Summary
Human glial progenitor cells (hGPCs) are promising cellular substrates to explore for the in situ production of new neurons for brain repair. Proof of concept for direct neuronal reprogramming of glial progenitors has been obtained in mouse models in vivo, but conversion using human cells has not yet been demonstrated. Such studies have been difficult to perform since hGPCs are born late during human fetal development, with limited accessibility for in vitro culture. In this study, we show proof of concept of hGPC conversion using fetal cells and also establish a renewable and reproducible stem cell-based hGPC system for direct neural conversion in vitro. Using this system, we have identified optimal combinations of fate determinants for the efficient dopaminergic (DA) conversion of hGPCs, thereby yielding a therapeutically relevant cell type that selectively degenerates in Parkinson's disease. The induced DA neurons show a progressive, subtype-specific phenotypic maturation and acquire functional electrophysiological properties indicative of DA phenotype.
Keywords: dopaminergic neurons, neuronal reprogramming, glial progenitor cells, hESC, Parkinson’s disease
Graphical Abstract
Highlights
-
•
Human glial progenitors (hGPCs) can be directly converted into functional neurons
-
•
Specific transcription factor combinations result in dopaminergic conversion
-
•
Reprogrammed neurons show subtype-specific and functional maturation over time
In this article, Parmar and colleagues show proof of concept that human glial progenitors (hGPCs) sourced from fetal brains can be converted into functional neurons. To allow for large-scale studies, a stem cell-based model system that mimics the fetal cell conversion was established and used to identify optimal combinations of fate determinants for the efficient dopaminergic conversion of hGPCs.
Introduction
Somatic cells can be directly converted into induced neurons (iNs) without a stem cell intermediate using defined sets of transcription factors, microRNAs, and/or through chemical manipulations. Since the original report (Vierbuchen et al., 2010), a number of studies have shown conversion of human fibroblasts into subtype-specific neurons, including dopaminergic (DA) neurons, striatal medium spiny neurons, peripheral sensory neurons, noradrenergic neurons, cholinergic neurons, and spinal motor neurons using different combinations of lineage specific transcription factors (Blanchard et al., 2015; Caiazzo et al., 2011; Li et al., 2019; Liu et al., 2013; Pfisterer et al., 2011; Son et al., 2011; Victor et al., 2014). Direct neural conversion opens up new possibilities to generate patient- and disease-specific neurons from somatic cells and is predicted to have a great impact on disease modeling, diagnostics, and other biomedical applications (Drouin-Ouellet et al., 2017b; Mertens et al., 2018).
Direct conversion could potentially also be used to generate neurons for transplantation-based cell replacement therapies. So far, a number of somatic cell types, including fibroblasts, hepatocytes, pericytes, and glia, have been successfully converted to neurons (Masserdotti et al., 2016). Since the direct conversion strategy does not rely on a proliferative intermediate, some of the safety concerns associated with the use of human pluripotent stem cells (hPSCs) are minimized, and both autologous and allogeneic strategies could be used (Fang et al., 2018; Grealish et al., 2016). As an extension of direct conversion in cell-based therapies, delivery of the conversion factors directly to the brain with the aim of converting endogenous glia is being developed as an alternative approach for the generation of therapeutic neurons in situ (Vignoles et al., 2019). In support of this concept, resident astrocytes and glial progenitor cells (GPCs, also called oligodendrocyte progenitor cells or NG2 cells) have been successfully converted into neurons in the mouse brain (Guo et al., 2014; Mattugini et al., 2019; Niu et al., 2015; Pereira et al., 2017; Qian et al., 2020; Rivetti di Val Cervo et al., 2017; Torper et al., 2015). For translational applications, it is important to document the conversion of human glia into subtype-specific and functional neurons. Initially, this can be done using in vitro cultures of human brain-derived cells and so far, human astrocytes and pericytes have been shown to generate functional and subtype-specific neurons after conversion in vitro (Guo et al., 2014; Karow et al., 2012). The possibility to convert human GPCs (hGPCs) in vitro has not yet been explored even though these cells may be more suitable than astrocytes for in situ conversion, because GPCs remain proliferative in the adult brain (Hughes et al., 2013; Simon et al., 2011). Proof of concept for GPC conversion has been obtained in mouse models in vivo (Guo et al., 2014; Heinrich et al., 2014; Pereira et al., 2017; Torper et al., 2015). However, studies of human GPC conversion are challenging because GPCs are not born until late in the second trimester, which limits their accessibility because it is difficult to source human embryos for cell isolation and subsequent in vitro cultures (Jakovcevski and Zecevic, 2005; Sim et al., 2011).
In this study, we show that hGPCs isolated from the human fetal brain can be efficiently converted into neurons in vitro, which provides important proof of concept for the approach. However, access to human fetal cells is limited, and we therefore sought to establish a human pluripotent stem cell (hPSC)-based experimental model system for hGPC reprogramming studies. To achieve this, we differentiated human embryonic stem cells (hESCs) into hGPCs, based on the protocol by Wang et al. (2013), and show that these hESC-derived hGPCs convert into iNs in a similar manner to the hGPCs isolated from the human fetal brain. We used stem cell-derived hGPCs to assess the effect of fate-determining genes with a focus on DA neurons, the principal neuronal subtype lost in Parkinson's disease (PD), and thus a therapeutically important cell type. We found that hGPCs can be directly converted into TH+ neurons using several different combinations of transcription factors and microRNAs. The resulting TH+ neurons have a gene and protein expression profile similar to mature midbrain DA neurons and adopt electrophysiological properties typical of functional DA neurons.
Thus, this study provides evidence that hGPCs isolated from both human fetal brain and differentiated from hESCs can be successfully reprogrammed into functional iNs, including induced DA neurons (iDANs).
