Abstract
The Harzianum clade of Trichoderma comprises many species, which are associated with a wide variety of substrates. In this study, four new species of Trichoderma, namely T. lentinulae, T. vermifimicola, T. xixiacum, and T. zelobreve, were encountered from a fruiting body and compost of Lentinula, soil, and vermicompost. Their colony and mycelial morphology, including features of asexual states, were described. For each species, their DNA sequences were obtained from three loci, the internal transcribed spacer (ITS) regions of the ribosomal DNA, the gene encoding the second largest nuclear RNA polymerase subunit (RPB2), the translation elongation factor 1-α encoding gene (TEF1-α). The analysis combining sequences of the three gene regions distinguished four new species in the Harzianum clade of Trichoderma. Among them, T. lentinulae and T. xixiacum clustered with T. lixii, from which these new species differ in having shorter phialides and smaller conidia. Additionally, T. lentinulae differs from T. xixiacum in forming phialides with inequilateral to a strongly-curved apex, cultural characteristics, and slow growth on PDA. Trichoderma vermifimicola is closely related to T. simmonsii, but it differs from the latter by producing phialides in verticillate whorls and smaller conidia. Trichoderma zelobreve is the sister species of T. breve but is distinguished from T. breve by producing shorter and narrower phialides, smaller conidia, and by forming concentric zones on agar plates. This study updates our knowledge of species diversity of Trichoderma.
Keywords: compost, fungicolous, Hypocreaceae , mycoparasite
Introduction
The genus Trichoderma Pers., introduced by Persoon (1794), is cosmopolitan, including saprotrophs and mycoparasites in a diversity of ecosystems, such as agricultural fields, prairies, forests, salt marshes, and fungal fruiting body (Gazis and Chaverri 2010; Chaverri et al. 2015; Qiao et al. 2018). Species of this genus have been widely used in the biocontrol of plant pathogens (Chaverri et al. 2015; Degenkolb et al. 2015; Bunbury-Blanchette and Walke 2019) and production of enzymes and bioactive compounds (Sun et al. 2016). Nevertheless, some of them are associated with green mold diseases in the commercial production of mushrooms (Innocenti et al. 2019; Sun et al. 2019a) . Morphologically, the asexual-morphs are similar in producing branched tree-like conidiophores with cylindrical to nearly subglobose phialides and ellipsoidal to globose conidia, but their variation is insufficient to differentiate the Trichoderma species (Chaverri et al. 2015; Qin and Zhuang 2017; Qiao et al. 2018). Multilocus molecular phylogeny, based on combined sequence data of the internal transcribed spacer (ITS) regions, RNA polymerase II subunit (RPB2), and the translation elongation factor 1-α gene (TEF1-α), enables rapid and accurate identification of the Trichoderma species (Druzhinina et al. 2005; Atanasova et al. 2013; Chaverri et al. 2015). Currently, the combination of multi-gene phylogenetic analysis and phenotypic characteristics is extensively applied in species delimitation of Trichoderma (du Plessis et al. 2018; Qiao et al. 2018; Innocenti et al. 2019).
Trichoderma harzianum Rifai is one of the most well-known Trichoderma species, due to its antifungal properties and effective bio-control ability, used to suppress soil-borne plant pathogens (Chaverri et al. 2015; Degenkolb et al. 2015; Bunbury-Blanchette and Walker 2019). As a cosmopolitan and ubiquitous fungus, it has been isolated from diverse substrates, such as soil, plant tissue, and mushrooms (Chaverri et al. 2015; Jaklitsch and Voglmayr 2015; Innocenti et al. 2019; Sun et al. 2019b). Since Chaverri et al. (2015) provided a systematic revision of species in the Harzianum clade, numerous new species have been described (Jaklitsch and Voglmayr 2015; Qin and Zhuang 2016a; Sun et al. 2016; Chen and Zhuang 2017b; Qiao et al. 2018). Currently, more than 60 species are placed in the Harzianum clade (Jaklitsch and Voglmayr 2015; Qin and Zhuang 2016a, b, 2017; Chen and Zhuang 2017b; Qiao et al. 2018; Phookamsak et al. 2019;) .
It is estimated that 136 new species of Trichoderma have been recognised since 2015 (www.indexfungorum.org 2020), with 84 among these reported from China (Sun et al. 2012; Qin and Zhuang 2016a, b, 2017; Chen and Zhuang 2017a, b; Qiao et al. 2018), which evidenced that China has a high species diversity of Trichoderma (Zhu and Zhuang 2015; Jiang et al. 2016). In our survey of Trichoderma, eighteen isolates were obtained from soil, mushroom substrates, and vermicompost from northern China. Four new species belonging to the Harzianum clade were identified based on morphological features and DNA sequence data at three loci: the genes encoding RNA polymerase II subunit (RBP2) and translation elongation factor 1-α gene (TEF1-α), and the internal transcribed spacer (ITS) regions of the nuclear ribosomal RNA gene.
Materials and methods
Sampling sites and strains isolation
Since Trichoderma is easily isolated from soil, mushroom substrates, and earthworm substrates, the soil, mushroom substrates, and earthworm were therefore collected from Yinchuan, Ningxia Hui Autonomous Region, and Chaoyang district, Beijing, China. All the samples were stored at 4 °C before fungal isolation. Trichoderma strains were isolated by gradient dilution and the spread plate method or directly from the mushroom substrates. Three dilutions (10-1, 10-2, and 10-3) were prepared with 1 g soil and sterile water, and 100 µl of each dilution was spread on a 9 cm diameter Petri dish of PDA agar with100 mg/L chloramphenicol added. The plates were then incubated at 25 °C. Each of the individual colonies was transferred to a new PDA dish after 1–3 days and incubated at 25 °C. Dried cultures from the single spore or specimens of new species were deposited in the Herbarium Mycologicum Academiae Sinicae (HMAS) and the ex-type strains were preserved in the China General Microbiological Culture Collection Center (CGMCC)
Morphological analysis
For morphological studies, we used three different media: cornmeal dextrose agar (CMD, Difco, BD Science, USA), PDA (Difco, BD Science, USA), and synthetic low nutrient agar (SNA, Difco, BD Science, USA) (Chaverri et al. 2015). Each strain was first cultured on an SNA plate for 3 days and a small agar piece of 0.5 cm diameter with mycelium was then transferred, respectively, to new CMD, PDA, and SNA plates. Strains were incubated in 9 cm diam with three replicates. Petri dishes at 25 °C with a 12 h natural light and 12 h darkness interval. Colony diameter at 25 °C was measured three days after inoculation, and the time when mycelium entirely covered the surface of the agar plate was also recorded. Micromorphological characters were examined from the cultures of one-week-old colonies on SNA (Chaverri et al. 2015). A Nikon Ellipse 80i light microscope, equipped with differential interference contrast (DIC) optics, was used to capture digital images.
DNA extraction, PCR and sequencing
Genomic DNA of each strain was extracted from fresh mycelium growing on PDA after 5 days of growth following the rapid “thermolysis” method described in Zhang et al. (2010). For the amplification of ITS, RPB2, and TEF1-α gene fragments, ITS4 and ITS5 for ITS (White et al. 1990), EF1-728F (Carbone and Kohn 1999) and TEF1LLErev (Jaklitsch et al. 2005) for TEF1, and RPB2-5F and RPB2-7R for rpb2 (Liu et al. 1999) were used. Each PCR reaction consisted of 12.5 μl T5 Super PCR Mix (containing Taq polymerase, dNTP, and Mg2+, Beijing TsingKe Biotech Co. Ltd., Beijing), 1.0 μl of forward primer (10 μM), 1.0 μl of reverse primer (10 μM), 0.5 μl DMSO, 3 μl DNA template and 7 μl double sterilized water. PCR reactions were in Eppendorf Mastercycler, following the protocols described by Sun et al. (2016). PCR products were purified with the PCR product purification kit (TIANGEN Biotech, Beijing, China), and sequencing was carried out in both directions on an ABI 3730 XL DNA sequencer (Applied Biosystems, Foster City, California) with primers used during PCR amplification.
Phylogenetic analyses
Preliminary BLAST searches with ITS, RPB2, and TEF1-α gene sequences of the new isolates against NCBI, TrichOKey (Druzhinina and Kopchinski 2006), and TrichoBlast (Kopchinskiy et al. 2005) databases identified species closely related to our isolates. Based on this information, sequences of ITS, RBP2, and TEF1-α of 133 strains, representing 59 species were downloaded from GenBank, following recent publications (Qin and Zhuang 2017; Qiao et al. 2018; Innocenti et al. 2019). Among them, 139 strains are belonging to the Harzianum clade, and Trichoderma ceramicum, T. parestonicum, and T. estonicum were chosen to represent the outgroup.
Tree alignment files were generated by using MAFFT version 7.03 with the Q-INS-I strategy (Katoh and Standley 2013). Conserved blocks were selected from the initial alignments with Gblocks 0.91 b (Castresana 2000). The appropriate nucleotide substitution model for each gene was determined by using MrModeltest v2.4 (Nylander 2004). HKY + I + G was estimated as the best-fit model for RPB2, and GTR + I + G was estimated as the best-fit model for TEF1-α and ITS under the output strategy of AIC. The partition homogeneity test (p = 0.01) indicated that the individual partitions were not significantly incongruent (Cunningham 1997), thus the aligned sequences of ITS, RPB2, and TEF1-α were combined for analyses. The multi-locus phylogenetic analyses included 1065 characters for RBP2, 587 characters for TEF1-α, and 555 characters for ITS. All characters were weighted equally and gaps were treated as missing characters.
