Abstract
Background: High risk human papillomavirus (hr-HPV)-associated oropharyngeal cancers (OPCs) are characterized by significantly better therapy responses. In order to implement a de-escalated treatment strategy for this tumor entity, it is highly crucial to accurately distinguish HPV-associated OPCs from non-HPV-associated ones. Methods: In this prospective study, 56 patients with histologically confirmed OPC were evaluated. A commercially available sandwich ELISA test system was used for the detection of hr-HPV E7 oncoprotein targeting the genotypes 16, 18 and 45. Results were presented as optical density. Positivity for HPV DNA and p16 immunohistochemistry (IHC) was taken as the reference method. Results: E7 positivity was significantly associated with the reference method (p = 0.048). The sensitivity, specificity, positive predictive value and negative predictive value for the E7 oncoptotein was 60.9% (95% CI 38.5 to 80.3%), 66.7% (95% CI 46% to 83.5%), 64.2% (95% CI 49.4 to 77.4%) and 63.01% (95% CI 48.9–75.2%), respectively, for the cutoff provided by the manufacturer. Conclusions: We found a significant association between E7 oncoprotein detection and the currently used combination. We believe that the use of the ELISA based E7 antigen test could be a valuable addition in cases of ambiguous findings and may be used in combination with other techniques to distinguish between HPV-driven and non-HPV-driven OPCs. However, the low sensitivity of the assay coupled with the small sample size in our study may represent a limitation. We recommend that future larger studies elucidate the diagnostic value of the E7 brush test.
Keywords: human papillomavirus, oropharyngeal cancer, E7 oncoprotein, brush test, p16 IHC
1. Introduction
An increase in oropharyngeal cancer (OPC) was first observed in the United States at the beginning of the 21st century [1,2]. Since then, a steady increase in OPCs in the USA and Europe has been described, caused by high risk human papillomavirus (hr-HPV) [3,4], while the number of smokers and the incidence of tobacco related head and neck cancers have declined [5]. Changes in sexual behavior in the last decade, like high numbers of oral sex partners, seem to play an important etiological role in the rising incidence of HPV-positive OPCs [6]. The HPV-positive OPC is a distinct tumor entity that can be distinguished from HPV-negative OPCs by its etiology, molecular characteristics and clinical presentation [7,8]. Patients with an HPV-positive OPC have a substantially better prognosis [2,9]. In a retrospective European study of 259 OPC patients, the HPV status was the most important parameter for overall survival, regardless of the treatment strategies. HPV association was shown to positively influence the survival more than, for example, the size of the primary tumor or smoking status [7]. Hence, precise distinction between HPV-driven and non-HPV-driven tumors is essential. The detection of HPV DNA in tumor tissue does not necessarily speak of an HPV-driven tumor as only a small proportion of HPV infections lead to a transforming lesion [10].
This highlights the need to incorporate a diagnostic surrogate marker which precisely distinguishes between a transient and a transforming hr-HPV infection. The biomarker currently in use for the diagnosis of HPV-driven OPC is the copresence of HPV DNA and overexpression of the cellular marker p16 protein [11,12].
P16 is highly expressed in tissues undergoing cell cycle deregulation, suggesting that the detected hr-HPV DNA in the tumor tissue may be the cause of OPC. However, being merely a cellular marker, p16 is also overexpressed in lesions with no HPV association. Rasmussen and coworkers, for example, observed in a cohort of 1243 OPC patients a group of p16-positive but HPV DNA-negative patients. These patients experienced a significantly higher hazard ratio (HR) for metastatic recurrence as compared to HPV+/p16+ patients (HR = 2.56) (p = 0.006) [12]. This may translate into lower reliability of this marker in correctly identifying HPV-induced OPC and justifies the need to search for a surrogate marker which is closely linked to hr-HPV oncogenesis. One possible alternative is the detection of upregulated expression of hr-HPV oncoproteins. Hr-HPV oncoproteins (E6 and E7) play a major role in HPV-associated malignant transformation since they are able to inactivate tumor suppressor proteins and consequently inhibit cell cycle control mechanisms [13,14]. The molecular mechanism of the E7 oncoprotein is the inactivation of the retinoblastoma (Rb) protein—a tumor suppressor cellular protein which controls key regulators of S-phase genes. Hr-HPV E7 oncoproteins interact with Rb at a higher efficiency than low-risk HPV E7 oncoproteins. The interaction of E7 with Rb causes disruption of the growth-suppressive Rb-E2F complexes, promoting G1-S cell cycle transition and uncontrolled cellular replication [15,16].
A recently published study evaluated ELISA-based detection of hr-HPV E7 oncoprotein as a screening method in cervical samples of healthy women. Agorastos and coworkers found in this study that E7 oncoprotein detection might be a promising marker for precisely distinguishing transformation-relevant hr-HPV infections from transient ones [17]. The ELISA-based procedure is easy to perform, less time-consuming and requires only a basic laboratory setup. Although this sandwich-based E7 antigen ELISA test has become available commercially within the last couple of years, no previous study ever evaluated this assay among patients with OPC.
