Highlights
-
•
The new antigen test is an indispensable tool in the control of the pandemic due to its adequate sensitivity and specificity.
-
•
The implementation of the point of care technique in primary care is feasible and has good results.
-
•
Results of the antigen techniques are determined by an onset of symptoms inferior to five days and a CT below 27 in PCR.
Abbreviations: COVID-19, Coronavirus disease 2019; CT, cycle threshold; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; NP, nucleocapsid protein; LFA, lateral flow assay; FDA, Food and Drug Administration; RT- qPCR, real-time reverse transcription polymerase chain reaction; RDT, rapid diagnostic test
Keywords: Rapid antigen detection test, SARS-CoV-2, RT-PCR, Point-of-care testing, Lateral flow assay, COVID19
Abstract
Background
RT-qPCR is the current recommended laboratory method to diagnose SARS-CoV-2 acute infection, several factors such as requirement of special equipment, time consuming, high cost and skilled staff limit the use of these techniques. A more rapid and high-throughput method is essential.
Methods
We analyzed clinical data and nasopharyngeal samples, collected during September 2020, from patients attended at the emergency department of a secondary hospital and in two primary healthcare centers in Madrid. The performance of the Panbio™ COVID-19 AG Rapid Test Device for the detection of SARS-CoV-2 antigen was compared to RT-qPCR.
Results
255 nasopharyngeal swabs, including 150 from the emergency department and 105 from primary helthcare centers, were tested. 184 patients were symptomatic (72.1 %). Amongst the 60 positive RT-qPCR samples, 40 were detected by the rapid antigen test, given an overall sensitivity of 73.3 %. All the samples detected positive with the rapid antigen test were also positive with RT-qPCR. The median cycle threshold was 23.28 (IQR 18.5–30.16). Patients with less than seven days onset of symptoms showed a higher viral load, and sensitivity for rapid antigen test (86.5 %), compared to those with more days (sensitivity of 53.8 %)(p < 0.004).
Conclusions
The rapid antigen test evaluated in this study showed a high sensitivity and specificity in samples obtained during the first week of symptoms and with high viral loads. This assay seems to be an effective strategy for controlling the COVID-19 pandemic for the rapid identification and isolation of SARS-CoV-2 infected patients.
1. Introduction
Ensuring accurate diagnosis is essential to limit the spread of SARS-CoV-2 and for the clinical management of COVID-19. Although real-time reverse transcription polymerase chain reaction (RT-qPCR) is the currently recommended laboratory method to diagnose SARS-CoV-2 acute infection, several factors such as requirement of special equipment, time consuming, high cost and skilled staff limit the use of these molecular techniques. A more rapid and high-throughput method is in growing demand [1,2].
Until now, the use of antigen detection tests alone had been ruled out and not recommended due to their low sensitivity [[3], [4], [5]]. Previously in the first wave, several easy to perform rapid antigen detection tests were developed as the first line of diagnostic. However, the results obtained were not good enough [[6], [7], [8], [9]].
2. Material and methods
2.1. Population and study period
Study was conducted between September 10, 2020 and September 15, 2020 at Prínice de Asturias Hospital in Alcalá de Henares, Madrid and its area of influence. The cumulative incidence rate of active cases in the last 14 days (Confirmed Cases per 100,000 inhabitants) in this area was 518.8 [10]. We included patients that were attended at our emergency department and in two of our primary healthcare centers:
Emergency department (ED) patients: we included 135 symptomatic patients that were admitted in our ED with clinical suspicion or COVID-19 and 17 asymptomatic patients with history of contact with another COVID-19 patient.
Primary healthcare (PH) patients: 50 symptomatic patients and 55 asymptomatic patients attendend in two of our primary healthcare centres.
A total of two consecutive nasopharyngeal swabs were obtained from each patient. One of them was employed to perform the antigenic rapid test and the other sample was employed to carry out the RT-PCR.
2.2. Diagnostic procedures
RT-PCR: one automatic extractor was employed to obtain viral RNA from clinical samples: Hamilton Microlab Starlet (Hamilton Company, Bonaduz, Switzerland). RNA amplification was made using Allplex SARS-CoV-2 assay (Seegene, Seoul, South Korea).
Antigenic rapid test: we applied the Panbio COVID-19 Ag Rapid Test Device (Abbott Rapid Diagnostic Jena GmbH, Jena, Germany). This test is a qualitative membrane-based immunoassay (immunochromatography) for the detection of Nucleocapsid protein of SARS-CoV-2 in nasopharyngeal samples.
