Abstract
Background
In early 2020, many scientists are rushing to discover novel drugs and vaccines against the coronavirus, and treatments for COVID-19, because coronavirus disease 2019 (COVID-19), a life-threatening viral disease, affected first in China and quickly spread throughout the world. In this article, in silico studies have been performed to explore the binding modes of chemical constituents for natural remedies like Curcuma longa (turmeric) and Andrographis paniculata against COVID-19 (PDB ID 5R82) targeting coronavirus using Schrodinger suit 2019-4. The molecular docking studies are performed by the Glide module, in silico ADMET screening was performed by the QikProp module, and binding energy of ligands was calculated using the Prime MM-GB/SA module.
Results
The chemical constituents from turmeric like cyclocurcumin and curcumin and from Andrographis paniculata like andrographolide and dihydroxy dimethoxy flavone are significantly binding with the active site of SARS CoV-2 main protease with Glide score more than − 6 when compared to the currently used drugs hydroxychloroquine (− 5.47) and nelfinavir (− 5.93). When compared to remdesivir (− 6.38), cyclocurcumin from turmeric is significantly more active. The docking results of the compounds exhibited similar mode of interactions with SARS CoV-2. Main protease and the residues THR24, THR25, THR26, LEU27, SER46, MET49, HIE41, GLN189, ARG188, ASP187, MET165, HIE164, PHE181, and THR54 play a crucial role in binding with ligands.
Conclusion
Based on in silico investigations, the chemical constituents from turmeric like cyclocurcumin and curcumin and from Andrographis paniculata like andrographolide and dihydroxy dimethoxy flavone, significantly binding with the active site of SARS CoV-2 main protease, may produce significant activity and be useful for further development.
Keywords: Coronavirus (COVID-19), Curcuma longa (turmeric), Andrographis paniculata, Docking studies, MM-GBSA
Background
Coronavirus disease 2019 (COVID-19) is a life-threatening disease which was affected first in China and quickly spread throughout the world [1–6]. According to the WHO data, as of the second week of April 2020, there are 21.5 lakhs peoples in the world affected by COVID-19, out of these more than 1.5 lakhs peoples died. With more asymptomatic infections being found among COVID-19 cases, it is worthy of consideration the detailed current evidence and understanding of the transmission of SARS CoV, MERS-CoV, and SARS CoV-2 and discussion on pathogen inactivation methods on coronaviruses is very important [7–12].
In this emergency situation, it is very difficult to discover novel drugs with all clinical trials and also determine the side effects, adverse effects, etc. So, it is important to find some natural remedies for the prevention and treatment of COVID-19. From the literatures, the natural products like Curcuma longa (turmeric) and Andrographis paniculata were reported for various biological activities and used traditionally for curing many diseases. Also, there is no or minimum side effects reported when compared to allopathic drugs.
The dried and powdered root Curcuma longa (turmeric) is belonging to the Zingiberaceae family, which is being cultivated in many countries worldwide. It has many uses such as textile dyes, herbal medicines, or food products. The biological properties of its chemical components were reported for inhibition of platelet aggregation [13], anti-diabetic [14], anti-tumor [15–17], anti-inflammatory effects [18], antioxidant effects [19], anti-platelet aggregation effects [20], gastro-protective effects [21], lipid-lowering effects [22], Alzheimer's effects [23], etc.
Andrographis paniculata was reported for the treatment of liver diseases [24], fever, common cold [25], acute diarrhea [26], hypertension [27], chicken pox, leprosy [28], hepatitis [29], malaria [30], anti-inflammatory effects [31], anti-cancer [32], diabetes [33], etc.
As part of our ongoing research on searching the potent biological molecules against various diseases by in silico and wet lab methods [34–44], we have designed and evaluated various heterocyclic compounds for their biological activities. Using different modules (Glide, QikProp, and Prime) of Schrödinger suite LLC various computational methods like molecular docking, ADMET screening, and binding-free energy, calculations were performed to find the interactions responsible for SARS CoV-2 main protease inhibition. These studies will provide the requirement of key structural features in the design of potential drug candidates.
Methods
The 3D crystal structure of COVID-19 protein called SARS CoV-2 main protease receptor co-crystallized with 6-(ethylamino) pyridine-3-carbonitrile (PDB ID 5R82, resolution 1.31 Å) was retrieved from the protein data bank. The protein was prepared using the protein preparation wizard of epic module of Schrödinger suite 2019-4. The protein structure retrieved from the RCSB protein data bank is a monomer with co-crystallized ligand. The protein was prepared by using the protein preparation wizard by refining bond orders, addition of hydrogens, and deleting water molecules beyond 5 Å, and missing chains are included by using the Prime module [45] of Schrödinger suite 2019-4. Protein minimization was performed using optimized potentials for liquid simulations (OPLS3) molecular force field with RMSD of crystallographic heavy atoms kept at 0.30 Å. A grid box was generated to define the centroid of the active site. All the compounds were docked into the catalytic pocket of SARS CoV-2 main protease by using the Glide module of Schrödinger suite 2019-4 in extra precision (XP) mode [46]. The ligands with significant Glide scores have more binding affinity towards SARS CoV-2 main protease enzyme. To predict the free energy of binding for the set of ligands in complex with a receptor, post-docking energy minimization studies were performed using Prime molecular mechanics-generalized Born surface area (MM-GB/SA) of Schrödinger 2019-4. The energy for minimized XP docked pose of ligand-receptor complex was calculated using the OPLS3 force field and generalized Born/surface area (GB/SA) continuum VSGB 2.0 solvent model [47, 48].