Confronted with the increasing need to establish a de-escalated therapy strategy explicitly for patients with HPV-driven OPC, we questioned whether this ELISA-based E7 oncoprotein test could be a reliable option in accurately distinguishing between HPV-driven and non-HPV-driven OPC.
2. Results
2.1. Study Population
During the study period, 56 patients with OPC were included. Of the study population, 46 (82.1%) patients were male, the mean age at diagnosis was 65.4 (standard deviation ± 10.12) years and 26 (46.4%) patients were positive for the E7 oncoprotein (Table 1). The mean follow-up time was 8.0 months ± 6 months.
Table 1.
Study population.
| Variables | N (%) |
|---|---|
| Male | 46 (82.1%) |
| Female | 10 (17.9%) |
| Mean age | 65.4 years (±10.12) |
| E7 positivity * | 26 (46.4%) |
* E7 oncoprotein for human papillomavirus 16, 18 and/or 45 without differentiation.
In 23 (41.1%) OPC patients, HPV DNA was detected, and the most common genotype was HPV 16 (83%) (Table 2).
Table 2.
HPV genotypes in oropharyngeal cancer (OPC) patients.
| HPV Subtypes | Number and Percent of HPV + OPC Patients |
|---|---|
| HPV 16 | 19 patients (83%) |
| HPV 18 | 2 patients (9%) |
| HPV 33 | 1 patient (4%) |
| HPV 58 | 1 patient (4%) |
The E7 oncoprotein was detected in 26 patients (46.4%) and was associated with the American Society of Anesthesiologists (ASA) classification (p = 0.04) and smoking was inversely and significantly associated with E7 oncoprotein positivity (p = 0.009). Further clinico-pathological parameters are shown in Table 3.
Table 3.
Clinico-pathological characteristics of OPC patients.
| Variables | E7 Positive (n = 26) |
E7 Negative (n = 30) |
p-Value |
|---|---|---|---|
| Sex | |||
| Male | 22 | 24 | p = 0.73 |
| Female | 4 | 6 | |
| Age | |||
| ≤65 years | 10 | 10 | p = 0.45 |
| >65 years | 16 | 20 | |
| ASA score | |||
| ASA I/II | 19 | 14 | p = 0.04 |
| ASA III/IV | 7 | 16 | |
| Smoking | |||
| Non-smokers | 16 | 8 | p = 0.009 |
| Smoker | 10 | 22 | |
| Alcohol consumption | |||
| Daily | 8 | 13 | p = 0.33 |
| Not daily | 18 | 17 | |
| Clinical T-stage | |||
| cT1/T2 | 14 | 16 | p = 1.0 |
| cT3/4 | 12 | 14 | |
| UICCC | |||
| Stage I | 1 | 3 | p = 0.84 |
| Stage II | 4 | 4 | |
| Stage III | 6 | 7 | |
| Stage IV | 15 | 16 | |
| Subsite oropharynx | |||
| Palatine tonsil | 17 | 19 | p = 0.14 |
| Base of tongue | 9 | 6 | |
| Uvula | 0 | 3 | |
| Lateral pharyngeal wall | 0 | 2 | |
| Therapy | |||
| Surgery only | 3 | 5 | p = 0.72 |
| Surgery and PORT | 3 | 4 | |
| Surgery and RCT/RIT | 1 | 0 | |
| Primary RCT/RIT | 14 | 13 | |
| Primary RT | 2 | 3 | |
| Chemo only | 2 | 3 | |
| p16 | |||
| Positive | 17 | 12 | p = 0.05 |
| Negative | 9 | 18 | |
| HPV DNA | |||
| Positive | 14 | 9 | p = 0.06 |
| Negative | 12 | 21 | |
| Follow up | |||
| No recurrence | 15 | 20 | p = 0.55 |
| Progression | 6 | 9 |
OPC, oropharyngeal cancer; UICC, Union for International Cancer Control; PORT, postoperative radiation; RCT, radiochemotherapy; RIT, radioimmunotherapy; RT, radiotherapy; ASA, American Society of Anesthesiologists.
2.2. Detection of E7 Oncoprotein, HPV DNA and p16
In 26/56 patients, E7 oncoprotein was detected; in 23/56 patients, HPV DNA was detected, and in 29/56 patients, p16 immunohistochemistry (IHC) positivity was detected. There was a high association between p16 and HPV DNA (p < 001) (Table 4). Patients being positive or negative for both test methods, HPV DNA and p16 IHC, served as reference method. The E7 oncoprotein was associated (p = 0.048) with the reference method, including 50 patients with concordant results for p16 and HPV DNA (Table 5). The two patients who were positive for HPV genotypes other than those detectable by the E7 oncoprotein ELISA tests were in one case positive (HPV 33) and in one case negative (HPV 58) for the E7 oncoprotein.
Table 4.
