Abstract
Ultrahigh-performance liquid chromatography Quadrupole-Orbitrap tandem mass spectrometry (UHPLC-Q-Orbitrap-MS/MS) was used to compare the composition of ginsenosides in white ginseng (WG) and extruded white ginseng (EWG). A total of 45 saponins, including original neutral ginsenosides, malonyl-ginsenosides, and chemical transformation of ginsenosides, were successfully identified in both WG and EWG. Multivariate statistical analyses including supervised orthogonal partial least squared discrimination analysis (OPLS-DA) and hierarchical clustering analysis (HCA) were used to analyze components of white ginseng before and after extrusion. As a result, three ginsenosides (malonyl (M)-Rb1, M-Rb2, and M-Rc) were found to be increased in WG, while three ginsenosides (Rb2, Rc, and Rg1) were elevated in EWG. In the OPLS-DA S-plot, the different compositions of ginsenoside that were distinguished between WG and EWG were screened out. Experimental results indicate that the UHPLC-Q-Orbitrap-MS/MS is a useful tool to characterize variations of ginsenosides in WG and EWG.
1. Introduction
The root of ginseng (Panax ginseng C. A. Mey.) has been widely used as traditional medicine and functional foods in China and other Asian countries and regions for over 2,000 years [1]. It is found that ginseng contains lots of bioactive and pharmacological effects ingredients such as polysaccharides, saponin, amino acids, vitamins, protein, and phenolic compounds [2]. In China, WG (white ginseng) and RG (red ginseng) are two main types of ginseng that are widely used in Chinese herbal medicine and food markets. WG is produced by drying fresh ginseng in the sun, and RG is manufactured by steaming fresh ginseng at 95–100°C for 2-3 h and then drying. Although both white ginseng and red ginseng were processed with fresh ginseng, they were used for different purposes in the clinical application of TCM (Traditional Chinese Medicine) practice [3]. Meanwhile, ginseng raw materials and products were popularized worldwide as a natural healthy food along with the global trends of preference for natural products [4]. Modern studies showed that the bioactive and pharmacological effects components are mainly recognized as ginsenosides [5], which exhibit anti-inflammatory, antioxidant, antiapoptotic, anticancer, and immune-stimulant pharmacological activities [6–15].
Extrusion is a continuous process with high temperature, high pressure, and short time. Many chemical and physical reactions occur during the extrusion process, such as starch gelatinization, protein denaturation, and Millard reaction. Meanwhile, functional properties are also changed [16]. Numerous studies have reported that the extruding process could increase some bioactive and pharmacological effects of ginseng products, compared with unprocessed WG [17–19]. The physical characteristics and chemical composition of ginseng are changed during the extrusion process, which further alters the functional properties of ingredients [20–24]. Many researchers have extruded white ginseng and red ginseng samples conducted to improve the chemical and physical properties [8, 16, 18, 19, 25–27]. The contents of crude saponin were higher in the extruded ginseng than that in unprocessed ginseng. In addition, the content of Rg2, Rh1, Rh2, and Rg3 in EWG was increased as the extrusion temperature was raised [28]. WG was more suitable for extrusion than RG because more significant increased antioxidant activity was obtained in extruded WG than that in RG [8]. Although studies on the physical, chemical, and functional properties of extrusion WG have been reported in the literature, the comparison of ginsenoside composition of WG before and after extrusion process has not yet been researched.
In the present study, we applied UHPLC-Q-Orbitrap-MS/MS combined with multivariate statistical analysis approach to assess the ginsenoside compositions of WG and EWG. We also investigated the changing chemical structures of WG and EWG and the possible reasons.
2. Materials and Methods
2.1. Materials and Reagents
HPLC-MS grade methanol, acetonitrile, and formic acid were purchased from TEDIA (Fairfield, OH, USA). Ultrapure water was filtered through a Milli-Q system (Millipore, Billerica, MA, USA). All other chemical reagents were of analytical grade. Ginsenoside Rb1, ginsenoside Rb2, ginsenoside Rb3, ginsenoside Rc, ginsenoside Rd, ginsenoside Re, ginsenoside Rf, ginsenoside Rg1, ginsenoside Rg2, ginsenoside Ro, and standard chemicals were purchased from Shanghai Standard Technology Co., Ltd. (purity ≥ 98%, Shanghai, China). The 5-year-old white ginseng power was obtained from Hongjiu Biotech Co., Ltd. (total saponins of Panax ginseng was 4.5%, moisture was 4.8%, Jilin, China).
2.2. Extrusion Process
The WG root powder was extruded with a co-rotating intermeshing twin-screw extruder (Fumach Food Stuff Engineering & Technology Co., Hunan, China). The parameters of extrusion were as follows: the moisture content of 20%, screw speed of 200 rpm, the feed rate of 10 kg/h, and die diameter of 3.0 mm. The temperature profile from the feed section to the die exit was set to 140°C. The extrudate was directly dried in the oven at 60°C for 12 h.
2.3. Extraction of Ginsenosides
The obtained powder was weighed (0.1 g) and extracted with 5 mL of 70% methanol in an ultrasonic water bath for 45 min, and the extract was filtered through a syringe filter (0.22 μm) and stored in a 4°C refrigerator for LC-MS analysis [29, 30].
