Abstract
High temperature during grain filling considerably reduces yield and quality in rice (Oryza sativa L.); however, how high temperature affects seed germination of the next generation is not yet well understood. Here, we report that seeds from plants exposed to high temperature during the grain filling stage germinated significantly later than seeds from unstressed plants. This delay remained even after dormancy release treatments, suggesting that it was not due to primary seed dormancy determined during grain filling. In imbibed embryos of heat-stressed seeds, expression of abscisic acid (ABA) biosynthesis genes (OsNCEDs) was higher than in those of control seeds, whereas that of ABA catabolism genes (OsABA8′OHs) was lower. In the aleurone layer, despite no change in GA signaling as evidenced by no effect of heat stress on OsGAMYB gene expression, the transcripts of α-amylase genes OsAmy1C, OsAmy3B, and OsAmy3E were significantly down-regulated in heat-stressed seeds in comparison with controls. Changes in promoter methylation levels were consistent with transcriptional changes of ABA catabolism-related and α-amylase genes. These data suggest that high temperature during grain filling results in DNA methylation of ABA catabolism-related and α-amylase gene promoters, delaying germination of heat-stressed seeds.
Subject terms: Plant physiology, Heat
Introduction
Environmental stresses, such as temperature, drought, salinity, and other abiotic stresses, strongly affect plant growth and development1. High temperature is one of the main environmental factors that cannot be avoided and cause losses in agricultural production worldwide. Ambient temperature strongly influences plant growth and development during both vegetative and reproductive stages2. Heat stress can shorten the period of grain development, resulting in insufficient grain filling and yield reduction in cereals3,4. Rice (Oryza sativa L.) is one of the most important crops for global food consumption, especially in Asia. It has been predicted that every 1 °C increase in average temperature would lead to about 10% reduction in rice yield5. Rice exposed to high temperature during grain filling develops chalky appearance, which reduces grain quality6–9. However, the effect of heat stress during grain filling on offspring growth and development remains to be elucidated.
Seed germination is the beginning of the plant’s life cycle. Seed dormancy and germination are strongly related to each other and are regulated by phytohormones, especially gibberellic acid (GA) and abscisic acid (ABA)10. After imbibition, GA biosynthesis and ABA catabolism are up-regulated to promote seed germination11,12. The transcription factor GAMYB induces α-amylase gene expression in a GA-dependent manner in the aleurone layers of barley, wheat, rice, and other cereals13,14. In response to GA from the embryo, GAMYB expression is induced and the encoded protein binds to the GARE boxes in the α-amylase gene promoters to induce α-amylase expression for starch degradation to fuel seed germination14,15. Environmental factors during seed development, especially temperature, strongly influence the level of primary dormancy16. A failure of a viable seed to germinate under favorable conditions is known as seed dormancy, which is controlled by several environmental factors such as light, temperature, and duration of seed storage17,18. In some barley (Hordeum vulgare L.) cultivars, moisture and low temperature during seed maturation are related to a lack of seed dormancy, resulting in preharvest sprouting19. In oilseed rape (Brassica napus L.), heat stress during seed filling decreases seed dormancy20. During seed development and maturation of Arabidopsis and wheat, temperature variations profoundly affect seed performance and dormancy16,21. In rice, high temperature during early endosperm development primes seed germination and seedling growth22.
