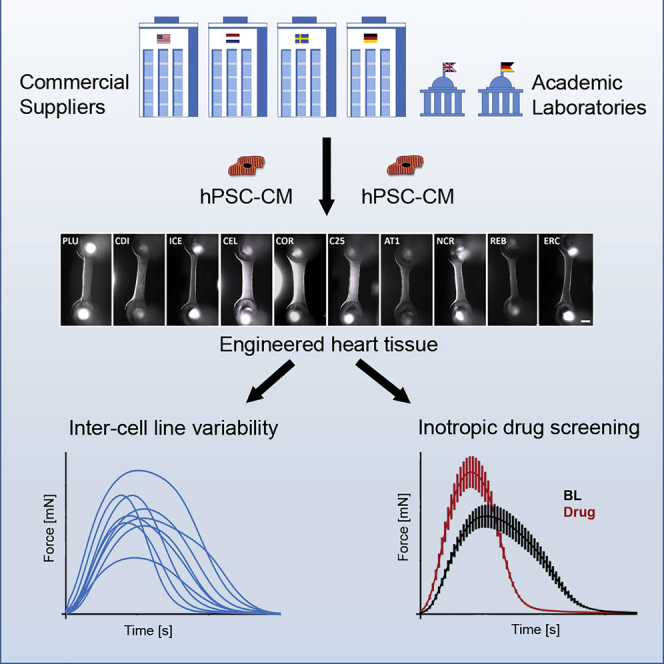
Summary
Human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM) are commercially available, and cardiac differentiation established routine. Systematic evaluation of several control hiPSC-CM is lacking. We investigated 10 different control hiPSC-CM lines and analyzed function and suitability for drug screening. Five commercial and 5 academic hPSC-CM lines were casted in engineered heart tissue (EHT) format. Spontaneous and stimulated EHT contractions were analyzed, and 7 inotropic indicator compounds investigated on 8 cell lines. Baseline contractile force, kinetics, and rate varied widely among the different lines (e.g., relaxation time range: 118-471 ms). In contrast, the qualitative correctness of responses to BayK-8644, nifedipine, EMD-57033, isoprenaline, and digoxin in terms of force and kinetics varied only between 80% and 93%. Large baseline differences between control cell lines support the request for isogenic controls in disease modeling. Variability appears less relevant for drug screening but needs to be considered, arguing for studies with more than one line.
Keywords: human induced pluripotent stem cells, cardiomyocytes, tissue engineering, drug safety pharmacology, variability
Graphical Abstract
Highlights
-
•
EHTs are stable from all tested hPSC-CM control lines
-
•
EHT baseline contractility shows high levels of variability between cell lines
-
•
Batch-to-batch variability is a large confounder of contractile parameters
-
•
Despite baseline variability, canonical drug responses were seen in all lines
In this article, Mannhardt and colleagues show that a systematic evaluation of 10 control hPSC-CM lines (commercial and academic) revealed high levels of variability regarding baseline contractility between the lines. In contrast, inotropic drug effects showed less prominent variability in canonical responses, arguing for a smaller relevance in drug screening.
Introduction
Developing a new drug is a very costly and time-consuming process, taking up to 10-15 years (Ashburn and Thor, 2004) and ∼1 billion Euro (Morgan et al., 2011). With cardiotoxicity (e.g., QTc interval prolongation, Torsade des Pointes arrhythmias) being a major cause of drug withdrawal and restrictions in the past (Onakpoya et al., 2016), new guidelines (ICH S7B and E14) for preclinical safety evaluation were established in 2004. The mandatory assessment of drug effects on single ion channels such as human ether-a-go-go (hERG) likely contributed to the elimination of potentially torsadogenic drugs, but also of potentially valuable safe drugs (Ewart et al., 2014). Moreover, single ion channel assays do not allow the analysis of inotropic effects or structural cardiotoxicity, another concern in preclinical drug development (Gintant et al., 2016). One key issue for achieving a higher level of predictivity is a testbed of human origin (Pang et al., 2019).
Cardiomyocytes (CMs) from human-induced pluripotent stem cells (hiPSC) offer great potential as a predictive human testbed for drug screening (Denning et al., 2016) and are routinely used in industrial and academic laboratories. Protocols for the differentiation of CMs have become highly efficient and robust, and hiPSC-derived CMs (hiPSC-CMs) are commercially available from several providers. While hiPSC-CMs replicate important features of native adult CMs with qualitatively normal responses to physiological and pharmacological stimuli, their immature developmental status remains a drawback (Sala et al., 2016). In general, hiPSC-CMs resemble fetal CMs in terms of morphology, contractility, electrophysiology, calcium handling, and metabolism (Scuderi and Butcher, 2017), associated with a hypersensitivity to calcium, a small response to beta-adrenergic stimulation, and a small contribution of the phosphodiesterase (PDE) 3 isoform compared with PDE4 (Guo et al., 2011; Mannhardt et al., 2017a).
Another obstacle is the variability between different control hPSC lines. A systematic comparison of 711 hiPSC lines from 301 donors showed that the variability of molecular signatures had a donor-dependent component, but that batches and unknown factors contributed to more than half of these (Kilpinen et al., 2017). Anecdotal reports described differences between batches, too (Huo et al., 2017; Mannhardt et al., 2017a; Nozaki et al., 2016). However, less is known about the functional variability between hiPSC-CM lines. In a small comparison of three batches of two commercial cardiomyocyte lines, Huo et al. (2017) saw indications that spontaneous beat rate and baseline field potential duration (FPD) affect the response to proarrhythmic stimuli. The Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative, a large study comparing the effects of many drugs on commercially available hiPSC-CM from two different lines, reported differences between the lines in detecting proarrhythmic effects (Blinova et al., 2017). A follow-up study with electrophysiological evaluation of 28 drugs across 10 different laboratories confirmed differences between the two commercial cell lines, but concluded that the cell line had minimal influence on drug categorization and their potential to detect drug-induced proarrhythmic effects (Blinova et al., 2018). Millard et al. (2018) investigated human stem cell-derived CMs provided by four different suppliers in three different platforms on multiple sites and saw differences with FPDc ranging from 271–577 ms. Comparison of four commercial cardiomyocyte lines in another study confirmed differences at baseline level with FPD ranging from 246–548 ms, highlighting the need for rate control during drug screening (Bot et al., 2018).
Systematic comparisons of hiPSC-CM contractility from larger numbers of lines are lacking. In this study we investigated CMs from 10 different hPSC control cell lines (4 commercial hiPSC-CM suppliers, 1 commercial hESC-CM supplier, and 5 academic hiPSC cell lines) in three-dimensional engineered heart tissue (EHT) format and compared their baseline phenotypes as well as their response to 7 drugs and detection of inotropic effects under electrical stimulation.
Results
EHT Formation and Baseline Contractility
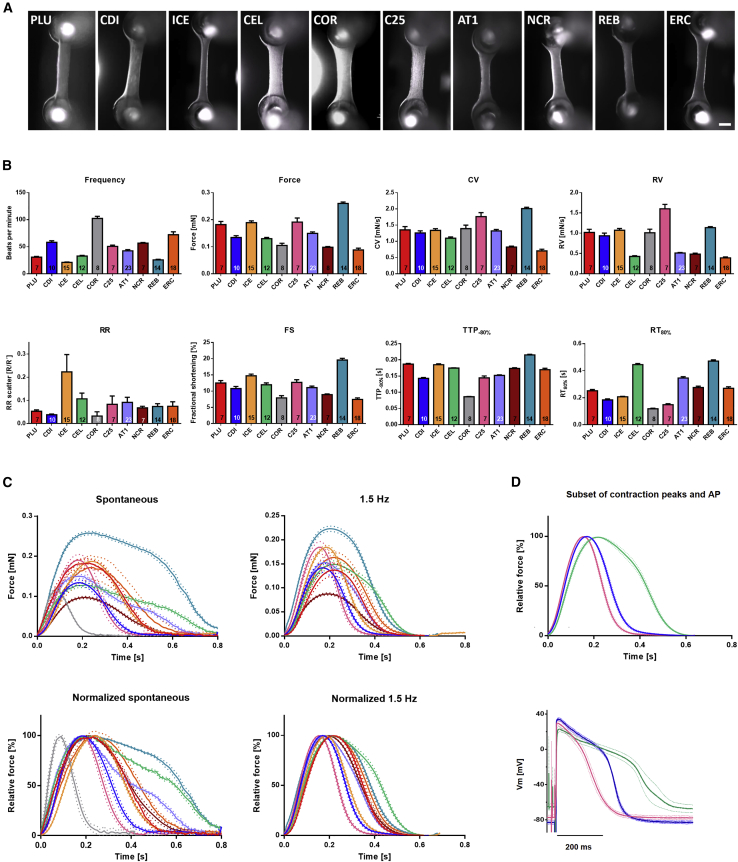
Human CMs from 10 control PSC lines (5 commercial, 5 academically generated lines from Hamburg, Nottingham, and the NIH) were used successfully to generate EHTs (Figure 1A). All tissues showed spontaneous macroscopic contractions that were analyzed with the video-optical EHT analysis system. Baseline contractility of EHT varied considerably between the different cell lines (Figure 1B). Spontaneous beating frequency (at culture day 23 ± 8) span from 21 ± 3 beats per minute (bpm; n = 15) in ICE-EHT to 102 ± 11 bpm (n = 8) in COR-EHT. Force varied from 0.09 ± 0.02 mN (n = 18) in ERC-EHT to 0.26 ± 0.02 mN (n = 14) in REB-EHT and fractional shortening from 7.5 ± 1.9% in ERC-EHT (n = 5) and 19.6 ± 2.0% (n = 14) in REB-EHT. Time to peak (TTP−80%; from 80% below peak; see also Figure S1) ranged from 86 ± 2 ms (n = 8) in COR-EHTs to 215 ± 7 ms (n = 14) in REB-EHTs. As contraction velocity (CV) depends on peak height/force, differences in CV varied from 0.70 ± 0.22 mN/s (n = 18) in ERC-EHTs to 2.01 ± 0.15 mN/s (n = 14) in REB-EHTs. Relaxation time (RT80%; from peak to 80% relaxation) showed the largest level of variability between the cell lines spanning from 118 ± 12 ms (n = 8) in COR-EHTs to 471 ± 33 ms (n = 14) in REB-EHTs. Relaxation velocity (RV) varied from 0.39 ± 0.09 mN/s (n = 18) in ERC-EHTs and maximal values of 1.60 ± 0.29 mN/s (n = 7) for C25-EHTs. EHTs from all lines beat rhythmically with an RR scatter between 0.03 ± 0.05 (n = 8) in the fast beating COR-EHTs and 0.22 ± 0.29 (n = 15) in slightly irregularly beating ICE-EHTs.