2.4. The Methods of UHPLC-Q-Orbitrap-MS/MS Analysis
Chromatographic separation was performed on an Ultimate 3000 ultrahigh-performance liquid chromatography system (Thermo Fisher, San Jose, CA, USA) coupled with the Supelco C18 column (3.0 × 50 mm, 2.7 μm; Sigma-Aldrich, USA). The column oven temperature was maintained at 35°C, and the mobile phases A and B were acetonitrile and water with 0.1% formic acid, respectively. The separation of experimental samples was programmed with the following gradient elution: the proportion of acetonitrile (A) was increased from 15% to 19% (0–5 min), 19–19% (5–10 min), 19–25% (10–13 min), 25–28% (13–15 min), 28–28% (15–18 min), 28–30% (18–22 min), 30–35% (22–25 min), 35–40% (25–30 min), 40–60% (30–35 min), 60–80% (35–38 min), 80–100% (38–40 min), 100–100% (40–45 min), and finally adjusted from 100% to 15% (45–50 min) and maintained at 15% for 10 minutes. The injection volume was 5 μL, and the flow rate was 0.4 mL/min.
Mass spectrometric detection was carried out on a Q-Orbitrap-MS/MS (Thermo Fisher, San Jose, CA, USA) equipped with electrospray ionization source operated in the negative ion mode. The parameters of ion source were set as follows: sheath gas flow of 35 Arb, aux gas flow of 10 Arb, and sweep gas flow of 1 Arb. S-Lens RF was 55%. The capillary voltage was set to −3.5 kV with a capillary temperature of 350°C. Full-scan MS data were acquired at the centroid mode from m/z 150 to 2000 Da, 70,000 resolution, automatic gain control (AGC), the target of 1 × 106, and maximum injection time (IT) of 100 ms. The parameters of dd-MS2 were set as follows: 17,000 resolution, automatic gain control (AGC) the target of 1 × 105, maximum injection time (IT) of 50 ms, Loop count 5, isolation window 4.0 m/z and NCE/stepped NCE.The MS/MS data were acquired in Full-MS/ddMS2 mode using the following settings: resolution 17,000 with AGC target of 1×105, maximum IT of 50 ms, and the normalized collision energy (NCE) of 25–55.
2.5. Data Processing and Multivariate Analysis
The SIEVE (version2.1, Thermo Fisher, San Jose, CA, USA) software was used to process the raw data of samples, which could detect the mass, retention time, and intensity of the peaks in each TIC. The max retention time shift was set at 0.20 min, and the m/z width was 10 ppm to align the features. The base peak min intensity and background were set at 105 and 3, respectively. After being aligned, the intensity of each ion was normalized by the total ion intensity of each TIC. The resultant dataset, containing m/z value @ retention time, the normalized intensity, and the sample code, was used to perform the multivariate statistical analysis. Then, the datasets were saved as .csv files and imported into SIMCA-P software 11.5 (Umetrics, Umea, Sweden) to conduct the multivariate statistical analysis, including orthogonal partial least squared discrimination analysis (OPLS-DA) and hierarchical clustering analysis (HCA). In the OPLS-DA model, ions with variable importance in projection VIP values larger than 1 were highlighted and were further filtered by t-test (SPSS19.0, Chicago, IL, USA). The components with p < 0.05 were considered significant and were selected as analytical markers.
3. Results and Discussion
3.1. UHPLC-Q-Orbitrap-MS/MS Analysis of White Ginseng and Extruded White Ginseng
The ultrahigh-performance liquid chromatography combined high-resolution mass spectrometry has been proved as an effective analytical tool for ginsenoside analysis in complex extracts of Chinese herb medicine [31–34]. Figure 1 shows the total ion chromatogram (TIC) of the extracts of WG and EWG samples by UHPLC-Q-Orbitrap/MS in the negative ion mode. The ginsenoside compounds were effectively separated in 45 min by the established method. The intensities of several peaks were different before and after extrusion. Compared to WG, the total ion chromatogram of EWG showed obvious lower or higher intensity of some peaks from 25 to 30 min. These demonstrated that the extrusion process changed the chemical composition of the ginseng sample. Meanwhile, the sample of ginseng contains water, and the determination of water content may be performed before extraction [35, 36], which contribute to accurate determination of polysaccharide in future research.
Figure 1.
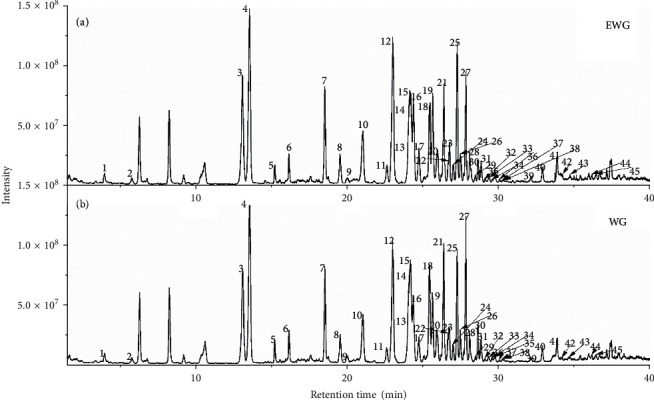
Total ion chromatogram (TIC) of extruded white ginseng (a) and white ginseng (b).
The Q-Orbitrap-MS was reliable and sensitive for measuring the exact mass values of the compounds from samples. Moreover, the retention time, accurate masses, and the characteristic fragment ions were compared with the standards to perform the compound identification.
