Abstract
Xanthomonas gardneri is one of the causal agents of bacterial spot (BS), an economically important bacterial disease of tomato and pepper. Field-deployable and portable loop-mediated isothermal amplification (LAMP)-based instruments provide rapid and sensitive detection of plant pathogens. In order to rapidly and accurately identify and differentiate X. gardneri from other BS-causing Xanthomonas spp., we optimized a new real-time monitoring LAMP-based method targeting the X. gardneri-specific hrpB gene. Specificity and sensitivity of real-time and colorimetric LAMP assays were tested on the complex of bacterial strains pathogenic to tomato and pepper and on plants infected by the pathogen. The assay detection limit was 1 pg/μL of genomic DNA with an assay duration of only 30 min. The use of portable and handheld instruments allows for fast analysis, reducing the diagnosis time, and can contribute to proper disease management and control of X. gardneri. Due to the high efficiency of this method, we suggest its use as a standard diagnostic tool during phytosanitary controls.
Keywords: Xanthomonas spp., bacterial spot, LAMP
1. Introduction
Xanthomonas gardneri is one of the species of the genus Xanthomonas that are currently considered to cause bacterial spot (BS) on tomato and pepper. It was first isolated in former Yugoslavia and named Pseudomonas gardneri [1]. Later, this name was suggested to be a synonym for X. vesicatoria [2]. However, DNA–DNA hybridization indicated that these organisms were genetically distinct [3]. Four distinct groups of Xanthomonas were differentiated based on various physiological and molecular tests [4]. Group A was named X. euvesicatoria; group B mostly included X. vesicatoria, group C X. perforans and group D X. gardneri [5]. Additionally, based on amplified fragment length polymorphism and multilocus sequence analysis (MLSA) of genes atpD, dnaK, efp and gyrB partial sequences, the four bacterial strains pathogenic for tomato and pepper were identified to the species level in Xanthomonas spp. [6]. The whole genomic sequence of X. euvesicatoria showed a 99.7% relationship with X. perforans [7]. It was suggested that the two species should be combined into one and subdivided into two pathovars: X. euvesicatoria pv. euvesicatoria and X. euvesicatoria pv. perforans [8]. Based on phylogenetic analysis and comparison of partial gyrase B gene sequences, X. gardneri and X. cynarae (an artichoke pathogen) were recognized as species closely related to X. hortorum [9]. New data from MLSA using four housekeeping genes (dnaK, fyuA, gyrB and rpoD) indicated that X. gardneri appears to be identical to X. cynarae but further investigation of the classification of Xanthomonas is still needed [10].
Various molecular, biochemical and physiological assays have been used to characterize Xanthomonas species. For Xanthomonas spp. causing BS, PCR-based molecular detection assays were used [11,12]. A region of hrpB operon as a potential source target for primers and probes for real-time TaqMan PCR assay for X. euvesicatoria, X. vesicatoria, X. gardneri and X. perforans was also evaluated [13]. PCR primer sets with TaqMan probes were developed to distinguish between X. gardneri and other Xanthomonas groups causing bacterial spot of tomato [14]. Currently, DNA-based techniques are the most used methods for molecular diagnostic of this group of pathogenic bacteria.
Nucleic acid amplification is one of the most valued methods in science but still requires sophisticated instruments for amplification and detection. A majority of the techniques for nucleic acid analysis utilize the polymerase chain reaction (PCR) amplification method, which requires repeated cycles of three temperature-dependent steps during the amplification of the target nucleic acid sequence [15,16]. Isothermal amplification techniques such as nucleic acid sequence-based amplification, helicase-dependent amplification, rolling circle amplification, multiple displacement amplification, recombinase polymerase amplification and loop-mediated isothermal amplification (LAMP) were developed to facilitate DNA amplification in simpler ways [17]. LAMP is a novel isothermal amplification method for amplifying a limited amount of DNA copies into millions of copies in less than an hour. It is a molecular technique of nucleic acid amplification where a set of four to six different primers binds to six to eight different regions on the target gene, providing high specificity [18]. A basic LAMP primer set consists of two outer primers (F3 and B3) and two inner primers (forward inner primer—FIP and backward inner primer—BIP). Loop primers may be used to accelerate the reaction [19]. LAMP methods utilize Bacillus smithii, Bacillus stearothermophilus or Geobacillus sp. DNA polymerases with high strand displacement activity at optimal working temperature ranged between 60 and 65 °C [18,20,21]. This method can be easily performed at the point-of-care thanks to its isothermal nature. It has been successfully used for rapid and specific detection of plant bacteria from infected plant tissues and soil. The objective of this study was to develop a real-time and colorimetric LAMP protocol for specific detection of X. gardneri that would be superior to the current PCR-based methods in its speed, ease of use and possibility of point-of-care application.
