Abstract
Testicular germ cell tumors (TGCTs) are highly prevalent in young men aged 20–40 years and are one of the most common lethal solid tumors in men of this age. Due to the current unclear mechanism of tumor development, there is a lack of effective treatment, and therefore in-depth research of the molecular mechanism of the occurrence and development of TGCT and the search for suitable and effective therapeutic targets and molecular markers are of great significance for achieving effective treatment. METTL3 is a very important methylase, which has been implicated in the progression of many cancers, but the role of METTL3 in TGCT has not been fully elucidated. In this article, we found that METTL3 expression was significantly downregulated in TGCT tissues, and patients with low expression levels had lower overall survival and relapse-free survival rates. After overexpressing METTL3, cell proliferation, invasion, and migration ability significantly increased, while influencing the expression of epithelial–mesenchymal transition (EMT)-related proteins. In addition, we observed that the expression level of METTL3 positively correlated with molecular markers and infiltration level of CD8+ and CD4+ T cells and natural killer cells. In sum, our findings identified that METTL3 can be used as an independent prognostic marker in patients with TGCT. METTL3 participates in the proliferation, migration, and invasion of TGCT cells by regulating the expression of EMT-related genes and may also play a role in activating the tumor immune response in TGCT.
Keywords: testicular germ cell tumors (TGCT), prognosis, METTL3, migration, invasion
Introduction
Testicular germ cell tumors (TGCTs) are a class of tumors that occur in the seminiferous epithelium and are generally classified into two types, seminoma and nonseminoma, accounting for about 1% of all male solid tumors, and 98% of all testicular tumors1. TGCTs are highly prevalent in young men aged 20–40 years and are one of the most common lethal solid tumors in men of this age. Studies have shown that the incidence of this tumor in China is about 1 in 100,0002, with recent data showing that the incidence of TGCT has increased worldwide3.
At present, the clinical treatment plan for TGCT is mainly testicular resection, accompanied by chemotherapy and radiotherapy, but still nearly 15% of patients face the status of recurrence and metastasis, so the prognosis is poor4. Due to the current unclear mechanism of tumor development, there is a lack of effective treatment, and therefore in-depth research of the molecular mechanism of the occurrence and development of TGCT and the search for suitable and effective therapeutic targets and molecular markers are of great significance for achieving effective treatment.
RNA is an important genetic material in organisms, with more than 100 modifications. N6-adenylate methylation (m6A), as the most common modification of eukaryotic mRNA5, can reduce the processing time of mRNA precursors, accelerate the speed of mRNA transport, and nuclear release in cells, ultimately regulating gene expression6. Many enzymes involved in m6A modification have been identified, including demethylases, methylases, and methylation recognition enzymes. These enzyme abnormalities can cause a variety of diseases, including tumors7, neurological diseases8, and embryonic retardation9,10. METTL3 is a very important methylase, which forms a heterocomplex with METTL14, and together with WTAP and other factors (such as KIAA1429) modifies adenylate11,12. Many studies have found that METTL3 is dysregulated in liver cancer13, gastric cancer14, colorectal cancer15, and other tumors, and is closely related to the occurrence and development of tumors. However, the expression, function, and mechanism of METTL3 in TGCT have not been fully elucidated. Herein, we analyzed the expression of METTL3 in TGCT and its relationship with prognosis through The Cancer Genome Atlas (TCGA) data, conducting cell experiments to verify the role of METTL3 in TGCT.
Materials and Methods
Online Database Data Analysis
Gene expression profiling and interactive analyses (GEPIA; http://gepia.cancer-pku.cn/) database is an interactive web version tool developed by the research team of Peking University. It uses a standard data processing process to integrate RNA sequencing data of 9736 tumor tissues and 8587 normal samples in the TCGA and GTEx database16 and hence is useful for our statistical analysis. We used the differential expression analysis tool and survival analysis tool of GEPIA to analyze the expression of METTL3 in TGCT and its correlation with the overall survival of TGCT patients. In addition, this database was also used to analyze the correlation between METTL3 and the expression of ZEB1, Vimentin (VIM), N-cadherin (CDH2), CD4, CD8A, and CD56. The TGCT microarray data set (GSE3218) was obtained from the Gene Expression Omnibus (GEO) database and used to analyze the differential expression of METTL3 in TGCT patients17. The Kaplan–Meier plotter (http://kmplot.com/analysis/) tool was used to analyze the relapse-free survival of TGCT patients with different METTL3 expressions; these data were obtained from TCGA by Kaplan–Meier plotter18. TIMER (http://timer.cistrome.org/) is a web server for the analysis of immune infiltrates based on TCGA data. We used this tool to evaluate the correlation between the expression level of METTL3 and the infiltration level of CD8+ and CD4+ T cells and natural killer cells19,20.
