Summary
Centrosome reduction and redistribution of pericentriolar material (PCM) coincides with cardiomyocyte transitions to a post-mitotic and matured state. However, it is unclear whether centrosome changes are a cause or consequence of terminal differentiation. We validated that centrosomes were intact and functional in proliferative human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs), consistent with their immature phenotype. We generated acentrosomal hPSC-CMs, through pharmacological inhibition of centriole duplication, and showed that centrosome loss was sufficient to promote post-mitotic transitions and aspects of cardiomyocyte maturation. As Hippo kinases are activated during post-natal cardiac maturation, we pharmacologically activated the Hippo pathway using C19, which was sufficient to trigger centrosome disassembly and relocalization of PCM components to perinuclear membranes. This was due to specific activation of Hippo kinases, as direct inhibition of YAP-TEAD interactions with verteporfin had no effect on centrosome organization. This suggests that Hippo kinase-centrosome remodeling may play a direct role in cardiac maturation.
Key words: cardiomyocytes, centrosomes, microtubules, Hippo kinase, maturation, polo-like kinase 4
Highlights
-
•
Centrosomes are intact and functional in immature human cardiomyocytes
-
•
Centrosome loss promotes maturation of human cardiomyocytes
-
•
Centrosomes are returned with cell cycle reinitiation in post-natal cardiomyocytes
-
•
Hippo kinases promote disassembly and redistribution of centrosomal proteins
In this article, Ng, Hudson, and colleagues demonstrated that the removal of centrosomes, through blocking centriole duplication, was sufficient to trigger post-mitotic transitions and differentiation of immature human cardiomyocytes. They also revealed that Hippo kinase activation triggered redistribution of centrosomal proteins to acentrosomal sites. This suggests that the regulated disassembly of centrosomes by Hippo kinases may promote cardiomyocyte maturation.
Introduction
The adult mammalian heart is a post-mitotic organ, with cardiomyocytes (CM) transitioning to a post-mitotic state during late embryonic development or shortly after birth (Porrello et al., 2011; Zhu et al., 2018). This transition occurs as part of a broader process of cardiac maturation, which includes alterations to cardiac metabolism, gene expression, electrophysiology, and cytoskeletal organization and coincides with the loss of regenerative potential (Porrello and Olson, 2014). The regulatory mechanisms that determine post-mitotic transition and lost proliferative potential of CMs may provide important insights into how cardiac regeneration can be re-engaged in adult tissue, specifically during disease contexts, and are thus of significant interest. Structural changes in centrosomes, non-membrane bound organelles with central functions in cytoskeletal organization, and signal transduction have been reported to coincide with the post-mitotic transition of mammalian CMs (Zebrowski et al., 2015).
The prototypical centrosome consists of two barrel-like centrioles that are linked via a proteinaceous tether and embedded in a dense pericentriolar matrix (PCM) (Nigg and Stearns, 2011). Centrosomes control cell morphology, division, polarity, and differentiation (Vertii et al., 2016). Moreover, the structure and protein constituents of the centrosome are highly dynamic and tightly regulated, which is necessary to prevent untimely duplication and to facilitate cell-cycle stage-specific formation of the primary cilium or the bipolar spindle (Gupta et al., 2015). In myocytes, centrosomes undergo a process of structural and functional reduction that is co-ordinated with skeletal myogenesis or terminal differentiation in the cardiac setting (Srsen et al., 2009; Zebrowski et al., 2015). This reflects changes in centrosome organization associated with differentiation in other cell types, with PCM and centriole disassembly to various degrees reported (Avidor-Reiss et al., 2015).
The process of centrosome reduction broadly encompasses (1) the breakdown and redistribution of the PCM resulting in lost microtubule nucleation and anchoring capacity, (2) the loss of centriole cohesion resulting in centrosome splitting, and (3) the partial to complete degradation of the centriole barrels (Manandhar et al., 2005; Zebrowski et al., 2015). During the maturation of rodent CMs, during late fetal and early post-natal stages, the PCM relocates from the centrioles to the nuclear membrane (Zebrowski et al., 2015). In addition, there is an increase in irreversible centriole splitting and loss of centrosome cohesion (Zebrowski et al., 2015). This coincides with a decrease in proliferation as the heart switches from hyperplasia to hypertrophic growth (Zebrowski et al., 2015). However, it remains undetermined whether the process is recapitulated in human CMs and if centrosome reduction is sufficient to trigger terminal differentiation. In this study, we utilized pharmacological targeting of centriole biogenesis in human pluripotent stem cell-derived CMs (hPSC-CMs), which are fetal-like and proliferative in nature, to demonstrate that centrosome loss is sufficient to trigger transition to a post-mitotic state and induce cytoskeletal and morphological characteristics of mature CMs. In addition, we demonstrate that Hippo pathway activation recapitulates features of centrosome reduction and post-mitotic transitions in hPSC-CMs. Our study indicates activation of Hippo kinases as a key regulatory mechanism required for the reorganization of centrosomes to induce human CM maturation.
