ABSTRACT
High ambient temperature attributable to global warming has a profound influence on plant growth and development at all stages of the life cycle. The response of plants to high ambient temperature, termed thermomorphogenesis, is characterized by hypocotyl and petiole elongation and hyponastic growth at the seedling stage. However, our understanding of the molecular mechanism of thermomorphogenesis is still rudimentary. Here, we show that a set of four SUPPRESSOR OF PHYA-105 (SPA) genes is required for thermomorphogenesis. Consistently, SPAs are necessary for global changes in gene expression in response to high ambient temperature. In the spaQ mutant at high ambient temperature, the level of SPA1 is unaffected, whereas the thermosensor phytochrome B (phyB) is stabilized. Furthermore, in the absence of four SPA genes, the pivotal transcription factor PIF4 fails to accumulate, indicating a role of SPAs in regulating the phyB-PIF4 module at high ambient temperature. SPA1 directly phosphorylates PIF4 in vitro, and a mutant SPA1 affecting the kinase activity fails to rescue the PIF4 level in addition to the thermo-insensitive phenotype of spaQ, suggesting that the SPA1 kinase activity is necessary for thermomorphogenesis. Taken together, these data suggest that SPAs are new components that integrate light and temperature signaling by fine-tuning the phyB-PIF4 module.
KEY WORDS: Arabidopsis, High ambient temperature, SUPPRESSOR OF PHYA-105 (SPA), PIF4, Phytochrome B
Highlighted Article: Four SUPPRESSOR OF PHYA-105 (SPA) genes destabilize the thermosensor phyB and are crucial for stabilizing PIF4 through direct phosphorylation, fine-tuning thermomorphogenesis in response to high ambient temperature.
INTRODUCTION
Among many facets of climate change, global warming, causing an elevated ambient temperature, has a profound influence on plant growth and development at all stages of the life cycle (Lippmann et al., 2019). Plants modify their body plan as an adaptive response to cope with the high ambient temperature. These changes in growth and development are termed thermomorphogenesis, which is characterized by hypocotyl and petiole elongation, hyponastic growth, reduced stomatal density and early flowering (Casal and Balasubramanian, 2019; Gil and Park, 2019; Quint et al., 2016).
Recent reports showed that the temperature and light perception mechanisms are tightly linked. For example, the red light photoreceptor phytochrome B (phyB) has been shown to function as a thermosensor, and the blue light photoreceptor Cryptochrome 1 (Cry1) was also shown to mediate a high ambient temperature response (Jung et al., 2016; Legris et al., 2016; Ma et al., 2016). In addition, an RNA thermoswitch within the 5′-untranslated region of the PHYTOCHROME INTERCTING FACTOR 7 (PIF7) has been shown to regulate the translational efficiency of PIF7 mRNA in response to high ambient temperature (Chung et al., 2020). Downstream of environmental sensors, a plethora of light and clock signaling components have been shown to regulate thermomorphogenesis. Among these, PIF4 acts as the main hub in regulating thermomorphogenesis (Koini et al., 2009). Both PIF4 and PIF7 are transcriptionally and post-translationally upregulated by elevated temperature, and the stabilized PIF4/7 controls downstream target genes, especially auxin biosynthesis and signaling genes, to promote hypocotyl elongation (Chung et al., 2020; Fiorucci et al., 2020; Franklin et al., 2011; Oh et al., 2012; Stavang et al., 2009). Furthermore, the DEETIOLATED 1 (DET1)-CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1)-ELONGATED HYPOCOTYL 5 (HY5) module controls PIF4 both transcriptionally and post-translationally (Delker et al., 2014; Gangappa and Kumar, 2017). Recently, HEMERA (HMR) has been shown to interact directly with PIF4 to stabilize PIF4 and also control PIF4 target genes to regulate thermomorphogenesis (Qiu et al., 2019).
Apart from light signaling genes, circadian clock and flowering time genes are also involved in regulating thermomorphogenesis. For example, EARLY FLOWERING 3 (ELF3) interacts physically with PIF4 and controls the activity of PIF4 to regulate thermomorphogenesis independent of the circadian clock (Box et al., 2015; Nieto et al., 2015). TIMING OF CAB EXPRESSION 1 (TOC1)-PIF4 interaction results in inhibition of PIF4 function to mediate circadian gating of thermomorphogenic growth at elevated temperature (Zhu et al., 2016). A GIGANTEA (GI)-REPRESSOR OF ga1-3 (RGA)-PIF4 signaling module enables daylength-dependent modulation of thermomorphogenesis (Park et al., 2020). Furthermore, transcription factors B-box 18 (BBX18) and B-box 23 (BBX23) promote thermomorphogenesis by inhibiting ELF3, resulting in activation of PIF4 function (Ding et al., 2018a). The RNA-binding protein FCA directly interacts with PIF4 and inhibits the promoter occupancy of PIF4 to attenuate auxin-induced stem elongation at elevated temperature (Lee et al., 2014). Finally, a group of TEOSINTE BRANCHED 1/CYCLOIDEA/PCF (TCP) transcription factors interacts physically with PIF4 and Cry1 to regulate thermomorphogenesis (Han et al., 2019; Zhou et al., 2019). Overall, a number of regulatory proteins impinge upon PIF4, highlighting the importance of PIF4 in regulating thermomorphogenesis.
The SUPPRESSOR OF PHYA-105 (SPA) family of genes were originally discovered as suppressors of phyA signaling pathways (Hoecker, 2017; Hoecker et al., 1999; Laubinger et al., 2004). They have been shown to function as part of a complex with COP1 as E3 Ubiquitin ligases and play both negative and positive roles in regulating photomorphogenesis (Hoecker, 2017; Pham et al., 2018). Very recently, SPA1 has been shown to function as a Ser/Thr kinase for the light-induced degradation of PHYTOCHROME INTERACTING FACTOR 1 (PIF1) (Paik et al., 2019). Although the COP1-SPA complex has been shown to function in thermomorphogenesis (Delker et al., 2014; Jang et al., 2015; Park et al., 2017), the molecular functions of SPA proteins and the significance of the SPA kinase activity in high ambient temperature signaling have not yet been demonstrated. Here, we show that the SPA proteins and SPA1 kinase activity are essential for regulating thermomorphogenesis by control of the phyB-PIF4 module.
