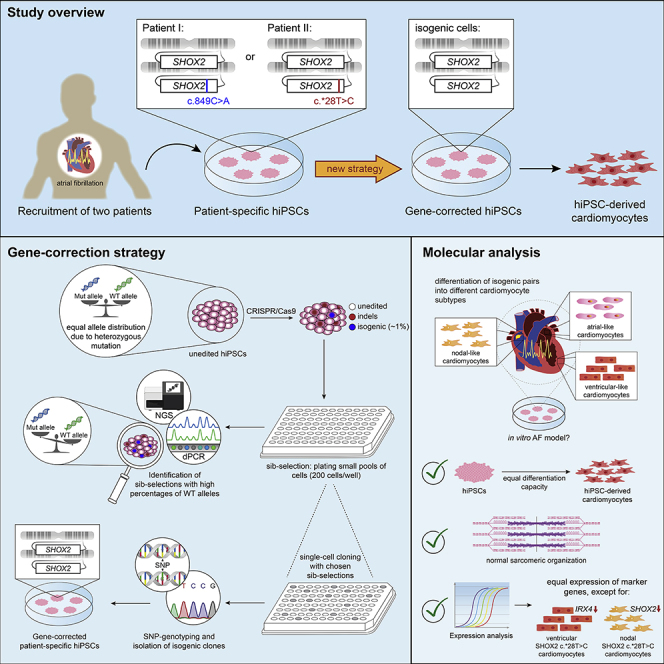
Summary
Patient-specific human induced pluripotent stem cells (hiPSCs) offer unprecedented opportunities for the investigation of multigenic disease, personalized medicine, and stem cell therapy. For heterogeneous diseases such as atrial fibrillation (AF), however, precise correction of the associated mutation is crucial. Here, we generated and corrected hiPSC lines from two AF patients carrying different heterozygous SHOX2 mutations. We developed a strategy for the scarless correction of heterozygous mutations, based on stochastic enrichment by sib selection, followed by allele quantification via digital PCR and next-generation sequencing to detect isogenic subpopulations. This allowed enriching edited cells 8- to 20-fold. The method does not require antibiotic selection or cell sorting and can be easily combined with base-and-prime editing approaches. Our strategy helps to overcome low efficiencies of homology-dependent repair in hiPSCs and facilitates the generation of isogenic control lines that represent the gold standard for modeling complex diseases in vitro.
Keywords: SHOX2, atrial fibrillation, precise gene editing, patient-derived iPSCs, isogenic control, sib selection
Graphical Abstract
Highlights
-
•
Model for atrial fibrillation using patient-specific and gene-corrected hiPSCs
-
•
Scarless gene correction of hiPSCs derived from patients with heterozygous mutations
-
•
Isolation of rare isogenic clones via sib selection and allele quantification
-
•
Strategy for difficult-to-target regions with low editing efficiency
In this study, Sumer, Hoffmann, et al., developed a strategy for the isolation of extremely rare hiPSC clones, suitable for scarless correction of heterozygous mutations by random enrichment of precisely edited cells and their detection via allele quantification. This strategy facilitates hiPSC-based gene correction regardless of the gene-editing approach.
Introduction
Research on the genetic basis of human cardiovascular disease has mainly relied on animal models in the past due to the limited access to human primary cardiac tissue. However, species-specific electrophysiological and transcriptional differences in cardiomyocytes can confound the translation of these findings to clinical relevance (Brandao et al., 2017; Moretti et al., 2013). Human induced pluripotent stem cells (hiPSCs) can help to overcome these limitations, as they provide an unlimited source of cardiomyocytes for the investigation of pathogenic mechanisms and the discovery of promising therapeutic targets (Abou-Saleh et al., 2018). Patient-derived hiPSCs represent a personalized drug-screening platform for precision medicine and individualized therapy (van Mil et al., 2018), Yet, the phenotype of the derived cardiomyocytes remains largely immature and the complex three-dimensional tissue structure can be mimicked only insufficiently in vitro (Moretti et al., 2013). In addition, substantial interline variability in cardiac differentiation capacity exists, emphasizing the need for reliable controls. Nevertheless, hiPSCs have been successfully used to model several cardiovascular diseases (Brandao et al., 2017; El-Battrawy et al., 2018; Goedel et al., 2017; Moretti et al., 2020).
The short stature homeobox 2 (SHOX2) transcription factor plays a crucial role in the development of the sinoatrial node, the native pacemaker of the heart. By antagonizing NKX2.5, it prevents the formation of working myocardial tissue, simultaneously activating the gene program of pacemaker cells, such as ISL1 and HCN4 (Blaschke et al., 2007; Hoffmann et al., 2013; Ye et al., 2015). We and others have shown that mutations in this gene are associated with early-onset and familiar forms of atrial fibrillation (AF) and sinus node dysfunction (Hoffmann et al., 2016, 2019; Li et al., 2018). In a previous screening of patients with an early-onset form of AF, we identified two mutations that interfere with the function of SHOX2: the coding mutation SHOX2 c.849C>A (= SHOX2 p.H283Q) was shown to impede the protein's function as a transcriptional activator, as demonstrated by the failed rescue of the bradycardia phenotype in a zebrafish model after knockdown of endogenous Shox2. Another SHOX2 variant resides in the 3′ UTR (SHOX2 c.∗28T>C), leading to a novel microRNA binding site for miR-92b-5p, which has been shown to be functional in vitro. AF patients harboring this variant have reduced microRNA expression levels in the plasma compared with AF-affected non-carriers and significantly prolonged PR intervals (Hoffmann et al., 2016). To develop an in vitro AF model, we recruited two patients with these SHOX2 mutations (SHOX2 c.849C>A or SHOX2 c.∗28T>C) for reprogramming of somatic cells to hiPSCs.
Recent advances in gene transduction and editing technologies have potentiated the applicability and versatility of hiPSCs (Brookhouser et al., 2017). Genome targeting has become feasible due to the development of site-specific nucleases, such as clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9. Following a Cas9-induced double-strand break (DSB), endogenous repair mechanisms are activated by the cell: the error-prone non-homologous end joining (NHEJ) or the precise homology-directed repair (HDR), which uses (partially) homologous DNA to restore the DNA sequence (Jackson and Bartek, 2009). In the presence of an exogenously introduced template, the alteration or insertion of specific DNA sequences, ranging from single-nucleotide exchanges to whole transgenes, can be achieved. Especially, the correction of disease-associated mutations in patient-specific hiPSCs holds great potential for precision disease modeling (Jang and Ye, 2016). These so-called isogenic controls differ only in single genetic variants, allowing for a direct genotype-to-phenotype comparison. This is of particular interest when studying multigenic diseases such as AF, where combinations of different variants with low impact are contributing to the observed phenotype.
A key limitation in the genome editing of hiPSCs is the fact that these cells preferentially choose the error-prone NHEJ pathway rather than the precise homology-directed approach (Guo et al., 2018). Under normal circumstances, frequencies of ∼1% for single-base substitutions or deletions are often reported (Miyaoka et al., 2014; Soldner et al., 2011). In recent years, substantial efforts have been made to develop strategies improving HDR efficiency, ranging from changes in the culturing conditions to suppressing NHEJ via small molecules or gene knockdown, modifications of the Cas9 enzyme, and optimization of the donor template (Anzalone et al., 2019; Chu et al., 2015; Guo et al., 2018; Komor et al., 2016; Okamoto et al., 2019; Yu et al., 2015).
Here, we propose a strategy to precisely correct heterozygous mutations adapted from a previously published method (Miyaoka et al., 2014), which was developed for the insertion of single-base substitutions into a wild-type (WT) background. It is based on the stochastic enrichment of precisely edited cells by subdivision of genome-edited cells into small pools and the detection of these subpopulations via allele quantification. With this approach, we were able to enrich our cells of interest 8- to 20-fold compared with the initial editing efficiency, allowing for a less work-intense screening for rare editing events, which can be combined with other optimization approaches. The corrected hiPSCs will help to elucidate the function of SHOX2 in the genetic network of atrial and nodal cardiomyocytes and its contribution to the development and progression of AF.