Results
Functional Centrosomes in Proliferating Human CMs
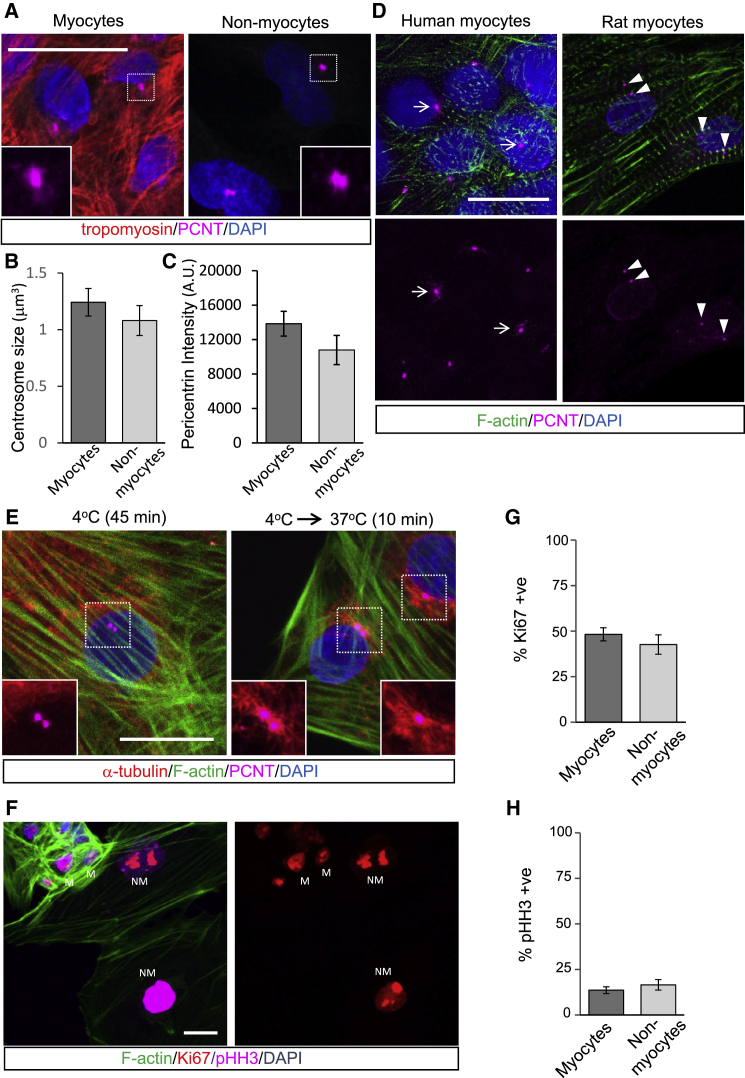
Neonatal rat ventricular CMs undergo centrosome reduction that coincides with lost proliferation capacity and terminal differentiation (Zebrowski et al., 2015). We reasoned that if this process were conserved in human CMs, the centrosomes of hPSC-CMs, validated to be fetal-like in nature by proteomic and transcriptomic analyses (Mills et al., 2017), would represent the primary microtubule-organizing center by nucleating and anchoring microtubules. hPSC-CMs were generated using previously published methods (Mills et al., 2017) and immunostained for pericentrin (PCNT) and tropomyosin as markers for the PCM and myocyte sarcomeres, respectively. The size and integrity of the PCM in hPSC-CMs, as indicated by volume and intensity of PCNT fluorescence, were comparable to those of non-myocytes within those cultures (Figures 1A–1C). In comparison to reduced PCM and split centrioles in post-natal rat CMs, centrosomes in hPSC-CMs exhibit intact centrosomes with expanded PCM (Figure 1D). To confirm that centrosomes in hPSC-CMs were functional, we depolymerized the microtubule array at 4°C and monitored regrowth of microtubules at centrosomes at a permissive temperature. Shortly upon return to 37°C we observed microtubules emanating from the PCM of hPSC-CMs and also from perinuclear regions near centrosomes (Figure 1E). This indicated that hPSC-CM centrosomes were functional and retained the capacity to nucleate and anchor microtubules. It is also likely that microtubules may be nucleated from Golgi or nuclear membranes. The morphology and functional capacity of centrosomes in hPSC-CMs are consistent with the proliferative capacity of immature CMs, which is comparable to that of non-myocytes in co-culture as indicated by staining with proliferation (Ki67) and mitotic (pHH3) markers (Figures 1F–1H). Thus, centrosomes in hPSC-CMs have yet to undergo a process of reduction, as they are morphologically and functionally indistinguishable from non-myocytes and this coincides with retained proliferative capacity.
Figure 1.
Immature hPSC-CMs Have Intact and Functional Centrosomes
F-actin and tropomyosin staining revealed myocytes within our hPSC-CM cultures.
(A) Immunofluorescence staining of centrosome morphology in CMs and non-myocytes in our hPSC-CM cultures.
(B and C) Centrosome size was quantified by measuring (B) volume and (C) intensity of pericentrin (PCNT) fluorescence.
(D) Centrosome morphology in hPSC-CMs compared with neonatal rat ventricular CMs. Arrows indicate intact centrosomes. Arrowheads show split/separated centroosmes.
(E) Microtubule regrowth assays indicated the capacity of hPSC-CMs to nucleate new microtubules at the PCM following shift from 4°C to 37°C.
(F) Immunofluorescence staining of hPSC-CMs for proliferative (Ki67) and mitotic (phospho-histone H3, pHH3) markers. M, myocytes, and NM, non-myocytes in culture differentiated by actin staining (phalloidin).
(G and H) Quantitation indicates CMs are actively cycling and dividing at levels comparable to those of non-myocytes. (G) Percentage Ki67 positive. (H) Percentage pHH3 positive.
Error bars represent SEM. Scale bars, 20 μm.