Figure 1.
Baseline Characterization of EHT
(A) Exemplary EHT of the 10 control cell lines used in this study. Scale bar, 1 mm.
(B) Baseline parameter of spontaneously beating in Tyrode's solution with 1.8 mM Ca2+. CV = contraction velocity, RV = relaxation velocity, RR‘ = regularity indicator, FS = fractional shortening, TTP-80% = time to peak starting at 20% above baseline, RT80% = relaxation time to 80% of total EHT relaxation (see also Figure S1). Replicate numbers are indicated in the respective column; data represent mean ± SEM.
(C) Average contraction peaks of spontaneously beating or electrically stimulated (1.5 Hz) EHTs. Red: PLU, n = 7; blue: CDI, n = 10; yellow: ICE, n = 15; green: CEL, n = 7; gray: COR, n = 8; pink: C25, n = 7; purple: AT1, n = 7; brown: NCR, n = 21; petrol: REB, n = 13; orange: ERC, n = 5. Data represent mean ± SEM of n EHTs (mean of 7–15 contraction peaks per EHT).
(D) Comparison of contraction peak shape and action potential shape at 1.5 Hz. Top: Subset of normalized average contraction peaks of electrically stimulated EHT displayed in C (green: CEL, blue: CDI, pink: C25). Bottom: Averaged action potentials from the three cell lines displayed in the top graph measured by sharp microelectrode (each n = 4/3; data represent mean ± SEM; see also Table S1).
To eliminate frequency-dependent effects on the contraction parameters, we compared baseline contractility also under electrical stimulation (1.5 Hz). Due to their high spontaneous beating frequency, COR-EHTs could not be captured with this slower pacing frequency but only at higher frequencies and were therefore not included in the average peak overlay (Figure 1C). Comparison of the other cell lines replicated the differences observed under spontaneous beating, but to a lower degree. Contraction kinetics, best seen in the overlay of the normalized average contraction peaks, varied maximally by a factor of two compared with a factor of four under spontaneous beating (Figure 1C).
Slow relaxation can have various reasons including differences in action potential (AP) duration and myofilament Ca2+ sensitivity. We therefore measured the AP in intact EHTs with a sharp microelectrode and compared the fastest (C25-EHTs) and a slow relaxing group (CEL-EHTs; Figure 1D). The AP results replicated the EHT contractility results confirming major cell line dependent differences (AP duration [APD]90: 222 ± 22 ms for C25-EHT; 292 ± 12 ms for CDI-EHTs; 435 ± 47 ms for CEL-EHTs; n/N = 4/3 each).
These data indicate that the EHT system is a robust platform for the functional evaluation of contractility working on all tested cell lines, but baseline values varied considerably.
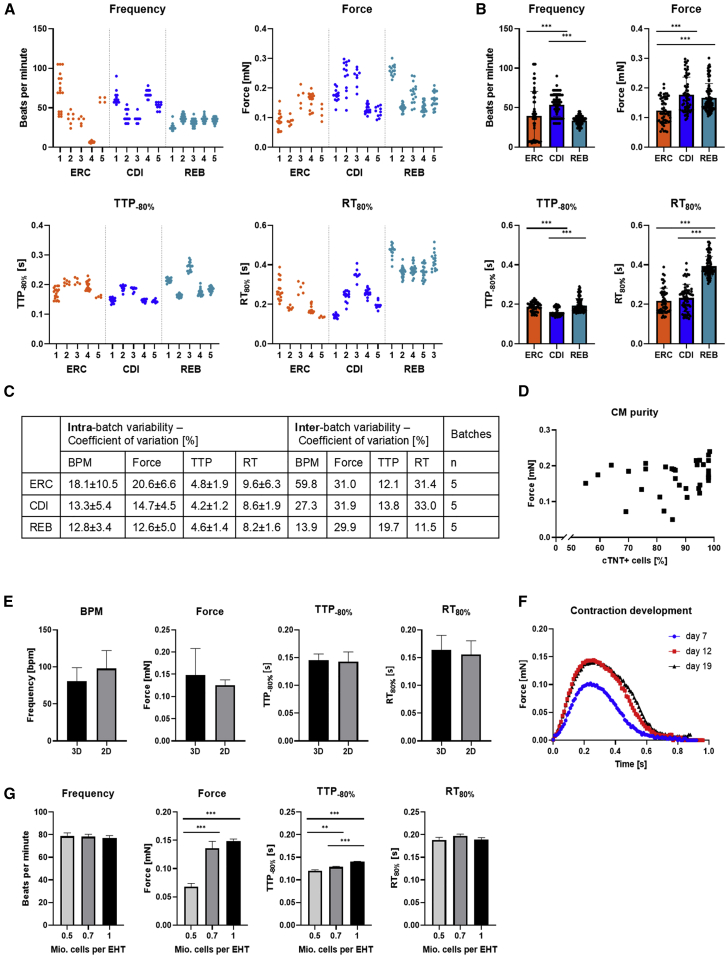
Batch-To-Batch Variability
Part of the variability could derive from differences between batches. We therefore tested 5 batches of independent cardiac differentiation runs of three cell lines and observed indeed strong variability between the batches (Figure 2A). Statistical analysis of the intra-batch variability showed that coefficients of variations (COV) were highest for force (20.6 ± 6.6% in ERC-EHT, 14.7 ± 4.5% in CDI-EHTs, and 12.6 ± 5.0% in REB-EHT). Intra-batch variability was smallest for contraction kinetics with COV amounting to 8.2%–9.6% for RT and 4.2–4.8 for TTP. As expected, inter-batch variability was higher than intra-batch variability with COV amounting to 13.9%–59.8% for spontaneous beating frequency, 29.9%–31.0% for force, 12.1%–19.7% for TTP and 11.5%–33% for RT (Figure 2C). Nevertheless, the much longer RT observed in REB-EHTs (Figure 1) was not an effect of batch-to-batch variability, but rather a true cell line-innate difference (Figure 2B).
Figure 2.
Batch-to-batch Variability and Other Variables Influencing EHT Contractility
(A) Contraction data (Frequency, force, TTP, and RT) for 5 batches of ERC, CDI, and REB-EHT. Scatterplot with each dot representing the mean of at least six peaks of one EHT.
(B) Pooled contraction data for three cell lines (ERC, CDI, REB) depicted in (A) 5 batches each, n = 52/5 EHTs (ERC), 59/5 EHTs(CDI), 96/5 EHTs (REB). Unpaired one-way ANOVA with Tukey's post-test, ∗∗∗p < 0.001.
(C) Intra-batch and inter-batch variability expressed as coefficients of variation for three cell lines.
(D) Correlation of cardiomyocyte purity and mean EHT force of 29 EHTs batches of five different cell lines at developmental plateau phase (stable force; see also Figure 2F) in EHT culture medium. Linear regression analysis showed no correlation of cardiomyocyte purity and respective force with a Pearson's correlation coefficient R2 of 0.04. Please note the cut x axis as all batches had a purity >50%.
(E) ERC-EHT casted with hiPSC-CM generated from either 3D-based or 2D-based differentiation protocols, mean of three batches each. Unpaired Student's t test: no significance.
(F) Changes in contraction peak shape in EHTs during development (day 7, 12, 19) indicating the plateau phase reached at day 19. Average contraction peaks of 3 peaks per n = 4 EHTs.
