Abstract
The systematic entry of SARS-CoV-2 into host cells, as mediated by its Spike (S) protein, is highly essential for pathogenicity in humans. Hence, targeting the viral entry mechanisms remains a major strategy for COVID-19 treatment. Although recent efforts have focused on the direct inhibition of S-protein receptor-binding domain (RBD) interactions with human angiotensin-converting enzyme 2 (hACE2), allosteric targeting remains an unexplored possibility. Therefore, in this study, for the first time, we employed an integrative meta-analytical approach to investigate the allosteric inhibitory mechanisms of SARS-CoV-2 S-protein and its association with hACE2. Findings revealed two druggable sites (Sites 1 and 2) located at the N-terminal domain (NTD) and S2 regions of the protein. Two high-affinity binders; ZINC3939013 (Fosaprepitant – Site 1) and ZINC27990463 (Lomitapide – Site 2) were discovered via site-directed high-throughput screening against a library of ~1500 FDA approved drugs. Interestingly, we observed that allosteric binding of both compounds perturbed the prefusion S-protein conformations, which in turn, resulted in unprecedented hACE2 displacement from the RBD. Estimated ΔGbinds for both compounds were highly favorable due to high-affinity interactions at the target sites. In addition, Site 1 residues; R190, H207, K206 and K187, I101, R102, I119, F192, L226, V126 and W104 were identified for their crucial involvement in the binding and stability of ZINC3939013. Likewise, energy contributions of Q957, N953, Q954, L303, Y313, Q314, L858, V952, N953, and A956 corroborated their importance to ZINC27990463 binding at the predicted Site 2. We believe these findings would pave way for the structure-based discovery of allosteric SARS-CoV-2 S-protein inhibitors for COVID-19 treatment.
Keywords: SARS-CoV-2, Spike protein, Allosteric targeting, Virtual high-throughput screening, Receptor binding domain, High-affinity binding
1. Introduction
The novel coronavirus disease also referred to as COVID-19 is caused by the SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2), with incidences first reported in Wuhan China in December 2019 [1]. This disease has, however, persisted till mid-2020, spreading across 212 countries with over 3,513,507 cases reported coupled with increasingly high casualties numbering over 245,544 globally [2]. SARS-CoV-2 belongs to a large group of coronaviruses which are known to cause respiratory infections and related complications. These RNA viruses are spherical, pleomorphic, positive-sensed, single-stranded and polyadenylated [3]. Of all known viruses, coronaviruses (CoVs) have the largest RNA genome [4], with diverse pathogenic effects in animals and humans. This virus class is divided into four genera namely: alpha-CoV, beta-CoV, gamma-CoV and delta CoV [[5], [6], [7]], with the beta-CoV class prominent for their disease-causing effects in humans (HCoVs). Seven HCoVs have been characterized to date [[6], [7], [8]]; among which four (HCoV-HKU1, HCoV-OC43, HCoV-NL63 and HCoV-229E) cause very mild respiratory symptoms [9,10]. On the other hand, MERS-CoV, SARS-CoV, and SARS-CoV-2 cause severe respiratory and gastrointestinal infections which, in most cases, can be fatal [11]. Although SARS-CoV-related infections were zoonotically transmitted into human populations [12,13], human to human transmissions has further contributed towards viral super-spread via respiratory aerosols [14].
The entry of SARS-CoV-2 coupled with its replication process in target human cells is achieved by the functionalities of a cohort of components, majorly non-structural and structural proteins, that make up the virus. Generally, about 16 non-structural proteins (NSPs) mediate diverse pro-pathogenic functions such as replication, processing and proof-reading of genomic frames, host immune evasion among many others, as previously reported [[15], [16], [17]]. More so, CoVs comprises of four major structural proteins that are integral to their pathogenesis [[18], [19], [20]]. These are the nucleocapsid (N), envelope (E), membrane (M) and spike (S) proteins. The N protein makes up the nucleocapsid and other viral genome-related processes [21] while the M protein is the most abundant of the four, playing major roles in maintaining viral structural integrity as well as coordinating other structural proteins [22]. E protein, on the other hand, is crucial to the maturation of the virus [[23], [24], [25], [26], [27]] while the trimeric S protein mediates viral entry into the host cell via the endosomal or non-endosomal route [28].
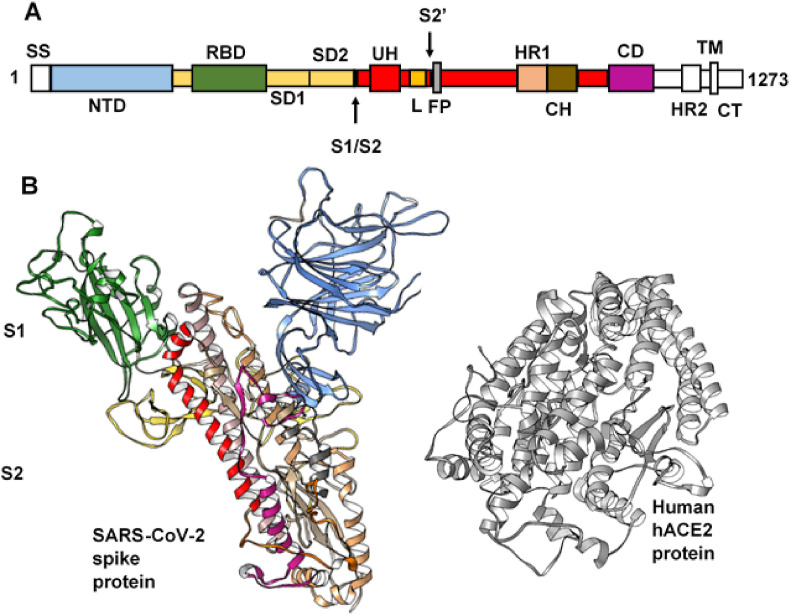
Two domains make up the S protein namely the N-terminal S1 domain and the C-terminal S2-membrane-anchored domain. The S2 region is extensively conserved in CoVs while constituent S1 region residues are highly diverge across the CoV strains [29]. These domains have been further characterized into subdomains due to specific functionalities with respect to host receptor recognition and binding (S1), coupled with membrane fusion and entry (S2) (Fig. 1 ). Similar to SARS-CoV architecture, some recent reports have sub-categorized the SARS-CoV-2 S1 ecto-domain into the N- terminal domain (NTD), a conserved receptor-binding domain (RBD) which recognizes the human angiotensin-converting enzyme 2 (hACE2) [30], and subdomains 1 and 2 (SD1 and SD2). During infection, proteolytic cleavage or priming of the S protein is crucial for viral fusion and entry into host cells, a process mediated by host cell proteases such as the transmembrane serine protease 2 (TMPRSS2) and Cathepsin L [[31], [32], [33]], at the S1/S2 (boundary between S1 and S2 subunits) and S2’ (immediately upstream S2 fusion peptide - FP) cleavage sites [[34], [35], [36]]. The S protein primarily exists in a metastable prefusion complex prior to cleavage, after which notable conformational arrangements occur in order to fuse the viral membrane into the host cell [[37], [38], [39]]. In addition, the RBD adopts disparate conformational motions to engage the host cell receptor [40,41].
Fig. 1.
Structural architecture of SARS-CoV-2 S-protein and target human ACE2 (protease domain). A. Structural topology of the SARS-CoV-2 S-protein showing its different components. NTD, N-terminal domain; RBD, receptor binding domain; SD1, subdomain 1, SD2, subdomain 2; S1/S2, furin cleavage site 1; UH, upstream helix; L, linker region; S2’, furin cleavage site 2; FP, fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane region, CT, cytoplasmic tail. B. 3D structure of the prefusion (S1/S2) S-protein and the interacting (protease) domain of the host hACE2 (grey).
These conformational states are transient, designated as up (open) and down (close) conformations [[42], [43], [44]]. The up conformation corresponds to the hACE2 accessible state while the down state cannot engage the host cell receptor [44]. The S2 domain, on the other hand, consists of the functionally important fusion peptide (FP), which is critical for viral fusion and formation of the post-fusion complex; heptad repeats 1 and 2 (HR1 and HR2); transmembrane domain (TM) and cytoplasmic tail (CT). The HRs of the S-protein trimer interact to form a fusion core of six-helical bundle which helps bring the membranes of the virus and host cell in close proximity for fusion and entry [42]. Therefore, the roles of SARS-CoV-2 S-protein present it as an important therapeutic target, which would enable the prevention of viral entry and fusion in host cells. Numerous studies have been reported over the past months with regards to the possibility of blocking direct interactions between SARS-CoV-2 S-protein and hACE2. Most of these studies were aimed at targeting the S protein RBD domain with antibodies, peptide-based or small molecule compounds that binds with a much higher affinity to block S-protein-hACE2 interactions [[45], [46], [47], [48], [49], [50]]. Also, targeting host proteases such as TMPRSS2 was explored in a recent study, with consequential impediments on SARS-CoV-2 entry [30].