Centrosome Loss Is Sufficient to Induce Post-mitotic Transitions in hPSC-CMs
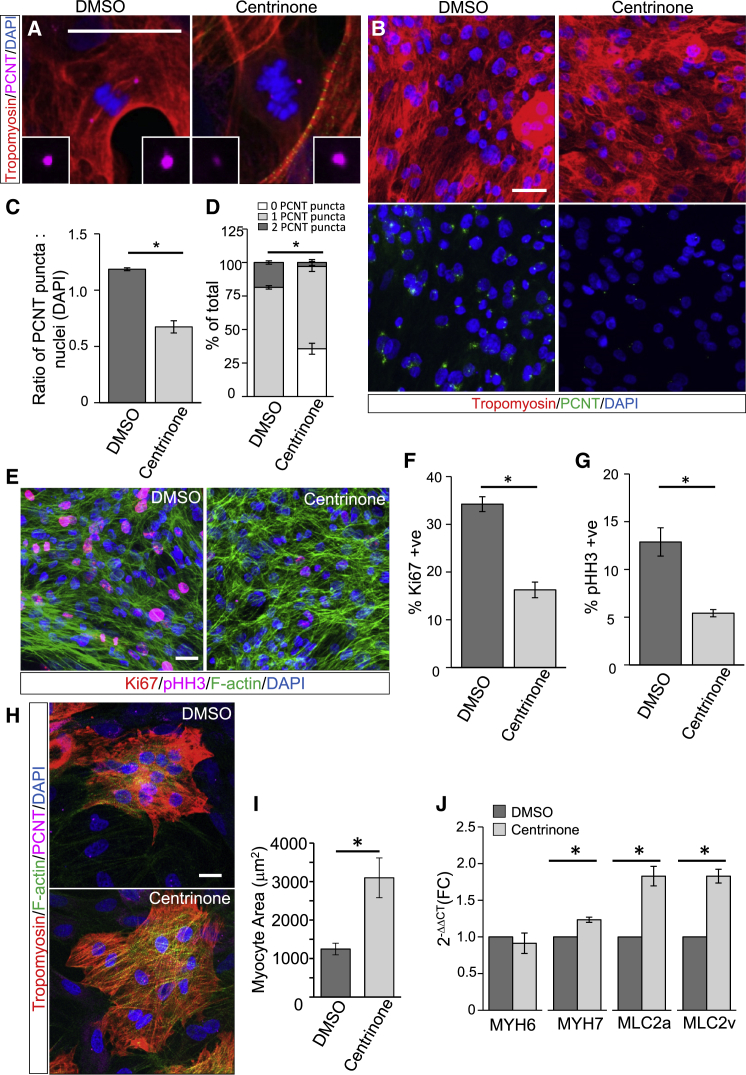
While the disassembly of centrosomes coincides with maturation of rodent CMs (Zebrowski et al., 2015), it is unclear if centrosome reduction is sufficient to induce CM terminal differentiation. To specifically target centrosomes, we utilized a highly selective and reversible small-molecule inhibitor of PLK4, a master regulatory centrosomal kinase that is essential for centriole duplication (Arquint and Nigg, 2016). Centrinone treatment results in centrosome loss in daughter cells following mitosis due to blocked centriole duplication (Wong et al., 2015). Centrinone treatment of hPSC-CMs resulted in mitotic myocytes with asymmetric inheritance of intact or reduced centrosomes into daughters (Figure 2A), leading to an overall decrease in cells with intact centrosomes (Figure 2B). An extended (3 day) exposure to centrinone resulted in a significant increase in acentrosomal CMs as indicated by a reduction in centrosomes-to-nuclei ratios and increased percentage of CMs with no PCNT staining (Figures 2C and 2D).
Figure 2.
Centrosome Loss Induces Post-mitotic Transition and Maturation of hPSC-CMs
(A) hPSC-CMs were treated with centrinone (0.5 μM, 72 h) to generate acentrosomal myocytes or a vehicle (DMSO) control. Insets show intact or centrosome remnants in mitotic myocytes inherited by daughter cells.
(B) Reduced PCNT staining in centrinone-treated hPSC-CMs.
(C) Reduced PCNT puncta-to-nuclei (DAPI) ratio in centrinone-treated hPSC-CMs.
(D) Increased percentage of CMs with <2 PCNT puncta following centrinone treatment.
(E) Decreased Ki67 and pHH3 staining in centrinone-treated hPSC-CMs.
(F and G) Centrinone-treatment reduced the percentages of (F) Ki67+ve and (G) pHH3+ve hPSC-CMs.
(H) Increased hPSC-CM size following centrinone treatment.
(I) Increased surface area (μm2) of centrinone-treated hPSC-CMs. See also Figure S1F.
(J) Centrinone treatment increased mRNA expression of sarcomeric proteins associated with CM maturation.
Error bars represent SEM. ∗p < 0.05. Scale bars, 20 μm.
Following centrinone exposure, we investigated the effect of centrosome loss on hPSC-CM proliferation by immunostaining for Ki67 and pHH3 (Figures 2E–2G). We observed a significant reduction in the proliferative capacity (Ki67+ve) of centrinone-treated CMs compared with vehicle-treated controls (Figures 2E and 2F). Similarly, the percentage of mitotic (pHH3+ve) hPSC-CMs was substantially reduced with centrinone treatment (Figures 2E and 2G). We repeated these studies using human CMs generated from induced pluripotent stem cells (iPSC-CMs) as an independent stem cell line. Centrinone treatment resulted in an increase in the proportion of acentrosomal myocytes (Figures S1A and S1B) and reduced the percentage of iPSC-CMs positively stained for Ki67 and pHH3 (Figures S1A–S1E). This indicates that the targeted loss of centrosomes is sufficient to trigger post-mitotic transition of immature human CMs.
To evaluate whether centrosome loss was sufficient to induce additional features of mature hPSC-CMs, we determined the impact of centrinone on hypertrophic responses and expression of maturation markers. We found that centrinone treatment increased the surface area of hPSC-CMs stained with tropomyosin and F-actin to mark myocytes and identify cell boundaries, respectively, which is indicative of hypertrophic growth (Figures 2H and S1F). Automated quantification indicated an ∼3-fold increase in cell area in response to centrinone treatment (Figure 2I). To determine if hPSC-CM hypertrophy coincided with maturation, we measured the expression of specific myosin heavy- and light-chain isoforms (MYH7, MLC2a, and MLC2v) as markers of CM maturation and found that mRNA levels were significantly increased following treatment with centrinone compared with a DMSO control (Figure 2J). Similarly, centrinone increased expression of mature sarcomeric markers in iPSC-CMs (Figure S1G). This shows that centrosome reduction with centrinone treatment was sufficient to recapitulate molecular and morphological aspects of CM maturation.
Our studies indicate that centrosome loss promotes maturation of human CMs in 2D culture. To evaluate centrosomes in human cardiac organoids (hCO) in 3D culture, we generated hCOs from hPSC-CMs as previously described (Mills et al., 2017). The hCO culture and subsequent incubation in medium optimized to promote maturation (MM) resulted in reduced proliferation and cell-cycle arrest as previously defined (Mills et al., 2017) (Figure S2A). When we investigated the morphology of centrosomes in hCOs, we observed that centrosome numbers (measured as a ratio of PCNT+ve puncta to nuclei) and size were not substantially altered by these culture conditions (Figures S2B–S2D). In support, mRNA expression profiles of centrosome components in hCOs cultured in MM were not substantially different from those of control conditions (Figure S2E). Moreover, the expression of centrosome components in MM hCOs was more similar to that of fetal human cardiac tissue and immature (20 days old) hPSC-CMs (Figure S3E). In contrast, the mRNAs encoding centrosome-associated scaffold proteins and kinases (including PLK4) were downregulated in adult human heart (Figure S2E). Although differences in cellular composition may account for differences in centrosomal protein expression in hCOs compared with adult ventricular tissues, our imaging analysis of centrosomes in NKX2.5+ve myocytes in hCOs revealed centrosomes with intact PCM (Figures S2F–S2H). This raises the intriguing question of whether centrosome targeting could promote the further maturation of hCOs. However, we found that centrinone treatment had limited effects on centrosome number or size in hCOs (Figures S2F–S2H). As cell division generates acentrosomal cells from PLK4 inhibition, we attributed the limited centrosome impact to hPSC-CM proliferation in 3D culture at the time of centrinone administration. Thus, an alternative approach to selective targeting of centrosomes is required to reveal their contribution to hCO maturation.