(G) ERC-EHT casted with different cell densities (n = 7-11). Unpaired one-way ANOVA with Dunnett's post-test, ∗∗p < 0.01, ∗∗∗p < 0.001. B, E-G: Data represent mean ± SEM
Confounders Influencing hiPSC-CM and EHT Contractility
To ensure that differences in contractility observed between the cell lines were not technical artifacts, we investigated possible confounders and their influence of hiPSC-CM and EHT contractility. Cardiomyocyte purity varied between cell lines (Table S1), but linear regression analysis of 29 EHT batches of five different cell lines showed no correlation with a Pearson's correlation coefficient R2 of 0.04 (Figure 2D). The method used for cardiac differentiation, either a 3D based protocol for large volumes in spinner flask suspension culture (Breckwoldt et al., 2017) or a 2D based protocol with adhesion culture in 6-well plates (Mosqueira et al., 2018) did not affect the contractile parameters of the EHT (Figure 2E). As shown in a previous publication (Mannhardt et al., 2017a), contractile parameters change during EHT development and their time in culture. After 15–20 days in culture, EHT development has reached a plateau phase with only minor changes in force and kinetics as indicated here in the overlay of paired average contraction peaks over time (Figure 2E). With age, spontaneous beating frequency is the only parameter that changes and slows down over time (data not shown). Because all our data were generated in the plateau phase of the EHT development, differences in EHT age are not likely the explanation for the large variability observed between the different cell lines. Reducing the cell concentration within the EHT to 70% or 50% of normal resulted in a significant reduction in TTP and, at 50%, also to a substantial (∼50%) reduction in force (Figure 2G). However, changes in cell concentration per EHT did not affect spontaneous beating frequency or RT.
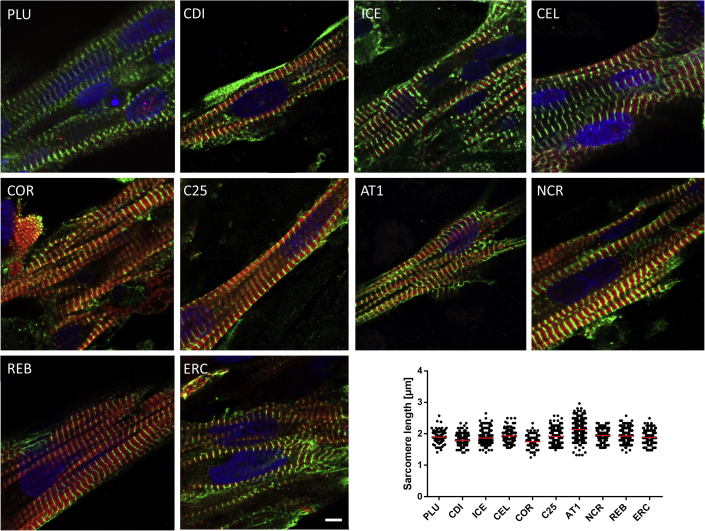
Histological Analysis
Analysis of sarcomeric structures was investigated by immunofluorescence analysis of whole EHTs (one per group) stained for alpha actinin and MLC2v. All EHTs showed CMs with longitudinal orientation and good sarcomeric organization, but the degree of MLC2v staining differed and ranged from rather low (PLU) to high (C25; Figure 3). Quantification of alpha actinin-positive Z-disk signals revealed significant differences in sarcomere length between the cell lines ranging from 1.75 ± 0.17 μm (n/N = 158/17) in COR-EHTs to 2.14 ± 0.27 μm (n/N = 344/28) in AT1-EHTs (see Table S2). For better overview, transversal sections of EHTs were stained with antibodies marking CM (dystrophin, MLC2v, MLC2a, alpha sarcomeric actin) and collagen or non-myocytes (smooth muscle actin, vimentin; see Figures S2 and S3). EHTs of all cell lines showed expression of mainly MLC2v-positive cells and less cells expressing MLC2a as a marker of immaturity. REB-EHTs were the only line in which MLC2a seemed to preside over MLC2v. Collagen was visible only sparsely in few EHT (PLU, ERC, C25) and absent in most investigated slides. Smooth muscle actin and vimentin-positive cells on the other hand confirmed the presence of non-myocytes within the population of commercial as well as academically generated cells used for the casting of EHT.
Figure 3.
Whole Mount Fluorescence Analysis
Immunofluorescence images of CM within EHT format. Green: Anti-alpha actinin. Red: Anti-MLC2v. Blue: DRAQ5. Scale bar, 5 μm. Quantification of sarcomere length indicated as mean ± SD. N = 158–358 sarcomeres; 10–20 cells; 1–2 EHTs. Two-way ANOVA with Tukey's post-test revealed numerous significant differences between the cell lines (see Table S2; see also Figures S2 and S3).
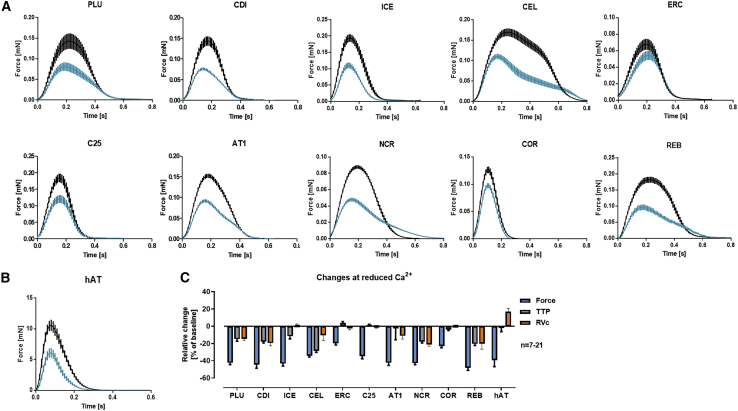
Response to Changes in Extracellular Calcium Concentrations
EHTs were exposed to a submaximal external calcium concentrations (0.5–1 mM), similar to previous studies (Mannhardt et al., 2016, 2017a) to study the effect of positive and negative inotropic drugs. Initial experiments revealed calcium-hypersensitivity (compared with adult heart tissues) of all tested cell lines with small differences between the lines. A threshold decrease in force of >20% (compared with baseline at 1.8 mM external calcium) was reached at external calcium concentrations ranging from 0.5 mM for PLU-, CEL-, and C25-EHTs and 0.6 mM for CDI-, ICE-, AT1-, NCR-, ERC-, and REB-EHTs to >1.0 mM for COR-EHTs (Figure 4). The calcium concentration in the medium not only reduced peak force, but also affected contraction and relaxation kinetics. 5/10 cell lines (PLU, CDI, CEL, NCR, REB) showed an unexpectedly large decrease in relaxation velocity (RVc) at lower external calcium levels. As the parallel decrease in force by itself already leads to a decrease in relaxation velocity, we corrected the RV for this change. Decreases in RVc are at variance with the response of human atrial trabeculae, which were studied for comparison and showed an increase in RVc (Figure 4C).
Figure 4.
Influence of External Calcium Concentration on Shape of Contraction Peak
(A and B) Average contraction peaks of paired EHTs (A) or human atrial trabeculae (hAT, [B] at high (1.8 mM; black) and low (0.5–1 mM; blue) external Ca2+ concentrations. PLU: 1 Hz, 0.5 mM Ca2+, n = 11. CDI: 1.5 Hz, 0.6 mM Ca2+, n = 10. ICE: 1.5 Hz, 0.6 mM Ca2+, n = 13. CEL: 1 Hz, 0.5 mM Ca2+, n = 7. ERC: 1.5 Hz, 0.6 mM Ca2+, n = 12. C25: 1.5 Hz, 0.5 mM Ca2+, n = 7. AT1: 1.5 Hz, 0.6 mM Ca2+, n = 8. NCR: 1.5 Hz, 0.6 mM Ca2+, n = 22. COR: 3 Hz, 1 mM Ca2+, n = 8. REB: 1 Hz, 0.6 mM Ca2+, n = 9. Please note differences in ordinate scales. (B) Average contraction peaks of paired human atrial trabeculae (hAT) at high (5 mM; black) and low (1.8 mM; blue) external Ca2+ concentrations; n = 8.
(C) Relative change in force, TTP and relaxation velocity RVc (corrected for force decrease) compared with baseline. All data are depicted as mean ± SEM.