RESULTS
SPAs are required for thermomorphogenesis
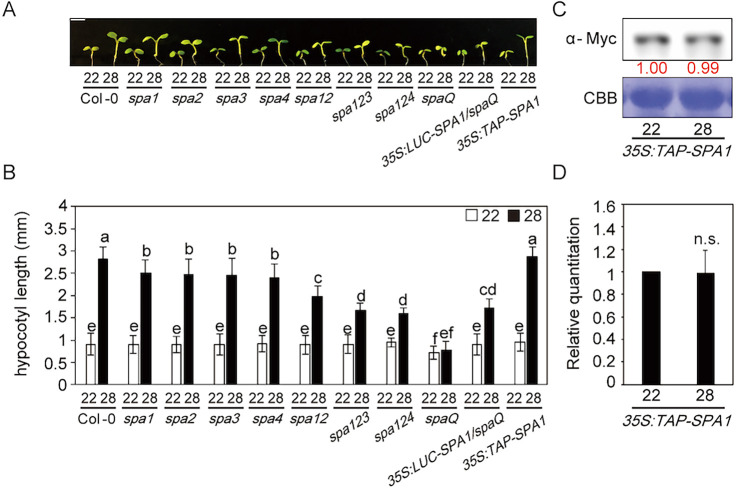
To examine whether the SPA genes are required for thermomorphogenesis, we tested high ambient temperature-mediated hypocotyl/petiole elongation and flowering time phenotypes using spa higher order mutants and TAP-tagged SPA1-overexpressing plants driven by the CaMV 35S promoter (Fig. 1A,B). To this end, we grew seedlings at 22°C under continuous white light for 2 days and then at either 22 or 28°C for an additional 4 days. Although we did not observe a statistically significant difference in hypocotyl growth at 22°C for spa double and triple mutants, the higher order spa mutants exhibited a shorter hypocotyl in high ambient temperature (28°C) conditions. Interestingly, the spaQ mutant, in which all four SPAs were mutated, displayed insensitivity to the high ambient temperature treatment, indicating that SPAs are necessary for thermomorphogenesis in a redundant manner. However, the overexpressed SPA1 (35S:TAP-SPA1) line showed a similar phenotype to the wild type (WT), indicating that an excessive amount of SPA1 might not necessarily enhance the high ambient temperature-mediated hypocotyl elongation.
Fig. 1.
SPAs are required for thermomorphogenesis. (A) Photograph showing seedling phenotypes of spa higher order mutants grown at normal or high ambient temperature. Seedlings were grown for 2 days in continuous white light at 22°C and then either kept at 22°C or transferred to 28°C for an additional 4 days before being photographed. More than 10 seedlings were measured for each experiment, and the experiment was repeated three times for one-way ANOVA with Tukey's HSD test. Scale bar: 2 mm. (B) Bar graph shows the hypocotyl lengths for seedlings grown in the conditions described in A. The letters a-f indicate statistically significant differences between means of hypocotyl lengths (P<0.05) based on one-way ANOVAs. Error bars indicate the s.d. (n=3). (C) Western blot shows the level of TAP-SPA1 from 35S:TAP-SPA1 whole seedlings grown for 5 days at 22°C and either kept at 22°C or transferred to 28°C for 4 h. TAP-SPA1 was detected using Myc antibody. The red numbers indicate the quantitation value from the anti-Myc band divided by the Coomassie Blue staining intensity. (D) Bar graph shows the relative amount of TAP-SPA1 (n=3).
We then examined whether SPA1 can complement the thermomorphogenic response in the spaQ mutant. We used previously reported LUC-SPA1/spaQ transgenic plants (Paik et al., 2019). As expected, SPA1 can partly rescue the spaQ mutant thermo-insensitive phenotype (Fig. 1A,B), indicating that SPA1 is required for thermomorphogenesis. Similar to the hypocotyl phenotype, higher order spa mutants also displayed reduced sensitivity for petiole elongation (Fig. S1A). However, flowering time was not affected at 28°C (Fig. S1B) in long-day conditions, which is similar to a previous report (Chung et al., 2020).
To investigate the potential temperature regulation on SPA1, we first examined the SPA1 protein level at 22 and 28°C using 35S:TAP-SPA1 plants (Fig. 1C,D). Intriguingly, the SPA1 protein level did not change at high ambient temperature, indicating that temperature does not regulate SPA1 protein stability post-translationally. We also examined the transcript level of SPAs at high ambient temperature (Fig. S2) and did not observe any difference in gene expression of SPAs, except for SPA1, which showed mildly decreased expression at high ambient temperature. Finally, we examined the transcript level of several thermo-responsive marker genes, such as IAA 29, PRE 5, SAUR 15 and CYP79B2. The expression of these genes was not upregulated in spaQ mutant, whereas obvious regulation was observed in the WT (Fig. S3). Taken together, our results showed that SPAs are required for thermomorphogenesis.
SPAs control global gene expression at high ambient temperature
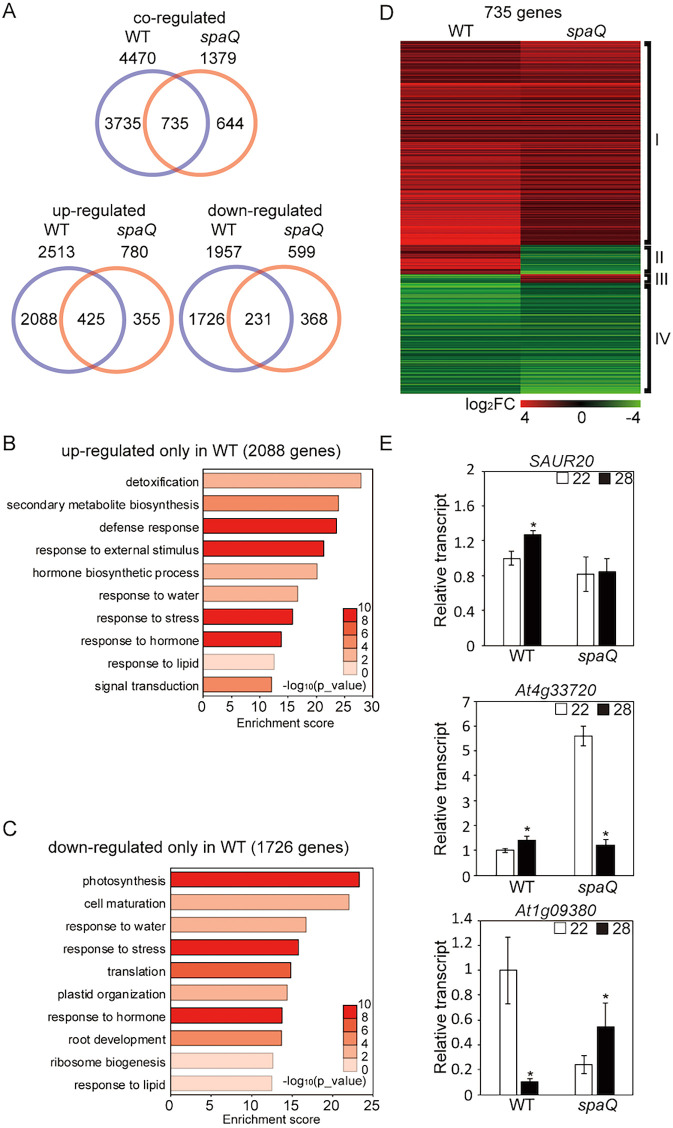
High ambient temperature induces global changes in gene expression to effect thermomorphogenesis (Jung et al., 2016). To test whether SPAs can regulate gene expression in response to high ambient temperature, RNA-seq was conducted using WT Col-0 and spaQ mutant at both 22 and 28°C. The results showed that 4470 genes were differentially regulated in the WT, whereas only 1379 genes were regulated in spaQ mutant in response to high ambient temperature (Fig. 2A). Moreover, 735 genes were shared between the WT and spaQ, which is >53% of genes regulated in spaQ. A closer examination showed that >54% of upregulated genes and 38.5% of downregulated genes in spaQ were shared with the WT (Fig. 2A), indicating that SPAs are crucial for transcriptional regulation in thermomorphogenesis.
Fig. 2.