The appropriate level of seed dormancy is a desirable trait for production of important cereal crops; understanding of how temperature during seed maturation affects seed dormancy and germination is necessary for crop production. Many studies have proposed the impacts of temperature during imbibition on seed germination23–25, but the effects of temperature during seed maturation have not been studied. In this study, we subjected rice plants to moderate heat stress from post-anthesis to harvest and show that high temperature during grain filling significantly delayed seed germination. We investigated the hormonal and epigenetic regulation of this phenomenon.
Results
Heat stress during grain filling significantly delays seed germination regardless of primary dormancy release
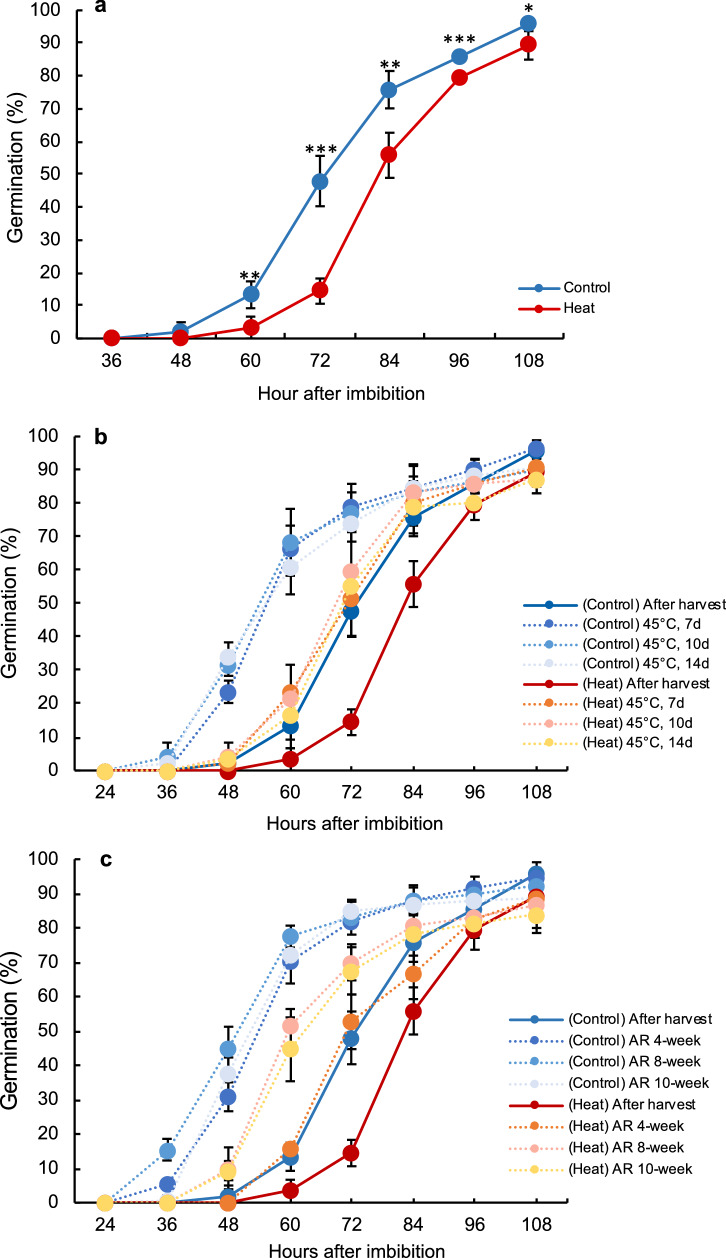
Seeds developed under heat stress germinated significantly later than control unstressed seeds (Fig. 1a). At 72 HAI (hours after imbibition), 48% of control seeds but only 14% of heat-stressed seeds germinated. Heat stress during grain filling did not affect seed viability, because germination rates of control and heat-stressed seeds at 108 HAI were similar. Similar delay was also observed in the other two cultivation years (Supplemental Fig. 1) and in other rice cultivars (Supplemental Fig. 2). To clarify whether this phenomenon was due to primary seed dormancy induced during grain development under stress, we subjected control and heat-stressed seeds to dormancy break treatment before germination test at 27 °C. Dormancy break treatment accelerated germination of both control and heat-stressed seeds (Fig. 1b). Although seed dormancy was released, control seeds still germinated faster than heat-stressed seeds. After-ripening treatment (26 °C) for 4, 8, or 10 weeks showed similar results (Fig. 1c). We calculated the average time required to reach 50% germination (T50) of seeds subjected or not to dormancy break or after-ripening treatment (Table 1). In both treatments, increasing treatment duration reduced T50 values, but T50 reached a plateau at 5 days (dormancy break) or 8 weeks (after-ripening treatment). Compared with after-harvest seeds, those subjected to dormancy-break or after-ripening treatment had lower T50 values regardless of heat stress; however, heat-stressed seeds still germinated significantly later in both treatments, as evidenced by significantly higher T50 values of heat-stressed seeds (Table 1). These results suggest that the delay in germination of heat-stressed seeds was not caused by primary dormancy.
Figure 1.
Delay in germination of seeds subjected to heat stress during grain filling stage. Germination rates of (a) freshly harvested seeds; (b) seeds subjected to 45 °C dormancy break treatments for 7, 10, or 14 days; and (c) seeds subjected to 26 °C after-ripening (AR) treatments for 4, 8, or 10 weeks. Significant differences are shown as P < 0.05*, P < 0.01**, and P < 0.001***, according to Student’s t-test (n = 3). Error bars, SD.
Table 1.
T50 values of after-harvest seeds and seeds subjected to dormancy-break or after-ripening treatment.
| Control | Heat | Student’s t-test | |
|---|---|---|---|
| Dormancy break | |||
| After-harvest | 71.28 ± 3.34 a | 79.72 ± 2.51 a | ** |
| 45 °C, 3 days | 61.17 ± 2.58 b | 73.70 ± 3.33 ab | *** |
| 45 °C, 5 days | 56.70 ± 6.03 bc | 69.67 ± 3.80 bc | ** |
| 45 °C, 7 days | 57.86 ± 3.87 bc | 67.42 ± 4.43 bc | ** |
| 45 °C, 10 days | 51.22 ± 5.29 c | 64.27 ± 8.25 c | * |
| 45 °C, 14 days | 55.74 ± 3.92 bc | 69.20 ± 3.28 bc | *** |
| After-ripening | |||
| After-harvest | 71.28 ± 3.34 a | 79.72 ± 2.51 a | ** |
| 26 °C, 2-week | 54.70 ± 2.96 b | 68.27 ± 8.53 b | * |
| 26 °C, 4-week | 52.02 ± 2.52 b | 69.04 ± 1.59 b | *** |
| 26 °C, 6-week | 52.35 ± 4.85 b | 60.95 ± 2.97 c | * |
| 26 °C, 8-week | 48.79 ± 2.19 b | 57.83 ± 1.27 c | *** |
| 26 °C, 10-week | 50.10 ± 3.92 b | 58.57 ± 3.58 c | ** |
All seeds were germinated at 27 °C (n = 5 per condition).Within each column, values followed by different letters are significantly different (P < 0.05). Within each row, the significance of differences between control and heat-stressed seeds is shown according to Student’s t-test (P < 0.05*, P < 0.01**, and P < 0.001***). SD values are shown after T50 values.