HPV DNA and p16.
| HPV DNA | p16 | |||
|---|---|---|---|---|
| Negative | Positive | Total | p-Value | |
| Negative | 27 | 6 | 33 | p < 0.001 |
| Positive | 0 | 23 | 23 | |
| Total | 27 | 29 | 56 | |
Table 5.
E7 oncoprotein and reference method.
| Reference Method | E7 Oncoprotein | |||
|---|---|---|---|---|
| Negative | Positive | Total | p-Value | |
| Negative | 18 | 9 | 27 | p = 0.048 |
| Positive | 9 | 14 | 23 | |
| Total | 27 | 23 | 50 | |
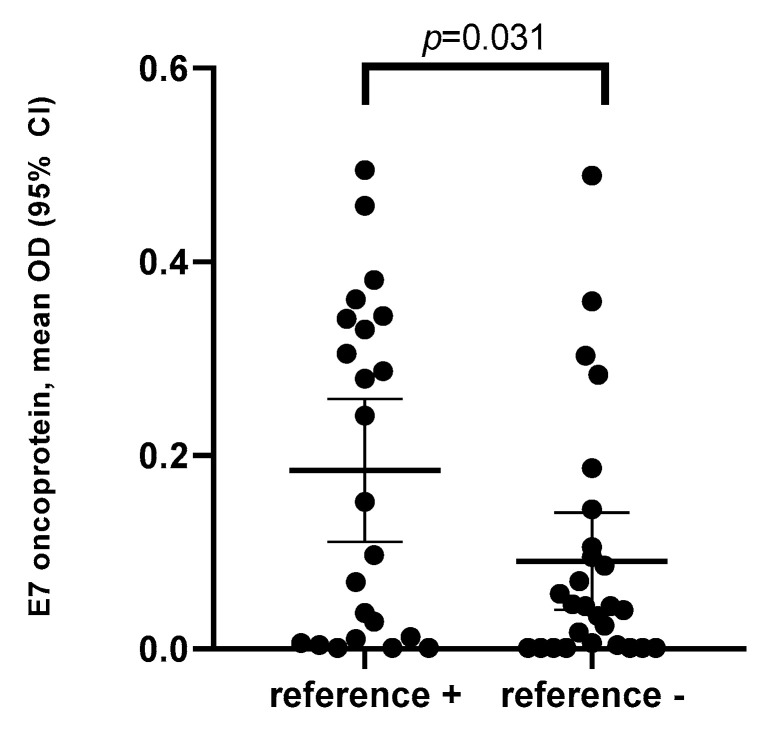
Mean optical density (OD) and 95% CI of the E7 oncoprotein ELISA was shown to be significantly higher among patients who were positive for the reference method (0.18, 95% CI 0.05–0.26) (median OD = 0.15) as compared to those who were negative for the reference method (0.09, 95% CI 0.05–1.13) (median OD = 0.04) (p = 0.031) (Figure 1).
Figure 1.
Mean optical density (OD) of the E7 oncoprotein level by the reference method (p16 and HPV DNA) among patients with histologically confirmed oropharyngeal cancer (OPC).
In addition to the cutoff value provided by the manufacturer, we also analyzed the performance of E7 oncoprotein using other arbitrarily selected cutoff points. The best agreement with the reference method is achieved when using a cutoff OD value of 0.2. With this cutoff value, the E7 oncoprotein ELISA shows the highest specificity (and the highest % agreement) as compared to the cutoff value provided by the manufacturer (Table S1).
2.3. Sensitivity, Specificity and Accuracy
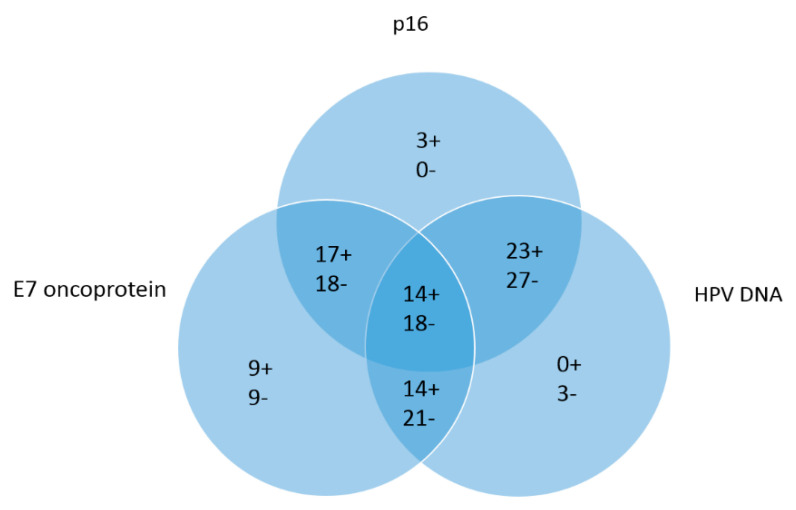
Sensitivity, specificity, positive predictive value and negative predictive value for the E7 oncoptotein was 60.9% (95% CI 38.5 to 80.3%), 66.7% (95% CI 46% to 83.5%), 64.2% (95% CI 49.4 to 77.4%) and 63.01% (95% CI 48.9–75.2%), respectively. The percent agreement between the standard approach and E7 method was 64%. Concordant results of the three test methods are shown in Figure 2.