Identification of other functional (allosteric) sites on the prefusion S protein could present another dynamic and effective approach of preventing SARS-CoV-2 infectivity relative to its interaction with the host cell ACE2 and proteases. This alternative target approach for SARS-CoV-2 S protein is important because its RBD (similar to other CoVs) has been associated with a high mutational propensity which may in turn alter the affinity of small molecule inhibitors or peptide designed to bind therein [51]. Allosteric targeting was explored in a recent study wherein the CoV-conserved S2 HR1 region was identified as an important target site for the development of broad-spectrum inhibitors of human CoVs. The resulting peptide inhibitor (EK1) was evaluated in vivo and exhibited desirable safety and efficacy [52]. More so, the Protein Contact Network (PCN) paradigm was used to map functional allosteric loci on SARS-CoV S protein [53].
Relatively, this study was implemented to (i) identify potential druggable sites across the S1 and S2 domains of the SARS-CoV-2 S protein other than the RBD-hACE2 interface (ii) perform high-throughput (virtual) screening of ~1500 FDA approved drugs against the most druggable site(s) (iii) investigate the binding dynamics and interaction mechanisms of the compounds and their consequential effects on the S-protein RBD-ACE2 complex.
We believe this systematic study will be able to provide structural and molecular insights into possible allosteric sites on SARS-CoV-2 S protein suitable for selective targeting and structure-based design. This would open up avenues for the development of allosteric inhibitors with the potential of disrupting hACE2 interactions at the S-protein RBD.
2. Computational methodologies
2.1. Protein retrieval and preparation
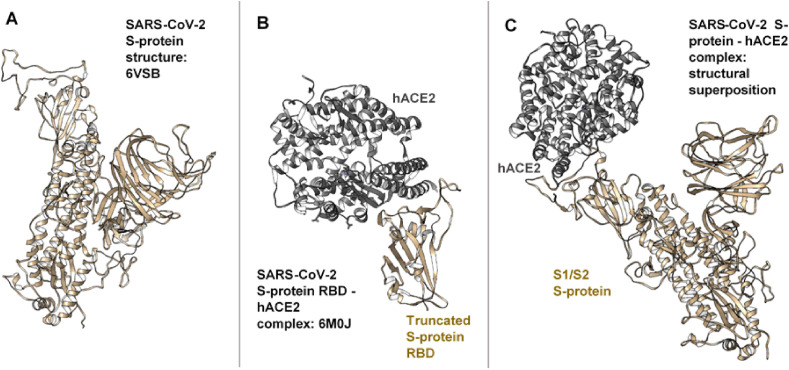
The three-dimensional structure of SARS-CoV-2 S-protein (prefusion) was retrieved from PDB with entry 6VSB [44]. This, as previously reported, represents the S-protein RBD conformation in its up (open) state, which is most suitable for hACE2 binding. Also, to model binding interactions between the prefusion SARS-CoV-2 S-protein (S1/S2) and the hACE2, a crystalized structure with PDB entry 6M0J [54] was separately retrieved. This complex depicts binding between the RBD domain (truncated) of SARS-CoV-2 S-protein and the protease domain (PD) of hACE2. Co-crystallized molecules not relevant to this study were removed while missing residues (gaps) in the structures were filled using the MODELLER algorithm [55]. This preparation was performed on the UCSF Chimera Graphic User Interface (GUI) [56]. Subsequently, using the structural superposition method, we were able to model a complex between prefusion S-protein (S1/S2) monomer (RBD - up conformation) and the hACE2 protein (Fig. 2 ).
Fig. 2.
Structural depiction of the modeling approach employed herein for obtaining the prefusion S-protein-hACE2 complex. A. 3D structure of the prefusion SARS-CoV-2 S-protein as retrieved from PDB (ID 6VSB)B. Retrieved 3D structure of the truncated S-protein RBD and hACE2 (PDB ID 6M0J) C. Modelled complex of prefusion SARS-CoV-2 S-protein and hACE2 as obtained via structural superposition of A and B, followed by removal of the truncated domain.
2.2. Predictive-mapping, cross-validation and characterization of allosteric sites
Possible druggable sites other than the SARS-CoV-2 RBD interface were predicted using approaches previously reported [[57], [58], [59], [60]]. Herein, we employed multiple tools for site identification and validation, which include SiteMap [61], Fpocket [62], Discovery studio 2016 Client [63] and Prankweb [64]. SiteMap is an exhaustive tool which ranks protein pockets based on properties such as druggability, surface exposure, hydrophobicity and hydrophilicity among others [[65], [66], [67]]. These details were then used to characterize the predicted pockets after which other predictive algorithms were used complementarily for cross-validation. Two highly ranked sites were then selected for further analyses.
2.3. High-throughput screening against predicted SARS-CoV-2 S-protein allosteric sites
Furthering on the rationale of the study, we mapped out the two most druggable sites on the target protein and virtually screened against them a large chemical library of FDA approved drugs (~1500 compounds) derived from the ZINC repository (http://zinc.docking.org/substances/subsets/fda/). This screening was performed using high-performance computing-integrated Autodock vina [68] prior to which coordinates of the predicted sites were mapped using gridboxes. Corresponding binding scores were retrieved from the resulting.pdbqt files and were used to filter down to the topmost 20 compounds for each predicted Sites 1 and 2. Subsequently, two compounds with the highest binding scores (most negative) were selected for the two predicted sites yielding complexes that were subjected to further simulation studies. As explained in 2.1, the prefusion S-proteins (ligand-bound and unbound) were superimposed with the RBD-hACE2 complex (6M0J) after which the single truncated RBD was removed. By so doing, we obtained models of allosterically-bound and unbound pre-fusion S-protein-ACE2 complex. This, as aimed in this study, would provide structural and dynamical insights into the mechanistic effects of allosteric targeting on SARS-CoV-2 host entry machinery.
2.4. Molecular dynamics (MD) simulations
Although computationally expensive (1673 residues), we proceeded with long-timescale MD simulation runs for the systems on AMBER18 Graphical Processing Unit (GPU) using its embedded modules [69]. Protein parameters were defined using the FF14SB forcefield while ligand parameters were generated with the antechamber and parmchk modules. Likewise, the LEAP program was used to define coordinate and topology files for the ligand-bound and unbound protein complexes. This program, also, was used to neutralize (addition of counter-ions; Na+ and Cl−) and solvate the systems in a TIP3P water box of size 10 Å. Structural minimization was first carried out partially for 5000steps with a restraint potential of 500 kcal/mol Å2 followed by another 100000 steps of full minimization with no restraints. A canonical (NVT) ensemble with a 5 kcal/mol Å2 harmonic restraints was used to heat the systems gradually from 0 to 300k for 50ps, after which the systems were equilibrated for 10000 ps at a constant 300k temperature without restraints in an NPT ensemble. Atmospheric pressure was maintained at 1 bar with a Berendsen barostat [70] while each protein system was subjected to a production run of 350ns. Studied systems include ZINC3939013-S-protein-hACE2 (allosteric Site 1), ZINC27990463-S-protein-hACE2 (allosteric Site 2), and unbound S-protein-hACE2. Corresponding trajectories were saved at every 1ps time-frame until the end of the simulation followed by data plot analyses using Microcal Origin software [71]. Snapshots were also taken and analyzed to monitor structural events and ligand interaction dynamics across the trajectories on the UCSF Chimera user interface (GUI) and Discovery Studio Client [63].
2.5. Calculations of binding free energies and per-residue decomposition
The Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) method was used to evaluate binding affinities of the predicted allosteric S-protein binders at their target sites. Binding energy profiles for both compounds, inclusive of their energy components, were estimated using 1000 snapshots from the terminal 30ns of MD trajectories where conformational stabilities were visible. This approach was important in order to minimize the effects of conformational disorder or entropy on ligand interactions. The equations below mathematically express binding energy calculations:
| ΔGbind = Gcomplex - (Greceptor + Ginhibitor) | (1) |
| ΔGbind = ΔGgas + ΔGsol - TΔS = ΔH - TΔS | (2) |
| ΔGgas = ΔEint + ΔEele + ΔEvdW | (3) |
| ΔGsol = ΔGele,sol(GB) - ΔGnp,sol | (4) |
| ΔGnp,sol = γSASA + β | (5) |
As shown, internal (ΔEint), electrostatic (ΔEele) and van der Waals (ΔEvdW) energies sum up the gas-phase energy (ΔGgas) while the solvation free energy (ΔGsol) is defined by the polar solvation (ΔGele,sol) and non-polar contribution to solvation (ΔGnp,sol) terms. The MM/GBSA method was used to estimate the Generalized Born (GB) for ΔGele,sol while the linear relationship between the surface tension proportionality constant (γ = 0.0072 mol−1 Å−2), solvent accessible surface area (SASA, Å2), and β constant was used to solve ΔGnp,sol. Furthermore, estimated ΔGbind was decomposed into individual residue energies, most especially those that constitute the predicted allosteric pockets where the ligands were bound. This method was essential to identify specific residues that contribute crucially to the stability and inhibitory activities of potential allosteric inhibitors.