Maximum Likelihood (ML) analyses were performed by RAxML (Stamatakis 2006), using the GTR-GAMMA-I model. The maximum likelihood bootstrap proportions (MLBP) were using 1000 replicates. Bayesian Inference (BI) analyses were conducted with MrBayes v3.2.6 (Ronquist et al. 2012). Metropolis-coupled Markov Chain Monte Carlo (MCMC) searches were calculated for 10,000,000 generations, sampling every 100th generation with the best best-fit model for each gene. Two independent analyses with six chains each (one cold and five heated) were carried out until the average standard deviation of the split frequencies dropped below 0.01. The initial 25% of the generations of MCMC sampling were discarded as burn-in. The refinement of the phylogenetic tree was used for estimating Bayesian inference posterior probability (PP) values. The Tree was viewed in FigTree v1.4 (Rambaut 2012), values of Maximum likelihood bootstrap proportions (MLBP) greater than 50% and Bayesian inference posterior probabilities (BIPP), greater than 95% at the nodes, are shown along branches. The final alignments and the trees obtained have been deposited in TreeBASE (TreeBASE accession number: 25400).
Results
Phylogeny
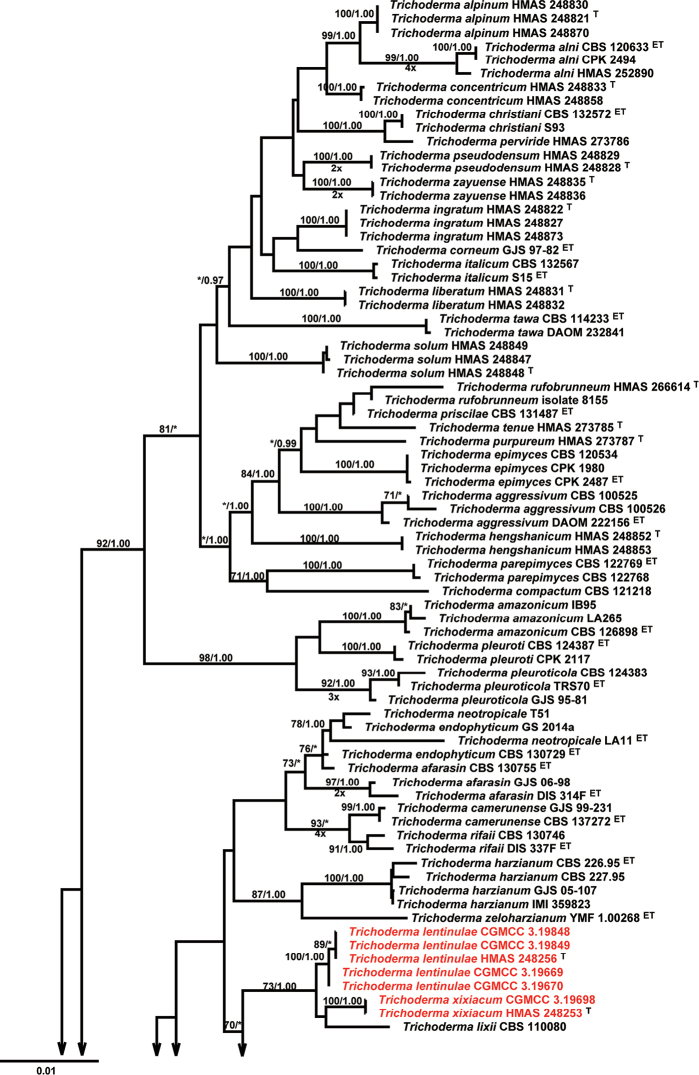
The preliminary BLAST searches with ITS, RPB2, and TEF1-α gene sequences of the new isolates suggest our isolates were highly similar to species from Trichoderma in the Harzianum-complex. Therefore, as the next step phylogenetic analyses were conducted by using a single gene of ITS, RPB2, TEF1-α, and multi-gene dataset of cascaded ITS, RPB2, and TEF1-α, respectively. The phylogenetic trees showed that our isolates were placed in the Harzianum clade (Fig. 1, Suppl. material 1: Fig. S1, Suppl. material 2: Fig. S2, Suppl. material 3: Fig. S3). In the phylogenetic tree conducted by a combined matrix of ITS, RPB2, and TEF1-α sequences, isolates of T. lentinulae, T. xixiacum, and T. lixii formed a well-supported clade (MLBP/BIBP = 73%/1.00). Within this clade, isolates of T. lentinulae and T. xixiacum formed a subclade with maximum support. Isolates of T. vermifimicola clustered together with T. simmonsii (BIBP = 1.00), both forming a subclade with maximum support (MLBP/BIBP = 100%/1.00, Fig. 1). Trichoderma zelobreve and T. breve, were distinguished by maximum support to respective clades while forming a highly supported clade (MLBP/BIBP = 100%/1.00, Fig. 1).
Figure 1.
Phylogenetic tree based on Maximum Likelihood analysis of a combined ITS, RPB2, and TEF1α sequence dataset. Trichoderma estonicum, Trichoderm parastinicum, Trichoderm ceramicum were chosen as the outgroup. Bootstrap Values higher than 70% from RAxML (BSML) (left) and Bayesian posterior probabilities greater than 0.95 (BYPP) (right) are given above the nodes. T indicates the type; ET indicates the ex-living type. Isolates obtained in this study are in red.
Figure 1.
Continued.
The ITS gene could not distinguish our isolates from other species within the Harzianum clade (Suppl. material 1: Fig. S1). In the phylogenetic tree resulted from the RPB2 gene, Trichoderma lentinulae, T. xixiacum, and T. lixii formed a highly supported clade (MLBP/BIBP = 100%/1.00), but within this clade, T. lentinulae, T. xixiacum were not distinguished (Suppl. material 2: Fig. S2). Isolates of T. vermifimicola formed a distinct clade (MLBP/BIBP = 100%/1.00) and grouped with T. simmonsii, T. guizhouense, and T. rugulosum but weakly supported (Suppl. material 3: Fig. S3). Trichoderma zelobreve and T. breve also formed a highly supported clade (MLBP/BIBP = 98%/1.00), but T. zelobreve and T. breve, were distinguished by maximum support to respective clades while forming a highly supported clade (MLBP/BIBP = 100%/1.00, Suppl. material 2: Fig. S2). In the phylogenetic tree resulted from the TEF1-α gene, T. zelobreve and T. breve also formed a highly supported clade (MLBP/BIBP = 98%/1.00), but were not distinct from each other (Suppl. material 3: Fig. S3). Isolates of T. lentinulae, T. xixiacum, T. vermifimicola, and T. simmonsii clustered together but this clade was not well-supported. Within this clade, isolates of T. lentinulae formed a well-supported subclade (MLBP/BIBP = 91%/1.00). Trichoderma xixiacum and T. vermifimicola formed a highly supported subclade (MLBP/BIBP = 100%/1.00). Within this group, isolates of T. vermifimicola clustered together with well-supported (MLBP/BIBP = 93%/1.00, Suppl. material 3: Fig. S3).
Taxonomy
Trichoderma lentinulae
Jing Z. Sun & X.Z. Liu sp. nov.
0AA38324-0ACC-5EC9-87D7-CD71AAE0401C
833233
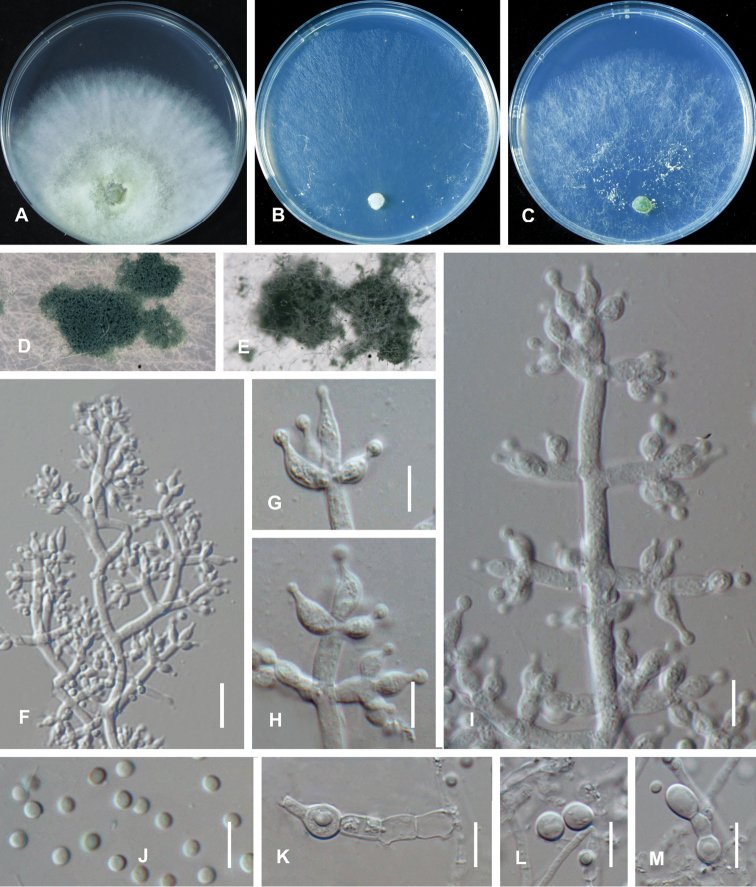
Figure 2.