SPAs regulate global gene expression at high ambient temperature. (A) Venn diagrams show co-regulated, upregulated and downregulated genes in the wild type (WT) versus the spaQ mutant at high ambient temperature. Six-day-old seedlings grown under white light were transferred to 22 or 28°C for an additional 24 h, and total RNA was extracted from three biological replicates for RNA-seq analyses. (B) Gene Ontology (GO) analysis of upregulated genes only in WT. (C) GO analysis of downregulated genes only in WT. (D) Hierarchical clustering, displaying 735 differentially expressed genes from co-regulated genes shared between WT and spaQ as shown in A. Co-regulated genes were identified with FDR<0.05. Each group represents genes upregulated in both WT and spaQ (I), genes upregulated in WT but downregulated in spaQ (II), genes downregulated in WT but upregulated in spaQ (III), and genes downregulated in both WT and spaQ (IV), respectively. (E) RT-qPCR analysis using SAUR20, already identified as a high ambient temperature marker gene, At4g33720 from group II in D, and At1g09380 from group III in D. RT-qPCR samples were from Col-0 whole seedlings grown for 5 days at 22°C and then either kept at 22°C or transferred to 28°C for 24 h. Three biological repeats were performed. Relative gene expression levels were normalized using expression levels of ACT7. Student's unpaired t-test: *P<0.05.
Using Gene Ontology (GO) analysis, we analyzed 2088 genes that were upregulated only in the WT but were not affected in the spaQ mutant, and also 1726 genes that were downregulated only in the WT but were not altered in the spaQ mutant (Fig. 2B,C). Many of these genes were responsible for abiotic responses, such as water and stress responses, in both upregulated and downregulated genes in the WT. Notably, many of the detoxification and secondary metabolite biosynthesis responsive genes were involved in thermomorphogenesis in the WT, but not in spaQ mutant. Also, many genes that are involved in metabolism or development, such as photosynthesis, cell maturation or root development, were downregulated in the WT but not in the spaQ mutant. Furthermore, 735 differentially expressed genes (DEGs) were shared in both WT and spaQ, which displayed four distinct patterns as shown in the heatmap analysis (Fig. 2D). Although the expression levels were different, two groups (I and IV) displayed similar patterns between the WT and spaQ mutant, whereas the other two (II and III) displayed opposite patterns of expression.
We conducted qPCR to confirm the RNA-seq data using three different genes, including genes from groups II and III (Fig. 2E). SAUR20, which is one of the 2088 genes in Fig. 2B, showed similar expression patterns in both qPCR and RNA-seq analyses, and was upregulated in the WT but did not change in the spaQ mutant at high ambient temperature. At4g33720 from group II and At1g09380 from group III also displayed similar patterns, showing opposite regulation between the WT and spaQ mutant. To investigate whether SPAs and PIF4 co-regulate gene expression in response to high ambient temperature, we compared our spaQ RNA-seq data with those of pif4 data from a previous report (Fig. S4) (Ding et al., 2018b). As expected, >30% of PIF4-regulated genes overlapped with SPA-regulated genes (Fig. S4A). Furthermore, most of the categories identified using GO analyses were temperature or stress response related, such as the cellular response to sulfur starvation, the cellular response to heat, heat acclimation and secondary metabolic processes (Fig. S4B). Overall, these data suggest that SPAs play a crucial role in thermomorphogenesis, in part through PIF4-mediated transcriptional regulation.
SPAs are essential for PIF4 stabilization at high ambient temperature
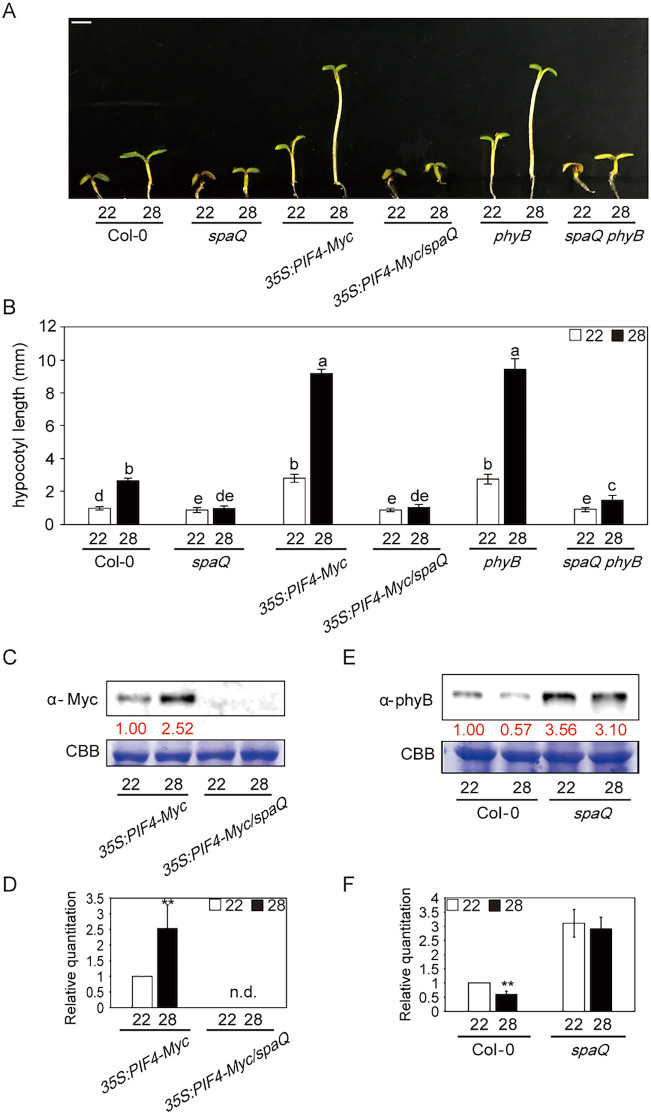
Given that the spaQ mutant did not respond to high ambient temperature-induced hypocotyl elongation and that SPA and PIF4 co-regulated gene expression, we examined whether SPAs regulated thermomophogenesis through PIF4 at high ambient temperature. To test the genetic relationship between PIF4 and SPAs, we generated an 35S:PIF4-Myc overexpression line on the spaQ mutant background and measured the phenotype at high ambient temperature (Fig. 3A,B). The results showed that the hypocotyl lengths of 35S:PIF4-Myc/spaQ and spaQ were similar at high ambient temperature, suggesting that spaQ is epistatic to the PIF4 overexpression phenotype. To examine whether SPA proteins regulated PIF4 protein stability, we performed immunoblot analyses from 35S:PIF4-Myc and 35S:PIF4-Myc/spaQ grown at 22 and 28°C. Strikingly, the PIF4-Myc level was not detectable in 35S:PIF4-Myc/spaQ compared with that in 35S:PIF4-Myc in WT (Fig. 3C,D), indicating that SPAs are essential for stabilization of PIF4 at both normal and elevated temperature. Taken together, these data suggest that SPAs might promote thermomorphogenesis at a high ambient temperature through stabilization of PIF4.
Fig. 3.