Responses to Drugs
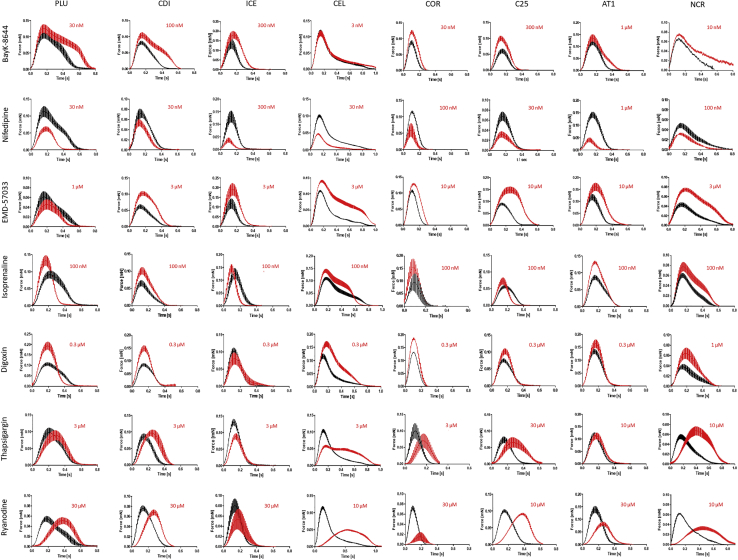
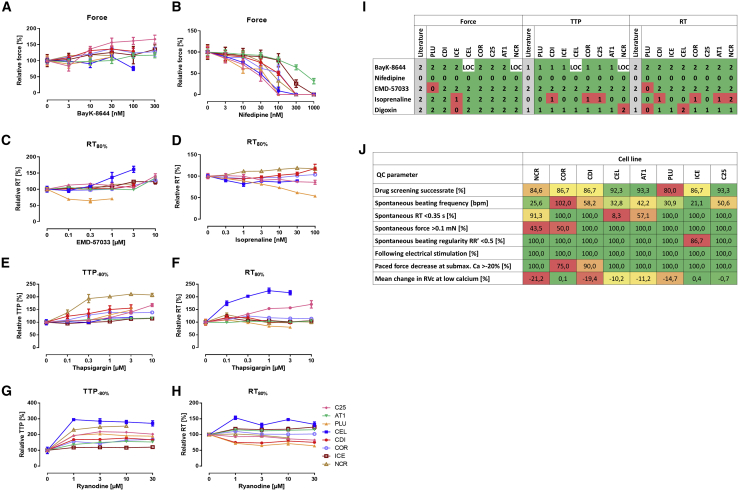
To evaluate the cell lines for their suitability for drug screening, EHTs of eight cell lines were exposed to seven inotropic indicator compounds (BayK-8644, nifedipine, EMD-57033, isoprenaline, digoxin, thapsigargin, ryanodine) in cumulative concentration-response curves under electric pacing (0.8–3 Hz; see Figure 5 legend). Figure 5 depicts the average contraction peaks at baseline (black) and at the indicated effective concentration of the respective drug (red). For full concentration-response curves with half-logarithmic steps see Figure S4 and Table S3. Exposure to the calcium channel activator BayK-8644 resulted in a positive inotropic and negative lusitropic effect (increase in RT). This effect was apparent in all cell lines (Figure 5) with differences in effect size as depicted in Figure 6A where most prominent differences in drug responses between lines are shown. CEL- and NCR-EHTs lost pacing capture due to the prolongation of relaxation on top of their long basal RT. The negative inotropic effect of the calcium channel blocker nifedipine was replicated in all cell lines (Figure 5), but the calculated half maximal inhibitory concentration (IC50) ranged from 20 nM in C25 to 497 nM in AT1 (Figure 6B). All cell lines, but PLU, showed a positive inotropic and negative lusitropic effect in response to the calcium sensitizer EMD-57033 (Figures 5 and 6C). A positive inotropic response to the beta-adrenergic agonist isoprenaline was also observed in all EHTs (Figure 5). The positive lusitropic effect (shortening of RT) of isoprenaline though, was not detected in most cell lines (Figure 6D). The Na+/K+-ATPase inhibitor digoxin led to a positive inotropic effect in all cell lines, but ICE (Figure 5). At higher concentrations, most cell lines showed decline in force amplitude, likely an indicator of toxicity. The SERCA blocker thapsigargin led to an increase in TTP (negative clinotropic effect) in most cell lines with variable effects on force and RT (Figures 5, 6E, and 6F). The ryanodine receptor antagonist ryanodine had a negative clinotropic effect on all lines, but ICE (Figures 5, 6G, and 6H). Fisher's exact test of the analyzed drug effects indicated an overall response rate of 87.9% (102/116) with the correct replication of canonical drug responses in the EHT. Because the effects of thapsigargin and ryanodine have not been studied well in human ventricular heart muscle preparations and show large inter-species differences (e.g., Sutko and Willerson, 1980), we decided to include only the five other drugs for the quantification of drug screening precision (Figure 6I). No single cell line replicated the entire canonical responses in all three parameters’ force, TTP, and RT. When regarding only the inotropic response, four cell lines (CDI, COR, AT1, C25) showed the expected change in force. Overall, the correctness varied between 80% (PLU) and 92%–93% (CEL, AT1, C25). Our findings suggest that differences in baseline contractility might be less relevant for drug screening, but at least two cell lines would be recommended to increase precision of the assay and detect all drug effects.
Figure 5.
Average Contraction Peaks of EHT Drug Responses
Average contraction peaks are depicted at baseline (black) and at the indicated concentration of the respective drug (red). Data were generated in Tyrode's solution with submaximal Ca2+ and electrical stimulation. Rows show effects of different drugs, columns separate different hiPSC-CM cell lines. PLU: 0.5 mM calcium, 1 Hz; CDI: 0.6 mM calcium, 1.5 Hz; ICE: 0.6 mM calcium, 1.5 Hz; CEL: 0.5 mM calcium, 0.8 Hz; COR: 1 mM calcium; 3 Hz/2.5 Hz; C25: 0.5 mM calcium, 1.5 Hz/2Hz; AT1: 0.6 mM calcium, 1.5 Hz; NCR: 0.6 mM calcium, 1 Hz; n = 2–6 each; data represent mean ± SEM; for details please see Table S2. Note: NCR data on BayK-8644 do not show electrically stimulated EHT peaks, as prolongation of RT led to failure of capture. Isoprenaline data paced with 1.5 Hz. Note that for visualization, contraction peaks of EHTs that ceased beating (e.g., COR at 0.3 μM digoxin) are not included in average peak illustration but in concentrations response curves (see also Figure S4 and Table S3).
Figure 6.
Cell Line-Specific Differences in Drug Responses
Examples of the most prominent differences in drug responses between cell lines.
(A) Relative change in force in response to BayK-8644 (C25: n = 5, AT1: n = 6, PLU: n = 4, CDI: n = 4, COR: n = 4, ICE, n = 6, CEL and NCR: n = 4, loss of after drug administration, see Figure S2). Note difference in effect size.
(B) Relative change in force in response to nifedipine (C25: n = 6, AT1: n = 6, PLU: n = 4, CDI: n = 3, COR: n = 4, ICE, n = 5, CEL: n = 4, NRC: n = 5). Note >10-fold differences in sensitivity.
(C) Relative change in RT80% in response to EMD-57033 (C25: n = 6, AT1: n = 6, PLU: n = 4, CDI: n = 3, COR: n = 4, ICE, n = 5, CEL: n = 2, NCR: n = 4). Note the unusual shortening of relaxation in PLU.
(D) Relative change in RT80% in response to isoprenaline (C25: n = 5, PLU: n = 7, CDI: n = 6, COR: n = 4, CEL: n = 6, NCR: n = 5). Note absence of lusitropic effect in most cell lines.
(E and F) Relative change TTP−80% and RT80% in response to thapsigargin (C25: n = 6, AT1: n = 6, PLU: n = 4, CDI: n = 3, COR: n = 4, ICE: n = 6, CEL: n = 4, NCR: n = 5). Note large differences in effect size ranging from no effect to >2-fold.
(G and H) Relative change in TTP−80% and RT80% in response to ryanodine (C25: n = 6, AT1: n = 5, PLU: n = 3, CDI: n = 3, COR: n = 3, ICE, n = 5, CEL: n = 3, NCR: n = 5). Note large differences in effect size and diverging effects on RT. Please note differences in scaling of the graph axes. For statistical analysis please see Table S3. A-H: All data represent mean ± SEM.
(I) Success of drug screening. Expected drug effects on force, TTP and RT are indicated in the respective first column (in gray, literature) and results observed for the respective cell lines in the following. Number coding: 2 = increase, 1 = no change, 0 = decrease of parameter, LOC = loss of capture – no results available. Color coding: green = canonical effect in congruence with literature (human ventricular muscle strips as gold standard), red = not-canonical response. Please note that “success” was visually evaluated from average contraction peaks shown in Figure 5 as they compile more detailed information than the narrow parameters listed in Table S3 alone.
(J) Classification of cell lines based on QC parameters and drug screening results with heatmap coloring indicating good performance in green and less ideal results in red.
Quality Control Parameter
As part of the internal EHT quality control (QC), we took into account a number of parameters that have proven critical in the past (Mannhardt et al., 2016, 2017a; Sala et al., 2018; Saleem et al., 2020). Figure 6J lists these QC parameters and their use for ranking of cell lines (see also Table S1). A high spontaneous beat rate obscures a positive force-frequency-relationship, which is typically only observed between 0.5 and 2 Hz, and can hinder the detection of positive inotropic effects (Saleem et al., 2020). Therefore, spontaneous beat rate should be low. In addition, a long RT (>0.35 s) is not physiological and can easily lead to loss of capture during electrical stimulation (e.g., CEL and NCR-EHT in BayK experiment, see also Figure 5). For detection of inotropic effects at submaximal calcium, a certain baseline force is required (threshold: 0.1 mN; Mannhardt et al., 2017a). Regularity of spontaneous beating is another QC parameter, as well as a reliable response to electrical stimulation. To enable detection of Ca-dependent inotropic effects, external calcium needs to be lowered to a half maximal effective concentration (EC50) concentration. We applied a QC threshold of more than −20% decrease in force and the absence of a relevant RVc in response to lowering of calcium (see also Figure 4).