Cell Culture
The human TGCT cell lines, NCCIT and Tcam-2, were used in this study, both of which were donated by the Institute of Reproductive and Stem Cell Engineering of Central South University. NCCIT cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS; Gibco, Gaithersburg, MD, USA) and 1% penicillin/streptomycin (PS; Gibco), and Tcam-2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing 10% FBS and 1% PS at 37 °C in 95% air and 5% carbon dioxide (CO2). Confluent cells were digested and passaged with 0.1% trypsin (Thermo Fisher Scientific, Shanghai, China).
Plasmid Construction and Transfection
METTL3 overexpression plasmid and control plasmid were constructed by GENECHEM (Shanghai, China). First, high-fidelity PCR amplification was performed to obtain the METTL3 cDNA, and then the cDNA was cloned into the Age I site of the GV208 plasmid. Finally, the purified plasmid was transformed into competent cells for amplification and used for subsequent plasmid extraction. There were two experimental groups: the overexpressing experimental group (METTL3) and the control group (Con). TGCT cells in the logarithmic growth phase were seeded in six-well plates at a density of 5 × 105/well, with three replicates per group. Plasmids were transfected when cells were grown to 60%–70% confluence. Plasmid transfection was performed using Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Methylthiotetrazole (MTT) Assay
NCCIT and Tcam-2 cells in the logarithmic growth phase were seeded in six-well plates at an appropriate density and grown to 60%–70% confluence for plasmid transfection. Cells were digested 24 h after transfection, and 1000 cells/well were seeded in 96-well plates, and cultured in a constant temperature incubator at 37 °C containing 5% CO2 for 6 h. After the cells adhered to the plate, 20 µl of MTT reagent (Sigma Aldrich, St. Louis, MO, USA) was added to each well, and the optical density (OD) was measured at a wavelength of 490 nm after incubation at 37 °C for 3 h in the dark. MTT reagent was added to the wells every day for 5 days thereafter, and the OD was measured and recorded.
Transwell Cell Migration and Invasion Experiments
Cells were collected 36 h after plasmid transfection, washed three times with phosphate-buffered saline (PBS; Gibco), and then resuspended in serum-free medium to adjust the cells to the appropriate concentration. Then, 800 µl of complete medium containing 15% FBS was added to the lower chamber of the Transwell chamber (a non-Matrigel-coated chamber for migration experiments and a Matrigel-coated chamber for invasion experiments; BD Biosciences, New Jersey, USA), and 200 µl of the cell suspension to the upper chamber. This cell suspension containing 20,000 cells was cultured in a 37 °C incubator for 36–48 h, then fixed with 10% paraformaldehyde for 20 min, and stained with crystal violet for 10 min. After washing the excess crystal violet with PBS, the chamber was allowed to dry before images were taken under a microscope and the number of cells counted.
Western Blot
Cell pellets were collected 48 h after transfection and lysed with radioimmunoprecipitation assay buffer on ice for 1 h. The cell lysates were then centrifuged at 12,000 rpm for 30 min, and the protein concentration was determined by the bicinchoninic acid method. Samples (30 µg protein) were electrophoresed on 8%–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA), and blocked with 5% nonfat milk at room temperature for 1.5 h. The PVDF membrane was then incubated with primary antibodies (Cell Signaling Technology, Danvers, MA, USA) at 4 °C overnight, before washing three times with phosphate-buffered saline with Tween (PBST, 20 min each time). Then, the secondary antibody was added, and the membrane incubated at 37 °C for 1 h, before washing three times with PBST (30 min each time). ECL luminescent solution was added to the membrane, and the protein bands were visualized using a gel imaging system.
Statistical Analysis
The experimental data were analyzed by GraphPad Prism software (GraphPad Software, San Diego, CA, USA). Count data were expressed as mean ± standard error. The comparison between the two groups was performed by Student’s t-test. The correlation analysis was performed by the Pearson’s correlation coefficient. The P (probability) <0.05 was considered as a statistically significant result.