Figure 2.
Venn diagram for E7 oncoprotein, p16 IHC and HPV DNA, (+) describing positive and (−) describing negative results. Field in the center describes concordant results for all 3 test methods (14 positive and 18 negative). The overlapping fields describe concordant results of E7 and HPV DNA (14 positive and 21 negative), E7 and p16 (17 positive and 18 negative) and HPV DNA and p16 (23 positive and 27 negative). The fields on the outside describe single positivity or negativity for the single test methods.
3. Discussion
To our knowledge, this is the first study evaluating the role of E7 oncoprotein detection in patients with OPC for a precise distinction of HPV-driven from non-HPV-driven OPCs with a simple tumor surface brush sample. This test was initially developed to triage HPV-positive women being screened for cervical cancer [17]. A positive result in this assay (values greater than 0.5 pg/well (OD > 0.076)) was defined to be consistent with upregulated E7 function, corresponding to a transcriptionally relevant hr-HPV infection. Compared to p16 IHC, this ELISA-based E7 oncoprotein assay is easy to perform and less time-consuming. As different de-escalation treatment strategies to reduce toxicity in HPV-positive OPC patients are in current development [18,19,20], the detection of HPV-induced OPCs is essential. As previously mentioned, p16 is not sufficient as a single test method, particularly because it is a cell cycle deregulation marker, which may be expressed also in other tumor setups with no HPV involvement. Rb loss through a non-HPV associated mutation can result likewise in p16 expression [21]. In this case, the presence of a high level of E7 oncoprotein, the only source of which is transcriptionally active hr-HPV infection detected, may be a more specific biomarker.
Despite the low sensitivity and specificity of the assay, E7 oncoprotein detection was significantly associated with the reference method (p = 0.048), consisting of concordant HPV DNA and p16 results.
E7 oncoprotein positivity was also associated with the ASA score (p = 0.04), indicating the lower rate of comorbidities [22] in HPV-positive patients [23]. This assumes that E7 oncoprotein detection may identify younger and healthier patients benefiting from a de-escalation therapy. Further findings were an association between the E7 oncoprotein and smoking status; the E7 oncoprotein was more often expressed in non-smokers (p = 0.009). This is in accordance with previous studies describing HPV-positive patients to be more commonly non-smokers, whereas smoking and alcohol consumption are the pathogenic mechanisms in non-HPV-driven OPCs [24,25,26]. There was no statistically significant difference regarding the subsite of the OPC; however, despite the low sample size of this subsite, no E7 oncoprotein expression was found in OPCs of the uvula or lateral pharyngeal wall.
Interestingly, in nine patients, a single positive result for the E7 oncoprotein was found, and also, in nine patients, a single negative result was found. The single negative ones may indicate an OPC definitely of non-HPV origin despite the presence of HPV DNA in the lesion, i.e., a non-transforming HPV co-infection.
The single negative result of E7 could indicate episomally latent hr-HPV in the cells of a tobacco smoker and alcohol-induced carcinoma without HPV being involved in the carcinogenesis. A further explanation may be the low sensitivity of ELISA-based antigen tests, with a consequence of failing to detect low levels but yet relevant amounts of E7 oncoprotein.
The single E7-positive result may also encourage us to consider resetting the cutoff value for E7 positivity. Since the test kit was originally approved for triaging in cervical cancer screening, it may be essential to re-evaluate the assay in the context of OPC to optimize an appropriate threshold for this entity. In our study, we observed much better performance characterized by the best percent agreement and high specificity when using a higher cutoff value than provided by the manufacturer (also supported by receiver operating characteristic analysis).
Data from cervical cancer studies show that, in some tumors, although definitely caused by HPV, tests are negative for HPV DNA [27,28]. A plausible explanation is the non-productive nature of the infection in the oncogenic setup, in which viral DNA is integrated in the host. This situation, being transcriptionally active, may be characterized by high expression of transforming proteins, with less virus being released. The fact that we are taking brush samples from the surface of the tumor may support this notion. We cannot explain with certainty the single E7 positive cases. They may be due to the non-productive nature of the infection in tumors. Such phenomena are known among patients with invasive cervical cancers [29].
Moreover, in this study, a certain proportion of patients (6 patients, 10.7%) positive for p16 IHC and negative for HPV DNA was observed. The importance of additional HPV DNA testing to identify HPV-positive OPCs was described in several studies [12,30,31]. As previously described, these patients seem to have a less favorable prognosis than HPV DNA and p16-positive patients [12,32]. Weinberger and coworkers classified 79 OPC patients into class I (HPV DNA negative/p16 low positive), class II (HPV DNA positive/p16 low positive) and class III (HPV DNA positive/ p16 high positive). Class III patients had improved overall survival (p = 0.0095) and disease-free survival (p = 0.03) in comparison to class I and class II OPC patients [33].