Changes in embryo ABA-metabolic gene expression and endogenous ABA content during seed germination
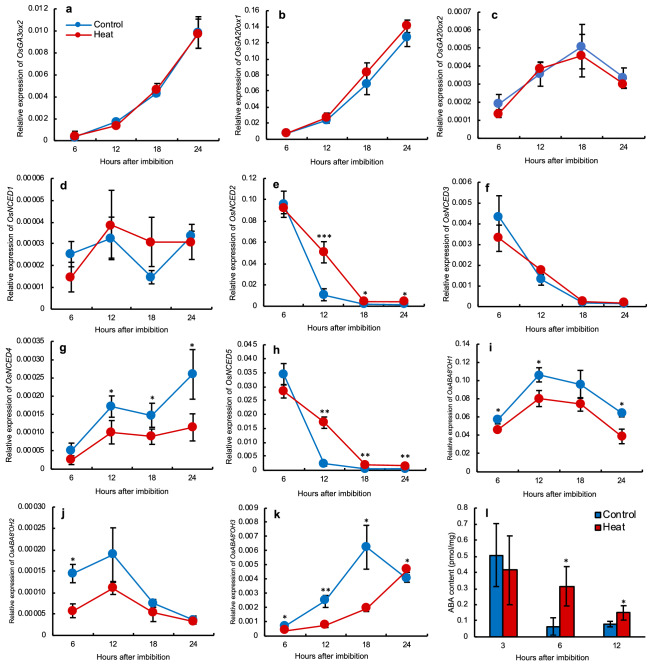
We examined the transcript levels of genes involved in the metabolism of GA and ABA, two major hormones regulating rice seed germination. In embryos, the expression of three GA biosynthesis genes, OsGA3ox2, OsGA20ox1, and OsGA20ox2, did not differ significantly between control and heat-stressed seeds (Fig. 2a–c), suggesting that GA biosynthesis has less effect on delayed germination. Among ABA biosynthesis genes, the expression of OsNCED1 fluctuated during germination, with no significant difference between treatments (Fig. 2d). The expression of OsNCED4 increased after imbibition and was higher in the control than in heat-stressed embryos (Fig. 2g). The expression of OsNCED2, OsNCED3, and OsNCED5 dropped dramatically upon imbibition from 6 to 18 HAI, suggesting their roles in seed germination (Fig. 2e,f,h). The expression of OsNCED2 was similar in control and heat-stressed embryos at 6 HAI, but was significantly higher in heat-stressed embryos at the later time points (4.7, 2.4 and 3.0-fold at 12, 18 and 24 HAI, respectively; Fig. 2e). Similarly, OsNCED5 expression was significantly higher in heat-stressed than in control embryos (7.6, 3.3 and 2.1-fold at 12, 18 and 24 HAI, respectively; Fig. 2h).
Figure 2.
Relative expression of GA and ABA metabolism-related genes in imbibed embryos and endogenous ABA content. (a) OsGA3ox2; (b) OsGA20ox1; (c) OsGA20ox2 (d) OsNCED1; (e) OsNCED2; (f) OsNCED3; (g) OsNCED4; (h) OsNCED5; (i) OsABA8′OH1; (j) OsABA8′OH2; (k) OsABA8′OH3; (l) endogenous ABA contents in embryos during imbibition. Significant differences are shown as P < 0.05*, P < 0.01**, and P < 0.001***, according to Student’s t-test (n = 3). Error bars, SD.
We also analysed transcriptional expression of three ABA catabolism genes. Upon imbibition, OsABA8′OH1 expression was down-regulated in heat-stressed embryos, and the difference from the control was significant at 6, 12, and 24 HAI (Fig. 2i). Significant down-regulation of expression in heat-stressed embryos was also observed at 6 HAI for OsABA8′OH2 (Fig. 2j), and from 6 to 18 HAI for OsABA8′OH3 (Fig. 2k). As a result of higher expression of ABA biosynthesis genes and lower expression of ABA catabolism genes during imbibition, endogenous ABA content was significantly higher in heat-stressed in embryos at 6 and 12 HAI (Fig. 2l). Overall, these data suggest that higher endogenous ABA content in the embryo after imbibition was involved in delayed germination of heat-stressed seeds.
Down-regulation of α-amylase gene expression in aleurone cells in response to exogenous GA
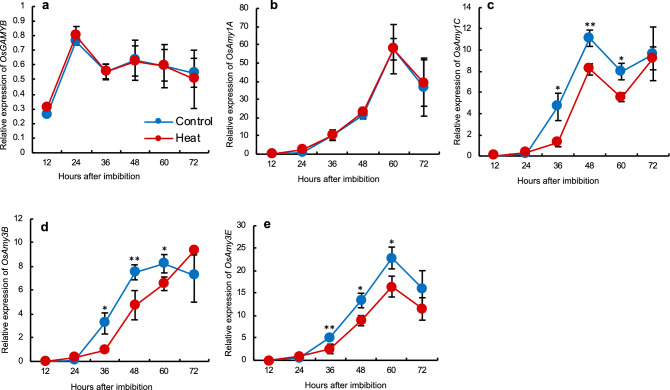
We also analysed transcriptional changes in aleurone cells during seed imbibition in response to GA. The expression of OsGAMYB, a GA-responsive transcription factor, in embryoless half-seeds imbibed with 1 μM GA changed similarly with time in control and heat-stressed seeds, with a peak at 24 HAI regardless of heat treatment (Fig. 3a). We measured the expression levels of OsAmy1A, OsAmy1C, OsAmy3B, and OsAmy3E because theses α-amylase genes are highly expressed in rice endosperm after seed imbibition41. OsAmy1A expression increased gradually, with the same pattern in control and heat-stressed seeds (Fig. 3b). The expression of OsAmy1C, OsAmy3B, and OsAmy3E was significantly lower in heat-stressed than in control seeds at 36–60 HAI (Fig. 3c–e), i.e., later than the GAMYB peak. These data suggest that down-regulated α-amylase gene expression in the aleurone layer contributed to the delayed germination of heat-stressed seeds.
Figure 3.
Expression of GAMYB transcription factor and α-amylase genes in embryoless imbibed seeds treated with exogenous 1 μM GA. (a) OsGAMYB; (b) OsAmy1A; (c) OsAmy1C; (d) OsAmy3B; (e) OsAmy3E. Significant differences are shown as P < 0.05* and P < 0.01** according to Student’s t-test (n = 3). Error bars, SD.