SPAs are necessary for controlling the phyB-PIF4 module at high ambient temperature. (A) Photograph shows the seedling phenotypes of Col-0, spaQ, 35S:PIF4-Myc, 35S:PIF4-Myc/spaQ, phyB and spaQ phyB plants at 22 or 28°C. Seedlings were grown for 2 days in continuous white light at 22°C and then either kept at 22°C or transferred to 28°C for an additional 4 days before being photographed. More than 10 seedlings were measured for each experiment, and the experiment was repeated three times for one-way ANOVA with Tukey's HSD test. Scale bar: 2 mm. (B) Bar graph shows the hypocotyl lengths for seedlings grown in the conditions described in A. The letters a-e indicate statistically significant differences between means of hypocotyl lengths (P<0.05) based on one-way ANOVA with Tukey's HSD test. Error bars indicate the s.d. (n=3). (C) Western blot shows PIF4-Myc protein level from either 35S:PIF4-Myc or 35S:PIF4-Myc/spaQ whole seedling grown for 5 days at 22°C and transferred to 22 or 28°C for 4 h. PIF4-Myc was detected using Myc antibody. Red numbers indicate the quantitation value from anti-Myc detection divided by Coomassie Blue staining intensity. (D) Bar graph shows the relative amount of PIF4-myc (n=3). n.d., not detectable. (E) Western blot shows the phyB protein level from either Col-0 or spaQ mutant at 22 or 28°C. (F) Bar graph shows the relative amount of phyB (n=3). Student's unpaired t-test: **P<0.01.
SPAs are essential for phyB degradation at high ambient temperature
Phytochrome B has been shown to function as a temperature sensor not only in diurnal conditions but also in continuous light (Jung et al., 2016; Legris et al., 2016; Qiu et al., 2019). Phytochromes are well-known red/far-red light receptors (Legris et al., 2019), and SPAs are one of the key components in light signaling pathways that bind to phyB (Lu et al., 2015; Paik et al., 2019; Sheerin et al., 2015). Given that phyB and SPA1 have been shown to interact directly with each other, we hypothesized that phyB and SPAs might also be linked tightly in the temperature signaling cascade. Therefore, we measured the phyB level in the WT and the spaQ mutant using phyB antibody. Interestingly, we first observed that the phyB level was decreased in the WT at high ambient temperature. However, unlike the WT, the phyB level was elevated at 22°C and remained similar in the spaQ mutant at high ambient temperature (Fig. 3E,F). These data suggest that high ambient temperature induces the thermomorphic response by decreasing the phyB protein level, possibly owing to receptor desensitization, and the SPAs are necessary for this reduction in phyB level. Furthermore, SPAs are required for phyB degradation at both normal and high ambient temperature. We also tested the spaQ phyB mutant phenotype to examine whether an absence/degradation of phyB has a role in thermomorphogenesis. Interestingly, the spaQ phyB mutant displayed a partial but weak recovery of phenotype compared with spaQ at high ambient temperature (Fig. 3A,B). These data indicate that the SPA-mediated regulation of the phyB level at high ambient temperature is essential for thermomorphogenic hypocotyl elongation.
SPA1 interacts with PIF4 and forms the SPA1-phyB-PIF4 complex in vitro
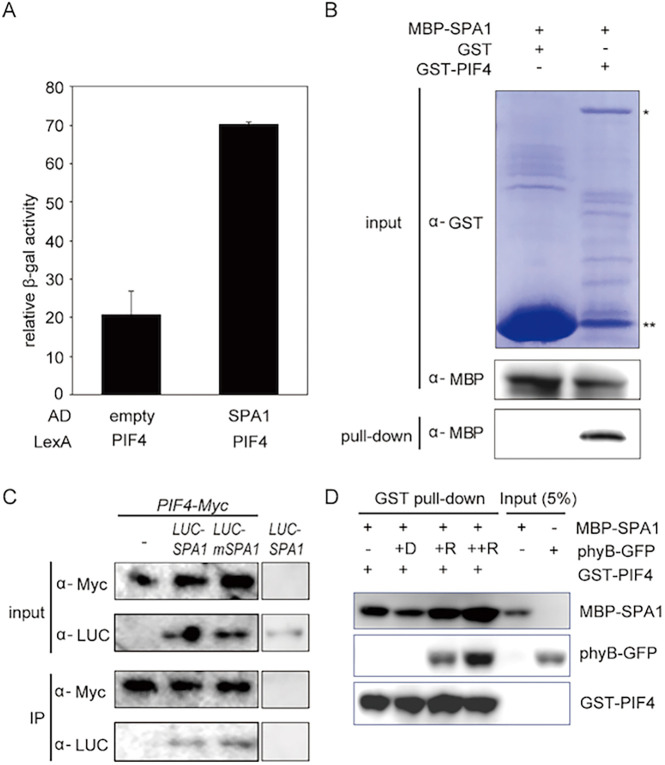
PIF4 is considered to be a main hub in high ambient temperature response pathways (Pham et al., 2018). The expression and stability of PIF4 are regulated at high ambient temperature (Foreman et al., 2011; Oh et al., 2012; Stavang et al., 2009). Stabilized PIF4 triggers transcriptional regulation in auxin responsive and brassinosteroid (BR) biosynthetic genes to promote thermomorphogenesis (Franklin et al., 2011; Jung et al., 2016; Koini et al., 2009; Legris et al., 2019; Martínez et al., 2018; Oh et al., 2012; Qiu et al., 2019). Given that SPAs regulate both PIF4 and phyB levels, we examined whether SPA1 can interact directly with PIF4. Initially, we performed yeast two-hybrid assays using PIF4 and SPA1. The results showed that PIF4 interacted with SPA1 in yeast (Fig. 4A). Next, we confirmed the physical interaction between SPA1 and PIF4 through in vitro pull-down assays using bacterially expressed MBP-tagged SPA1 and GST-tagged PIF4 (Fig. 4B). To confirm that these two proteins interact in plants, we performed in vivo co-immunoprecipitation assays using double transgenic plants expressing PIF4-myc and LUC-SPA1 or LUC-mSPA1. The results showed that PIF4 could interact with both LUC-SPA1 and LUC-mSPA1 (Fig. 4C). These data suggest that SPA1 might regulate PIF4 via physical interaction.
Fig. 4.
SPA1 interacts with PIF4 and forms an SPA1-PIF4-phyB ternary complex. (A) SPA1 interacts with PIF4 in yeast two-hybrid assays. LexA-PIF4 was co-transformed with empty AD or AD-SPA1. The error bars represent the s.d. Three biological replicates were used in this experiment. (B) SPA1 interacts with PIF4 in in vitro pull-down assays. GST-PIF4 and GST only (as a control) was used to pull down MBP-SPA1. Proteins were detected using anti-MBP and anti-GST antibodies. *GST-PIF4 band; **GST-only protein. (C) SPA1 interacts with PIF4 in vivo. Double transgenic plants expressing 35S:PIF4-myc with 35S:LUC-SPA1 or LUC-mSPA1 were used for this assay. 35S:PIF4-myc and 35S:LUC-SPA1 only were used as controls. PIF4-myc was used as a bait to co-immunoprecipitate LUC-SPA1 or LUC-mSPA1 and then detected using anti-LUC and anti-myc antibodies. (D) SPA1 forms a ternary complex with PIF4 and phyB in vitro. The in vitro pull-down assay shows that phyB enhances PIF4-SPA1 interaction in a light- and concentration-dependent manner. GST-PIF4 was used as a bait to pull-down MBP-SPA1 without and with an increasing amount of phyB. The samples were exposed to red light or kept in the dark. Precipitated proteins were detected using anti-MBP, anti-GFP or anti-GST HRP-conjugated antibodies.