Pharmacologically Enhanced Proliferation of Rat Ventricular CMs Coincides with the Return of Centrosomes
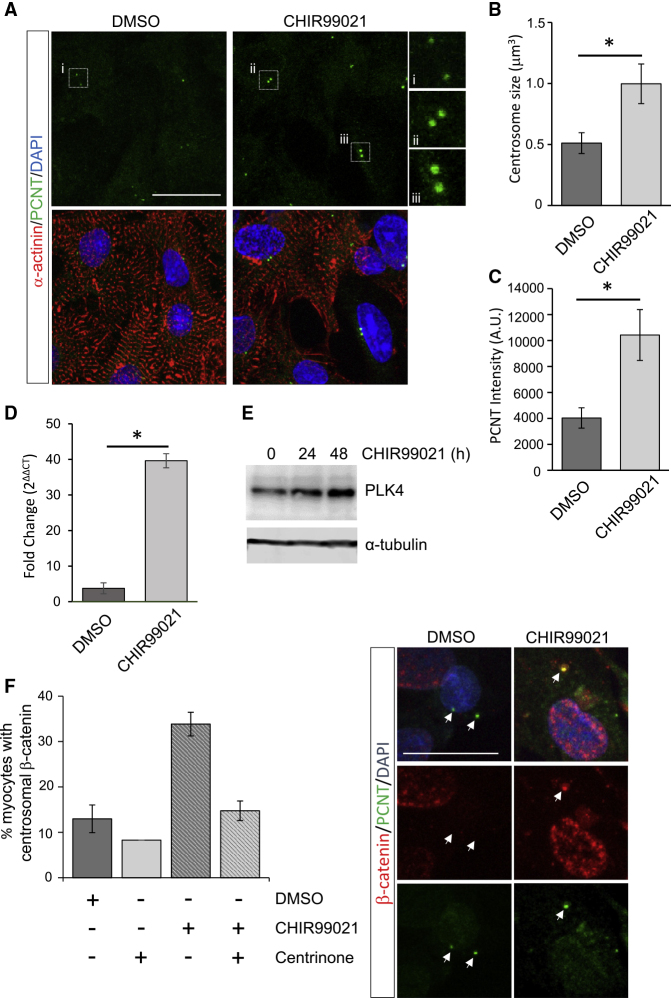
The post-mitotic state of post-natal CMs can be reversed to some degree by manipulation of molecular pathways involved in cell cycle or redox regulation (Cheng et al., 2011; Matrone et al., 2017). Specifically, inhibition of GSK-3β and subsequent β-catenin stabilization has been shown to increase proliferation of post-natal rat ventricular CMs (Woulfe et al., 2010). We confirmed that treatment with a GSK-3β inhibitor, CHIR99021, significantly increased the proliferation (Ki67) and mitotic index (phospho-histone H3) of primary-isolated 3 day old rat ventricular CMs (Figures S3A–S3C). The increased CM proliferation coincided with β-catenin stabilization and nuclear localization (Figure S3D). In response to CHIR99021 treatment, we found that a proportion of rat ventricular CMs (α-actinin+ve) presented centrosomes with increased PCM size and resembled centrosomes of proliferating non-myocytes (Figure 3A). A quantitative analysis indicated that centrosome size and PCM staining intensity were substantially increased following CHIR99021 treatment compared with DMSO (Figures 3B and 3C). Centrosome return following CHIR99021 was also accompanied by increased PLK4 levels (Figures 3D and 3E). This indicated that re-established organization of centrosomes may be mediated by increased PLK4 and coincides with increased CM proliferation.
Figure 3.
GSK-3 Inhibition Returns Centrosome Integrity to Post-natal Rat CMs
(A) Post-mitotic 3 day old rat CMs were treated with CHIR99021 (5 μM, 24 h) or a vehicle control and immunostained for PCNT and α-actinin.
(B and C) (B) Quantitation of centrosome volume or (C) PCNT intensity revealed significant increases in centrosome size following CHIR99021 treatment.
(D and E) (D) CHIR99021 increased PLK4 mRNA and (E) protein levels in 3 day old rat CMs.
(F) Immunofluorescence staining of β-catenin at centrosomes in post-natal rat cardiomyocytes is increased with CHIR99021 (5 μM, 24 h) treatment. Centrinone (0.5 μM) co-treatment prevented CHIR99021-induced increased β-catenin at centrosomes. Arrows indicate centrosome position.
Error bars represent SEM. ∗p < 0.05. Scale bars, 20 μm. See also Figure S3.
A distinct centrosomal pool of β-catenin is known to regulate centrosome organization independent of transcriptional activity in non-cardiac contexts (Bahmanyar et al., 2010; Yang et al., 2018). Here we show that, in addition to increased nuclear levels, CHIR99021 treatment increased β-catenin localization to centrosomes (Figure 3F) in post-natal rat ventricular CMs. Notably, CHIR99021-mediated increase in centrosomal β-catenin, but not nuclear β-catenin, was blocked with PLK4 activity inhibition (Figures 3F and S3D). Our findings suggest that, in response to GSK-3β inhibition, the stabilization and increased levels of centrosomal β-catenin require PLK4 and may contribute to the return of functional centrosomes required for CM proliferation.