DNA methylation at the promoters of ABA catabolism-related and α-amylase genes in heat-stressed seeds
To test whether epigenetic changes affect the expression of the above genes, we analysed their promoters for possible DNA methylation regions. We found predicted CpG islands in the promoters of the ABA metabolism genes OsNCED5, OsABA8′OH1, and OsABA8′OH3, and of the α-amylase genes OsAmy1C, OsAmy3B, and OsAmy3E, but not in those of OsNCED2, OsABA8′OH2, or OsAmy1A. Relative methylation levels in the CG-context in dry seeds are shown in Fig. 4. The methylation level at OsABA8′OH1:proP4, the predicted region nearest to the transcription start site (Fig. 4a), in heat-stressed seeds was 3.8 times higher than that in control seeds (Fig. 4b). Methylation of the OsABA8′OH3:proP1 region was also significantly higher in heat-stressed than in control seeds, but that of the OsNCED5:proP1 region was lower but not significant (Fig. 4b). OsAmy1C:proP1 and OsAmy3B: proP1 regions showed significant hyper-methylation (1.3 and 2.5-fold, respectively) in heat-stressed seeds in comparison with the control (Fig. 5). Hyper-methylations of OsABA8′OH1:proP4, OsABA8′OH3:proP1, OsAmy1C:proP1 and OsAmy3B:proP1 promoters were also observed by using MeDIP-qPCR identifying methylation levels, suggesting the similar results (Supplemental Fig. 5). The methylation levels of OsAmy3B:proP2 and OsAmy3E:proP1 were not affected by heat treatment. Thus, heat stress significantly increased methylation levels in promoters of some genes.
Figure 4.
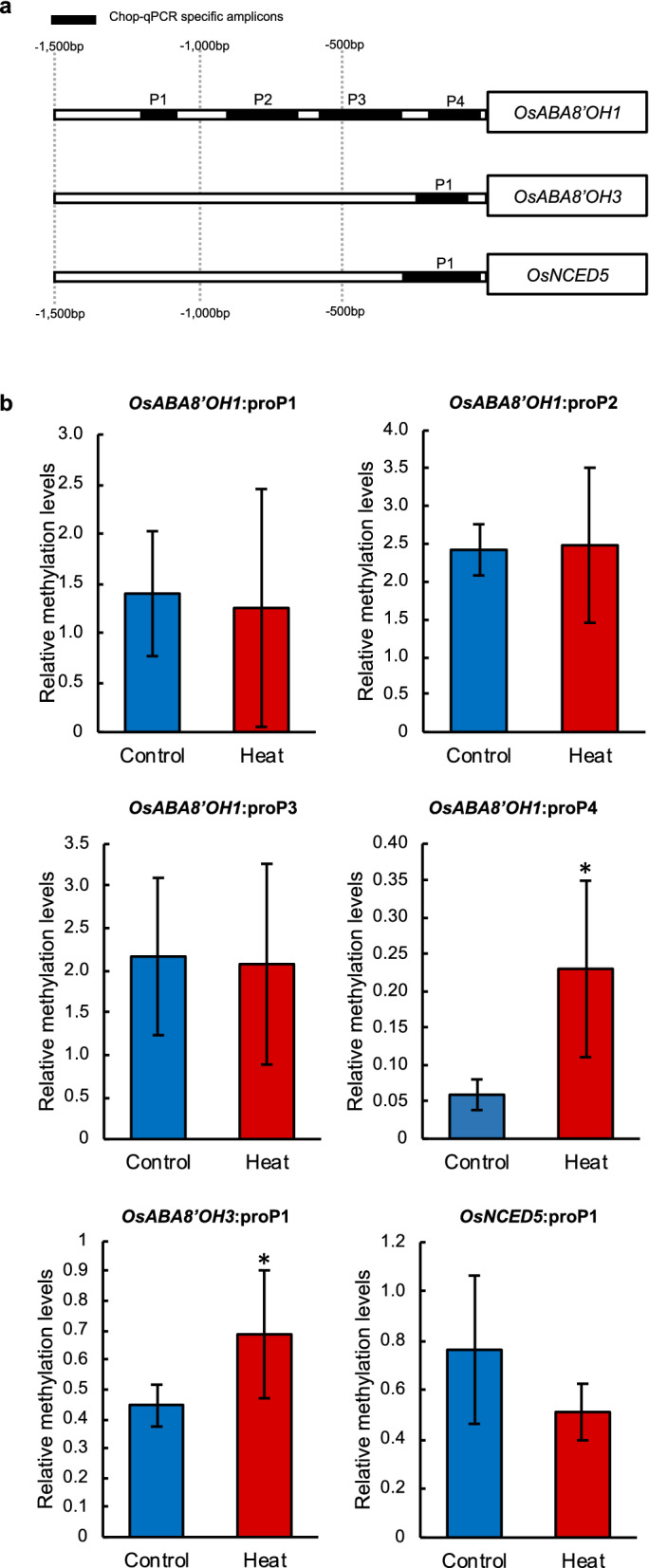
Relative methylation levels of OsABA8′OH1, OsABA8′OH3 and OsNCED5 promoter regions measured by Chop-qPCR. (a) Map of PCR amplicons of CpG islands predicted by MethPrimer. (b) Relative DNA methylation levels of OsABA8′OH1, OsABA8′OH3, and OsNCED5 promoter regions in after-harvest control and heat-stressed seeds. Significant differences are shown as P < 0.05* according to Student’s t-test (n = 6).
Figure 5.