To reveal whether SPA1-phyB-PIF4 could form a ternary complex, we performed in vitro pull-down assays using GFP-tagged full-length phyB protein purified from Pichia pastoris, MBP-tagged SPA1 and GST-tagged PIF4 purified from bacteria in dark and red light conditions. The data showed that SPA1, phyB and PIF4 could form a ternary complex in vitro in response to red light (Fig. 4D). Thus, SPAs might regulate PIF4 and phyB by direct interaction.
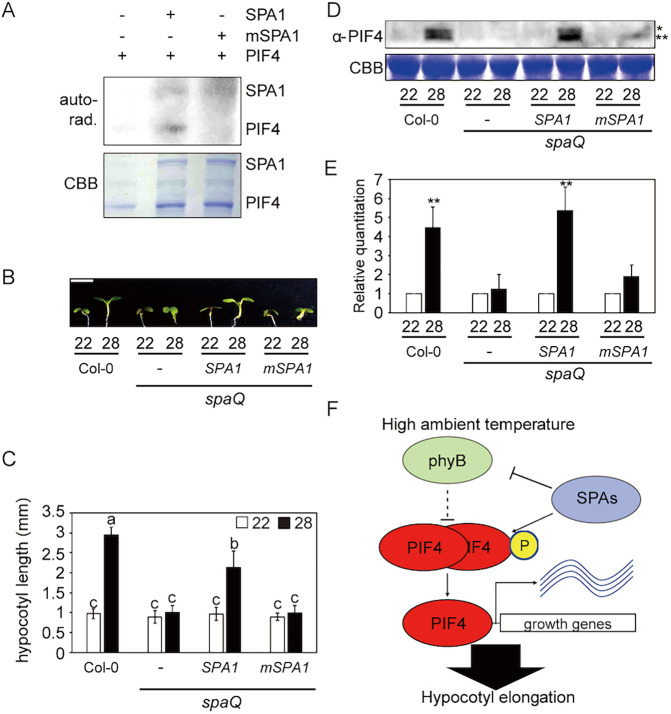
Phosphorylation of PIF4 by SPA1 is necessary for thermomorphogenesis
Recently, we reported that SPA1 acts as a Ser/Thr kinase that phosphorylates PIF1 and controls PIF1 stability in response to light (Paik et al., 2019). Given that PIF4 also interacts with SPA1, we hypothesized that SPA1 might phosphorylate PIF4. Therefore, we performed an in vitro kinase assay to examine whether SPA1 could phosphorylate PIF4 directly (Fig. 5A). A GFP-tagged full-length SPA1 protein purified from Pichia pastoris was used for the kinase assay along with bacterially expressed GST-PIF4. As expected, PIF4 was phosphorylated by SPA1 in vitro. A conserved amino acid mutation (R517E) in the kinase domain of SPA1 greatly reduces its kinase activity in vitro (Paik et al., 2019). In vitro kinase assays also showed that mSPA1 displayed significantly reduced kinase activity toward PIF4 (Fig. 4A), suggesting that PIF4 might be a bona fide substrate of SPA1 kinase.
Fig. 5.
Phosphorylation of PIF4 by SPA1 is essential for thermomorphogenesis. (A) The autoradiograph (top panel) shows that SPA1 directly phosphorylates PIF4, whereas mSPA1 significantly reduces PIF4 phosphorylation in vitro. In vitro kinase assay performed using purified SPA1 or mSPA1 from Pichia pastoris and GST-PIF4 from Escherichia coli. The Coomassie Blue-stained gel (bottom panel) shows the amount of protein used in the assay. (B) Photograph shows the seedling phenotypes of Col-0, spaQ, 35S:LUC-SPA1/spaQ and 35S:LUC-mSPA1/spaQ at 22 or 28°C. Three spaQ background plants are indicated with a dash for spaQ, SPA1 for 35S:LUC-SPA1/spaQ, and mSPA1 for 35S:LUC-mSPA1/spaQ, respectively. Seedlings were grown for 2 days in continuous white light at 22°C and then either kept at 22°C or transferred to 28°C for an additional 4 days before being photographed. Scale bar: 2 mm. (C) Bar graph shows the hypocotyl lengths for seedlings grown in the conditions described in B. More than 10 seedlings were measured for each experiment, and the experiment was repeated three times for one-way ANOVA with Tukey's HSD test. The letters a-c indicate statistically significant differences between means of hypocotyl lengths (P<0.05) based on one-way ANOVA with Tukey's HSD test. Error bars indicate the s.d. (n=3). (D) Western blot shows native PIF4 protein level from Col-0, spaQ, 35S:LUC-SPA1/spaQ and 35S:LUC-mSPA1/spaQ whole seedlings grown for 5 days at 22°C and then either kept at 22°C or transferred to 28°C for 4 h. PIF4 was detected using native PIF4 antibody. *Phosphorylated PIF4; **unphosphorylated PIF4. (E) Bar graph shows the relative amount of PIF4 (n=3). Student's unpaired t-test: **P<0.01. (F) Simplified model shows the role of SPA proteins in regulating thermomorphogenesis at elevated temperature. After exposure to high ambient temperature, SPAs promote the degradation of phyB, but stabilize PIF4 by direct phosphorylation and/or other unknown mechanisms. Stabilized PIF4 then promotes hypocotyl elongation through direct promoter binding to hypocotyl elongation-responsive genes.
To examine whether the kinase activity of SPA1 is necessary for thermomorphic hypocotyl elongation, we measured the phenotype of 35S:LUC-mSPA1/spaQ at high ambient temperature (Fig. 5B,C). In contrast to WT 35S:LUC-SPA1/spaQ, 35S:LUC-mSPA1/spaQ did not rescue the thermo-insensitive phenotype of spaQ at high ambient temperature, indicating that the kinase activity of SPA1 is necessary for thermomorphogenesis. Moreover, an immunoblot using anti-PIF4 antibody showed that both the phosphorylated and unphosphorylated forms of native PIF4 are unstable on the spaQ background compared with WT (Fig. 5D,E). Strikingly, the PIF4 level was not rescued in the 35S:LUC-mSPA1/spaQ, whereas the expression of 35S:LUC-SPA1/spaQ largely rescued both the phosphorylated and unphosphorylated forms of PIF4, consistent with the phenotype of the 35S:LUC-SPA1/spaQ seedlings. In addition, treatment with a proteasome inhibitor (bortezomib) stabilized both the unphosphorylated and phosphorylated forms of PIF4 on the 35S:LUC-SPA1/spaQ background (Fig. S5). However, only the unphosphorylated form of PIF4 accumulated on the 35S:LUC-mSPA1/spaQ background after bortezomib treatment (Fig. S5), suggesting that phosphorylation of PIF4 by SPA1 kinase is necessary for the stabilization of PIF4 at high ambient temperature. The PIF4 transcript level remained unchanged on the 35S:LUC-SPA1/spaQ and 35S:LUC-mSPA1/spaQ backgrounds (Fig. S6), suggesting that the PIF4 protein level is likely to be regulated post-translationally. Taken together, these data suggest that SPAs regulate thermomorphogenesis through direct phosphorylation of PIF4 (Fig. 5F).