2.3. Clinical data
Demographic (age and sex) and clinical variables of the study population were obtained from the medical records. We also recorded the time from the onset of symptoms and the story of prior contact with COVID-19 patients.
2.4. Statistical analysis
Specificity and sensitivity with 95 % confidence intervals (95 %CI) were calculated using the RT-PCR results as gold standard. Sensitivity was evaluated globally and also according to the time from the onset of symptoms (≤ 5days, < 7 days and > 7 days). Agreement between techniques was evaluated using Cohen's kappa score [11].
Continuous variables were presented as median and interquartile range (IQR) and categorical variables as proportions. We used the Mann-Whitney U-test, χ2 test, or Fisher's exact test to compare differences between survivors and non-survivors where appropriate. For these comparisons, a p value of 0.05 or below was considered significant. Statistical analysis was performed with SPSS v20.0 (IBM Corp., Armonk, NY, USA) (Table 1 ).
Table 1.
Patients.
| Characteristics | Emergency department | Primary care | p-value |
|---|---|---|---|
| No. patients | 150 (58.8 %) | 105 (41.2 %) | |
| Demographic | |||
| Gender (female) | 75 (50.0 %) | 56 (53.3 %) | 0.596 |
| Age (years) | 515 (37,0–71,8) | 39,0 (25,0–56,0) | <0.001 |
| Asymptomatic patients with close contact | 13 (8.6 %) | 54 (51.4 %) | <0.001 |
| Days from close contact* | 2 (1–3) | 6 (4–8) | <0.001 |
| Symptomatic patients | 134 (89.3 %) | 50 (47.6 %) | ≤0.001 |
| Days from the onset of symptoms | 2 (1–5) | 4 (2–7) | ≤0.001 |
| Fever (> 38.0 °C) | 41(27.3 %) | 15(14.3 %) | 0.013 |
| Myalgia | 3(2.0 %) | 7(6.7 %) | 0.552 |
| Headache | 14(9.3 %) | 17(16.2 %) | 0.099 |
| Sore throat | 9(6.0 %) | 17(16.2 %) | 0.008 |
| Cough | 23(7 %) | 19(18.1 %) | 0.558 |
| Dyspnoea | 40(26.7 %) | 1(1.0 %) | <0.001 |
| Anosmia and/or ageusia | 0 | 3(2.8 %) | 0.037 |
| Diarrhea | 13(8.7 %) | 4(3.8 %) | 0.126 |
| confusional syndrome | 6(4.0 %) | 1(1.0 %) | 0.143 |
Only asymptomatic patients. Days from close contact with a confirmed COVID-19 case*.
3. Results
255 nasopharyngeal swabs collected from symptomatic and asymptomatic patients were tested. 60 (23,5 %) positive RT-qPCR samples, 44 (17,2 %) were detected by the rapid antigen test. In our study population, the overall sensitivity was 73.3 % (95 % IC: 62.2–83.8).
Considering only symptomatic patients with <5 days, <7 days or ≥7days since onset, the sensitivity was 85.3 % (95 % IC: 73.4–97.2) (Cohen's kappa score = 0.897), 86.5 % (95 % IC: 75.5–97.5) (Cohen's kappa score = 0.904) and 53.8 % (95 % IC: 26.7–80.9) (Cohen's kappa score = 0.617)(Fisher's exact test was p < 0.001 independently of group of patients) respectively. Specificity was always 1.0 % (Table 2 ).
Table 2.
Agreement between antigenic test and PCR.
| ≼ 5 days |
< 7 days |
≽ 7 days |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | PCR | PCR | |||||||||
| + | – | + | – | + | – | ||||||
| Antigen | + | 29 | 0 | Antigen | + | 32 | 0 | Antigen | + | 7 | 0 |
| – | 5 | 102 | – | 5 | 104 | – | 6 | 29 | |||
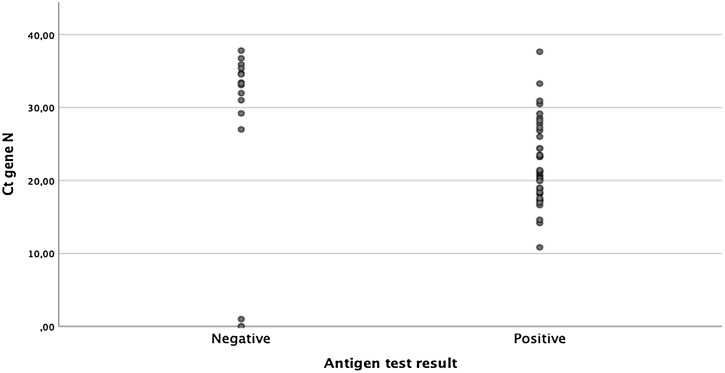
The RDT evaluated in this study showed a high sensitivity and specificity in samples mainly obtained during the first week of symptoms and with high viral loads (Tables 2 and 3 ). The range of cycle threshold (Ct) values was 0–37.8 (median 23.28 IQR 18.5–30.16). There were statistically significant differences when comparing the Ct gene N of patients with <7 days onset compared to the rest (p < 0.004) (Fig. 1 , Table 4 ).
