Summary
Cerebral organoids (COs) are rapidly accelerating the rate of translational neuroscience based on their potential to model complex features of the developing human brain. Several studies have examined the electrophysiological and neural network features of COs; however, no study has comprehensively investigated the developmental trajectory of electrophysiological properties in whole-brain COs and correlated these properties with developmentally linked morphological and cellular features. Here, we profiled the neuroelectrical activities of COs over the span of 5 months with a multi-electrode array platform and observed the emergence and maturation of several electrophysiologic properties, including rapid firing rates and network bursting events. To complement these analyses, we characterized the complex molecular and cellular development that gives rise to these mature neuroelectrical properties with immunohistochemical and single-cell transcriptomic analyses. This integrated approach highlights the value of COs as an emerging model system of human brain development and neurological disease.
Keywords: brain organoids, cerebral organoids, cerebral cortex, electrophysiology, multi-electrode array, MEA, neural network, single cell RNA sequencing
Highlights
-
•
CO electrophysiology can be quantified with a multi-electrode array method
-
•
CO electrophysiological trajectories correlate with molecular and cellular development
-
•
The neurotrophin/TRK signaling pathway is active in COs by 5 months in culture
Cerebral organoids (COs) have a unique advantage in modeling human-specific features of early brain development. Here, we profile their neuroelectrical activities with an MEA platform over time and correlate these activities with their increasingly complex molecular and cellular features. This integrated approach highlights the value of COs as an emerging model system of human brain development and neurological disease.
Introduction
Our understanding of mammalian brain development has largely been derived from rodent models. Although these models have provided invaluable mechanistic insight, they are significantly limited in that they do not fully recapitulate the molecular, structural, and genetic complexity of human conditions. For example, human brains exemplify unique anatomical features, such as an expanded outer subventricular zone (SVZ), that are not present in mice (Sousa et al., 2017). This zone contains basal radial glia (bRG) that are critical for human cortical expansion (Fietz et al., 2010; Hansen et al., 2010; Johnson et al., 2015). Furthermore, our current knowledge of human brain development is largely based on analyses of postmortem or pathological specimens. Although they have provided important fundamental knowledge, these tools are not amenable to experimental manipulation (Hartlaub et al., 2019; Stiles and Jernigan, 2010).
Recent advances in stem cell and three-dimensional tissue culture techniques led to the advent of cerebral organoids (COs) that can model early periods of human fetal brain development (Camp et al., 2015; Hester and Hood, 2017; Lancaster et al., 2013; Lancaster and Knoblich, 2014; Qian et al., 2016; Quadrato et al., 2017; Yin et al., 2016). COs, which are generated from human pluripotent stem cells, are three-dimensional structures that can self-organize to form discrete regions of the human brain, such as the cerebral cortex. Several CO protocols have been developed that rely either on intrinsic patterning molecules or on exposure to morphogens under directed differentiation. Protocols that rely on spontaneous cues generate COs with diverse cell populations and multiple regional brain identities at the expense of increased interorganoid variability compared with directed differentiation approaches (Pasca, 2018).
Despite significant advances in discerning the morphological and cellular complexity of COs, very little work has been done to characterize their electrophysiological (EP) properties. A previous report showed the presence of spontaneous activity arising from photosensitive, retinal-like cells using a 64-channel neural probe implanted within 8-month-old whole-brain COs (Quadrato et al., 2017). Other studies performed EP measurements on organoid slices using whole-cell patch-clamp techniques (Birey et al., 2017; Xiang et al., 2017) or measured network activity in COs via imaging of synchronized calcium activities (Sakaguchi et al., 2019). Most recently, Trujillo et al. used a forebrain organoid model to compare organoid EP features to EEG signatures from premature infants (Trujillo et al., 2019).
Here we examine the EP development of COs using a 64-channel multi-electrode array (MEA) array platform. This culture method enabled us to detect weak spontaneous electrical activity in COs after as early as 1 month in culture. We noted the gradual evolution of EP features in COs within 3 months in culture, as evidenced by elevated mean spike rates and characteristic spontaneous action potentials with discrete depolarizations. In addition, we characterized the simultaneous development of morphological and transcriptomic features in COs with immunohistochemistry (IHC) and single-cell RNA sequencing (scRNA-seq). Maturation of electrical features correlated with dynamic changes in the development of cell types within COs, such as the emergence of astrocytes and diverse neuronal populations. Last, as COs transition into increased cellular and morphological complexity, we observed activation of the neurotrophin (NTR)/TRK receptor signaling pathway. Our findings highlight the use of COs as a model not only for recapitulating complex morphological features of human brain development, but also as a functional testing platform to investigate how genetic and environmental factors perturb the development and maturation of neural circuits. Further, our results provide an impetus to use COs as a system to screen and functionally characterize potential pharmaceutical neuromodulators and gene therapeutics that would restore proper EP function in neurodevelopmental disorders.
Results
Generation of COs from Human Induced Pluripotent Stem Cells
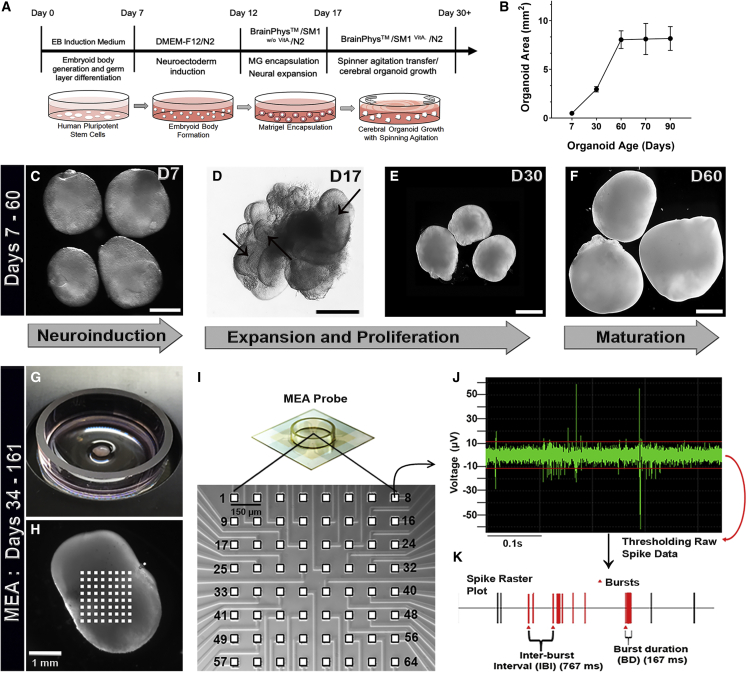
To investigate the contribution of diverse human brain cell types to the developmental maturation of electrical properties in COs, we used an undirected differentiation approach to generate human whole-brain COs (Di Lullo and Kriegstein, 2017; Hester and Hood, 2017; Lancaster et al., 2013; Pasca, 2018). Specifically, we used our previously described methods to generate human whole-brain COs based on a modified protocol from Lancaster et al. (Hester and Hood, 2017; Lancaster et al., 2013) (Figure 1A). We incorporated BrainPhys medium in our methodology, which has been shown to enhance neuronal synaptic function and promote the EP maturation of neuronal monolayer cultures (Bardy et al., 2015) and has not been previously used for long-term culture of COs (Figure 1A). We generated COs from a well-characterized human induced pluripotent stem cell (hiPSC) line (Burridge et al., 2011) that exhibits molecular characteristics of pluripotency (Figures S1A–S1E). These COs showed similar growth properties compared with previously established protocols (Figure 1B). Of note, the hiPSC-derived neuroepithelial (NE) bodies that give rise to the COs had translucent borders (Figure 1C) and expanded into NE buds at day 17 in culture (Figure 1D), similar to the developmental timing seen in conventional culture methods (Figures S1F–S1H). When switched to constant agitation culture conditions, COs showed steady growth characteristics (Figures 1B, 1E, and 1F) and did not significantly increase in growth area after day 60 in culture. Molecular analyses of early-stage CO cultures at day 25 revealed abundant proliferative SOX2+ neural progenitor cells (NPCs) organized into neuroepithelium-like structures as shown by the expression of the cell-cycle marker Ki67 and the mitosis-specific cell-cycle marker pHH3 (Figures S1I and S1J).