Trichoderma lentinulae (CGMCC 3.19847). Cultures at 25 °C after 3 days (A on PDA B on CMDC on SNA) D conidiation pustules on CMD after 10 days E conidiation pustules on CMD after 10 d F conidiophores G–I Conidiophores and phialides J conidia K–M chlamydospores. Scale bars: 25 µm (F); 10 µm (G–M).
Etymology.
Latin, lentinulae, refers to the host from which the fungus was isolated.
Type.
China. Haidian District, Beijing, 39°57'40"N, 116°19'40"E, ca. 27 m elev., from a fruiting body and mushroom spawn of Lentinula edodes, 19 Oct 2018, Jing Z. Sun (HMAS 248256, holotype), ex-type culture CGMCC 3.19847.
Description.
On CMD after 72 h, colony radius 57–58 mm at 25 °C, covering the plate at 30 °C, 4–5 mm at 35 °C. Colony hyaline, weak, indistinctly radial. Aerial hyphae short, inconspicuous. No diffusing pigment noted, odor indistinct (Fig. 2B). Conidial production noted after 3 days, scant, effuse in aerial hyphae, becoming blue-green after 7 days. Chlamydospores not observed.
On PDA after 72 h, colony radius 45–46 mm at 25 °C, mycelium covering the plate at 30 °C, 11–12 mm at 35 °C. Colony white to yellowish-white, regularly circular, indistinctly zonate; mycelium dense and radial. No diffusing pigment, not distinct odor (Fig. 2A). Conidial production noted after 3 days, starting around the original inoculum, effuse in the aerial hyphae, first white, turning green after 3 d. Chlamydospores unobserved.
On SNA after 72 h, colony radius 51–52 mm at 25 °C, 52–53 mm at 30 °C, 4–5 mm at 35 °C. Colony hyaline, indistinctly zonate; mycelium loose, especially at the margin. Aerial hyphae loose. No diffusing pigment, not distinct odor (Fig. 2C). Conidial production noted after 2 days, starting around the inoculum, effuse in the aerial hyphae. Small pustules formed around the inoculum, first white, turning green after 3 d, with hairs protruding beyond the surface. Conidiophores pyramidal with opposing branches, less frequently solitary, closely-spaced branches, each branch, and the main axis terminating in 2–5 cruciately to nearly verticillately disposed phialides (Fig. 2F, H, I). Phialides ampulliform, typically strongly constricted below the tip, less frequently lageniform and then usually apex and inequilateral to strongly curved, hyaline, (3.5–)4.0–6.0(–6.5) × (2.0–)2.5–3.0(–3.5) µm (x̄= 4.5 × 3.0 μm, n = 30), length/width ratio (1.5–)2.0–3.0(–5.0) (x̄= 2.0, n = 30), base 1.0–2.5 μm (x̄= 1.5 μm)(Fig. 2G, H, I). Conidia ovoid to globose, smooth, hyaline when young, becoming green to dark green with age, (2.0–)2.5–3.0(–3.5) × (1.5–)2.0–2.5(–3.0) µm (x̄= 2.5 × 2.2 μm, n = 50), length/width ratio (1.0–)1.1–1.4 (–1.5) (x̄= 1.2, n = 50) (Fig. 2J). Chlamydospores common, apex or intercalary, ellipsoid or subglobose, (3.5–)5.0–6.5(–7.0) × (3.0–)4.0–5.0(–6.0) µm (x̄= 5.5 × 4.5 μm, n = 30), length/width ratio (1.0–)1.2–1.5 (–1.7) (x̄= 1.2, n = 30) (Fig. 2K–M).
Additional specimen examined.
China. Haidian District, Beijing, 39°57'40"N, 116°19'40"E, ca. 27 m elev., From a fruiting body and mushroom spawn of Lentinula edodes, 19 Oct 2018, Jing Z. Sun, living culture CGMCC 3.19848; Xixia District, Yinchuan, Ningxia Hui Autonomous Region, 38°38'52"N, 106°9'33"E, ca. 1127 m elev., from rhizosphere soil of Lycium chinois, 17 Oct 2018, Jing Z. Sun, living culture CGMCC 3.19699; ibid., living culture CGMCC 3.19670.
Teleomorph.
Undetermined.
Note.
The species is characterized by tree-like conidiophores, phialides verticillate or in whorls of 3–4, spindle-like to fusiform phialides (4.0–6.0 × 2.5–3.0 μm) and ovoid to subglobose conidia. Differs from T. lixii by shorter and wider phialides and smaller conidia. Differs from Trichoderma xixiacum by compact, relatively smaller phialides, and the pustules not forming distinctly zonate of pustules on SNA.
Trichoderma vermifimicola
Jing Z. Sun & X.Z. Liu sp. nov.
F20EA222-A753-5CD4-ABBF-A000DCDE66A2
833234
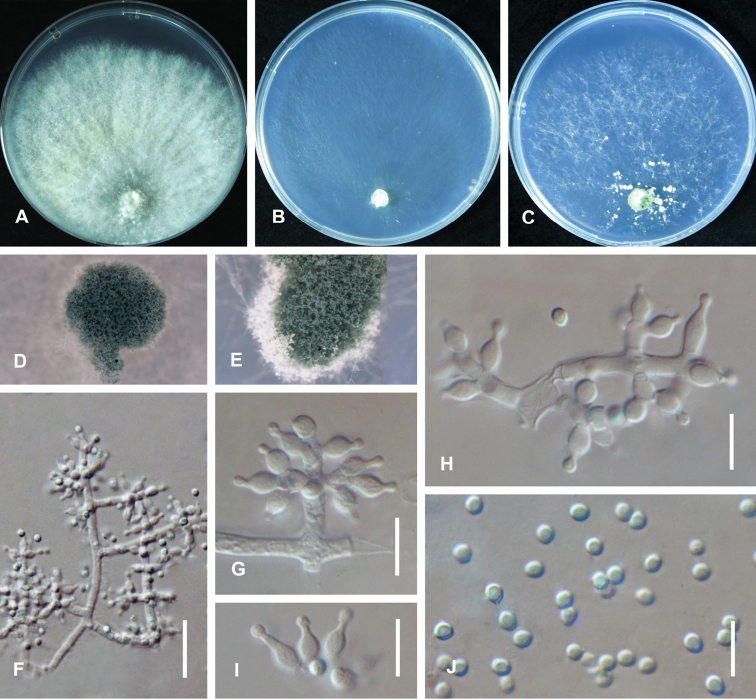
Figure 3.
Trichoderma vermifimicola (CGMCC 3.19694). Cultures at 25 °C after 3 days (A on PDA B on CMDC on SNA) D conidiation pustules on CMD after 10 days E conidiation pustules on SNA after 10 d F, H conidiophores G, J, K conidiophores and phialides I conidia. Scale bars: 25 µm (F, H); 10 µm (G, J–K).
Etymology.
Latin, vermifimicola, refers to the habitat of the type species.
Type.
China. Yongning, Yinchuan, the Ningxia Hui Autonomous Region, 40°0'41"N, 116°23'37"E, ca. 1678 m elev., from the substrates for earthworm cultivation, 18 Oct 2018, Jing Z. Sun (HMAS 248255, holotype), ex-type culture CGMCC 3.19694.
Description.
On CMD after 72 h, colony radius 49–51 mm at 25 °C, 51–52 mm at 30 °C, 4–5 mm at 35 °C. Colony hyaline, irregularly circular, indistinctly zonate; mycelium loose. Aerial hyphae short, inconspicuous. No diffusing pigment, not distinct odor. Conidial production noted after 3 days, starting around the inoculum (Fig. 3B). Small pustules formed at the colony margin, first white, turning blue-green after 7 d, with hairs protruding beyond the surface. Chlamydospores unobserved.
On PDA after 72 h, colony radius 55–58 mm at 25 °C, 55–56 mm at 30 °C, 5–6 mm at 35 °C. Colony white-green to bright green, regularly circular, distinctly zonate; mycelium dense and radial. Aerial hyphae short, inconspicuous. No diffusing pigment, not distinct odor. Conidial production noted after 2 days, starting around the inoculum, effuse in the aerial hyphae, first white, turning green after 2 d (Fig. 3A). Chlamydospores unobserved.