Detection of the HPV oncoproteins in the clinical routine of OPC patients is still challenging. The brush test is very feasible and easily applied; however, further studies based on our results and further developments may be necessary. The brush is designed for the cervix, so, in the oropharyngeal region, a smaller brush would be more applicable in order to ensure that the tumor surface can be brushed more precisely without touching the surrounding tissue. A major limitation of the study is the lack of data on E7 mRNA expression. Taken by some as the gold standard for the classification of transcriptionally active hr-HPV, results of mRNA PCR may have supplemented the low sensitivity of this ELISA-based antigen assay. However, we used the combination of HPV DNA and p16 detection as a reference method, since this combination is the standard method for the diagnosis HPV-driven tumor according to many guidelines [11,12]. The role of routine E7 mRNA detection should, however, be elucidated in a future study. Since the great majority (>95%) of HPV driven tumors are due to HPV 16 and HPV 18 [34], the fact that our ELISA assay was limited to the three genotypes (16, 18 and 45) is a negligible limitation. A possible cross-reaction between other genotypes genetically closely linked to these three genotypes cannot be excluded. A further limitation of the study is the fact that this ELISA assay detects only E7 protein and not E6, another oncogenic protein owned by HPV which is of carcinogenic significance in a different pathway. The additional detection of E6 oncoprotein might have added valuable information to the data already generated by this study. Further studies need to elaborate on the diagnostic value of this method to additionally determine E6 oncoprotein for these hr-HPV types in order to accurately identify HPV-driven OPCs.
4. Materials and Methods
This was a prospectively designed study, and patients presenting with OPC between January 2018 and June 2020 at the Department of Otorhinolaryngology, Medical University Innsbruck, Austria were included. Each patient who agreed to participate in this study gave informed consent. The study was conducted in full accordance with the principles expressed in the Declaration of Helsinki and was approved by the ethics committee of the Medical University Innsbruck. The respective reference number was 1147/2018.
Patients were included only when the histology confirmed squamous cell carcinoma of the oropharynx and the patient’s age was at least 18 years. Patients with other diagnoses than OPC in the histopathological examination were excluded. With the exception of 3 patients, all patients had a primary tumor in the oropharyngeal regions, and 3 patients presented with a recurrence or second primary in the oropharynx. In all patients, the E7 brush test was performed before treatment.
4.1. Specimen Harvest and Handling
Details of specimen harvesting and handling has been described in a previous work [35]. In short, each patient suspected to have OPC received a panendoscopy under anesthesia. In this procedure, two different cytology brush tests for HPV DNA and E7 oncoprotein detection were conducted (digene® HC2 DNA Collection Device, Qiagen, Hilden, Germany and ThinPrep® PreservCyt Solution, Hologic, Manchester, UK) by gently brushing the tumor surface. The brushes were then placed in a sterile container and sent to the Institute of Virology, Medical University Innsbruck. Furthermore, tumor biopsies were obtained and fixed in formalin for routine histopathological examination at the Department of Pathology, Medical University Innsbruck. Additionally, a tumor biopsy for p16 IHC detection was kept in cell culture medium and immediately sent to the Laboratory for Molecular Biology and Oncology, Department of Otorhinolaryngology.
4.2. E7 Oncoprotein Detection by Brush Test
Detection of E7 oncoprotein for genotypes of HPV 16, 18 and/or 45 was conducted using a sandwich ELISA test system (recomWell HPV 16/18/45, Mikrogen, Neurid, Germany) developed and validated (CE-labelled) initially to support diagnostic and therapeutic decisions in cervical cancer screening [17]. The ELISA microtiter plates (MTPs) were coated with rabbit monoclonal antibodies (RabMabs) specific for the three genotypes mentioned above. The remaining oropharyngeal swab samples, after cytology and HPV genotyping, were centrifuged and the pellets were incubated with the RabMabs after a couple of lysis steps. Biotinylated polyclonal goat-anti-E7 antibodies were used to detect E7 antigens which remained bound to the monoclonal antibodies after final washing steps. Results were provided as optical density (OD), with a limit of detection of 0.5 pg of protein per well, with the corresponding cutoff value of the OD being 0.076. We also evaluated the association between the reference method and E7 oncoprotein positivity using further arbitrarily selected cutoff values.
4.3. DNA Amplification and HPV Genotyping
For HPV DNA detection, real-time PCR was used based on the amplification of the L1 open reading frames (ORF). As internal control for the availability of cellular material, a PCR for the housekeeping gene beta globin was performed. The HPV DNA was considered positive if the fluorescence signal appeared before the fortieth cycle [36]. Further genotyping was performed on all HPV-positive samples using reverse line blot hybridization on nitrocellulose membrane strips containing genotype specific probes (AmpliQuality HPV-TYPE EXPRESS, AB Analitica®, Padova, Italy) [37]. With this genotyping kit, it is possible to identify 40 different HPV types, including all hr and several low-risk HPV types.