3. Results and discussion
3.1. Potential allosteric sites on the prefusion SARS-CoV-2 S-protein – identification, cross-validation and characterization
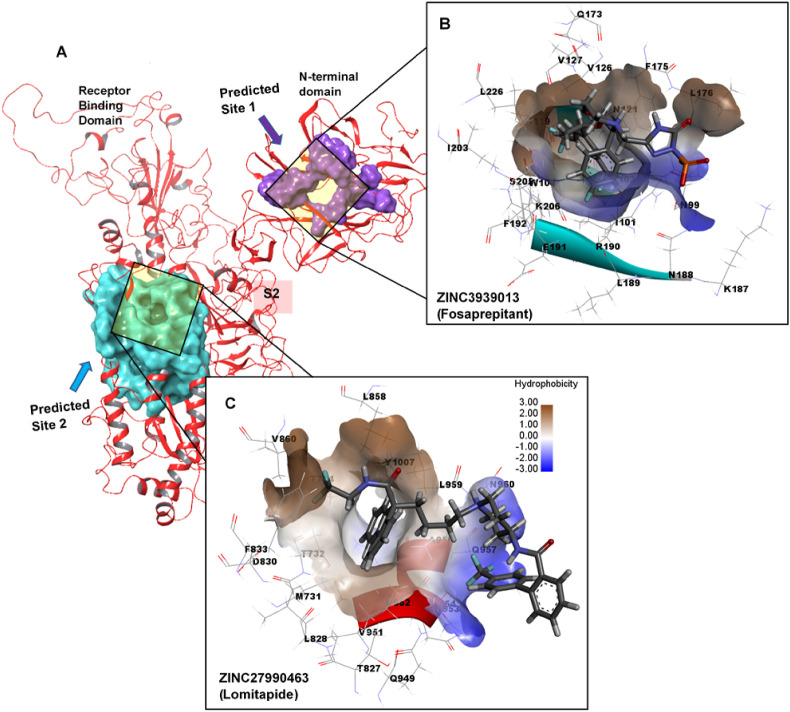
Based on the study rationale, we set out to identify possible sites for drugging the target protein other than the RDB domain where interaction with hACE2 reportedly occurs. Prediction and cross-validation were done using aforementioned tools with results showing corresponding residues (domain). This represents a combinatorial method for identifying, validating and cross-validating possible allosteric sites that are druggable in SARS-CoV-2 S-protein. From the multiple predictions, Site 1 entails residues that make up the NTD (7–307) of SARS-CoV-2 S-protein while Site 2 residues overlap across the FP (816–831), HR1 (898–985) and CR (838–897) (Table 1 ). The architectures of these pockets are shown in Fig. 3 . Furthermore, defining the druggability of a site on target proteins depends on the size (volume) and hydrophobicity (with minimal hydrophilicity) while, on the other hand, high hydrophilicity, reduced hydrophobicity, small pocket size and shallowness characterize “difficult-to-drug” and undruggable pockets [61,[65], [66], [67]]. While large hydrophilicity could have repulsive effects on ligand mobility at the binding site, a small or shallow cavity would impede ligand access, fitness, optimal binding and stability.
Table 1.
Identification and cross validation of potential allosteric sites on SARS-CoV-2 prefusion S-protein using multiple predictive algorithms.
| Predicted sites | Binding site prediction and cross-validation |
Corresponding S-protein domain | |||
|---|---|---|---|---|---|
| SiteMap | Fpocket | DS 2016 Client | Prankweb | ||
| Site 1 | 65, 66, 95–115, 121–136, 202, 201, 203, 204, 205, 206, 211, 221, 222, 223 | 101, 103, 104, 108, 109, 142, 143, 144, 145, 146, 147, 178 | 92,93, 94, 95,96, 97, 98, 99, 101, 102, 103, 104, 105, 106, 117–129, 170–175, 189–194, 203–207, 223, 224, 225 | 99, 101, 102, 104, 119, 120, 121, 126, 172, 173, 175, 177, 190, 192, 203, 205 | NTD |
| Site 2 | 731-737, 791–806, 815,819, 823–828, 833–840, 951, 954, 959, 962-968 | 705-713, 720, 789, 792, 793, 794, 807, 832, 833, 834, 835, 836, 837, 840, 848, 852–863, 904, 908–910, 947-960 | 726, 727, 822–837, 845–854, 856–861, 944-963 | 828, 833, 837, 854, 856, 858, 859, 860, 956, 959, 960, 963 | FP, HR1, CD |
| Site 3 | 18, 19, 20, 21, 22, 23, 53–61, 288–293, 585–593, 623-635 | 13, 28, 47, 50, 51, 53, 54, 57, 317, 318, 372 | 37, 38, 39, 40, 41, 42, 51, 52,53, 54, 55, 195-204 | 50, 51, 52, 273, 274, 291, 292, 298, 301, 302, 304, 315, 317, 631, 632 | NTD, RBD, SD1 |
| Site 4 | 363, 364, 365, 366, 367, 368, 369, 370, 382, 383, 385, 423–434, 512, 513, 514, 515 | 355, 380, 396, 398, 412, 423, 425, 426, 429, 430, 431, 432, 433, 464, 512, 513, 514, 515, 516, 518 | 326, 327, 328, 329, 330, 331, 357, 358, 359, 360, 361, 362, 363, 380–396, 428, 429, 430, 431, 432, 513–526, 541–548, 559-580 | 355-360, 396, 398, 425, 426, 429, 430, 431, 433, 464, 466, 514, 515, 516 | RBD, SD1 |
| Site 5 | 596-603, 660–674, 697–702, 710,711, 712, 713, 714, 715, 716, 717, 718, 719, 775-781 | 769, 774, 797, 798, 800, 882, 883, 895, 896, 897, 898, 899, 902, 916, 920, 923 | 719-734, 769–785, 833, 860–875, 1069 | 729, 730, 731, 774, 775, 778, 782, 830, 863, 865, 867, 870, 1056, 1058, 1059 | SD1, CR, HR1 |
| Site 6 | 1068, 1069, 1077, 1078, 1079, 1080, 1089, 1090, 1091, 1092, 1093, 1094, 1095, 1105, 1106, 1107 | 1010, 1012, 1015, 1019, 1108, 1119, 1208, 1210, 1211, 1212, 1214, 1215, 1218 | 909. 910, 911, 1089, 1090, 1091, 1092, 1093, 1094, 1104, 1105, 1106, 1107, 1108, 1109, 1207-1216 | 1081, 1083, 1084, 1088, 1115, 1116, 1117, 1122, 1123, 1136, 1137, 1138, 1201 | SD2, βH, TM, CH |
NTD, N-terminal domain; FP, fusion peptide; HR1, heptad repeat 1; CD, connector domain; RBD, receptor binding domain; SD2, subdomain 2; SD1, subdomain 1; CH, central helix; βH – β hairpin; TM, transmembrane region.
Fig. 3.
Predicted allosteric sites and their locations on SARS-CoV-2 S-protein. A. 3D structure of the SARS-CoV-2 prefusion S-protein showing surface representation of the predicted Sites 1 and 2. B. Closer look at the predicted Site 1, constituent residues and starting orientation of ZINC3939013. C. Inset showing the predicted Site 2, constituent residues and binding orientation of ZINC27990463.
More so, a >0.83 (Halgren Dscore) threshold applies for druggable sites [61] while a 0.8 cut-off point was set for the SiteScore, which, altogether are parameters used to distinguish between sites that are druggable and non-druggable.
From our results, four prediction algorithms employed herein commonly identified and ranked specific regions on the target protein, indicating their possibilities as allosteric sites. As earlier stated, constituent residues of the top two ranked sites correspond to the fusion peptide (FP), Heptad repeat 1 (HR1), connecting region/domain (CR/CD) and N-terminal domain (NTD). From Table 2 , Sites 1 → 3 ranks above the 0.83 Halgren Dscore threshold making them suitable for therapeutic targeting. Relatively, Site 1 appears to be highly surface-exposed with a score of 0.933 while a large pocket size and volume for Site 2 could favor the use of large-molecule compounds.
Table 2.