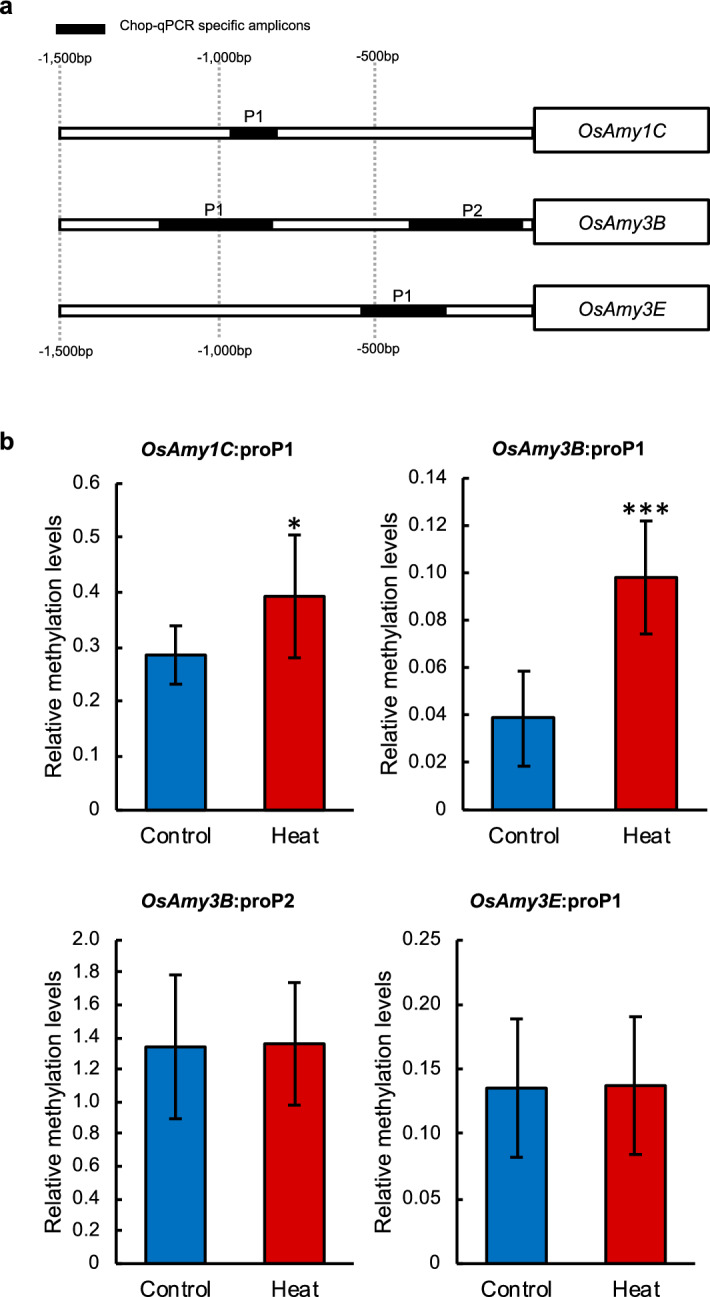
Relative methylation levels of α-amylase promoter regions measured by Chop-qPCR. (a) Map of PCR amplicons of CpG islands predicted by MethPrimer. (b) Relative DNA methylation levels of OsAmy1C, OsAmy3B, and OsAmy3E promoter regions in dry control and heat-stressed seeds. Significant differences are shown as P < 0.05* and P < 0.001*** according to Student’s t-test (n = 6). Error bars, SD.
Discussion
We found that heat stress during grain filling significantly delayed seed germination, but that this phenomenon was not due to primary dormancy. Germination of the seeds of Nipponbare and the other rice cultivars tested (one heat stress-sensitive and five tolerant) was delayed to different extents by heat stress during grain filling. Under heat stress, sensitive cultivars develop more grain chalkiness than do tolerant cultivars8. Therefore, this phenomenon is not cultivar specific and is not associated with the level of grain chalkiness under heat stress.
High temperature during maturation increases rice seed dormancy26,27. After harvest rice seed dormancy is broken by high temperature28,29, and after-ripening30,31 are commonly used to release seed dormancy. In this study, the dormancy levels of both control and heat-stressed seeds were significantly decreased after dormancy release by dormancy-break or after-ripening treatment. Yet heat-stressed seeds still germinated significantly slower than control seeds. Therefore, the delay in germination by exposure to high temperature during maturation is not due to primary dormancy. Delayed of germination1 (DOG1), is a well-known master key controlling primary dormancy through maternal environmental factors during seed maturation32. Although of some rice japonica-type DOG1-like genes33 were up-regulated during grain filling stage under heat stress (Supplemental Fig. 3a), a previous study in Arabidopsis thaliana shows that after-ripening treatment changes the protein structure of DOG1 and abolishes its activity34. Also, OsDOG1-like gene expression in imbibed embryos was not strongly induced or inhibited by heat stress during grain filling (Supplemental Fig. 3b). Taking together, primary dormancy and DOG1 were not involved in delayed germination of heat stressed seeds.
During seed imbibition, GA content increases14 and ABA content drops dramatically35 to allow germination. In imbibed embryos, the expression of the GA biosynthesis genes OsGA3ox2, OsGA20ox1, and OsGA20ox2 gradually increased toward germination. OsGA3ox2 is highly expressed during seed germination to induce the expression of α-amylase, whereas the expression of its homolog, OsGA3ox1, is lower14. OsGA20ox1 and OsGA20ox2 are involved in GA biosynthesis during seed germination36,37. In this study, no difference in GA biosynthesis gene expression was observed, but higher expression of ABA biosynthesis genes and lower expression of ABA catabolism genes were observed in heat-stressed seeds. OsNCED2 and OsABA8′OH3 play predominant roles in ABA metabolism in imbibed rice seeds38. The changes in the transcription of these genes in embryos during imbibition resulted in a significantly higher ABA content in heat-stressed seeds. At the very early phase of imbibition, control and heat-stressed seeds had the same ABA levels. However, the ABA levels were significantly higher in heat-stressed embryos at later time points. GAMYB is a GA-responsive transcription factor essential for α-amylase induction in the aleurone39. A GARE box in the α-amylase promoter is essential for direct binding of GAMYB and for transcriptional induction40. We showed that the OsGAMYB gene expression level was indistinguishable between control and heat-stressed seeds, indicating that the response to exogenous was not affected by heat stress during grain filling. Among eight rice α-amylase isozymes, promoters of genes encoding OsAmy1A, OsAmy1C, OsAmy3B, and OsAmy3E contain GARE boxes41. The expression of α-amylase genes started to increase after the GAMYB expression had peaked. Among the α-amylase genes examined, OsAmy1A was particularly highly induced by GA in the aleurone. OsAmy1A, the most predominant GA responsive gene, showed no difference between the two treatments, while OsAmy1C, OsAmy3B, and OsAmy3E expression was significantly lower in heat-stressed than in control seeds at 36–60 HAI. This might result in non-drastic delayed seed germination. These observations suggest that changes in the expression of ABA metabolism genes in the embryo and of α-amylase genes in the aleurone layer after imbibition delayed germination of heat-stressed seeds.