Results
Lower METTL3 Expression in TGCT Correlates with Prognosis
To determine the expression of METTL3 in TGCT, TGCT data in GEPIA online database was used to statistically analyze the expression of METTL3, showing that METTL3 expression was significantly downregulated in TGCT tissues (Fig. 1A). Based on another TGCT dataset GSE3218 from the GEO database, we found that METTL3 was also significantly downregulated in TGCT tissues (Fig. 1B). Furthermore, low METTL3 expression was significantly associated with shorter overall survival and relapse-free survival in TGCT patients (Fig. 1C, D). So METTL3 can be used as an independent prognostic indicator for TGCT patients. Taken together, these data suggested that METTL3 may function as a tumor suppressor gene in TGCT.
Figure 1.
METTL3 correlates with the prognosis of TGCT patients. (A) METTL3 expression data in TGCT from the GEPIA dataset. (B) Comparison of METTL3 expression levels in tumor tissues and normal tissues from the GEO dataset in accession GSE3218. (C) Correlation between METTL3 and overall survival in TGCT patients. (D) Correlation between METTL3 and relapse-free survival in TGCT patients. *P < 0.05. GEO: Gene Expression Omnibus; GEPIA: Gene expression profiling and interactive analyses; N: normal; T: tumor; TGCT: testicular germ cell tumors.
Overexpression of METTL3 Promotes TGCT Cell Proliferation
To determine whether METTL3 can inhibit the progress of TGCT, we constructed a METTL3 overexpression vector and successfully transfected it into NCCIT and Tcam-2 cells (Fig. 2A, B). The proliferation ability of the transfected cells was then assessed by the MTT assay. Unexpectedly, overexpression of METTL3 significantly promoted the proliferation of TGCT cells on the second day, and this effect was more obvious on the third day (Fig. 3A, B).
Figure 2.
TGCT cells successfully overexpress METTL3. (A) Microscopic and fluorescent pictures of NCCIT cells transfected with METTL3 overexpression plasmid. (B) Microscopic and fluorescent pictures of Tcam-2 cells transfected with METTL3 overexpression plasmid. Scale bars=100 μm. TGCT: testicular germ cell tumors.
Figure 3.
Effect of overexpressed METTL3 on the proliferation capacity of TGCT cells. (A) Cell growth curve of NCCIT cells transfected with METTL3 overexpression plasmid. (B) Cell growth curve of Tcam-2 cells transfected with METTL3 overexpression plasmid. **P < 0.01. TGCT: testicular germ cell tumors.
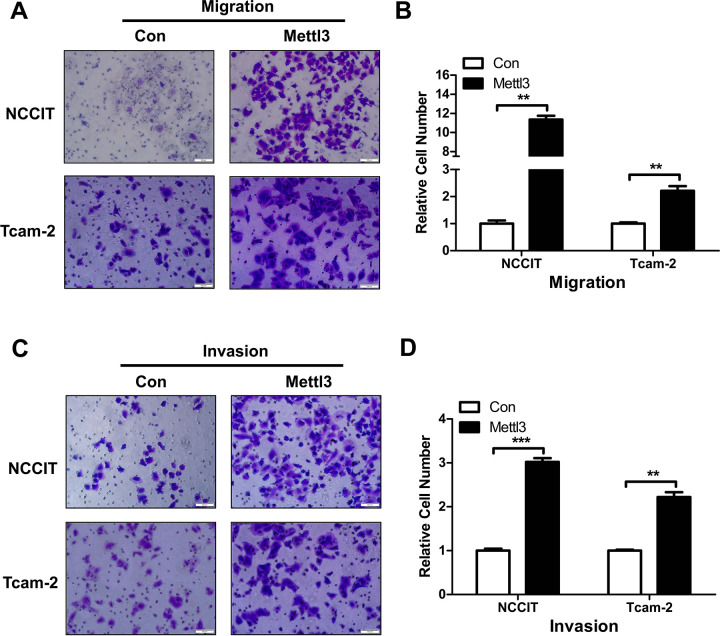
Overexpression of METTL3 Promotes Migration and Invasion of TGCT Cells
The effect of overexpressed METTL3 on TGCT cell migration and invasion ability was investigated using the Transwell assay, showing that the migration and invasion ability of both groups of transfected cells increased more than twofold after METTL3 overexpression (Fig. 4A–D). This indicates that METTL3 plays a very important role in the migration and invasion of TGCT, warranting further investigation.
Figure 4.