Results
Human Fetal Glial Progenitors Efficiently Convert to Functional iDANs In Vitro
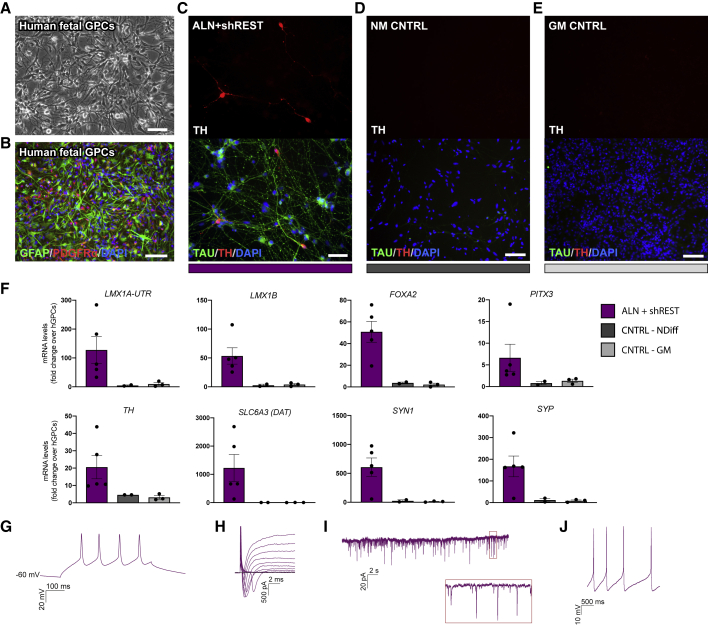
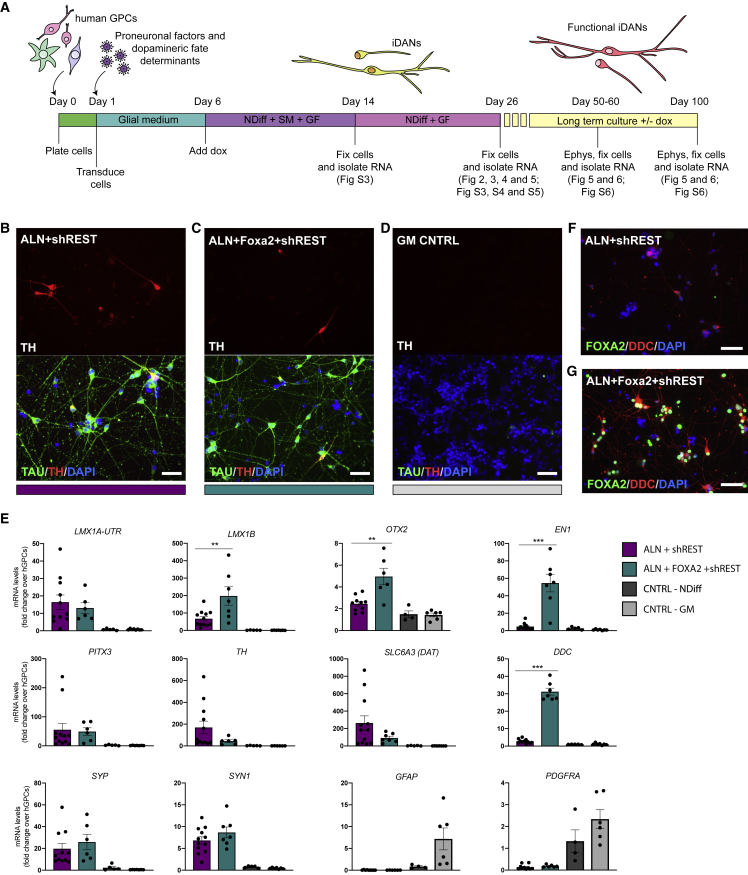
To establish the potential of hGPCs to convert into neurons in vitro, we used hGPCs isolated from the fetal brain (Figures 1A, 1B and S1A–S1AC) (Benraiss et al., 2016) and transduced them with doxycycline (dox)-inducible vectors carrying the conversion factors Ascl1, Lmx1a, and Nr4a2 (Nurr1), (together refered to as ALN), a combination of factors that has previously been used to successfully reprogram human fetal fibroblasts into iDANs (Caiazzo et al., 2011; Torper et al., 2013) together with short hairpin (sh) RNA against the RE1-silencing transcription factor (REST) complex. Already at 1 week after transgene activation, glial markers were downregulated, and some TH+/MAP2+ cells could be detected in the cultures (Figure S1D). After an additional 2 weeks, the reprogrammed cells showed a more pronounced neuronal morphology and expressed many DA-related genes, including TH, SLC6A3 (DAT), FOXA2, LMX1A, and PITX3 (Figures 1C, 1F, and S1E). Control cultures of untransduced cells that were kept in parallel in either glial medium or neuronal conversion medium (containing small molecules and growth factors) continued to express GFAP and PDGFRα (Figures S1F and S1G) and did not give rise to any TH+ neurons (Figures 1D and 1E).
Figure 1.
Fetal hGPCs Can Be Converted to Induced DA Neurons In Vitro
(A and B) Fetal hGPCs visualized by bright field imaging (A) or stained for GFAP and PDGFRα (B).
(C) Reprogrammed TH+/TAU+ neurons 3 weeks after transgene activation.
(D and E) hGPCs kept in parallel for 3 weeks in neuronal conversion medium (D) or glial medium (E) do not give rise to any TH+/TAU+ neurons.
(F) qPCR analysis of reprogrammed cells 3 weeks after transgene activation shows an upregulation of pan-neuronal and DA genes.
(G) Representative trace of voltage responses from the whole-cell patch-clamp technique showing multiple induced APs.
(H) Representative trace of inward sodium- and outward potassium-rectifying currents triggered by stepwise depolarization of the cell.
(I and J) The reprogrammed fetal hGPCs show spontaneous postsynaptic currents (I) and spontaneous firing at their resting membrane potential (J).
In Figure 1F, data are presented as means ± SEM, and all data points have been visualized in the graphs. Each data point represents a replicate from an independent experiment (n = 5 for ALN + shREST; n = 2 for CNTRL-NDIFF; n = 3 for CNTRL-GM). Scale bars: (A, B, D, and E) 100 μm; (C) 50 μm. See also Figure S1.
Whole-cell patch-clamp recordings were performed after 80–120 days in vitro and confirmed functional maturation of the hGPC-derived iNs. At this point, the reprogrammed neurons (n = 37) exhibited the ability to fire evoked action potentials (APs) in the form of single and multiple APs (multiple APs, 14%; single APs, 22%; n = 37) (Figure 1G and Table S1). The cells also displayed fast-inactivated inward and outward currents characteristic of sodium and delayed-rectifier potassium currents (Figure 1H), as well as spontaneous postsynaptic activity (Figure 1I), suggestive of synaptic engagement. Moreover, a proportion of the converted neurons developed spontaneous firing at resting membrane potential (7/25 converted neurons; Figure 1J), a characteristic typical of DA neurons.
Taken together, this establishes the ability of fetal hGPCs to be reprogrammed into functional iNs, with a gene and protein expression profile and functionality indicative of a DA identity using ALN + shREST. However, hGPCs arise late in the gestational time period, following the neuron-to-glial cell switch, and are therefore difficult to obtain from the limited amounts of fetal brain tissue available at this late gestational time point (Rowitch and Kriegstein, 2010; Sim et al., 2011). To enable larger experimental studies of hGPC conversion, we set out to develop a more accessible experimental in vitro system.
Establishing a Pluripotent Stem Cell-Based Model System for hGPC Reprogramming
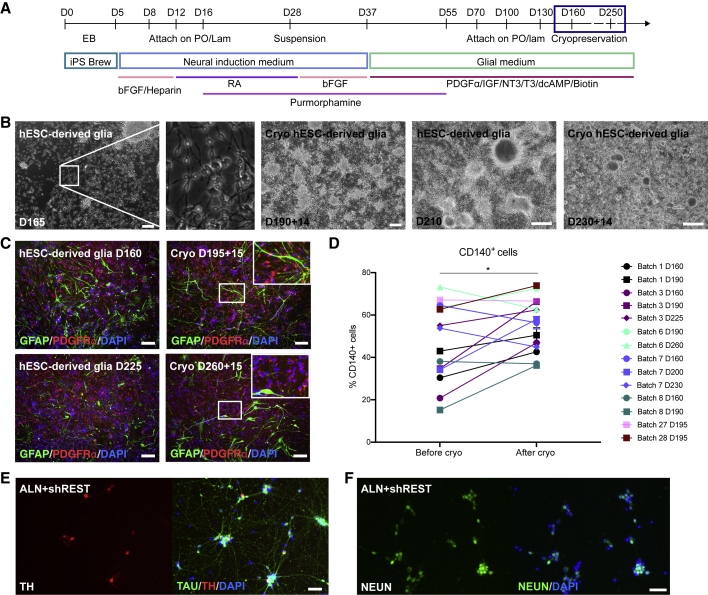
To establish an in vitro model system for direct neuronal conversion using cells that are as close as possible to the bona fide hGPCs in the brain, we used a previously published protocol for generating hGPCs from hPSCs (Wang et al., 2013). The protocol was adapted to a defined and xeno-free hESC culture system for maintenance of hPSCs on recombinant human laminin, in iPS Brew medium. We also established a cryopreservation step in order to improve the logistics and reproducibility of the protocol (Figure 2A). During the glial differentiation of the hESCs, the proportion of cells positive for CD140 (PDGFRα), which identifies myelinogenic and migration-competent human oligodendrocyte progenitor cells (Sim et al., 2011), increases with time in culture (Figure S2A). Cells generated from two different hESC lines (RC17 and HS1001) were expanded and cryopreserved from day 160 up to day 250 (Figures 2A and 2B). At this point. the majority of the cells were positive for CD140, and we confirmed that this phenotype was maintained following cryopreservation (Figures 2C, 2D, and S2A–S2E). The glial marker CD44 labels astrocyte biased progenitors, or when co-expressed with CD140, bipotential glial progenitors. In contrast to CD140, the proportion of CD44+ cells did not increase with time in culture (Figure S2A), and the cryopreservation procedure did not significantly affect the relative proportion of CD44+ cells in the culture (p = 0.952; paired, two-tailed t test). Under these culture conditions, very few if any cells expressed neuronal TAU (HT7) or MAP2a/MAP2b (Figures S2F and S2G), whereas MAP2c, an isoform of MAP2 that is expressed in developing progenitors, both neuronal and oligodendroglial (Garner et al., 1988; Vouyiouklis and Brophy, 1995), was abundantly expressed (Figure S2F). Rare TH+ cells could also be found in both fetal- and hESC-derived hGPC cultures, but co-labeling with glial and neuronal markers confirmed that these cells were of a glial identity, since TH was only found co-labeled with glial markers and not with neuronal markers (Figures S1C, S2G, and S2H).