2. Materials and Methods
2.1. Bacterial Strains and Culturing
Bacterial strains (Table 1) were obtained from Belgian Co-ordinated Collections of Microorganisms—Bacteria Collection, Gent (BCCM/LMG); French Collection of Plant associated bacteria, Beaucouze Cedex (CFBP); Czech Collection of Microorganisms, Brno (CCM); Crop Research Institute Collection, Czech Republic, Prague—Ruzyně (CRI); Deutsche Sammlung Microorganismen und Zellkulturen GmbH, Germany (DSMZ); National Collection of Plant Pathogenic Bacteria, UK, York (NCPPB); National Collection of Agricultural and Industrial Microorganisms—(NCAIM); and Horticulture Research International, Wellesbourne, UK, (HRI-W). All Clavibacter spp. were grown on nutrient broth yeast extract (NBY) [22] at 27 °C for 3 to 7 days, depending on the subspecies. Pantoea agglomerans was grown on King’s B medium [23] at 25 °C for 24 to 48 h. Other bacteria were cultured on MPAg medium (meat–peptone agar with glucose: 20 g of nutrient agar no. 2, 2.6 g of yeast extract, 5 g of glucose, 10 g of agarose—added to 500 mL of distilled H2O, pH adjusted to 7.2 with 1 M NaOH and solidified) at 28 °C for 24–48 h, or 3–7 days (only X. fragariae).
Table 1.
1 Bacteria listed in collection; 2 Type; 3 Pathotype; 4 not specified (N.S.) by collection; 5 (+) for positive detection, (−) for negative detection; BCCM/LMG—Belgian Co-ordinated Collections of Microorganismse Bacteria Collection; CFBP—French Collection of Plant Pathogenic Bacteria; CCM—Czech Collection of Microorganisms; CRI—Crop Research Institute Collection; NCPPB—National Collection of Plant Pathogenic Bacteria; DSMZ—Deutsche Sammlung von Mikroorganismen und Zellkulturen; NCAIM—National Collection of Agricultural and Industrial Microorganisms; HRI-W—Horticulture Research International, Wellesbourne, UK.
| Name 1 | Collection | Number in Collection | Geographic Origin | LAMP Result 5 |
|---|---|---|---|---|
| Xanthomonas gardneri | ||||
| X. gardneri 2 | DSMZ | 19127 | Yugoslavia | + |
| X. gardneri | CFBP | 8588 | France (Réunion) | + |
| X. gardneri | CFBP | 7992 | France (Réunion) | + |
| X. gardneri | CFBP | 8120 | Costa Rica | + |
| Other (non-gardneri) Xanthomonas | ||||
| X. alfalfae subsp. alfalfae | CFBP | 3836 | Sudan | − |
| X. arboricola pv. pruni | BCCM/LMG | 854 | New Zealand | − |
| X. axonopodis pv. allii | CFBP | 6369 | Not specified (N.S.) 4 | − |
| X. axonopodis pv. carotoae | NCPPB | 3440 | Brazil | − |
| X. axonopodis pv. vesicatoria | CRI | 1013 | Czech Republic | − |
| X. campestris pv. incanae | HRI-W | 6377 | UK | − |
| X. campestris pv. phaseoli | NCAIM | B.01695 | Hungary | − |
| X. campestris pv. pisi | NCAIM | B.01393 | N.S. 4 | − |
| X. campestris pv. raphani | HRI-W | 8305 | UK | − |
| X. campestris pv. vesicatoria | BCCM/LMG | 934 | Brazil | − |
| X. campestris pv. vesicatoria | BCCM/LMG | 921 | USA (Long Island) | − |
| X. euvesicatoria | BCCM/LMG | 918 | India | − |
| X. euvesicatoria | BCCM/LMG | 922 | USA (Florida) | − |
| X. euvesicatoria | BCCM/LMG | 921 | USA (Long Island) | − |
| X. fragariae 2 | CFBP | 6766 | USA | − |
| X. oryzae pv. Oryzicola 3 | CFBP | 2286 | N.S. 4 | − |
| X. perforans 9.2 2 | CFBP | 7293 | USA (Florida) | − |
| X. perforans 9.2 | CFBP | 8122 | Thailand | − |
| X. perforans 2 | DSMZ | 18975 | USA | − |
| X. vesicatoria | BCCM/LMG | 925 | Hungary | − |
| X. vesicatoria 2 | CFBP | 2537 | New Zealand | − |
| X. vesicatoria | BCCM/LMG | 920 | Italy | − |
| Other species | ||||
| Agrobacterium tumefaciens | CCM | 2835 | Czech Republic | − |
| Burkholderia glumae | BCCM/LMG | 20138 | Philippines (province Jalajala Riza) | − |
| B. glumae 2 | BCCM/LMG | 2196 | Japan (Ehime) | − |
| Clavibacter michiganensis subsp. michiganensis | CFBP | 1460 | France | − |
| C. michiganensis subsp. sepedonicus | NCPPB | 3467 | Poland | − |
| Erwinia amylovora | CRI | Ea10/96 | Czech Republic | − |
| Pantoea agglomerans 2 | CFBP | 3845 | N.S. 4 | − |
| Pseudomonas syringae pv. phaseolicola | CRI | 186/2 | Czech Republic | − |
| P. syringae pv. pisi | NCPPB | 3496 | USA | − |
| P. syringae pv. syringae | NCPPB | 2306 | Italy | − |
| P. syringae pv. tomato | CRI | 8119 | Czech Republic | − |
| Ralstonia pseudosolanacearum | CFBP | 3936 | China (Guangdong) | − |
| R. solanacearum | NCPPB | 2505 | Sweden | − |
| Stenotrophomonas sp. | NCPPB | 2859 | Turkey | − |
2.2. DNA Isolation
Freshly grown bacteria (5–10 mg) were taken from plates. Total DNA was extracted using NucleoSpin® Microbial DNA kit (Macherey-Nagel, Dylan, Germany) according to the manufacturer’s protocol. For agitation, a Retsch® Mixer Mill MM400 was used for 4 min at maximal frequency (30 Hz). Concentration of extracted DNA was measured with a BioSpec-nano spectrophotometer (Shimadzu, Kyoto, Japan).