Results
Sib Selection Stochastically Enriches Isogenic Subpopulations That Are Detected by Allele Quantification
We generated hiPSCs derived from peripheral blood mononuclear cells of two AF patients harboring heterozygous SHOX2 mutations (SHOX2 c.849C>A and SHOX2 c.∗28T>C) using non-integrating Sendai viruses encoding KLF4, OCT3/4, SOX2, or c-MYC (Figure S1A) (for detailed patient characteristics see Table S1). The presence of the patient-specific SHOX2 c.849C>A and SHOX2 c.∗28T>C mutations was confirmed by Sanger sequencing (Figure S1B). All derived clones (two clones from patient I and three clones from patient II) displayed stem cell-like features and pluripotency capacity, as shown by high activities of alkaline phosphatase (Figure S1C), had lost the viral transgenes after 10–20 passages (Figure S1D), expressed pluripotency markers on protein and RNA level (Figures S1E and S1F), and differentiated spontaneously into all three germ layers (Figure S1G). Moreover, they were karyotypically normal (Figure S1H) and shared their respective patient origin (Table S2). Clones no. 1 from patient I and patient II were used for gene correction.
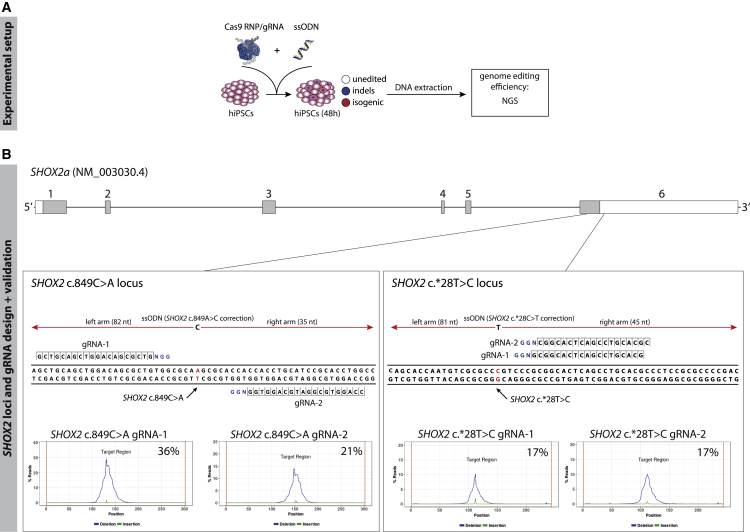
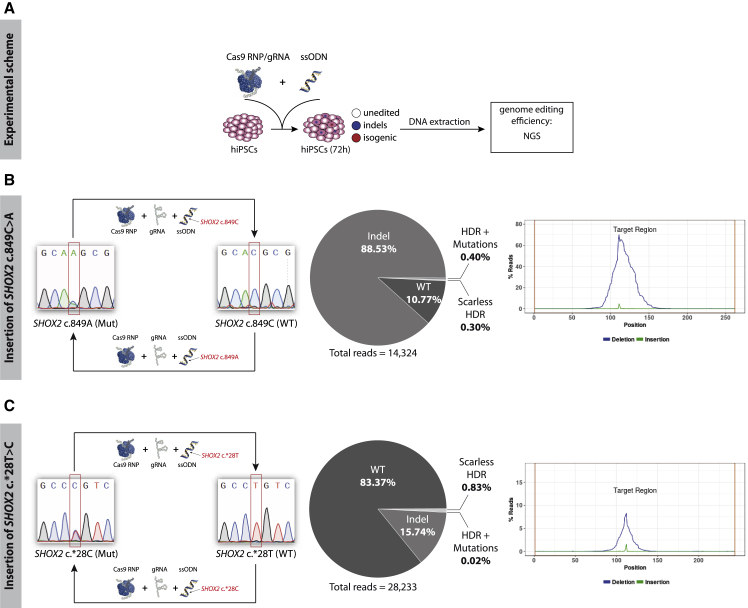
To target the heterozygous mutations, two single guide RNAs (gRNAs) per locus with cutting sites close to the mutations were selected (Figure 1). The gRNAs were predicted to range from moderately to highly efficient, with a substantial number of off-targets. However, as gene conversion tracks are relatively short in mammalian cells (Elliott et al., 1998), the distance between the Cas9 cut site and the targeted DNA sequence has to be minimized to achieve high HDR efficiency. Cells were transfected with Cas9 ribonucleoprotein (RNP)/gRNA complexes and a single-stranded oligodesoxynucleotide (ssODN) as the HDR template (Figure 1A). After 48 h the genome-targeting efficiency of each gRNA was determined by next-generation sequencing (NGS) for a precise estimation of indel size, frequency, and sequence identity. For the SHOX2 c.849C>A locus, the two selected gRNAs were moderately effective, producing indel frequencies of 36% and 21%, respectively. For the SHOX2 c.∗28T>C locus, gRNA-1 and gRNA-2 were less effective, producing indel frequencies of 17% in hiPSCs (Figure 1B). Due to the heterozygous nature of the SHOX2 mutations, the frequency of HDR events could not be precisely determined in these large cell pools.
Figure 1.
gRNA Design and Validation for the SHOX2 c.849C>A and the SHOX2 c.∗28T>C Loci
(A) Experimental overview of gRNA validation. hiPSCs were transfected with Cas9 RNP/gRNA complexes and ssODNs. After 48 h, the Cas9-targeted region was amplified and analyzed via NGS.
(B) Boxes, top: The SHOX2 c.849C>A and SHOX2 c.∗28T>C loci with selected gRNA binding sites. PAM sequences are depicted in blue, the mutated base pair is in red. Boxes, bottom: Indel frequency for each gRNA in hiPSCs 48 h after transfection. The targeted region is centered. The cumulative frequency of deletions (blue) and insertions (green) at each position is depicted in percentage of reads (% Reads). Abbreviations: RNP, ribonucleoprotein; gRNA, single guide RNA; ssODN, single-stranded oligodesoxynucleotide; NGS, next-generation sequencing.
The fractionation of a heterogeneous population of genome-edited stem cells can randomly enrich desired subpopulations, such as isogenic cells. A population of cells containing a small number of cells of interest is subdivided into small pools (“sib selection”). Of these, the one with the highest percentage of target cells is selected and subjected to a new round of subdivision. After the enrichment of target cells to a reasonable amount, single-cell cloning can be performed to achieve a pure cell population. We hypothesized that—by quantifying the ratio of WT and Mut alleles in these sib selections—we would be able to detect cells that had precisely corrected the heterozygous mutation. A present subpopulation of isogenic cells contributes two WT alleles to the DNA pool, leading to a shift in the WT/Mut allele ratio. Sib selections with an overaverage abundance of WT alleles potentially contain enriched amounts of isogenic cells and can, therefore, be selected for further analysis and single-cell cloning.
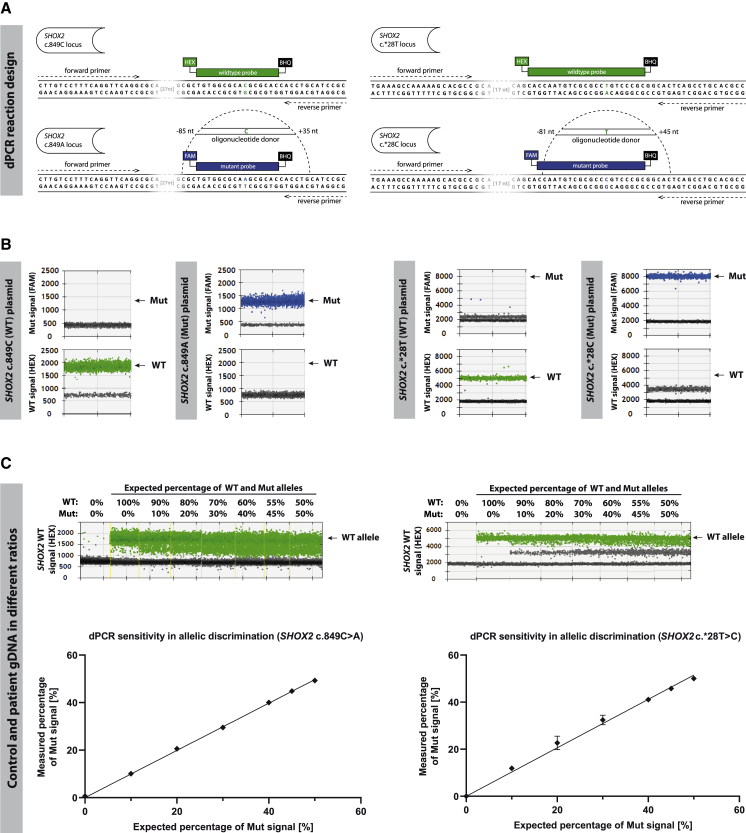
We utilized digital PCR (dPCR) to precisely quantify and identify DNA molecules to determine WT/Mut allele ratios within individual sib selections (Figure 2A). Using plasmids containing either the mutated or the unmutated SHOX2 gene, we confirmed the specificity of our TaqMan probes (Figure 2B). The sensitivity and specificity of the system were tested by mixing different ratios of genomic DNA from healthy donors with DNA from patient-specific hiPSCs and comparing detected allele ratios to actual ratios. The strong correlation between calculated and measured Mut alleles indicated a high precision of allelic quantification as well as a specificity of WT and Mut TaqMan probes for their respective alleles (Figure 2C).