Results
Results are summarized in Tables 1, 2, and 3 and Figs. 1, 2, 3, 4, 5, 6, and 7. The results revealed that the SARS CoV-2 main protease inhibitory property of the compounds isolated from some natural products like Curcuma longa (turmeric) and Andrographis paniculata greatly depended on the chemical nature of the substituents. The chemical structures of selected major bioactive constituents of Curcuma longa (turmeric) and Andrographis paniculata are given in Fig. 1a and b. The anti-malarial drug which was currently recommended in many countries like the USA, India, etc. [49] for the treatment of COVID-19 is hydroxychloroquine (Fig. 1c).
Table 1.
Docking studies for phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata with SARS CoV-2 main protease (5R82)
| Cpd | Glide score | Lipophilic EvdW |
Phob En | H bond | XP electro | Low MW | Rot Penal | XP penalties |
|---|---|---|---|---|---|---|---|---|
| T4_Cyclocurcumin | − 6.77 | − 4.12 | 0 | − 2.36 | − 0.35 | − 0.27 | 0.18 | 0 |
| N1_Andrographolide | − 6.26 | − 1.27 | 0 | − 4.01 | − 1.3 | − 0.33 | 0.2 | 0 |
| N7_dihydroxydimethoxyflavone | − 6.23 | − 2.69 | 0 | − 2.44 | − 0.84 | − 0.45 | 0.08 | 0 |
| T1_Curcumin | − 6.13 | − 4.14 | 0 | − 1.46 | − 0.72 | − 0.27 | 0.37 | 0 |
| T3_Bisdemethoxycurcumin | − 5.36 | − 4.13 | 0 | − 0.7 | − 0.6 | − 0.47 | 0.5 | 0 |
| T2_Demethoxycurcumin | − 5.25 | − 3.69 | 0 | − 1.06 | − 0.62 | − 0.37 | 0.42 | 0 |
| T7_Curcuphenol | − 5.13 | − 4.34 | 0 | − 0.7 | − 0.31 | − 0.5 | 0.6 | 0 |
| N3_14deoxy12hydroxyandrographolide | − 5.11 | − 2.89 | 0 | − 2.12 | − 0.27 | − 0.33 | 0.27 | 0 |
| T6_Curlone | − 3.89 | − 4.05 | 0 | 0 | − 0.07 | − 0.5 | 0.6 | 0 |
| N2_14deoxyandrographolide | − 3.88 | − 1.96 | 0 | − 2.07 | − 0.54 | − 0.39 | 0.29 | 0 |
| T5_Turmerone | − 3.78 | − 3.87 | 0 | 0 | − 0.01 | − 0.5 | 0.6 | 0 |
| N8_cinnamateester | − 3.31 | − 3.55 | 0 | 0 | − 0.02 | − 0.5 | 0.76 | 0 |
| N5_Stigmasterol | − 2.28 | − 3.6 | 0 | 0 | 0.02 | − 0.12 | 0.15 | 1 |
| N6_βSitosterylfattyacidesters | − 1.89 | − 2.66 | 0 | − 0.3 | − 0.02 | 0 | 0.32 | 0 |
| N4_betaSitosterol | − 1.36 | − 1.86 | 0 | 0 | − 0.06 | − 0.12 | 0.2 | 0 |
| Hydroxychloroquine (Std) | − 5.47 | − 3.15 | 0 | − 1.75 | − 0.69 | − 0.38 | 0.5 | 0 |
Table 2.