2.3. Primer Design
Partial sequence of the hrpB gene (GenBank ID: KX437681.1) was used for X. gardneri primer design [13]. One set of five primers (external primers F3 and B3; internal primers FIP and BIP and one loop primer LoopB) selected from total of three prospective primer sets was designed in PrimerExplorer software (Eiken Chemical Company, Tokyo, Japan; https://primerexplorer.jp/e/) and synthesized by Macrogen (Republic of Korea). Oligonucleotide sequences of the best primer set used for further testing are listed in Table 2. A sixth primer (LoopF) could not be designed due to the configuration of the template binding sites.
Table 2.
Loop-mediated isothermal amplification (LAMP) primers.
| Primer Name | Primer Length (nt) | Tm (°C) | Sequence (5′–3′) |
|---|---|---|---|
| F3 | 16 | 61.60 | CGGGGTGCAGGTCAGC |
| B3 | 15 | 61.13 | ACCGGCACCGCCAAG |
| FIP | 37 | - | CCACCTCGGCACGTTGCAGGCGAGGTATGCGAGTTGC |
| BIP | 35 | - | GCCGCCATCTCGCCTTGCGCCCCGATCCGATCACG |
| LB | 17 | 61.26 | CGAGCTGGTGGGCTTGT |
2.4. Real-Time LAMP
The LAMP reactions were performed in a QuantStudio™ 6 Flex Real-Time PCR System (Thermofisher, Waltham, MA, USA) and BioRangerTM (Diagenetix, Honolulu, HI, USA). The reaction mixture contained 12.5 µL of Isothermal master mix (Optigene, Horsham, UK), 0.2 μM of each of outer primers (F3 and B3), 1.6 μM of each of inner primers (FIP and BIP) and 0.4 μM of loop primer (only LoopB). Lastly, 3 µL of template genomic DNA (10 ng/µL) was added and the reaction was brought to a final volume of 25 µL with nuclease-free H2O. The LAMP reaction mixtures were incubated for 60 min at 65 °C, followed by heating at 98 °C for 2 min to terminate the reactions. Melting analysis followed at temperature range from 70 °C to 99 °C with increments of 0.1 °C per second that allow for the generation of derivative melting curves. All LAMP assays were replicated at least three times, and all experiments included negative (no-template) controls.
2.5. Electrophoresis of LAMP
LAMP reaction products (5 and 10 µL, respectively) were analyzed via electrophoresis on a 1.5% agarose gel made of 1× TBE buffer (Tris/Borate/EDTA: 89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.3) stained with ethidium bromide (SigmaAldrich, St. Louis, MO, USA) at 90 V (4 V/cm) for 1 h. DirectLoad™ Wide Range DNA Marker was used for LAMP samples as a molecular standard for comparing molecular weight. To avoid contamination among samples, different laboratories were used for DNA extraction and for reaction mixture preparation, PCR Clean™ (Minerva Biolabs, Berlin, Germany, DE) was used for surface cleaning and only filtered pipette tips were used. After electrophoresis, the gel was visualized under UV illumination on GeneSys (Syngene, Cambridge, UK).
2.6. Colorimetric LAMP
Colorimetric LAMP reactions were carried out in a 25 μL volume containing 1.6 μM of each of inner primers FIP and BIP, 0.2 μM of each of outer primers F3 and B3, 0.4 μM of LoopB primer, 12.5 μL of 2× Colorimetric LAMP Master Mix (Cat. No. M1800, New England Biolabs) and 5 μL of DNA template (10 ng/µL). Reactions were incubated at 65 °C for 15 and 30 min in a heat block before results were recorded by naked eye.
2.7. Sensitivity and Specificity (Real-Time and Colorimetric LAMP)
Serial 10-fold dilutions of 10 ng/μL X. gardneri DNA ranging from 10 ng to 10 fg were used as a template for sensitivity test of DNA amplification assays. Specificity of assays was tested using Xanthomonas spp. strains and other related bacteria pathogenic for tomato and pepper (Table 1). No-template control (NC; water) was included in each LAMP run.
2.8. LAMP Assay on Plant Tissues
For testing of the LAMP assay with an infected plant tissue, 50 tomato and pepper plants were grown in growth chamber (Sanyo MLR-351H, Osaka, Japan) in temperature of 24 °C and 72% humidity for 4 weeks. Inoculation was done according to ISTA (2015) [24] methodology. Two youngest fully developed leaves were pierced in the area surrounding the major veins by six needles dipped in the suspension of X. gardneri type strain DSMZ 19127. First symptoms were observed after 3–4 weeks. Samples for subsequent testing were taken from the boundary of healthy and infected tissue and transferred to 50 µL of TE buffer. DNA extraction was performed as described above.
3. Results
In this study, we developed a LAMP-based real-time and colorimetric assay for specific, sensitive, reliable and robust detection and differentiation of X. gardneri. For primer design, we selected the hrpB gene due to its high species specificity.