Gene Expression Analysis
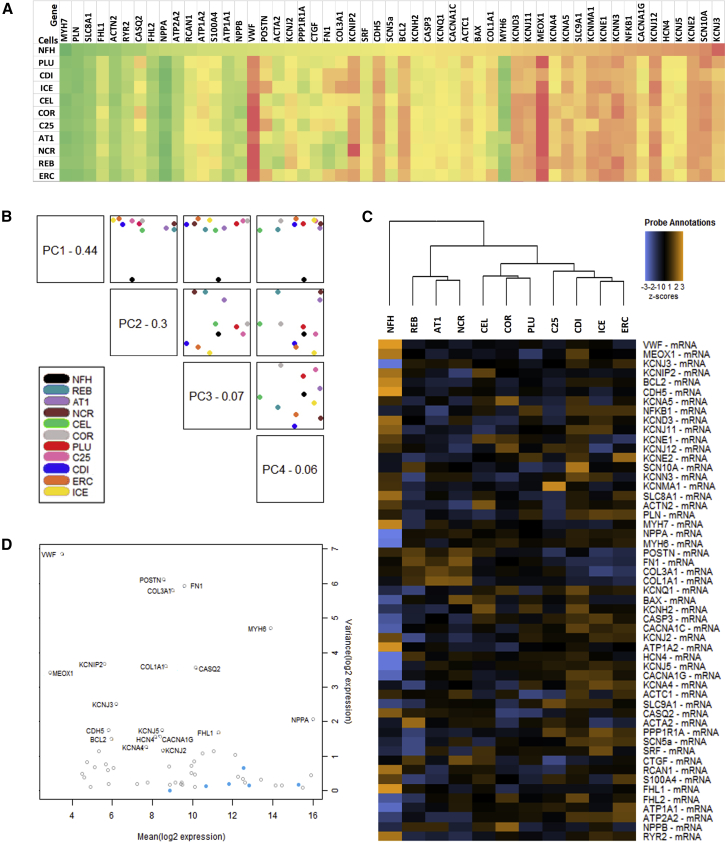
To evaluate whether differences between the EHT groups were also apparent on the transcript level, we performed Nanostring gene expression analysis in EHTs from the 10 different hPSC control cell lines and, for direct comparison, a sample from left ventricular myocardium from a non-failing human heart (NFH). We investigated 54 genes coding for proteins that are involved in cardiac excitation-contraction coupling or are dysregulated in heart failure (Figure 7). Gene expression in the hPSC lines was overall very similar without apparent difference between the nine hiPSC and the one hESC line or in general between commercial or in-house differentiated CM. Higher levels of variability (coefficient of variation >100%) were observed for genes associated with matrix proteins and non-CMs (COL1A/3A1, FN1, MEOX1, POSTN, VWF, KCNIP2, KCNMA1) and lesser variability (coefficient of variation <30%) for cardiac-specific genes or markers of apoptosis (ACTN2, ATP1A1/2A2, CACNA1C, FHL2, KCNE2, KCNQ1, NPPA, PLN, SLC8A1/9A1, SRF, BAX, CASP3). Interestingly, transcript abundance of many important cardiac genes (e.g., MYH7, PLN, RYR2, ATP1A1/2A2, KCNH2, CACNA1C) was similar in EHTs of all lines and NFH. Nevertheless, the immaturity of the hPSC-CM was apparent by higher MYH6 and HCN4 and lower CSQ abundance in EHTs than in NFH. As expected, all EHT samples—casted without the addition of endothelial cells or fibroblasts—showed much lower levels of markers of these cell types (VWF, CDH5, POSTN, COL3A1; Figure 7A). Principal-component analysis as well as hierarchical clustering indicated clear discrimination between NFH and the hPSC-EHT samples. Interestingly, hPSC-CMs from the same “house”/provider (e.g., REB/AT1 from Nottingham or COR/PLU from Pluriomics or ICE/CDI from CDI or ERC/C25 from Hamburg) clustered closer together than others (Figures 7B and 7C), indicating effects of local technical procedures.
Figure 7.
Gene Expression Analysis of hiPSC-EHTs
Transcriptome analysis (Nanostring) of non-failing human heart (NFH) and the 10 different hiPSC-CM control cell lines in EHT format.
(A) Heatmap of gene expression (log 10) normalized to housekeeping genes and ordered according to NFH levels. Gene expression is color coded with high expression levels marked in green and low gene expression levels in red. See also Figure S5.
(B) Principal-component analysis plotting the first four principal components of the gene expression data against each other.
(C) Hierarchical clustering and dendrogram for the full gene panel. Heatmap of the Z scores of the normalized data, scaled to give all genes equal variance, generated via unsupervised clustering.
(D) Internal quality control plotting variance versus mean normalized signal plot across all lines. Blue dots: housekeeping genes used for normalization. Clear circles: Endogenous genes.
Analysis of genes marking atrial or ventricular CM revealed overall higher expression of atrial genes in hiPSC-CM as compared with NFH sample taken from the left ventricle. In between the cell lines, lower expression of atrial genes was observed in C25 or PLU-EHTs, whereas higher expression of atrial genes was measured in ERC or COR-EHTs (Figure S5A). Ventricular genes were similarly expressed to the level in NFH for PLU, COR, AT1 and ERC-EHT, but lower expression was shown in REB- and C25-EHT (Figure S5A). Transcriptomic differences in expression of atrial or ventricular genes did not correlate with EHT contraction data and could not explain the differences in contractile phenotype of the cell lines. Markers of fibrosis and extracellular matrix proteins were both higher (AT1, REB, NCR) and lower expressed (ICE, ERC, CDI, C25) than in NFH (Figure S5B). A significant positive correlation with EHT RT was detected for CTGF and a similar trend was seen for COL1A, COL3A, FN1, and POSTN, but without reaching statistical significance (Figure S5C).
Correlation analysis between EHT contraction parameters and transcript levels (Figure S6 and Table S4) confirmed well-known groups of coupling partners (e.g., CACNA1C/PLN/RYR2/ATP2A2 with force/fractional shortening). It also revealed interesting new possible interaction partners that might be worth further investigation in the future (e.g., beat rate/TTP with KCNA5/KCNJ12, and MYH7 with RV, and FHL1 with RT).
Discussion
Biological variability is a natural phenomenon that needs to be dealt with in scientific research. Even with highly standardized manufacturing processes and QC, a certain degree of variability at baseline level is apparent also in commercial cardiomyocyte cell lines (Huo et al., 2017; Mannhardt et al., 2017a; Sala et al., 2016). This study investigated the baseline differences of 10 different control hPSC-CM lines and their consequences for drug screening in 8 lines. We found that baseline phenotypes of healthy control cell lines differ considerably, nevertheless canonical drug responses were observed in most EHTs of the tested cell lines. Variability appears less relevant for drug screening, but needs to be considered, arguing for drug testing to be done in more than one line.
Baseline Variability
Even though we and others have demonstrated variability of commercial hiPSC-CM due to cell batches (Huo et al., 2017; Mannhardt et al., 2017a), the profound level of variability between cell lines (amounting to coefficients of variation of 50.1% for beat rate, 34.8% for force, 21.2% for TTP and 43.6% for RT) exceeded our initial expectations, but are well in line with literature. Sala et al. (2016) showed APD90 to vary between cell lines from 120-600 ms at 1 Hz. Blinova et al. (2018) observed a higher beat rate with shorter APD90c in COR than ICE cells (299 ± 17 ms versus 463 ± 31 ms) with similar results across different platforms. These data match the differences in contraction duration (TTP + RT; COR: 205 ± 14 ms versus ICE: 392 ± 25 ms) observed in our study between these two lines. Platform and lab independence is further supported by the iCell cardiomyocyte (1.generation) APD80 values of 452 ± 10 ms at 1.5 Hz (Herron et al., 2016) matching the 435 ± 47 ms we measured for the CDI-EHT. These and our present data clearly indicate that “unrelated control lines” are unsuitable for disease modeling studies and confirm the request for isogenic controls to substantiate often subtle phenotypic differences between mutation-carrier and non-carrier lines (Sala et al., 2016). Taken into account the batch-to-batch variability, multiple independent batches are mandatory and effect sizes should exceed inter- and intra-batch coefficient of variation to allow for discrimination between scatter and phenotypic differences. The lack of these appropriate controls may lead to conflicting results between models which impairs further understanding of cardiac disease progression (Mosqueira et al., 2019).