Figure 2.
Pretests for Allele Quantification via dPCR
(A) Primer/probe design for detection of WT and Mut alleles with specific probes. The mutation-spanning oligonucleotide donor is depicted above the mutant allele.
(B) dPCR result for different ratios of control and patient genomic DNA represented as HEX channel 1D amplitude (top) and plotted against the expected percentage (bottom); n = 3, error bars represent ±SD of the mean.
(C) Probe specificity test with plasmids containing WT (SHOX2c.849C, SHOX2 c.∗28T) and Mut (SHOX2 c.849A, SHOX2 c.∗28C) alleles. Data are expressed as mean ± SD of three independent experiments. Abbreviations: dPCR, digital PCR; WT, wild type; Mut, mutant; gDNA, genomic DNA; FAM, 6-carboxyfluorescein; HEX, hexachloro-fluorescein; BHQ, Black Hole Quencher.
See also Table S5.
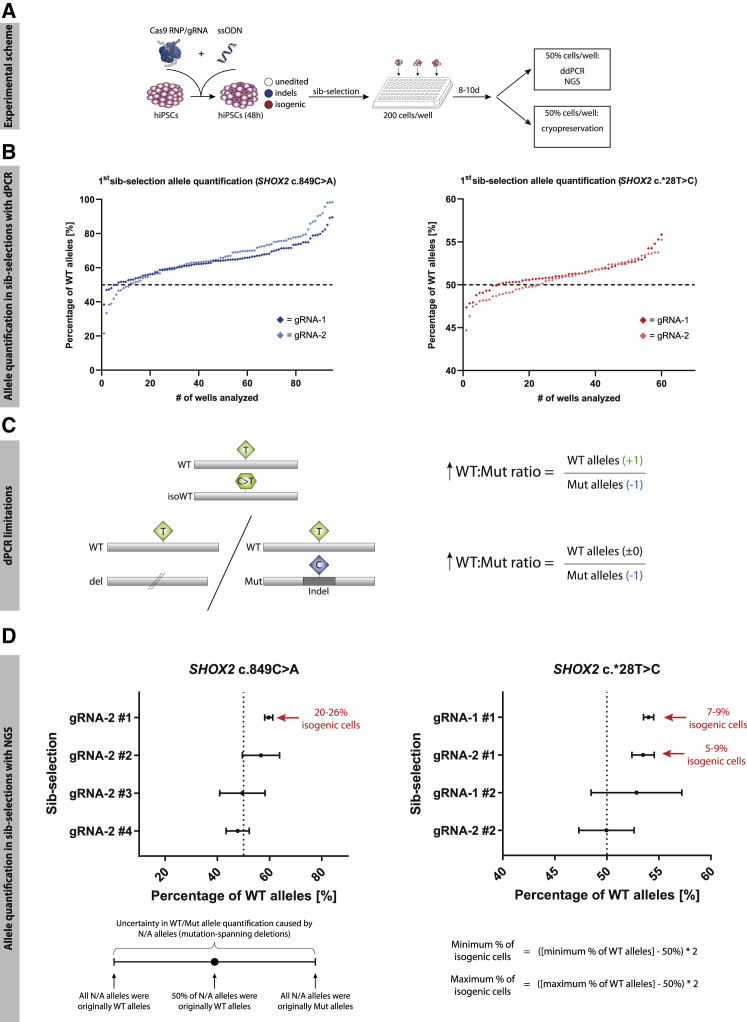
Cas9 RNP/gRNA/ssODN-transfected hiPSCs were seeded into small cell pools of 200 cells per well on a 96-well plate, 48 h after transfection and grown until confluency (∼8–10 days) (Figure 3A). From each well, 50% of the cells were cryopreserved, while the other 50% were subjected to DNA extraction and subsequent allele quantification with dPCR (Figure 3B). Most of the samples after the sib selection still showed a nearly equal allele distribution of 50:50 (dotted line), which is characteristic of a heterozygous mutation and indicates no enrichment of targeted cells. However, a higher abundance of WT alleles was seen in some of the samples that were selected for further analysis.
Figure 3.
Allele Quantification in Sib Selections via dPCR and NGS
(A) Experimental scheme of gene-editing approach.
(B) WT and Mut alleles were quantified in each sib selection 10 days after transfection via dPCR. Each dot represents the result for one sib selection. The dotted line marks the 50% WT allele percentage expected in unedited hiPSCs. Sib selections with the highest abundance of WT alleles were thawed and re-analyzed.
(C) Limitations of dPCR in allele quantification: due to the PCR-based allele detection, non-amplifiable Mut alleles can lead to shifts in the WT/Mut ratio, similar to what is caused by isogenic subpopulations.
(D) Allele quantification via NGS: alleles with SHOX2 c.849- and SHOX2 c.∗28-spanning deletions cause an uncertainty in allele quantification that is addressed by defining those alleles as all WT or all Mut. The resulting span of possible allele ratios is represented as error bars. Sib selections in which an increased WT/Mut allele ratio is not solely explicable by a loss of detectable Mut alleles were chosen for single-cell cloning. The percentage of isogenic cells was calculated with the given formula. Abbreviations: Indel, insertion/deletion; N/A, non-assignable.
Isogenic Subpopulations in Sib Selections Are Re-quantified via Next-Generation Sequencing
The major limitation of allele quantification via dPCR is its dependence on a functional PCR reaction. Mutations or the complete loss of primer/probe binding sites in alleles can prevent a successful amplification of the DNA strands and therefore their identification. If large populations of hiPSCs in a sib selection contain WT alleles and non-detectable mutant alleles, it will generally lead to a shift in the WT/Mut allele ratio similar to what is caused by isogenic cells (Figure 3C). Consequently, sequences had to be analyzed in detail via NGS to confirm the presence of isogenic cells in the chosen sib selections. A total of 11 sib selections were deep sequenced. To determine the percentage of isogenic cells from WT/Mut allele ratios, we made two assumptions: first, we considered the probability of SHOX2 copy number variations (CNVs), for example, due to trisomy or gene duplications, to be very low. CNVs could lead to changes in allele ratios that are also not caused by isogenic subpopulations. Second, we neglected the probability of an imprecise correction of the SHOX2 mutations (mutation lost, but other mutations introduced) for that moment. Under normal circumstances, HDR leads to a complete restoration of the sequence. This allowed us to directly calculate the percentage of isogenic subpopulations from the WT and Mut alleles (Figure 3D). We classified NGS reads into three categories, WT alleles, Mut alleles, and non-assignable (N/A) alleles, in which the base of interest (SHOX2 c.849 or SHOX2 c.∗28) was deleted (for detailed sequences see Table S3). This classification was carried out independent of additional mutations in the respective alleles. N/A alleles caused uncertainty in allele quantification, as their origin from either WT or Mut alleles could not be determined. This uncertainty was addressed by defining all N/A alleles as either WT or Mut alleles when calculating the WT/Mut allele ratio. The result was a range of possible ratios, spanning the two extreme scenarios in which N/A alleles were counted as either all WT or all Mut (Figure 3D). Using gRNA-1 to correct the SHOX2 c.849C>A mutation led to a high percentage of reads with deletions spanning the targeted mutation, which made the precise quantification of isogenic sib selections impossible (Figure S2). For the SHOX2 c.849C>A sib selections gRNA-2 nos. 2–4, a large fraction of N/A alleles led to a wide range of potential allele ratios (Figure 3D). As this included a scenario in which the higher percentage of WT alleles could be explained solely by a loss of detectable Mut alleles, a subpopulation of isogenic cells was possible, but not guaranteed. On the other hand, sib selection SHOX2 c.849C>A gRNA-2 no. 1 had a strong and robust increase in WT alleles, indicating a large fraction of isogenic clones. For SHOX2 c.∗28T>C sib selections, gRNA-1 no. 1 and gRNA-2 no. 1 showed similar shifts and were therefore selected for single-cell cloning together with SHOX2 c.849C>A gRNA-2 no. 1. Allelic distributions were used to calculate the percentage of isogenic cells in these cell pools, estimating frequencies of 20%–26% for SHOX2 c.849C>A gRNA-2 no. 1, as well as 7%–9% and 5%–9% in sib selection gRNA-1 no. 1 and gRNA-1 no. 2, respectively (Figure 3D). With a supposed initial efficiency of ∼1% for precise genome editing, the sib-selection process led to a significant enrichment of target cells.