Table 3.
Color scale stepped from white to gray of median of Cycle threshold (Ct), number of positive results for antigen test and PCR test and sensitivity of antigen test considering days since onset of a) symptoms or contact and b) only symptoms. *Number of positive results.
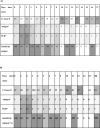 |
Fig. 1.
COVID-19 antigen results according to viral load.
Table 4.
Sensitivity of antigen test and percentage of patients with less of seven days from the onset of symptoms according to the viral loads represented by Cycle Threshold Values (Ct) of N gene.
| Ct N group | Symptomatic patients<7days* | Sensitivity |
|---|---|---|
| Ct < 25 (n = 34) | 26 (76.5 %) | 97.1 % |
| Ct<30 (n = 9) | 9 (1.0 %) | 77.8 % |
| Ct<35 (n = 10) | 1 (10 %) | 30 % |
| Ct<40 (n = 7), | 1 (14 %) | 14 % |
Days from the onset of symptoms<7days.
Considering asymptomatic patients with close contact the overall sensitivity was 54.5 % (95 % IC: 0,25-0,84) (Cohen's kappa score = 0.667). However in asymptomatic patients with less of seven days from close contact with a confirmed COVID-19 case it was 1.0 %, although only three positive RT-qPCR samples were included.
4. Discussion
As of September 18, 2020, the Food and Drug Administration (FDA) has approved more tan 150 SARS-CoV-2 RNA detection kits [12,13]. The lateral flow assay (LFA) is user-friendly, cheap, and easily mass-produced. In addition, it may be the optimal method for in-field detection of SARS-CoV-2 antigens if accurate, easy-to-use, rapid, and cost-effective [14]. In fact this is the first study carried out in primary care and shows the usability of the technique at this level of care.
Our results suggest that Panbio™ COVID-19 AG Rapid Test Device can rapidly identify SARS-CoV-2-infected individuals with moderate to high viral loads. Antigenic tests have shown 1.0 % specificity in all types of patients and can be a powerful tool of high positive predictive value to control the COVID pandemia. It could be used as a substitute technique for PCR in this type of patients to shortcut delays and intensive labour costs generated by the massive use of PCRs.
In the group of symptomatic patients with less than 7 days of evolution, the test reaches a sensitivity of 86.5 %, low to that described in the technical data sheet of the test. It is necessary to carry out studies with more patients to assess the usefulness that this technique could have in evaluating the infective potential of close contacts with PCR positive patients [15,16]. When testing with antigen tests, it must be considered that the infection prevalence and the clinical context of the recipient of the test affects at the pretest probability of the result being correct. Also, rapid antigen tests could be used for screening in high-risk clusters settings to identify quickly persons with a SARS-CoV-2 infection and prevent the transmission by repeat testing.
The role for this antigen-detecting rapid test can be considered in areas that are experiencing widespread community transmission, where the health system may be overburdened and where it may not be possible to test all suspect cases by RT- qPCR. In these situations, primary care may be the appropriate place to use it.
The RDT evaluated in this study showed a high sensitivity and specificity in samples mainly obtained during the first week of symptoms and with high viral loads (Ct<25). Its massive use, as suggested by the new protocols [5,17], can change the course of the pandemic. This assay seems to be an effective strategy for controlling the COVID-19 for the rapid identification and isolation of SARS-CoV-2 infected patients [5,17,18].
Informed consent
We also made sure that oral informed consent was obtained from each patient to participate on a voluntary basis in the study and a questionnaire with complete epidemiological and clinical questions was filled out. Oral informed consent was recorded in each questionnaire and was obtained from the next of kin, caretakers, or guardians on behalf of the minors/children enrolled in this study. No economic compensation was granted for participating in the study. All the data were treated confidentially and anonymized.