Figure 1.
CO Protocol Timeline and Early-Stage Morphological Features
(A) Timeline of major developmental features and technical milestones in generating human COs. EB, embryoid body; MG, Matrigel encapsulation; VitA, vitamin A.
(B) Growth curve of organoid areas shows a plateau at day 60 (minimum of four independent organoids per time point). Data are presented as mean ± SEM.
(C) Encapsulated NE body that has been induced toward neuroectoderm. Scale bar, 200 μm.
(D) Encapsulated NE body with expanding neuroepithelial folds (black arrows). Scale bar, 100 μm.
(E) Day 30 COs. Scale bar, 1 mm.
(F) Day 60 COs. Scale bar, 1 mm.
(G and H) Spontaneous electrical activity can be detected from COs using a multi-electrode array (MEA) system. (G) A transferred CO on an MEA probe during acquisition of spontaneous electrical activity. (H) CO recording setup with 64 planar microelectrodes.
(I) Microelectrode array at high magnification.
(J) Local field potential trace from an individual microelectrode.
(K) Spike raster plot depicting electrical activities derived from a CO. Bursts are represented as red triangles.
Probing Electrical Activities in COs Using an MEA Platform
To probe the electrical activities of maturing whole-brain COs, we established a method to monitor their spontaneous electrical activity with a 64-channel MEA platform to detect and record extracellular field potential change activity (Figures 1G–1K). Planar MEA systems have a non-invasive, multi-site, electrical recording interface to functionally assay the electrophysiology of neuronal cultures. MEAs have been widely used with monolayer neuronal cultures to study their neurophysiology (Kizner et al., 2019; Mehta et al., 2018), although the application of MEAs to the study of EP features in three-dimensional whole-brain COs is underexplored. Since CO cultures require agitation in suspension to enhance nutrient diffusion and maintain optimal three-dimensional growth conditions, we developed a non-destructive method to functionally assay their surface electrical activities (Figures 1G–1K). Specifically, in this method, COs are transferred onto an MEA probe to record spontaneous electric activities. This protocol allowed us to identify the precise developmental timing of EP property emergence in COs. Since the recording distances of MEAs are not expected to be greater than 60 μm (Delgado Ruz and Schultz, 2014), we expected to measure electrical activities not only from superficial layer neurons positioned up to three cell layers from the surface layer Cajal-Retzius (CR) cells. In addition, based on the developmental time points at which we performed the MEA analyses, we expected to record from a diverse set of excitatory and inhibitory neuronal populations.
Evolution of EP Properties in COs
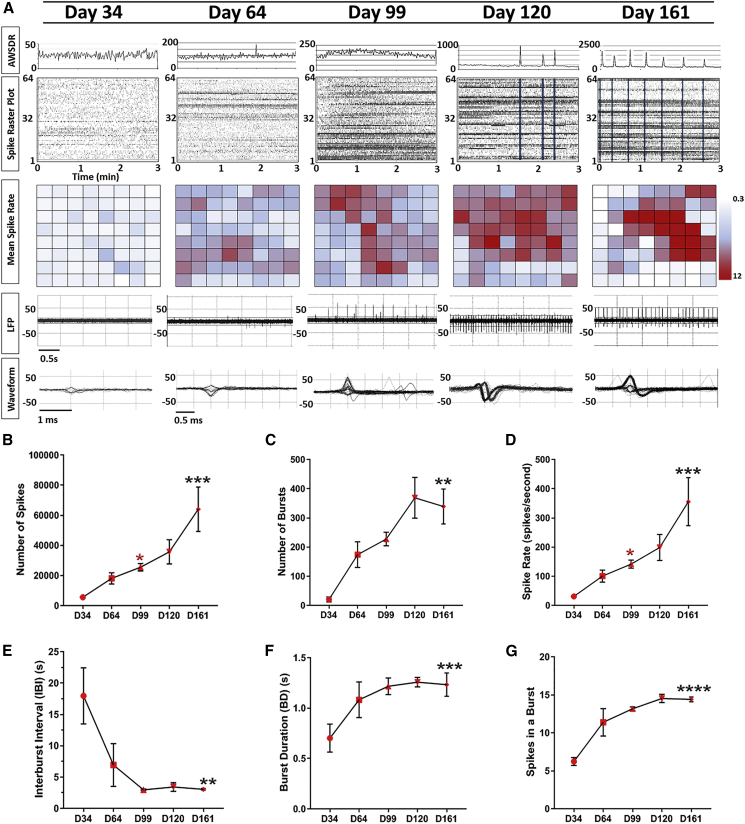
Using a 64-channel MEA array, we investigated the earliest time point at which electrical activity emerges in COs. We detected weak spiking activity (Figure 2) at day 34 in culture, which is expected, as this time point is correlated to a developmental window of rapid expansion of neuroepithelium and the beginning of early neurogenesis within COs that precedes neuronal maturation (Figures 1D, S1I, and S1J). After day 64 in culture, COs exhibited increases in both mean spike rate and field spike amplitude (local field potential), with both properties continuing to increase thereafter into day 99 of culture (Figures 2A and 2D). In addition, cortical neurons in COs showed characteristic spontaneous action potentials with discrete depolarizations (waveforms in days 120 and 161) that coincided with the observed expansion of astrocytes (Figures 4F, 4G, and 2A). At day 120, as indicated by a significant increase in the array-wide spike detection rate, COs exhibited synchronized burst firings (SBFs), which are prominent characteristic features of functional cortical neural networks in the developing mammalian cortex and a hallmark of synaptic signal propagation (Figure 2A). Day 161 COs showed further increases in the number of SBFs, which suggests the COs formed highly interconnected neural networks (Figure 2). Network burst metrics were analyzed in day 161 COs and showed an average network burst duration of 985 ± 152 ms, an average spike number of 1700 ± 300 in a network burst, and an average network burst interval of 25.3 ± 4.2 s. We observed a significant increase in the number of spikes, bursts, and spike rates over time in culture, which reflects the neuronal synaptic maturation of electrical transmission within COs (Figures 2B–2G). Altogether, these results demonstrate that COs show a gradual evolution of EP properties over development that resembles hallmark features of the developing neonatal brain. This trajectory begins in day 30–64 culture when COs have immature electrical activities as evidenced by random, low-amplitude, spike events and matures as more complex network bursting events or SBFs emerge by day 120 and increase to day 161.
Figure 2.
Developmental Trajectories of Electrophysiological Properties in COs
(A) Detection of spontaneous electrical activity in day-34 COs and approximately every month thereafter shows a progressive increase in and complexity of EP properties. Top: Graphical representation of the array-wide spike detection rate (AWSDR) above every spike raster plot with a 3-min recording interval. Synchronized burst firing (SBF) activity is observed in late-stage CO cultures and is indicated by light blue columns within raster plots of days 120 and 161. Note the corresponding peaks in the AWSDR above each SBF. Middle: Heatmap plots of the mean spike rates of neurons within COs recorded by each individual microelectrode. Bottom: Local field potential (LFP) traces and a corresponding waveform taken from a shorter time interval showing a progressive increase in voltage amplitude and spiking rate at successive developmental time points. Note the characteristic action potential waveforms shown in days 99, 120, and 161.