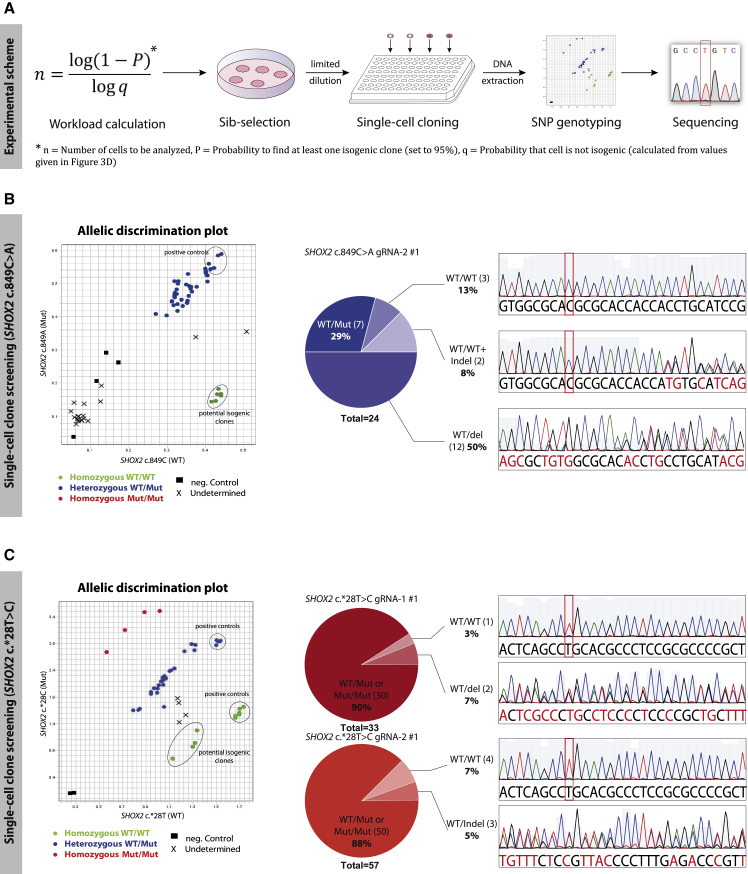
The number of cells that had to be screened to find at least one isogenic clone with a given probability was determined by negative binominal distribution. The applied parameters were the calculated frequency of target cells (∼23% or ∼8%), the desired number of positive clones to be found (≥1 clone), and a self-defined chance of success to find one (95%) (Figure 4A). Twelve clones for sib selection SHOX2 c.849C>A gRNA-2 no. 1 and 35–40 clones for SHOX2 c.∗28T>C sib selections were generated to find at least one clone of interest with a 95% probability. Compared with the nearly 300 cells that would have to be screened for the same chances of success—if the target cell population was only ∼1%—still, this represented a substantial reduction in screening workload.
Figure 4.
Single-Cell Cloning with Sib Selections and Screening for Isogenic Clones
(A) Experimental scheme for single-cell cloning and screening: the number of clones needed to be analyzed was calculated with a binomial distribution function. Sib selections with isogenic subpopulations were thawed for single-cell cloning via limited dilution. Clones were screened via TaqMan probe-based SNP genotyping, and potential homozygous WT clones were confirmed with Sanger sequencing.
(B) Screening for isogenic clones derived from heterozygous SHOX2 c.849C>A cells: 24 single-cell clones were genotyped and sequenced. In 5/24 clones (21%) the mutation was corrected back to WT, with 3 clones showing no additional mutations several hundred nucleotides up- and downstream.
(C) Screening for isogenic clones derived from heterozygous SHOX2 c.∗28T>C cells: single-cell-derived clones were genotyped. Ten annotated homozygous WT clones were sequenced to confirm the loss of the SHOX2 c.∗28T>C mutation. In 5/10 clones the mutation was repaired precisely back to WT, in the other 5/10 clones, deletions on the Mut allele explained the false annotation. Abbreviations: SNP, single-nucleotide polymorphism, here, SHOX2 c.849C>A and SHOX2 c.∗28T>C; Indel, insertion/deletion.
See also Figures S3 and S4, Tables S3 and S5.
Monoclonal cell populations were obtained via limiting dilution (Figure S3). Both dPCR primers and probes were reused for TaqMan SNP genotyping of SHOX2 c.849C>A and of SHOX2 c.∗28T>C to preselect potentially homozygous WT clones. From the allele discrimination plot, all annotated homozygous WT clones were identified (Figures 4B and 4C). Subsequent sequencing confirmed the precise correction of the heterozygous SHOX2 c.849C>A or SHOX2 c.∗28T>C mutation and was used to screen for additional mutations up- and downstream of the Cas9 target site. For SHOX2 c.849C>A, five of 24 sequenced clones (∼21%) were confirmed to have lost the patient mutation, matching the expected frequency of 20%–26%. However, in two of them, additional mutations were introduced during the editing process. For SHOX2 c.∗28T, five of the annotated WT clones (1× from sib selection gRNA-1 no. 1 and 4× gRNA-2 no. 1) were confirmed to be isogenic with no additional detectable mutations neither ∼600 bp upstream nor ∼260 bp downstream of the Cas9 cut site. For the rest of the clones that were predicted to be homozygous WT via genotyping, a loss of primer/probe binding sites on the mutant allele explained the false annotation (Figures 4B and 4C).
For subsequent detailed re-characterization, we selected one isogenic clone for SHOX2 c.849C>A and SHOX2 c.∗28T>C. Both clones had maintained their stem cell-like morphology and pluripotent capacity and exhibited high alkaline phosphatase activity, showed expression of pluripotency markers on the RNA/protein level, and spontaneously differentiated into derivates of all three germ layers (Figure S4A–D). Classical cytogenetic analysis on Giemsa-stained chromosomes revealed a normal male karyotype in 30 investigated metaphases each (Figure S4E). Cell line authentication confirmed their patient-specific origin and excluded cross-contamination with the control line (Table S2). We sequenced 12 and 11 highly scored off-targets for SHOX2 c.849C>A gRNA-2 and SHOX2 c.∗28T>C gRNA-1 consisting of exonic regions as well as intronic and intergenic regions with potential regulatory relevance, but found no additional mutations introduced in these sites (Table S4).
The Initial HDR Frequency Is Determined by Re-introducing the Patient-Specific Mutations into the Isogenic Control Lines
To determine if the sib-selection process indeed enriched the targeted cells, the initial HDR efficiency had to be quantified. Therefore, SHOX2 c.849C>A or SHOX2 c.∗28T>C mutations were re-introduced into the generated cell line using the same Cas9 enzyme batch and gRNAs but replacing the ssODNs with analogous versions that would lead to the insertion of the point mutations rather than to their repair. Seventy-two hours after transfection, the frequency of HDR events was determined by NGS. The SHOX2 c.849C>A and SHOX2 c.∗28T>C mutations could be detected in 0.70% and 0.85% of the reads, respectively, correlating with often-reported frequencies of ∼1% (Figure 5) (Miyaoka et al., 2014).
Figure 5.
Reverse Genome Editing to Determine the Initial HDR Efficiency
(A) Experimental scheme for reverse gene editing. The newly generated isogenic clones were used to determine the initial HDR efficiency. For this, the same Cas9 RNP and gRNA were used in combination with an oligonucleotide donor differing only in the one base analogous to SHOX2 c.849 (B) or SHOX2 c.∗28 (C). In consequence, HDR events would lead to the introduction of the SHOX2 c.849C>A or SHOX2 c.∗28T>C mutation. The frequency of HDR was determined by NGS 72 h after isogenic cells were transfected with Cas9/gRNA/ssODNs. Abbreviations: HDR, homology-directed repair.