In silico ADMET screening for phytochemical constituents of Curcuma longa (turmeric) and Andrographis paniculata
| Compounds | Mol. Wt. | Dipole | Donor HB | Accpt HB | QPlog o/w | #metab | Rule of five | %Human oral absorption |
|---|---|---|---|---|---|---|---|---|
| T1_Curcumin | 368.385 | 8.366 | 2 | 7 | 3.301 | 5 | 0 | 88.976 |
| T2_Demethoxycurcumin | 338.359 | 9.291 | 2 | 6.25 | 2.821 | 4 | 0 | 85.615 |
| T3_Bisdemethoxycurcumin | 308.333 | 8.477 | 2 | 5.5 | 2.585 | 3 | 0 | 81.091 |
| T4_Cyclocurcumin | 368.385 | 5.335 | 2 | 5.75 | 3.488 | 6 | 0 | 90.504 |
| T5_Turmerone | 218.338 | 3.649 | 0 | 2 | 4.036 | 6 | 0 | 100 |
| T6_Curlone | 218.338 | 3.147 | 0 | 2 | 3.991 | 5 | 0 | 100 |
| T7_Curcuphenol | 218.338 | 1.472 | 1 | 0.75 | 4.419 | 6 | 0 | 100 |
| N1_Andrographolide | 350.454 | 6.319 | 3 | 8.1 | 1.455 | 6 | 0 | 77.655 |
| N2_14deoxyandrographolide | 334.455 | 4.004 | 2 | 6.4 | 2.46 | 6 | 0 | 91.184 |
| N3_14deoxy12OH_andrographolide | 350.454 | 4.508 | 2 | 7.1 | 2.04 | 6 | 0 | 83.156 |
| N4_betaSitosterol | 414.713 | 2.542 | 1 | 1.7 | 7.643 | 3 | 1 | 100 |
| N5_Stigmasterol | 412.698 | 2.464 | 1 | 1.7 | 7.473 | 5 | 1 | 100 |
| N6_βSitosterylfattyacidesters | 526.885 | 3.304 | 0 | 2 | 9.625 | 3 | 2 | 100 |
| N7_dihydroxydimethoxyflavone | 314.294 | 3.726 | 1 | 4.5 | 2.682 | 4 | 0 | 93.829 |
| N8_cinnamateester | 218.295 | 4.054 | 0 | 2 | 3.983 | 0 | 0 | 100 |
| Hydroxychloroquine (std) | 335.876 | 6.854 | 2 | 5.7 | 3.369 | 5 | 0 | 93.213 |
| Recommended values | 130–725 | 1–12.5 | 0–6 | 2–20 | − 2–6.5 | 1–8 | max 4 |
> 80% is high < 25% is poor |
Mol. Wt. molecular weight of the molecule, Dipole computed dipole moment, Donor HB estimated number of hydrogen bonds that would be donated by the solute to water molecules in an aqueous solution, Accpt HB estimated number of hydrogen bonds that would be accepted by the solute from water molecules in an aqueous solution, QPlog o/w predicted octanol/water partition coefficient, #metab number of likely metabolic reactions, Rule of five number of violations of Lipinski’s rule of five, %Human oral absorption predicted human oral absorption on 0 to 100% scale
Table 3.
Binding free energy calculation using Prime/MM-GBSA approach
| Compd | MMGBSA_dG_Bind | MMGBSA _dG_Bind_ Coulomb |
MMGBSA _dG_Bind_ Covalent |
MMGBSA_dG_Bind Hbond |
MMGBSA _dG_Bind_ Lipo |
MMGBSA_dG_Bind_vdW |
|---|---|---|---|---|---|---|
| T4_Cyclocurcumin | − 36.0315 | − 31.6404 | 8.0570 | − 0.1385 | − 11.6120 | − 28.1951 |
| N1_Andrographolide | − 34.6766 | − 28.4227 | 6.9893 | − 1.9077 | − 4.4014 | − 28.5162 |
| N7_diOHdiOMeflavone | − 50.6953 | − 41.1895 | 1.6877 | − 1.5594 | − 8.5302 | − 26.2631 |
| T1_Curcumin | − 50.3408 | − 18.0802 | 1.5801 | − 2.6563 | − 11.9153 | − 42.2597 |
| T3_BisdeOMecurcumin | − 43.5559 | − 22.2542 | − 1.599 | 0.5231 | − 11.2505 | − 35.4184 |
| T2_Demethoxycurcumin | − 38.7071 | − 9.1482 | 6.6291 | − 1.3511 | − 11.0807 | − 42.2478 |
| T7_Curcuphenol | − 26.1402 | − 11.4548 | 14.9959 | 0.0274 | − 17.0435 | − 27.3193 |
| N3_14deoxy12OHandrographolide | − 29.3622 | − 5.8525 | − 9.0786 | 0.1215 | − 10.4797 | − 33.9966 |
| T6_Curlone | − 22.3669 | − 23.7502 | 2.6632 | − 0.3572 | − 9.3057 | − 17.1282 |
| N2_14deoxyandrographolide | − 39.6148 | − 17.6018 | 7.7512 | − 1.4333 | − 14.1630 | − 35.8594 |
| T5_Turmerone | − 24.1033 | 14.8710 | − 12.9950 | 3.1283 | − 8.2392 | − 37.1469 |
| N8_cinnamateester | − 30.7961 | 6.1548 | 1.5202 | 0.4830 | − 10.6485 | − 41.1941 |
| N5_Stigmasterol | − 31.5416 | 18.6211 | − 6.1436 | 2.5054 | − 15.3476 | − 41.7355 |
| N6_βSitosterylfattyacidesters | 1.1272 | 27.1428 | − 0.7667 | 3.0617 | − 5.4991 | − 29.3366 |
| N4_betaSitosterol | − 25.0588 | − 5.7182 | 1.8587 | 1.9414 | − 10.8267 | − 31.8647 |
| Hydroxychloroquine (std) | − 26.9975 | − 4.9621 | 2.1824 | 0.0011 | -9.2894 | − 33.0622 |
Fig. 1.