Ethical approval
The study was conducted according to the ethical requirements established by the Declaration of Helsinki. The Ethics Committee of Hospital Universitario Príncipe de Asturias (Madrid) approved the study.
Author contributions
Study concept and design: ML, RPT and JCG.
Patients’ selection and clinical data acquisition: ML, RPT, AC, JR, FPG, PGH and JCG.
Sample processing: ML, RPT, AC, JR, FPG, PGH, TA and JCG.
Statistical analysis and interpretation of data: ML and RPT.
Writing of the manuscript: ML, RPT, JR, FPG, PGH, TA and JCG.
Critical revision of the manuscript for relevant intellectual content: ML, RPT, AC, JR, FPG, PGH, TA and JCG.
Supervision and visualization: JCG.
All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
References
- 1.Loeffelholz M.J., Tang Y.W. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg. Microbes Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieberman J.A., Pepper G., Naccache S.N., Huang M.L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J. Clin. Microbiol. 2020;58(8):e00821–20. doi: 10.1128/JCM.00821-20. Published 2020 Jul 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephen M., Hahn M.D. Commisioner of Food and Drugs; 2020. Coronavirus (COVID-19) Update: FDA Authorizes First Antigen Test to Help in the Rapid Detection of the Virus That Causes COVID-19 in Patients. May 09. [Google Scholar]
- 4.Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagura-Ikeda M., Imai K., Tabata S. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J. Clin. Microbiol. 2020;58(9):e01438–20. doi: 10.1128/JCM.01438-20. Published 2020 Aug 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vashist S.K. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics (Basel) 2020;10(April (4)):202. doi: 10.3390/diagnostics10040202. PMID: 32260471; PMCID: PMC7235801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das D., Kammila S., Suresh M.R. Development, characterization, and application of monoclonal antibodies against severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Vaccine Immunol.: CVI. 2010;17(12):2033–2036. doi: 10.1128/CVI.00293-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kyosei Y., Namba M., Yamura S. Proposal of de novo antigen test for COVID-19: ultrasensitive detection of spike proteins of SARS-CoV-2. Diagnostics (Basel) 2020;10(8):E594. doi: 10.3390/diagnostics10080594. Published 2020 Aug 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mak G.C., Cheng P.K., Lau S.S. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.2020. Informe vigilancia de COVID-19. Infección por el nuevo coronavirus COVID-19. Semana 38 (Datos provisionales). Red de Vigilancia Epidemiológica Martes, 22 de septiembre de 2020.https://www.comunidad.madrid/sites/default/files/doc/sanidad/epid/informe_epidemiologico_semanal_covid.pdf Retrieved Sep 24, 2020. [Google Scholar]
- 11.McHugh M.L. Interrater reliability: the kappa statistic. Biochem. Med. (Zagreb) 2012;22:276–282. [PMC free article] [PubMed] [Google Scholar]
- 12.FDA . 2020. Coronavirus Disease 2019 (COVID-19) Emergency Use Authorizations for Medical Devices.https://www.fda.gov/medical-devices/emergency-use-authorizations-medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices Retrieved Sep 18, 2020. [Google Scholar]
- 13.2020. Information for Laboratories About Coronavirus (COVID-19)https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html Retrieved Sep 18, 2020, from. [Google Scholar]
- 14.Dinnes J., Deeks J.J., Adriano A. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020;8 doi: 10.1002/14651858.CD013705. Published 2020 Aug 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDC . 2020. Discontinuation of Transmission-based Precautions and Disposition of Patients with COVID-19 in Healthcare Settings (Interim Guidance)https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html Retrieved Sep 18, 2020. [Google Scholar]
- 16.CDC . 2020. Duration of Isolation and Precautions for Adults with COVID-19.https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html#:-:text=Recommendations,-Duration%20of%20isolation&text=For%20most%20persons%20with%20COVID,with%20improvement%20of%20other%20symptoms Retrieved Sep 18, 2020. [Google Scholar]
- 17.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., Kakizaki Y., Miyashita Y., Yagi S., Kojima S., Omata M. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int. J. Infect. Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. Aug 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.2020. Estrategia de detección precoz, vigilancia y control de COVID-19 Instituto de Salud Carlos III and Ministerio de Sanidad (Gobierno de España)https://www.mscbs.gob.es/en/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos.htm September 22. [Google Scholar]