Hippo Kinase Activation Promotes Centrosome Reduction in Human CMs
The endogenous process of centrosome reduction involves PCM redistribution to perinuclear sites (Zebrowski et al., 2015). Although our findings with PLK4 targeting indicate that centrosome loss was sufficient to induce CM maturation, this approach does not recapitulate the acentrosomal redistribution of PCM components. In exploring potential pathways involved in endogenous centrosome disassembly, we investigated the Hippo kinases, MST and LATS, that have been reported to localize to centrosomes and have established functions regulating CM proliferation (Bolgioni and Ganem, 2016; Wackerhage et al., 2014). Hippo kinase activity is dramatically increased post-natally, as well as in heart failure patients, and could potentially trigger centrosomal loss during maturation (Leach et al., 2017).
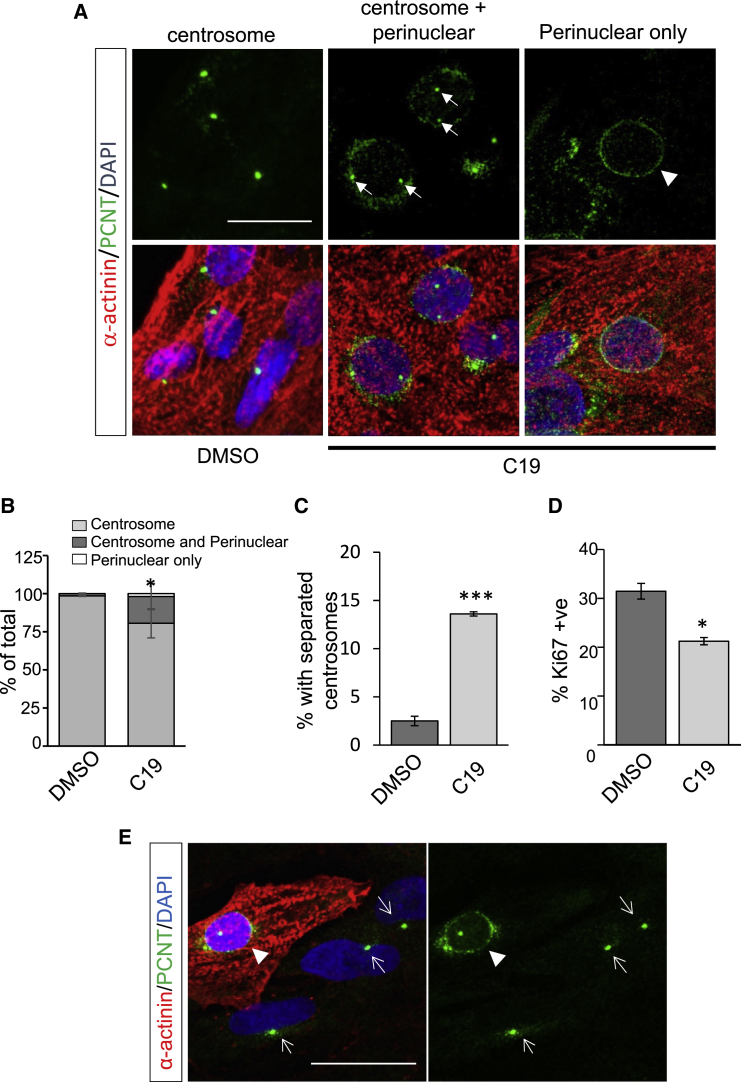
We find that treatment with the C19 compound, previously shown to activate MST/LATS (Basu et al., 2014), resulted in PCNT redistribution from centrosomes to perinuclear regions in a subset of hPSC-CMs (Figures 4A and 4B). A significant proportion (∼20%) of C19-treated CMs exhibited both centrosome and perinuclear PCNT localization, while a smaller proportion (∼4%) of CMs exhibited PCNT exclusively at the perinuclear region, reminiscent of mature myocytes (Figure 4B). In addition, following C19 treatment, we observed a significant increase in hPSC-CMs with split centrosomes (Figure 4C), indicative of precocious centriole disjunction and reduced centrosome cohesion (Mardin et al., 2010; Zebrowski et al., 2015). This coincided with a decrease in proliferation (Figure 4D). C19-mediated redistribution of PCNT to acentrosomal sites was observed only in α-actinin+ve cells and never in α-actinin−ve non-myocytes (Figure 4E), reinforcing centrosome remodeling as a myocyte-specific occurrence. Interestingly, the redistribution of PCNT to acentrosomal sites was not observed following treatment with verteporfin, a benzoporphyrin derivative that directly inhibits YAP function by disrupting YAP-TEAD interactions (Figures S4A and S4B) (Liu-Chittenden et al., 2012). In addition, the proportion of hPSC-CMs with split centrosomes and PCNT intensity at the PCM was unchanged with verteporfin treatment (Figures S4C and S4D). This suggests that centrosome remodeling is independent of YAP-TEAD-regulated transcription and may represent an additional response to Hippo kinase activation. Our results suggest that the activation of Hippo kinases MST/LATS leads to centrosome disassembly and redistribution of PCM to acentrosomal sites, which is associated with CM maturation.
Figure 4.
MST1 Induces Centrosome Reduction in Human CMs
(A) C19 (10 μM, 48 h) treatment induced translocation of PCNT to a perinuclear region (arrowhead) in hPSC-CMs. C19 treatment also resulted in split centrosomes, indicative of centriole separation (arrows).
(B) Proportion of hPSC-CMs with perinuclear PCNT in response to C19 treatment.
(C) Proportion of C19-treated hPSC-CMs with split centrosomes.
(D) Reduced proliferation of C19-treated hPSC-CMs.
(E) The relocalization of PCNT to perinuclear regions in response to C19 was observed in α-actinin+ve myocytes (arrowhead) and not in non-myocytes (α-actinin−ve, arrows).
Error bars represent SEM. ∗p < 0.05, ∗∗∗p < 0.001. Scale bars, 20 μm. See also Figure S4.