4.4. Immunohistochemistry
First, five-micrometer thin paraffin sections were dewaxed and then antigens were retrieved in an automated staining system (Ventana, Discovery, Tucson, AZ, USA). For p16 detection, a commercial diagnostic assay was used (CINtec® Histology V-Kit, Roche diagnostics, Basel, Switzerland). The staining was completed by using a universal secondary antibody solution, the DAB MAP Kit and hematoxylin counterstaining (both Ventana products). One experienced observer evaluated the tumor cell areas, and specimens were considered p16-positive if ≥66% of the cells in the tumor areas revealed immunohistochemical reaction products.
4.5. Data Analysis
Patient clinical data were presented in tabular form. A comparison of HPV DNA, E7 detection and p16 IHC in samples obtained from OPC patients was performed. For each investigated variable, a binary outcome (positive/negative) was obtained. Combined hr-HPV DNA positivity and p16 positivity served as the reference method. Contingency tables were analyzed with Fisher exact test or Pearson chi-square. Diagnostic accuracy parameters including sensitivity and specificity were calculated using the diagnostic test routines of MedCalc. For data analysis, SPSS Statistics 24 software (IBM Corporation, Armonk NY, USA) was used.
5. Conclusions
We do not have strong evidence that confirms that ELISA-based E7 oncoprotein replaces the reference method for diagnosis of HPV-associated OPC. However, looking at the significant association with the reference method of diagnosis, it could be a valuable addition in the case of ambiguous findings. We strongly recommend further investigation and optimization of the test in large prospective studies.
Acknowledgments
The authors thank the biomedical analysts at the Institute of Virology, Bettina Hofer and Agnes Mayr, for supporting this work by conducting E7 ELISA.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/9/2388/s1, Table S1: Comparing the performance of hr-HPV E7 oncoprotein ELISA in detecting HPV-driven OPC* (n = 50).
Author Contributions
Methodology, W.B., J.D.; software, V.H.S.; validation, W.B., B.K., J.L.; formal analysis, V.H.S.; investigation, B.K., W.B., T.B.S.; resources, H.R.; data curation, V.H.S., H.S.; writing—original draft preparation, B.K.; writing—review and editing, W.B., J.D., J.L.; visualization, J.I., M.C.G.; supervision, W.B.; project administration, W.B., B.K.; funding acquisition, H.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chaturvedi A.K., Engels E.A., Pfeiffer R.M., Hernandez B.Y., Xiao W., Kim E., Jiang B., Goodman M.T., Sibug-Saber M., Cozen W., et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahal B.A., Catalano P.J., Haddad R.I., Hanna G.J., Kass J.I., Schoenfeld J.D., Tishler R.B., Margalit D.N. Incidence and Demographic Burden of HPV-Associated Oropharyngeal Head and Neck Cancers in the United States. Cancer Epidemiol. Biomark. Prev. 2019;28:1660–1667. doi: 10.1158/1055-9965.EPI-19-0038. [DOI] [PubMed] [Google Scholar]
- 3.Wittekindt C., Wagner S., Bushnak A., Prigge E.S., von Knebel Doeberitz M., Wurdemann N., Bernhardt K., Pons-Kuhnemann J., Maulbecker-Armstrong C., Klussmann J.P. Increasing Incidence rates of Oropharyngeal Squamous Cell Carcinoma in Germany and Significance of Disease Burden Attributed to Human Papillomavirus. Cancer Prev. Res. (Phila) 2019;12:375–382. doi: 10.1158/1940-6207.CAPR-19-0098. [DOI] [PubMed] [Google Scholar]
- 4.Dahlstrom K.R., Bell D., Hanby D., Li G., Wang L.E., Wei Q., Williams M.D., Sturgis E.M. Socioeconomic characteristics of patients with oropharyngeal carcinoma according to tumor HPV status, patient smoking status, and sexual behavior. Oral Oncol. 2015;51:832–838. doi: 10.1016/j.oraloncology.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sturgis E.M., Cinciripini P.M. Trends in head and neck cancer incidence in relation to smoking prevalence: An emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110:1429–1435. doi: 10.1002/cncr.22963. [DOI] [PubMed] [Google Scholar]
- 6.Schnelle C., Whiteman D.C., Porceddu S.V., Panizza B.J., Antonsson A. Past sexual behaviors and risks of oropharyngeal squamous cell carcinoma: A case-case comparison. Int. J. Cancer. 2017;140:1027–1034. doi: 10.1002/ijc.30519. [DOI] [PubMed] [Google Scholar]