On SNA after 72 h, colony radius 48–50 mm at 25 °C, 51–52 mm at 30 °C, 3–4 mm at 35 °C. Colony hyaline, regularly circular, distinctly zonate; mycelium loose, especially at the margin. Aerial hyphae short, inconspicuous. No diffusing pigment, not distinct odor. Conidial production noted after 2 days, starting around the inoculum, effuse in the aerial hyphae. Small pustules formed along with two concentric rings, first white, turning yellow-green after 3 d, with hairs protruding beyond the surface (Fig. 3C). Conidiophores pyramidal with opposing branches, the distance between branches relatively large, each branch terminating in a whorl of 2–3 phialides, phialides sometimes solitary on the main axis (Fig. 3F, H, K); whorls typically cruciate, but often nearly verticillate (Fig. 3K); rarely conidiophores nodose and phialides disposed in more or less botryose clusters (Fig. 3H). Phialides ampulliform to lageniform, often constricted below the tip to form a narrow neck, hyaline, (4.4–)5.0–10.5(–11.2) × (2.0–)2.5–3.0(–3.5) µm (x̄= 6.6 × 2.7 μm, n = 30), length/width ratio (1.5–)1.8–2.8(–5.3) (x̄= 2.4, n = 30), base 1.6–2.5 μm (x̄= 1.9 μm) (Fig. 3G, I, K). Conidia ovoid to subglobose, smooth, hyaline when young, becoming green to dark green with age, (2.0–)2.3–2.6(–3.0) × (1.5–)2.0–2.4(–2.8) µm (x̄= 2.4 × 2.2 μm, n = 50), length/width ratio (1.0–)1.1–1.4(–1.7) (x̄= 1.2, n = 50) (Fig. 3J). Chlamydospores unobserved. No odor; no diffusing pigment observed.
Additional specimen examined.
China. Xixia District, Yinchuan, Ningxia Hui Autonomous Region, 38°38'52"N, 106°9'33"E, ca. 1127 m elev., from rhizosphere soil of Lycium chinois, 17 Oct 2018, Jing Z. Sun, living CGMCC 3.19697.
Teleomorph.
Undetermined.
Note.
Characterized by tree-like conidiophores, verticillate or in whorls of 3–4, ampulliform to lageniform phialides (5.0–10.5 × 2.5–3.0 μm), ovoid to subglobose conidia (2.4–2.6 × 2.0–2.5 μm). Differs from Trichoderma simmonsii by forming loose branches in whorls, relatively longer and thinner phialides, smaller conidia, and the fewer pustules on SNA.
Trichoderma xixiacum
Jing Z. Sun & X.Z. Liu sp. nov.
E6B880C9-D090-5DE6-A633-F72AFF924701
833235
Figure 4.
Trichoderma xixiacum (CGMCC 3.19697). Cultures at 25 °C after 3 d (A on PDA B on CMDC on SNA) D conidiation pustules on CMD after 10 d E conidiation pustules on SNA after 10 d F, G, I conidiophores and phialides H conidia. Scale bars: 10 µm (F, G); 10 µm (H, I).
Etymology.
Latin, xixiacum, refers to the type locality.
Type.
China. Xixia District, Yinchuan, Ningxia Hui Autonomous Region, 38°38'52"N, 106°9'33"E, ca. 1127 m elev., from rhizosphere soil of Lycium chinois, 17 Oct 2018, Jing Z. Sun (HMAS 248253, holotype), ex-type culture CGMCC 3.19697.
Description.
On CMD after 72 h, colony radius 55–56 mm at 25 °C, covering the plate at 30 °C, 9–11 mm at 35 °C. Colony hyaline, indistinctly zonate, mycelia loose. Aerial hyphae short, inconspicuous. No diffusing pigment, not distinct odor (Fig. 4B). Conidial production noted after 3 days, effuse in aerial hyphae, becoming blue-green after 4 days. Chlamydospores unobserved.
On PDA after 72 h, colony radius 59–60 mm at 25 °C, covering the plate at 30 °C, 7–8 mm at 35 °C. Colony white to yellow-white, regularly circular, indistinctly zonate; mycelium dense and radial. Aerial hyphae conspicuous. No diffusing pigment, not distinct odor (Fig. 4A). Conidial production noted after 3 days, starting around the original inoculum, effuse in the aerial hyphae, first white, turning blue-green after 7 d. Chlamydospores unobserved.
On SNA after 72 h, colony radius 51–52 mm at 25 °C, 52–53 mm at 30 °C, 4–5 mm at 35 °C. Colony hyaline, indistinctly zonate; mycelium loose, especially at the margin. Aerial hyphae short. No diffusing pigment, not distinct odor (Fig. 4C). Conidial production noted after 2 days, starting around the inoculum, effuse in the aerial hyphae. Small pustules formed around the inoculum, first white, turning green after 3 d, with hairs protruding beyond the surface. Conidiophores pyramidal with opposing branches, less frequently solitary, closely-spaced branches, each branch, and the main axis terminating in 2–5 cruciately to nearly verticillately disposed phialides (Fig. 4F, G, I). Phialides ampulliform to lageniform, often constricted below the tip to form a narrow neck, hyaline, (3.2–)3.5–7.0(–9.3) × (2.3–)2.6–3.3(–3.6) µm (x̄= 5.0 × 3.0 μm, n = 50), length/width ratio (1.2–)1.5–2.5(–4) (x̄= 1.8, n = 50), base 1.6–2.2 μm (x̄= 1.8 μm, n = 50) (Fig. 4I). Conidia subglobose to globose, smooth, hyaline when young, becoming green to dark green with age, (2.0–)2.3–2.7(–3.0) × (1.6–)2.0–2.6(–3.0) µm (x̄= 2.5 × 2.2 μm, n = 50), length/width ratio 1.0–1.3(–1.7) (x̄= 1.1, n = 50) (Fig. 4H). Chlamydospores unobserved. No odor; no diffusing pigment observed.
Additional specimen examined.
China. Xixia District, Yinchuan, Ningxia Hui Autonomous Region, 38°38'52"N, 106°9'33"E, ca. 1127 m elev., from rhizosphere soil of Lycium chinois, 17 Oct 2018, Jing Z. Sun, living CGMCC 3.19697.
Teleomorph.
Undetermined.
Note.
Characterized by tree-like conidiophores, verticillate or in whorls of 3–4, ampulliform to lageniform phialides (3.5–7.0 × 2.6–3.4 μm), subglobose to globose conidia (2.2–2.6 × 2.0–2.4 μm). Differs from Trichoderma lentinulae by compact, relatively smaller phialides, and the character of pustules on SNA. Differs from Trichoderma lixii by shorter and wider phialides and smaller conidia.
Trichoderma zelobreve
Jing Z. Sun & X.Z. Liu sp. nov.
6CE22E31-B776-5834-97A4-36AAE0236F30
833236
Figure 5.
Trichoderma zelobreve (CGMCC 3.19695). Cultures at 25 °C after 3 days (A on PDA B on CMDC on SNA) D conidiation pustules on CMD after 10 days E conidiation pustules on SNA after 10 d F conidiophores G, I conidiophores and phialides H phialides with conidia J conidia. Scale bars: 25 µm (F); 10 µm (G–J).
Etymology.
Greek zelo, meaning emulation + breve, referred to Trichoderma breve.
Type.
China. Chaoyang District, Beijing, 40°0'41"N, 116°23'37"E, ca. 35 m elev., 19 Oct 2018, isolated from soil, Jing Z. Sun (HMAS 248254, holotype), ex-type culture CGMCC 3.19695.
Description.
On CMD after 72 h, colony radius covering the plate at 25 °C and 30 °C, 11–12 mm at 35 °C. Colony hyaline, indistinctly radial; Aerial inconspicuous. No diffusing pigment, not distinct odor (Fig. 5B). Conidial production noted after 5 days, starting around the original inoculum. Small pustules formed at the colony margin, first white, olivaceous after 6 d, with hairs protruding beyond the surface. Chlamydospores unobserved.
On PDA after 72 h, colony radius 55–58 mm at 25 °C, covering the plate at 30 °C, 8–9 mm at 35 °C. Colony white to yellow-white; mycelium dense and radial. Aerial conspicuous. No diffusing pigment, not distinct odor (Fig. 5A). Conidial production noted after 3 days, starting around the inoculum, effuse in the aerial hyphae, first white, turning green after 4 d. Chlamydospores unobserved.
On SNA after 72 h, colony radius 62–63 mm at 25 °C, covering the plate at 30 °C, 7–8 mm at 35 °C. Colony hyaline, regularly circular; mycelium loose. Aerial conspicuous. No diffusing pigment, not distinct odor (Fig. 5A). Conidial production noted after 2 days, starting around the inoculum, effuse in the aerial hyphae. Small pustules formed along with two concentric rings, first white, turning yellow-green after 3 d, with hairs protruding beyond the surface. Conidiophores pyramidal with opposing branches, the distance between branches relatively large (Fig. 5F). Phialides, sometimes solitary, often paired or in whorls of 2–3 (Fig. 5F); whorls typically cruciate but often nearly verticillate; rarely conidiophores nodose and phialides disposed in more or less botryose clusters (Fig. 5G, H). Phialides ampulliform to lageniform, often constricted below the tip to form a narrow neck, hyaline (Fig. 5G, H, I), (3.5–)4.0–6.0(–7.0) × (2.2–)2.6–3.2(–3.5) µm (x̄= 4.8× 2.9 μm, n = 30), length/width ratio (1.1–)1.4–2.1(–2.5) (x̄= 1.5, n = 30), base 1.4–2.1 μm (x̄= 1.7 μm). Conidia ovoid to subglobose, smooth, hyaline when young, becoming green to dark green with age, (2.0–)2.3–2.6(–2.9) × (1.5–)1.8–2.2(–2.5) µm (x̄= 2.4 × 2.0 μm, n = 30), length/width ratio (0.8–)1.1–1.4(–1.7) (x̄= 1.2, n = 30) (Fig. 5J). Chlamydospores unobserved.