Figure 2.
Establishing a Renewable and Reproducible Source of hGPCs from hESCs for In Vitro Modeling of Direct Neuronal Conversion
(A) Schematic overview of the protocol for generating hESC-derived hGPCs, with an indication of the time frame used for cryopreservation.
(B) Bright-field images of the hESC-derived hGPCs cultures at different stages and before and after cryopreservation.
(C) GFAP/PDGFRα immunostaining of the hESC-derived hGPC cultures at different stages and before and 15 days after cryopreservation. Insets show close-up morphology of cells in the boxed areas.
(D) Percentage of CD140+ cells within the cultures before and after cryopreservation as determined by flow cytometry.
(E and F) TH+/TAU+ cells (E) and NEUN+ cells (F) 3 weeks after activation of the transgenes ALN + shREST in hESC-derived hGPCs.
The proportions of CD140+ cells in Figure 2D were compared using a paired two-tailed t test; ∗p < 0.05 (p = 0.0327). Scale bars: 100 μm. See also Figures S2 and S3.
We next assessed whether cryopreserved hESC-derived hGPCs could be converted into iDANs using the same protocol and the same combination of reprogramming factors (ALN + shREST) as was used for the fetal hGPCs. In accord with our observations in converting fetal progenitors, some TH+ neurons could be detected 1 week after conversion (Figure S3A). Three weeks after activation of ALN + shREST, the glial markers were efficiently downregulated, and the cultures contained few if any GFAP+ or PDGFRα+ cells (Figure S3B). Control cells that were not transduced but cultured in neuronal conversion medium (Figures S3C and S3D), as well as control glial cultures that were analyzed in parallel (Figure S3E), maintained their glial identity (Figures S3D and S3E), and no TH+ neurons were detected (Figure S3C). At this time point, reprogrammed cells with a clear neuronal morphology that were positive for TH/TAU and NEUN were observed only after reprogramming with ALN + shREST (Figures 2E, 2F, and 5G). Together, these data establish that both fetal hGPCs and hESC-derived hGPCs can be converted to TH+ iNs, using the same factors and with comparable timing and efficiencies (Figures S1E and 5G). No significant differences were noted between fetal- and hESC-derived cells in the relative proportions of either neurons (p = 0.102) or TH+ neurons (p = 0.139; both determined using unpaired, two-tailed t tests). These data support the applicability of our stem cell-based model system for neuronal conversion studies.
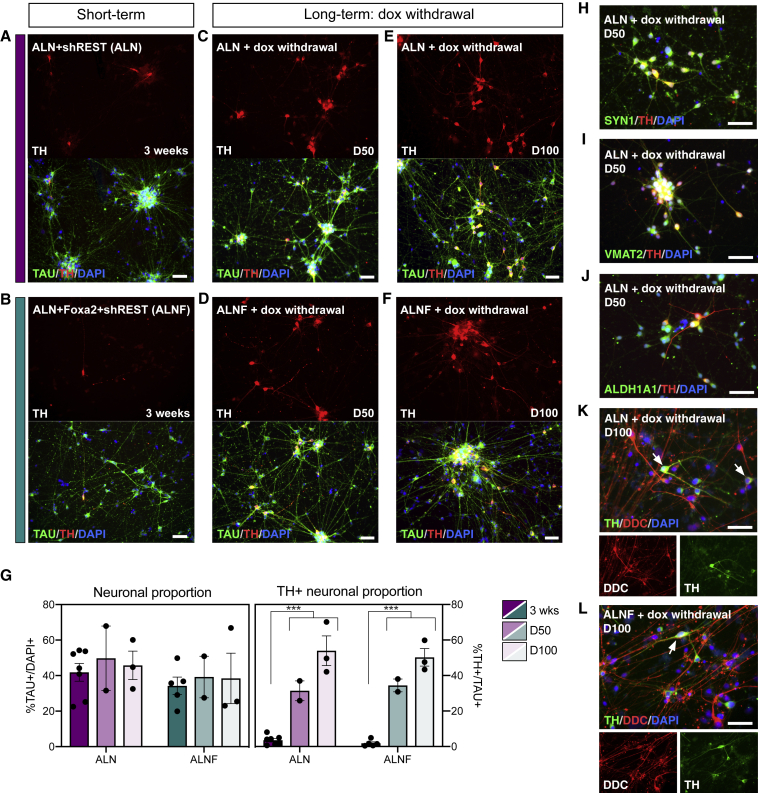
Figure 5.
Progressive Subtype-Specific Maturation of the iDANs over Time
(A, C, and E) The proportion of TH+ iDANs increase when doxycycline is withdrawn after 3 weeks and the ALN + shREST reprogrammed neurons are kept in culture for 50 days (C) and 100 days (E), compared with the 3-week time point (A).
(B, D, and F) The proportion of TH+ iDANs increases when doxycycline is withdrawn after 3 weeks and the ALN + Foxa2 + shREST reprogrammed neurons are kept in culture for 50 days (D) and 100 days (F), compared with the 3-week time point (B).
(G) Quantifications of neuronal proportion and TH+ neuronal proportion over time. TH+ neuronal proportion (ALN + shREST): 3 weeks, 3.6% ± 1.0%, n = 7; 50 days, 31.4% ± 5.6%, n = 2; 100 days, 53.9% ± 8.3%, n = 3. TH+ neuronal proportion (ALN + Foxa2 + shREST): 3 weeks, 1.8% ± 0.8%, n = 5; 50 days, 34.5% ± 3.6%, n = 2; 100 days, 50.2% ± 4.9%, n = 3. Neuronal proportion (ALN + shREST): 3 weeks, 41.8% ± 5.0%, n = 7; 50 days, 49.8% ± 18.1%, n = 2; 100 days, 45.8% ± 8.0%, n = 3. Neuronal proportion (ALN + Foxa2 + shREST): 3 weeks, 34.3% ± 5.0%, n = 5; 39.2% ± 11.7%, n = 2; 100 days, 38.4% ± 14.2%, n = 3.
(H–J) The generated iDANs are positive for SYN1 (H), VMAT2 (I), and ALDH1A1 (J).
(K and L) TH+/DDC+ double-labeled cells are observed in long-term cultures of both ALN + shREST (K) and ALN + Foxa2 + shREST (L) reprogrammed cells. White arrows indicate examples of double-labeled cells.
Data are presented as means ± SEM and all data points have been visualized in the graphs. Each data point represents a replicate from an independent experiment (n = 7 for ALN + shREST at the 3-week time point; n = 5 for ALN + Foxa2 + shREST at the 3-week time point; n = 2 for both conditions at the D50 time point and n = 3 for both conditions at the D100 time point). In (G), the proportion of TAU+ and TAU+/TH+ cells at the 3-week time point was compared with the late time points day 50 and day 100 combined using an unpaired, two-tailed t test. ∗∗∗p < 0.001 (p < 0.0001 for TH+ cells in ALN + shREST, p < 0.0001 for TH+ cells in ALN + Foxa2 + shREST); n.s. (p = 0.5307 for TAU+ cells in ALN + shREST, p = 0.6637 for TAU+ cells in ALN + Foxa2 + shREST). Scale bars: 50 μm. See also Figure S6.