(B–G) EP quantifications of developing COs at approximately monthly intervals initiating at day 34 (n = 4/time point). EP data include (B) number of spikes, (C) number of bursts, (D) spike rate (number of spikes/s), (E) interburst interval, (F) burst duration, and (G) number of spikes in a burst. Data are presented as mean ± SEM and statistical analyses were performed using one-way ANOVA with Tukey's multiple comparisons test (n = 4 independent organoids per time point recorded and 2 independent experiments performed). Differences between day 34 and day 161 or between day 99 and day 161 time points are shown by black or red asterisks, respectively. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001. All statistical analyses are shown in Table S3.
Figure 4.
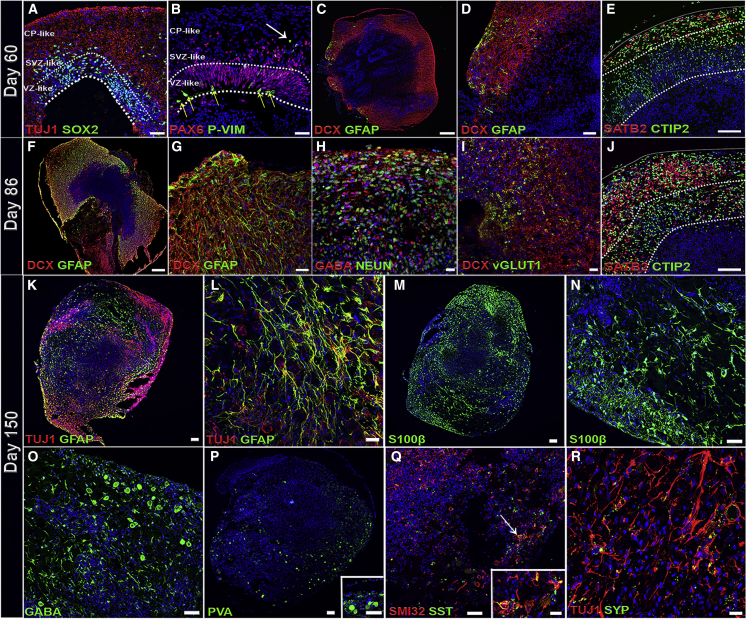
Cell-Type Diversity in COs
(A) Immunostaining of day 60 COs shows SOX2+ NPCs lining the ventricle-like structures within COs. At day 60, the majority of SOX2+ NPCs are restricted to the VZ-like areas within the area of dashed lines. SOX2+ NPCs are also observed migrating into SVZ-like areas and rarely observed in CP-like zones as shown by TUJ1 immunostaining at this developmental stage. Scale bar, 50 μm.
(B) Immunostaining for the radial glial (RG) marker PAX6, showing dense expression primarily within the VZ-like area, in addition to the mitotic RG marker phospho-Vimentin (P-VIM), showing RG preferentially dividing at the apical surface (yellow arrows, lower dashed line). PAX6+ RGs observed outside of the VZ-like zone representbasal RG cells, which also remain mitotically active to a lesser extent, as shown by P-VIM staining (white arrow). Scale bar, 50 μm.
(C) Day 60 COs show the emergence of astroglia as shown by GFAP staining. DCX, a neuronal migratory marker, demarcates the CP-like zone. Scale bar, 200 μm.
(D) High-magnification image of (E) showing the presence of GFAP+ astrocytes. Scale bar, 50 μm.
(E) Immunostaining for the late-born and superficial CP marker SATB2 and the deep-layer CP marker CTIP2. Dashed lines indicate upper SATB2+ cortical layer zone with deeper layer CTIP2+ zone. Scale bar,100 μm.
(F) Day 86 COs show significant expansion of GFAP+ astrocytes within the CP-like zone, which also contains DCX+ neurons. Scale bar, 200 μm.
(G) High-magnification image of (I) showing the increased presence of GFAP+ astrocytes compared with day 60. Scale bar, 50 μm.
(H) Immunostaining for the inhibitory neuronal marker GABA and the mature neuronal marker NEUN. Scale bar, 20 μm.
(I) Presence of excitatory neurons is also observed as shown by staining for the vesicular glutamate transporter 1, vGLUT1, which co-localized with DCX in the CP-like zone. Scale bar, 50 μm.
(J–N) (J) Day 86 COs show an expansion of the superficial SATB2+ cortical layer zone compared with day 60 COs. Dashed lines indicate more defined upper SATB2+ cortical layer zone and deeper layer CTIP2+ zone. Scale bar, 100 μm. (K–N) Immunostaining of day 150 cerebral COs shows presence of neuronal (K and L, TUJ1) and astrocytic cell lineages (K and L, GFAP, and M and N, S100β). Scale bars, (K and M) 100 μm, (L) 20 μm, and (N) 50 μm.
(O–R) Maturation of day 150 COs exhibits GABAergic neurons as shown by (O) GABA staining and interneuron subtypes, (P) parvalbumin (PVALB), and (Q) somatostatin (SST). Scale bars, (O) 50 μm; (P) 100 μm and inset, 50 μm; (Q) 50 μm and inset, 25 μm. (R) Synaptic maturation is also observed in day 150 COs as shown by expression of the pre-synaptic marker, synaptophysin (SYP). Scale bar, 25 μm. All images were stained with DAPI (blue) (n = 4 independent organoids, all time points).
Increased Cellular Diversity in COs and Correlation with Their Electrophysiological Trajectories
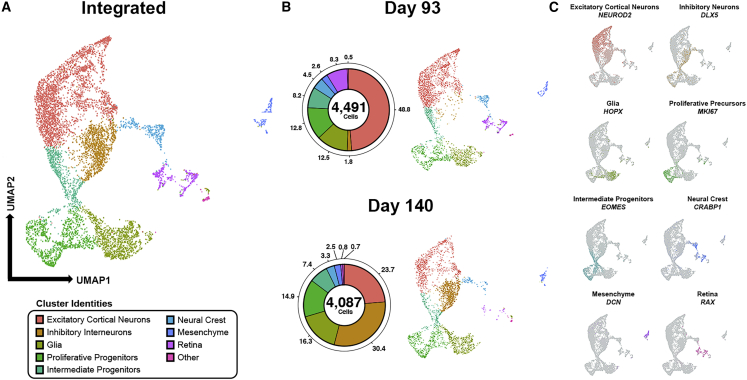
To correlate the trajectories of electrical activities we observed in COs to their morphological and molecular properties, we performed scRNA-seq in COs and IHC analyses of neuronal and glial lineages. We first subjected COs to scRNA-seq at late developmental time points, including days 93 and 140, to characterize the dynamic transcriptome of their diverse cell types (Figures 3 and S2A). We performed unsupervised clustering on gene expression profiles from an integrated dataset of 8,578 cells (Figures 3A and 3B) and identified eight major clusters using known cell-type-specific marker genes (Figures 3C and S2A). Excitatory cortical neurons (NEUROD4+) and inhibitory interneurons (GAD2+ and DLX5+) comprised approximately 50% and 2% of all cells in day 93 COs, respectively, and approximately 24% and 30% of all cells in day 140 COs, respectively. The presence of inhibitory neurons in late-stage COs may indicate the maturation of local cortical circuitry within neuronal networks, which correlates with the maturation of EP properties we observed, such as the high degree of SBFs observed in day 161 COs (Figure 2). This developmental link supports their previously described role in the establishment and control of neural network activity (Kuijlaars et al., 2016; Mann and Paulsen, 2007). We also identified several non-neuronal populations, including proliferative progenitors (MKI67+, NUSAP1+), glia (GFAP+, HOPX+), and intermediate progenitors (EOMES+, NHLH1+), that had approximately equal representation in both day 93 and day 140 COs. The role of glia and progenitor populations in neurogenesis, neuronal maturation, and neural network activity has been extensively characterized in the developing human brain (Felix et al., 2020; Wilton et al., 2019). Their stable presence in early- and late-stage COs is likely a reflection of their similar functions in developing COs.