Additional specimen examined.
China. Chaoyang District, Beijing, 40°0'41"N, 116°23'37"E, ca. 35 m elev., isolated from soil, 19 Oct 2018, Jing Z. Sun, living culture CGMCC 3.19696.
Teleomorph.
Undetermined.
Note. Characterized by tree-like conidiophores, branches paired or in whorls of 3–4, ampulliform to lageniform (4.0–6.0 × 2.6–3.2 μm), ovoid to subglobose conidia (2.2–2.6 × 1.8–2.2 μm). Differs from Trichoderma breve by shorter phialides and smaller conidia, as well as the cultural characteristics and growth rates.
Discussion
A combination of phylogenetic, morphological, ecological, and biogeographical data has robustly resolved the taxonomy of Trichoderma (Jaklitsch and Voglmayr 2015; Qin and Zhuang 2016a; Sun et al. 2016; Chen and Zhuang 2017b; Qiao et al. 2018). In this study, phylogenetic analysis based on a single gene of ITS could not distinguish species of Trichoderma in the Harzianum clade from each other (Suppl. material 1: Fig. S1), which confirmed that the ITS region is not suitable for species delimitation of Trichoderma (Jaklitsch et al. 2012; Qin et al. 2018). Sequences of RPB2 and TEF1-α were powerful due to their suitable interspecific variations (Jaklitsch and Voglmayr 2015), and these have extensively been used in solving the taxonomy of Trichoderma (Jaklitsch and Voglmayr 2015; Qin and Zhuang 2016a; Chen and Zhuang 2017a, b; Qiao et al. 2018). Despite the phylogenetic analyses based on the single gene of RPB2 and TEF1-α generally revealed the phylogenetic relationship within the Harzianum clade (Suppl. material 1: Fig. S2, Suppl. material 3: Fig. S3), but the relationships among T. lentinulae, T. xixiacum, T. vermifimicola, T. zelobreve, and their closed taxa were not well distinct. Consideration of the universality and reliability of barcodes for species in the Trichoderma genus (Qiao et al. 2018), combined ITS, RPB2, and TEF1-α dataset was used for phylogenetic analysis in this study, revealing phylogenetic relationship among species in Harzianum clades, and suggesting that T. lentinulae, T. xixiacum, T. vermifimicola, and T. zelobreve are distinguishable from each other and species within and outside of Harzianum clade as well.
Table 1.
Species, strains and their corresponding GenBank accession numbers of sequences used for phylogenetic analyses.
| Species | Voucher/ culture Nos. | Origin | Substrate | GenBank accession No. | ||
|---|---|---|---|---|---|---|
| ITS | RPB2 | TEF1-a | ||||
| Trichoderma afarasin | CBS 130755 ET | Cameroon | Soil | AY027784 | AF348093 | |
| DIS 314F | Cameroon | Wood | FJ442259 | FJ442778 | FJ463400 | |
| GJS 06 98 | Cameroon | Soil | FJ442630 | FJ463327 | ||
| Trichoderma afroharzianum | CBS 124620 ET | Peru | Moniliophthora roreri | FJ442265 | FJ442691 | FJ463301 |
| CBS 466.94 | Netherlands | KP009262 | KP009150 | KP008851 | ||
| GJS 04-193 | Cameroon | Soil | FJ442233 | FJ442709 | FJ463298 | |
| Trichoderma aggressivum | CBS 100525 | UK | Mushroom compost | AF057600 | AF545541 | AF348095 |
| DAOM 222156 ET | Mushroom compost | AF456924 | FJ442752 | AF348098 | ||
| CBS 100526 | Ireland | Mushroom compost | FJ442607 | KP009166 | KP008993 | |
| Trichoderma alni | CBS 120633 ET | UK, England | Alnus glutinosa | EU518651 | EU498349 | EU498312 |
| CPK 2494 | EU518652 | EU498350 | EU498313 | |||
| HMAS 252890 | KT343763 | KT343758 | ||||
| Trichoderma alpinum | HMAS 248821 T | China, Sichuan | Soil | KY687906 | KY687958 | KY688012 |
| HMAS 248830 | KY687912 | KY687961 | KY688015 | |||
| HMAS 248870 | KY687953 | KY687963 | KY688017 | |||
| Trichoderma amazonicum | CBS 126898 ET | Peru | Hevea brasiliensis | HM142358 | HM142367 | HM142376 |
| IB95 | HM142359 | HM142368 | HM142377 | |||
| LA265 | HM142360 | HM142369 | HM142379 | |||
| Trichoderma atrobrunneum | GJS 05-101 | FJ442677 | FJ442745 | FJ463392 | ||
| GJS 90-254 | AF443926 | FJ442735 | AF443943 | |||
| Trichoderma atrogelatinosum | BMCC LU498 | New Zealand | KJ871087 | |||
| CBS 237.63 ET | New Zealand | MH858272 | KJ842201 | |||
| DAOM 167632 | KJ871083 | |||||
| Trichoderma bannaense | HMAS 248840 T | China, Yunan | Soil | KY687923 | KY687979 | KY688037 |
| HMAS 248865 | KY687948 | KY688003 | KY688038 | |||
| Trichoderma breve | HMAS 248844 T | China, Beijing | Soil | KY687927 | KY687983 | KY688045 |
| HMAS 248845 | KY687928 | KY687984 | KY688046 | |||
| Trichoderma brevicrassum | HMAS 248871 T | Soil | KY687954 | KY688008 | KY688064 | |
| HMAS 248872 | Soil | KY687955 | KY688009 | KY688065 | ||
| Trichoderma brunneoviride | CBS 120928 | EU518661 | EU498358 | EU498318 | ||
| CBS 121130 ET | EU518659 | EU498357 | EU498316 | |||
| Trichoderma camerunense | CBS 137272 ET | Cameroon | Soil | AY027780 | – | AF348107 |
| GJS 99 231 | AY027783 | AF348108 | ||||
| Trichoderma catoptron | DAOM 232830 | KJ842166 | KJ871245 | |||
| GJS 02 76 ET | Sri Lanka | Wood | AY737766 | AY737726 | ||
| Trichoderma ceramicum | CBS 114576 | FJ860743 | FJ860531 | FJ860628 | ||
| Trichoderma cerinum | BMCC LU784 | KJ871244 | ||||
| DAOM 230012 ET | Nepal | KC171336 | KJ842184 | KJ871242 | ||
| Trichoderma christiani | CBS 132572 ET | Spain | KJ665244 | KJ665439 | ||
| S93 | KJ665245 | KJ665442 | ||||
| Trichoderma cinnamomeum | GJS 96-128 | AY391916 | AY391977 | |||
| GJS 97-233 | AY391919 | AY391978 | ||||
| Trichoderma cinnamomeum | GJS 97-237 ET | USA, Missour | Decaying wood | AY737759 | AY391920 | AY737732 |
| Trichoderma compactum | CBS 121218 | AY941822 | KF134789 | KF134798 | ||
| Trichoderma concentricum | HMAS 248833 T | China, Hubei | Soil | KY687915 | KY687971 | KY688027 |
| HMAS 248858 | KY687941 | KY687997 | KY688028 | |||
| Trichoderma corneum | GJS 97-82 ET | Thailand | KJ665252 | KJ665455 | ||
| Trichoderma endophyticum | CBS 130729 ET | Ecuador | Theobroma gileri | FJ442243 | FJ463319 | |
| GS 2014a | FJ884177 | FJ967822 | ||||
| Trichoderma epimyces | CBS 120534 ET | Austria | EU518663 | EU498360 | EU498320 | |
| CPK 1980 | EU518662 | EU498359 | EU498319 | |||
| CPK 2487 ET | EU518665 | EU498361 | EU498322 | |||
| Trichoderma estonicum | GJS 96-129 | AY737767 | AF545514 | AF534604 | ||
| Trichoderma guizhouense | DAOM 231435 | EF191296 | EF191321 | |||
| HGUP0038 T | JN191311 | JQ901400 | JN215484 | |||
| S628 | KJ665273 | KJ665511 | ||||
| Trichoderma harzianum | CBS 226.95 ET | U.K. | Soil | AJ222720 | AF545549 | AF348101 |
| CBS 227.95 | AF057605 | AF348100 | ||||
| GJS 05 107 | FJ442679 | FJ442708 | FJ463329 | |||
| IMI 359823 | EF113587 | AF348092 | ||||
| Trichoderma hausknechtii | CBS 133493 | France | KJ665276 | KJ665515 | ||
| Trichoderma helicolixii | CBS 133499 ET | Spain | KJ665278 | KJ665517 | ||
| Trichoderma helicolixii | CBS 135583 | KJ665277 | KJ665516 | |||
| Trichoderma hengshanicum | HMAS 248852 T | China, Hubei | Soil | KY687935 | KY687991 | KY688054 |
| HMAS 248853 | KY687936 | KY687992 | KY688055 | |||
| Trichoderma hirsutum | HMAS 248834 T | China, Hubei | Soil | KY687916 | KY687972 | KY688029 |
| HMAS 248859 | KY687942 | KY687998 | KY688030 | |||
| Trichoderma hunanense | HMAS 248841 T | China, Hunan | Soil | NR_154571 | KY687980 | KY688039 |
| HMAS 248867 | KY687950 | KY688005 | KY688040 | |||
| Trichoderma ingratum | HMAS 248822 T | China, Sichuan | Soil | KY687917 | KY687973 | KY688018 |
| HMAS 248827 | KY687909 | KY687966 | KY688021 | |||
| HMAS 248873 | KY687956 | KY688010 | KY688022 | |||
| Trichoderma inhamatum | CBS 273.78 ET | Colombia | Soil | FJ442680 | FJ442725 | AF348099 |
| Trichoderma italicum | CBS 132567 | KJ665282 | KJ665525 | |||
| S15 ET | Italy | KJ665283 | KJ665526 | |||
| Trichoderma lentiforme | CBS 100542 ET | French Guiana | Decorticated wood | AF469189 | – | AF469195 |
| DIS 253B | FJ442619 | FJ442756 | FJ851875 | |||
| DIS 94D | FJ442615 | FJ442749 | FJ463379 | |||
| Trichoderma lentinulae | HMAS 248256 T | China | Lentinula | MN594469 | MN605867 | MN605878 |
| CGMCC 3.19848 | China | Lentinula | MN594470 | MN605868 | MN605879 | |
| CGMCC 3.19849 | China | Lentinula | MN594471 | MN605869 | MN605880 | |