Purified Populations of hESC-Derived hGPCs Can Be Converted to TH+ INs
We initiated conversion from hESC-derived hGPCs at different time points, ranging from day 160 to day 270 in culture (corresponding to passage 2–6 of the hGPCs; see Table S2) and established that the time in culture did not affect the efficiency of the hGPCs to convert into iNs, based on the comparable induction of TH and SYNAPSIN 1 (SYN1) as assessed 3 weeks after initiation of reprogramming (Figure S3F). The differentiation protocol used here results in primarily oligodendrocyte biased progenitors (CD140+/CD44−) but also produces a minority of bipotential (CD140+/CD44+) and astrocyte biased (CD140−/CD44+) progenitors. By analyzing neuronal conversions from different glial differentiation runs containing varying ratios of these three subtypes of glial progenitors (Table S2), we found that the induction of SYN1 in the reprogrammed cells correlated positively with a high CD140+ starting population, and negatively with a high astrocyte progenitor population, while TH expression did not vary in correlation with the composition of cells at the start of neuronal conversion (Figure S3G).
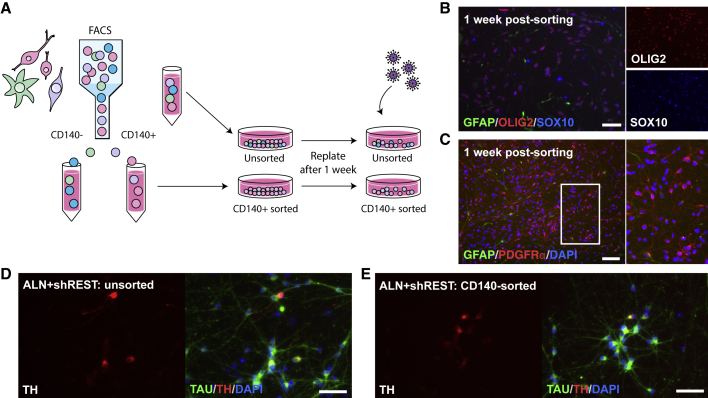
To assess experimentally if a purified population of CD140+ hGPCs can be converted into TH+ iNs, we next used fluorescence-activated cell sorting to extract the CD140+ fraction from the hESC-derived hGPCs cultures (Figures 3A–3C and S4). Importantly, by converting unsorted cells (Figure 3D) and CD140-sorted cells (Figure 3E) from the same batch of hESC-derived hGPCs side by side, we confirmed that using our differentiation strategy, CD140+ hGPCs are indeed amenable to DA conversion in vitro.
Figure 3.
Direct Conversion of Purified CD140-expressing Progenitors
(A) Schematic overview of the strategy for neuronal conversion of CD140-sorted and unsorted cells side by side. Astrocyte biased progenitors are depicted in green, oligodendrocyte biased progenitors are depicted in pink, bipotent glial progenitors are depicted in purple, and cells of an unknown identity are depicted in blue.
(B and C) CD140-sorted hGPCs are positive for OLIG2/SOX10 (B) and PDGFRα (C). A subset of the hGPCs are positive for GFAP (B and C), representing a bipotent progenitor state.
(D and E) TH+/TAU+ cells 3 weeks after reprogramming, starting from either unsorted (neuronal proportion, 53.7%; TH+ neuronal proportion, 1.7%; D) or CD140-sorted (neuronal proportion, 57.0%; TH+ neuronal proportion, 1.3%; E) hESC-derived hGPCs.
Scale bars: (C) 100 μm; (B, D, and E) 50 μm. See also Figure S4.
Timed Delivery of Foxa2 Augments DA Reprogramming of Fetal- and Stem Cell-Derived hGPCs
Previous efforts from our lab and others have identified Otx2, Foxa2, Nurr1, NeuroD1, Ascl1, Lmx1a, and mir218 as factors that can improve the DA reprogramming of human fibroblasts and human astrocytes when expressed in specific combinations (Pereira et al., 2014; Rivetti di Val Cervo et al., 2017; Torper et al., 2013). Here, we tested the effect of these additional DA fate determinants when converting hGPCs to DA neurons (Figure 4A). Gene expression analysis 3 weeks after initiation of reprogramming showed that the addition of Otx2 to the original reprogramming cocktail did not add any beneficial effects in terms of reprogramming efficiency and DA phenotype (Figures S5A–S5C). The NeuroD1, Ascl1, Lmx1a, and mir218 (NeAL218) factor combination used in Rivetti di Val Cervo et al. (2017) resulted in a comparable number of TH+ iDANs and a similar gene expression profile as ALN + shREST reprogrammed cells, when assessed 3 weeks after transgene activation (Figures S5D and S5E). In contrast, when Foxa2 was added to the initial cocktail (Figures 4B and 4C), the gene expression analysis showed a higher endogenous expression of the midbrain DA genes LMX1B (p = 0.0098), EN1 (p = 0.0001), and OTX2 (p = 0.0075) (Figure 4E). The addition of Foxa2 also resulted in increased expression of the DA synthesis-related enzyme DDC (p < 0.0001) (Figure 4E), which was confirmed at the protein level using immunocytochemistry (Figures 4F, 4G, and S5F). At the same time, the proportion of TH+ neurons in the 3-weeks culture did not differ significantly between the ALN + shREST and ALN + Foxa2 + shREST transduced cells (p = 0.224 as determined using an unpaired, two-tailed t test and see Figure 5G). We also tested the NeAL218 and ALN + Foxa2 + shREST factor combinations in the fetal hGPCs and confirmed that while both these combinations successfully generate iNs with a DA phenotype (Figures S5G and S5H), the addition of Foxa2 again resulted in a higher expression of LMX1B and DDC (Figure S5I).
Figure 4.
Direct Conversion of hESC-Derived hGPCs Using Different Combinations of DA Fate Determinants
(A) Schematic overview of the neuronal conversion protocol indicating the different time points of analysis and references to the figures related to each time point.
(B and C) TH+/TAU+ neurons generated with either ALN + shREST (B) or ALN + Foxa2 + shREST (C) 3 weeks after transgene activation starting from hESC-derived hGPCs.
(D) No TH+/TAU+ neurons are generated when the hESC-derived hGPCs are kept for 3 weeks in parallel in glial medium.
(E) qPCR analysis of gene expression of iDANs 3 weeks after transgene activation comparing cells reprogrammed with either ALN + shREST or ALN + Foxa2 + shREST.
(F and G) Immunocytochemical analysis of DDC and FOXA2 in cells reprogrammed using either ALN + shREST (F) or ALN + Foxa2 + shREST (G).
Data are presented as means ± SEM, and all data points have been visualized in the graphs. Each data point represents a replicate from an independent experiment (n = 9–12 for ALN + shREST; n = 6–7 for ALN + Foxa2 + shREST; n = 4–5 for CNTRL-NDIFF; n = 6–8 for CNTRL-GM). The gene expression levels for the conditions ALN + shREST and ALN + Foxa2 + shREST were compared using a Mann-Whitney test; ∗∗p < 0.01 (p = 0.0098 for LMX1B, p = 0.0075 for OTX2); ∗∗∗p < 0.001 (p = 0.0001 for EN1, p < 0.0001 for DDC). Scale bars: 50 μm. See also Figure S5.