Figure 3.
scRNA-seq Reveals Dynamic Cell Type Diversity Throughout the Developmental Trajectory of COs
(A) Uniform-manifold approximation map (UMAP) of single-cell RNA-sequencing data from two COs with 8,578 cells total analyzed in one independent experiment. Each point on the UMAP represents a single cell. Color map specifies cell type, as outlined in the legend.
(B) Donut plots and UMAPs for day 93 and day 140 capture of the cell type diversity within COs as a function of time. Day 93: 4,491 cells shown, including 2,190 excitatory cortical neurons (48.8%), 79 inhibitory interneurons (1.8%), 560 glia (12.5%), 575 proliferative progenitors (12.8%), 370 intermediate progenitors (8.2%), 203 neural crest (4.5%), 116 mesenchyme (2.6%), 373 retina (8.3%), and 25 other (0.5%). Day 140: 4,087 cells shown, including 968 excitatory cortical neurons (23.7%), 1,242 inhibitory interneurons (30.4%), 667 glia (16.3%), 608 proliferative progenitors (14.9%), 304 intermediate progenitors (7.4%), 133 neural crest (3.3%), 104 mesenchyme (2.6%), 34 retina (0.8%), and 27 other (0.7%).
(C) UMAPs depict canonical markers for each cell cluster type, excluding other. Additional markers are provided in Figure S4.
To complement our scRNA-seq analyses, we also performed a comprehensive IHC analysis of key cell-type-specific markers during CO development. At day 40, COs showed expansion of neuroepithelium as revealed by expression of the telencephalic markers FOXG1 and LHX2 (Figure S2B). LHX2 displayed a more restrictive expression pattern in day 40 COs compared with FOXG1 expression, coinciding with its predominate expression in the dorsal telencephalon (Godbole et al., 2018). LHX2 maintained expression at day 93 in proliferative progenitors and a subset of excitatory neurons, although expression diminished at day 140 (Figure S2C). In contrast, FOXG1 maintained robust expression in both excitatory and inhibitory neurons at days 93 and 140 (Figure S2C). In addition, the mitotic NPC marker SOX2 was expressed in the proliferative neuroepithelium at day 40 (Figure S2B) and absent in postmitotic neurons, as expected (Figure S2C). Further, IHC analysis of SOX2 at day 60 revealed expression in NPCs, which lined ventricle-like structures within COs and also expanded minimally in SVZ-like zones (Figure 4A). Similarly, PAX6, a dorsal cortical marker, which is characteristically expressed in radial glial (RG) cells, was also expressed predominately at ventricular zone (VZ)-like zones (Figure 4B). We also performed a more extensive analysis of markers for neuronal subtypes within progenitor zone-like and cortical plate (CP)-like regions at day 60. As shown in Figure S3B, CTIP2+ and TBR1+ neurons are observed extensively within CP-like regions, and TBR2+ neurons are restricted predominately to the SVZ-like zones. We also observed SOX2+/HOPX+ progenitors within VZ-like zones and the cortical layer 1 marker, REELIN, expressed at the most upper cortical layer that contains CR cells (Figure S3B). We and others also observe transient survival of CR neurons in COs (Renner et al., 2017), as indicated by low levels of RELN expression in day 140 COs (Figure S5A).
We further investigated the cellular composition of day 60 COs and observed a minor population of astrocytes emerging, as shown by GFAP expression (Figures 4C and 4D), that expanded significantly after an additional 26 days in day 86 COs (Figures 4F, 4G, and S3A). We found that our transcriptomic data were in agreement with this observation, as we detected substantial expression of astroglia markers GFAP, S100β, AGT, and AQP4 (Figure S6). It is plausible that the progressive electrical maturation of COs during this developmental period could be driven in part by the expansion of astrocytes as these cell types have been shown to promote the functional maturation of cortical neurons in vivo and in vitro (Kuijlaars et al., 2016; Sloan et al., 2017).
The inhibitory marker GABA was initially apparent at very low levels at day 40 and increased significantly as GABAergic neurons appeared in later-stage COs, from day 60 to day 120 (Figures 3B, 4H, 4O, and S3A). To complement these analyses, we stained for the glutamatergic neuronal marker vesicular glutamate transporter 1 (VGLUT1), and observed the typical punctate staining pattern indicating the presence of this excitatory pre-synaptic marker (Figure 4I). At day 86, a large increase in SATB2+ neurons was observed within expanded upper CP layers compared with day 60 COs (Figure 4E), which began to more clearly define upper and deeper CP layers (Figure 4J). Thus, it is probable that our MEA recordings at days 64 and 99 comprise electrical activities arising from CR and CTIP2+ and SATB2+ neurons, whereas day 120 and 161 recordings also include the electrical activities of GABAergic neurons. These data, which are consistent with previous reports, indicate that whole-brain COs can develop for an extended duration in culture and generate diverse brain cell lineages.
COs Exhibit a Diversity of Apical and Basal Radial Glial Subtypes
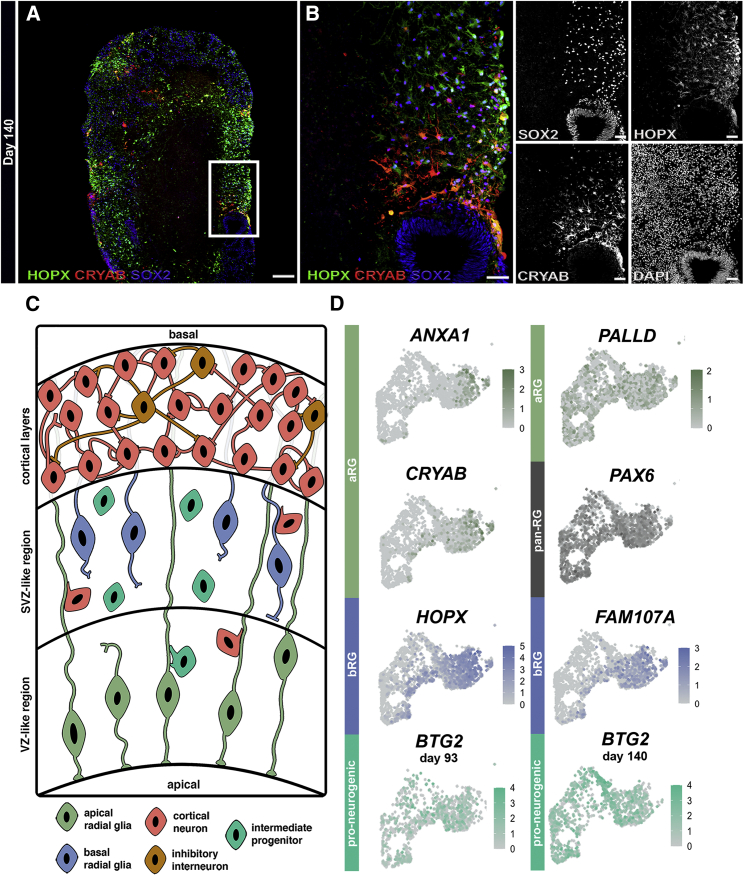
Next, we examined whether our whole-brain COs generated the diversity of RG cell types that are present in the developing human brain. Given the instrumental role that apical radial glia (aRG) and bRG play in the structural scaffold and maturation of the developing human brain, we first confirmed their presence in the transcriptome of late-stage COs with our scRNA-seq data (Figure 5). We observed substantial RG marker expression in both the glia and the proliferative progenitor clusters (Figure 5D), including the aRG markers ANXA1, PALLD, and CRYAB; the late bRG markers HOPX and FAM107A; and the pan-radial glia markers SOX2 and PAX6 (Thomsen et al., 2016). We also noted a remarkable increase in the anti-proliferative and pro-neurogenic BTG2 (also known as TIS21) from day 93 to day 140 (Figure 5D), which indicates a significant increase in progenitor division fates destined to become neurons (Attardo et al., 2010; Iacopetti et al., 1999). We then characterized their development from day 60 to day 140 by IHC. At day 60, we noted pVIM+ mitotic RG cells within VZ-like and radially distant, SVZ-like regions comprising early progenitors found in the scaffold of the embryonic brain (Figure 4B). At day 90, the VZ- and SVZ-like regions primarily contained SOX2+/HOPX+ progenitors (Figures S6A–S6C). By day 140, CRYAB+ progenitors appeared at the apical surface, whereas HOPX+ progenitors appeared in a distant outer VZ-like zone (Figures 5A and 5B). This patterning suggests that COs contain regionally defined aRG and bRG populations that become readily apparent by day 140. Recently, a study described the emergence of truncated radial glia (tRG) that marks the beginning stages of the transition from a continuous to a discontinuous RG scaffold in late human brain development (Di Lullo and Kriegstein, 2017; Nowakowski et al., 2016). tRG, which express the CRYAB marker, are formed when aRG retract their basal fibers and terminate them in the outer SVZ, thus resembling a truncated morphology. Interestingly, we identified CRYAB+ progenitors in COs, although future studies are necessary to further validate whether they represent tRG as observed in vivo.