Transwell experiments to investigate the effects of METTL3 overexpression on TGCT cell migration and invasion. (A) NCCIT and Tcam-2 cells overexpress METTL3 cell migration pictures. (B) Statistics of relative numbers of migrated NCCIT and Tcam-2 cells after METTL3 overexpression. (C) NCCIT and Tcam-2 cells overexpress METTL3 cell invasion pictures. (D) Statistics of relative invasion of NCCIT and Tcam-2 cells after METTL3 overexpression. **P < 0.01; ***P < 0.001. Scale bars = 100 μm. TGCT: testicular germ cell tumors.
METTL3 Regulates Expression of EMT-Related Proteins
Many studies have shown that the EMT signaling pathway is involved in the migration and invasion of many tumors21,22, and METTL3 has also been reported to be associated with EMT in gastric cancer23. In addition, we analyzed the correlation between METTL3 and EMT markers (ZEB1, VIM, and CDH2) using the GEPIA database and found that METTL3 was significantly positively correlated with these EMT markers (Fig. 5A–C). It was hypothesized that METTL3 may participate in the process of TGCT by regulating EMT signals, and therefore the expression of EMT-related proteins after overexpression of METTL3 was assessed by western blotting. The expression of CDH2, VIM, and ZEB1 proteins was significantly upregulated, while the expression of E-cadherin was significantly downregulated (Fig. 5D), suggesting that the METTL3-mediated EMT pathway may be an important mechanism involved in TGCT migration and invasion.
Figure 5.
Effects of METTL3 overexpression on EMT-related proteins detected by western blot. (A–C) GEPIA database analysis of the correlation between METTL3 and EMT markers. (D) After overexpressing METTL3, the expression of E-cadherin, a marker of epithelial cells, was significantly downregulated, and the expression of VIM, CDH2, ZEB-1, markers of mesenchymal cells, was significantly upregulated. Red arrow: upregulated; green arrow: downregulated. CDH2: N-cadherin; GEPIA: gene expression profiling and interactive analyses; VIM: vimentin.
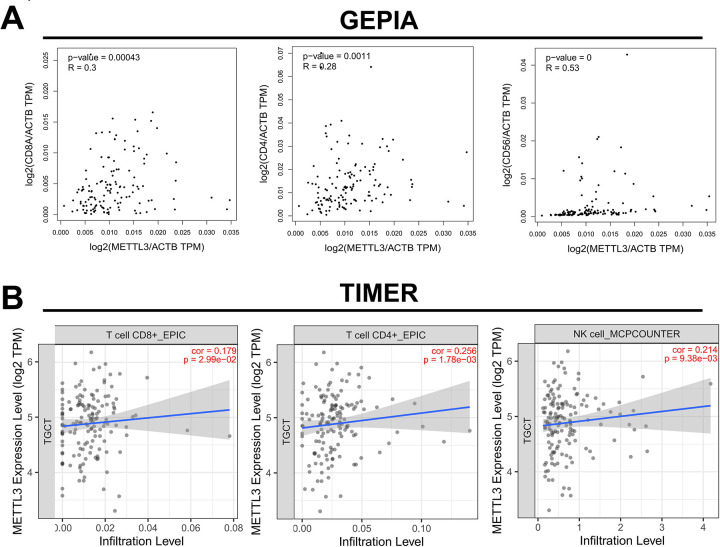
METTL3 Correlates with Molecular Marker Expression in Immune Cells
Studies have shown that m6A modification can participate in tumor immune response24, and hence it was hypothesized that METTL3 may play a role in the regulation of tumor immunity in addition to the EMT pathway. Spearman correlation analysis revealed that the expression level of METTL3 in TGCT was significantly positively correlated with the expression of molecular markers CD8A, CD4, and CD56 of CD8-positive T cells, CD4-positive T cells, and NK cells (Fig. 6A). Furthermore, we analyzed the correlation between METTL3 and the infiltration of these three immune cells using the TIMER tool. We found that METTL3 was positively correlated with the infiltration of these three immune cells in TGCT (Fig. 6B). These data indicate that the higher the METTL3 expression, the greater the number of these tumor infiltration-related inflammatory cells. Therefore, METTL3 may also participate in the immune response of tumors by regulating the immune microenvironment of TGCT.
Figure 6.
Online database analysis of the correlation between METTL3 and immune cells. (A) Correlation between METTL3 and the expression of CD8+ T cell marker CD8A, CD4+ T cell marker CD4, and NK cell marker CD56 (based on GEPIA database). (B) Correlation between METTL3 and the infiltration level of CD8+ and CD4+ T cells and NK cells (based on TIMER database). GEPIA: gene expression profiling and interactive analyses; NK: natural killer.