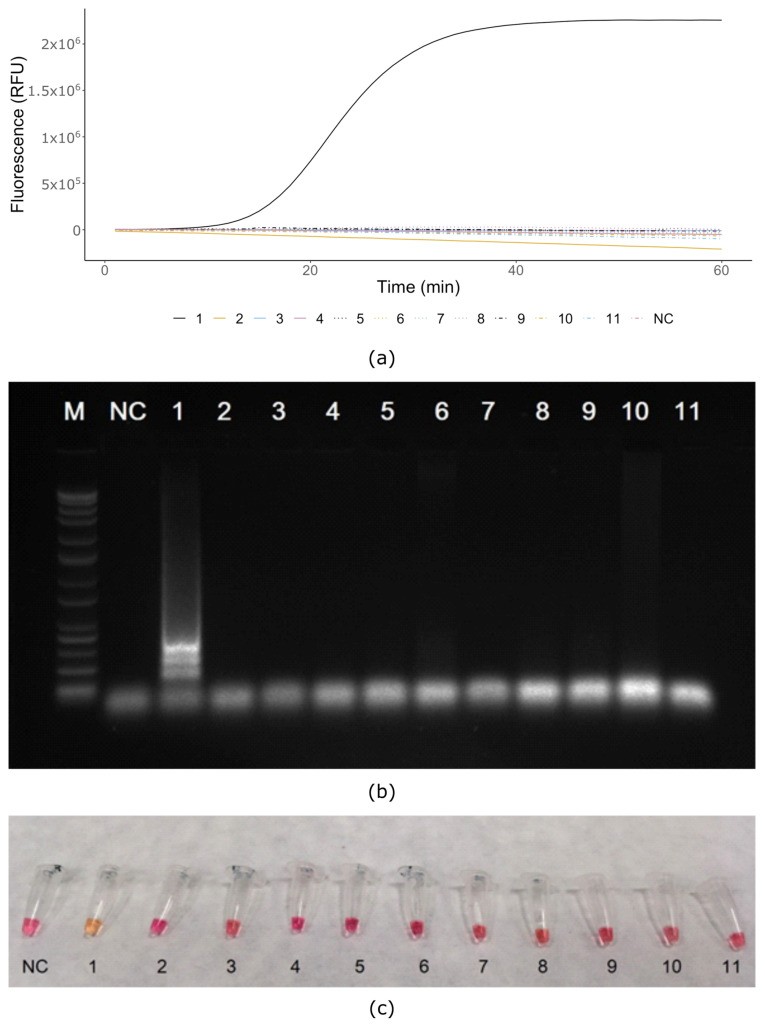
LAMP assays distinctly amplified X. gardneri DNA, but none of the nontarget Xanthomonas species, including strains closely related to the X. gardneri, produced amplicons within 60 min (Figure 1a, Table 1). All the real-time LAMP assays were performed using the Isothermal master mix in both a QuantStudio™ 6 Flex Real-Time PCR System and a BioRangerTM (Diagenetix, USA). Amplification of X. gardneri was first observable after 15 min of isothermal amplification. All reactions were subsequently analyzed on electrophoresis gel after 60 min of amplification, to confirm the results (Figure 1b). The X. gardneri reaction product displayed the typical ladder-like appearance of LAMP products, while no amplicons were detectable from the other reactions. Colorimetric LAMP assays were performed using 2× Colorimetric LAMP Master Mix in a heat block. No amplification was observed for any sample after 15 min. After 30 min, amplification was only visible in test tubes with X. gardneri DNA (Figure 1c).
Figure 1.
Specificity of selected bacterial species: (a) real-time LAMP assay (QuantStudio™ 6 Flex Real-Time PCR System); (b) gel electrophoresis of LAMP products confirms results of real-time LAMP; (c) Colorimetric Master Mix assay after 30 min reaction time; M—ladder 100 bp (NEB, Hitchin, UK), NC—negative control water, 1—Xanthomonas gardneri DSMZ 19127, 2—Xanthomonas axonopodis pv. vesicatoria CRI 1013, 3—Xanthomonas perforans DSMZ 18975, 4—Xanthomonas vesicatoria BCCM/LMG 920, 5—Erwinia amylovora CRI Ea10/96, 6—Burkholderia glumae BCCM/LMG 20138, 7—Clavibacter michiganensis susp. michiganensis CFBP 1460, 8—Clavibacter michiganensis subsp. sepedonicus NCPPB 3467, 9—Pseudomonas syringae pv. syringae NCPPB 2306, 10—Pseudomonas syringae pv. tomato CRI 8119, 11—Ralstonia solanacearum NCPPB 2505.