Because the negative ion mode gave a much clearer fragmentation pattern for structure identification of ginsenoside compounds, we detected ginsenoside and confirmed the molecular weight by full-scan MS in negative ion mode. The [M-H]− and [M + HCOO]− ions of ginsenoside were formed in the negative ion mode. Compared to the theoretical value, all molecular ions were measured within the mass accuracy of 10 ppm. Table 1 shows the characteristic fragment ions of ginsenosides. The aglycone type and the sequence of ginsenosides were confirmed by the tandem MS information. Figure 2 shows the tandem MS spectra of protopanaxadiol saponins (Rg1 and Rb1), malonyl saponins (mRb1), and oleanolic acid saponins (Ro). As shown in Figures 2(a)–2(c), the [M-H]− ion produced fragment ions at m/z 475, 459, and 455, which were the characteristic ions of ginsenosides. The fragment ion at m/z 475 was produced by loss of two glucose residues (162 Da + 162 Da). The fragment ion at m/z 459 was subtracted from four glucose residues (162 Da + 162 Da + 162 Da + 162 Da). Figures 2(c) and 2(d) show the MS/MS spectra of ginsenoside Ro and malonyl-ginsenoside Rb1. The fragment ion at m/z 455 was produced by loss of two glucose residues and β-d-glucuronic acid (162 Da + 162 Da + 176 Da). In Figure 2(d), the [M-H]− ion produced fragment ions at m/z 1107, representing malonyl-ginsenoside cleavage by loss of one malonyl residue. Similar to Rb1, the fragment ion at m/z 459 is produced by loss of four glucose residues (162 Da + 162 Da + 162 Da + 162 Da). The experimental results show that the tandem mass spectrum fragmentation provided structural rich information for the elucidation of ginsenosides. By comparing the retention time, full scan, and the tandem MS spectra with the standard, ginsenoside structure could be identified. The components without standard could be tentatively assigned by comparing the data with the literature record. As a result of our analysis, a total of 45 ginsenosides were identified from WG and EWG. The identification information of ginsenosides is listed in Table 1.
Table 1.
Compounds identified from white ginseng and extruded white ginseng.
| No. | Rt (min) | Formula | Detected mass (Da) | Mass error (ppm) | MS/MS fragment ions | Identification |
|---|---|---|---|---|---|---|
| 1 | 3.37 | C42H72O15 | 861.4844a | 1.0 | 650.2734, 542.4258, 415.3136 | Ginsenoside Re5 |
| 2 | 5.81 | C22H32O13 | 503.1771b | 0.2 | 473.1600, 415.2855, 390.2661 | Cistanoside H |
| 3 | 13.14 | C42H72O14 | 845.4894a | 1.2 | 799.4832, 637.4314, 475.3788 | Ginsenoside Rg1 |
| 4 | 13.57 | C48H82O18 | 991.5472a | 1.1 | 945.5403, 799.4826, 637.4302, 475.3785 | Ginsenoside Re |
| 5 | 15.24 | C45H74O17 | 885.4991b | 1.4 | 841.4911, 781.4704, 637.4300, 475.3782 | Malonyl-ginsenoside Rg1 |
| 6 | 16.2 | C51H84O21 | 1031.5425b | 0.7 | 945.5348, 783.4891, 637.4325, 475.3785 | Malonyl-ginsenoside Re |
| 7 | 18.55 | C42H72O14 | 845.4893a | 1.3 | 799.4816, 637.4309, 475.3787 | Ginsenoside Rf |
| 8 | 19.57 | C41H70O13 | 815.4793a | 0.6 | 637.4122,475.2591 | Noto-ginsenoside R2 |
| 9 | 20.04 | C59H100O27 | 1239.6353b | 0.9 | 1077.5878, 945.5434, 783.4840, 681.4325 | Noto-ginsenoside R4/ginsenoside Ra3 |
| 10 | 21.03 | C42H72O13 | 829.4858a | 0.4 | 637.4292, 475.3785 | Ginsenoside Rg2 |
| 11 | 22.64 | C58H98O26 | 1255.6304a | 2.2 | 1239.6354, 1209.6183, 1007.5946, 945.5434, 799.4820, 650.1565, 475.3796 | Ginsenoside Ra2 |
| 12 | 23.04 | C54H92O23 | 1153.5977a | 1.2 | 1107.5927,945.5379,783.4874, 621.4355, 459.3850 | Ginsenoside Rb1 |
| 13 | 24.06 | C48H76O19 | 955.4892b | 1.7 | 793.4366, 613.3745, 569.3832, 455.3529 | Ginsenoside Ro |