DISCUSSION
In this study, we provide genetic and biochemical evidence supportive of the hypothesis that SPA proteins play an essential role in promoting thermomorphogenesis. Phenotypic analyses of a series of spa single and higher order mutants showed that all four SPA genes are necessary for promoting hypocotyl elongation at high ambient temperature. In addition, the expression of a large number of genes is defective on the spaQ background compared with WT at high ambient temperature. The exaggerated phenotype of the 35S:PIF4-myc overexpression line is also suppressed on the spaQ background. These phenotypic and gene expression analyses support the conclusion that SPAs represent new components in the regulation of thermomorphogenesis. Thus, SPAs and COP1 play a positive role in regulating thermomorphogenesis (Delker et al., 2014; Gil and Park, 2019; Jang et al., 2015; Park et al., 2017).
Although phyB acts as a thermosensor (Jung et al., 2016; Legris et al., 2016), many factors impinge upon the central transcription factor PIF4 to regulate thermomorphogenesis (Casal and Balasubramanian, 2019; Gil and Park, 2019; Koini et al., 2009). ELF3, TOC1 and FCA interact directly with PIF4 and inhibit PIF4 function to negatively regulate thermomorphogenesis (Lee et al., 2014; Nieto et al., 2015; Zhu et al., 2016). GI stabilizes DELLA proteins that inhibit PIF4 activity (Park et al., 2020), whereas BBX18 and BBX23 inhibit ELF3 to promote PIF4 function (Ding et al., 2018a). In addition, the TCP transcription factors control the expression and activity of PIF4 to promote thermomorphogenesis (Han et al., 2019; Zhou et al., 2019). HMR and HOOKLESS also interact with PIF4 and control the activity of PIF4 to promote thermomorphogenesis (Jin et al., 2020; Qiu et al., 2019).
The biochemical data presented here suggest that SPAs regulate thermomorphogenesis by controlling the level of the thermosensor, phyB, in addition to the central transcription factor, PIF4. Although SPAs are necessary for stabilizing PIF4, they induce destabilization of phyB at high ambient temperature. In fact, SPA1 interacts physically with PIF4 in vitro and in vivo and forms a SPA1-PIF4-phyB ternary complex in vitro. The high ambient temperature-induced destabilization of phyB appears to be similar to the red light-mediated destabilization of phyB, because phyB acts as a receptor for both red light and high ambient temperature. Previously, PIFs have been shown to induce degradation of phyB under red light in a mutually destructive manner (Leivar et al., 2008; Ni et al., 2014). Thus, the increased level of PIF4 at high ambient temperature might induce degradation of phyB as part of a receptor desensitization, as previously hypothesized (Zhu and Huq, 2014).
The SPA-induced stabilization of PIF4 appears to be linked to the recently described kinase activity of SPA proteins (Paik et al., 2019). Previously, the BR signaling kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2) has also been shown to phosphorylate PIF4 directly, and the BIN2-mediated phosphorylation destabilizes PIF4 (Bernardo-García et al., 2014). By contrast, SPA1 directly phosphorylates PIF4 in vitro, and the phosphorylation of PIF4 is necessary for the stability of PIF4. Thus, multiple antagonistic kinases might fine-tune the PIF4 level to regulate seedling development at ambient and elevated temperatures. Moreover, these data also highlight the difference between two PIFs, PIF1 versus PIF4. Both PIFs are phosphorylated directly by SPA1. The SPA1-mediated phosphorylation of PIF1 is necessary for PIF1 degradation (Paik et al., 2019), whereas the PIF4 phosphorylation by the same kinase is necessary for its stabilization. It is possible that a certain phosphosite or a combination of sites might signal for degradation, whereas other phosphosites might signal for stabilization. Identification of SPA1-mediated phosphorylation sites in both PIF1 and PIF4 will help us to understand these differences.
In summary, our data uncovered a new role of SPA proteins, which act as positive regulators of thermomorphogenesis by controlling the phyB-PIF4 module (Fig. 5F). Although SPAs destabilize the thermosensor, phyB, they are crucial for stabilizing PIF4 through direct phosphorylation. In contrast to SPAs, ELF3, TOC1, FCA, HMR, TCP and others interact with PIF4 and control the activity of PIF4 to regulate thermomorphogenesis (Lee et al., 2014; Nieto et al., 2015; Paik et al., 2017; Qiu et al., 2019; Zhou et al., 2019; Zhu et al., 2016). Thus, SPAs in concert with other factors fine-tune thermomorphogenesis in response to high ambient temperature.
MATERIALS AND METHODS
Plant materials, growth conditions and phenotypic analyses
Seeds from the Col-0 ecotype of Arabidopsis thaliana were used in this study. Seeds were surface-sterilized and plated on Murashige and Skoog (MS) medium containing no sucrose. After 3 days at 4°C for stratification, seedlings were placed in 22°C for 2 days and transferred to 22 or 28°C for an additional 4 and 12 days for measurement of hypocotyl and petiole length, respectively. The number of leaves required for bolting was measured in long-day (16 h light:8 h dark) conditions. Hypocotyl length (n>10), petiole length (n=5) and the number leaves at flowering (n>5) were measured using ImageJ software and analyzed statistically using one-way ANOVA with Tukey's HSD test.
Protein extraction and western blot analyses
Total protein of 50 seedlings for each sample was extracted using 50 µl urea extraction buffer [8 M urea, 0.35 M Tris-Cl (pH 7.5), 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protease inhibitor cocktail]. Subsequently, 6× SDS buffer was added, followed by 5 min of denaturation at 95°C. Then, samples were centrifuged at 16,000 g for 15 min. Supernatant from the samples was loaded onto 8% SDS-PAGE gels for separation. A PVDF membrane (Millipore) was used for transfer, and western blots were detected using anti-Myc (Cell Signaling, 2276), anti-PIF4 (AS16 3955; Agrisera) or anti-phyB antibody (Dr Peter Quail; Table S2) at dilutions of 1:5000, 1:1000 and 1:500, respectively. Coomassie Blue staining was also used for the loading control.
RNA extraction, complementary DNA synthesis and qRT-PCR
RNA-seq was performed using 6-day-old seedlings grown under white light, with three independent biological replicates (n=3). Seeds were kept at 22°C under continuous white light for 6 days. After 6 days, seedlings were either kept at 22°C or transferred to 28°C for an additional 24 h under continuous white light. Total RNA was extracted from these seedlings using the Plant RNA purification kit (Sigma-Aldrich) according to the manufacturer's protocols. For complementary DNA synthesis, 2 µg of total RNA was used for reverse transcription with M-MLV Reverse Transcriptase (Thermo Fisher Scientific). SYBR Green PCR master mix (Thermo Fisher Scientific) and gene-specific oligonucleotides (Table S1) were used to conduct qPCR analyses. Finally, the relative transcription level was calculated using 2ΔCt with ACT2 normalization.