Characterization of the predicted allosteric sites on SARS-CoV-2 prefusion S-protein based on site attributes.
| Predicted sites | Druggability score (Dscore) | SiteScore | Surface-exposure | Pocket Size | Pocket Volume (A [3]) | Hydrophobicity | Hydrophilicity | Hydrogen donor/acceptor |
|---|---|---|---|---|---|---|---|---|
| Site 1 | 0.893 | 0.935 | 0.933 | 482 | 245.82 | 0.168 | 0.336 | 0.875 |
| Site 2 | 1.046 | 0.899 | 0.794 | 503 | 294.31 | 0.243 | 0.203 | 1.922 |
| Site 3 | 0.842 | 0.823 | 0.862 | 319 | 208.6 | 0.142 | 0.822 | 1.886 |
| Site 4 | 0.803 | 0.817 | 0.852 | 263 | 214.30 | 0.111 | 0.613 | 2.539 |
| Site 5 | 0.738 | 0.801 | 0.800 | 218 | 148.8 | 0.353 | 0.705 | 1.294 |
| Site 6 | 0.711 | 0.786 | 0.696 | 267 | 237.64 | 0.392 | 0.884 | 0.634 |
Taken together, high surface-exposure coupled with relatively large volumes, hydrophobicity and favorable donor/acceptor properties for Sites 1 and 2 could account for their suitability as targetable allosteric regions on the S-protein other than the RBD (Fig. 3). These presumptions are also reflected by the estimated Dscore and SiteScore values. In addition, since these predicted sites are highly functional, particularly the overlapping FP, HR1 and CR, targeting them could disrupt structural events associated with SARS-CoV-2 fusion into the host-cell as explicitly reported in recent literatures [39,41,44,54,72].
3.2. High-throughput screening and identification of potential allosteric binders to the predicted sites 1 and 2
High-throughput screening using a library of ~1500 FDA approved drug compounds (http://zinc.docking.org/substances/subsets/fda/) were performed against the two predicted allosteric sites.
Autodock vina grid boxes were centered on coordinates (center, size, x = 107.1, 14.45; y = 99.65, 16.33, z = 114.74, 15.82) for Site 1 and (center, size, x = 129.2, 17.84; y = 85.23, 24.15, z = 139.11, 18.47) for Site 2.
Results for the top 20 compounds with the highest binding scores are presented in Supplementary Table S1 and Supplementary Table S2 for Sites 1 and 2 respectively.
From the screening results, overall highest scores were estimated for ZINC3939013 (−10 kcal/mol) at Site 1 and ZINC27990463 (−9.3 kcal/mol) at Site 2.
3.3. Allosteric targeting disrupts S-protein conformation and hACE2 association at the RBD
As highlighted in our methods, MD simulations were performed for the prefusion S-protein-hACE2 complexes bound distinctly at two potential allosteric sites.
This approach was essential to investigate the likely effects of allosteric targeting on the entry/fusion mechanisms of SARS-CoV-2 via host hACE2.
Firstly, we monitored conformational events associated with the allosteric binding of Compds A and B at the predicted Sites 1 and 2 of SARS-CoV-2 S-protein. This was done by obtaining snapshots from the MD trajectories from selected time-frames which were then visually compared with their starting structures.
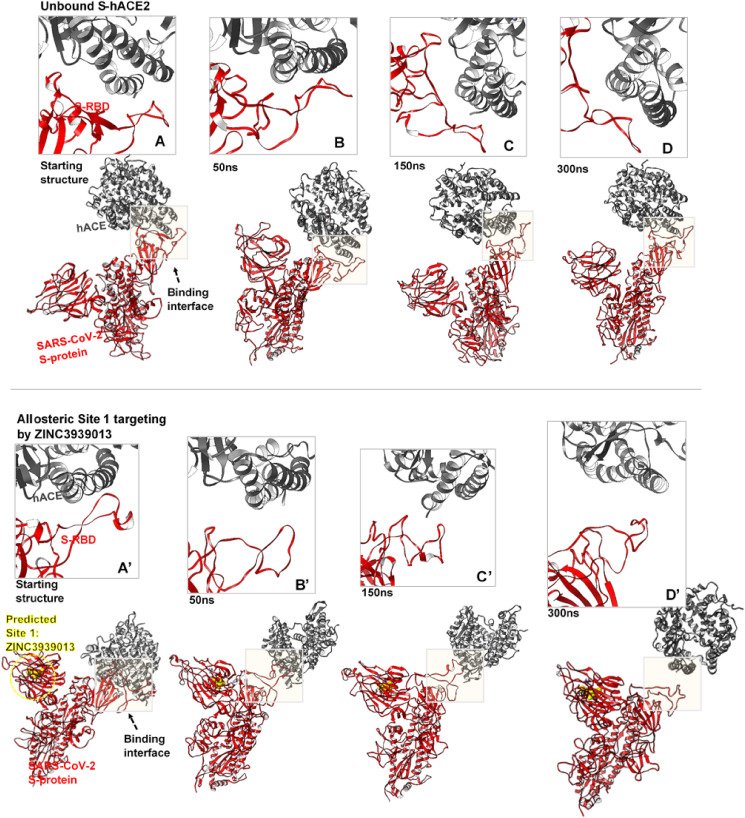
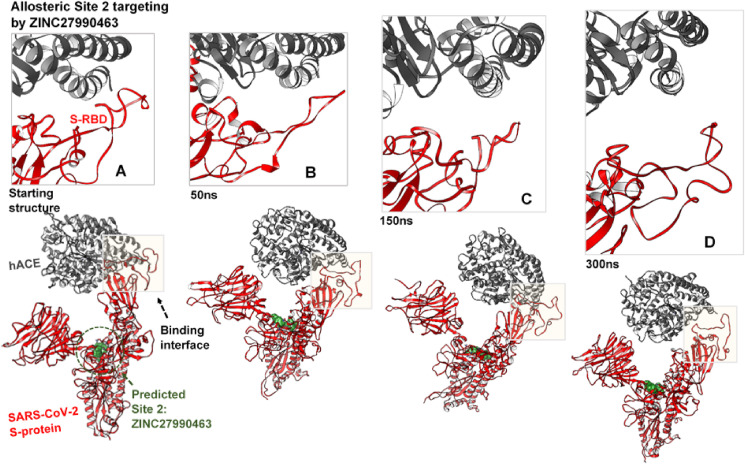
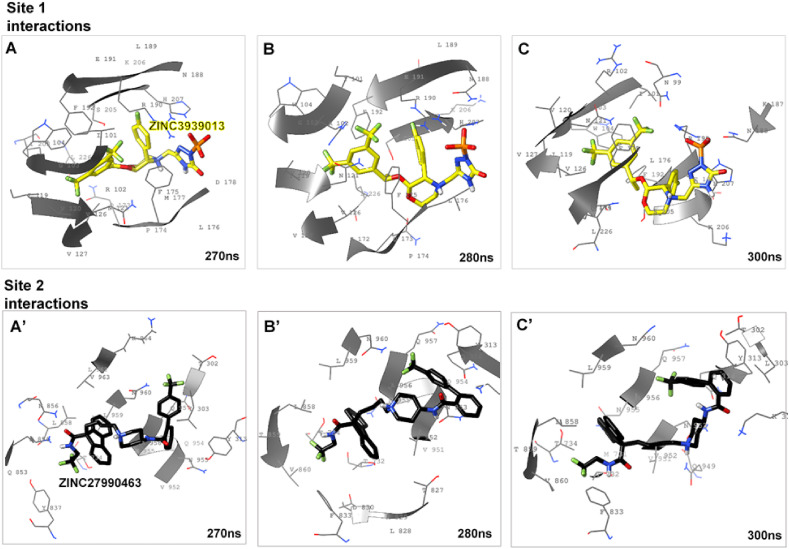
Our findings reveal that the unbound S-hACE2 complex remained intact over the simulation time while the hACE2, after about 50ns, was gradually displaced from the RBD interfaces in S-proteins bound by ZINC3939013 (Fosaprepitant) at Site 1 and ZINC27990463 (Lomitapide) at Site 2 (Fig. 4, Fig. 5 ). These variations, as induced by the allosteric binders, proceeded more prominently until the end of the simulation. As shown, RBD for the unbound S-protein exhibited relative ‘elongated’ or ‘out-stretched’ conformations which most likely favored hACE2 binding. However, this conformation appeared distorted the allosterically-bound S-proteins and could account for displacement motions of the interacting hACE2 from the RBD interface. Therefore, the allosteric-mediated disruption of SARS-CoV-2 S-protein RBD and its interaction with hACE2, as reported herein, is a major finding that could indicate the viability of allosteric targeting in SARS-CoV-2 therapy.
Fig. 4.
Allosteric disruption of S-protein hACE2 binding at the RBD. Inset A-D shows the trajectorial motions of the RBD (red) and its associated hACE2 (grey) along the simulation period from the starting to the ultimate structure at 300ns. Inset A’-D’ shows systematic perturbations of the RBD and time-based displacement of the hACE2 as induced by the binding of ZINC3939013 (yellow surface) at the predicted Site 1.
Fig. 5.
Systematic perturbations of the S-protein RBD. Inset A’-D’ shows time-based alterations of the RBD and displacement of the hACE2 as induced by the allosteric binding of ZINC27990463 (green surface) at the predicted Site 2.