Discussion
TGCT occurs in young men, posing a huge threat to the life and reproductive health of young male patients, but the pathogenesis has not been fully elucidated. Many studies have shown that genetic factors and epigenetic factors are closely related to the occurrence and development of TGCT25–27. METTL3 is an important molecule that regulates m6A modification in epigenetics and can initiate meiosis by promoting the differentiation process of spermatogenesis28. METTL3 may be related to the survival of TGCT cells29, playing an important role in spermatogenesis. However, the expression, function, and molecular mechanism of METTL3 in TGCT are still unclear.
In this study, we performed data mining using public databases to show that METTL3 was significantly downregulated in TGCT tissues and that low METTL3 expression was associated with poor prognosis in TGCT patients. This is similar to the findings of another research team in colorectal cancer, who found that patients with low METTL3 expression had poorer overall and disease-free survival30. Based on the above analysis, it was hypothesized that METTL3 plays a similar role as a tumor suppressor gene in both tumors. Interestingly, the overexpression of METTL3 promoted cell proliferation, migration, and invasion in vitro, contrary to our previous hypothesis. However, most current studies have reported that METTL3 mainly plays a tumor-promoting role in tumors, promoting the progression and metastasis of bladder cancer31, lung cancer10, and gastric cancer23. This suggests that METTL3 may have multiple and different functions in different tumors, which is also worthy of further research.
Many studies have shown that EMT is involved in the invasion and metastasis of various tumors21,22,32. We also observed at the molecular level that METTL3 upregulated the expression of CDH2, VIM, and ZEB1 that promote EMT-related molecules and downregulated the expression of E-cadherin that suppresses EMT-related molecules. These results are consistent with the previous experimental results of cell invasion and migration, suggesting that METTL3 can indeed participate in tumor progression by regulating the EMT pathway in TGCT. Previously, a research team found that METTL3 participates in the invasion and migration process by inhibiting the transcription process of the EMT core molecule, E-cadherin, in gastric cancer23. However, whether METTL3 participates in the EMT process in TGCT remains to be further confirmed.
The tumor microenvironment (TME) comprises tumor cells, resident and recruited host cells (fibroblasts and immune cells associated with cancer, respectively), products secreted by cells (cytokines and chemokines), and noncellular components in the extracellular matrix33. Infiltrating inflammatory cells in the TME play an important role in determining tumor survival and patient prognosis34. Analysis of the GEPIA data revealed that the expression of METTL3 positively correlated with the molecular markers and infiltration level of CD8+ T cells, CD4+ T cells, and NK cells, indicating that METTL3 may affect the prognosis of TGCT patients by regulating the immune response of tumors and that the regulation of tumor immunity is more important and extensive. However, this hypothesis should be tested by further in vivo experiments. This is also the drawback of the study.
In summary, our study found that METTL3 can promote the migration and invasion of TGCT cells in vitro and can be used as a prognostic molecular marker for TGCT patients. METTL3 positively correlated with the markers of tumor-infiltrating inflammatory cells and might be related to tumor immunity. This study identified a new mode of action of METTL3, contrary to its in vitro and in vivo functions, which has enriched our understanding of the mechanism of TGCT development.
Footnotes
Author Contributions: YL and YM designed the study. YS and LL performed the cell culture, MTT assay, and Transwell cell migration and invasion experiments. YL conducted Western blot and plasmid construction and transfection tests. YM made the data analysis and the statistical analysis of the data in this article. YL and YM wrote and edited the manuscript. All the authors reviewed the article and agreed to publish it.
Ethical Approval: Ethical approval was obtained for all experimental procedures by the Ethics Committee of Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Statement of Human and Animal Rights: This article does not contain any studies with human and animals.
Statement of Informed Consent: Informed consent is not applicable for this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the National Natural Science Foundation of China (No. 81701414).