| 14 | 24.19 | C57H94O26 | 1193.5942b | 1.6 | 1107.5925, 945.5403, 783.4891, 621.3830, 459.3846 | Malonyl-ginsenoside Rb1/isomer |
| 15 | 24.39 | C53H90O22 | 1123.5885a | 1.9 | 1077.5790, 945.5386,7 83.4573, 621.2311,459.3891 | Ginsenoside Rc |
| 16 | 24.75 | C58H98O26 | 1255.6209a | 1.5 | 1239.6340, 1209.6176, 1007.5927, 945.5438, 799.4827, 650.1566, 475.3743 | Ginsenoside Ra1 |
| 17 | 25.25 | C36H62O9 | 683.4382a | 0.9 | 475.3733 | Ginsenoside F1 |
| 18 | 25.46 | C56H92O25 | 1163.5841b | 1.2 | 1077.5323,945.5419,783.4745, 621.42631,459.3257 | Malonyl-ginsenoside Rc |
| 19 | 25.66 | C53H90O22 | 1123.5887a | 1.7 | 1077.5820, 945.5396, 783.4877, 621.4354, 459.3839 | Ginsenoside Rb2 |
| 20 | 25.95 | C57H94O26 | 1193.5946b | 1.3 | 1149.6058, 1177.5963, 945.5458, 783.4924, 621.4364, 459.3817 | Malonyl-ginsenoside Rb1/isomer |
| 21 | 26.03 | C53H90O22 | 1123.5873a | 2.9 | 1077.5817, 945.5402, 783.4875, 621.4353, 459.3840 | Ginsenoside Rb3 |
| 22 | 26.4 | C56H92O25 | 1163.584b | 1.3 | 1077.5225,945.5573,783.4354, 621.4602, 459.3924 | Malonyl-ginsenoside Rb2/isomer |
| 23 | 26.75 | C56H92O25 | 1163.584b | 1.3 | 1077.5347, 945.5523, 783.4616, 621.4473, 459.3248 | Malonyl-ginsenoside Rb2/isomer |
| 24 | 27.02 | C42H66O14 | 793.4374b | 0.8 | 631.3840, 569.3831, 455.3528 | Chikusetsusaponin IVa |
| 25 | 27.27 | C48H82O18 | 991.5472a | 1.5 | 783.4174, 621.3566, 459.3527 | Ginsenoside Rd |
| 26 | 27.48 | C56H92O25 | 1163.5837b | 1.5 | 1077.5360, 945.5741, 783.4784, 621.4541 | Malonyl-ginsenoside Rb3/isomer |
| 27 | 27.83 | C51H84O21 | 1031.5424b | 0.8 | 945.5354, 783.4422, 637.4179, 475.3627 | Malonyl-ginsenoside Rd |
| 28 | 28.1 | C50H84O19 | 1033.5565a | 2.3 | 945.2658, 783.2891, 637.4630, 475.2571 | Acetylginsenoside Re |
| 29 | 28.22 | C55H92O23 | 1165.6007a | 0.3 | 1077.5847, 945.5405, 783.4880, 621.4357, 459.3843 | GinsenosideRs1/Rs2/isomer |
| 30 | 28.65 | C55H92O23 | 1119.5957b | 0.5 | 1077.5479, 945.5371, 783.4766, 621.4918, 459.3737 | Ginsenoside Rs1/Rs2/isomer |
| 31 | 28.86 | C51H84O21 | 1031.5422b | 0.1 | 945.52647, 783.4873, 621.4560, 459.7781 | Malonyl-ginsenoside Rd/isomer |
| 32 | 29.11 | C51H84O21 | 1031.5421b | 0.2 | 945.5331, 783.4842, 621.4179, 459.3679 | Malonyl-ginsenoside Rd/isomer |
| 33 | 29.34 | C54H86O24 | 1117.5417b | 1.7 | 945.5157, 783.4358, 621.4479, 459.3297 | PPD-Glc-(Glc-Glc)-2malonyl |
| 34 | 29.41 | C47H80O17 | 961.8364a | 1.5 | 915.5361, 785.9601, 621.4492, 459.3247 | Noto-ginsenoside Fe/vinaginsenoside R16 |
| 35 | 29.58 | C54H86O24 | 1117.5422b | 1.3 | 945.5359, 927.6159, 783.4793, 621.6795 | PPD-Glc-(Glc-Glc)-2malonyl |
| 36 | 29.77 | C50H84O19 | 1033.5579a | 1.0 | 987.5502,945.5998,783.4875, 459.3838 | Pseudo-ginsenoside Rc1 |
| 37 | 30.08 | C47H80O17 | 961.5368a | 1.0 | 915.5049, 783.2958, 621.4138, 459.3660 | Noto-ginsenoside Fe/vinaginsenoside R16 |
| 38 | 30.26 | C50H82O20 | 1001.5317b | 1.0 | 915.5296, 783.4793, 622.4159, 459.3647 | Malonylnoto-ginsenoside Fe |
| 39 | 32.16 | C42H72O13 | 829.4955a | 0.8 | 621.42671, 459.3097 | Ginsenoside F2 |
| 40 | 32.89 | C42H66O14 | 793.4374b | 0.8 | 631.3840, 569.3831, 497.3645, 455.3528 | Zingibroside R1 |
| 41 | 33.86 | C42H72O13 | 829.4947a | 1.0 | 637.4154, 475.3612 | Ginsenoside Rg3/isomer |
| 42 | 34.12 | C42H72O13 | 829.4946a | 1.1 | 637.4304,475.3143 | Ginsenoside Rg3/isomer |
| 43 | 34.72 | C42H72O14 | 799.4841b | 0.8 | 734.8464, 621.4331, 542.4661, 481.0309 | Ginsenoside Rg7 |
| 44 | 36.44 | C42H70O12 | 811.4843a | 0.7 | 765.4680, 649.9458, 545.0465, 432.8917, 304.9154 | Ginsenoside Rg5 |
| 45 | 36.70 | C36H62O8 | 667.4419a | 1.2 | 631.3836, 455.5328, 304.9154 | Rh2 |
a[M+COOH]−; b[M − H]−.
Figure 2.