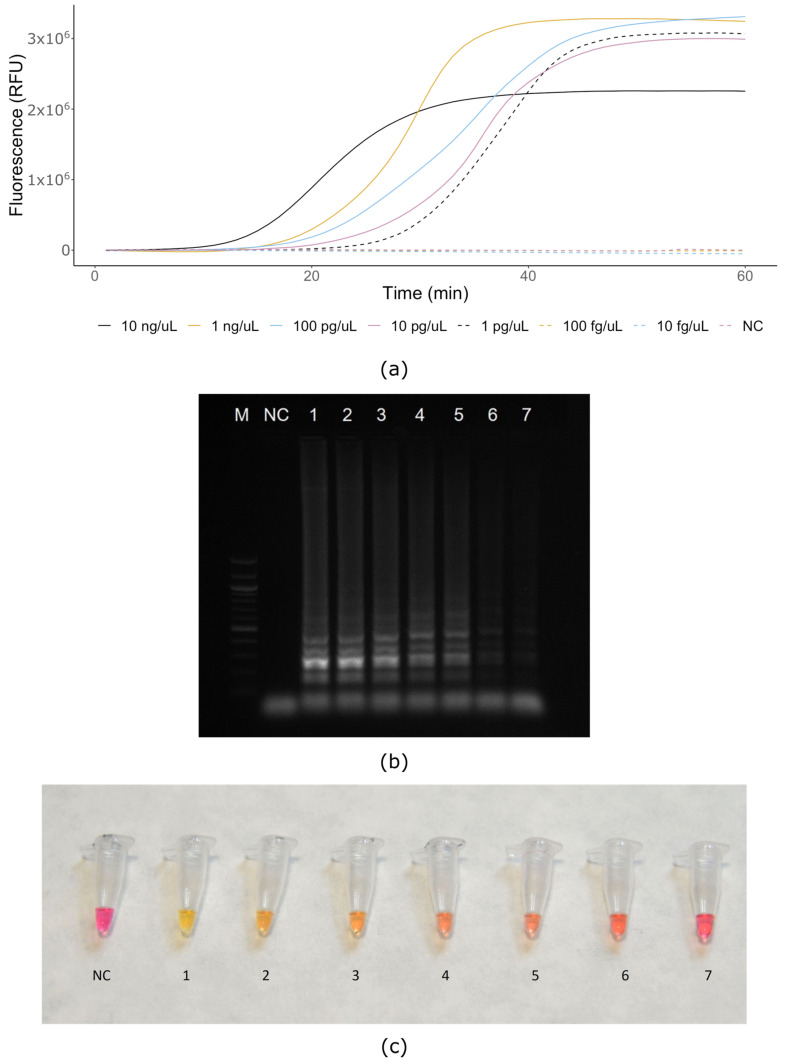
To determine the sensitivity of real-time and colorimetric assays, genomic DNA diluted to concentrations ranging from 10 ng to 10 fg was used. A no-template control (water) was included in each experimental replication. The lowest concentration limit of detection was 1 pg/μL for X. gardneri (Figure 2a). However, very weak amplification was also observable via electrophoresis with concentrations of 100 fg and 10 fg (Figure 2b). Considering all the variants of the assay (real-time, electrophoresis, colorimetry), we defined the lowest amount of bacterial genomic DNA required to reliably detect the bacterium to 1 pg/μL after 30 min of amplification (Figure 2a–c) as an aggregate detection limit.
Figure 2.
Serial 10-fold dilutions: (a) real-time LAMP assay (QuantStudio™ 6 Flex Real-Time PCR System); (b) gel electrophoresis of LAMP products confirms results of real-time LAMP; (c) Colorimetric Master Mix assay after 30 min reaction time; M—ladder 100 bp (NEB, UK), NC—negative control water, 1—10 ng/μL, 2—1 ng/μL, 3—100 pg/μL, 4—10 pg/μL, 5—1 pg/μL, 6—100 fg/μL, 7—10 fg/μL.
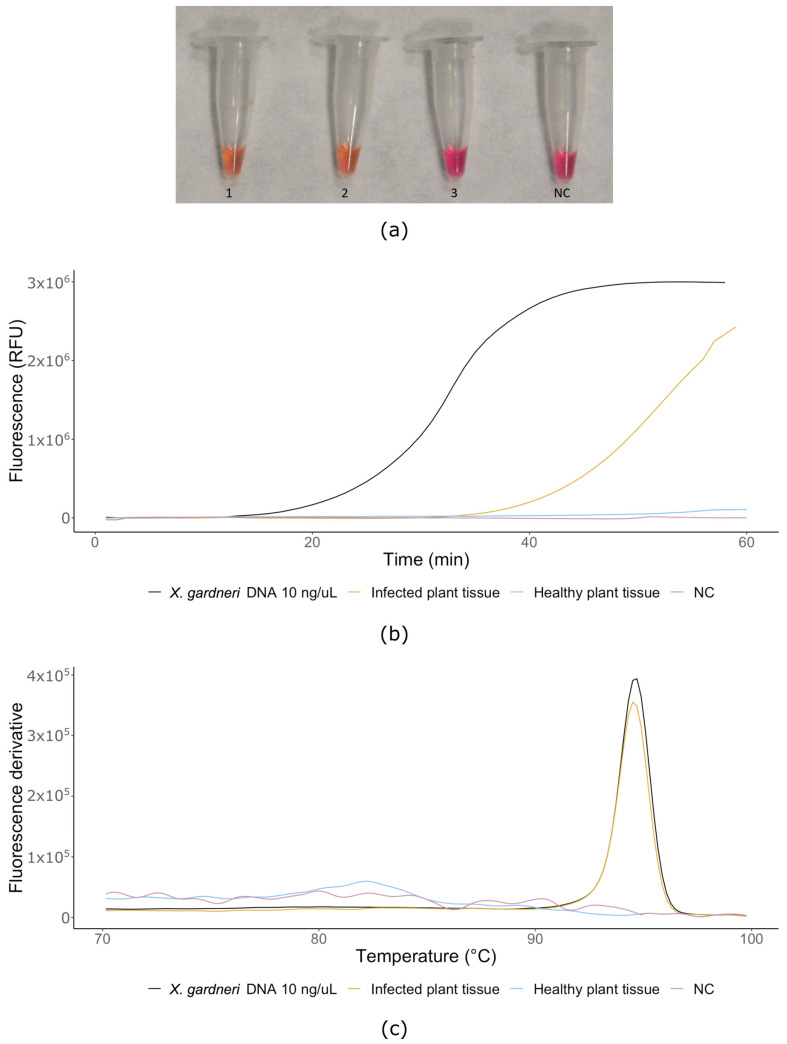
All X. gardneri infected tomato and pepper plant samples were positive for X. gardneri. No positive amplification was observed when LAMP primers were tested with healthy tomato plants (Figure 3a,b). The melting curves obtained for all X. gardneri isolates and from infected tomato plant showed the same melting temperature (Tm) 94.27 °C, indicating similar sequences, and hence similar amplicons (Figure 3c). The possibility of using the LAMP assay in the field was tested on the BioRangerTM platform using tomato plants inoculated by X. gardneri (Figure 3b). Amplification was detected by 30 min (45 min on BioRangerTM), suggesting that with our primers, this platform is a convenient and reliable method for point-of-care detection of X. gardneri if technological limitations of mobile devices are carefully considered.