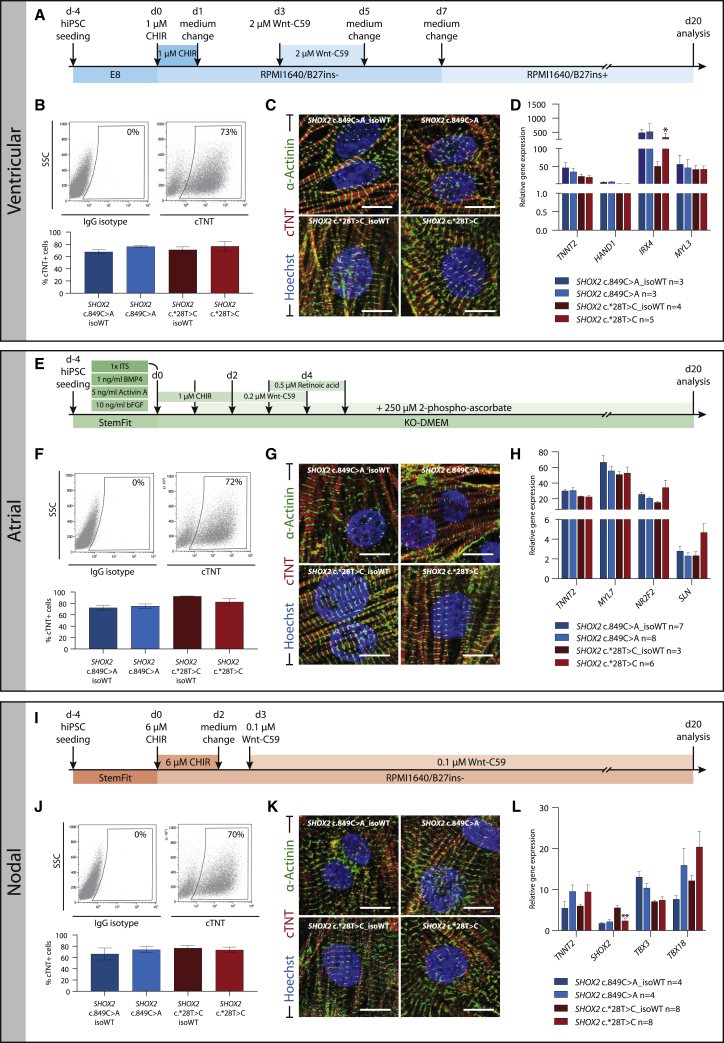
Patient-Specific and Isogenic hiPSCs Were Differentiated into Different Cardiomyocyte Subtypes
To prove the suitability of these lines as a cardiac disease model, patient-derived hiPSCs and their isogenic counterparts were differentiated into ventricular-, atrial-, and nodal-like cardiomyocytes using previously described protocols (Foo et al., 2018; Liang et al., 2019; Zhang et al., 2015a) (Figures 6A, 6E, and 6I). Flow-cytometry analysis of the pan-cardiac marker cardiac Troponin T showed comparable differentiation capacities between isogenic pairs (Figures 6B, 6F, and 6J). Immunostaining of cardiac Troponin T (A band) and sarcomeric α-actinin (Z band) of 24-day-old cardiomyocytes revealed normal sarcomeric organization in all lines (Figures 6C, 6G, and 6K). We analyzed the expression of several subtype-specific marker genes on the RNA level and found no significant differences between patient and isogenic lines, with the exception of SHOX2 expression that was reduced in nodal-like cells and a reduced expression of the ventricular homeobox transcription factor IRX4 in the presence of the c.∗28T>C mutation (Figures 6D, 6H, and 6L). We conclude that these lines are well suited to future investigations on how these SHOX2 mutations contribute to the prevalence of AF.
Figure 6.
Differentiation of Patient-Specific and Isogenic Control hiPSCs into Cardiomyocyte Subtypes
(A, E, and I) hiPSCs were differentiated into ventricular- (A–D), atrial- (E–H), and nodal-like (I–L) cardiomyocytes using the given differentiation schemes. (B, F, and I) Flow-cytometry analysis revealed similar numbers of cTNT+ cells in isogenic pairs, indicating an unimpaired differentiation capacity in patient lines. Top: Representative flow-cytometry dot plots for IgG isotype control (left) and cTNT (right) of differentiated cells at day 20. Bottom: Percentage of cTNT+ cells in n = 3 independent experiments. (C, G, and K) Immunostaining for cardiac Troponin T (sarcomere A band) and α-actinin A (sarcomere Z band) showed normal sarcomeric organization in patient and control lines; scale bar, 50 μm. (D, H, and L) qRT-PCR analysis of subtype-specific markers showed equal gene expression between patient and control hiPSC-derived cardiomyocytes, with the exception of SHOX2 and IRX4, which were significantly downregulated in the SHOX2 mutant c.∗28T>C line. Data are expressed as mean ± SD of three independent experiments. ∗p ≤ 0.05, ∗∗p ≤ 0.01. Abbreviation: cTNT, cardiac Troponin T.
Discussion
The concept of using stochastic enrichment of cells by sib selection to introduce precise mutations into hiPSCs derived from a healthy donor has been proposed before (Miyaoka et al., 2014). The insertion of disease-linked variants into a WT background helps to interrogate the influence of these mutations on the onset and progression of a disease by direct comparison of mutated cells to their isogenic counterpart. Yet, this approach is limited to monogenic diseases or variances with a high impact on the phenotype. Here, we demonstrate that this approach can be used to correct heterozygous mutations as well. This is of particular interest when using patient-derived hiPSCs that already harbor putative disease-causing variants. These patient models play a central role in the investigation of sporadic or idiopathic diseases, where a combination of multiple risk alleles with low effect size is thought to be the genetic basis and individual risk variants might not be sufficient to cause a disease-associated phenotype.
AF is a multifactorial disease with a strong genetic component and a complex heritability (Feghaly et al., 2018). Multiple genetic loci have been associated with this disease (Roselli et al., 2018) and mutations in potassium and sodium channels as well as mutations in transcription factors and structural proteins have been identified (Feghaly et al., 2018). However, AF models based on human stem cell-derived cardiomyocytes are sparse (Benzoni et al., 2019; Laksman et al., 2017) and to date, linking specific mutations to an AF phenotype in hiPSC-cardiomyocytes has not been achieved. The correction of putative disease-contributing variants in hiPSCs could unravel subtle phenotypic changes, even if the disease phenotype overall persisted.
Our proposed strategy can be applied to a precise repair of heterozygous mutations that does not require the use of selection marker integration, its transient expression, or an enrichment of nuclease-expressing cells by fluorescence-activated cell sorting (Lonowski et al., 2017; Mitzelfelt et al., 2017; Steyer et al., 2018). This does not only abolish the need to optimize the selection or sorting process, but also allows the use of unlabeled nuclease proteins or gene constructs. The enrichment of cells with a high expression of Cas9 via puromycin or cell sorting has led to concerns regarding off-target effects, as prolonged expression or high concentrations of nucleases tend to increase the probability of unwanted DSBs (Chen et al., 2016; Zhang et al., 2015b). This can be countered by the use of Cas9 RNPs, which reportedly show less off-targeting due to the shortened activity span by immediate DNA cleavage after delivery into cells, followed by a rapid degradation (Kim et al., 2014; Ramakrishna et al., 2014). Avoiding delivery vectors that have the potential to integrate into the genome also opens possibilities for gene editing in clinical settings under good manufacturing practice conditions. In addition, high-fidelity gRNAs with few predicted off-targets should be preferentially chosen (Hsu et al., 2013). However, to increase the frequency of HDR events, gRNAs must be selected according to the distance from the induced DSB to the HDR-targeted DNA section, rather than off-target scores or predicted efficiency. Nevertheless, to completely rule out additional editing, a genome-wide analysis via whole-genome sequencing would be required. Enriching cells of interest before single-cell seeding is especially beneficial for cell lines that behave poorly during clonal expansion. In our experience, keeping cells in small pools of 200 cells per well of a 96-well plate has significantly improved survival rates upon splitting. This avoids the selective enrichment of cell populations with abnormal survival advantages or growth rates caused by chromosomal aberrations. Subsequently, low clonability rates do not lead to an immense increase in time and material consumption due to large-scale cloning efforts as only a handful of clones have to be analyzed to find an isogenic one. In fact, even with the low clonability rates of 10%–20% that we achieved with our patient lines, only two or three 96-well plates per line were sufficient to find several isogenic populations.