| CGMCC 3.19699 | China | Soil | MN594478 | MN605876 | MN605887 | |
| CGMCC 3.19670 | China | Soil | MN594479 | MN605877 | MN605888 | |
| Trichoderma liberatum | HMAS 248831 T | China,Hubei | Soil | KY687913 | KY687969 | KY688025 |
| Trichoderma liberatum | HMAS 248832 | KY687914 | KY687970 | KY688026 | ||
| Trichoderma linzhiense | HMAS 248846 T | China, Tibet | Soil | KY687929 | KY687985 | KY688047 |
| HMAS 248874 | KY687957 | KY688011 | KY688048 | |||
| Trichoderma lixii | CBS 110080 ET | Thailand | Decayed Ganoderma | AF443920 | KJ665290 | AF443938 |
| Trichoderma neotropicale | LA11 ET | HQ022407 | HQ022771 | |||
| T51 | FJ884180 | FJ967825 | ||||
| Trichoderma parestonicum | CBS 120636 ET | FJ860803 | FJ860565 | |||
| Trichoderma parepimyces | CBS 122768 | FJ860801 | FJ860563 | FJ860665 | ||
| CBS 122769 ET | Austria | Wood | MH863234 | FJ860562 | FJ860664 | |
| Trichoderma perviride | HMAS 273786 | China,Hubei | Wood | KX026962 | KX026954 | |
| Trichoderma pinicola | KACC 48486 ET | Korea | root of Pinus densiflora | MH050354 | MH025993 | MH025981 |
| SFC20130926-S014 | MH025991 | MH025978 | ||||
| SFC20130926-S111 | MH025992 | MH025980 | ||||
| Trichoderma pleuroti | CBS 124387 ET | Korea | Pleurotus substrate | HM142363 | HM142372 | HM142382 |
| CPK 2117 | EU279975 | |||||
| Trichoderma pleuroticola | CBS 124383 ET | Korea | Pleurotus substrate | HM142362 | HM142371 | HM142381 |
| GJS 95 81 | AF345948 | AF348102 | ||||
| TRS70 ET | KP009264 | KP009172 | KP008951 | |||
| Trichoderma polypori | HMAS 248855 T | Hunan | Soil | KY687938 | KY687994 | KY688058 |
| HMAS 248861 | KY687944 | KY688000 | KY688059 | |||
| Trichoderma polysporum | S72 | KJ665685 | ||||
| Trichoderma priscilae | CBS 131487 ET | Spain | KJ665333 | KJ665691 | ||
| Trichoderma pseudodensum | HMAS 248828 T | Hubei | Soil | KY687910 | KY687967 | KY688023 |
| HMAS 248829 | KY687911 | KY687968 | KY688024 | |||
| Trichoderma pseudogelatinosum | CNUN309 ET | Japan | Shiitake mushroom | HM769754 | HM920173 | HM920202 |
| Trichoderma purpureum | HMAS 273787 T | China,Hubei | KX026961 | KX026953 | ||
| Trichoderma pyramidale | CBS 135574 ET | Italy | Olea europaea | KJ665334 | KJ665699 | |
| Trichoderma rifaii | CBS 130746 | Ecuador | Theobroma gileri | FJ442663 | FJ463324 | |
| DIS 337F ET | FJ442621 | FJ442720 | FJ463321 | |||
| Trichoderma rufobrunneum | HMAS 266614 T | China,Jilin | Rotten wood | KF729998 | KF730010 | KF729989 |
| isolate 8155 | KF730007 | KF729992 | ||||
| Trichoderma rugulosum | SFC20180301-001 T | MH050353 | MH025986 | MH025984 | ||
| SFC20180301-002 | MH025987 | MH025985 | ||||
| Trichoderma simmonsii | CBS 130431 | USA, Maryland | Decaying wood bark | AF443917 | FJ442757 | AF443935 |
| S297 | KJ665711 | |||||
| S7 | KJ665337 | KJ665719 | ||||
| Trichoderma simplex | HMAS 248842 T | China, Guangxi | Soil | KY687925 | KY687981 | KY688041 |
| HMAS 248860 | KY687943 | KY687999 | KY688042 | |||
| Trichoderma solum | HMAS 248847 | KY687930 | KY687986 | KY688049 | ||
| HMAS 248848 T | China, Hubei | Soil | KY687931 | KY687987 | KY688050 | |
| HMAS 248849 | KY687932 | KY687988 | KY688051 | |||
| Trichoderma stramineum | CBS 114248 ET | Sri Lanka | Decaying wood | AY737765 | AY391945 | AY737746 |
| TAMA 0425 | AB856609 | AB856748 | AB856675 | |||
| Trichoderma tawa | CBS 114233 ET | Thailand | Decaying bark | AY737756 | AY391956 | FJ463313 |
| DAOM 232841 | KJ842187 | EU279972 | ||||
| Trichoderma tenue | HMAS 273785 ET | China,Hubei | Wood | KX026960 | KX026952 | |
| Trichoderma tomentosum | DAOM 171918 | AY605715 | AY605759 | |||
| DAOM 178713a ET | Canada, Ontario | Ulmus wood | EU330958 | AF545557 | AY750882 | |
| DAOM 234236 | EU280083 | EU279971 | ||||
| Trichoderma velutinum | DAOM 230013 ET | Nepal | Soil | AF149873 | JN133569 | AY937415 |
| HMAS 273865 T | China, Heilongjiang | Soil | KX026965 | KX026957 | ||
| Trichoderma vermifimicola | CGMCC 3.19850 | China | Compost | MN594472 | MN605870 | MN605881 |
| HMAS 248255 T | China | Compost | MN594473 | MN605871 | MN605882 | |
| Trichoderma xixiacum | HMAS 248253 T | China | Soil | MN594476 | MN605874 | MN605885 |
| CGMCC 3.19698 | China | Soil | MN594477 | MN605875 | MN605886 | |
| Trichoderma zayuense | HMAS 248835 T | China,Tibet | Soil | KY687918 | KY687974 | KY688031 |
| HMAS 248836 | KY687919 | KY687975 | KY688032 | |||
| Trichoderma zelobreve | HMAS 248254 T | China | Mushroom | MN594474 | MN605872 | MN605883 |
| CGMCC 3.19696 | China | Soil | MN594475 | MN605873 | MN605884 | |
| Trichoderma zeloharzianum | YMF 1.00268 ET | China,Yunan | Soil | MH113932 | MH158996 | MH183181 |
Trichoderma lentinulae was phylogenetically close to T. xixiacum and T. lixii but represents a taxon (Fig. 1). Morphologically, it differed from T. xixiacum in producing less frequently lageniform phialides with inequilateral to a strongly-curved apex. The conidia of T. lentinulae are usually more slender (x̄= 2.0), than those of T. xixiacum (x̄= 1.8). In addition, the conidia of T. lentinulae (length/width ratio, x‒= 1.2) are slightly more slender than T. xixiacum (length/width ratio, x‒= 1.1). The two species also differ from each other in their cultural characteristics and growth rates (Figs 2A–C, 4 A–C). Trichoderma lentinulae differed from T. lixii in producing less frequently lageniform phialide with inequilateral to a strongly-curved apex. Additionally, T. lentinulae forms 2–5 apex phialides on the main axis (Fig. 2F, I) in contrast to 2–4 apex phialides of T. lixii (Chaverri et al. 2015). Trichoderma lentinulae is also clearly distinguished from T. lixii (phialides, 6.5–3.5 μm; conidia, 3.0–2.7 μm)(Chaverri et al. 2015) in producing shorter phialides (x̄= 4.5 × 3.0 μm) and smaller conidia (x̄= 2.5 × 2.2 μm). Trichoderma vermifimicola was phylogenetically associated with T. simmonsii (Fig. 1). Morphologically, it is hard to distinguish T. vermifimicola from T. simmonsii , because both form similar tree-like conidiophores, ampulliform to lageniform phialides and ovoid to subglobose conidia, but phialide whorls of T. vermifimicola were often nearly verticillate rather than cruciate in T. simmonsii (Chaverri et al. 2015). Furthermore, T. simmonsii grew fast (PDA 25–55 mm, SNA 10–35 mm) at 35 °C than T. vermifimicola. Additionally, the length/width ratio phialide of T. vermifimicola is larger (x̄= 2.4) than that of T. simmonsii (x̄= 1.9) (Chaverri et al. 2015), and T. vermifimicola also produces smaller conidia (x̄= 2.4 × 2.2 μm) (Fig. 3) than T. simmonsii (3.0–2.7 μm) (Chaverri et al. 2015). Trichoderma zelobreve was closely related to Trichoderma breve in the multi-gene phylogenetic analysis (Fig. 1). Morphologically, both fungi have short phialides, however, T. zelobreve differs from T. breve by producing shorter and narrower phialides (4.0–6.0 × 2.6–3.2 μm) than that of T. breve (6.7–10.0 × 2.8–3.9 μm) (Chen and Zhuang 2017a). The conidia of T. zelobreve are smaller (x̄= 2.4 × 2.0 μm) than those of T. breve (x̄= 3.0 × 2.8 μm). Additionally, T. zelobreve does not form a zonate colony on CMD, PDA, and SNA, whereas the colony of T. breve presents concentric zones on CMD and PDA and finely concentric zones on SNA (Chen and Zhuang 2017a). In a previous study, the phylogenetic analysis indicated that T. breve was a sister taxon of T. bannaense, but morphologically more similar to T. harzianum (Chen and Zhuang (2017a). Herein, our phylogenetic analyses presented T. breve was associated with T. zelobreve (Fig. 1), resulted from the little genetic variation of sequences of ITS and TEF1-α between them. The phylogenetic analysis in Chaverri et al. (2015) presented that T. simmonsii was associated with T. camerunense. In this study, our phylogenetic analysis presented that T. simmonsii was phylogenetically closed to T. vermifimicola, and T. camerunense phylogenetic to T. rifaii (Fig. 1, Suppl. material 3: Fig. S3). In a previous study, these species were recognized as the cryptic species in under T. harzianum (Chaverri et al. (2015).