Figure 5.
Presence of Diverse RG Cells in COs
(A) Low-power composite image of HOPX, CRYAB, and SOX2 expression in day 140 COs. White frame indicates reference for (B).
(B) High-power merged and single-channel images from inset in (A).
(C) Schematic depicting progenitor zones in the CO model.
(D) Feature UMAPs of integrated glia and proliferative progenitor clusters (note the retained shape of clusters from Figure 3). From top to bottom: expression of aRG-specific markers PALLD, ANXA1, and CRYAB; bRG-specific markers HOPX and FAM107A; pan-RG markers PAX6 and SOX2 in integrated scRNA-seq dataset; and anti-proliferative/pro-neurogenic marker BTG2 shows increased expression over time (left, day 93, and right, day 140) (n = 4 independent organoids). Scale bar (A), 200 μm. All scale bars in (B), 50 μm.
Neuronal and Synaptic Maturation in COs
Neuronal and synaptic development is indispensable to the formation of neural circuits and drives EP maturation of COs. Therefore, we sought to identify evidence of this maturation at the protein and transcriptome levels around days 120–150 in culture. We first noted the formation of synapses in day 120 COs with expression of the mature pre-synaptic marker synaptophysin (SYP) that partially co-localized with the mature neuronal dendritic marker MAP2 (Figure S4A). We further examined the expression levels of a more comprehensive gene panel of dendritic, presynaptic, and postsynaptic markers in our scRNA-seq data in day 140 COs (Figure S4B). We evaluated synaptic markers enriched within either inhibitory or excitatory neurons. These markers may be critical for synaptic propagation, as they correlate with the increased spiking activity and SBF that we detected in late-stage COs. For example, GAP43, SYT4, SYP, and SLC17A7 (Figure S4B, highlighted) are pre-synaptic markers enriched in excitatory neurons. Conversely, NRXN3 and SLC32A1 (Figure S4B, highlighted) are pre-synaptic markers enriched in inhibitory neurons. In some cases, these genes encode well-documented functions for these classes of neurons. For example, SLC17A7 encodes vesicular glutamate transporter 1 (VGLUT1) and SLC32A1 encodes for VIAAT, which is a membrane protein associated with the uptake of GABA and glycine into synaptic vesicles. In addition to pre-synaptic markers, we also observed expression of the post-synaptic marker HOMER1, GPHN, NLGN1, NLGN2, DLG3, DLG4 (also known as PSD95), and SHANK gene family members in both neuronal classes, although most markers showed enrichment in excitatory neurons.
Given this evidence of synaptic maturation, we extended our analyses in day 150 COs for additional support of neuronal maturation. Of note, we observed a significant increase in GFAP+ and S100β+ astrocytes by day 150 in culture (visualized in Figures 4K–4N, quantified in S3A). These astrocytes exhibited extensive, ramified processes that indicate their maturation and association with neuronal populations (Figures 4K–4N). We also found that from day 86 to day 150, our COs exhibited significantly increased levels of GABA expression (visualized in Figure 4O, quantified in Figure S3A). GABAergic neurons in day 150 COs showed significantly larger cell bodies compared with those in earlier-stage COs (Figure 4O). We were able to identify specific GABAergic neuron subtypes, including those that were immunoreactive to either parvalbumin or somatostatin, which further provides evidence for GABAergic neuron maturation (Figures 4P and 4Q). Finally, we noted the pan-neuronal marker TUJ1 to localize with the pre-synaptic marker SYP, which depicts the development of mature CO networks (Figure 4R).
Gene Expression Profiling in Mature COs Reveals Activation of the NTR/Trk Receptor Signaling Pathway
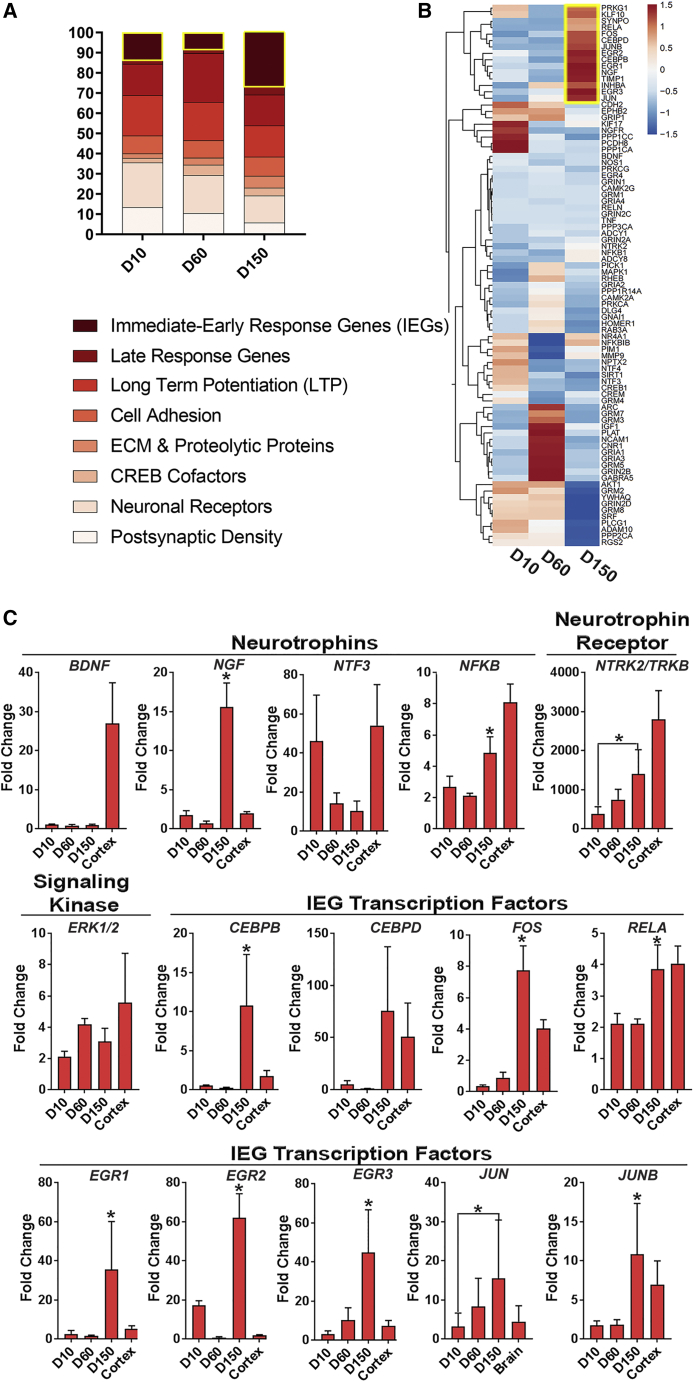
To further investigate whether the observed EP maturation of COs correlated with concomitant changes in gene expression patterns, we performed qRT-PCR and profiled the expression of 84 key genes associated with human synaptic plasticity at three additional time points during CO development (day 10, day 60, and day 150) (Figure 6). For this analysis, we selected a wide range of time points to identify synaptic gene expression changes that more clearly delineated immature (day 60) from mature (day 150) neuroelectrical properties in COs. As shown in Figure 6A, these genes encode a diverse set of protein functions that are associated with synaptic activity and were functionally categorized into 10 groups, such as neuronal receptors, cell adhesion molecules, or immediate-early response gene (IEG) groups. Genes that were upregulated 3-fold or higher relative to hiPSC gene expression levels were collated into these 10 groups and their percentage abundance was determined from each organoid time point. Interestingly, this analysis revealed increased expression of the TRKB receptor (Figure 6C) during organoid development without a concomitant increase in the expression of one of its ligands, BDNF. These data agree with a previous study showing that activation of TrkB in newborn cortical neurons occurs through EGFR signaling rather than through BDNF, which is expressed at low levels during brain development but increases in expression after birth (Puehringer et al., 2013). In contrast, we also observed a relative increase in the levels of NGF at day 150, which may reflect the expansion of inhibitory neurons at this time point and agrees with a report demonstrating that this NTR is primarily produced in GABAergic neurons (Biane et al., 2014). In addition, in agreement with our IHC analysis, we observed an increase in AMPA/NMDA receptor expression at day 60, which signifies the maturation of excitatory neurons (Figure 6B). The relative expression of these genes appears to decrease by day 150, although this observation most likely reflects the increased population of inhibitory neurons at this time point, which artificially dilutes AMPA/NMDA expression levels (Figures 3 and S5B). We did not observe a significant change in the expression levels of these genes specifically in excitatory neurons in our scRNA-seq dataset, which supports this conclusion (Figure S5B).