Although hESC-derived hGPCs could be reprogrammed into TAU+ iNs at 3 weeks after transgene activation with an efficiency of 35%–40%, the relative proportion of TH+ iDANs remained low at this point (Figures 5A, 5B, and 5G). Gene expression analysis at this time point suggested that other subtypes of neurons, such as glutamatergic and GABAergic neurons, could also be present in the cultures (Figure S6A). To investigate if the proportion of TH+ neurons increases with time, we maintained the ALN + shREST and ALN + Foxa2 + shREST reprogrammed cells in long-term culture, with either maintained transgene expression (i.e., continued dox administration to the medium) or with transgene de-activation via dox withdrawal after 3 weeks (Figure 4A). Quantification at day 50 and day 100 (24 and 74 days after dox withdrawal, respectively) showed that the proportion of TH+ iDANs in both culture conditions was significantly increased over time, reaching as high as 50%, while the neuronal content did not change significantly from the 3-week time point (Figures 5C–5G). The observations that the total number of cells did not change (Figure S6F) and the neuronal proportions were comparable between time points, while the relative proportion of TH+ neurons increased, suggested that the converted cells undergo progressive DA maturation; this was later confirmed with electrophysiology (see Figure 6). Interestingly, when ALN + shREST was used for conversion, a high number of TH+ iDANs was generated both with and without dox withdrawal (Figures 5E and S6B), but when Foxa2 was added to the reprogramming mix, a high number of TH+ iDANs was only observed when dox was withdrawn from the medium (Figures 5F and S6C). This indicates that while the addition of Foxa2 to the conversion factor cocktail increased DA-specific conversion initially, maintenance of the exogenous Foxa2 expression had a negative effect of the subtype identity and maturation of the iDANs. Indeed, the unphysiologically high and/or prolonged expression of Foxa2 seemed to directly interfere with the expression of TH, since none of the cells that were strongly positive for FOXA2 co-labeled with TH, and the TH+ neurons only showed weak nuclear labeling of FOXA2 by immunocytochemistry (Figure S6D). Immunocytochemistry confirmed the expression of the mature neuronal- and DA neuron markers SYN1, VMAT2, and ALDH1A1 in long-term cultures of ALN + shREST reprogrammed cells (Figures 5H–5J), and at this late time point, DDC+ neurons were present in equal quantities in ALN + shREST and ALN + Foxa2 + shREST reprogrammed cultures (Figures 5K, 5L, and S6G).
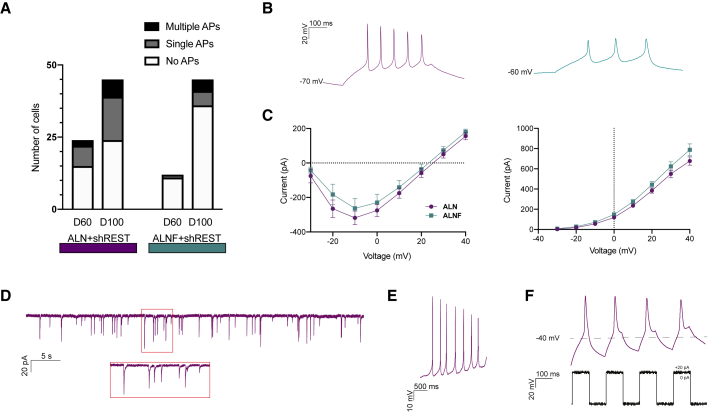
Figure 6.
hESC-Derived hGPCs Reprogrammed iDANs Become Functionally Mature
(A) Graph showing the number of cells exhibiting multiple, single, or no AP responses evoked by current steps at 60 and 100 days after reprogramming start with ALN + shREST and ALN + Foxa2 + shREST.
(B) Representative trace of voltage responses from the whole-cell patch-clamp technique showing multiple induced APs in cells reprogrammed with ALN + shREST (left, purple) ALN + Foxa2 + shREST (right, green).
(C) Inward sodium current peak (left) and outward potassium current (right) plotted against voltage steps of cells reprogrammed with ALN + shREST (purple) and ALN + Foxa2 + shREST (green).
(D) Spontaneous postsynaptic currents in an ALN + shREST reprogrammed neuron.
(E) Spontaneous firing at resting membrane potential in an ALN + shREST reprogrammed neuron.
(F) Repetitive induced AP at small current injections in an ALN + shREST reprogrammed neuron.
See also Figure S6.
The Reprogrammed iDANs Become Functionally Mature
To better characterize the maturation and functional properties of iDANs at later stages of reprogramming, we focused on ALN + shREST and ALN + Foxa2 + shREST with dox withdrawal from the medium after the first 3 weeks of conversion. Comparing the gene expression profile of the reprogrammed iDANs at 3 weeks with the expression profile at later time points (day 50 and day 100) showed that while the endogenous expression of the DA progenitor markers LMX1A, FOXA2, and LMX1B decreased over time, the expression of the post-mitotic DA markers TH and NURR1 increased (Figure S6E).
To assess the electrophysiological properties of iDANs that had been reprogrammed from hESC-derived hGPCs, whole-cell patch-clamp recordings were performed at day 60 and day 100 of reprogramming. At these time points, no detectable differences in intrinsic membrane properties were observed between the groups (see Table S3). After 60 days (34 days after dox withdrawal), cells reprogrammed with ALN + shREST showed induced APs in higher proportion (9/24 cells firing), compared with ALN + Foxa2 + shREST (1/12 cells firing; Figure 6A). At day 100, the iDANs had matured further, with APs in 21/45 cells for ALN + shREST and 9/45 cells for ALN + Foxa2 + shREST. There was a trend of a greater number of cells displaying multiple APs in ALN + shREST condition compared ALN + Foxa2 + shREST condition (Figures 6A and 6B). The induced APs could be selectively blocked by tetrodotoxin (TTX, 1 μM) (Figure S6H). Both ALN + shREST and ALN + Foxa2 + shREST iDANs displayed fast-inactivated inward and outward currents characteristic of sodium and delayed-rectifier potassium currents (Figure 6C and Table S3) as well as spontaneous postsynaptic activity indicative of synaptic integration (Figures 6D and S6I, d100: n = 13/43 for ALN + shREST and 10/41 for ALN + Foxa2 + shREST). iDANs from both conditions also showed the presence of spontaneous firing at resting membrane potential (Figure 6E) and repetitive APs at small current injections (Figure 6F), indicative of a DA phenotype. The proportion of cells exhibiting spontaneous firing was 11% for ALN + shREST and 8% ALN + Foxa2 + shREST at day 60 with a higher proportion at day 100 for the ALN + shREST group (37%). Moreover, the iDANs also showed repetitive AP at small current injections already at day 60 (11% for ALN + shREST and 10% for ALN + Foxa2 + shREST, Figure 6F), a feature that was further increased in ALN + shREST condition at day 100 (ALN + shREST 37%, ALN + Foxa2 + shREST 10%).
Taken together, these results show that hESC-derived hGPCs convert into functional neurons with DA properties that achieve mature profiles after long-term culture.
Discussion
The potential of cell replacement therapy as a means of treating PD has been established through clinical trials using human fetal ventral midbrain tissue (Barker et al., 2015), and efforts are currently underway with the aim of assessing hPSCs as a source of transplantable DA progenitors (Barker et al., 2017; Doi et al., 2020; Schweitzer et al., 2020). In vivo direct reprogramming of resident glia into DA neurons is similarly based on DA neuronal replacement but circumvents cell transplantation, as the new neurons are generated by reprogramming resident cells in situ (Grealish et al., 2016; Vignoles et al., 2019).
Several studies have shown that it is possible to obtain subtype-specific and functional neurons via transcription factor-mediated reprogramming of mouse GPCs in vivo (Guo et al., 2014; Heinrich et al., 2014; Pereira et al., 2017; Torper et al., 2015) or via inhibition of PTB (Qian et al., 2020), but analogous studies using human cells have been lacking. In this study, we therefore established an in vitro model of direct neural conversion from hESC-derived hGPCs. This model has a distinct advantage over existing models that use fetal tissue-derived glia, since generation from stem cells of highly expandable hGPCs provides us with the opportunity to reproducibly assess the biology of neuronal reprogramming from human cells, while doing so at scale using a preferentially permissive but otherwise scarce cell type.