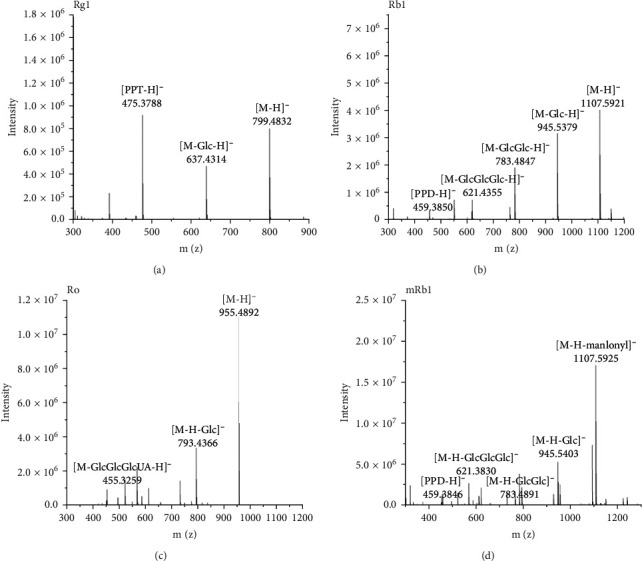
The tandem mass spectrum of ginsenosides in the negative ion mode: (a) Rg1; (b) Rb1; (c) Ro; (d) mRb1.
3.2. Multivariate Statistical Analysis of White Ginseng and Extruded White Ginseng
Some researchers obtained chemical profiling data using high-resolution mass spectrometry combined with multivariate statistical analysis [37–41]. The antioxidant activity of EWG was better than that of WG because of the chemical composition changes after extruding [16, 18]. Thus, we used statistical methods to explore the differences in composition. The extract of WG and EWG was analyzed by the UHPLC-Q-Orbitrap-MS/MS. After data preprocessing, the dataset was generated and used to conduct multivariate statistical analysis.
Multivariate data analyses were performed to characterize the distinct composition of various chemicals from WG and EWG in detail. As shown in Figure 3(a), the score plot of OPLS-DA were effectively used todistinguishbetween WG and EWG samples. No overfitting was found because the permutation R2 (0.993) and Q2 (0.989) values on the right are higher than those on the left (Figure 3(b)). In the S-plot, the points of the chemical composition that importantly contributed to the variance between WG and EWG were plotted farther along the x-axis and y-axis (Figure 3(c)). In the comparison between WG and EWG, the increased components of WG were shown in the lower left quadrant of S-plot, while the upper right quadrant showed the increased components of EWG. The results showed that three ginsenosides (malonyl (M)-Rb1, M-Rb2, and M-Rc) and three ginsenosides (Rb2, Rc, and Rg1) were elevated in WG and EWG, respectively. In the OPLS-DA and S-plot, different compositions of the chemical were distinguished between WG and EWG.
Figure 3.
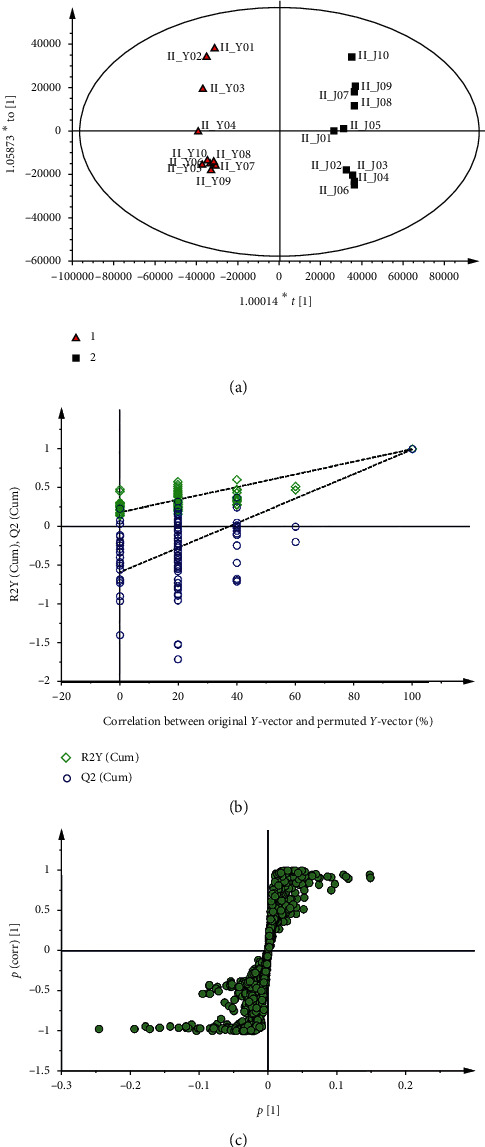
The OPLS-DA/permutation test/S-plot of white ginseng and extruded white ginseng in negative ion mode.
In order to visualize the tendency of the variation of the chemical markers of WG and EWG, a heat map was constructed based on the relative intensity of each compound. As shown in Figure 4, color differences indicated the composition change in the WG group and the EWG group. Between the WG and EWG groups , the contents ofginsenoside-Rh2, -Rg3, -Rg1, -F1, -Rb3, -Ra1, -Rg7, -Rc, and Rd in EWG samples were significantly higher. However, the contents of malonylginsenoside-Rb1, -Rc, -Rb3, -Rd, -Rb2, -Rg1, and -Re, and ginsenoside-Re5 in all the WG samples were significantly higher.. This was illustrated in the heating trial, in which the concentration of ginsenosides was affected by the thermal processing condition and the degree of conversion between malonyl and neutral ginsenosides [37, 42, 43]. The results suggested that malonyl-ginsenoside was thermally unstable and especially susceptible to hydrolysis by extruding treatment. The malonyl residue was well preserved in WG because its dehydration was conducted in sunlight without extruding treatment.