ORCID iD: Yang Luo  https://orcid.org/0000-0002-9931-6579
https://orcid.org/0000-0002-9931-6579
Reference
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. [DOI] [PubMed] [Google Scholar]
- 2. Shanmugalingam T, Soultati A, Chowdhury S, Rudman S, Van Hemelrijck M. Global incidence and outcome of testicular cancer. Clin Epidemiol. 2013;5:417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Le Cornet C, Lortet-Tieulent J, Forman D, Beranger R, Flechon A, Fervers B, Schuz J, Bray F. Testicular cancer incidence to rise by 25% by 2025 in Europe? Model-based predictions in 40 countries using population-based registry data. Eur J Cancer. 2014;50(4):831–839. [DOI] [PubMed] [Google Scholar]
- 4. Batool A, Karimi N, Wu XN, Chen SR, Liu YX. Testicular germ cell tumor: a comprehensive review. Cell Mol Life Sci. 2019;76(9):1713–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu BB, Wang XY, Gu XY, Zou C, Gao ZJ, Zhang H, Fan Y. N(6)-methyladenosine (m(6)A) RNA modification in gastrointestinal tract cancers: roles, mechanisms, and applications. Mol Cancer. 2019;18(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, Riggs AD, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, Di W, Hu B, An J, Kong L, Pan L, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38(1):393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sorci M, Ianniello Z, Cruciani S, Larivera S, Ginistrelli LC, Capuano E, Marchioni M, Fazi F, Fatica A. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018;9(8):796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. [DOI] [PubMed] [Google Scholar]
- 14. Lin S, Liu J, Jiang W, Wang P, Sun C, Wang X, Chen Y, Wang H. METTL3 promotes the proliferation and mobility of gastric cancer cells. Open Med (Wars). 2019;14:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, Chen D. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Korkola JE, Houldsworth J, Chadalavada RS, Olshen AB, Dobrzynski D, Reuter VE, Bosl GJ, Chaganti RS. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66(2):820–827. [DOI] [PubMed] [Google Scholar]
- 18. Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bo H, Fan L, Li J, Liu Z, Zhang S, Shi L, Guo C, Li X, Liao Q, Zhang W, Zhou M. High expression of lncRNA AFAP1-AS1 promotes the progression of colon cancer and predicts poor prognosis. J Cancer. 2018;9(24):4677–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bo H, Fan L, Gong Z, Liu Z, Shi L, Guo C, Li X, Liao Q, Zhang W, Zhou M, Xiang B. Upregulation and hypomethylation of lncRNA AFAP1AS1 predicts a poor prognosis and promotes the migration and invasion of cervical cancer. Oncol Rep. 2019;41(4):2431–2439. [DOI] [PubMed] [Google Scholar]
- 23. Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, Zhao G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, Aplin AE, Lu Z, Hwang S, He C, He YY. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10(1):2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Z, McGlynn KA, Rajpert-De Meyts E, Bishop DT, Chung CC, Dalgaard MD, Greene MH, Gupta R, Grotmol T, Haugen TB, Karlsson R. Meta-analysis of five genome-wide association studies identifies multiple new loci associated with testicular germ cell tumor. Nat Genet. 2017;49(7):1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Costa AL, Moreira-Barbosa C, Lobo J, Vilela-Salgueiro B, Cantante M, Guimaraes R, Lopes P, Braga I, Oliveira J, Antunes L, Henrique R. DNA methylation profiling as a tool for testicular germ cell tumors subtyping. Epigenomics. 2018;10(12):1511–1523. [DOI] [PubMed] [Google Scholar]
- 27. Bo H, Cao K, Tang R, Zhang H, Gong Z, Liu Z, Liu J, Li J, Fan L. A network-based approach to identify DNA methylation and its involved molecular pathways in testicular germ cell tumors. J Cancer. 2019;10(4):893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, Chen YS, Zhang XX, Wang CX, Jiang LY, Liu C. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27(9):1100–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nettersheim D, Berger D, Jostes S, Kristiansen G, Lochnit G, Schorle H. N6-Methyladenosine detected in RNA of testicular germ cell tumors is controlled by METTL3, ALKBH5, YTHDC1/F1/F2, and HNRNPC as writers, erasers, and readers. Andrology. 2019;7(4):498–506. [DOI] [PubMed] [Google Scholar]
- 30. Deng R, Cheng Y, Ye S, Zhang J, Huang R, Li P, Liu H, Deng Q, Wu X, Lan P, Deng Y. m(6)A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther. 2019;12:4391–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, Wei JF. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He B, Li W, Wu Y, Wei F, Gong Z, Bo H, Wang Y, Li X, Xiang B, Guo C, Liao Q. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7(9):e2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. [DOI] [PubMed] [Google Scholar]
- 34. Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B, Wu X, Ma J, Zhou M, Li X, Li Y. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]