- 7.Wagner S., Wittekindt C., Sharma S.J., Wuerdemann N., Juttner T., Reuschenbach M., Prigge E.S., von Knebel Doeberitz M., Gattenlohner S., Burkhardt E., et al. Human papillomavirus association is the most important predictor for surgically treated patients with oropharyngeal cancer. Br. J. Cancer. 2017;116:1604–1611. doi: 10.1038/bjc.2017.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kofler B., Laban S., Busch C.J., Lorincz B., Knecht R. New treatment strategies for HPV-positive head and neck cancer. Eur. Arch. Oto-Rhino-Laryngol. 2014;271:1861–1867. doi: 10.1007/s00405-013-2603-0. [DOI] [PubMed] [Google Scholar]
- 9.Mirghani H., Bellera C., Delaye J., Dolivet G., Fakhry N., Bozec A., Garrel R., Malard O., Jegoux F., Maingon P., et al. Prevalence and characteristics of HPV-driven oropharyngeal cancer in France. Cancer Epidemiol. 2019;61:89–94. doi: 10.1016/j.canep.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Jung A.C., Briolat J., Millon R., de Reynies A., Rickman D., Thomas E., Abecassis J., Clavel C., Wasylyk B. Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int. J. Cancer. 2010;126:1882–1894. doi: 10.1002/ijc.24911. [DOI] [PubMed] [Google Scholar]
- 11.Mena M., Taberna M., Tous S., Marquez S., Clavero O., Quiros B., Lloveras B., Alejo M., Leon X., Quer M., et al. Double positivity for HPV-DNA/p16(ink4a) is the biomarker with strongest diagnostic accuracy and prognostic value for human papillomavirus related oropharyngeal cancer patients. Oral Oncol. 2018;78:137–144. doi: 10.1016/j.oraloncology.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen J.H., Gronhoj C., Hakansson K., Friborg J., Andersen E., Lelkaitis G., Klussmann J.P., Wittekindt C., Wagner S., Vogelius I.R., et al. Risk profiling based on p16 and HPV DNA more accurately predicts location of disease relapse in patients with oropharyngeal squamous cell carcinoma. Ann. Oncol. 2019;30:629–636. doi: 10.1093/annonc/mdz010. [DOI] [PubMed] [Google Scholar]
- 13.Narisawa-Saito M., Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: Roles of E6 and E7 proteins. Cancer Sci. 2007;98:1505–1511. doi: 10.1111/j.1349-7006.2007.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.zur Hausen H. Papillomaviruses causing cancer: Evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- 15.Moody C.A., Laimins L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat Rev. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez S.L., Stremlau M., He X., Basile J.R., Munger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 2001;75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agorastos T., Chatzistamatiou K., Moysiadis T., Kaufmann A.M., Skenderi A., Lekka I., Koch I., Soutschek E., Boecher O., Kilintzis V., et al. Human papillomavirus E7 protein detection as a method of triage to colposcopy of HPV positive women, in comparison to genotyping and cytology. Final results of the PIPAVIR study. Int. J. Cancer. 2017;141:519–530. doi: 10.1002/ijc.30761. [DOI] [PubMed] [Google Scholar]
- 18.Foster C.C., Seiwert T.Y., MacCracken E., Blair E.A., Agrawal N., Melotek J.M., Portugal L., Brisson R.J., Gooi Z., Spiotto M.T., et al. Dose and Volume De-Escalation for Human Papillomavirus-Positive Oropharyngeal Cancer is Associated with Favorable Post-Treatment Functional Outcomes. Int. J. Radiat. Oncol. Biol. Phys. 2020 doi: 10.1016/j.ijrobp.2020.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Bigelow E.O., Seiwert T.Y., Fakhry C. Deintensification of treatment for human papillomavirus-related oropharyngeal cancer: Current state and future directions. Oral Oncol. 2020;105:e104652. doi: 10.1016/j.oraloncology.2020.104652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel R.R., Ludmir E.B., Augustyn A., Zaorsky N.G., Lehrer E.J., Ryali R., Trifiletti D.M., Adeberg S., Amini A., Verma V. De-intensification of therapy in human papillomavirus associated oropharyngeal cancer: A systematic review of prospective trials. Oral Oncol. 2020;103:e104608. doi: 10.1016/j.oraloncology.2020.104608. [DOI] [PubMed] [Google Scholar]
- 21.Liang C., Marsit C.J., McClean M.D., Nelson H.H., Christensen B.C., Haddad R.I., Clark J.R., Wein R.O., Grillone G.A., Houseman E.A., et al. Biomarkers of HPV in head and neck squamous cell carcinoma. Cancer Res. 2012;72:5004–5013. doi: 10.1158/0008-5472.CAN-11-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riechelmann H., Neagos A., Netzer-Yilmaz U., Gronau S., Scheithauer M., Rockemann M.G. The ASA-score as a comorbidity index in patients with cancer of the oral cavity and oropharynx. Laryngo-Rhino-Otol. 2006;85:99–104. doi: 10.1055/s-2005-870291. [DOI] [PubMed] [Google Scholar]
- 23.Grisar K., Dok R., Schoenaers J., Dormaar T., Hauben E., Jorissen M., Nuyts S., Politis C. Differences in human papillomavirus-positive and -negative head and neck cancers in Belgium: An 8-year retrospective, comparative study. Oral Surg., Oral Med. Oral Pathol. Oral Radiol. 2016;121:456–460. doi: 10.1016/j.oooo.2015.10.035. [DOI] [PubMed] [Google Scholar]