Drug Screening
While canonical drug responses were observed for most drugs in almost all lines (87.9%), some notable exceptions were apparent (Figure 6) and could theoretically be due to differences in gene expression of drug targets. However, no differences in RNA abundance were found to correlate with different drug responses. The lack of positive lusitropic response to isoprenaline in some lines (CDI, CEL, AT1, NCR, COR) could not be correlated to differences in gene expression. PLN expression levels were similar to NFH for PLU, CDI, ICE, ERC and ∼60% for CEL, COR, C25, AT1, REB, NCR. ATP2A2 (encoding SERCA2a) gene expression was similar to NFH or up to 2-fold higher for some hPSC-EHT (CDI, ICE, ERC). Differences between lines did not explain differences in inotropic response to isoprenaline or the SERCA inhibitor thapsigargin. The latter had surprisingly moderate negative inotropic effects in a rabbit working heart preparation (Elliott et al., 2012). Similarly, the SERCA inhibitor cyclopiazonic acid (CPA) did not reduce peak force in human ventricular muscle strips even when combined with ryanodine, indicating that, at least for some time, the heart can compensate for the loss of SR function (Chung et al., 2018). Given this complexity and the above-mentioned species differences in the response to a single application of ryanodine (Sutko and Willerson, 1980), we decided to exclude the responses to thapsigargin and ryanodine from our correctness analysis.
Expression levels of the Na+/K+-ATPase (ATP1A1) were 4-fold and for NCX (SLC8A1) ∼2-fold lower in all hPSC-EHT (incl. ICE) than in NFH, which could explain the quantitatively rather small positive inotropic effects of digoxin. Ryanodine receptor (RYR2) transcript levels were lower in all hPSC-EHT than in NFH. Slightly higher expression levels in ICE could be an indicator for the relatively small negative clinotropic effect of ryanodine in these EHTs. BayK-8644 and nifedipine, both interacting with the L-type calcium channel encoded by the CACNA1C gene, provoked clear effects in EHTs matching the high gene transcript levels. The reason for the unusual prolongation of relaxation at low external calcium concentrations in some lines (CDI, CEL, NCR, PLU, REB) is unknown and did not clearly correlate with any of the analyzed genes (e.g., PLN/ATP2A2 ratio [Biesiadecki et al., 2014]). As cardiomyocyte relaxation is not only governed by the rate of calcium transient decline, but also deactivation of the thin filaments and actin-myosin interaction, quantification of myofilament protein expression as well as post-translational modifications might contribute to the observed deceleration of relaxation. A limitation of the study is that not enough material was available for a proteome analysis which may have provided a more comprehensive picture of the actual biology than the transcript analysis.
With regard to the variability, comparison of relative changes to the respective baseline has proven most appropriate for drug screening (Abi-Gerges et al., 2017; Blinova et al., 2018, 2017; Mannhardt et al., 2017a). In a MEA-based study the observed lack of correlation between beat rate and FPD (Abi-Gerges et al., 2017) resulted in the requirement for a beat rate correction model to interpret direct drug effects on FPD. Huo et al. (2017) saw indications that spontaneous beat rate and baseline FPD affect the response to proarrhythmic stimuli arguing for rate control during drug screening. We circumvented this obstacle with electrical pacing of the tissues during the experiment and highly recommend analysis under frequency-controlled conditions for drug screening. As incorporation of an electrical pacing system is technically not always feasible, emerging technologies for optical stimulation could be of value (Lemme et al., 2020; Molokanova et al., 2017). Of note, pacing frequency might interfere with the results as prolongation in FPD was only visible at certain pacing frequencies (Lapp et al., 2017) and many positive inotropic effects are only observed at low pacing rate (Saleem et al., 2020). In addition, positive chronotropic effects (e.g., isoprenaline) or extensive APD- and relaxation-prolonging effects might cause failure of capture (see also Figure 5: BayK-8644 on NCRM5) and require adjustment of the experimental protocol. These considerations let us define low spontaneous beat rate and an RT of <0.35 s as quality criteria (Figure 6J).
Confounder—Cell Handling and Genetic Background
The high level of variability between cell lines observed in our study could be attributed to several factors. Prior to casting of EHTs and culture in this three-dimensional format, culture conditions and cell treatment differed between the various cell lines. For the commercial CM, detailed conditions regarding culture and upbringing prior to arrival in the laboratory are proprietary and hence not disclosed. (1) The origin of the somatic cell used for the generation of iPSC could cause differences between cell lines as it is has been reported to influence the molecular and functional properties of mouse iPSCs (Polo et al., 2010). Differences in gene expression between hiPSC derived from fibroblasts, keratinocytes and adipose tissue have been shown and discussed as indication of an “epigenetic memory” (Gaborit et al., 2010; Ghosh et al., 2010; Huo et al., 2019). (2) The generation of hiPSC was achieved by different reprogramming techniques (retrovirus: C25, AT1, CDI, ICE, CEL; Sendai virus: ERC, REB; episomal plasmids: NCR, non-integrative: PLU). As it turned out recently, Cor4U is in fact hESC-derived (Ncardia 04-2019). While Bock et al. (2011) have shown transcriptional differences between hESC and hiPSC lines, the impact on the phenotype of hPSC-derived CM is unknown. Further variabilities between cell lines could very well arise from (3) different protocols regarding stem cell culture (e.g., physical handling, media renewal, or passage number) and (4) cardiac differentiation with confidential protocols for commercial hiPSC-CM versus growth factor and Wnt-inhibitor based cardiac differentiation protocols in suspension or adhesion culture for the in-house differentiated cell lines (C25, ERC, NCR, REB, AT1 [Breckwoldt et al., 2017; Mosqueira et al., 2018]). These factors could explain the clustering of AT1, REB, and NCR or C25 and ERC or COR and PLU cells seen in transcriptome analysis, as the respective cell lines were all cultivated in the same laboratories or obtained from one supplier. (5) Purity of the CM is another confounder of this study. Whereas the in-house differentiated cell lines as well as PLU and CEL were used with a rather similar cardiomyocyte content of >70% (see also Table S1), COR, ICE and CDI are supposed to be essentially free of non-CM. The real purity may be lower as FACS analyses reported only 91.4% ± 4.4% CM for iCell (CDI) and 89.2% ± 7.6% for Cor4U (COR; Huo et al., 2017) and histological analysis of our EHT indicated vimentin- and smooth muscle actin-positive cells also in the latter lines (see Figure S3). As the native heart muscle contains 70%–80% non-CM, the benefit and/or necessity for supporting stromal cells were discussed (Kawatou et al., 2017; Kim et al., 2010; Ravenscroft et al., 2016; Soong et al., 2012). In line with our findings that do not indicate correlation of purity and force, purity positively correlated with contraction velocity, but not with maximum active force of hESC-CM based cardiac patches (Iseoka et al., 2018; Zhang et al., 2013). Though all cell preparations in our study resulted in spontaneously contracting EHT, the genetically sorted, highly pure COR cells did result in EHTs of limited stability that got very thin and ruptured after 3 weeks in culture. While this results was compatible with the idea of the necessity of stromal cells for good cardiac tissue formation, the phenomenon could not be recapitulated with CDI cells of similar purity (see above) or with EHT from in-house-differentiated hiPSC-CM of high purity (92%) that remained strong (0.16 ± 0.06 mN; n = 6) and stable until experiment termination beyond day 180 (data not shown). (6) Non-myocytes as byproducts of the cardiac differentiation are predominantly myofibroblast-like cells (Mannhardt et al., 2016; see also Figure S3). Even though mRNAs for some markers of fibrosis and extracellular matrix were more abundant in some cell lines (AT1, NCR, REB), increased fibrosis could not be confirmed histologically and was absent in all investigated slides with the exception of a few collagen fibrils on the surface of ERC-EHT. However, this issue should be taken into consideration when working with multicellular EHT preparations or different matrix protein such as collagen in the future (Giacomelli et al., 2020; Li et al., 2017; Mills et al., 2017; Nagaraju et al., 2019). (7) Predominance of cells with atrial or ventricular-like phenotype could theoretically explain differences in baseline contractility. EHT from atrial CM exhibited higher spontaneous beating frequency, lower force and faster kinetics than their ventricular counterparts (Goldfracht et al., 2020; Lemme et al., 2018). Comparison of our contraction data with gene expression (Figure S5) and histological markers (Figures S2 and S3) however, did not indicate a clear correlation, arguing against this idea. (8) Cell storage and transport of differentiated CM in a frozen state (e.g., ICE, CDI, PLU, CEL) or shipment of live cells (e.g., COR, AT1, REB) are other variables potentially affecting the response of the cells. We thawed frozen cells with a drop-wise protocol in accordance to the CDI application note, but used the respective thawing medium of each company provided with their cells, if available. Living CM were dissociated with collagenase according to a previous protocol (Breckwoldt et al., 2017). The final step of EHT casting and culture of the tissues was done according to the same protocol (Breckwoldt et al., 2017; Mannhardt et al., 2017b) for all cell lines. An accurate head-to-head comparison of cell lines in search of the ideal candidate for drug testing would not only require the exact same culture conditions and protocols starting at the isolation of the somatic cells (Kilpinen et al., 2017), but also a certain number of replications to eliminate batch-to-batch variability (Huo et al., 2017; Mannhardt et al., 2017a) within one protocol itself.