Figure 6.
Gene Expression Profiling Reveals Activation of an IEG Cluster in Day 150 COs
(A) Functional categorization of upregulated genes involved in synaptic plasticity. The highlighted boxed segments in the bar graph at each time point refer to IEGs.
(B) Heatmap of gene expression dynamics associated with synaptic activity at days 10, 60, and 150 during CO development.
(C) Quantitative expression analyses of NTR/TRK signaling and IEG activation genes regulating synaptic plasticity at days 10, 60, and 150 during CO development. Profiled gene members include NTRs, the TRKB receptor, the ERK 1/2 signaling kinase, and multiple IEG transcription factors. Gene expression values that showed significant differences (p < 0.05) between day 60 and day 150 time points are indicated by an asterisk. Adult human brain cortex was included as a positive control (n = 5 independent organoids pooled together and 3 independent experiments performed).
In addition, 14 IEG genes were upregulated in day 150 COs (Figure S5C). IEGs have been shown to play critical roles in promoting synaptic formation, transmission, and maturity (Kim et al., 2018; Lanahan and Worley, 1998), which suggests that these genes perform similar functions to promote EP maturation in late-stage COs. Clustering analysis confirmed this finding and revealed 33 genes that were upregulated in day 150 COs that consisted of IEGs, neuronal receptor genes, postsynaptic density genes, and others. In addition, the highest upregulated gene cluster within the day 150 group comprised primarily IEGs (11/15 genes). We next determined whether these upregulated genes functioned within a common signaling pathway and used the Ingenuity Pathway Analysis (IPA) software tool to perform this bioinformatics analysis. This analysis revealed 205 canonical pathways with a default threshold (−log(p value) > 1.3). We focused on those canonical pathways that were both enriched in the developing human brain and were top ranked according to their statistical significance. Based on these criteria, we identified IL17A and the NTR/TRK signaling pathway as the top two ranked canonical pathways (−log(p value) = 13.2 and 13.1, respectively) (Table S2). We further verified whether the receptors mediating the activation of these downstream molecules were relevant to brain development. We noted a significant increase in the expression of the TRKB receptor at day 140 compared with day 90 (Table S2 and Figure S5E), which suggests NTR/TRK signaling is enriched as COs mature. Conversely, we did not note significant changes in the expression of IL-17RA or the related IL-17RC over time (Figure S5D and Table S4) and believe the apparent enrichment of IL17A signaling in our IPA is a reflection of its common downstream targets with the NTR/TRK signaling pathway. Taken together, these findings suggest that NTR/TRK signaling contributes to promoting synaptic maturation in COs and may drive complex network bursting activities.
Discussion
COs have shown significant promise as three-dimensional model systems of human brain development and have shaped our current understanding of key neurodevelopmental milestones (Birey et al., 2017; Camp et al., 2015; Hartlaub et al., 2019; Lancaster et al., 2017; Pasca, 2018; Qian et al., 2016; Yin et al., 2016). Here we expanded upon these studies to correlate molecular and morphological features in COs with their EP trajectories. To accomplish this analysis, we used our MEA methodology to evaluate the transition of neural circuits from spontaneous activity to more robust electrical features that give rise to network bursting. This EP trajectory indicates the development of highly connected neural networks, which is reminiscent of the functional maturation of early brain development. We simultaneously characterized the morphological development of the progenitor and cortical populations that give rise to this neuroelectrical maturation with IHC and transcriptomic analyses. For instance, we found that the emergence of GABAergic neurons in our transcriptomic and IHC data from late-stage COs correlated with the onset of mature network bursting observed in our MEA data. We also noted the increased incidence of neurogenic progenitor divisions, supported by increased BTG2 expression in the transcriptome of day 140 COs, which coincided with the rapid development of neural networks and subsequent functional maturation.
Our findings are congruent with reports by Giandomenico et al. and Trujillo et al. that describe network bursting in CO slices and intact forebrain COs, respectively. These bursting events become prominent between 4 and 6 months in culture and continue to develop nested oscillatory network events by 8–10 months in culture. The work presented here expands these findings by identifying the morphological characteristics and cellular diversity that emerge with EP maturation in whole-brain, undirected COs. It is apparent that the organoid culture conditions are pertinent to these developmental phenotypes, as we observed relatively high representation of inhibitory neurons as early as 4 months in organoid development compared with other studies (Quadrato et al., 2017; Tanaka et al., 2020; Trujillo et al., 2019).
Alternative methods that rely on calcium imaging to evaluate neuronal network activity in COs have been recently developed (Bershteyn et al., 2017; Lancaster et al., 2017; Mansour et al., 2018; Sakaguchi et al., 2019; Watanabe et al., 2017) and would greatly complement the analyses we report here. These methods permit the quantitation of changes in intracellular calcium levels in neuronal and glial cultures (Bonifazi and Massobrio, 2019; Wong, 1998) and have been adapted for use in dissociated organoid cultures by Sakaguchi et al. The primary advantage of calcium-imaging-based methods is their ability to measure functional activity with spatial resolution. A combined approach that incorporates both calcium imaging and MEA techniques would therefore provide the advantage of analyzing functional activities with both high temporal and spatial resolution and allow a more accurate assessment of neural network dynamics in COs.