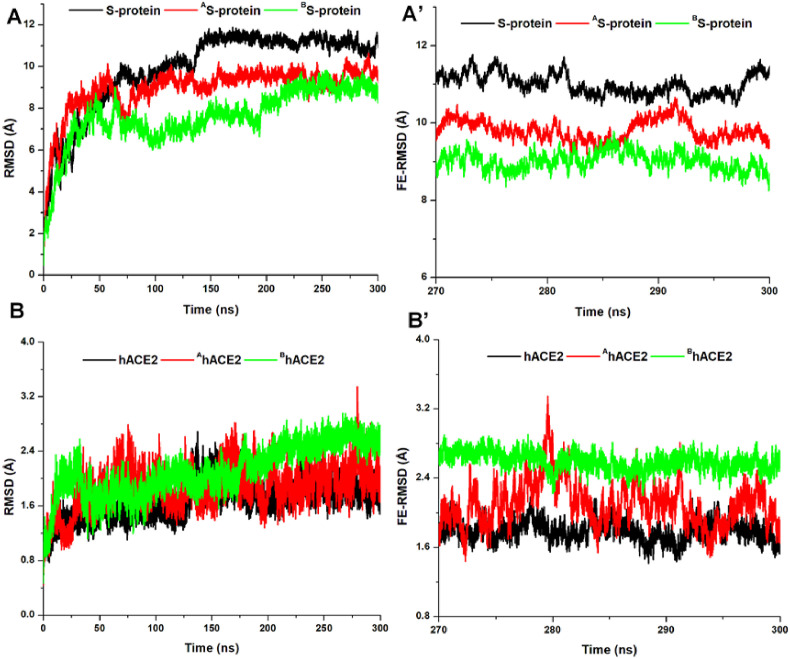
Furthermore, we measured structural stabilities across the ligand-protein complexes relative to the unbound system using the RMSD metrics. As shown in Fig. 6 , structural instability was highest in the unbound S-protein while its associated hACE2 was relatively stable compared to the allosterically-bound S-proteins which exhibited reductions in Cα deviations across the simulation time (RMSD <2 Å). Interestingly, hACE2s for the bound proteins showed higher structural motions and could correlate with their displacement motions away from the RBD as earlier revealed. Comparatively, allosteric ligand binding at both sites distinctly lowered RMSD in the S-protein and induced high RMSDs in the hACE2.
Fig. 6.
Relative structural stabilities of the S-protein and corresponding hACE2 among the unbound and bound (red and green) systems. A and A’ shows the overall RMSD and FE-RMSD for unbound (black), ZINC3939013-bound (red), and ZINC27990463-bound (green) S-protein. B and B’ shows overall RMSD and FE-RMSD for hACE2s associated to unbound (black), ZINC3939013-bound (red), and ZINC27990463-bound (green) S-proteins.
This could indicate the structural effects of allosteric targeting on the S-protein and its interaction with hACE2. Estimated mean RMSDs, as presented in Table 3, corroborates conformational variations among the unbound and bound protein complexes.
Table 3.
Estimations of relative conformational variations across the studied systems.
| Structural analysis (Å) | Unbound Spike (hACE2) | ZINC393013Spike (hACE2) | ZINC27990463Spike (hACE2) |
|---|---|---|---|
| Whole RMSD | 17.09 ± 1.1 (1.77 ± 0.23) | 13.31 ± 0.9 (1.93 ± 0.32) | 11.00 ± 2.0 (2.2 ± 0.4) |
| FE-RMSD | 11.05 ± 0.3 (1.66 ± 0.26) | 9.57 ± 0.4 (1.87 ± 0.31) | 7.61 ± 1.4 (2.6 ± 0.1) |
| FE-RMSF | 5.40 ± 1.9 (0.73 ± 0.32) | 4.87 ± 1.7 (1.39 ± 0.45) | 3.79 ± 1.2 (3.0 ± 0.82) |
| FE-RoG | 41.08 ± 0.41 (24.49 ± 0.1) | 38.4 ± 0.36 (25.01 ± 0.16) | 36.9 ± 0.61 (26.0 ± 0.22) |
| Receptor binding domain (RBD) | |||
| FE-RMSD | 4.1 ± 0.1 | 5.6 ± 0.3 | 6.1 ± 0.2 |
| FE-RoG | 15.3 ± 0.2 | 16.1 ± 0.3 | 16.2 ± 0.2 |
To minimize the effects of structural disorderliness (entropy) in our calculations, we selected, from the MD trajectories, terminal time-frames (270–300ns) from which the systems appeared to relatively stabilize. These were defined as the finally equilibrated (FE) time-frames and were used for subsequent structural analyses (Table 3 ).
From the resulting FE-RMSD plots, unbound S-protein was highly unstable while its associated hACE2 exhibited low structural motion in line with the RMSD calculations, which could also imply that the binding of S-protein stabilized hACE2. In contrast, the allosterically-bound S-proteins (Sites 1 and 2) were notably stable while their corresponding hACE2 showed high structural instability that could correlate with their systemic motions at the S-protein RBD as earlier mentioned.
Furthermore, we measured the Cα mobilities of the proteins in their bound and unbound forms by calculating their relative FE-RoGs. From the plots in Supplementary Figure S1, we observed that atomistic mobilities were distinctly high in the unbound S-protein while its corresponding hACE2 maintained stable motions which on the contrary was prominently high in the allosterically-bound systems. This also agrees with the FE-RMSD analysis which, altogether indicate that high Cα mobility in hACE2s associated with the allosterically-bound S-protein could be as a result of their motions at the S-protein RBD.
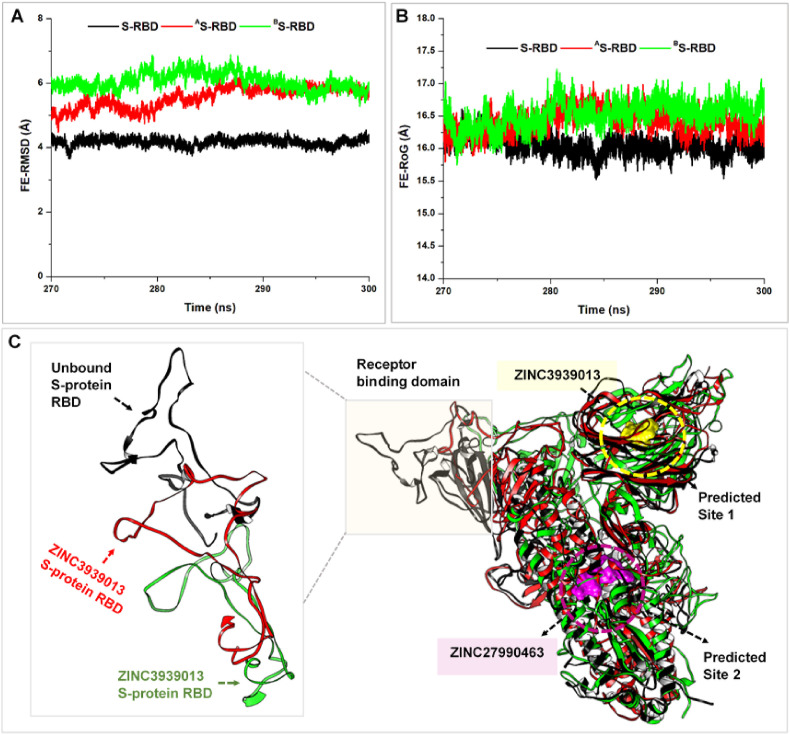
We then investigated Cα motions at the S-protein RBDs since this region is reportedly critical for hACE2 binding [54,72]. FE-RMSD calculation revealed distinct stability (RMSD < 2 Å) at the RBD of unbound S-protein while high structural motions or instability characterized RBDs of the allosterically-bound S-proteins. This corroborated high distortions earlier reported at the RBDs of Compd A and B-bound S-proteins.
Presumably, allosteric targeting or inhibition of SARS-CoV-2 S-protein perturbs the RBD conformation essential for interactions with the host ACE2, leading to eventual dissociation. More so, we determined the degree of Cα mobility at the respective RBDs using the FE-RoG metrics. From the resulting plot, the unbound S-protein RBD exhibited low Cα motions, which were increased in the allosterically-bound S-proteins (Fig. 7 ). This observation agrees with the estimated FE-RMSD which could altogether indicate that allosteric binding of the compounds induced conformational variations that may have altered hACE2 binding and stability at the RBD.
Fig. 7.
Estimations of Cα motions at the RBD domain of unbound and allosterically-bound S-proteins relative to hACE2 interactions. A. Comparative Cα RMSD plot of unbound (black), ZINC3939013-bound (red), and ZINC27990463-bound (green) S-protein RBD. B. Comparative Cα RoG plot of unbound (black), ZINC3939013-bound (red), and ZINC27990463-bound (green) S-protein RBD. C. Visual analyses of structural alterations that occurred differentially at the RBDs of unbound (black), ZINC3939013-bound (red), and ZINC27990463-bound (green) S-protein. These depictions were obtained via structural superposition of their resulting average structures.