In regions that are difficult to target, the initial HDR efficiency may still be low even after incorporation of improvement strategies such as optimizing the DNA template (Okamoto et al., 2019; Richardson et al., 2016), modifying the Cas9 enzyme itself (Anzalone et al., 2019; Komor et al., 2016), or applying small molecules to increase HDR (Yu et al., 2015). Our proposed strategy can be combined with any of these approaches as it is locus independent and relies solely on a stochastic enrichment rather than the modulation of biological processes. Especially, emerging techniques like base editing and prime editing (Anzalone et al., 2019; Komor et al., 2016) can profit from this approach by complementing their increased efficiency in targeted editing with an additional enrichment of correctly edited cells. We purposely avoided the insertion of silent blocking mutations, which prevent re-cutting by Cas9 after HDR-mediated editing (Okamoto et al., 2019), together with the mutation correction in the ssODN sequence. We highly recommend restricting their application to coding mutations, as unwanted side effects in UTRs or non-coding regulatory regions by altered posttranscriptional regulation or transcription factor binding cannot be ruled out completely. An example of this is the SHOX2 c.∗28T>C mutation, which resides within the 3′ UTR and mediates its detrimental effect by the generation of a novel microRNA binding site (Hoffmann et al., 2016).
However, despite the clear advantages of this method, several limitations remain. Laboratories are required to have fast and easy access to NGS and potentially dPCR. We used dPCR to preselect sib selections with potentially high fractions of isogenic cells, despite the stated limitations of this method to quantify such subpopulations. In addition, some dPCR primer/probe combinations seem to systematically favor the amplification of WT or Mut alleles in dPCRs, thus allowing only a relative comparison between sib selections. The preselection of a few samples made singleplex NGS possible, which could be analyzed with freely available online tools such as Cas-analyzer (Park et al., 2017). Nevertheless, a more straightforward approach would be to quantify alleles in all sib selections at once with deep sequencing. Although this is more cost intense and requires amplicon-NGS analysis knowledge due to sample multiplexing, it would speed up the process significantly and allow one to find the one sib selection that truly has the highest percentage of isogenic cells. With recent and future advances in NGS, we expect this method to become more feasible and affordable (Levy and Myers, 2016; Park and Kim, 2016). The additional passaging and cryopreservation required in the sib-selection process is a potential source for acquiring mutations and chromosomal translocations in the extended culturing periods (Martins-Taylor and Xu, 2012; Merkle and Eggan, 2013). On the other hand, stressful processes like cell sorting and antibiotic selection are not required. Yet, its applicability to primary cells and cell types that cannot be extensively passaged is limited. Furthermore, the additional culturing periods are time consuming. Even under ideal circumstances, the isolation of isogenic clones takes several weeks to months. However, most of the time is spent waiting for the cells to grow, and during allele quantification procedures, sib selections are cryopreserved, making the process easily interruptible. The quantification of HDR events after each sib selection allows precise workload calculations for the next step and prevents tedious single-cell cloning, even if no isogenic cells are present.
In conclusion, we provide a strategy for the scarless correction of heterozygous mutations by random enrichment of precisely edited cells and their detection via allele quantification. We propose that the frequency of isogenic cells can be determined by comparing WT/Mut allele ratios with two assumptions: no CNV and an error-free correction of the mutation via HDR. This approach can facilitate the generation of isogenic control cells, which represent the gold standard of controls when investigating the influence of putative disease-causing variants on the disease phenotype. Future detailed electrophysiological and molecular analysis of this in vitro disease model will aid in refining the paradigm of how SHOX2 mutations influence the onset and progression of AF by direct genotype-phenotype comparison and even has the potential to serve as a platform for drug discovery and personalized medicine. Compared with already existing AF models (Benzoni et al., 2019; Marczenke et al., 2017), this would be one using patient-specific and gene-corrected hiPSCs.
Experimental procedures
Cell Culture and Reagents
Standard cultivation of hiPSCs was performed with 5% CO2 at 37°C in a humidified atmosphere. Cells were grown in StemFit, supplemented with 100 ng/mL basic fibroblast growth factor (Peprotech) on a Geltrex LDEV-free, hESC-qualified, Reduced Growth Factor Basement Membrane matrix (1:100, Thermo Fisher) according to the manufacturer's instructions. Cells were dissociated with TrypLE Express (Gibco).
CRISPR/Cas9 gRNA Design, Off-Target Analysis, and gRNA Synthesis
gRNA design was performed with the CCTop-CRISPR/Cas9 target online predictor (https://crispr.cos.uni-heidelberg.de/) (Stemmer et al., 2015) with spCas9, a target-site length of 20 nucleotides, and standard settings as input (Table S5). Off-targets were predicted with CCTop using standard settings and the Homo sapiens GRCh37/hg19 assembly as a reference genome. The top 20 off-target sites were further evaluated: exonic off-target sites were automatically included into downstream analysis. For intronic and intergenic off-target sites, the target sequence coordinates were analyzed in the UCSC Genome Browser (https://genome.ucsc.edu/). The inclusion criteria for downstream analysis were conservation among species, DNase clustering, expressed sequence tags, and active chromatin marks. If a combination of these criteria indicated a potential regulatory relevance of this DNA segment, the off-target site was sequenced (12 and 11 off-targets per gRNA) (Table S3). gRNAs were in vitro synthesized with the GeneArt Precision gRNA Synthesis Kit (Invitrogen) according to the provided protocol and quantified with the Qubit RNA BR Assay Kit (Invitrogen) on the Qubit 2.0 fluorometer (Invitrogen).
Design of Primers/Probes and Oligonucleotide Donors
ssODNs were designed as previously described (Bollen et al., 2018; Richardson et al., 2016). The template spanned the Cas9 cut site asymmetrically with a longer 5′ homology arm for improved hybridization (Table S5). The DNA oligo was synthesized and purified via desaltation by Integrated DNA Technologies. Primers and probes for the dPCR were designed via Primer3plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and further evaluated with OligoAnalyzer (http://eu.idtdna.com/calc/analyzer) and UCSC in silico PCR (http://rohsdb.cmb.usc.edu/GBshape/cgi-bin/hgPcr). Primers were designed by default for an annealing temperature of 60°C and TaqMan probes for 65°C. The PCR product length was set to be 90–120 nucleotides for dPCR or 150–450 nucleotides for NGS (Table S5). For quantitative PCR, primers were designed with the Universal ProbeLibrary Assay Design Center (Roche) (https://lifescience.roche.com/en_de/brands/universal-probe-library.html#assay-design-center) (Table S5).
Electroporation and Sib Selection
iPSCs were dissociated with TrypLE Express and transfected with the 4D-Nucleofector System (Lonza) and the P3 Primary Cell 4D-Nucleofector X Kit S according to manufacturer's guidelines. For genome editing, 1 μg of Platinum or Truecut v.2 Cas9 RNP (Invitrogen), 250 ng of in vitro synthesized gRNA, and 50 pmol of ssODN were electroporated into 200,000 hiPSCs using program DN-100. Cells were seeded at high density posttransfection (50,000 cells/cm2) and cultured in medium containing 10 μM Y-27632 (ROCK inhibitor) for the first 24 h Forty-eight hours after transfection, hiPSCs were dissociated and seeded at 200 cells/well on a precoated 96-well plate (sib selection). The amount of 200 cells per sib selection was chosen as a compromise between cell viability upon splitting and throughput (the more cells per well, the better) and a cell pool size that is small enough to also detect smaller isogenic subpopulations. In a sib selection of 200 cells, the minimum allele frequency is 0.25%. If an overaverage abundance of isogenic cells (>1% or >2/200 cells) is present, the shift in the allele frequency would be >0.5%, which we considered to be detectable in dPCR and NGS. These sib selections were cultured until they reached ∼80% confluency (8–10 days) and then dissociated with 30 μL TrypLE Express per well. Fifteen microliters of the cell suspension was directly used for DNA extraction with the Quick-DNA 96 Plus Kit (Zymo Research), the other 15 μL was mixed with 85 μL Bambanker (Nippon Genetics) and frozen in a Styrofoam container at −80°C until further use.
dPCR
The dPCR consisted of 250 nM allele-specific TaqMan probes labeled with FAM (Mut allele) or HEX (WT allele) and 900 nM forward + reverse primers in 1× dPCR Supermix for Probes (Bio-Rad), containing 50–150 ng of genomic DNA or 50,000 plasmid copies in a total volume of 22 μL before the generation of droplets was performed with a QX200 droplet generator according to the manufacturer's instructions (Bio-Rad). Droplets were immediately pipetted into 96-well PCR plates (Bio-Rad), which were heat sealed and transferred into a C1000 thermal cycler (Bio-Rad). The conducted thermal cycling program consisted of step 1, 95°C 10 min; step 2, 94°C 30 s; step 3, 57°C or 60°C 60 s, repetition of step 2 + 3 39 times, 98°C 10 min. After completion, the droplets were analyzed on a QX200 droplet reader (Bio-Rad) using the Quantasoft software (version 1.7.4.0917). To calculate the percentage of WT alleles, WT allele copies/mL was divided by WT plus Mut copies/mL. We chose sib selections for NGS based on the percentage of WT alleles and the number of detectable copies/μL (value given by the droplet reader). We hypothesized that a high value here indicates a reliable result and a dPCR that was not inhibited by non-detectable DNA alleles, thus giving a small range of possible allele ratios.