Figure 3.
Naturally infected plants: (a) colorimetric LAMP assay, 1—X. gardneri bacterial culture DNA (10 ng/μL, strain DSMZ 19127), 2—infected plant sample, 3—healthy plant tissue, NC—non-template control (water); (b) real-time LAMP in BioRangerTM device; (c) melting curves of LAMP products.
Although 30 min was enough to reliably detect the pathogen in most cases, we recommend 60 min of amplification due to possible variations in detection setup in different laboratories. The real-time nature of the LAMP assay may suggest its use for quantification of X. gardneri as well. However, our assay was not designed with quantification in mind, and all data resulting from following our recommendations should only be treated as detection, or not, of X. gardneri.
4. Discussion
Here, we developed a LAMP assay to detect X. gardneri, one of the causal agens of BS of tomato. LAMP assays are available for some other Xanthomonas spp. like X. arboricola pv. pruni, the causal agent of Stone Fruit Bacterial Spot; X. oryzae pv. oryzae the causal agent of Bacterial Blight disease; X. oryzae pv. oryzicola, the causal agent of Bacterial Leaf Streak disease; X. campestris pv. musacearum, the causal agent of Banana Xanthomonas Wilt; X. citri subsp. citri and X. fuscans subsp. aurantifolii, the causal agents of Citrus Bacterial Canker; and X. translucens, causal agent of cereal leaf streak [25,26,27,28,29]. LAMP assay for detection of X. euvesicatoria, the causal agent of BS of tomato, was also developed [30]. Our X. gardneri-specific assay substantially improves the detection of the bacteria associated with BS of tomato detection.
Different assays for detection of all the BS of tomato associated Xanthomonas were developed before. Most notably, many PCR based assays are available for single pathogens, and several multiplex-PCR assays for detection of all four species has also been developed [12,13,14,31,32]. In general, specificity of PCR based assays compared to LAMP seems to be similar. Sensitivity varies, and sometimes is not directly comparable (in some tests of sensitivity assessment bacterial cultures are diluted before DNA extraction, other tests use diluted DNA). With a detection limit 1 pg/μL of target DNA, our LAMP assay performs considerably better than the most sensitive X gardneri molecular detection assay by Araújo et al. [31] with detection limit of 50 pg/μL or 5 × 104 CFU/mL.
The time to obtain results with LAMP is about 30 min, compared to one or more hours using real-time PCR or even more time-consuming end-point PCR. Although single LAMP reactions are usually slightly more expensive than PCR-based alternatives due to additional primers and more expensive polymerase, LAMP reactions can be performed on considerably cheaper and simpler devices such as the BioRangerTM or Genie® II (Optigene, Horsham, UK). This allows the use of LAMP assays in field conditions, which also makes LAMP more suitable for environments where high initial investment is not possible or desirable. Unlike PCR, LAMP does not directly provide information about size of amplified DNA, which is usually determined by performing melting analysis. For routine pathogen detection, this is not an issue, if the assay is well designed and tested. LAMP assays are, however, more difficult to design compared to PCR-based methods due to the higher complexity of the reaction and lower availability of software for LAMP assay design. Additionally, quantification is less accurate than real-time PCR [33]. Lower precision amplification devices (such as mobile devices like BioRangerTM), DNA extraction method, in-field contaminants and other factors may further decrease the precision of quantification, so we suggest only qualitative interpretation of results resulting from our assay. When preparing the assay, end users should carefully consider prolonging the time of amplification with mobile devices and for less efficient DNA extraction methods with possible contamination of sample DNA. For evaluation of the data, we suggest using the multi-operator validation test [30], which provides results without further statistical analysis and is therefore suitable for point-of-care detection.
The assay developed in this work is based on the gene sequence of hrpB gene. The hrp gene cluster was successfully used before as a target for molecular detection of phytopathogenic bacteria [13,34,35]. Other common targets include genes for ribosomal RNA, atpD, gumD, gyrB, rpfB and many others, or other non-annotated sequences obtained by whole genome analysis [28,36,37,38]. As expected from the literature, our own DNA alignment and BLAST search, the hrpB gene sequence of X. gardneri is conserved within the BS of tomato related Xanthomonas (with some variable regions within the group, allowing distinction of the four species), while being distinct enough from other related or tomato specific bacteria. This allowed us to design a LAMP primer set, which is specific only for X. gardneri, enabling improvement of current BS-related Xanthomonas detection and distinction between X. gardneri and other species. To date, no other LAMP assay for detection of this bacteria was published.