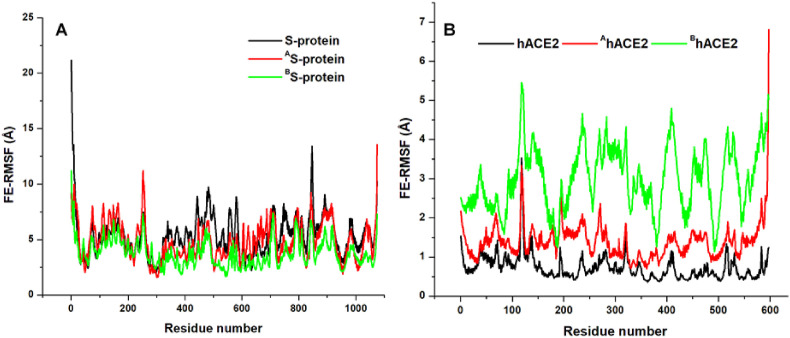
Systemic fluctuations of constituent residues across the protein units were then monitored by calculating the FE-RMSF values using the stable time-frames (Fig. 8 ). Results showed that the S-protein was, on the average, less flexible in the bound systems relative to unbound while high conformational flexibility characterized the corresponding hACE2 proteins in the bound systems.
Fig. 8.
Cα FE-RMSF plots showing disparate per-residue motions and conformational flexibility among unbound and ligand-bound S-proteins together with their corresponding hACE2s A. Comparative per-residue fluctuations in S-proteins of unbound (black), ZINC3939013-bound (red), and ZINC27990463-bound (green) S-protein B. Comparative per-residue fluctuations in their corresponding hACE2s.
In line with earlier results, high structural fluctuations manifested by hACE2 in the bound S-proteins could indicate the degree of dynamical variations relative to their starting positions at the RBD. We also mapped out equilibrated fluctuations of the receptor-binding domain (RBD), a crucial component of the S-protein that interacts directly with the hACE2. Likewise, residual fluctuations across the CTD2 and CTD3 regions were investigated in addition to the S2 subunit (Table 4 ). As shown in Supplementary Figure S2, the RBD region of unbound S-protein exhibited the least fluctuation while allosteric ZINC27990463 binding induced the highest fluctuation at this region. The posterior S2 domain of the S-protein was highly fluctuated in the unbound form while allosteric binding lowered residual fluctuation in this region, particularly by ZINC27990463 at Site 2. Residual fluctuations were also lowered at the SD1 and SD2 domains of ZINC3939013 and ZINC27990463-bound S-proteins which further indicates the allosteric inhibitory effects were extended into crucial parts of the protein. This could present allosteric SARS-CoV-2 S-protein targeting as a viable strategy for disrupting viral entry mechanisms into human hosts.
Table 4.
FE-RMSF calculations of the structural components of SARS-CoV-2 S-protein in their unbound and allosterically-bound forms.
| Residual fluctuation (Å) | |||
|---|---|---|---|
| Structural domains | Unbound Spike | ZINC3939013-Spike | ZINC27990463-Spike |
| NTD | 1.35 ± 1.25 | 1.64 ± 0.52 | 1.26 ± 0.6 |
| RBD | 1.75 ± 0.7 | 1.9 ± 1.24 | 2.14 ± 0.44 |
| SD1 | 1.67 ± 0.39 | 1.53 ± 0.44 | 1.35 ± 0.3 |
| SD2 | 1.85 ± 0.49 | 1.72 ± 0.67 | 1.19 ± 0.31 |
| S2 | 1.39 ± 0.89 | 1.30 ± 0.66 | 1.15 ± 0.43 |
Also from the table above, we could deduce possible allostery communication mechanisms wherein ligand binding at Site 1 altered the corresponding Site 2 region (FP, HRI and CR) which altogether constitute the S2. This could provide an avenue to identify and characterize allosteric communication networks within the target protein [73]. Accordingly, while structural fluctuation was slightly difference among the NTD and S2 domain of unbound S-protein (~0.04 Å), systematic fluctuation increased by 0.34 Å and 0.11 Å respectively in the Site 1 and Site 2-bound S-protein. In other words, ligand binding at the NTD reduced structural perturbations at the S2 which opposes structural occurrence in the unbound protein.
3.4. Interaction dynamics of Compds A and B at the predicted SARS-CoV-2 S-protein allosteric sites
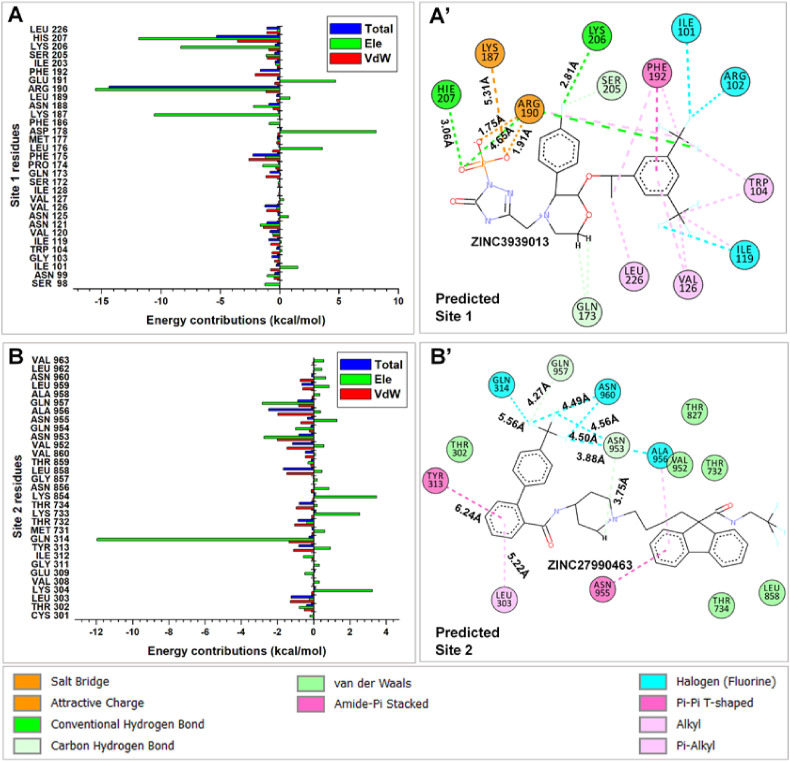
Structural analyses of ligand orientations at the respective allosteric sites of SARS-CoV-2 S-protein were performed using averaged structures from the MD trajectories (Fig. 9 ). Findings reveal that the allosteric binding of ZINC3939013 (Fosaprepitant) was stabilized at the NTD. Fosaprepitant contains a terminal triphosphate group that orients towards residues such as N99, K187, N188, R190 and H207. Likewise, its trifluoromethyl group oriented towards D102 while constituent –O and –NH groups mediate interactions with Q173 and N121, among others. These altogether could facilitate high-affinity interactions accountable for its stability and allosteric inhibitory effects against the SARS-CoV-2 and associated hACE2.
Fig. 9.
Binding orientations of the allosteric binders at the predicted S-protein sites over the terminal post-equilibrated time-frames. A-C. show ZINC3939013 orientations at Site 1 along time-frames 270-300ns A’-C’. shows the trans-domain orientations of Site 2-bound ZINC27990463 from 270-300ns of the simulated trajectory.
On the other hand, systematic orientations of ZINC27990463 show that it transverses the predicted Site 2 and adjacent NTD-RBD linker (L) at the initial time-frames. Chemically, ZINC27990463 (Lomitapide) consists of two trifluoromethyl groups at its two terminal ends, and as observed, while a terminal trifluoromethyl moiety (together with amino and hydroxyl substituents) was oriented into the hydrophobic pocket formed by T732, T734, D830, F833, Y837, Q853, K854, N856, L858, T859, V860, the second trifluoromethyl moiety was extended towards T302, L303, K310, I312 and Y313 of the NTD-RBD linker subdomain.
Also, HR1 residues (N955, A956, L959, N960, L962, V963 and K964) were involved in ligand binding. This trans-domain binding activity of ZINC27990463 could account for its prominent disruptive activities on the S-protein structure, particularly at the RBD.
3.5. Estimations of binding free energies and per-residue contributions
Binding affinities of the compounds were determined using the MM/PBSA technique, which also allowed us to measure the energy contributions of interactive residues at the predicted allosteric sites. Energy calculations, as presented in Table 5 were performed using relatively stable time-frames (270–300ns) to minimize entropical effects that may interfere with ligand binding activities.
Table 5.