Structures of phytochemical constituents. a Chemical structures of selected major bioactive constituents of Curcuma longa (turmeric). b Chemical structures of selected major bioactive constituents of Andrographis paniculata (Burm.f.) Nees. c Structure of hydroxychloroquine (Std)
Fig. 2.
Docked poses of all compounds with SARS CoV-2 main protease (5R82)
Fig. 3.
a Ligand interaction of compound T4_Cyclocurcuminwith SARS CoV-2 main protease (5R82). b Ligand interaction of compound N1_Andrographolidewith SARS CoV-2 main protease (5R82). c Ligand interaction of compound N7_dihydroxydimethoxyflavonewith SARS CoV-2 main protease (5R82). d Ligand interaction of compound T1_Curcumin with SARS CoV-2 main protease (5R82). e Ligand interaction of compound hydroxychloroquine (Std) with SARS CoV-2 main protease (5R82)
Fig. 4.
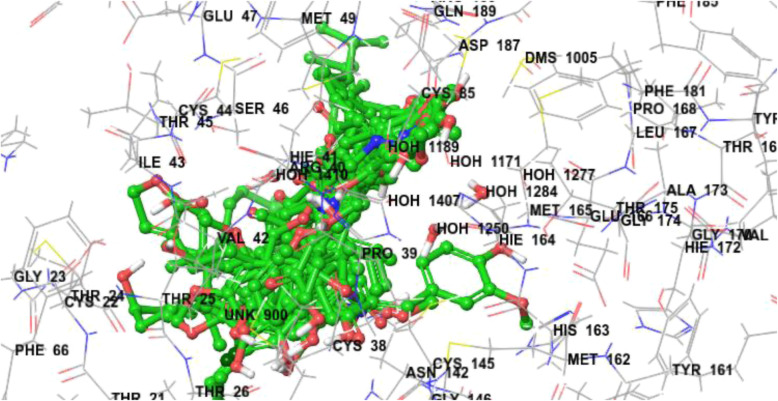
Best affinity mode of docked compounds with SARS CoV-2 main protease (5R82)
Fig. 5.
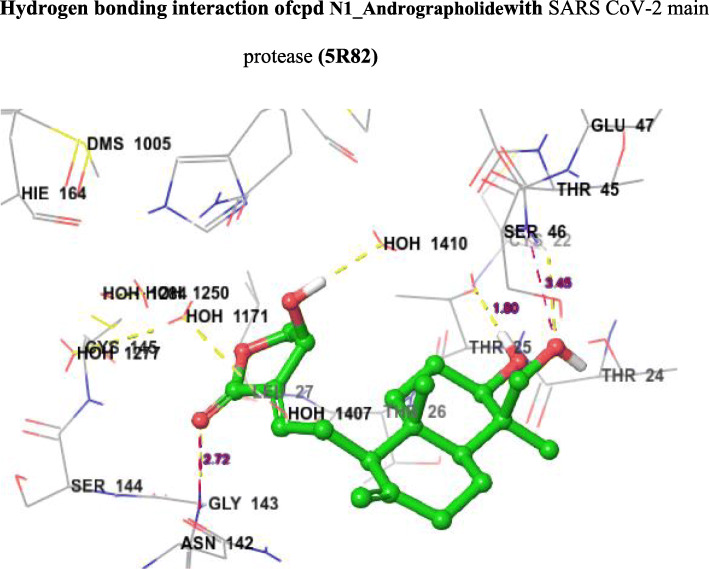
Hydrogen bonding interaction of cpd N1_Andrographolide with SARS CoV-2 main protease (5R82)
Fig. 6.
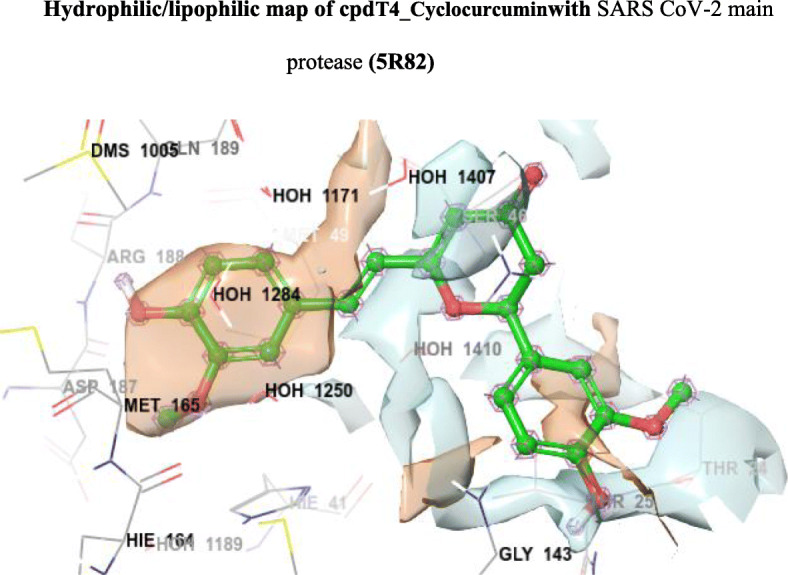
Hydrophilic/lipophilic map of cpd T4_Cyclocurcumin with SARS CoV-2 main protease (5R82)
Fig. 7.