Currently, the Harzianum clade contains more than 60 species which were isolated from soil, plant tissues, and other fungi (Jaklitsch and Voglmayr 2015; Qin and Zhuang 2016a; Chen and Zhuang 2017b; Qiao et al. 2018; Sun et al. 2019a, b). Several studies have confirmed that species in this clade are important because of their mycoparasitism (Chaverri et al. 2015; Chen and Zhuang 2017a; Sun et al. 2019). When numerous biological control agents were explored deriving from species in the Harzianum clade (Chaverri et al. 2015, several taxa, such as T. atrobrunneum T. pleuroti, and T. pleuroticola were recognized as causing agents of “Green mold” disease of cultivated mushroom (Innocenti et al. 2019; Sun et al. 2019a, b). In this study, T. lentinulae was isolated from a fruiting body and the cultivated substrates of L. edodes, causing the decay of the host as well. How T. lentinulae affect the cultivation of Lentinula edodes is worthy of further studies. Since T. lentinulae was isolated from mushroom, T. lentinulae and T. vermifimicola were isolated from the mushroom spawn and substrates for earthworm cultivation, T. xixiacum and T. zelobreve were isolated from soil, confirming that species in the Harzianum clade have flexible nutrition modes (Chaverri and Samuels 2013; Zhang et al. 2018). The new species introduced here are not only potential candidates for biological agent exploration, but also improve our understanding of the diversity of Trichoderma, especially of the Harzianum clade in China.
Supplementary Material
Acknowledgements
This research was jointly supported by Key Research and Development Programs in Ningxia Hui Autonomous Region (2018BBF02004) and the Natural Science Foundation of China (no. 31600024).
Citation
Gu X, Wang R, Sun Q, Wu B, Sun J-Z (2020) Four new species of Trichoderma in the Harzianum clade from northern China. MycoKeys 73: 109–132. https://doi.org/10.3897/mycokeys.73.51424
Funding Statement
Key Research and Development Programs in Ningxia Hui Autonomous Region (2018BBF02004);Natural Science Foundation of China (no. 31600024)
Supplementary materials
Figure S1
Xin Gu, Rui Wang, Quan Sun, Bing Wu, Jing-Zu Sun
Data type
phylogenetic tree
Explanation note
Phylogenetic tree based on Maximum Likelihood analysis of ITS sequence dataset. Trichoderm ceramicum, Trichoderma estonicum, and Trichoderm parastinicum were chosen as the outgroup. Bootstrap Values higher than 70% from RAxML (BSML) (left) and Bayesian posterior probabilities greater than 0.95 (BYPP) (right) are given above the nodes. T indicates the type; ET indicates the ex-living type. Isolates obtained in this study are in red.
Figure S2
Xin Gu, Rui Wang, Quan Sun, Bing Wu, Jing-Zu Sun
Data type
phylogenetic tree
Explanation note
Phylogenetic tree based on Maximum Likelihood analysis of RPB2 sequence dataset. Trichoderm ceramicum, Trichoderma estonicum, and Trichoderm parastinicum were chosen as the outgroup. Bootstrap Values higher than 70% from RAxML (BSML) (left) and Bayesian posterior probabilities greater than 0.95 (BYPP) (right) are given above the nodes. T indicates the type; ET indicates the ex-living type. Isolates obtained in this study are in red
Figure S3
Xin Gu, Rui Wang, Quan Sun, Bing Wu, Jing-Zu Sun
Data type
phylogenetic tree
Explanation note
Phylogenetic tree based on Maximum Likelihood analysis of TEF1α sequence dataset. <em>Trichoderm ceramicum</em> was chosen as the outgroup. Bootstrap Values higher than 70% from RAxML (BSML) (left) and Bayesian posterior probabilities greater than 0.95 (BYPP) (right) are given above the nodes. <sup>T</sup> indicates the type; <sup>ET</sup> indicates the ex-living type. Isolates obtained in this study are in red.
References
- Atanasova L, Druzhinina IS, Jaklitsch WM, Mukherjee P, Horwitz B, Singh U. (2013) Two hundred Trichoderma species recognized on the basis of molecular phylogeny. Trichoderma: Biology and Applications CABI, Wallingford: 10–42. 10.1079/9781780642475.0010 [DOI]
- Bunbury-Blanchette AL, Walker AK. (2019) Trichoderma species show biocontrol potential in dual culture and greenhouse bioassays against Fusarium basal rot of onion. Biological Control 130: 127–135. 10.1016/j.biocontrol.2018.11.007 [DOI] [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chaverri P, Branco-Rocha F, Jaklitsch W, Gazis R, Degenkolb T, Samuels GJ. (2015) Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 107: 558–590. 10.3852/14-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaverri P, Samuels GJ. (2013) Evolution of habitat preference and nutrition mode in a cosmopolitan fungal genus with evidence of interkingdom host jumps and major shifts in ecology. Evolution 67: 2823–2837. 10.1111/evo.12169 [DOI] [PubMed] [Google Scholar]
- Chen K, Zhuang WY. (2016) Trichoderma shennongjianum and Trichoderma tibetense, two new soil-inhabiting species in the Strictipile clade. Mycoscience 57: 311–319. 10.1016/j.myc.2016.04.005 [DOI] [Google Scholar]
- Chen K, Zhuang WY. (2017a) Discovery from a large-scaled survey of Trichoderma in soil of China. Scientific Reports 7, 9090. 10.1038/s41598-017-07807-3 [DOI] [PMC free article] [PubMed]
- Chen K, Zhuang WY. (2017b) Three new soil-inhabiting species of Trichoderma in the Stromaticum clade with test of their antagonism to pathogens. Current Microbiology 74: 1049–1060. 10.1007/s00284-017-1282-2 [DOI] [PubMed] [Google Scholar]
- Cunningham CW. (1997) Can three incongruence tests predict when data should be combined? Molecular Biology and Evolution 14: 733–740. 10.1093/oxfordjournals.molbev.a025813 [DOI] [PubMed]
- Degenkolb T, Nielsen KF, Dieckmann R, Branco-Rocha F, Chaverri P, Samuels GJ, Thrane U, von Dohren H, Vilcinskas A, Bruckner H. (2015) Peptaibol, secondary-metabolite, and hydrophobin pattern of commercial biocontrol agents formulated with species of the Trichoderma harzianum Complex. Chemistry & Biodiversity 12: 662–684. 10.1002/cbdv.201400300 [DOI] [PubMed] [Google Scholar]
- Druzhinina IS, Kopchinskiy AG. (2006) TrichOKEY v. 2 – A DNA Oligonucleotide BarCode Program for the Identification of Multiple Sequences of Hypocrea and Trichoderma In: Meyer W, Pearce C (Eds) International Proceedings of the 8th International Mycological Congress. Cairns, Australia, Medimond, Bologna, Italy.