Discussion
During the organization of microtubules, the transition from centrosomal to acentrosomal juxtanuclear sites is characteristic of muscle development (Muroyama and Lechler, 2017). In skeletal muscle, centrosome reduction and release of PCM components occur early during myoblast differentiation and prior to fusion into multinucleated myotubes (Tassin et al., 1985). In cardiac muscle, centrosomes remain functional in proliferating contractile myocytes but are disassembled during cardiac maturation at late gestational or neonatal stages (Zebrowski et al., 2015). In both cases, centrosome disassembly coincides with a transition to a post-mitotic state. In this study we investigated centrosome contribution to the terminal differentiation of myocytes. We utilized pharmacological targeting of PLK4 to generate acentrosomal human CMs and found that this resulted in post-mitotic transitions, increased CM area, and the expression of matured sarcomeric proteins. This suggests that centrosome loss was sufficient to promote molecular and morphological aspects of CM maturation. The 3D culture of hCOs in defined medium similarly promotes maturation (Mills et al., 2017). Interestingly, we find here that CM centrosomes remained largely intact in hCOs. This indicates that centrosome reduction was not necessarily required to initiate maturation, but raises the intriguing possibility that centrosome targeting may augment the process or trigger irreversible transitions. Centrosomes integrate signal transduction and cytoskeletal elements to regulate multiple cellular processes, including ciliogenesis, intracellular trafficking, cell-cycle progression, cell shape, and organelle positioning (Nigg and Raff, 2009). For example, in skeletal myotubes, PCM components are redistributed to nuclear membranes for nuclear positioning and normal skeletal muscle development (Espigat-Georger et al., 2016; Gimpel et al., 2017). It is likely that several of the processes described above would be altered with centrosome loss and may variously contribute to CM maturation. The specific molecular and cellular consequences of centrosome loss involved in CM maturation remain to be fully defined.
In our studies we targeted PLK4 with a highly specific inhibitor (centrinone) to generate acentrosomal cells (Wong et al., 2015). PLK4 is restricted to centrosomes, and the described functions of PLK4 are principally in mediating centriole duplication and centriole assembly (Arquint and Nigg, 2016; Galletta et al., 2016). Therefore, our reported CM maturation effects in response to PLK4 inhibition would logically be the result of lost centrosomes. However, it remains a possibility that inhibition of unidentified centrosome-independent functions of PLK4 may contribute to CM differentiation. The loss of centrosomes in hPSC-CMs also appeared to be irreversible, as drug washout following prolonged centrinone treatment (5 days) did not increase or return centrosome numbers (data not shown). In contrast, GSK-3β inhibition in post-natal rat CMs demonstrated that centrosome reduction could be reversed to coincide with reinitiated proliferation. These findings likely reflect the difference seen in the complete loss of centrioles in response to prolonged centrinone treatment compared with post-natal myocytes that have undergone an endogenous process of centrosome reduction but retained centrioles in a seemingly non-functional state. Centrosomes undergo an analogous process of reduction during animal and insect sperm formation with PCM removal and disassembly of the distal centriole to result in an atypical structure initially thought dispensable (Fishman et al., 2018; Khire et al., 2016). Recent studies, however, have revealed that sperm atypical centrioles are functional, required for fertilization, and seemingly reassembled to facilitate zygotic cell divisions (Fishman et al., 2018). This suggests that the reduced/remnant centrosomes in post-natal myocytes may retain functionality and raises the intriguing notion that post-natal myocytes may be enticed to re-enter the cell cycle with centrosome reassembly. Future studies will determine if the capacity to reassemble centrosomes is progressively lost as CMs age.
The mechanisms underlying centrosome remodeling during muscle development remain mysterious. Here we report that pharmacological activation of Hippo kinases MST1/2 and LATS1/2 decreased hPSC-CM proliferation and recapitulated the perinuclear redistribution of PCM proteins typical of mature CMs. The mammalian STE20-like protein kinases (MST1/2) and large tumor suppressor kinases (LATS1/2) are core components of the Hippo pathway that has important functions in development and human pathologies, including heart failure (Harvey et al., 2013; Irvine and Harvey, 2015; Leach et al., 2017). MST1/2 and its scaffold protein, Salvador, promotes the activation of LATS1/2, which subsequently phosphorylates Yes-associated proteins (YAP/TAZ) to promote their nuclear exclusion. YAP/TAZ interacts with TEAD transcriptional repressors in the nucleus to derepress target genes that promote growth and proliferation and attenuate apoptosis (Irvine and Harvey, 2015). Thus, Hippo signaling reduces proliferation and is tumor suppressive. In skeletal and cardiac tissues, MST/LATS activation, and subsequent inhibition of YAP/TAZ, promotes differentiation and reduced proliferation (Wackerhage et al., 2014). Moreover, the conditional deletion of Salvador or LATS or overexpression of constitutively active YAP promotes CM proliferation, while YAP knockout promotes apoptosis (Leach et al., 2017; Monroe et al., 2019; Wackerhage et al., 2014). Thus, our findings are consistent with a regulatory role for Hippo signaling in promoting CM terminal differentiation.
Our findings raise the question as to how activation of Hippo kinases promotes centrosome disassembly. The direct targeting of YAP-TEAD interactions with verteporfin did not trigger equivalent centrosome remodeling, which suggests that centrosomal effects of MST/LATS may not involve downstream components of the canonical Hippo pathway. In contrast, several studies suggest that MST and LATS may exert their actions on centrosomal components more directly. MST1/2 have been reported to regulate centriole duplication (Hergovich et al., 2009). In addition, LATS1/2 are known to be localized to centrosomes in an Aurora-A-dependent manner to regulate spindle organization and mitosis (Abe et al., 2006; Toji et al., 2004). Importantly, MST1/2 interactions with the centrosomal kinase NEK2 are involved in the phosphorylation of CGNAP and rootletin, which are required for centrosome cohesion (Mardin et al., 2010). This triggers centrosome disjunction, the detachment of mother and daughter centrioles, at the onset of mitosis (Mardin et al., 2010). Interestingly, centrosome disjunction and splitting of centrosomes are similarly observed as CMs mature (Zebrowski et al., 2015). This suggests that analogous mechanisms, mediated by Hippo pathway kinases, may be at play in the remodeling of centrosomes in CMs. In summary, our studies indicate that centrosome reduction is not simply a consequence of terminal differentiation, but rather centrosome remodeling may actively promote their morphological and functional maturation.