Next-Generation Sequencing and Analysis
For amplicon-NGS, the target regions spanning the SHOX2 mutations and the Cas9 cut site were amplified using the Q5 High-Fidelity DNA polymerase (New England Biolabs) or the HotStarTaq polymerase (QIAGEN) according to the manufacturer's instructions. Five hundred nanograms of the PCR product in 25 μL was sent to GENEWIZ, Germany, for single-sample paired-end amplicon NGS on the HiSeq system (Illumina). To determine gRNA efficiencies and the frequency of HDR upon reverse gene editing, the provided SNP/indel analysis from GENEWIZ was used. For allele quantification in sib selections, the .fastq files were uploaded to the JavaScript-based assessment tool Cas-Analyzer (http://www.rgenome.net/cas-analyzer/) with the following analysis parameters: comparison range 100 nucleotides, minimum frequency 0.25% of total reads containing both indicator sequences (determined by setting it to 1 at first, doing the analysis, and then calculating 0.25% from the total number of reads containing both indicator sequences) and no WT marker. The resulting sequence table was used for manual allele classification (Table S4).
Single-Cell Cloning
Thawed sib selections were dissociated and a cell suspension with 10 cells/mL was generated by serial dilution. The suspension was mixed 1:50 with ice-cold Geltrex and 100 μL per well (=1 cell/well) was immediately pipetted on a 96-well plate using multichannel pipets. Seventy-two hours after seeding, plates were screened for single cell-derived clones (Figure S3). Medium was changed every other day until day 8, when hiPSC colonies were split onto 2× precoated 96 wells each. After reaching confluency, one well was subjected to cell lysis with directPCR lysis reagent (VWR), the other was used for expansion and cryopreservation.
Genotyping
To identify potential isogenic WT clones, single-cell-derived clones were genotyped in 96-well plates via TaqMan probe-based SNP genotyping using 1× TaqPath ProAmp Master Mix (Thermo Fisher) according to the manufacturer's instructions. DNA from control and patient-specific cells served as positive control for homozygous and heterozygous WT annotations. Potential isogenic clones were identified in the allelic discrimination plot.
Subtype-Specific Cardiomyocyte Differentiation
Differentiation into ventricular- (Foo et al., 2018), atrial- (Zhang et al., 2015a), and nodal-like cardiomyocytes (Liang et al., 2019) was done as described previously. At day 20, differentiating myocytes were dissociated with papain (Worthington Biochemical Corporation) and analyzed for cTNT+ cells by flow cytometry or further purified with the human PSC-derived Cardiomyocyte Isolation Kit (Miltenyi Biotech) followed by RNA isolation for qPCR analyses or plating on fibronectin-coated dishes for immunohistological analysis (detailed methods are described in the Supplemental Information).
Data and Code Availability
Unprocessed sequencing files (.fastq) have been uploaded to the NCBI Sequence Read Archive and are available via the accession no. PRJNA659248 or under https://www.ncbi.nlm.nih.gov/bioproject/PRJNA659248/.
Author Contributions
Conceptualization, S.A.S. and S.H.; Methodology, S.A.S. and S.H.; Validation, S.A.S. and S.H.; Formal Analysis, S.A.S. and S.H.; Investigation, S.A.S., S.H., K.R., S.L., B.C., K.R., V.F., T.D., J.W.G.J., and A.J.; Resources, S.C., S.K., J.W.G.J., A.J., A.M., and G.A.R.; Writing – Original Draft, S.A.S.; Writing – Review & Editing, S.H., T.D., A.M., and G.A.R.; Supervision, A.M. and G.A.R.; Funding Acquisition, S.H., A.M., K-L.L., and G.A.R.
Acknowledgments
The authors declare no conflict of interest. This work was funded by the German Research Foundation (DFG) RA 380/14-4 (to G.A.R.); DZHK (German Centre for Cardiovascular Research) 81X2500133 and 81X2500113 (to S.H.); the European Research Council, ERC 788381 (to A.M.); the German Research Foundation (DFG), Transregio Research Unit 152 (to A.M., K-L.L.) and 267 (to A.M., K-L.L.); and DZHK (German Centre for Cardiovascular Research) 81X2600603 and 81X2600607 (to A.M.).
Published: September 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.08.015.
Supplemental Information
Detailed sequences and read numbers for each sib selection analyzed via NGS are listed here. For allele quantification, only reads with a frequency higher than 0.25% of total reads (minimum allele frequency in 200 cells) were considered. These reads were classified into three categories: wild type (“WT,” no detectable SHOX2 c.849C>A or SHOX2 c.∗28T>C mutations), mutant (“Mut,” detectable SHOX2 c.849C>A or SHOX2 c.∗28T>C mutations), or non-assignable (“N/A,” where position SHOX2 c.849 or SHOX2 c.28∗ was deleted).
All primer sequences used for in vitro synthesis, dPCR, NGS, Sanger sequencing, and qRT-PCR of pluripotency and cardiomyocyte subtype marker genes are listed here. In addition, the sequences of single-stranded oligodeoxynucleotides that were transfected together with Cas9 RNP/gRNA complexes to serve as a repair template are given.
References
- Abou-Saleh H., Zouein F.A., El-Yazbi A., Sanoudou D., Raynaud C., Rao C., Pintus G., Dehaini H., Eid A.H. The march of pluripotent stem cells in cardiovascular regenerative medicine. Stem Cell Res. Ther. 2018;9:201. doi: 10.1186/s13287-018-0947-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzoni P., Campostrini G., Landi S., Bertini V., Marchina E., Iascone M., Ahlberg G., Olesen M.S., Crescini E., Mora C. Human iPSC modeling of a familial form of atrial fibrillation reveals a gain of function of if and ICaL in patient-derived cardiomyocytes. Cardiovasc. Res. 2019;116:1147–1160. doi: 10.1093/cvr/cvz217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke R.J., Hahurij N.D., Kuijper S., Just S., Wisse L.J., Deissler K., Maxelon T., Anastassiadis K., Spitzer J., Hardt S.E. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- Bollen Y., Post J., Koo B.K., Snippert H.J.G. How to create state-of-the-art genetic model systems: strategies for optimal CRISPR-mediated genome editing. Nucleic Acids Res. 2018;46:6435–6454. doi: 10.1093/nar/gky571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandao K.O., Tabel V.A., Atsma D.E., Mummery C.L., Davis R.P. Human pluripotent stem cell models of cardiac disease: from mechanisms to therapies. Dis. Model. Mech. 2017;10:1039–1059. doi: 10.1242/dmm.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookhouser N., Raman S., Potts C., Brafman D.A. May i cut in? gene editing approaches in human induced pluripotent stem cells. Cells. 2017;6:5. doi: 10.3390/cells6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu X., Zhang Y., Wang H., Ying H., Liu M., Li D., Lui K.O., Ding Q. A self-restricted CRISPR system to reduce off-target effects. Mol. Ther. 2016;24:1508–1510. doi: 10.1038/mt.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- El-Battrawy I., Zhao Z., Lan H., Li X., Yucel G., Lang S., Sattler K., Schunemann J.D., Zimmermann W.H., Cyganek L. Ion channel dysfunctions in dilated cardiomyopathy in limb-girdle muscular dystrophy. Circ. Genom Precis Med. 2018;11:e001893. doi: 10.1161/CIRCGEN.117.001893. [DOI] [PubMed] [Google Scholar]
- Elliott B., Richardson C., Winderbaum J., Nickoloff J.A., Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feghaly J., Zakka P., London B., Macrae C.A., Refaat M.M. Genetics of atrial fibrillation. J. Am. Heart Assoc. 2018;7:e009884. doi: 10.1161/JAHA.118.009884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo K.S., Lehtinen M.L., Leung C.Y., Lian X., Xu J., Keung W., Geng L., Kolstad T.R.S., Thams S., Wong A.O. Human ISL1(+) ventricular progenitors self-assemble into an in vivo functional heart patch and preserve cardiac function post infarction. Mol. Ther. 2018;26:1644–1659. doi: 10.1016/j.ymthe.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedel A., MY I., Sinnecker D., Moretti A. Perspectives and challenges of pluripotent stem cells in cardiac arrhythmia research. Curr. Cardiol. Rep. 2017;19:23. doi: 10.1007/s11886-017-0828-z. [DOI] [PubMed] [Google Scholar]