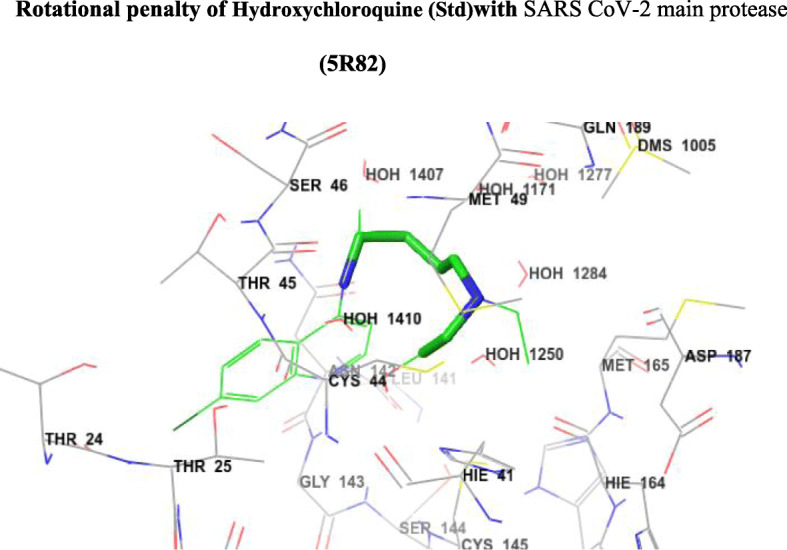
Rotational penalty of hydroxychloroquine (Std) with SARS CoV-2 main protease (5R82)
The docking studies of the ligands to protein active sites were performed by an advanced molecular docking program Glide module of Schrodinger suite 2019 Maestro-12.2 version for determining the binding affinities of the compounds. The designed analogues were docked towards the SARS CoV-2 main protease (PDB ID 5R82) in order to ascertain their inhibitory activity. The analogues show best fit root mean square difference (RMSD) value of 0.2.
The results are summarized in Table 1. Almost all the compounds are docked in the same binding pocket.
The 2D-ligand interaction diagrams of T4_Cyclocurcumin, N1_Andrographolide, N7_dihydroxydimethoxyflavone, and T1_Curcumin with SARS CoV-2 main protease (PDB ID 5R82) are given in Fig. 3a–d. The 2D-ligand interaction diagram of hydroxychloroquine is given in Fig. 3e.
From the molecular docking study, it was revealed that the ligands have shown agreeable Glide G score values from − 6.13 kcal/mol (T1_Curcumin) to − 6.77 kcal/mol (T4_Cyclocurcumin) when compared to the currently recommended drugs for COVID-19 hydroxychloroquine (G score − 5.47) and nelfinavir (− 5.93). When compared to remdesivir (− 6.38), cyclocurcumin from turmeric is significantly more active. From the obtained binding modes, it was illustrated that the ligands formed hydrophobic interactions and hydrogen bonding interactions with different residues THR24 to GLN192 surrounding the active pocket which was shown in Fig. 4. The ligand N1_Andrographolide exhibited hydrogen bonding interaction with some amino acid residues and with some water molecules which are shown in Fig. 5. The presence of aromatic features and different heterocyclic rings majorly contributed towards lipophilic factors (Fig. 6).
The Glide score of the standard compound hydroxychloroquine was decreased because of the rotational penalty of the side alkyl chain which was shown in Fig. 7.
Molecular docking was additionally assessed with MM-GBSA free restricting vitality which is identified with the post-scoring approach for SARS CoV-2 main protease (PDB ID 5R82) target and the values are shown in Table 3.
Discussion
From the docking results, as shown in Table 1, it is clearly demonstrated that some of the chemical constituents from turmeric like cyclocurcumin and curcumin and from Andrographis paniculata like andrographolide and dihydroxy dimethoxy flavone significantly bind with the active site of COVID-19 main protease with Glide score more than − 6 when compared to the currently recommended drug hydroxychloroquine (G score − 5.47) and significantly inhibit SARS CoV-2 main protease and may be active against COVID-19 on further process. The above compounds have good affinity to the receptor due to more lipophilic character and also due to hydrogen bonding. From the 2D-ligand interaction diagrams, almost all the compounds exhibited similar mode of interactions with SARS CoV-2 main protease and the residues THR24, THR25, THR26, LEU27, SER46, MET49, HIE41, GLN189, ARG188, ASP187, MET165, HIE164, PHE181, and THR54 play a crucial role in binding with ligands.