DNA methylation is a well-known epigenetic response for gene regulation, including silencing42. A recent study has shown that DNA methylation in the promoter of the gene for ALLANTOINASE, a negative regulator of dormancy, is stimulated by cold experienced by the mother plant and is passed to the seeds43, suggesting that DNA methylation in the mother plant is important for seed germination. We found that OsABA8′OH1 and OsABA8′OH3 promoters were highly methylated and the OsNCED5 promoter tended to be hypo-methylated in heat-stressed seeds, which corresponded to the lower expression of the former two genes and higher expression of OsNCED5. Promoter analysis revealed that only OsAmy1A contained no predicted CpG island for DNA methylation, at least within − 1500 bp, whereas OsAmy1C and OsAmy3B promoters were highly methylated in heat-stressed seeds and their methylation levels corresponded to down-regulation of gene expression. The presence of CpG islands and hyper-methylation in α-amylase promoters play a role in heat stress-induced transcriptional regulation during seed imbibition. In conclusion, we propose that heat stress during grain filling changes the expression of genes involved in ABA metabolism and α-amylase genes during imbibition via DNA methylation of the respective gene promoters, delaying seed germination.
Other factors such as histone modifications are involved in epigenetic regulation. In Arabidopsis, the HUB1 (HISTONE MONOUBIQUITINATION 1) gene is involved in H2B monoubiquitination, which regulates the methylation of this histone44, and the hub1 mutant shows reduced expression of NCED9 and ABI4 (ABSCISIC ACID INSENSITIVE4) but increased expression of ABA8′OH245. In Arabidopsis, acetylation of H3K9K19 also regulates ABA8′OH expression and seed dormancy during maturation46. In rice, H3K9 deacetylation by OsSRT1 (Sirtuin1) mediates starch metabolism genes such as OsAmy3B, OsAmy3E, OsBmy4 and OsBmy947.
These data suggest that other epigenetic changes together with DNA methylation might regulate germination in heat-stressed seeds.
Materials and methods
Plant materials and growth conditions
Three-week-old seedlings of rice (Oryza sativa L.) ‘Nipponbare’ were transplanted into 1/5000-a Wagner pots (10 plants per pot) with 8.75 g of basal dressing compound fertilizer (N–P2O–K2O: 4%–4%–4%) and 0.85 g of sigmoid-type controlled-release coated urea in 2015. In addition, 0.5 g of ammonium sulfate (N: 21%) was applied twice, at the tiller development stage and at the panicle booting stage. All fully developed tillers were removed and only the main stems were used in this study. Plants were grown under natural conditions at Kyushu University (33°67′N, 130°42′E) until spikelets located on the upper primary rachis branches flowered in more than 50% of the whole population. That day was set as the day of flowering (0 days after flowering: DAF) and the plants were transferred into two different temperature regimes (25 °C, control; or 30 °C, heat treatment) in a growth chamber with natural light until harvest at 42 DAF. Harvested seeds were dried at room temperature for 1 week and stored at − 0 °C to maintain seed dormancy. Same cultivation methods were also repeated in 2016 and 2017. In some experiments, seeds were incubated at 45 °C (dormancy-break treatment) for 3, 5, 7, 10 and 14 days, or at 26 °C (after-ripening treatment) for 2, 4, 6, 8, 10 weeks in the dark prior to germination tests.
Germination test
Seeds were rested at room temperature for 1 h and sterilized with 0.2% NaClO for 20 min. Seeds were allowed to germinate on filter paper in 9-cm Petri dishes filled with 10 mL distilled water at 27 °C in the dark. One Petri dish contained 30 seeds (one replication). Germination rates (emergence of 1-mm shoots) were checked every 12 h. The T50 (the average time to reach 50% germination) of each sample was calculated48.
RNA extraction and quantitative real-time PCR
Seeds were germinated as above and embryos from each treatment group were sampled at 6, 12, 18, or 24 h after imbibition (HAI). Embryoless half-seeds were placed upside down in Petri dishes with filter paper soaked with 6 mL of 1 μM GA and were sampled at 12, 24, 36, 48, 60, and 72 HAI. Total RNA from embryos and embryoless half-seeds was extracted from frozen materials using the SDS/phenol/LiCl method49. cDNA was synthesised from extracted RNA using ReverTra Ace reverse transcriptase (Toyobo) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using a MiniOpticon real-time PCR detection system (Bio-Rad) with SYBR Green (Toyobo) as described in the manufacturers’ instructions. Primers used for qRT-PCR are listed in Table S1. PCR thermal cycling conditions were as follows: initial denaturation, 94 °C for 2 min; 40 cycles of denaturation 94 °C for 20 s, annealing at primer-specific temperature for 20 s, and extension at 72 °C for 20 s; followed by melting and plate reading. The results were normalized to the expression level of the OsActin gene.
Endogenous ABA content in embryos
Embryos isolated from 30 imbibed seeds (1 replicate; 30–50 mg) at 3, 6, and 12 HAI were crushed thoroughly with a pestle in a mortar with 2 mL 80% methanol and centrifuged at 10,000 rpm at 4 °C for 20 min. Supernatants were evaporated overnight. ABA content was measured as absorbance at 405 nm using a Phytodek competitive ELISA kit (Agdia) according to the manufacturer’s instructions.