Experimental Procedures
Human CM Differentiation
Ethical approval for the use of human embryonic stem cells was obtained from The University of Queensland's Medical Research and QIMR Berghofer's Ethics Committee (2014000801 and P2385) and experiments were carried out in accordance with the National Health and Medical Research Council regulations. Human CMs were differentiated from embryonic hPSCs or iPSCs according to previously described protocols (Voges et al., 2017). The human iPSC line without known cardiovascular disease (CW30382A) was obtained from the California Institute of Regenerative Medicine hPSC Repository through FUJIFILM. Briefly, HES3 hPSCs (WiCell) and iPSCs were differentiated in medium (RPMI, 100 units/mL penicillin-streptomycin, 200 mM ascorbic acid, B27 supplement) supplemented with 9 ng/mL Activin A, 5 ng/mL BMP, 5 ng/mL bFGF, and 1 μM CHIR99021 for 3 days followed by cardiac fate specification for 3 days in B27 medium (minus insulin) containing 5 μM Wnt inhibitor, IWP4, and a further 7 days in B27 medium (with insulin) containing 5 μM IWP4. hPSC-CMs and iPSC-CMs were maintained on coated glass coverslips in αMEM supplemented with GlutaMAX, 10% (v/v) fetal bovine serum, and 100 units/mL penicillin/streptomycin. hCOs were fabricated and analyzed as previously described (Mills et al., 2017).
Immunofluorescence
At the completion of cell treatments, coverslips were washed in cold PBS and fixed in pre-chilled MeOH (−20°C) for 3 min or 4% (w/v) paraformaldehyde for 20 min as appropriate. Cells were permeabilized with 0.2 (v/v) Triton X-100 in PBS and blocked with 10% (v/v) fetal calf serum in PBS. Primary antibody staining was performed by diluting antibodies in 1% (w/v) BSA in PBS followed by Alexa-conjugated secondary antibodies diluted in 1% BSA in PBS. Antibodies used included anti-PCNT (Abcam, ab4448), α-actinin (Sigma, A7811), γ-tubulin (Sigma, T5326), Ki67 (BD Bioscience, 550609), pHH3 (Abcam, ab47297), tropomyosin (Sigma, T2780), and β-catenin (CST, 9582). Co-staining with phalloidin conjugated to Alexa 488 was performed concurrent with secondary antibodies. Coverslips were finally stained with DAPI, mounted in ProLong Diamond Antifade mountant, and imaged on a confocal microscope (Leica TCS SP8) using a 63×/1.3 NA glycerol or 100×/1.4 NA oil immersion objective. Images were collected using LAS X and minimally processed with ImageJ and Adobe Photoshop. Unless indicated otherwise, fluorescence images are maximum intensity projections of confocal z-stack images and inset images within a panel utilize the same magnification. Image analyses and quantifications were performed using Imaris and Metamorph.
Statistical Analysis
Statistics were calculated with GraphPad Prism 7. Immunofluorescence and qRT-PCR values were analyzed using a t test with Welch correction or one/two-way ANOVA followed by a post hoc multiple comparisons t test with Tukey correction. Mean values from n = 3 experimental replicates (>150 total cells from a minimum of eight randomly selected images/condition/experimental replicate) are depicted.
Additional methodological details can be found in the Supplemental Information.
Author Contributions
Conceptualization, D.C.H.N., D.K.R., and J.E.H.; Investigation, D.K.R., R.J.M., H.L.P., U.Y.H., H.K.V., and C.R.T.; Data Analysis, D.C.H.N., D.K.R., H.L.P., R.J.M., U.Y.H., C.R.T., J.K.P., and J.E.H.; Writing – Original Draft, D.C.H.N. and D.K.R.; Writing – Review & Editing, D.C.H.N., J.K.P., and J.E.H.; Funding Acquisition, Resources & Supervision, D.C.H.N., J.K.P., and J.E.H.
Conflict of Interests
R.J.M. and J.E.H. are listed as co-inventors on pending patents held by The University of Queensland and QIMR Berghofer Medical Research Institute that relate to cardiac organoid maturation and putative cardiac regeneration therapeutics. J.E.H. is a co-inventor on licensed patents held by the University of Goettingen. R.J.M. and J.E.H. are co-founders, scientific advisors, and stockholders of Dynomics.
Acknowledgments
D.N. acknowledges funding support from the National Health and Medical Research Council (GNT1046032, GNT1162652), Australian Research Council (FT120100193), and Cancer Council (GNT1101931).
Published: September 17, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.08.007.
Supplemental Information
References
- Abe Y., Ohsugi M., Haraguchi K., Fujimoto J., Yamamoto T. LATS2-Ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 2006;580:782–788. doi: 10.1016/j.febslet.2005.12.096. [DOI] [PubMed] [Google Scholar]
- Arquint C., Nigg E.A. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem. Soc. Trans. 2016;44:1253–1263. doi: 10.1042/BST20160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T., Khire A., Fishman E.L., Jo K.H. Atypical centrioles during sexual reproduction. Front. Cell Dev. Biol. 2015;3:21. doi: 10.3389/fcell.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar S., Guiney E.L., Hatch E.M., Nelson W.J., Barth A.I. Formation of extra centrosomal structures is dependent on beta-catenin. J. Cell Sci. 2010;123(Pt 18):3125–3135. doi: 10.1242/jcs.064782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D., Lettan R., Damodaran K., Strellec S., Reyes-Mugica M., Rebbaa A. Identification, mechanism of action, and antitumor activity of a small molecule inhibitor of hippo, TGF-β, and Wnt signaling pathways. Mol. Cancer Ther. 2014;13:1457–1467. doi: 10.1158/1535-7163.MCT-13-0918. [DOI] [PubMed] [Google Scholar]
- Bolgioni A.F., Ganem N.J. The interplay between centrosomes and the Hippo tumor suppressor pathway. Chromosome Res. 2016;24:93–104. doi: 10.1007/s10577-015-9502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Woodgett J., Maamari M., Force T. Targeting GSK-3 family members in the heart: a very sharp double-edged sword. J. Mol. Cell. Cardiol. 2011;51:607–613. doi: 10.1016/j.yjmcc.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espigat-Georger A., Dyachuk V., Chemin C., Emorine L., Merdes A. Nuclear alignment in myotubes requires centrosome proteins recruited by nesprin-1. J. Cell Sci. 2016;129:4227–4237. doi: 10.1242/jcs.191767. [DOI] [PubMed] [Google Scholar]