- 24.Beynon R.A., Lang S., Schimansky S., Penfold C.M., Waylen A., Thomas S.J., Pawlita M., Waterboer T., Martin R.M., May M., et al. Tobacco smoking and alcohol drinking at diagnosis of head and neck cancer and all-cause mortality: Results from head and neck 5000, a prospective observational cohort of people with head and neck cancer. Int. J. Cancer. 2018;143:1114–1127. doi: 10.1002/ijc.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deschler D.G., Richmon J.D., Khariwala S.S., Ferris R.L., Wang M.B. The "new" head and neck cancer patient-young, nonsmoker, nondrinker, and HPV positive: Evaluation. Otolaryngol. Head Neck Surg. 2014;151:375–380. doi: 10.1177/0194599814538605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lassen P., Lacas B., Pignon J.P., Trotti A., Zackrisson B., Zhang Q., Overgaard J., Blanchard P., Group M.C. Prognostic impact of HPV-associated p16-expression and smoking status on outcomes following radiotherapy for oropharyngeal cancer: The MARCH-HPV project. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2018;126:107–115. doi: 10.1016/j.radonc.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Kaliff M., Karlsson M.G., Sorbe B., Bohr Mordhorst L., Helenius G., Lillsunde-Larsson G. HPV-negative Tumors in a Swedish Cohort of Cervical Cancer. Int. J. Gynecol. Pathol. 2020;39:279–288. doi: 10.1097/PGP.0000000000000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arroyo Muhr L.S., Lagheden C., Eklund C., Lei J., Nordqvist-Kleppe S., Sparen P., Sundstrom K., Dillner J. Sequencing detects human papillomavirus in some apparently HPV-negative invasive cervical cancers. J. Gen. Virol. 2020;101:265–270. doi: 10.1099/jgv.0.001374. [DOI] [PubMed] [Google Scholar]
- 29.Mills A.M., Dirks D.C., Poulter M.D., Mills S.E., Stoler M.H. HR-HPV E6/E7 mRNA In Situ Hybridization: Validation Against PCR, DNA In Situ Hybridization, and p16 Immunohistochemistry in 102 Samples of Cervical, Vulvar, Anal, and Head and Neck Neoplasia. Am. J. Surg. Pathol. 2017;41:607–615. doi: 10.1097/PAS.0000000000000800. [DOI] [PubMed] [Google Scholar]
- 30.Rietbergen M.M., Snijders P.J., Beekzada D., Braakhuis B.J., Brink A., Heideman D.A., Hesselink A.T., Witte B.I., Bloemena E., Baatenburg-De Jong R.J., et al. Molecular characterization of p16-immunopositive but HPV DNA-negative oropharyngeal carcinomas. Int. J. Cancer. 2014;134:2366–2372. doi: 10.1002/ijc.28580. [DOI] [PubMed] [Google Scholar]
- 31.Parfenov M., Pedamallu C.S., Gehlenborg N., Freeman S.S., Danilova L., Bristow C.A., Lee S., Hadjipanayis A.G., Ivanova E.V., Wilkerson M.D., et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc. Natl. Acad. Sci. USA. 2014;111:15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrone F., Gloghini A., Cortelazzi B., Bossi P., Licitra L., Pilotti S. Isolating p16-positive/HPV-negative oropharyngeal cancer: An effort worth making. Am J Surg Pathol. 2011;35:774–777. doi: 10.1097/PAS.0b013e3182116a45. [DOI] [PubMed] [Google Scholar]
- 33.Weinberger P.M., Yu Z., Haffty B.G., Kowalski D., Harigopal M., Brandsma J., Sasaki C., Joe J., Camp R.L., Rimm D.L., et al. Molecular classification identifies a subset of human papillomavirus--associated oropharyngeal cancers with favorable prognosis. J. Clin. Oncol. 2006;24:736–747. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- 34.LeConte B.A., Szaniszlo P., Fennewald S.M., Lou D.I., Qiu S., Chen N.W., Lee J.H., Resto V.A. Differences in the viral genome between HPV-positive cervical and oropharyngeal cancer. PLoS ONE. 2018;13:e0203403. doi: 10.1371/journal.pone.0203403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kofler B., Borena W., Manzl C., Dudas J., Wegscheider A.S., Jansen-Durr P., Schartinger V., Riechelmann H. Sensitivity of tumor surface brushings to detect human papilloma virus DNA in head and neck cancer. Oral Oncol. 2017;67:103–108. doi: 10.1016/j.oraloncology.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Abreu A.L., Souza R.P., Gimenes F., Consolaro M.E. A review of methods for detect human Papillomavirus infection. Virol. J. 2012;9:e262. doi: 10.1186/1743-422X-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason A.G.S., Vettorato M., Negri G., Mian C., Brusauro F., Bortolozzo K. Detection of high-risk HPV genotypes in cervical samples: A comparison study of a novel real time pcr/reverse line blot-based technique and the digene HC2 assay. Pathologica. 2012;6:104–146. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.