Once established, this hPSC-based experimental model system for hGPC reprogramming studies allowed us to conduct detailed in vitro studies of the timeline of reprogramming, the effect of different reprogramming factors, and the phenotypic profile and electrophysiological properties of the reprogrammed iDANs. Using several combinations of transcription factors, including ALN, ALN + Foxa2, and NeAL218, which have previously been shown to successfully reprogram human fibroblasts and astrocytes into DA neurons (Caiazzo et al., 2011; Rivetti di Val Cervo et al., 2017; Torper et al., 2013), we confirmed that the same factors could successfully reprogram hGPCs as well. Knockdown of REST, which acts to suppress neuronal identity, has previously been shown to greatly increase the efficiency of neuronal reprogramming, starting from adult human fibroblasts (Drouin-Ouellet et al., 2017a). REST is expressed by both GPCs and differentiating astrocytes, where it restricts the neuronal program, and its knockdown has been shown to result in an induction of neuronal genes (Dewald et al., 2011; Kohyama et al., 2010; Liu et al., 2019). Accordingly, the combined knockdown of REST together with ALN resulted in iNs with a DA neuron profile when reprogramming hGPCs. The addition of Foxa2 to the reprogramming cocktail resulted in a stronger endogenous expression of midbrain DA progenitor genes, such as OTX2, LMX1B, and EN1 after 3 weeks of conversion, but sustained expression directly interfered with the acquisition of a TH+ phenotype.
Long-term assessment of the reprogrammed iDANs established that the cells were stably converted and did not rely on continuous expression of conversion factors. Phenotypic and functional assessment using patch-clamp electrophysiology revealed that the reprogrammed neurons matured over time, resulting in an approximately 15-fold increase of TH+ iDANs in the culture and an adoption of functional electrophysiological properties in line with a DA neuron identity. However, just as when fibroblasts are converted to DA neurons (Caiazzo et al., 2011; Pfisterer et al., 2011; Torper et al., 2013), not all of the reprogrammed cells adopt a DA neuron identity. In this study, we also detected expression of markers suggesting the presence of other subtypes of neurons, including GABAergic and glutamatergic neurons, and the exact phenotype of these neurons remains to be established. Furthermore, while glial markers are rapidly suppressed in these cells, not all glia convert into neurons. This may be due to a technical limitation, in that the conversion factors are delivered on separate vectors, with the result that not all cells are transduced to co-express all factors, and the stoichiometry of gene expression may vary across the transduced cell population. Going forward, such incompletely transduced or reprogrammed cells will need to be removed before iDANs can be evaluated for clinical use. The reprogrammed neurons matured functionally after long-term culture. However, even at the late time points, a proportion of the neurons were not functionally mature. This can be due to initial asynchrony in reprogramming, culture conditions that may not be ideal for maturation, or insufficient support of mature cells in 2D cultures, and future studies are needed to develop culture conditions more optimal for long-term maintenance of functionally mature cells.
Together, these observations suggest that our hPSC-based system for hGPC reprogramming is effective at reproducibly producing iDANs, while providing a platform for broader studies aiming at converting hGPCs into other subtypes of neurons as well, because it provides scalable populations of reprogramming-permissive human cells. In the current study, we identified combinations of transcription factors that were able to successfully convert both fetal- and stem cell-derived hGPCs directly into functional iDANs. Thus, this study provides an important proof of principle that hPSC-derived hGPCs comprise especially permissive targets for efforts aimed at converting endogenous cells into context-appropriate neurons, in both the diseased and damaged brain, including DA neuronal replacement in PD.
Experimental Procedures
hESC Culture and Generation of Human GPCs from hESCs
Glial differentiation was initiated from two different hESC lines: RC17 (RCe021-A, p26-30) and HS1001 (KIe055-A, p8). The undifferentiated hESCs were maintained on LN521 (0.5 μg/cm2; Biolamina)-coated tissue culture plates in iPS Brew XF medium (StemMACS; Milteny) and passaged weekly with EDTA (0.5 mM).
For differentiation of hESCs into hGPCs, the protocol developed in the Goldman lab (Wang et al., 2013) was employed with some minor modifications. For detailed information, see Supplemental Experimental Procedures.
Culturing of Human Fetal GPCs
Human fetal brain tissue from aborted fetuses (20 weeks gestational age) was obtained with donor consent and under protocols approved by the University of Rochester-Strong Memorial Hospital Research Subjects Review Board. The isolation of glial progenitors from the fetal brain tissue was performed as previously described (Benraiss et al., 2016). The cells were subsequently grown on suspension plates (Greiner Bio-One) in DMEM/F12/N1 medium supplemented with bFGF (10 ng/mL) and 2% PD-FBS at 2.0 × 105 cells/mL and passaged every 10–14 days.
Viral Vectors and Transduction
Doxycycline-regulated lentiviral vectors expressing mouse cDNA and the Tet-On trans-activator (FuW.rtTA-SM2; Addgene), have been described previously (Pereira et al., 2014; Pfisterer et al., 2011; Rivetti di Val Cervo et al., 2017). REST shRNA sequences were either expressed in lentiviral constructs under control of a U6 promotor or expressed in a single doxycycline vector construct containing REST shRNA sequences and Ascl1 cDNA. Third-generation lentiviral vectors were produced as previously described (Zufferey et al., 1997) and titrated by quantitative PCR analysis (Georgievska et al., 2004). All viruses used in this study titered between 3 × 108 and 8 × 109. Both fetal- and hESC-derived hGPCs were quite sensitive to the viral transduction, resulting in relatively high levels of cell loss during the early stage of reprogramming compared with starting from fibroblasts for example. This effect showed some cell-batch dependency, and the later inclusion of additional reprogramming factors to the conversion cocktail resulted in an increased viral load, which in general resulted in slightly higher cell death. Therefore, conversion of fetal- and hESC-derived hGPCs required lower multiplicities of infections (MOI; 0.5–2) compared with studies using fibroblasts (Drouin-Ouellet et al., 2017a).
Direct Neuronal Conversion of Human GPCs
Fetal hGPCs or hESC-derived hGPCs were seeded in their normal culturing medium on plates that had been serially coated with polyornithine (PO), laminin (Lam), and fibronectin (FN), at a density of 50,000 cells/cm2. For electrophysiological analysis, the cells were seeded onto PO/Lam/FN-coated coverslips that had been pre-treated according to Richner et al. (2015). One day after seeding, the cells were transduced at an MOI of 0.5–1 per vector (fetal hGPCs) or MOI of 1–2 per vector (hESC-derived hGPCs), and the medium was changed the next day to remove the viral vectors. Doxycycline (2 μg/mL) was added to the culture medium 5 days after transduction to activate the transgenes, and 2 days later the medium was switched to neural differentiation medium (NDiff227; Takara-Clontech) containing the following small molecules (SMs): CHIR99021 (2 μM; Axon), SB-431542 (10 μM; Axon), noggin (0.5 μg/mL; R&D Systems), LDN-193189 (0.5 μM; Axon), and valproic acid sodium salt (VPA 1 mM; Merck Millipore); and the following growth factors (GFs): LM-22A4 (2 μM; R&D Systems), GDNF (2 ng/mL; R&D Systems), NT3 (10 ng/mL; R&D Systems), and db-cAMP (0.5 mM; Sigma). Two-thirds of the medium was changed every 2–3 days. Two weeks post transduction, the SMs were withdrawn from the neuronal differentiation medium, and the medium was supplemented with only the GFs until the end of the experiment. For long-term assessment of the maturation of the reprogrammed cells, doxycycline was either kept in the medium until the end or withdrawn 3 weeks post transgene activation.