RNA-seq analyses
For the RNA-seq analysis, the 3′Tag-Seq method was used in this study (Lohman et al., 2016). Raw read quality was accessed using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). The raw reads were aligned to the Arabidopsis genome using Bowtie2 (Langmead and Salzberg, 2012) and TopHat (Trapnell et al., 2012). The annotation of the Arabidopsis genome was obtained from TAIR10 (www.arabidopsis.org/). Read count data were performed by HTseq (Anders et al., 2015; https://htseq.readthedocs.io/en/master/). Differentially expressed genes in spaQ/WT were identified using the EdgeR (Robinson et al., 2010). The differential gene expression was defined using a cut-off of ≥2-fold with adjusted P-value (FDR) ≤0.05. Venn diagrams were generated using the website (http://bioinformatics.psb.ugent.be/webtools/Venn/). A heatmap was generated using Morpheus (https://software.broadinstitute.org/morpheus/). We used the options as Hierarchical clustering with one minus cosine similarity metric combined with the average linkage method for the heatmap. Also, GO enrichment analyses were performed using TAIR (https://www.arabidopsis.org/tools/go_term_enrichment.jsp). GO bar graphs were generated based on the significantly enriched terms with the lowest P-value and FDR (≤0.05) for GO terms. Raw data and processed data for RNA-Seq in Col-0 and spaQ can be accessed from the Gene Expression Omnibus database under accession number GSE142354.
Yeast two-hybrid analyses
Cloning of SPA1 in pJG4-5 has been described previously (Xu et al., 2014). PIF4 was cloned into the pEG202 vector using EcoRI and XhoI restriction enzyme sites. Different combinations of AD-LexA and BD-fusion plasmids were introduced into the yeast strain EGY48-0 and selected on -His, -Ura, -Trp minimal synthetic medium. The β-galactosidase assay was performed according to the manufacturer's protocol (Matchmaker Two–Hybrid System; Takara, https://www.takarabio.com).
In vitro pull-down and in vivo co-immunoprecipitation assays
For in vitro pull-down assays, MBP-SPA1 (Xu et al., 2014) and GST-PIF4 were used. GST-PIF4 was expressed from the pGEX4T-1 vector. The bacterial extract expressing GST-PIF4 was incubated with glutathione resin in the binding buffer (50 mM Tris-Cl pH 7.5, 200 mM NaCl and 0.1% NP-40) for 2 h. Resin was washed more than five times with wash buffer (1× PBS and 0.1% Tween 20). Samples were boiled and analyzed using western blot. Anti-MBP (E8032S; New England Biolabs) and anti-GST-HRP conjugate (RPN1236; GE Healthcare Bio-Sciences) were used to detect MBP-SPA1 and GST or GST-PIF4, respectively.
For in vivo co-immunoprecipitation assays, 35S:LUC-SPA1, 35S:LUC-mSPA1 and 35S:PIF4-myc were crossed for each combination, and double transgenic plants expressing both proteins were selected. Immunoprecipitation was conducted using 50 seedlings for each sample with Dynabeads protein A and anti-Myc (Abcam). Western blots using anti-Myc (Cell Signaling) or anti-LUC antibody (Abcam) were used to detect the proteins.
For the ternary complex in vitro pull-down assay, GST-PIF4 and MBP-SPA1 were purified from bacteria, and phyB-GFP was purified from Pichia pastoris. One microgram of purified GST-PIF4 was bound to GST resin and incubated with 500 ng of MBP-SPA1 and an increasing amount of phyB-GFP. Co-precipitated SPA1-GFP, phyB-GFP and GST-PIF4 were immunoblotted using anti-GFP (Thermo Fisher Scientific) and anti-GST-HRP conjugate (RPN1236; GE Healthcare Bio-Sciences).
In vitro kinase assay
For the SPA1 kinase assay, ∼500 ng of SPA1-GFP or mSPA1-GFP and 1 µg of GST-PIF4 were used with the kinase buffer (50 mM Tris, pH 7.5, 4 mM β-mercaptoethanol, 1 mM EDTA and 10 mM MgCl2). 32P radio-labeled γ-ATP (BLU502A; Perkin Elmer) was added to the reaction and incubated at 28°C for 1 h. The reaction was stopped using 6× SDS sample buffer, and boiled proteins were loaded onto a 7.8% SDS-PAGE gel. After separation, gels were dried, exposed to a phosphor screen and scanned using Typhoon FLA 9500 (GE Healthcare).
Supplementary Material
Acknowledgements
We thank Dr Sibum Sung and the members of the Huq laboratory for critical reading of the manuscript, Dr Hong-Quan Yang for sharing spaQ phyB seeds, Dr Peter Quail for sharing anti-phyB antibody and Dr Taeyoung Lee for comments analyzing RNA-seq. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing high-performance computing, visualization and database resources that have contributed to the research results reported in this paper.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.L., I.P., E.H.; Methodology: S.L., I.P., E.H.; Software: S.L.; Validation: S.L., I.P.; Formal analysis: S.L., I.P., E.H.; Data curation: S.L., I.P.; Writing - original draft: S.L.; Writing - review & editing: I.P., E.H.; Supervision: E.H.; Project administration: E.H.; Funding acquisition: E.H.
Funding
This work was supported by grants from the National Science Foundation (MCB-2014408) and the National Institutes of Health (NIH) (GM-114297) to E.H., and by Integrative Biology (IB) Startup Fellowship grant from the University of Texas at Austin to S.L. Deposited in PMC for release after 12 months.
Data availability
Raw data and processed data for RNA-Seq in Col-0 and spaQ have been deposited in GEO under accession number GSE142354.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.189233.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.189233.reviewer-comments.pdf
References
- Anders S., Pyl P. T. and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-García S., de Lucas M., Martínez C., Espinosa-Ruiz A., Davière J.-M. and Prat S (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28, 1681-1694 10.1101/gad.243675.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box M. S., Huang B. E., Domijan M., Jaeger K. E., Khattak A. K., Yoo S. J., Sedivy E. L., Jones D. M., Hearn T. J., Webb A. A. R. et al. (2015). ELF3 Controls thermoresponsive growth in Arabidopsis. Curr. Biol. 25, 194-199. 10.1016/j.cub.2014.10.076 [DOI] [PubMed] [Google Scholar]
- Casal J. J. and Balasubramanian S (2019). Thermomorphogenesis. Annu. Rev. Plant Biol. 70, 321-346. 10.1146/annurev-arplant-050718-095919 [DOI] [PubMed] [Google Scholar]
- Chung B. Y. W., Balcerowicz M., Di Antonio M., Jaeger K. E., Geng F., Franaszek K., Marriott P., Brierley I., Firth A. E. and Wigge P. A (2020). An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat. Plants 6, 522-532. 10.1038/s41477-020-0633-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C., Sonntag L., James G. V., Janitza P., Ibañez C., Ziermann H., Peterson T., Denk K., Mull S., Ziegler J. et al. (2014). The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Reports 9, 1983-1989. 10.1016/j.celrep.2014.11.043 [DOI] [PubMed] [Google Scholar]
- Ding L., Wang S., Song Z. T., Jiang Y., Han J. J., Lu S. J., Li L. and Liu J. X (2018a). Two B-Box domain proteins, BBX18 and BBX23, interact with ELF3 and regulate thermomorphogenesis in Arabidopsis. Cell Rep. 25, 1718-1728.e4. 10.1016/j.celrep.2018.10.060 [DOI] [PubMed] [Google Scholar]
- Ding Z., Wang C., Feng G. and Zhang X (2018b). Thermo-responsive fluorescent polymers with diverse LCSTs for ratiometric temperature sensing through FRET. Polymers (Basel) 10, 283 10.3390/polym10030283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci A.-S., Galvão V. C., Ince Y. Ç., Boccaccini A., Goyal A., Allenbach Petrolati L., Trevisan M. and Fankhauser C (2020). Phytochrome interacting factor 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol. 226, 50-58. 10.1111/nph.16316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J., Johansson H., Hornitschek P., Josse E. M., Fankhauser C. and Halliday K. J (2011). Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 65, 441-452. 10.1111/j.1365-313X.2010.04434.x [DOI] [PubMed] [Google Scholar]