Figure 4.
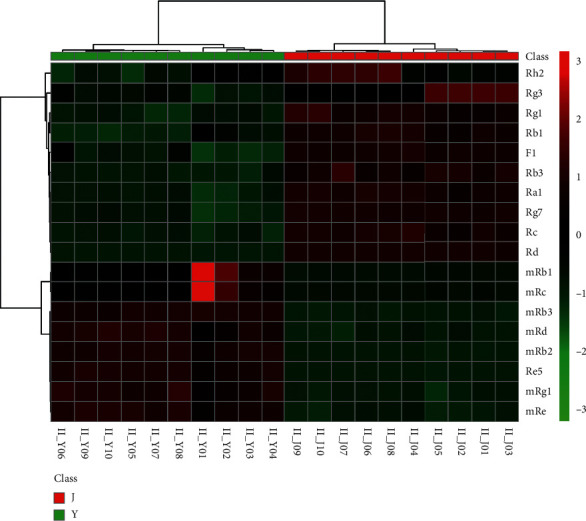
The heat maps visualizing the intensities of the 17 ginsenoside datasets from white ginseng and extruded white ginseng (n = 10) and extruded white ginseng (n = 10).
In the case of crude saponin (some ginsenosides such as Rb1, Rg1, and Rc) and rare saponin (some ginsenosides such as Rh2 and Rg3) content, there was a slight increase after extrusion. The malonyl-ginsenoside is prone to be transformed into ginsenoside due to heat and pressure treatment; the conversion efficiency of the ginsenosides was increased by the extrusion process. The amount of several ginsenosides was increased by the extrusion process because the cell or tissues of ginseng underwent transformations, which allowed easier extraction of these components through the extrusion process. Meanwhile, this result was caused by the weakening of molecular bonds and increased water absorption at the high temperature, pressure, and shear forces involved in the extrusion process.
4. Conclusions
In this study, we perform the chemical comparison of white ginseng before and after extrusion by UHPLC-Q-Orbitrap-MS/MS combined with multivariate statistical analysis. Multivariate statistical analyses were applied to differentiate the chemical components of WG and EWG, and 6 ginsenosides were found as the characteristic chemical markers of the analytical sample. The experimental information will help to supervise the production of each type of processed ginseng product in the herbal medicine market or food industry. Finally, this study verified the application value of this method in the quality evaluation of WG and EWG. It is expected that the established method will be useful for evaluating processed products not only by ginseng samples but also other agricultural and food product processing such as tea, grape, meat, and so on.
Acknowledgments
The authors gratefully acknowledge the financial support provided by the National Key Research and Development Program of China (no. 2016YFD0400702), Scientific Research Program of Education Department of Jilin Province of China (no. JJKH20180654KJ), and Scientific Research Startup Fund Program of Jilin Agricultural University (no. 201715), The National Grain Industry Leading Talent Program of China (no. LL2018201).
Contributor Information
Yu-Zhu Wu, Email: 604198532@qq.com.
Jing-Sheng Liu, Email: liujingshengname@163.com.
Data Availability
The data used to support the results of this study are included within the article. Any further information is available from the authors upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
All authors have given approval to the final version of the manuscript. Yun-Long Guo, Yang Wang, Yi-Lin Zhao, Xiu-Ying Xu, Hao Zhang, and Cheng-Bin Zhao conceived the experiments. Ming-Zhu Zheng, Shu-Ying Liu, Yu-Zhu Wu, and Jing-Sheng Liu supervised the study, provided financial support via projects, and wrote the paper.
References
- 1.Zhou Y., Yang Z., Gao L., et al. Changes in element accumulation, phenolic metabolism, and antioxidative enzyme activities in the red-skin roots of Panax ginseng. Journal of Ginseng Research. 2017;41(3):307–315. doi: 10.1016/j.jgr.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin Y., Kim Y.-J., Jeon J.-N., et al. Effect of white, red and black ginseng on physicochemical properties and ginsenosides. Plant Foods for Human Nutrition. 2015;70(2):141–145. doi: 10.1007/s11130-015-0470-0. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H.-M., Li S.-L., Zhang H., et al. Holistic quality evaluation of commercial white and red ginseng using a UPLC-QTOF-MS/MS-based metabolomics approach. Journal of Pharmaceutical and Biomedical Analysis. 2012;62:258–273. doi: 10.1016/j.jpba.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Kim B.-G., Choi S.-Y., Suh H. J., Park H. J. Bitterness reduction and enzymatic transformation of ginsenosides from Korean red ginseng (panax ginseng) extract. Journal of Food Biochemistry. 2011;35(4):1267–1282. doi: 10.1111/j.1745-4514.2010.00450.x. [DOI] [Google Scholar]
- 5.Hwang C. R., Lee S. H., Jang G. Y., et al. Changes in ginsenoside compositions and antioxidant activities of hydroponic-cultured ginseng roots and leaves with heating temperature. Journal of Ginseng Research. 2014;38(3):180–186. doi: 10.1016/j.jgr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rai D., Bhatia G., Sen T., Palit G. Anti-stress effects of Ginkgo biloba and Panax ginseng: a comparative study. Journal of Pharmacological Sciences. 2003;93(4):458–464. doi: 10.1254/jphs.93.458. [DOI] [PubMed] [Google Scholar]