Binding free energy profiles of the ligand-protein complexes.
| Energy Components (kcal mol−1) | ZINC3939013-Site 1 | ZINC27990463-Site 2 |
|---|---|---|
| ΔEvdW | −49.93 ± 0.21 | −43.19 ± 0.15 |
| ΔEele | −213.0 ± 1.54 | 34.68 ± 0.54 |
| ΔGgas | −262.9 ± 1.49 | −8.51 ± 0.56 |
| ΔGele,sol(GB) | 214.2 ± 1.38 | −21.65 ± 0.51 |
| ΔGnp,sol | −7.5 ± 0.01 | −6.4 ± 0.02 |
| ΔGsol | 206.7 ± 1.38 | −28.05 ± 0.51 |
| ΔH | −56.2 ± 0.19 | −36.56 ± 0.26 |
| TΔS | −3.5 ± 0.02 | 3.99 ± 0.1 |
| ΔGbind | −52.7 ± 0.26 | −32.57 ± 0.17 |
Findings revealed that both targets sites are highly favorable for ligand binding as both compounds exhibited favorable ΔG binds of −52.7 kcal/mol (ZINC3939013 at Site 1) and −32.57 kcal/mol (ZINC27990463 at Site 2). These estimations further suggest both predicted sites as high-affinity regions for achieving the allosteric inhibition of SARS-CoV-2 S-protein. Also from the estimations, we could deduce that entropical effects on the ΔG binds were minimal.
In addition, we observed that electrostatic effects contributed most notably to the allosteric binding of ZINC3939013 at the NTD region while van der Waals contributions had the highest effect on the binding of ZINC27990463 at the predicted Site 2 pocket. Electrostatic contributions at Site 1 could be due to the high number of electropositive residues that constitute the pocket, as shown in Fig. 10 , which may form high-affinity interactions with electronegative moieties of the compound. Calculations further revealed that ΔE vdW and ΔE ele were more favorable in the gas phase for ZINC3939013 while polar solvation energies were more favorable for ZINC27990463 at the S2 region of the S-protein. This could imply that while the former was buried in the deep hydrophobic pocket of the NTD, the latter was surface exposed due to its trans-domain binding activity as earlier reported. To understand the mechanistic binding of the compounds at both predicted sites, we decomposed the binding free energies into individual contributions of the interacting residues. These were juxtaposed with structural analysis that showed the type and nature of interactions involved. From the per-residue plot, we observed high ΔE ele contributions by R190, H207, K206 and K187 with values of −15.5 kcal/mol, −11.90 kcal/mol, −8.35 kcal/mol and −10.55 kcal/mol indicative of their crucial roles in stabilizing ZINC3939013 at the predicted Site 1. R190 and H207 had notably high total energy contributions of −14.42 kcal/mol and −5.3 kcal/mol respectively while vdW contributions were also high in H207, F175 and F192 with values of −3.5 kcal/mol, −2.58 kcal/mol and −2.06 kcal/mol. Unfavorable ΔE ele interactions, which could be due to steric clashes, were mediated by L176, D178 and E191 but were however compensated by the high negative energies. Coherent with our calculations, 2D analysis of the ligand-residue interactions revealed that K187, R190, K206, H207 formed high-affinity salt bridges, ionic and hydrogen interactions with the phosphoryl group of ZINC3939013. The salt bridge interaction was unique to R190 which could account for its high ΔE tot contribution as estimated. Also, H–F bonds mediated by I101, R102, I119 coupled with aromatic (π) interactions by F192, L226, V126, and W104 could further contribute to the stability of the inhibitor at the predicted Site 1. It is also important to mention that these bonds have short distances ranging from 1.2 Å - 6 Å indicative of their strengths and stabilities. At Site 2, ΔE ele contribution was highest in Q314 (−11.9 kcal/mol) followed by Q957, N953, and Q954 which had estimated energy values of −2.83 kcal/mol, −2.73 kcal/mol and −1.00 kcal/mol respectively. These energies were however insufficient to compensate unfavorable ΔE ele contributions by K304 (3.24 kcal/mol), K733 (2.54 kcal/mol), K854 (3.48 kcal/mol) and N955 (1.3 kcal/mol). However, vdW contributions by L303 (−1.28 kcal/mol), Y313 (−1.1 kcal/mol), Q314 (−1.37 kcal/mol), L858 (−1.47 kcal/mol), V952 (−1.46 kcal/mol), N953 (−2.0 kcal/mol) and A956 (−1.96 kcal/mol) complemented ligand binding and stability. Overall, total energies above the −1 kcal/mol threshold were contributed by A956 (−2.48 kcal/mol), L858 (−1.658), L303 (−1.24 kcal/mol) and V952 (−1.16 kcal/mol) indicative of their importance to ligand binding at Site 2. Structural-interaction analysis revealed that halogen (H–F) bond predominated ZINC27990463 binding as mediated by Q314, A956 and N953. In addition, Q957 and N953 engaged two F atoms via hydrogen bonds while L303, L858 and A956 interacted with the constituent rings via aromatic (π-alkyl) interactions. More so, π-π stacked interaction between Y313 and a benzene ring (of the 4-tri-fluoromethyl-1,1′-biphenyl group) could be highly crucial for the stability of the compound.
Fig. 10.
Ligand-residue interactions and energy contributions at the predicted allosteric sites of SARS-CoV-2 S-protein. A. Per-residue energy decomposition plot for ZINC3939013 at S-protein Site 1 A’. Complementary interaction pattern mediated by ZINC3939013 at S-protein Site 1 with constituent residues. Bond distances are also shown for crucial residues indicative of their strength B. Energy plot showing contributions of individual residues of the predicted Site 2 towards the stability of ZINC27990463. Corresponding interactions are showed in B’.
Taken together, electrostatic energies favored the binding of ZINC3939013 at Site 1 while vdW energies favored ZINC27990463 binding at Site 2, which consequentially, were able to perturb the S-protein RBD and allosterically disrupt hACE2 interactions.
4. Conclusion
The systemic entry of SARS-CoV-2 into the human host cell is a crucial process that underlies its virulence and pathogenicity in humans and other animals it infects. This mechanism is mediated by its interaction with the host ACE2 (hACE2) via attachment and fusion. Potential intervention approaches in SARS-CoV-2 treatment include therapeutic strategies that could prevent SARS-CoV-2 S-protein binding to hACE2. In this study, we implemented an exhaustive approach to identify drug molecules that could potentially bind to SARS-CoV-2 S-protein at other sites other than the RBD. Pertinent to the allosteric targeting approach implemented herein was the identification of highly druggable sites inherent in the S-protein (S1/S2), which was carried out using multiple pocket prediction algorithms for identification and validation of possible allosteric sites. Predicted pockets were then characterized based on their attributes after which two highly probable pockets were selected. These were then screened distinctly against a library of ~1500 FDA approved drugs retrieved from the ZINC database. Amongst all, ZINC3939013 (Fosaprepitant) and ZINC27990463 (Lomitapide) were obtained for both Sites 1 and 2. All atom-MD simulation methods were then used to investigate their interaction dynamics and possible inhibitory effects on the S-protein (S1/S2)-hACE2 complex. Findings revealed that the allosteric binding of the compounds induced varying degrees of conformational alterations across the protein structures, including the RBD where hACE2 interactions reportedly occur. Time-based analysis revealed a systematic displacement of the hACE2 away from the allosterically-bound S-protein RBD domain, which on the other hand exhibited reduced Cα motions. Conformational variations across the different components of the S1/S2 protein provided further structural insights into the inhibitory mechanisms of the allosteric binders; ZINC3939013 and ZINC27990463.
Analyses of interaction dynamics revealed that while Compd A (ZINC3939013) was optimally buried within the deep hydrophobic pocket of Site 1, Compd B (ZINC27990463) exhibited trans-domain binding across Site 2 and its adjacent NTD-RBD linker (L) region. This binding activity demonstrated by Compd B could account for its prominent disruptive effects on the S-protein-hACE2 complexes. ΔG binds of −52.7 kcal/mol and −32.57 kcal/mol were estimated for ZINC3939013 and ZINC27990463 respectively. Per-residue energy decomposition revealed the crucial contributions of R190, H207, K206 and K187, I101, R102, I119, F192, L226, V126, and W104 to the binding of ZINC3939013. Also, energy contributions of Q957, N953, Q954, L303, Y313, Q314, L858, V952, N953, and A956 to ZINC27990463 binding and stability were reported. While de novo binding site characterization performed herein were based on computational modeling methods, experimental methods such as Surface Plasmon Resonance (SPR), Isothermal Titration Calorimetry (ITC), Biolayer Interferometry (BLI), and Microscale Thermophoresis (MST) can be employed for further validation. These implementations will provide additional insights into the targetability and suitability of these pockets for novel COVID-19 therapeutics. Findings from this study paves way for novelty in the structure-based design of high-affinity allosteric inhibitors or disruptors of SARS-CoV-2 association with host hACE2 thereby preventing viral entry.