Acknowledgments
The authors would like to thank Eliška Peňázová and the mentioned bacterial strain collections for providing the strains for testing.
Author Contributions
All authors participated in the conception and design of the experiments and analyzed the resulting data; D.S. and P.B. performed the experiments; D.S., P.B., S.P.C. and V.Č. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the Ministry of Education, Youth and Sports of the Czech Republic within INTER-EXCELLENCE/INTER-COST programme, grant: LTC19014 (MSMT-15739/2019-8). This article is based on work from European Cooperation in Science and Technology (COST) action CA16107 EuroXanth, supported by COST. S.P.C. is supported by the AFRI Education and Workforce Development Postdoctoral Fellowship (grant no. 2018-08122) from the U.S. Department of Agriculture, National Institute of Food and Agriculture.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Šutic D.D. Bakterioze crvenog patlidžana = Bacteriosis of tomatoes. Institut za zaštitu bilja; Belgrade, Serbia: 1957. pp. 1–65. [Google Scholar]
- 2.Dye D.W. Cultural and biochemical reactions of additional Xanthomonas spp. N. Z. J. Sci. 1966;9:913. [Google Scholar]
- 3.De Ley J., Segers P., Gillis M. Intra- and Intergeneric Similarities of Chromobacterium and Janthinobacterium Ribosomal Ribonucleic Acid Cistrons. Int. J. Syst. Bacteriol. 1987;28:154–168. doi: 10.1099/00207713-28-2-154. [DOI] [Google Scholar]
- 4.Jones J.B., Bouzar H., Stall R.E., Almira E.C., Roberts P.D., Bowen B.W., Sudberry J., Strickler P.M., Chun J. Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. Int. J. Syst. Evol. Microbiol. 2000;50:1211–1219. doi: 10.1099/00207713-50-3-1211. [DOI] [PubMed] [Google Scholar]
- 5.Jones J.B., Lacy G.H., Bouzar H., Stall R.E., Schaad N.W. Reclassification of the Xanthomonads Associated with Bacterial Spot Disease of Tomato and Pepper. Syst. Appl. Microbiol. 2004;27:755–762. doi: 10.1078/0723202042369884. [DOI] [PubMed] [Google Scholar]
- 6.Hamza A.A., Robène-Soustrade I., Jouen E., Gagnevin L., Lefeuvre P., Chiroleu F., Pruvost O. Genetic and Pathological Diversity Among Xanthomonas Strains Responsible for Bacterial Spot on Tomato and Pepper in the Southwest Indian Ocean Region. Plant Dis. 2010;94:993–999. doi: 10.1094/PDIS-94-8-0993. [DOI] [PubMed] [Google Scholar]
- 7.Barak J.D., Vancheva T., Lefeuvre P., Jones J.B., Timilsina S., Minsavage G.V., Koebnik R. Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of type 3 effectors. Front. Plant Sci. 2016;7:1805. doi: 10.3389/fpls.2016.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Constantin E.C., Cleenwerck I., Maes M., Baeyen S., Van Malderghem C., De Vos P., Cottyn B. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol. 2016;65:792–806. doi: 10.1111/ppa.12461. [DOI] [Google Scholar]
- 9.Parkinson N., Aritua V., Heeney J., Cowie C., Bew J., Stead D. Phylogenetic analysis of Xanthomonas species by comparison of partial gyrase B gene sequences. Int. J. Syst. Evol. Microbiol. 2007;57:2881–2887. doi: 10.1099/ijs.0.65220-0. [DOI] [PubMed] [Google Scholar]
- 10.Young J.M., Park D.C., Shearman H.M., Fargier E. A multilocus sequence analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 2008;31:366–377. doi: 10.1016/j.syapm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Araújo E.R., Costa J.R., Ferreira M.A.S.V., Quezado-Duval A.M. Widespread distribution of Xanthomonas perforans and limited presence of X. gardneri in Brazil. Plant Pathol. 2017;66:159–168. doi: 10.1111/ppa.12543. [DOI] [Google Scholar]
- 12.Koenraadt H., Van Betteray B., Germain R., Hiddink G., Jones J.B., Oosterhof J. Development of specific primers for the molecular detection of bacterial spot of pepper and tomato. Acta Hortic. 2009;808:99–102. doi: 10.17660/ActaHortic.2009.808.13. [DOI] [Google Scholar]
- 13.Strayer A.L., Jeyaprakash A., Minsavage G.V., Timilsina S., Vallad G.E., Jones J.B., Paret M.L. A Multiplex Real-Time PCR Assay Differentiates Four Xanthomonas Species Associated with Bacterial Spot of Tomato. Plant Dis. 2016;100:1660–1668. doi: 10.1094/PDIS-09-15-1085-RE. [DOI] [PubMed] [Google Scholar]
- 14.Cuppels D.A., Louws F.J., Ainsworth T. Development and Evaluation of PCR-Based Diagnostic Assays for the Bacterial Speck and Bacterial Spot Pathogens of Tomato. Plant Dis. 2006;90:451–458. doi: 10.1094/PD-90-0451. [DOI] [PubMed] [Google Scholar]
- 15.Fang X., Li J., Chen Q. One new method of nucleic acid amplification—Loop-mediated isothermal amplification of DNA. Virol. Sin. 2008;23:167–172. doi: 10.1007/s12250-008-2929-8. [DOI] [Google Scholar]