- 7.Jia L., Zhao Y. Current evaluation of the millennium phytomedicine-ginseng (I): etymology, pharmacognosy, phytochemistry, market and regulations. Current Medicinal Chemistry. 2009;16(19):2475–2484. doi: 10.2174/092986709788682146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gui Y., Gil S.-K., Ryu G.-H. Effects of extrusion conditions on the physicochemical properties of extruded red ginseng. Preventive Nutrition and Food Science. 2012;17(3):203–209. doi: 10.3746/pnf.2012.17.3.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi S. Epidermis proliferative effect of thePanax ginseng Ginsenoside Rb2. Archives of Pharmacal Research. 2002;25(1):71–76. doi: 10.1007/bf02975265. [DOI] [PubMed] [Google Scholar]
- 10.Lu J.-M., Yao Q., Chen C. Ginseng compounds: an update on their molecular mechanisms and medical applications. Current Vascular Pharmacology. 2009;7(3):293–302. doi: 10.2174/157016109788340767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attele A. S., Wu J. A., Yuan C.-S. Ginseng pharmacology. Biochemical Pharmacology. 1999;58(11):1685–1693. doi: 10.1016/s0006-2952(99)00212-9. [DOI] [PubMed] [Google Scholar]
- 12.Lee D. G., Lee J., Kim K.-T., et al. High-performance liquid chromatography analysis of phytosterols in Panax ginseng root grown under different conditions. Journal of Ginseng Research. 2018;42(1):16–20. doi: 10.1016/j.jgr.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surh Y., Na H. K., Lee J. Y., Keum Y. S. Molecular mechanisms underlying anti-tumor promoting activities of heat-processed Panax ginseng C.A. Meyer. Journal of Korean Medical Science. 2001;16:38–41. doi: 10.3346/jkms.2001.16.s.s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu W., Sun L., Zhang Z., Guo Y., Liu S. Profiling and multivariate statistical analysis of Panax ginseng based on ultra-high-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis. 2015;107:141–150. doi: 10.1016/j.jpba.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 15.Cho J.-G., In S.-J., Jung Y.-J., et al. Re-evaluation of physicochemical and NMR data of triol ginsenosides Re, Rf, Rg2, and 20-gluco-Rf from Panax ginseng roots. Journal of Ginseng Research. 2014;38(2):116–122. doi: 10.1016/j.jgr.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui Y., Ryu G.-H. Effects of extrusion cooking on physicochemical properties of white and red ginseng (powder) Journal of Ginseng Research. 2014;38(2):146–153. doi: 10.1016/j.jgr.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C.-Z., Aung H., Ni M., et al. Red American ginseng: ginsenoside constituents and antiproliferative activities of heat-processed Panax quinquefolius roots. Planta Medica. 2007;73(7):669–674. doi: 10.1055/s-2007-981524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma X., Jin Z., Jin T. Effects of extrusion conditions on chemical properties of extruded white ginseng root hair. Journal of the Science of Food and Agriculture. 2019;99(6):3186–3191. doi: 10.1002/jsfa.9535. [DOI] [PubMed] [Google Scholar]
- 19.Sun S., Wang C.-Z., Tong R., et al. Effects of steaming the root of Panax notoginseng on chemical composition and anticancer activities. Food Chemistry. 2010;118(2):307–314. doi: 10.1016/j.foodchem.2009.04.122. [DOI] [Google Scholar]
- 20.Singh S., Gamlath S., Wakeling L. Nutritional aspects of food extrusion: a review. International Journal of Food Science & Technology. 2007;42(8):916–929. doi: 10.1111/j.1365-2621.2006.01309.x. [DOI] [Google Scholar]
- 21.Lazou A., Krokida M. Structural and textural characterization of corn-lentil extruded snacks. Journal of Food Engineering. 2010;100(3):392–408. doi: 10.1016/j.jfoodeng.2010.04.024. [DOI] [Google Scholar]
- 22.Leblanc N., Saiah R., Beucher E., Gattin R., Castandet M., Saiter J.-M. Structural investigation and thermal stability of new extruded wheat flour based polymeric materials. Carbohydrate Polymers. 2008;73(4):548–557. doi: 10.1016/j.carbpol.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 23.Thymi S., Krokida M. K., Pappa A., Maroulis Z. B. Structural properties of extruded corn starch. Journal of Food Engineering. 2005;68(4):519–526. doi: 10.1016/j.jfoodeng.2004.07.002. [DOI] [Google Scholar]
- 24.Kumar R., Xavier K. A. M., Lekshmi M., Balange A., Gudipati V. Fortification of extruded snacks with chitosan: effects on techno functional and sensory quality. Carbohydrate Polymers. 2018;194:267–273. doi: 10.1016/j.carbpol.2018.04.050. [DOI] [PubMed] [Google Scholar]
- 25.Chang Y. H., Ng P. K. W. Effects of extrusion process variables on extractable ginsenosides in Wheat−Ginseng extrudates. Journal of Agricultural and Food Chemistry. 2009;57(6):2356–2362. doi: 10.1021/jf8031827. [DOI] [PubMed] [Google Scholar]
- 26.Gui Y., Ryu G. H. The effect of extrusion conditions on the acidic polysaccharide, ginsenoside contents and antioxidant properties of extruded Korean red ginseng. Journal of Ginseng Research. 2013;37(2):219–226. doi: 10.5142/jgr.2013.37.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha D. C. Chemical components of red, white and extruded root ginseng. Journal of The Korean Society of Food Science and Nutrition. 2005;34(2):247–254. doi: 10.3746/jkfn.2005.34.2.247. [DOI] [Google Scholar]