Importantly, our results highlight the translational potential of COs to model neurodevelopmental disorders that arise from aberrant neural network formation. To date, COs have provided critical insights into the structural etiology of several neurodevelopmental disorders, including autism (Hou et al., 2020; Mariani et al., 2015; Papariello and Newell-Litwa, 2020; Wang et al., 2020; Zhang et al., 2020) and epilepsy (Niu and Parent, 2020; Sun et al., 2019). However, the characterization of neural network activity is indispensable in several of these disorders, including epilepsy. The combined functional, morphological, and transcriptomic analyses of COs may be implemented to characterize the developmental trajectory of disease phenotypes. Further refinement in correlating EP properties in COs with clinically relevant measures in the neonatal brain using EEG analysis as studied by Trujillo et al. represents an important step in validating COs as a functional testing model to investigate environment and genetic insults to the neonatal brain.
In sum, our work shows that EP maturation and network activity arising in COs is coupled with dynamic developmental changes in molecular and cellular complexity. The characterization of how EP features progress in COs with their developmental properties is key in validating this model for investigating molecular and cellular mechanisms underlying neuroelectrical deficits associated with neurological disorders. As an extension to this work, this functional testing platform may be used in parallel with morphological and transcriptomic analyses to characterize disease-specific features of early brain development.
Experimental Procedures
CO Culture
The human episomal-derived iPS-1 line was maintained under feeder-free and defined, serum-free medium conditions (Burridge et al., 2011). iPSCs were cultured in E8 medium (Thermo Fisher) and on vitronectin-coated plates. Whole-brain COs were generated according to our previously described protocol (Hester and Hood, 2017). Half of the medium within tissue culture dishes was replaced every 4 days.
MEA
Spontaneous extracellular field potentials in whole-brain COs were acquired with a 64-channel MEA system (MED64 system, Alpha MED Scientific) at a sampling rate of 20 kHz/channel and at 37°C using a temperature controller device. MEA probes (Alpha MED Scientific, MED-P515A) were treated with 0.005% polyethyleneimine and 2 μg/mL laminin prior to recording. Three-minute recordings were captured from four healthy COs per time point. COs were transferred from culture to the center of the probe containing the 8 × 8 channel array with 150 μL conditioned organoid culture medium. Spike data were imported into MED64 Mobius Offline software (version 1.4.5) and EP metrics were determined.
scRNA-seq
Approximately 10,000 live cells were used to generate scRNA-seq libraries with the 10× chromium single-cell 3′ V2 library kit (10× Genomics) according to the manufacturer's instructions, and sequencing was performed on a HiSeq 4000 System (Illumina).
Statistical Analyses
All statistical analyses were performed in GraphPad Prism v.8.1.2 or R v.3.6.1 and data are shown as the mean ± the SEM. Statistical significance was analyzed using one-way ANOVA with Tukey's multiple comparisons tests. Symbols of statistical significance were used as follows: ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001. All statistical analyses of EP metrics and scRNA-seq of COs are shown in Tables S3 and S4, respectively.
See Supplemental Experimental Procedures for detailed methods for MEA (also see Table S5 for burst detection parameters), IHC, and our scRNA-seq data handling pipeline.
Data and Code Availability
The accession number for the single cell RNA sequencing data reported in this paper is GEO: GSE157019.
Author Contributions
S.R.F., D.J., A.M.H., S.T.P., G.M., T.B., and M.E.H. conceived and designed the experiments. S.R.F., D.J., A.M.H., M.E.H., S.T.P., G.M., T.L.S., E.W.H., A.B.H., W.E.A., M.A., T.B., and G.Z. performed the research. S.R.F., D.J., A.M.H., M.E.H., T.B., A.B.H., J.I., S.T.P., G.M., N.L.M., C.A.M., and I.A.B. analyzed the data. S.R.F., A.M.H., M.E.H., D.J., A.B.H., E.W.H., J.I., C.A.M., N.L.M., and I.A.B. wrote the manuscript. All authors critically reviewed and approved the final manuscript for publication. This team works in the Steve and Cindy Rasmussen Institute for Genomic Medicine at Nationwide Children's Hospital. The institute is generously supported by the Nationwide Foundation Pediatric Innovation Fund.
Conflicts of Interest
Dr. Hester does not declare a conflict of interest related to this study. Dr. Imitola has been a consultant for Biogen and Novartis and has received funds from Biogen for another study. All other authors do not declare a conflict of interest.
Acknowledgments
We thank all laboratory members for their expert advice and constructive critiques in this study. We are also grateful to Dr. Susan Reynolds for critically reading and providing insights and edits to the manuscript and to Dr. Kathrin Meyer for providing support and guidance with the single-cell RNA sequencing experiments. This work was supported by research funds from The Abigail Wexner Research Institute at Nationwide Children's Hospital.
Published: September 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.08.017.
Supplemental Information
Please see associated Excel file.
References
- Attardo A., Fabel K., Krebs J., Haubensak W., Huttner W.B., Kempermann G. Tis21 expression marks not only populations of neurogenic precursor cells but also new postmitotic neurons in adult hippocampal neurogenesis. Cereb. Cortex. 2010;20:304–314. doi: 10.1093/cercor/bhp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy C., van den Hurk M., Eames T., Marchand C., Hernandez R.V., Kellogg M., Gorris M., Galet B., Palomares V., Brown J. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl. Acad. Sci. U S A. 2015;112:E2725–E2734. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershteyn M., Nowakowski T.J., Pollen A.A., Di Lullo E., Nene A., Wynshaw-Boris A., Kriegstein A.R. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell. 2017;20:435–449.e34. doi: 10.1016/j.stem.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biane J., Conner J.M., Tuszynski M.H. Nerve growth factor is primarily produced by GABAergic neurons of the adult rat cortex. Front. Cell. Neurosci. 2014;8:220. doi: 10.3389/fncel.2014.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifazi P., Massobrio P. Reconstruction of functional connectivity from multielectrode recordings and calcium imaging. Adv. Neurobiol. 2019;22:207–231. doi: 10.1007/978-3-030-11135-9_9. [DOI] [PubMed] [Google Scholar]
- Burridge P.W., Thompson S., Millrod M.A., Weinberg S., Yuan X., Peters A., Mahairaki V., Koliatsos V.E., Tung L., Zambidis E.T. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Brauninger M., Lewitus E., Sykes A., Hevers W., Lancaster M. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado Ruz I., Schultz S.R. Localising and classifying neurons from high density MEA recordings. J. Neurosci. Methods. 2014;233:115–128. doi: 10.1016/j.jneumeth.2014.05.037. [DOI] [PubMed] [Google Scholar]
- Di Lullo E., Kriegstein A.R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix L., Stephan J., Rose C.R. Astrocytes of the early postnatal brain. Eur. J. Neurosci. 2020 doi: 10.1111/ejn.14780. [DOI] [PubMed] [Google Scholar]