Consent
Not applicable to this study.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Fisayo A. Olotu: Conceptualization, Formal analysis, Writing - review & editing, conceptualized, implemented, analyzed, interpreted and wrote the manuscript. Kehinde F. Omolabi: performed molecular dynamics simulation. Mahmoud E.S. Soliman: Writing - review & editing, revised and approved the manuscript for submission.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Authors thank the college of health sciences, University of KwaZulu-Natal, South Africa for providing infrastructural support and we also acknowledge the Center for High Performance Computing (CHPC), Capetown, South Africa, for providing computational resources.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100451.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Worldometer . Worldometer; 2020. Coronavirus cases. [Google Scholar]
- 3.González J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navas-Martín S., Weiss S.R. Coronavirus replication and pathogenesis: implications for the recent outbreak of severe acute respiratory syndrome (SARS), and the challenge for vaccine development. J Neurovirol. 2004;10:75–85. doi: 10.1080/13550280490280292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong L.Y.R., Lui P.Y., Jin D.Y. Virol. Sin. Science Press; 2016. February) A molecular arms race between host innate antiviral response and emerging human coronaviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Elsevier Ltd; 2016. June) epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microb Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J C., F L., Zl S. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17 doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bucknall R.A., King L.M., Kapikian A.Z., Chanock R.M. Studies with human coronaviruses II. Some properties of strains 229E and OC43. Proc. Soc. Exp. Biol. Med. 1972;139:722–727. doi: 10.3181/00379727-139-36224. [DOI] [PubMed] [Google Scholar]
- 10.Woo P.C.Y., Lau S.K.P., Tsoi H., Huang Y., Poon R.W.S., Chu C., Lee R.A., Luk W., Wong G.K.M., Wong B.H.L., Cheng V.C.C., Tang B.S.F., Wu A.K.L., Yung R.W.H., Chen H., Guan Y., Chan K., Yuen K. Clinical and molecular epidemiological features of coronavirus HKU1–associated community‐acquired pneumonia. J Infect Dis. 2005;192:1898–1907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Z.W., Yuan S., Yuen K.S., Fung S.Y., Chan C.P., Jin D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. NLM (Medline) 2020 doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan J.F.W., To K.K.W., Tse H., Jin D.Y., Yuen K.Y. October) Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. Trends Microbiol. 2013 doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X., Lou Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R., Di Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adhikari, S. P., Meng, S., Wu, Y.-J., Mao, Y.-P., Ye, R.-X., Wang, Q.-Z., Sun, C., Sylvia, S., Rozelle, S., Raat, H., and Zhou, H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. [DOI] [PMC free article] [PubMed]
- 15.Astuti I., Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehr A.R., Perlman S. Coronaviruses: methods and protocols. Springer; New York: 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Liu Q., Guo D. April) Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. John Wiley and Sons Inc; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. Adv Virus Res. 2006 doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortola E., Roy P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 2004;576:174–178. doi: 10.1016/j.febslet.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C., Zheng X., Gai W., Zhao Y., Wang H., Wang H., Feng N., Chi H., Qiu B., Li N., Wang T., Gao Y., Yang S., Xia X. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget. 2017;8:12686–12694. doi: 10.18632/oncotarget.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Haan C.A.M., Rottier P.J.M. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. Adv Virus Res. 2005 doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeDiego M.L., Álvarez E., Almazán F., Rejas M.T., Lamirande E., Roberts A., Shieh W.-J., Zaki S.R., Subbarao K., Enjuanes L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo L., Masters P.S. The small envelope protein E is not essential for murine coronavirus replication. J Virol. 2003;77:4597–4608. doi: 10.1128/JVI.77.8.4597-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortego J., Ceriani J.E., Patiño C., Plana J., Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368:296–308. doi: 10.1016/j.virol.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.Y. Coronaviruses-drug discovery and therapeutic options. Nat Rev Drug Discov. 2016 doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012 doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosch B.J., Bartelink W., Rottier P.J.M. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian Z., Dominguez S.R., Holmes K.V. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim Y., Ng Y., Tam J., Liu D. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016 doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017 doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017 doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;80– doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan A., Benthin C., Zeno B., Albertson T.E., Boyd J., Christie J.D., Hall R., Poirier G., Ronco J.J., Tidswell M., Hardes K., Powley W.M., Wright T.J., Siederer S.K., Fairman D.A., Lipson D.A., Bayliffe A.I., Lazaar A.L. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit Care. 2017;21 doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020:1–4. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Othman, H., Bouslama, Z., Brandenburg, J.-T., Da Rocha, J., Hamdi, Y., Ghedira, K., Abid, N.-S., and Hazelhurst, S. In silico study of the spike protein from SARS-CoV-2 interaction with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. [DOI] [PMC free article] [PubMed]
- 49.Wu Yan, Wang Feiran, Shen Chenguang, Peng Weiyu, Li Delin, Zhao Cheng, Li Zhaohui, Li Shihua, Bi Yuhai, Yang Yang, Gong Yuhuan, Xiao Haixia, Fan Zheng, Tan Shuguang, Wu Guizhen, Tan Wenjie, Lu Xuancheng, Fan Changfa, Wang Qihui, Liu Yingxia, Chen Zh, L L., Wu Yan, Wang Feiran, Shen Chenguang, Peng Weiyu, Li Delin, Zhao Cheng, Li Zhaohui, Li Shihua, Bi Yuhai, Yang Yang, Gong Yuhuan, Zheng Fan H.X., Tan Shuguang, Wenjie Tan G.W., Lu Xuancheng, Fan Changfa, Wang Qihui, Chen Zhang Y.L., George Fu Gao J.Q., G F., L L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;80 doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020 doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu G., Wang Q., Gao G.F. August) Bat-to-human: spike features determining “host jump” of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. Elsevier Ltd; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.T.K., Wang Q., Du L., Tan W., Wilson I.A., Jiang S., Yang B., Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paola, L. Di, and Giuliani, A. Mapping active allosteric loci SARS-CoV spike proteins by means of protein Contact networks.
- 54.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 55.Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M.-Y., Pieper U., Sali A. Comparative protein structure modeling using MODELLER. Curr Protein Pept Sci. 2007 doi: 10.1002/0471140864.ps0209s50. [Chapter 2], Unit 2.9. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z., Lasker K., Schneidman-Duhovny D., Webb B., Huang C.C., Pettersen E.F., Goddard T.D., Meng E.C., Sali A., Ferrin T.E. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adeniji E.A., Olotu F.A., Soliman M. Exploring the lapse in druggability: sequence Analysis, Structural Dynamics and binding site characterization of K-RasG12C variant, a feasible oncotherapeutics target. Anticancer. Agents Med. Chem. 2018;18 doi: 10.2174/1871520618666180718110231. [DOI] [PubMed] [Google Scholar]
- 58.Balmith M., Soliman M.E.S. Potential Ebola drug targets – filling the gap: a critical step forward towards the design and discovery of potential drugs. Biologia (Bratisl). 2017;72:1–13. [Google Scholar]
- 59.Chetty S., Soliman M.E.S. Possible allosteric binding site on Gyrase B, a key target for novel anti-TB drugs: homology modelling and binding site identification using molecular dynamics simulation and binding free energy calculations. Med Chem Res. 2015;24:2055–2074. [Google Scholar]
- 60.Broomhead N.K., Soliman M.E. Can we rely on computational predictions to correctly identify ligand binding sites on novel protein drug targets? Assessment of binding site prediction methods and a protocol for validation of predicted binding sites. Cell Biochem Biophys. 2017 doi: 10.1007/s12013-016-0769-y. [DOI] [PubMed] [Google Scholar]
- 61.Halgren T.A. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- 62.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinf. 2009 doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Biovia D.S. San Diego Dassault Systèmes; 2016. Discovery studio 2016 client. [Google Scholar]
- 64.Jendele L., Krivak R., Skoda P., Novotny M., Hoksza D. PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Halgren T. New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- 66.Patschull A.O.M., Segu L., Nyon M.P., Lomas D.A., Nobeli I., Barrett T.E., Gooptu B. Therapeutic target-site variability in α 1-antitrypsin characterized at high resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:1492–1497. doi: 10.1107/S1744309111040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patschull A.O.M., Gooptu B., Ashford P., Daviter T., Nobeli I. In silico assessment of potential druggable pockets on the surface of ??1-antitrypsin conformers. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Case D.A. Univ. California; San Fr: 2018. Amber 18. [Google Scholar]
- 70.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 71.Seifert E. OriginPro 9.1: scientific data analysis and graphing software—software review. J Chem Inf Model. 2014;54:1552. doi: 10.1021/ci500161d. –1552. [DOI] [PubMed] [Google Scholar]
- 72.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;80 doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J., Jain A., McDonald L.R., Gambogi C., Lee A.L., Dokholyan N.V. Mapping allosteric communications within individual proteins. Nat Commun. 2020 doi: 10.1038/s41467-020-17618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.