- 28.Ryu B.-H., Choi M.-J., Chung K.-C., Lee S.-K. Effect of extrusion process on the change of components in ginseng. Korean Journal of Food Science and Technology. 2012;44(4):411–416. doi: 10.9721/kjfst.2012.44.4.411. [DOI] [Google Scholar]
- 29.Li L., Wang Y., Xiu Y., Liu S. Chemical differentiation and quantitative analysis of different types of Panax genus stem-leaf based on a UPLC-Q-exactive orbitrap/MS combined with multivariate statistical analysis approach. Journal of Analytical Methods in Chemistry. 2018;2018:16. doi: 10.1155/2018/9598672.9598672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L., Ma L., Guo Y., Liu W., Wang Y., Liu S. Analysis of oligosaccharides from Panax ginseng by using solid-phase permethylation method combined with ultra-high-performance liquid chromatography-Q-Orbitrap/mass spectrometry. Journal of Ginseng Research. 2019 doi: 10.1016/j.jgr.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuzzati N., Gabetta B., Jayakar K., Pace R., Peterlongo F. Liquid chromatography-electrospray mass spectrometric identification of ginsenosides in Panax ginseng roots. Journal of Chromatography A. 1999;854(1-2):69–79. doi: 10.1016/s0021-9673(99)00463-x. [DOI] [PubMed] [Google Scholar]
- 32.Van Breemen R. B., Huang C.-R., Lu Z.-Z., Rimando A., Fong H. H. S., Fitzloff J. F. Electrospray liquid chromatography/mass spectrometry of ginsenosides. Analytical Chemistry. 1995;67(21):3985–3989. doi: 10.1021/ac00117a026. [DOI] [Google Scholar]
- 33.Wu W., Song F., Guo D., et al. Mass spectrometry-based approach in ginseng research: a promising way to metabolomics. Current Analytical Chemistry. 2012;8(1):43–66. doi: 10.2174/157341112798472189. [DOI] [Google Scholar]
- 34.Wang X., Sakuma T., Asafu-Adjaye E., Shiu G. K. Determination of ginsenosides in plant extracts fromPanax ginsengandPanax quinquefolius L. By LC/MS/MS. Analytical Chemistry. 1999;71(8):1579–1584. doi: 10.1021/ac980890p. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M., Zhao C., Zhao C., et al. Determination of water content in corn stover silage using near-infrared spectroscopy. International Journal of Agricultural and Biological Engineering. 2019;12(6):143–148. doi: 10.25165/j.ijabe.20191205.4914. [DOI] [Google Scholar]
- 36.Zhao C., Cao Y., Ma Z., Shao Q. Optimization of liquid ammonia pretreatment conditions for maximizing sugar release from giant reed (Arundo donax L.) Biomass & Bioenergy. 2017;98:61–69. doi: 10.1016/j.biombioe.2017.01.001. [DOI] [Google Scholar]
- 37.Lin H., Zhu H., Tan J., et al. Non-targeted metabolomic analysis of methanolic extracts of wild-simulated and field-grown American ginseng. Molecules. 2019;24(6):p. 1053. doi: 10.3390/molecules24061053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Zhao H., Liu Y., et al. GC-MS-Based metabolomics to reveal the protective effect of gross saponins of tribulus terrestris fruit against ischemic stroke in rat. Molecules. 2019;24(4):p. 793. doi: 10.3390/molecules24040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W.-Z., Shi X.-J., Yao C.-L., et al. A novel neutral loss/product ion scan-incorporated integral approach for the untargeted characterization and comparison of the carboxyl-free ginsenosides from Panax ginseng, Panax quinquefolius, and Panax notoginseng. Journal of Pharmaceutical and Biomedical Analysis. 2020;177 doi: 10.1016/j.jpba.2019.112813.112813 [DOI] [PubMed] [Google Scholar]
- 40.Zhang C., Zuo T., Wang X., et al. Integration of data-dependent acquisition (DDA) and data-independent high-definition MSE (HDMSE) for the comprehensive profiling and characterization of multicomponents from Panax japonicus by UHPLC/IM-QTOF-MS. Molecules. 2019;24(15):p. 2708. doi: 10.3390/molecules24152708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao T., Yang Z., Mei X., Xu L., Fan Y. Metabolic disturbance in Korean red ginseng-induced “Shanghuo” (excessive heat) Journal of Ethnopharmacology. 2020;253:p. 112604. doi: 10.1016/j.jep.2020.112604. [DOI] [PubMed] [Google Scholar]
- 42.Lee J., Ji S.-H., Choi B.-R., et al. UPLC-QTOF/MS-Based metabolomics applied for the quality evaluation of four processed Panax ginseng products. Molecules. 2018;23(8):p. 2062. doi: 10.3390/molecules23082062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin M.-S., Song J. H., Choi P., et al. Stimulation of innate immune function by Panax ginseng after heat processing. Journal of Agricultural and Food Chemistry. 2018;66(18):4652–4659. doi: 10.1021/acs.jafc.8b00152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the results of this study are included within the article. Any further information is available from the authors upon request.