DNA methylation analysis by Chop-qPCR
Genomic DNA from 0.1 g dry seeds was extracted using a DNeasy Plant Maxi Kit (Qiagen) as described in the manufacturer’s instructions. Genomic DNA (0.5 μg) was digested (total reaction volume, 20 μL) with the methylation-sensitive restriction enzyme50 HpaII (New England Biolabs) at 37 °C for 1 h, followed by enzyme inactivation at 80 °C for 20 min. Digested DNA (25 ng) was subjected to qRT-PCR using SYBR Green (Toyobo). Relative methylation levels were calculated from ΔCt values and were standardized to undigested control DNA50. Specific primer sets (listed in Supplemental Table 1) were designed for putative methylation regions predicted in MethPrimer software (https://www.urogene.org/methprimer/).
DNA methylation analysis by MeDIP-qPCR
Genomic DNA from 40 dry seeds per sample was extracted using DNeasy Plant Kit (Qiagen) and was sheared to about 500 bp by sonication. Sheared DNA was immunoprecipitated using MagMeDIP Methylated DNA Immunoprecipitation Kit as described by the manufacturer’s protocol and a previous study applied to plant samples51. Immunoprecipitated DNA was subjected to qRT-PCR using SYBR using the same specific primers as Chop-qPCR. Percent recovery was calculated as described in manufacturer’s protocol (% recovery = 2^[Ct(10% input) − 3.32 − Ct(IP sample)] × 100).
All data generated or analysed during this study are included in this published article and its “Supplementary information” files.
Supplementary information
Acknowledgements
This work was supported by JSPS KAKENHI Grant numbers 15K14639, 18J20784, 20H02971 and Takahashi Industrial and Economic Research Foundation.
Author contributions
C.S., M.I. and Y.I. designed the experiments; C.S., Y.O., N.H. and Y.I. performed the experiments; C.S., T.I., K.M. and S.Y. performed plant hormone measurement. N.H. and Y.I. performed data analysis; C.S. and Y.I. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-74281-9.
References
- 1.Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005;16:123–132. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhury SI, Wardlaw IF. The effect of temperature on kernel development in cereals. Aust. J. Agric. Res. 1978;29:205–223. doi: 10.1071/AR9780205. [DOI] [Google Scholar]
- 3.Wardlaw IF. Interaction between drought and chronic high temperature during kernel filling in wheat in a controlled environment. Ann. Bot. 2002;90:469–476. doi: 10.1093/aob/mcf219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altenbach SB, et al. Temperature, water and fertilizer influence the timing of key events during grain development in a US spring wheat. J. Cereal Sci. 2003;37:9–20. doi: 10.1006/jcrs.2002.0483. [DOI] [Google Scholar]
- 5.Peng S, et al. Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA. 2004;101:9971–9975. doi: 10.1073/pnas.0403720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamakawa H, Hirose T, Kuroda M, Yamaguchi T. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 2007;144:258–277. doi: 10.1104/pp.107.098665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakata M, et al. Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol. J. 2012;10:1110–1117. doi: 10.1111/j.1467-7652.2012.00741.x. [DOI] [PubMed] [Google Scholar]
- 8.Tanamachi K, et al. Differential responses to high temperature during maturation in heat-stress-tolerant cultivars of Japonica rice. Plant Prod. Sci. 2016;19:300–308. doi: 10.1080/1343943X.2016.1140007. [DOI] [Google Scholar]
- 9.Suriyasak C, et al. Reactive oxygen species induced by heat stress during grain filling of rice (Oryza sativa L.) are involved in occurrence of grain chalkiness. J. Plant Physiol. 2017;216:52–57. doi: 10.1016/j.jplph.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Hilhorst HWM, Karssen CM. Seed dormancy and germination: The role of abscisic acid and gibberellins and the importance of hormone mutants. Plant Growth Regul. 1992;11:225–238. doi: 10.1007/BF00024561. [DOI] [Google Scholar]
- 11.Liu Y, Ye N, Liu R, Chen M, Zhang J. H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 2010;61:2979–2990. doi: 10.1093/jxb/erq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishibashi Y, et al. A role for reactive oxygen species produced by NADPH oxidases in the embryo and aleurone cells in barley seed germination. PLoS ONE. 2015;10:1–17. doi: 10.1371/journal.pone.0143173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubler F, et al. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 1999;17:1–9. doi: 10.1046/j.1365-313X.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko M, Itoh H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M. The alpha-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 2002;128:1264–1270. doi: 10.1104/pp.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodger FJ, Millar A, Murray F, Jacobsen JV, Gubler F. The role of GAMYB transcription factors in GA-regulated gene expression. J. Plant Growth Regul. 2003;22:176–184. doi: 10.1007/s00344-003-0025-8. [DOI] [Google Scholar]
- 16.He H, et al. Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis. J. Exp. Bot. 2014;65:6603–6615. doi: 10.1093/jxb/eru378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bewley J. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Curr. Opin. Plant Biol. 2002;5:33–36. doi: 10.1016/S1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 19.Gubler F, Millar AA, Jacobsen JV. Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 2005;8:183–187. doi: 10.1016/j.pbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Brunel-Muguet S, et al. Heat stress during seed filling interferes with sulfur restriction on grain composition and seed germination in oilseed rape (Brassica napus L.) Front. Plant Sci. 2015;6:213. doi: 10.3389/fpls.2015.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker-Simmons M, Sesing J. Temperature effects on embryonic abscisic acid levels during development of wheat grain dormancy. J. Plant Growth Regul. 1990;9:51–56. doi: 10.1007/BF02041941. [DOI] [Google Scholar]