From Fig. 5, the docking score of the ligand N1_Andrographolide is increased due to hydrogen bonding interaction with SER46 (H-bond length 3.45 Å), GLY143 (H-bond length 2.72 Å), and THR25 (H-bond length 1.90 Å) residues and with some water molecules. From Fig. 6, it is clearly demonstrated that most of the aromatic features are covered in the lipophilic region (red color) which contributed towards lipophilic factors.
From Fig. 7, the Glide score of the standard hydroxychloroquine is decreased because of the rotational penalty due to the rotation of the side alkyl side chain present at the fourth position of quinoline.
From the results of MM-GB/SA studies, the dG bind values were observed in the range of − 34.6766 (N1_Andrographolide) to − 50.69 kcal/mol (N7_dihydroxy dimethoxy flavone) for significantly active compounds and also dG vdw values, dG lipophilic values, and the energies are positively contributing towards total binding energy. The accuracy of docking is confirmed by examining the lowest energy poses predicted by the scoring function. The Glide score and MM-GBSA free energy obtained by the docking of ligands into the coupling pocket are more stable.
Conclusion
From the results of the docking study, the chemical constituents of Curcuma longa (turmeric) and Andrographis paniculata demonstrated better arrangement at a dynamic site. The in silico structuring strategy embraced in the present investigation helped for recognizing some lead molecules and furthermore may somewhat clarify their useful impact for further determinations like in vitro and in vivo assessments. Results from the in silico study exhibited that many of the chemical constituents from Curcuma longa (turmeric) and Andrographis paniculata family may be useful against COVID-19 by inhibiting SARS CoV-2 main protease enzyme. Based on in silico studies, the chemical constituents such as cyclocurcumin and curcumin from turmeric and andrographolide and dihydroxy dimethoxy flavone from Andrographis paniculata are significantly active against COVID-19 by inhibiting SARS CoV-2 main protease enzyme with remedial possibilities and are probably going to be helpful after further refinement. In conclusion, consuming turmeric in our diet regularly may be a useful remedy in the prevention of the coronavirus.
Acknowledgements
The authors express their sincere gratitude to JSS Academy of Higher Education & Research, Mysuru and also thank the principal Dr. S.P. Dhanabal, JSS College of Pharmacy, Ooty, for the technical support.
Abbreviations
- COVID-19
Coronavirus disease 2019
- MM-GBSA
Molecular mechanics-generalized Born surface area
- PDB
Protein data bank
- OPLS3
Optimized potentials for liquid simulations
- XP
Extra precision
Authors’ contributions
The authors KR and GB contributed to the technical and preparation of the manuscript. PV and BA contributed to the collection of literature and preparation of the manuscript. All authors have read and approved the manuscript and ensure that this is the case.
Funding
Not applicable/no funding was received.
Availability of data and materials
All data and material are available upon request.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors have no competing interests to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kalirajan Rajagopal, Email: rkalirajan@ymail.com, Email: rkalirajan@jssuni.edu.in.
Potlapati Varakumar, Email: p.varakumar778@gmail.com.
Aparma Baliwada, Email: aparna.baliwada@gmail.com.
Gowramma Byran, Email: gowrammab@rediffmail.com.
References
- 1.Wang D, Hu B, Hu C, Xiong, Zhao, Li, Wang X, Peng Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holshue ML, DeBolt C, Lindquist S, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Sara Wilkerson MN, Tural A, Diaz G, Cohn A, Fox LA, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK (2020) First case of 2019 novel coronavirus in the United States. N Engl J Med. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed]
- 3.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu J, Han B, Wang J (2020) COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 10.1053/j.gastro.2020.02.054 [DOI] [PMC free article] [PubMed]
- 5.Calisher C, Carroll D, Colwell R, Corley RB, Daszak P, Drosten C, Enjuanes L, Farrar J, Field H, Golding J. Statement in support of the scientists, public health professionals, and medical professionals of China combatting COVID-19. Lancet. 2020;395(10226):e42–e43. doi: 10.1016/S0140-6736(20)30418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.To KK-W, Tsang OT-Y, Yip CC-Y, Chan K-H, Wu T-C, Chan JM-C, Leung W-S, Chik TS-H, Choi CY-C, Kandamby DH. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Liang W, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Yang L, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed]