Immunocytochemistry
The cells were fixed in 4% paraformaldehyde solution for 15 min at room temperature (RT) before staining. The cells were pre-incubated in a blocking solution containing 0.1 M phosphate buffered saline with potassium (KPBS) + 0.1% Triton + 5% serum (of secondary antibody host species) for 1–3 h before the primary antibody solution was added (see Table S4). The cells were incubated with the primary antibodies over night at 4°C, and washed with KPBS the following day before adding the secondary antibody solution containing fluorophore-conjugated antibodies (1:200; Jackson ImmunoResearch Laboratories) and DAPI (1:500). The cells were incubated with the secondary antibodies for 2 h at RT and finally washed with KPBS.
qRT-PCR
RNA was isolated from the cells using the RNeasy Micro kit (QIAGEN). Reverse transcription was performed with a Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Fisher Scientific), using 0.2–0.5 μg of RNA per sample. The cDNA (1 μL) was pipetted together with LightCycler 480 SYBR Green I Master (5 μL; Roche) and relevant primers (4 μL, see Table S5; Integrated DNA Technologies) using the Bravo pipetting robot instrument (Agilent) and analyzed by qRT-PCR on a LightCycler 480 II instrument (Roche) using a 40× cycle two-step protocol with a 60°C, 1-min annealing/elongation step and a 95°C, 30-s denaturation step. All samples were run in technical triplicates, and the average CT values were used for calculating the relative gene expression using the ΔΔCT method. All fold changes were calculated as the average fold change based on two different housekeeping genes (b-actin and GAPDH) relative to the starting cells before conversion was initiated.
High Content Analysis
Cells were imaged at 10× magnification using a Cellomics Array Scan VTI HCS reader (Thermo Fisher Scientific). The total number of cells per well positive for DAPI, TAU, and TH were quantified using HCS Studio 3.0 Scan software (Thermo Fischer), which is an automated process enabling unbiased assessment of fluorescence intensity and distribution. Applying the program “Target Activation,” data from 289 fields were collected from the entire well (corresponding to an average number of 21,049 cells). TAU+ neurons were defined as cells containing a DAPI+ nuclei and a Cy2-TAU average fluorescence intensity above background of internal negative control cells. TH+ neurons were defined as TAU+ neurons with a Cy3-TH average fluorescence intensity above background of internal negative control cells.
Statistical Analysis
All data are expressed as means ± standard error of the mean (SEM). For gene expression analysis and cellomics analysis, n represent replicates from independent experiments. A Shapiro-Wilk normality test was used to assess the normality of the distribution, and parametric or nonparametric tests were performed accordingly.
For comparing the proportions of CD140+ cells before and after cryopreservation in Figure 2D, a paired, two-tailed t test was used. For qPCR data related to Figure 4E, outliers were identified using Grubbs's test and removed from the dataset. Since normality could not be proven, a Mann-Whitney test was used to compare the gene expression levels of ALN + shREST and ALN + Foxa2 + shREST. To compare the number of TAU+ and TAU+/TH+ at early and late time point in Figure 5G, the 3-week time point was compared with day 50 and day 100 combined using an unpaired, two-tailed t test. In Figures S3F and S3G, correlations were assessed using Pearson's r. In Figures S5F and S6H, the proportions of DDC+ cells were compared using unpaired, two-tailed t tests.
Statistical analyses were conducted using GraphPad Prism 8.0.
Author Contributions
S.N. conceived the project, designed and performed the conversion experiments with hESC-derived and fetal-derived hGPCs, interpreted data, and analyzed results; J.G. performed experiments, interpreted data, and analyzed results; D.B.H. designed and performed conversion experiments with fetal hGPCs and analyzed the results; A.B. and M.B. performed, analyzed, and interpreted patch-clamp experiments and wrote part of the paper; M. Pereira designed and performed conversion experiments with human fetal GPCs; D.R.O. designed, performed, analyzed, and interpreted patch-clamp experiments; S.A.G. designed experiments and interpreted results; M. Parmar conceived the project, designed experiments, and interpreted results. S.N. and M. Parmar wrote the paper with input from all authors.
Acknowledgments
We thank Anna Hammarberg, Sol Da Rocha Baez, Jenny Johansson, Ulla Jarl, and Marie Persson Vejgården for excellent technical assistance; members of Steve Goldman's group at the University of Rochester for help with transferring the protocol for hESC-based generation of hGPCs to Lund; and Ernest Arenas for kindly providing virus plasmids. The research leading to these results has received funding from the New York Stem Cell Foundation (M. Parmar), the European Research Council (ERC grant agreement no. 771427, M. Parmar), the Swedish Research Council (grant agreements 2016-00873, M. Parmar; 2017-01234, D.R.O.), Swedish Parkinson Foundation (Parkinsonfonden, M. Parmar), Jeansson Foundation (Jeansson stiftelser, D.R.O.), Swedish Brain Foundation (Hjärnfonden, M. Parmar), the Strategic Research Area at Lund University Multipark (Multidisciplinary research in Parkinson's disease), Knut och Alice Wallenberg Foundation (2018.0040, M. Parmar), National Institute of Neurological Disorders and Stroke (NINDS, S.A.G.), National Institute of Mental Health (NIMH, S.A.G.), ADELSON Medical Research Foundation (S.A.G.), Novo Nordisk Foundation (S.A.G.), Lundbeckfonden (S.A.G.), Oscine Corp. and Sana Biotechnology (S.A.G.). M. Parmar is a New York Stem Cell Foundation Robertson Investigator.
Published: September 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.08.013.
Supplemental Information
References
- Barker R.A., Drouin-Ouellet J., Parmar M. Cell-based therapies for Parkinson disease-past insights and future potential. Nat. Rev. Neurol. 2015;11:492–503. doi: 10.1038/nrneurol.2015.123. [DOI] [PubMed] [Google Scholar]
- Barker R.A., Parmar M., Studer L., Takahashi J. Human trials of stem cell-derived dopamine neurons for Parkinson's disease: dawn of a new era. Cell Stem Cell. 2017;21:569–573. doi: 10.1016/j.stem.2017.09.014. [DOI] [PubMed] [Google Scholar]
- Benraiss A., Wang S., Herrlinger S., Li X., Chandler-Militello D., Mauceri J., Burm H.B., Toner M., Osipovitch M., Jim Xu Q. Human glia can both induce and rescue aspects of disease phenotype in Huntington disease. Nat. Commun. 2016;7:11758. doi: 10.1038/ncomms11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J.W., Eade K.T., Szucs A., Lo Sardo V., Tsunemoto R.K., Williams D., Sanna P.P., Baldwin K.K. Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 2015;18:25–35. doi: 10.1038/nn.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M., Dell'Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Dewald L.E., Rodriguez J.P., Levine J.M. The RE1 binding protein REST regulates oligodendrocyte differentiation. J. Neurosci. 2011;31:3470–3483. doi: 10.1523/JNEUROSCI.2768-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi D., Magotani H., Kikuchi T., Ikeda M., Hiramatsu S., Yoshida K., Amano N., Nomura M., Umekage M., Morizane A. Pre-clinical study of induced pluripotent stem cell-derived dopaminergic progenitor cells for Parkinson’s disease. Nat. Commun. 2020;11:3369. doi: 10.1038/s41467-020-17165-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin-Ouellet J., Lau S., Brattas P.L., Rylander Ottosson D., Pircs K., Grassi D.A., Collins L.M., Vuono R., Andersson Sjoland A., Westergren-Thorsson G. REST suppression mediates neural conversion of adult human fibroblasts via microRNA-dependent and -independent pathways. EMBO Mol. Med. 2017;9:1117–1131. doi: 10.15252/emmm.201607471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin-Ouellet J., Pircs K., Barker R.A., Jakobsson J., Parmar M. Direct neuronal reprogramming for disease modeling studies using patient-derived neurons: what have we learned? Front Neurosci. 2017;11:530. doi: 10.3389/fnins.2017.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., El Wazan L., Tan C., Nguyen T., Hung S.S.C., Hewitt A.W., Wong R.C.B. Potentials of cellular reprogramming as a novel strategy for neuroregeneration. Front. Cell. Neurosci. 2018;12:460. doi: 10.3389/fncel.2018.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner C.C., Brugg B., Matus A. A 70-kilodalton microtubule-associated protein (MAP2c), related to MAP2. J. Neurochem. 1988;50:609–615. doi: 10.1111/j.1471-4159.1988.tb02954.x. [DOI] [PubMed] [Google Scholar]