- Fishman E.L., Jo K., Nguyen Q.P.H., Kong D., Royfman R., Cekic A.R., Khanal S., Miller A.L., Simerly C., Schatten G. A novel atypical sperm centriole is functional during human fertilization. Nat. Commun. 2018;9:2210. doi: 10.1038/s41467-018-04678-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta B.J., Fagerstrom C.J., Schoborg T.A., McLamarrah T.A., Ryniawec J.M., Buster D.W., Slep K.C., Rogers G.C., Rusan N.M. A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat. Commun. 2016;7:12476. doi: 10.1038/ncomms12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpel P., Lee Y.L., Sobota R.M., Calvi A., Koullourou V., Patel R., Mamchaoui K., Nédélec F., Shackleton S., Schmoranzer J. Nesprin-1α-dependent microtubule nucleation from the nuclear envelope via Akap450 is necessary for nuclear positioning in muscle cells. Curr. Biol. 2017;27:2999–3009. doi: 10.1016/j.cub.2017.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G.D., Coyaud E., Goncalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Cheung S.W. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Hergovich A., Kohler R.S., Schmitz D., Vichalkovski A., Cornils H., Hemmings B.A. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr. Biol. 2009;19:1692–1702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Irvine K.D., Harvey K.F. Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 2015;7:a019224. doi: 10.1101/cshperspect.a019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khire A., Jo K.H., Kong D., Akhshi T., Blachon S., Cekic A.R., Hynek S., Ha A., Loncarek J., Mennella V., Avidor-Reiss T. Centriole remodeling during spermiogenesis in Drosophila. Curr. Biol. 2016;26:3183–3189. doi: 10.1016/j.cub.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach J.P., Heallen T., Zhang M., Rahmani M., Morikawa Y., Hill M.C., Segura A., Willerson J.T., Martin J.F. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature. 2017;550:260–264. doi: 10.1038/nature24045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y., Huang B., Shim J.S., Chen Q., Lee S.J., Anders R.A., Liu J.O., Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar G., Schatten H., Sutovsky P. Centrosome reduction during gametogenesis and its significance. Biol. Reprod. 2005;72:2–13. doi: 10.1095/biolreprod.104.031245. [DOI] [PubMed] [Google Scholar]
- Mardin B.R., Lange C., Baxter J.E., Hardy T., Scholz S.R., Fry A.M., Schiebel E. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 2010;12:1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrone G., Tucker C.S., Denvir M.A. Cardiomyocyte proliferation in zebrafish and mammals: lessons for human disease. Cell. Mol. Life Sci. 2017;74:1367–1378. doi: 10.1007/s00018-016-2404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. U S A. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe T.O., Hill M.C., Morikawa Y., Leach J.P., Heallen T., Cao S., Krijger P.H.L., de Laat W., Wehrens X.H.T., Rodney G.G., Martin J.F. YAP partially reprograms chromatin accessibility to directly induce adult cardiogenesis in vivo. Dev. Cell. 2019;48:765–779.e7. doi: 10.1016/j.devcel.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroyama A., Lechler T. Microtubule organization, dynamics and functions in differentiated cells. Development. 2017;144:3012–3021. doi: 10.1242/dev.153171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., Raff J.W. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Nigg E.A., Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., Sadek H.A. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello E.R., Olson E.N. A neonatal blueprint for cardiac regeneration. Stem Cell Res. 2014;13(3 Pt B):556–570. doi: 10.1016/j.scr.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srsen V., Fant X., Heald R., Rabouille C., Merdes A. Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol. 2009;10:28. doi: 10.1186/1471-2121-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A.M., Maro B., Bornens M. Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 1985;100:35–46. doi: 10.1083/jcb.100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toji S., Yabuta N., Hosomi T., Nishihara S., Kobayashi T., Suzuki S., Tamai K., Nojima H. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells. 2004;9:383–397. doi: 10.1111/j.1356-9597.2004.00732.x. [DOI] [PubMed] [Google Scholar]
- Vertii A., Hehnly H., Doxsey S. The centrosome, a multitalented renaissance organelle. Cold Spring Harb. Perspect. Biol. 2016;8:a025049. doi: 10.1101/cshperspect.a025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges H.K., Mills R.J., Elliott D.A., Parton R.G., Porrello E.R., Hudson J.E. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development. 2017;144:1118–1127. doi: 10.1242/dev.143966. [DOI] [PubMed] [Google Scholar]
- Wackerhage H., Del Re D.P., Judson R.N., Sudol M., Sadoshima J. The Hippo signal transduction network in skeletal and cardiac muscle. Sci. Signal. 2014;7:re4. doi: 10.1126/scisignal.2005096. [DOI] [PubMed] [Google Scholar]
- Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348:1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woulfe K.C., Gao E., Lal H., Harris D., Fan Q., Vagnozzi R., DeCaul M., Shang X., Patel S., Woodgett J.R. Glycogen synthase kinase-3beta regulates post-myocardial infarction remodeling and stress-induced cardiomyocyte proliferation in vivo. Circ. Res. 2010;106:1635–1645. doi: 10.1161/CIRCRESAHA.109.211482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Tylkowski M.A., Huber D., Contreras C.T., Hoyer-Fender S. ODF2 maintains centrosome cohesion by restricting beta-catenin accumulation. J. Cell Sci. 2018;131:jcs220954. doi: 10.1242/jcs.220954. [DOI] [PubMed] [Google Scholar]
- Zebrowski D.C., Vergarajauregui S., Wu C.C., Piatkowski T., Becker R., Leone M., Hirth S., Ricciardi F., Falk N., Giessl A. Developmental alterations in centrosome integrity contribute to the post-mitotic state of mammalian cardiomyocytes. Elife. 2015;4:e05563. doi: 10.7554/eLife.05563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Zhang E., Zhao M., Chong Z., Fan C., Tang Y., Hunter J.D., Borovjagin A.V., Walcott G.P., Chen J.Y. Regenerative potential of neonatal porcine hearts. Circulation. 2018;138:2809–2816. doi: 10.1161/CIRCULATIONAHA.118.034886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.