- Guo Q., Mintier G., Ma-Edmonds M., Storton D., Wang X., Xiao X., Kienzle B., Zhao D., Feder J.N. 'Cold shock' increases the frequency of homology directed repair gene editing in induced pluripotent stem cells. Sci. Rep. 2018;8:2080. doi: 10.1038/s41598-018-20358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S., Berger I.M., Glaser A., Bacon C., Li L., Gretz N., Steinbeisser H., Rottbauer W., Just S., Rappold G. Islet1 is a direct transcriptional target of the homeodomain transcription factor Shox2 and rescues the Shox2-mediated bradycardia. Basic Res. Cardiol. 2013;108:339. doi: 10.1007/s00395-013-0339-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S., Clauss S., Berger I.M., Weiss B., Montalbano A., Roth R., Bucher M., Klier I., Wakili R., Seitz H. Coding and non-coding variants in the SHOX2 gene in patients with early-onset atrial fibrillation. Basic Res. Cardiol. 2016;111:36. doi: 10.1007/s00395-016-0557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S., Paone C., Sumer S.A., Diebold S., Weiss B., Roeth R., Clauss S., Klier I., Kaab S., Schulz A. Functional characterization of rare variants in the SHOX2 gene identified in sinus node dysfunction and atrial fibrillation. Front. Genet. 2019;10:648. doi: 10.3389/fgene.2019.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y.Y., Ye Z. Gene correction in patient-specific iPSCs for therapy development and disease modeling. Hum. Genet. 2016;135:1041–1058. doi: 10.1007/s00439-016-1691-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laksman Z., Wauchop M., Lin E., Protze S., Lee J., Yang W., Izaddoustdar F., Shafaattalab S., Gepstein L., Tibbits G.F. Modeling atrial fibrillation using human embryonic stem cell-derived atrial tissue. Sci. Rep. 2017;7:5268. doi: 10.1038/s41598-017-05652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S.E., Myers R.M. Advancements in next-generation sequencing. Annu. Rev. Genomics Hum. Genet. 2016;17:95–115. doi: 10.1146/annurev-genom-083115-022413. [DOI] [PubMed] [Google Scholar]
- Li N., Wang Z.S., Wang X.H., Xu Y.J., Qiao Q., Li X.M., Di R.M., Guo X.J., Li R.G., Zhang M. A SHOX2 loss-of-function mutation underlying familial atrial fibrillation. Int. J. Med. Sci. 2018;15:1564–1572. doi: 10.7150/ijms.27424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Han P., Kim E.H., Mak J., Zhang R., Torrente A.G., Goldhaber J.I., Marban E., Cho H.C. Canonical Wnt signaling promotes pacemaker cell specification of cardiac mesodermal cells derived from mouse and human embryonic stem cells. Stem Cells. 2019;38:352–368. doi: 10.1002/stem.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonowski L.A., Narimatsu Y., Riaz A., Delay C.E., Yang Z., Niola F., Duda K., Ober E.A., Clausen H., Wandall H.H. Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat. Protoc. 2017;12:581–603. doi: 10.1038/nprot.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczenke M., Fell J., Piccini I., Ropke A., Seebohm G., Greber B. Generation and cardiac subtype-specific differentiation of PITX2-deficient human iPS cell lines for exploring familial atrial fibrillation. Stem Cell Res. 2017;21:26–28. doi: 10.1016/j.scr.2017.03.015. [DOI] [PubMed] [Google Scholar]
- Martins-Taylor K., Xu R.H. Concise review: genomic stability of human induced pluripotent stem cells. Stem Cells. 2012;30:22–27. doi: 10.1002/stem.705. [DOI] [PubMed] [Google Scholar]
- Merkle F.T., Eggan K. Modeling human disease with pluripotent stem cells: from genome association to function. Cell Stem Cell. 2013;12:656–668. doi: 10.1016/j.stem.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Mitzelfelt K.A., Mcdermott-Roe C., Grzybowski M.N., Marquez M., Kuo C.T., Riedel M., Lai S., Choi M.J., Kolander K.D., Helbling D. Efficient precision genome editing in iPSCs via genetic co-targeting with selection. Stem Cell Rep. 2017;8:491–499. doi: 10.1016/j.stemcr.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y., Chan A.H., Judge L.M., Yoo J., Huang M., Nguyen T.D., Lizarraga P.P., So P.L., Conklin B.R. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods. 2014;11:291–293. doi: 10.1038/nmeth.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A., Fonteyne L., Giesert F., Hoppmann P., Meier A.B., Bozoglu T., Baehr A., Schneider C.M., Sinnecker D., Klett K. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat. Med. 2020;26:207–214. doi: 10.1038/s41591-019-0738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A., Laugwitz K.L., Dorn T., Sinnecker D., Mummery C. Pluripotent stem cell models of human heart disease. Cold Spring Harb. Perspect. Med. 2013;3:a014027. doi: 10.1101/cshperspect.a014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S., Amaishi Y., Maki I., Enoki T., Mineno J. Highly efficient genome editing for single-base substitutions using optimized ssODNs with Cas9-RNPs. Sci. Rep. 2019;9:4811. doi: 10.1038/s41598-019-41121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lim K., Kim J.S., Bae S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33:286–288. doi: 10.1093/bioinformatics/btw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.T., Kim J. Trends in next-generation sequencing and a new era for whole genome sequencing. Int. Neurourol J. 2016;20:S76–S83. doi: 10.5213/inj.1632742.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S., Kwaku Dad A.B., Beloor J., Gopalappa R., Lee S.K., Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C.D., Ray G.J., Dewitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- Roselli C., Chaffin M.D., Weng L.C., Aeschbacher S., Ahlberg G., Albert C.M., Almgren P., Alonso A., Anderson C.D., Aragam K.G. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018;50:1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F., Laganiere J., Cheng A.W., Hockemeyer D., Gao Q., Alagappan R., Khurana V., Golbe L.I., Myers R.H., Lindquist S. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J.L. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyer B., Bu Q., Cory E., Jiang K., Duong S., Sinha D., Steltzer S., Gamm D., Chang Q., Saha K. Scarless genome editing of human pluripotent stem cells via transient puromycin selection. Stem Cell Rep. 2018;10:642–654. doi: 10.1016/j.stemcr.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mil A., Balk G.M., Neef K., Buikema J.W., Asselbergs F.W., Wu S.M., Doevendans P.A., Sluijter J.P.G. Modelling inherited cardiac disease using human induced pluripotent stem cell-derived cardiomyocytes: progress, pitfalls, and potential. Cardiovasc. Res. 2018;114:1828–1842. doi: 10.1093/cvr/cvy208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Wang J., Song Y., Yu D., Sun C., Liu C., Chen F., Zhang Y., Wang F., Harvey R.P. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development. 2015;142:2521–2532. doi: 10.1242/dev.120220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., Liu H., La Russa M., Xie M., Ding S., Qi L.S. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Schulte J.S., Heinick A., Piccini I., Rao J., Quaranta R., Zeuschner D., Malan D., Kim K.P., Ropke A. Universal cardiac induction of human pluripotent stem cells in two and three-dimensional formats: implications for in vitro maturation. Stem Cells. 2015;33:1456–1469. doi: 10.1002/stem.1964. [DOI] [PubMed] [Google Scholar]
- Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed sequences and read numbers for each sib selection analyzed via NGS are listed here. For allele quantification, only reads with a frequency higher than 0.25% of total reads (minimum allele frequency in 200 cells) were considered. These reads were classified into three categories: wild type (“WT,” no detectable SHOX2 c.849C>A or SHOX2 c.∗28T>C mutations), mutant (“Mut,” detectable SHOX2 c.849C>A or SHOX2 c.∗28T>C mutations), or non-assignable (“N/A,” where position SHOX2 c.849 or SHOX2 c.28∗ was deleted).
All primer sequences used for in vitro synthesis, dPCR, NGS, Sanger sequencing, and qRT-PCR of pluripotency and cardiomyocyte subtype marker genes are listed here. In addition, the sequences of single-stranded oligodeoxynucleotides that were transfected together with Cas9 RNP/gRNA complexes to serve as a repair template are given.
Data Availability Statement
Unprocessed sequencing files (.fastq) have been uploaded to the NCBI Sequence Read Archive and are available via the accession no. PRJNA659248 or under https://www.ncbi.nlm.nih.gov/bioproject/PRJNA659248/.