In conclusion, baseline phenotypes of hiPSC-CM from apparently healthy control lines differ considerably. The extent of variability exceeds differences between patient- and healthy-proband-derived parameters that often have been interpreted as a “disease phenotype”. Therefore, the data provide support for the request of isogenic controls in disease modeling studies. For drug screening though, baseline variability appears less relevant, as drug responses were qualitatively similar. Nevertheless, non-response as well as responses opposite to the canonical response (e.g., shortening of RT with the calcium sensitizer EMD57033) argue for careful selection of a “predictive cell line” and/or the routine testing with multiple cell lines after thorough QC evaluation for the respective screening assay.
Experimental Procedures
Generation of EHT
Human PSC-derived CM were obtained from 4 commercial hiPS cell lines (PLU = Pluricyte CM from Pluriomics (now Ncardia); CDI = iCell CM, and ICE = iCell2 CM both Cellular Dynamics International; CEL = Cellartis CM from Takara Bio), 1 commercial hES cell line (COR = Cor4U CM from Axiogenesis) and differentiated from 3 Hamburg hiPS cell lines including 1 NIH-registered iPS cell line NCRM5 (C25, ERC = UKEi003-C, NCR = ND50031), and 2 Nottingham hiPS cell lines (AT1, REB = REBL-PAT). EHTs were generated from fresh or frozen human PS-derived CM using 1 × 106 cells per 100 μL tissue (see also Figure S1). There were no additional non-CM added to the master mix to test the hiPSC-CM alone as “of the shelf”-product. EHTs showed spontaneous macroscopic contractions, deflecting the silicone posts, after 7-14 days.
For detailed description of the Experimental Procedures please see the Supplemental Information.
Author Contributions
I.M. and T.E. designed the project and wrote the manuscript. I.M., U.S., M.F.L., B.M.U., and M.D.L. performed experiments and analyzed the data. D.M., C.L., C.A., T.K., and M.LH.V. provided cells. All authors discussed the project and results and commented on the manuscript.
Conflict of Interests
I.M., A.H., and T.E. are co-founders of EHT Technologies GmbH.
Acknowledgments
We would like to thank our colleagues of the CRACK-IT challenge for their most valuable input and lively discussions. We greatly appreciate the assistance of Kristin Hartmann from the UKE HEXT mouse pathology core facility. We thank the cardiac differentiation team at the UKE group of A.H. for their support, especially Mirja Schulze, Aya Domke-Shibamiya, Thomas Schulze, and Birgit Klampe. We are grateful to Alessandra Moretti (Department of Cardiology, Klinikum rechts der Isar, Technische Universität München) for providing the control hiPSC cell line C25. Many thanks as well to Steven Schulze (UKE, AG Cuello) for IT support and technical advice.
The work with the hiPSC lines was supported by the British National Centre for the Replacement Refinement & Reduction of Animals in Research (NC3Rs CRACK-IT grant 35911-259146), the entire study was additionally funded by the Freie und Hansestadt Hamburg, the DFG (German Research Foundation; DFG Es 88/12-1), the European Research Council (ERC-AG Indivuheart), the British Heart Foundation (BHF; grant numbers: SP/15/9/31605, PG/14/59/31000, RG/14/1/30588, RM/13/30157, P47352/CRM), Britain Israel Research and Academic Exchange Partnership (BIRAX; 04BX14CDLG), Medical Research Council (MRC; MR/M017354/1), Engineering and Physical Sciences Research Council (EPSRC; DM's Doctoral Prize Research Fellowship), as well as Heart Research UK and the DZHK (German Center for Cardiovascular Research).
Published: October 13, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.09.002.
Contributor Information
Ingra Mannhardt, Email: i.mannhardt@uke.de.
Thomas Eschenhagen, Email: t.eschenhagen@uke.de.
Supplemental Information
References
- Abi-Gerges N., Pointon A., Oldman K.L., Brown M.R., Pilling M.A., Sefton C.E., Garside H., Pollard C.E. Assessment of extracellular field potential and Ca2+ transient signals for early QT/pro-arrhythmia detection using human induced pluripotent stem cell-derived cardiomyocytes. J. Pharmacol. Toxicol. Methods. 2017;83:1–15. doi: 10.1016/j.vascn.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Biesiadecki B.J., Davis J.P., Ziolo M.T., Janssen P.M.L. Tri-modal regulation of cardiac muscle relaxation; intracellular calcium decline, thin filament deactivation, and cross-bridge cycling kinetics. Biophys. Rev. 2014;6:273–289. doi: 10.1007/s12551-014-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinova K., Stohlman J., Vicente J., Chan D., Johannesen L., Hortigon-Vinagre M.P., Zamora V., Smith G., Crumb W.J., Pang L. Comprehensive translational assessment of human-induced pluripotent stem cell derived cardiomyocytes for evaluating drug-induced arrhythmias. Toxicol. Sci. 2017;155:234–247. doi: 10.1093/toxsci/kfw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinova K., Dang Q., Millard D., Smith G., Pierson J., Guo L., Brock M., Lu H.R., Kraushaar U., Zeng H. International multisite study of human-induced pluripotent stem cell-derived cardiomyocytes for drug proarrhythmic potential assessment. Cell Rep. 2018;24:3582–3592. doi: 10.1016/j.celrep.2018.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z.D., Ziller M., Croft G.F., Amoroso M.W., Oakley D.H. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bot C.T., Juhasz K., Haeusermann F., Polonchuk L., Traebert M., Stoelzle-Feix S. Cross - site comparison of excitation-contraction coupling using impedance and field potential recordings in hiPSC cardiomyocytes. J. Pharmacol. Toxicol. Methods. 2018;93:46–58. doi: 10.1016/j.vascn.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckwoldt K., Letuffe-Brenière D., Mannhardt I., Schulze T., Ulmer B., Werner T., Benzin A., Klampe B., Reinsch M.C., Laufer S. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017;12:1177–1197. doi: 10.1038/nprot.2017.033. [DOI] [PubMed] [Google Scholar]
- Chung J.-H., Canan B.D., Whitson B.A., Kilic A., Janssen P.M.L. Force-frequency relationship and early relaxation kinetics are preserved upon sarcoplasmic blockade in human myocardium. Physiol. Rep. 2018;6:e13898. doi: 10.14814/phy2.13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning C., Borgdorff V., Crutchley J., Firth K.S.A., George V., Kalra S., Kondrashov A., Hoang M.D., Mosqueira D., Patel A. Cardiomyocytes from human pluripotent stem cells: from laboratory curiosity to industrial biomedical platform. Biochim. Biophys. Acta. 2016;1863:1728–1748. doi: 10.1016/j.bbamcr.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott E.B., Kelly A., Smith G.L., Loughrey C.M. Isolated rabbit working heart function during progressive inhibition of myocardial SERCA activity. Circ. Res. 2012;110:1618–1627. doi: 10.1161/CIRCRESAHA.111.262337. [DOI] [PubMed] [Google Scholar]
- Ewart L., Aylott M., Deurinck M., Engwall M., Gallacher D.J., Geys H., Jarvis P., Ju H., Leishman D., Leong L. The concordance between nonclinical and phase I clinical cardiovascular assessment from a cross-company data sharing initiative. Toxicol. Sci. 2014;142:427–435. doi: 10.1093/toxsci/kfu198. [DOI] [PubMed] [Google Scholar]
- Gaborit N., Varro A., Le Bouter S., Szuts V., Escande D., Nattel S., Demolombe S. Gender-related differences in ion-channel and transporter subunit expression in non-diseased human hearts. J. Mol. Cell. Cardiol. 2010;49:639–646. doi: 10.1016/j.yjmcc.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Ghosh Z., Wilson K.D., Wu Y., Hu S., Quertermous T., Wu J.C. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E., Meraviglia V., Campostrini G., Cochrane A., Cao X., van Helden R.W.J., Krotenberg Garcia A., Mircea M., Kostidis S., Davis R.P. Human-iPSC-Derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell. 2020;26:862–879.e11. doi: 10.1016/j.stem.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gintant G., Sager P.T., Stockbridge N. Evolution of strategies to improve preclinical cardiac safety testing. Nat. Rev. Drug Discov. 2016;15:457–471. doi: 10.1038/nrd.2015.34. [DOI] [PubMed] [Google Scholar]
- Goldfracht I., Protze S., Shiti A., Setter N., Gruber A., Shaheen N., Nartiss Y., Keller G., Gepstein L. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 2020;11:1–15. doi: 10.1038/s41467-019-13868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Abrams R.M.C., Babiarz J.E., Cohen J.D., Kameoka S., Sanders M.J., Chiao E., Kolaja K.L. Estimating the risk of drug-induced proarrhythmia using human induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2011;123:281–289. doi: 10.1093/toxsci/kfr158. [DOI] [PubMed] [Google Scholar]
- Herron T.J., Da Rocha A.M., Campbell K.F., Ponce-Balbuena D., Willis B.C., Guerrero-Serna G., Liu Q., Klos M., Musa H., Zarzoso M. Extracellular matrix–mediated maturation of human pluripotent stem cell–derived cardiac monolayer structure and electrophysiological function. Circ. Arrhythmia Electrophysiol. 2016;9:e003638. doi: 10.1161/CIRCEP.113.003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Kamalakar A., Yang X., Word B., Stockbridge N., Lyn-Cook B., Pang L. Evaluation of batch variations in induced pluripotent stem cell-derived human cardiomyocytes from 2 major suppliers. Toxicol. Sci. 2017;156:25–38. doi: 10.1093/toxsci/kfw235. [DOI] [PubMed] [Google Scholar]