- 8.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Q, Herrmann A (2020) Fast assessment of human receptor-binding capability of 2019 novel coronavirus (2019-nCoV). Bio Rxiv. 10.1101/2020.02.01.930537
- 10.Zhang H, Kang Z, Gong H, Xu D, Wang J, Li Z, Cui X, Xiao J, Meng T, Wang Z, Liu J, Xu H (2020) The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv:927806. 10.1101/2020.01.30.927806
- 11.Chang, L., Yan,Y., Wang, L. (2020) Coronavirus disease 2019: coronaviruses and blood safety. Transfus Med Rev. 10.1016/j.tmrv.2020.02.003. [DOI] [PMC free article] [PubMed]
- 12.Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed]
- 13.Prakash P, Misra A, Surin WR, Jain M, Bhatta RS, Pal R. Anti-platelet effects of Curcuma oil in experimental models of myocardial ischemia-reperfusion and thrombosis. Thromb Res. 2011;127:111–118. doi: 10.1016/j.thromres.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda M, Mimaki Y, Nishiyama T, Mae T, Kishida H, Tsukagawa K. Hypoglycemic effects of turmeric (Curcuma longa L. Rhizomes) on genetically diabetic KK-Ay mice. Biol Pharm Bull. 2005;5:937–939. doi: 10.1248/bpb.28.937. [DOI] [PubMed] [Google Scholar]
- 15.Aggarwal BB, Kumar A, Bharti AC. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res. 2003;23(1A):363–398. [PubMed] [Google Scholar]
- 16.Parthasarathy VA, Chempakam B, Zachariah TJ (2008) Chemistry of spices. United Kingdom. CABI:445–447
- 17.Haddad M, Sauvain M, Deharo E. Curcuma as a parasiticidal agent: a review. Planta Med. 2010;77:672–678. doi: 10.1055/s-0030-1250549. [DOI] [PubMed] [Google Scholar]
- 18.Abe Y, Hashimoto S, Horie T. Curcumin inhibition of inflammatory cytokine production by human peripheral blood monocytes and alveolar macrophages. Pharmacol Res. 1999;39(1):41–47. doi: 10.1006/phrs.1998.0404. [DOI] [PubMed] [Google Scholar]
- 19.Gupta B, Ghosh B. Curcuma longa inhibits TNF-alpha induced expression of adhesion molecules on human umbilical vein endothelial cells. Int J Immunopharmacol. 1999;21(11):745–757. doi: 10.1016/S0192-0561(99)00050-8. [DOI] [PubMed] [Google Scholar]
- 20.Srivastava R, Dikshit M, Srimal RC, Dhawan BN. Anti-thrombotic effect of curcumin. Thromb Res. 1985;40(3):413–417. doi: 10.1016/0049-3848(85)90276-2. [DOI] [PubMed] [Google Scholar]
- 21.Bhatia A, Singh GB, Khanna NM. Effect of curcumin, its alkali salts and Curcuma longa oil in histamine-induced gastric ulceration. Indian J Exp Biol. 1964;2:158–160. [Google Scholar]
- 22.Ramirez-Tortosa MC, Mesa MD, Aguilera MC, Quiles JL, Baro L, Ramirez-Tortosa CL, Martinez-Victoria E, Gil A. Oral administration of a turmeric extract inhibits LDL oxidation and has hypocholesterolemic effects in rabbits with experimental atherosclerosis. Atherosclerosis. 1999;147(2):371–378. doi: 10.1016/S0021-9150(99)00207-5. [DOI] [PubMed] [Google Scholar]
- 23.Kim DS, Park SY, Kim JK. Curcuminoids from Curcuma longa L. (Zingiberaceae) that protect PC12 rat pheochromocytoma and normal human umbilical vein endothelial cells from betaA(1-42) insult. Neurosci Lett. 2001;303(1):57–61. doi: 10.1016/S0304-3940(01)01677-9. [DOI] [PubMed] [Google Scholar]
- 24.Boopathi C. Andrographis spp.: a source of bitter compounds for medicinal use. Anc Sci Life. 2000;19(3-4):164–168. [PMC free article] [PubMed] [Google Scholar]
- 25.Jarukamjorn K, Nemoto N. Pharmacological aspects of Andrographis paniculata on health and its major diterpenoid constituent andrographolide. J Health Sci. 2008;54(4):370–381. doi: 10.1248/jhs.54.370. [DOI] [Google Scholar]
- 26.Kabir MH, Hasan N, Rahman MM, Khan JA, Hoque NT, Quddus Bhuiyan MR, Mou SM, Jahan R, Rahmatullah M. A survey of medicinal plants used by the Deb barma clan of the Tripura tribe of Moulvibazar district, Bangladesh. J Ethnobiol Ethnomed. 2014;10(1):19. doi: 10.1186/1746-4269-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borhanuddin M, Shamsuzzoha M, Hussain AH. Hypoglycaemic effects of Andrographis paniculata Nees on nondiabetic rabbits. Bangladesh Med Res Counc Bull. 1994;20(1):24–26. [PubMed] [Google Scholar]
- 28.Akbar S. Andrographis paniculata: a review of pharmacological activities and clinical effects. Altern Med Rev. 2011;16(1):66–77. [PubMed] [Google Scholar]
- 29.Chaturvedi GN, Tomar GS, Tiwari SK, Singh KP. Clinical studies on Kalmegh (Andrographis paniculata Nees) in infective hepatitis. J Int Inst Ayurveda. 1983;2:208–211. [PMC free article] [PubMed] [Google Scholar]