- Georgievska B., Jakobsson J., Persson E., Ericson C., Kirik D., Lundberg C. Regulated delivery of glial cell line-derived neurotrophic factor into rat striatum, using a tetracycline-dependent lentiviral vector. Hum. Gene Ther. 2004;15:934–944. doi: 10.1089/hum.2004.15.934. [DOI] [PubMed] [Google Scholar]
- Grealish S., Drouin-Ouellet J., Parmar M. Brain repair and reprogramming: the route to clinical translation. J. Intern. Med. 2016;280:265–275. doi: 10.1111/joim.12475. [DOI] [PubMed] [Google Scholar]
- Guo Z., Zhang L., Wu Z., Chen Y., Wang F., Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Bergami M., Gascon S., Lepier A., Vigano F., Dimou L., Sutor B., Berninger B., Gotz M. Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem Cell Reports. 2014;3:1000–1014. doi: 10.1016/j.stemcr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes E.G., Kang S.H., Fukaya M., Bergles D.E. Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat. Neurosci. 2013;16:668–676. doi: 10.1038/nn.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski I., Zecevic N. Sequence of oligodendrocyte development in the human fetal telencephalon. Glia. 2005;49:480–491. doi: 10.1002/glia.20134. [DOI] [PubMed] [Google Scholar]
- Karow M., Sanchez R., Schichor C., Masserdotti G., Ortega F., Heinrich C., Gascon S., Khan M.A., Lie D.C., Dellavalle A. Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell. 2012;11:471–476. doi: 10.1016/j.stem.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Kohyama J., Sanosaka T., Tokunaga A., Takatsuka E., Tsujimura K., Okano H., Nakashima K. BMP-induced REST regulates the establishment and maintenance of astrocytic identity. J. Cell Biol. 2010;189:159–170. doi: 10.1083/jcb.200908048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Shi Y., Yao X., Wang X., Shen L., Rao Z., Yuan J., Liu Y., Zhou Z., Zhang Z. Conversion of astrocytes and fibroblasts into functional noradrenergic neurons. Cell Rep. 2019;28:682–697.e7. doi: 10.1016/j.celrep.2019.06.042. [DOI] [PubMed] [Google Scholar]
- Liu M.L., Zang T., Zou Y., Chang J.C., Gibson J.R., Huber K.M., Zhang C.L. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Osipovitch M., Benraiss A., Huynh N.P.T., Foti R., Bates J., Chandler-Militello D., Findling R.L., Tesar P.J., Nedergaard M. Dysregulated glial differentiation in schizophrenia may be relieved by suppression of SMAD4- and REST-dependent signaling. Cell Rep. 2019;27:3832–3843.e6. doi: 10.1016/j.celrep.2019.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masserdotti G., Gascon S., Gotz M. Direct neuronal reprogramming: learning from and for development. Development. 2016;143:2494–2510. doi: 10.1242/dev.092163. [DOI] [PubMed] [Google Scholar]
- Mattugini N., Bocchi R., Scheuss V., Russo G.L., Torper O., Lao C.L., Gotz M. Inducing different neuronal subtypes from astrocytes in the injured mouse cerebral cortex. Neuron. 2019;103:1086–1095.e5. doi: 10.1016/j.neuron.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J., Reid D., Lau S., Kim Y., Gage F.H. Aging in a dish: iPSC-derived and directly induced neurons for studying brain aging and age-related neurodegenerative diseases. Annu. Rev. Genet. 2018;52:271–293. doi: 10.1146/annurev-genet-120417-031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W., Zang T., Smith D.K., Vue T.Y., Zou Y., Bachoo R., Johnson J.E., Zhang C.L. SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem Cell Reports. 2015;4:780–794. doi: 10.1016/j.stemcr.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira M., Birtele M., Shrigley S., Benitez J.A., Hedlund E., Parmar M., Ottosson D.R. Direct reprogramming of resident NG2 glia into neurons with properties of fast-spiking parvalbumin-containing interneurons. Stem Cell Reports. 2017;9:742–751. doi: 10.1016/j.stemcr.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira M., Pfisterer U., Rylander D., Torper O., Lau S., Lundblad M., Grealish S., Parmar M. Highly efficient generation of induced neurons from human fibroblasts that survive transplantation into the adult rat brain. Sci. Rep. 2014;4:6330. doi: 10.1038/srep06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A., Bjorklund A., Lindvall O., Jakobsson J., Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Kang X., Hu J., Zhang D., Liang Z., Meng F., Zhang X., Xue Y., Maimon R., Dowdy S.F. Reversing a model of Parkinson's disease with in situ converted nigral neurons. Nature. 2020;582:550–556. doi: 10.1038/s41586-020-2388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner M., Victor M.B., Liu Y., Abernathy D., Yoo A.S. MicroRNA-based conversion of human fibroblasts into striatal medium spiny neurons. Nat. Protoc. 2015;10:1543–1555. doi: 10.1038/nprot.2015.102. [DOI] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P., Romanov R.A., Spigolon G., Masini D., Martin-Montanez E., Toledo E.M., La Manno G., Feyder M., Pifl C., Ng Y.H. Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson's disease model. Nat. Biotechnol. 2017;35:444–452. doi: 10.1038/nbt.3835. [DOI] [PubMed] [Google Scholar]
- Rowitch D.H., Kriegstein A.R. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Schweitzer J.S., Song B., Herrington T.M., Park T.Y., Lee N., Ko S., Jeon J., Cha Y., Kim K., Li Q. Personalized iPSC-derived dopamine progenitor cells for Parkinson's disease. N. Engl. J. Med. 2020;382:1926–1932. doi: 10.1056/NEJMoa1915872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim F.J., McClain C.R., Schanz S.J., Protack T.L., Windrem M.S., Goldman S.A. CD140a identifies a population of highly myelinogenic, migration-competent and efficiently engrafting human oligodendrocyte progenitor cells. Nat. Biotechnol. 2011;29:934–941. doi: 10.1038/nbt.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C., Gotz M., Dimou L. Progenitors in the adult cerebral cortex: cell cycle properties and regulation by physiological stimuli and injury. Glia. 2011;59:869–881. doi: 10.1002/glia.21156. [DOI] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torper O., Ottosson D.R., Pereira M., Lau S., Cardoso T., Grealish S., Parmar M. In vivo reprogramming of striatal NG2 glia into functional neurons that integrate into local host circuitry. Cell Rep. 2015;12:474–481. doi: 10.1016/j.celrep.2015.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torper O., Pfisterer U., Wolf D.A., Pereira M., Lau S., Jakobsson J., Bjorklund A., Grealish S., Parmar M. Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. U S A. 2013;110:7038–7043. doi: 10.1073/pnas.1303829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor M.B., Richner M., Hermanstyne T.O., Ransdell J.L., Sobieski C., Deng P.Y., Klyachko V.A., Nerbonne J.M., Yoo A.S. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84:311–323. doi: 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignoles R., Lentini C., d'Orange M., Heinrich C. Direct lineage reprogramming for brain repair: breakthroughs and challenges. Trends Mol. Med. 2019;25:897–914. doi: 10.1016/j.molmed.2019.06.006. [DOI] [PubMed] [Google Scholar]
- Vouyiouklis D.A., Brophy P.J. Microtubule-associated proteins in developing oligodendrocytes: transient expression of a MAP2c isoform in oligodendrocyte precursors. J. Neurosci. Res. 1995;42:803–817. doi: 10.1002/jnr.490420609. [DOI] [PubMed] [Google Scholar]
- Wang S., Bates J., Li X., Schanz S., Chandler-Militello D., Levine C., Maherali N., Studer L., Hochedlinger K., Windrem M. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12:252–264. doi: 10.1016/j.stem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.