- Franklin K. A., Lee S. H., Patel D., Kumar S. V., Spartz A. K., Gu C., Ye S., Yu P., Breen G., Cohen J. D. et al. (2011). PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl Acad. Sci. USA 108, 20231-20235. 10.1073/pnas.1110682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S. N. and Kumar S. V (2017). DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Reports 18, 344-351. 10.1016/j.celrep.2016.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil K.-E. and Park C.-M (2019). Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 221, 1215-1229. 10.1111/nph.15518 [DOI] [PubMed] [Google Scholar]
- Han X., Yu H., Yuan R., Yang Y., An F. and Qin G (2019). Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience 15, 611-622. 10.1016/j.isci.2019.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U. (2017). The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 37, 63-69. 10.1016/j.pbi.2017.03.015 [DOI] [PubMed] [Google Scholar]
- Hoecker U., Tepperman J. M. and Quail P. H (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284, 496-499. 10.1126/science.284.5413.496 [DOI] [PubMed] [Google Scholar]
- Jang K., Gil Lee H., Jung S.-J., Paek N.-C. and Joon Seo P (2015). The E3 ubiquitin ligase COP1 regulates thermosensory flowering by triggering GI degradation in Arabidopsis. Sci. Rep. 5, 12071 10.1038/srep12071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Lin J. and Zhu Z (2020). PIF4 and HOOKLESS1 impinge on common transcriptome and isoform regulation in thermomorphogenesis. Plant Commun. 1, 100034 10.1016/j.xplc.2020.100034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.-H., Domijan M., Klose C., Biswas S., Ezer D., Gao M., Khattak A. K., Box M. S., Charoensawan V., Cortijo S. et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354, 886-889. 10.1126/science.aaf6005 [DOI] [PubMed] [Google Scholar]
- Koini M. A., Alvey L., Allen T., Tilley C. A., Harberd N. P., Whitelam G. C. and Franklin K. A (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408-413. 10.1016/j.cub.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Fittinghoff K. and Hoecker U (2004). The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16, 2293-2306. 10.1105/tpc.104.024216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-J., Jung J.-H., Cortés Llorca L., Kim S.-G., Lee S., Baldwin I. T. and Park C.-M (2014). FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat. Commun. 5, 5473 10.1038/ncomms6473 [DOI] [PubMed] [Google Scholar]
- Legris M., Ince Y. Ç. and Fankhauser C (2019). Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 10, 5219 10.1038/s41467-019-13045-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M., Klose C., Burgie E. S., Costigliolo C., Neme M., Hiltbrunner A., Wigge P. A., Schäfer E., Vierstra R. D. and Casal J. J (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897-900. 10.1126/science.aaf5656 [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J. M., Ecker J. R. and Quail P. H (2008). The Arabidopsis Phytochrome-Interacting Factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20, 337-352. 10.1105/tpc.107.052142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann R., Babben S., Menger A., Delker C. and Quint M (2019). Development of wild and cultivated plants under global warming conditions. Curr. Biol. 29, R1326-R1338. 10.1016/j.cub.2019.10.016 [DOI] [PubMed] [Google Scholar]
- Lohman B. K., Weber J. N. and Bolnick D. I (2016). Evaluation of TagSeq, a reliable low-cost alternative for RNAseq. Mol. Ecol. Resour. 16, 1315-1321. 10.1111/1755-0998.12529 [DOI] [PubMed] [Google Scholar]
- Lu X.-D., Zhou C.-M., Xu P.-B., Luo Q., Lian H.-L. and Yang H.-Q (2015). Red-light-dependent interaction of phyB with SPA1 Promotes COP1–SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8, 467-478. 10.1016/j.molp.2014.11.025 [DOI] [PubMed] [Google Scholar]
- Ma D., Li X., Guo Y., Chu J., Fang S., Yan C., Noel J. P. and Liu H (2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 113, 224-229. 10.1073/pnas.1511437113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C., Espinosa-Ruíz A., de Lucas M., Bernardo-García S., Franco-Zorrilla J. M. and Prat S (2018). PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 37, e99552 10.15252/embj.201899552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.-L., Tepperman J. M., Stanley D. J., Maltby D. A., Gross J. D., Burlingame A. L., Wang Z.-Y. and Quail P. H (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160-1164. 10.1126/science.1250778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C., Lopez-Salmeron V., Daviere J. M. and Prat S (2015). ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr. Biol. 25, 187-193. 10.1016/j.cub.2014.10.070 [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.-Y. and Wang Z.-Y (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802-809. 10.1038/ncb2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I., Chen F., Ngoc Pham V., Zhu L., Kim J.-I. and Huq E (2019). A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 10, 4216 10.1038/s41467-019-12110-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I., Kathare P. K., Kim J.-I. and Huq E (2017). Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10, 1035-1046. 10.1016/j.molp.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.-J., Kim J. Y., Lee J.-H., Lee B.-D., Paek N.-C. and Park C.-M (2020). GIGANTEA shapes the photoperiodic rhythms of thermomorphogenic growth in Arabidopsis. Mol. Plant 13, 459-470. 10.1016/j.molp.2020.01.003 [DOI] [PubMed] [Google Scholar]
- Park Y. J., Lee H. J., Ha J. H., Kim J. Y. and Park C. M (2017). COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol. 215, 269-280. 10.1111/nph.14581 [DOI] [PubMed] [Google Scholar]
- Pham V. N., Kathare P. K. and Huq E (2018). Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025-1038. 10.1104/pp.17.01384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Li M., Kim R. J.-A., Moore C. M. and Chen M (2019). Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA. Nat. Commun. 10, 140 10.1038/s41467-018-08059-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M., Delker C., Franklin K. A., Wigge P. A., Halliday K. J. and van Zanten M (2016). Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2, 15190 10.1038/nplants.2015.190 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. and Smyth G. K (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin D. J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.-D., Huq E. and Hiltbrunner A. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27, 189-201. 10.1105/tpc.114.134775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang J. A., Gallego-Bartolomé J., Gómez M. D., Yoshida S., Asami T., Olsen J. E., García-Martínez J. L., Alabadí D. and Blázquez M. A (2009). Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 60, 589-601. 10.1111/j.1365-313X.2009.03983.x [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L. and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562-578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Bu Q., Huang X., Deng X. W. and Huq E (2014). PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of constitutive photomorphogenic1 to synergistically repress Photomorphogenesis in Arabidopsis. Plant Cell 26, 1992-2006. 10.1105/tpc.114.125591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Xun Q., Zhang D., Lv M., Ou Y. and Li J (2019). TCP transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate Thermomorphogenesis in Arabidopsis thaliana. iScience 15, 600-610. 10.1016/j.isci.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. and Huq E (2014). Suicidal co-degradation of the Phytochrome Interacting Factor 3 and phytochrome B in response to light. Mol. Plant 7, 1709-1711. 10.1093/mp/ssu108 [DOI] [PubMed] [Google Scholar]
- Zhu J. Y., Oh E., Wang T. and Wang Z.-Y (2016). TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 7, 13692 10.1038/ncomms13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.