- 30.Dua VK, Ojha VP, Roy R, Joshi BC, Valecha N, Devi CU, et al. Anti-malarial activity of some xanthones isolated from the roots of Andrographis paniculata. J Ethnopharmacol. 2004;95:247–251. doi: 10.1016/j.jep.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 31.Parichatikanond W, Suthisisang C, Dhepakson P, Herunsalee A. Study of anti-inflammatory activities of the pure compounds from Andrographis paniculata (Burm.f.) Nees and their effects on gene expression. Int Immunopharmacol. 2010;10:1361–1373. doi: 10.1016/j.intimp.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Jade SR, Suburb GS, Matthews C, Hamzah AS, Lapis NH, Saad MS. Semisynthesis and in vitro anticancer activities of andrographolide analogues. Phytochem. 2007;68:904–912. doi: 10.1016/j.phytochem.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 33.Husen R, Pihie AH, Nallappan M. Screening for antihyperglycaemic activity in several local herbs of Malaysia. J Ethnopharmacol. 2004;95:205–208. doi: 10.1016/j.jep.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Kalirajan R, Mohammed rafick MH, Sankar S, Gowramma B. Green synthesis of some novel chalcone and isoxazole substituted 9-anilinoacridine derivatives and evaluation of their antimicrobial and larvicidal activities. Indian J Chem. 2018;57B:583–590. [Google Scholar]
- 35.Kalirajan R, Muralidharan V, Jubie S, Sankar S. Microwave assisted synthesis, characterization and evaluation for their antimicrobial activities of some novel pyrazole substituted 9-anilino acridine derivatives. Int J Health Allied Sci. 2013;2(2):81–87. doi: 10.4103/2278-344X.115682. [DOI] [Google Scholar]
- 36.Kalirajan R, Muralidharan V, Jubie S, Gowramma B, Gomathy S, Sankar S, Elango K. Synthesis of some novel pyrazole substituted 9-anilinoacridine derivatives and evaluation for their antioxidant and cytotoxic activities. J Heterocyclic Chem. 2012;49:748–754. doi: 10.1002/jhet.848. [DOI] [Google Scholar]
- 37.Kalirajan R, Mohammed rafick MH, Jubie S, Sankar S (2012) Docking studies, synthesis, characterization and evaluation of their antioxidant and cytotoxic activities of some novel isoxazole substituted 9-anilinoacridine derivatives. Sci World J:165258. 10.1100/2012/165258 [DOI] [PMC free article] [PubMed]
- 38.Kalirajan R, kulshrestha V, Sankar S, Jubie S. Docking studies, synthesis, characterization of some novel oxazine substituted 9-anilinoacridine derivatives and evaluation for their anti-oxidant and anticancer activities as topo isomerase II inhibitors. Eur J Med Chem. 2012;56:217–224. doi: 10.1016/j.ejmech.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Kalirajan R, Rathore L, Jubie S, Gowramma B, Gomathy S, Sankar S. Microwave assisted synthesis of some novel pyrazole substituted benzimidazoles and evaluation of their biological activities. Indian J Chem. 2011;50B:1794–1801. [Google Scholar]
- 40.Kalirajan R, Sankar S, Jubie S, Gowramma B. Molecular docking studies and in-silico ADMET screening of some novel oxazine substituted 9-anilinoacridines as topoisomerase II inhibitors. Indian J Pharm Educ Res. 2017;51(1):110–115. doi: 10.5530/ijper.51.1.15. [DOI] [Google Scholar]
- 41.Kalirajan R, Gowramma B, Jubie S, Sankar S. Molecular docking studies and in silico ADMET screening of some novel heterocyclic substituted 9-anilinoacridines as topoisomerase II inhibitors. JSM Chem. 2017;5(1):1039–1044. [Google Scholar]
- 42.Kalirajan R, Gaurav K, Pandiselvi A, Gowramma B, Sankar S. Novel thiazine substituted 9-anilinoacridines: synthesis, antitumour activity and structure-activity relationships. Anti-Cancer Agents Med Chem. 2019;11:1350–1358. doi: 10.2174/1871520619666190408134224. [DOI] [PubMed] [Google Scholar]
- 43.Kalirajan R, Vivek kulshrestha, Sankar, S. Synthesis, characterization and evaluation for antitumour activity of some novel oxazine substituted 9-anilinoacridines and their 3D-QSAR studies. Indian J Pharm Sci. 2018;80(5):921–929. doi: 10.4172/pharmaceutical-sciences.1000439. [DOI] [Google Scholar]
- 44.Kalirajan R, Pandiselvi A, Gowramma B, Balachandran P. In-silico design, ADMET screening, MM-GBSA binding free energy of some novel isoxazole substituted9-anilinoacridines as HER2 inhibitors targeting breast cancer. Curr Drug Res Rev. 2019;11(2):118–128. doi: 10.2174/2589977511666190912154817. [DOI] [PubMed] [Google Scholar]
- 45.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comp Aided Mol Design. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 46.Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 47.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein–ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Abel R, Zhu K, Cao Y, Zhao S, Friesner RA. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modelling. Proteins. 2011;79:2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cross R (2020) Chloroquine’s use to treat COVID-19 is backed by US government, but many questions remain. Drug Dev 98(12)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and material are available upon request.