- Fietz S.A., Kelava I., Vogt J., Wilsch-Brauninger M., Stenzel D., Fish J.L., Corbeil D., Riehn A., Distler W., Nitsch R. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- Godbole G., Shetty A.S., Roy A., D'Souza L., Chen B., Miyoshi G., Fishell G., Tole S. Hierarchical genetic interactions between FOXG1 and LHX2 regulate the formation of the cortical hem in the developing telencephalon. Development. 2018;145:dev154583. doi: 10.1242/dev.154583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D.V., Lui J.H., Parker P.R., Kriegstein A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hartlaub A.M., McElroy C.A., Maitre N.L., Hester M.E. Modeling human brain circuitry using pluripotent stem cell platforms. Front. Pediatr. 2019;7:57. doi: 10.3389/fped.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester M.E., Hood A.B. Generation of cerebral organoids derived from human pluripotent stem cells. In: Srivastava A., Snyder E., Teng Y., editors. Vol. 126. Humana Press; 2017. pp. 123–134. (Stem Cell Technologies in Neuroscience. Neuromethods: Springer Protocols). [Google Scholar]
- Hou P.S., hAilin D.O., Vogel T., Hanashima C. Transcription and beyond: delineating FOXG1 function in cortical development and disorders. Front. Cell. Neurosci. 2020;14:35. doi: 10.3389/fncel.2020.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacopetti P., Michelini M., Stuckmann I., Oback B., Aaku-Saraste E., Huttner W.B. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. Proc. Natl. Acad. Sci. U S A. 1999;96:4639–4644. doi: 10.1073/pnas.96.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.B., Wang P.P., Atabay K.D., Murphy E.A., Doan R.N., Hecht J.L., Walsh C.A. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim H., Um J W. Synapse development organized by neuronal activity-regulated immediate-early genes. Exp. Mol. Med. 2018;50:11. doi: 10.1038/s12276-018-0025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizner V., Fischer S., Naujock M. Multielectrode array (MEA)-Based detection of spontaneous network activity in human iPSC-derived cortical neurons. Methods Mol. Biol. 2019;1994:209–216. doi: 10.1007/978-1-4939-9477-9_19. [DOI] [PubMed] [Google Scholar]
- Kuijlaars J., Oyelami T., Diels A., Rohrbacher J., Versweyveld S., Meneghello G., Tuefferd M., Verstraelen P., Detrez J.R., Verschuuren M. Sustained synchronized neuronal network activity in a human astrocyte co-culture system. Sci. Rep. 2016;6:36529. doi: 10.1038/srep36529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan A., Worley P. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Corsini N.S., Wolfinger S., Gustafson E.H., Phillips A.W., Burkard T.R., Otani T., Livesey F.J., Knoblich J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E.O., Paulsen O. Role of GABAergic inhibition in hippocampal network oscillations. Trends Neurosci. 2007;30:343–349. doi: 10.1016/j.tins.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Mansour A.A., Goncalves J.T., Bloyd C.W., Li H., Fernandes S., Quang D., Johnston S., Parylak S.L., Jin X., Gage F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J., Coppola G., Zhang P., Abyzov A., Provini L., Tomasini L., Amenduni M., Szekely A., Palejev D., Wilson M. FOXG1-Dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S.R., Tom C.M., Wang Y., Bresee C., Rushton D., Mathkar P.P., Tang J., Mattis V.B. Human Huntington's disease iPSC-derived cortical neurons display Altered transcriptomics, morphology, and maturation. Cell Rep. 2018;25:1081–1096.e6. doi: 10.1016/j.celrep.2018.09.076. [DOI] [PubMed] [Google Scholar]
- Niu W., Parent J.M. Modeling genetic epilepsies in a dish. Dev. Dyn. 2020;249:56–75. doi: 10.1002/dvdy.79. [DOI] [PubMed] [Google Scholar]
- Nowakowski T.J., Pollen A.A., Sandoval-Espinosa C., Kriegstein A.R. Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron. 2016;91:1219–1227. doi: 10.1016/j.neuron.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papariello A., Newell-Litwa K. Human-derived brain models: windows into neuropsychiatric disorders and drug therapies. Assay Drug Dev. Technol. 2020;18:79–88. doi: 10.1089/adt.2019.922. [DOI] [PubMed] [Google Scholar]
- Pasca S.P. The rise of three-dimensional human brain cultures. Nature. 2018;553:437–445. doi: 10.1038/nature25032. [DOI] [PubMed] [Google Scholar]
- Puehringer D., Orel N., Luningschror P., Subramanian N., Herrmann T., Chao M.V., Sendtner M. EGF transactivation of Trk receptors regulates the migration of newborn cortical neurons. Nat. Neurosci. 2013;16:407–415. doi: 10.1038/nn.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Min Yang S., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M., Lancaster M.A., Bian S., Choi H., Ku T., Peer A., Chung K., Knoblich J.A. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36:1316–1329. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi H., Ozaki Y., Ashida T., Matsubara T., Oishi N., Kihara S., Takahashi J. Self-organized synchronous calcium transients in a cultured human neural network derived from cerebral organoids. Stem Cell Reports. 2019;13:458–473. doi: 10.1016/j.stemcr.2019.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan S.A., Darmanis S., Huber N., Khan T.A., Birey F., Caneda C., Reimer R., Quake S.R., Barres B.A., Pasca S.P. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron. 2017;95:779–790.e6. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A.M.M., Meyer K.A., Santpere G., Gulden F.O., Sestan N. Evolution of the human nervous system function, structure, and development. Cell. 2017;170:226–247. doi: 10.1016/j.cell.2017.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles J., Jernigan T.L. The basics of brain development. Neuropsychol. Rev. 2010;20:327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A.X., Yuan Q., Fukuda M., Yu W., Yan H., Lim G.G.Y., Nai M.H., D'Agostino G.A., Tran H.D., Itahana Y. Potassium channel dysfunction in human neuronal models of Angelman syndrome. Science. 2019;366:1486–1492. doi: 10.1126/science.aav5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Cakir B., Xiang Y., Sullivan G.J., Park I.H. Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep. 2020;30:1682–1689.e3. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen E.R., Mich J.K., Yao Z., Hodge R.D., Doyle A.M., Jang S., Shehata S.I., Nelson A.M., Shapovalova N.V., Levi B.P. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods. 2016;13:87–93. doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo C.A., Gao R., Negraes P.D., Gu J., Buchanan J., Preissl S., Wang A., Wu W., Haddad G.G., Chaim I.A. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019;25:558–569.e7. doi: 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhang L., Gage F.H. Modeling neuropsychiatric disorders using human induced pluripotent stem cells. Protein Cell. 2020;11:45–59. doi: 10.1007/s13238-019-0638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Buth J.E., Vishlaghi N., de la Torre-Ubieta L., Taxidis J., Khakh B.S., Coppola G., Pearson C.A., Yamauchi K., Gong D. Self-organized cerebral organoids with human-specific features predict effective drugs to combat zika virus infection. Cell Rep. 2017;21:517–532. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton D.K., Dissing-Olesen L., Stevens B. Neuron-glia signaling in synapse elimination. Annu. Rev. Neurosci. 2019;42:107–127. doi: 10.1146/annurev-neuro-070918-050306. [DOI] [PubMed] [Google Scholar]
- Wong R.O. Calcium imaging and multielectrode recordings of global patterns of activity in the developing nervous system. Histochem. J. 1998;30:217–229. doi: 10.1023/a:1003251504594. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Tanaka Y., Patterson B., Kang Y.J., Govindaiah G., Roselaar N., Cakir B., Kim K.Y., Lombroso A.P., Hwang S.M. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21:383–398.e7. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Mead B.E., Safaee H., Langer R., Karp J.M., Levy O. Engineering stem cell organoids. Cell Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Ma L., Yang M., Shao Q., Xu J., Lu Z., Zhao Z., Chen R., Chai Y., Chen J.F. Cerebral organoid and mouse models reveal a RAB39b-PI3K-mTOR pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Genes Dev. 2020;34:580–597. doi: 10.1101/gad.332494.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please see associated Excel file.
Data Availability Statement
The accession number for the single cell RNA sequencing data reported in this paper is GEO: GSE157019.