- 22.Begcy K, Sandhu J, Walia H. Transient heat stress during early seed development primes germination and seedling establishment in rice. Front. Plant Sci. 2018;9:1–13. doi: 10.3389/fpls.2018.01768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonai T, et al. Abscisic acid in the thermoinhibition of lettuce seed germination and enhancement of its catabolism by gibberellin. J. Exp. Bot. 2004;55:111–118. doi: 10.1093/jxb/erh023. [DOI] [PubMed] [Google Scholar]
- 24.Dong T, Tong J, Xiao L, Cheng H, Song S. Nitrate, abscisic acid and gibberellin interactions on the thermoinhibition of lettuce seed germination. Plant Growth Regul. 2012;66:191–202. doi: 10.1007/s10725-011-9643-5. [DOI] [Google Scholar]
- 25.Huo H, Dahal P, Kunusoth K, McCallum CM, Bradford KJ. Expression of 9-cis-epoxycarotenoid dioxygenase4 is essential for thermoinhibition of lettuce seed germination but not for seed development or stress tolerance. Plant Cell. 2013;25:884–900. doi: 10.1105/tpc.112.108902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikehashi H. Induction and test of dormancy of rice seeds by temperature condition during maturation. Japan J. Breed. 1972;22:209–216. doi: 10.1270/jsbbs1951.22.209. [DOI] [Google Scholar]
- 27.Takahashi N. Effect of environmental factors during seed formation on pre-harvest sprouting. Cereal Res. Commun. 1980;8:175–183. [Google Scholar]
- 28.Doku GD, Glover MK, Glover EK, Dartey KPA. The effect of the use of temperature on the breakage of dormancy and the subsequent performance of rice (Oryza spp.) Int. J. Plant Sci. Ecol. 2016;2:1–9. [Google Scholar]
- 29.Naredo MEB, Juliano AB, Lu BR, De Guzman F, Jackson MT. Responses to seed dormancy-breaking treatments in rice species (Oryza L.) Seed Sci. Technol. 1998;26:675–689. [Google Scholar]
- 30.Du W, et al. Physiological characteristics and related gene expression of after-ripening on seed dormancy release in rice. Plant Biol. 2015;17:1156–1164. doi: 10.1111/plb.12371. [DOI] [PubMed] [Google Scholar]
- 31.Finch-Savage WE, Cadman CSC, Toorop PE, Lynn JR, Hilhorst HWM. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 2007;51:60–78. doi: 10.1111/j.1365-313X.2007.03118.x. [DOI] [PubMed] [Google Scholar]
- 32.Carrillo-Barral N, Rodríguez-Gacio MC, Matilla AJ. Delay of germination-1 (DOG1): A key to understanding seed dormancy. Plants. 2020;9:480. doi: 10.3390/plants9040480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashikawa I, Abe F, Nakamura S. DOG1-like genes in cereals: Investigation of their function by means of ectopic expression in Arabidopsis. Plant Sci. 2013;208:1–9. doi: 10.1016/j.plantsci.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 34.Nakabayashi K, et al. The time required for dormancy release in Arabidopsis is determined by delay of germination1 protein levels in freshly harvested seeds. Plant Cell. 2012;24:2826–2838. doi: 10.1105/tpc.112.100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millar AA, et al. Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 2006;45:942–954. doi: 10.1111/j.1365-313X.2006.02659.x. [DOI] [PubMed] [Google Scholar]
- 36.Guo X, et al. The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J. 2013;75:403–416. doi: 10.1111/tpj.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye H, et al. Map-based cloning of seed dormancy1–2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 2015;169:2152–2165. doi: 10.1104/pp.15.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu G, Ye N, Zhang J. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 2009;50:644–651. doi: 10.1093/pcp/pcp022. [DOI] [PubMed] [Google Scholar]
- 39.Washio K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 2003;133:850–863. doi: 10.1104/pp.103.027334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaneko M, et al. Loss-of-function mutations of the rice GAMYB gene impair the plant cell. Plant Cell. 2004;16:33–44. doi: 10.1105/tpc.017327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen PW. Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of -amylase genes. Plant Cell Online. 2006;18:2326–2340. doi: 10.1105/tpc.105.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huh JH, Bauer MJ, Hsieh TF, Fischer R. Endosperm gene imprinting and seed development. Curr. Opin. Genet. Dev. 2007;17:480–485. doi: 10.1016/j.gde.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwasaki M, Hyvarinen L, Piskurewicz U, Lopez-Molina L. Non-canonical RNA-directed DNA methylation participates in maternal and environmental control of seed dormancy. eLife. 2019;8:e37434. doi: 10.7554/eLife.37434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chinnusamy V, Gong Z, Zhu JK. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant Biol. 2008;50:1187–1195. doi: 10.1111/j.1744-7909.2008.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Koornneef M, Soppe WJJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell Online. 2007;19:433–444. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, et al. Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1. Nat. Commun. 2016;7:1–14. doi: 10.1038/ncomms13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Lu Y, Zhao Y, Zhou D. Plant Science OsSRT1 is involved in rice seed development through regulation of starch metabolism gene expression. Plant Sci. 2016;248:28–36. doi: 10.1016/j.plantsci.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 48.Farooq M, Basra SMA, Ahmad N, Hafeez K. Thermal hardening: A new seed vigor enhancement tool in rice. J. Integr. Plant Biol. 2005;47:187–193. doi: 10.1111/j.1744-7909.2005.00031.x. [DOI] [Google Scholar]
- 49.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H, et al. Protocol: A beginner’s guide to the analysis of RNA-directed DNA methylation in plants. Plant Methods. 2014;10:1–9. doi: 10.1186/1746-4811-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding CJ, Liang LX, Diao S, Su XH, Zhang BY. Genome-wide analysis of day/night DNA methylation differences in Populus nigra. PLoS ONE. 2018;13:1–14. doi: 10.1371/journal.pone.0190299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.