- Druzhinina IS, Kopchinskiy AG, Komoń M, Bissett J, Szakacs G, Kubicek CP. (2005) An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genetics and Biology 42: 813–828. 10.1016/j.fgb.2005.06.007 [DOI] [PubMed] [Google Scholar]
- du Plessis IL, Druzhinina IS, Atanasova L, Yarden O, Jacobs K. (2018) The diversity of Trichoderma species from soil in South Africa, with five new additions. Mycologia 110: 559–583. 10.1080/00275514.2018.1463059 [DOI] [PubMed] [Google Scholar]
- Gazis R, Chaverri P. (2010) Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecology 3: 240–254. 10.1016/j.funeco.2009.12.001 [DOI] [Google Scholar]
- Innocenti G, Montanari M, Righini H, Roberti R. (2019) Trichoderma species associated with green mould disease of Pleurotus ostreatus and their sensitivity to prochloraz. Plant Pathology 68: 392–398. 10.1111/ppa.12953 [DOI] [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS. (2005) Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97: 1365–1378. 10.1080/15572536.2006.11832743 [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Voglmayr H. (2015) Biodiversity of Trichoderma (Hypocreaceae) in southern Europe and Macaronesia. Studies in Mycology 80: 1–87. 10.1016/j.simyco.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang JL, Chen J, Mao LJ, Feng XX, Zhang CL, Lin FC. (2016) Trichoderma biodiversity of agricultural fields in East China reveals a gradient distribution of species. PLoS ONE 11(8): e0160613. 10.1371/journal.pone.0160613 [DOI] [PMC free article] [PubMed]
- Katoh K, Standley DM. (2013) MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopchinskiy A, Komoń M, Kubicek CP, Druzhinina IS. (2005) TrichoBLAST: a multilocus database for Trichoderma and Hypocrea identifications. Mycological Research 109: 658–660. 10.1017/S0953756205233397 [DOI] [PubMed] [Google Scholar]
- Liu YJJ, Whelen S, Benjamin DH. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. https://github.com/nylander/MrModeltest2
- Phookamsak R, Hyde KD, Jeewon R, Bhat DJ, Jones EBG, Maharachchikumbura SSN, Raspé O, Karunarathna SC, Wanasinghe DN, Hongsanan S, Doilom M, Tennakoon DS, Machado AR, Firmino AL, Ghosh A, Karunarathna A, Mešić A, Dutta AK, Thongbai B, Devadatha B, Norphanphoun C, Senwanna C, Wei D, Pem D, Ackah FK, Wang GN, Jiang HB, Madrid H, Lee HB, Goonasekara ID, Manawasinghe IS, Kušan I, Cano J, Gené J, Li J, Das K, Acharya K, Raj KNA, Latha KPD, Chethana KWT, He MQ, Dueñas M, Jadan M, Martín MP, Samarakoon MC, Dayarathne MC, Raza M, Park MS, Telleria MT, Chaiwan N, Matočec N, de Silva NI, Pereira OL, Singh PN, Manimohan P, Uniyal P, Shang QJ, Bhatt RP, Perera RH, Alvarenga RLM, Nogal-Prata S, Singh SK, Vadthanarat S, Oh SY, Huang SK, Rana S, Konta S, Paloi S, Jayasiri SC, Jeon SJ, Mehmood T, Gibertoni TB, Nguyen TTT, Singh U, Thiyagaraja V, Sarma VV, Dong W, Yu XD, Lu YZ, Lim YW, Chen Y, Tkalčec Z, Zhang ZF, Luo ZL, Daranagama DA, Thambugala KM, Tibpromma S, Camporesi E, Bulgakov T, Dissanayake AJ, Senanayake IC, Dai DQ, Tang LZ, Khan S, Zhang H, Promputtha I, Cai L, Chomnunti P, Zhao RL, Lumyong S, Boonmee S, Wen TC, Mortimer PE, Xu J. (2019) Fungal diversity notes 929–1036: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 95: 1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Qiao M, Du X, Zhang Z, Xu JP, Yu ZF. (2018) Three new species of soil-inhabiting Trichoderma from southwest China. Mycokeys: 63–80. 10.3897/mycokeys.44.30295 [DOI] [PMC free article] [PubMed]
- Qin WT, Zhuang WY. (2016a) Four new species of Trichoderma with hyaline ascospores from central China. Mycological Progress 15: 811–825. 10.1007/s11557-016-1211-y [DOI] [Google Scholar]
- Qin WT, Zhuang WY. (2016b) Two new hyaline-ascospored species of Trichoderma and their phylogenetic positions. Mycologia 108: 205–214. 10.3852/15-144 [DOI] [PubMed] [Google Scholar]
- Qin WT, Zhuang WY. (2017) Seven new species of Trichoderma (Hypocreales) in the Harzianum and Strictipile clades. Phytotaxa 305: 121–139. 10.11646/phytotaxa.305.3.1 [DOI] [Google Scholar]
- Rambaut A. (2012) FigTree v1. 4. Molecular evolution, phylogenetics and epidemiology. Edinburgh: University of Edinburgh, Institute of Evolutionary Biology.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Sun JZ, Liu XZ, Jeewon R, Li YL, Lin CG, Tian Q, Zhao Q, Xiao XP, Hyde KD, Nilthong S. (2019a) Fifteen fungicolous Ascomycetes on edible and medicinal mushrooms in China and Thailand. Asian Journal of Mycology 2(1): 129–169. 10.5943/ajom/2/1/7 [DOI] [Google Scholar]
- Sun JZ, Liu XZ, McKenzie EH, Jeewon R, Liu JK, Zhang XL, Zhao Q, Hyde KD. (2019b) Fungicolous fungi: terminology, diversity, distribution, evolution, and species checklist. Fungal Diversity 95(1): 337–430. 10.1007/s13225-019-00422-9 [DOI] [Google Scholar]
- Sun RY, Liu ZC, Fu KH, Fan LL, Chen J. (2012) Trichoderma biodiversity in China. Journal of Applied Genetics 53: 343–354. 10.1007/s13353-012-0093-1 [DOI] [PubMed] [Google Scholar]
- Sun JZ, Pei YF, Li EW, Li W, Hyde KD, Yin WB, Liu XZ. (2016) A new species of Trichoderma hypoxylon harbours abundant secondary metabolites. Scientific Reports 6, 37369. 10.1038/srep37369 [DOI] [PMC free article] [PubMed]
- White T, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (Eds) PCR protocols: a guide to methods and applications, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Zhang WW, Zhang XL, Li K, Wang CS, Cai L, Zhuang WY, Xiang M, Liu XZ. (2018) Introgression and gene family contraction drive the evolution of lifestyle and host shifts of hypocrealean fungi. Mycology 9: 176–188. 10.1080/21501203.2018.1478333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Zhang S, Liu XZ, Wen HA, Wang M. (2010) A simple method of genomic DNA extraction suitable for analysis of bulk fungal strains. Letters in applied microbiology 51(1): 114–118. 10.1111/j.1472-765X.2010.02867.x [DOI] [PubMed] [Google Scholar]
- Zhu ZX, Zhuang WY. (2015) Trichoderma (Hypocrea) species with green ascospores from China. Persoonia 34: 113–129. 10.3767/003158515X686732 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Xin Gu, Rui Wang, Quan Sun, Bing Wu, Jing-Zu Sun
Data type
phylogenetic tree
Explanation note
Phylogenetic tree based on Maximum Likelihood analysis of ITS sequence dataset. Trichoderm ceramicum, Trichoderma estonicum, and Trichoderm parastinicum were chosen as the outgroup. Bootstrap Values higher than 70% from RAxML (BSML) (left) and Bayesian posterior probabilities greater than 0.95 (BYPP) (right) are given above the nodes. T indicates the type; ET indicates the ex-living type. Isolates obtained in this study are in red.
Figure S2
Xin Gu, Rui Wang, Quan Sun, Bing Wu, Jing-Zu Sun
Data type
phylogenetic tree
Explanation note
Phylogenetic tree based on Maximum Likelihood analysis of RPB2 sequence dataset. Trichoderm ceramicum, Trichoderma estonicum, and Trichoderm parastinicum were chosen as the outgroup. Bootstrap Values higher than 70% from RAxML (BSML) (left) and Bayesian posterior probabilities greater than 0.95 (BYPP) (right) are given above the nodes. T indicates the type; ET indicates the ex-living type. Isolates obtained in this study are in red
Figure S3
Xin Gu, Rui Wang, Quan Sun, Bing Wu, Jing-Zu Sun
Data type
phylogenetic tree
Explanation note
Phylogenetic tree based on Maximum Likelihood analysis of TEF1α sequence dataset. <em>Trichoderm ceramicum</em> was chosen as the outgroup. Bootstrap Values higher than 70% from RAxML (BSML) (left) and Bayesian posterior probabilities greater than 0.95 (BYPP) (right) are given above the nodes. <sup>T</sup> indicates the type; <sup>ET</sup> indicates the ex-living type. Isolates obtained in this study are in red.