- Huo J., Wei F., Cai C., Lyn-Cook B., Pang L. Sex-Related differences in drug-induced QT prolongation and torsades de Pointes: a new model system with human iPSC-CMs. Toxicol. Sci. 2019;167:360–374. doi: 10.1093/toxsci/kfy239. [DOI] [PubMed] [Google Scholar]
- Iseoka H., Miyagawa S., Fukushima S., Saito A., Masuda S., Yajima S., Ito E., Sougawa N., Takeda M., Harada A. Pivotal role of non-cardiomyocytes in electromechanical and therapeutic potential of induced pluripotent stem cell-derived engineered cardiac tissue. Tissue Eng. Part A. 2018;24:287–300. doi: 10.1089/ten.tea.2016.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawatou M., Masumoto H., Fukushima H., Morinaga G., Sakata R., Ashihara T., Yamashita J.K. Modelling Torsade de Pointes arrhythmias in vitro in 3D human iPS cell-engineered heart tissue. Nat. Commun. 2017;8:1078. doi: 10.1038/s41467-017-01125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H., Goncalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F.P., Culley O.J. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Majdi M., Xia P., Wei K.A., Talantova M., Spiering S., Nelson B., Mercola M., Chen H.-S.V., Al K.I.M.E.T. Non-cardiomyocytes influence the electrophysiological maturation of human embryonic stem cell-derived cardiomyocytes during differentiation. Stem Cells Dev. 2010;19:783–795. doi: 10.1089/scd.2009.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapp H., Bruegmann T., Malan D., Friedrichs S., Kilgus C., Heidsieck A., Sasse P. Frequency-dependent drug screening using optogenetic stimulation of human iPSC-derived cardiomyocytes. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-09760-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemme M., Ulmer B.M., Lemoine M.D., Zech A.T.L., Flenner F., Ravens U., Reichenspurner H., Rol-Garcia M., Smith G., Hansen A. Atrial-like engineered heart tissue: an in vitro model of the human atrium. Stem Cell Reports. 2018;11:1378–1390. doi: 10.1016/j.stemcr.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemme M., Braren I., Prondzynski M., Aksehirlioglu B., Ulmer B.M., Schulze M.L., Ismaili D., Meyer C., Hansen A., Christ T. Chronic intermittent tachypacing by an optogenetic approach induces arrhythmia vulnerability in human engineered heart tissue. Cardiovasc. Res. 2020;116:1487–1499. doi: 10.1093/cvr/cvz245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Asfour H., Bursac N. Age-dependent functional crosstalk between cardiac fibroblasts and cardiomyocytes in a 3D engineered cardiac tissue. Acta Biomater. 2017;55:120–130. doi: 10.1016/j.actbio.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhardt I., Breckwoldt K., Letuffe-brenière D., Schaaf S., Schulz H., Neuber C., Benzin A., Werner T., Eder A., Schulze T. Human engineered heart tissue: analysis of contractile force. Stem Cell Reports. 2016;7:29–42. doi: 10.1016/j.stemcr.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhardt I., Eder A., Dumotier B., Prondzynski M., Krämer E., Traebert M., Söhren K.-D., Flenner F., Stathopoulou K., Lemoine M.D. Blinded contractility analysis in hiPSC-cardiomyocytes in engineered heart tissue format: comparison with human atrial trabeculae. Toxicol. Sci. 2017;158:164–175. doi: 10.1093/toxsci/kfx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhardt I., Saleem U., Benzin A., Schulze T., Klampe B., Eschenhagen T., Hansen A. Automated contraction analysis of human engineered heart tissue for cardiac drug safety screening. J. Vis. Exp. 2017:e55461. doi: 10.3791/55461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard D., Dang Q., Shi H., Zhang X., Strock C., Kraushaar U., Zeng H., Levesque P., Lu H.-R., Guillon J. Cross-Site reliability of human induced pluripotent stem cell-derived cardiomyocyte based safety assays using microelectrode arrays: results from a blinded CiPA pilot study. Toxicol. Sci. 2018;164:550–562. doi: 10.1093/toxsci/kfy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. U S A. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molokanova E., Mercola M., Savchenko A. Bringing new dimensions to drug discovery screening: impact of cellular stimulation technologies. Drug Discov. Today. 2017;22:1045–1055. doi: 10.1016/j.drudis.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan S., Grootendorst P., Lexchin J., Cunningham C., Greyson D. The cost of drug development : a systematic review. Health Policy (New. York) 2011;100:4–17. doi: 10.1016/j.healthpol.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Mosqueira D., Mannhardt I., Bhagwan J.R., Lis-Slimak K., Katili P., Scott E., Hassan M., Prondzynski M., Harmer S.C., Tinker A. CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. Eur. Heart J. 2018;44:3879–3892. doi: 10.1093/eurheartj/ehy249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosqueira D., Smith J.G.W., Bhagwan J.R., Denning C. Modeling hypertrophic cardiomyopathy: mechanistic insights and pharmacological intervention. Trends Mol. Med. 2019;25:775–790. doi: 10.1016/j.molmed.2019.06.005. [DOI] [PubMed] [Google Scholar]
- Nagaraju C.K., Dries E., Gilbert G., Abdesselem M., Wang N., Amoni M., Driesen R.B., Sipido K.R. Myofibroblast modulation of cardiac myocyte structure and function. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-45078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki Y., Honda Y., Watanabe H., Saiki S., Koyabu K., Itoh T., Nagasawa C., Nakamori C., Nakayama C., Iwasaki H. CSAHi study: validation of multi-electrode array systems (MEA60/2100) for prediction of drug-induced proarrhythmia using human iPS cell-derived cardiomyocytes -assessment of inter-facility and cells lot-to-lot-variability- Regul. Toxicol. Pharmacol. 2016;77:75–86. doi: 10.1016/j.yrtph.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Onakpoya I.J., Heneghan C.J., Aronson J.K. Post-marketing withdrawal of 462 medicinal products because of adverse drug reactions: a systematic review of the world literature. BMC Med. 2016;14:10. doi: 10.1186/s12916-016-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang L., Sager P., Yang X., Shi H., Sannajust F., Brock M., Wu J.C., Abi-Gerges N., Lyn-Cook B., Berridge B.R. Workshop report: FDA workshop on improving cardiotoxicity assessment with human-relevant platforms. Circ. Res. 2019;125:855–867. doi: 10.1161/CIRCRESAHA.119.315378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Liu S., Figueroa M.E., Kulalert W., Eminli S., Tan K.Y., Apostolou E., Stadtfeld M., Li Y., Shioda T. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft S.M., Pointon A., Williams A.W., Cross M.J., Sidaway J.E. Cardiac non-myocyte cells show enhanced pharmacological function suggestive of contractile maturity in stem cell derived cardiomyocyte microtissues. Toxicol. Sci. 2016;152:kfw069. doi: 10.1093/toxsci/kfw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala L., Bellin M., Mummery C.L. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: has the time come? Br. J. Pharmacol. 2016;174:1–17. doi: 10.1111/bph.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala L., van Meer B.J., Tertoolen L.G.J., Bakkers J., Bellin M., Davis R.P., Denning C., Dieben M.A.E., Eschenhagen T., Giacomelli E. MUSCLEMOTION: a versatile open software tool to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Circ. Res. 2018;122:e5–e16. doi: 10.1161/CIRCRESAHA.117.312067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem U., van Meer B.J., Katili P.A., Mohd Yusof N.A.N., Mannhardt I., Garcia A.K., Tertoolen L., de Korte T., Vlaming M.L.H., McGlynn K. Blinded, multicenter evaluation of drug-induced changes in contractility using human-induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2020;176:103–123. doi: 10.1093/toxsci/kfaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuderi G.J., Butcher J. Naturally engineered maturation of cardiomyocytes. Front Cell Dev Biol. 2017;5:1–28. doi: 10.3389/fcell.2017.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong P.L., Tiburcy M., Zimmermann W. Cardiac differentiation of human embryonic stem cells and their assembly into engineered heart muscle. Curr. Protoc. Cell Biol. 2012:1–21. doi: 10.1002/0471143030.cb2308s55. Chapter 23:Unit23.8.1-23.8.21. [DOI] [PubMed] [Google Scholar]
- Sutko J.L., Willerson J.T. Ryanodine alteration of the contractile state of rat ventricular myocardium. Comparison with dog, cat, and rabbit ventricular tissues. Circ. Res. 1980;46:332–343. doi: 10.1161/01.res.46.3.332. [DOI] [PubMed] [Google Scholar]
- Zhang D., Shadrin I.Y., Lam J., Xian H.Q., Snodgrass H.R., Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials. 2013;34:5813–5820. doi: 10.1016/j.biomaterials.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.