- 16.Parida M., Sannarangaiah S., Dash P.K., Rao P.V.L., Morita K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008;18:407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zanoli L.M., Spoto G. Isothermal Amplification Methods for the Detection of Nucleic Acids in Microfluidic Devices. Biosensors. 2013;3:18–43. doi: 10.3390/bios3010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 20.Aliotta J.M., Pelletier J.J., Ware J.L., Moran L.S., Benner J.S., Kong H. Thermostable Bst DNA polymerase I lacks a 3′ → 5′ proofreading exonuclease activity. Genet. Anal. Biomol. Eng. 1996;12:185–195. doi: 10.1016/S1050-3862(96)80005-2. [DOI] [PubMed] [Google Scholar]
- 21.Hawwa R., Aikens J., Turner R.J., Santarsiero B.D., Mesecar A.D. Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus. Arch. Biochem. Biophys. 2009;488:109–120. doi: 10.1016/j.abb.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaad N.W., Jones J.B., Chun W. Laboratory Guide for the Identification of Plant. Pathogenic Bacteria. 3rd ed. American Phytopathological Society, APS Press; St. Paul, MN, USA: 2001. [Google Scholar]
- 23.King E.O., Ward M.K., Raney D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954;44:301–307. doi: 10.5555/uri:pii:002221435490222X. [DOI] [PubMed] [Google Scholar]
- 24.Roberts S.J., Koenraadt H. 7-019a: Detection of Xanthomonas Campestris pv. Campestris on Brassica spp. 4th ed. International Seed Testing Association; Bassersdorf, Switzerland: 2015. International Rules for Seed Testing, Chapter 7: Validated Seed Health Testing Methods. [Google Scholar]
- 25.Bühlmann A., Pothier J.F., Tomlinson J.A., Frey J.E., Boonham N., Smits T.H.M., Duffy B. Genomics-informed design of loop-mediated isothermal amplification for detection of phytopathogenic Xanthomonas arboricola pv. pruni at the intraspecific level. Plant Pathol. 2013;62:475–484. doi: 10.1111/j.1365-3059.2012.02654.x. [DOI] [Google Scholar]
- 26.Hodgetts J., Hall J., Karamura G., Grant M., Studholme D., Boonham N., Karamura E., Smith J.J. Rapid, specific, simple, in-field detection of Xanthomonas campestris pathovar musacearum by loop-mediated isothermal amplification. J. Appl. Microbiol. 2015;119:1651–1658. doi: 10.1111/jam.12959. [DOI] [PubMed] [Google Scholar]
- 27.Lang J.M., Langlois P., Nguyen M.H.R., Triplett L.R., Purdie L., Holton T.A., Djikeng A., Vera Cruz C.M., Verdier V., Leach J.E. Sensitive Detection of Xanthomonas oryzae Pathovars oryzae and oryzicola by Loop-Mediated Isothermal Amplification. Appl. Environ. Microbiol. 2014;80:4519–4530. doi: 10.1128/AEM.00274-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langlois P.A., Snelling J., Hamilton J.P., Bragard J.P., Koebnik R., Verdier V., Triplett L.R., Blom J., Tisserat N.A., Leach J.E. Characterization of the Xanthomonas translucens Complex Using Draft Genomes, Comparative Genomics, Phylogenetic Analysis, and Diagnostic LAMP Assays. Phytopathology. 2017;107:519–527. doi: 10.1094/PHYTO-08-16-0286-R. [DOI] [PubMed] [Google Scholar]
- 29.Rigano L.A., Marano M.R., Castagnaro A.P., Do Amaral A.M., Vojnov A.A. Rapid and sensitive detection of Citrus Bacterial Canker by loop-mediated isothermal amplification combined with simple visual evaluation methods. BMC Microbiol. 2010;10:176. doi: 10.1186/1471-2180-10-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larrea-Sarmiento A., Dhakal U., Boluk G., Fatdal L., Alvarez A., Strayer-Scherer A., Paret M., Jones J., Jenkins D., Arif M. Development of a genome-informed loop-mediated isothermal amplification assay for rapid and specific detection of Xanthomonas euvesicatoria. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-32295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Araújo E.R., Costa J.R., Ferreira M.A.S.V., Quezado-Duval A.M. Simultaneous detection and identification of the Xanthomonas species complex associated with tomato bacterial spot using species-specific primers and multiplex PCR. J. Appl. Microbiol. 2012;113:1479–1490. doi: 10.1111/j.1365-2672.2012.05431.x. [DOI] [PubMed] [Google Scholar]
- 32.Obradovic A., Mavridis A., Rudolph K., Janse J.D., Arsenijevic M., Jones J.B., Minsavage G.V., Wang J.F. Characterization and PCR-based typing of Xanthomonas campestris pv. vesicatoria from peppers and tomatoes in Serbia. Eur. J. Plant Pathol. 2004;110:285–292. doi: 10.1023/B:EJPP.0000019797.27952.1d. [DOI] [Google Scholar]
- 33.Wang H., Turechek W.W. A loop-mediated isothermal amplification assay and sample preparation procedure for sensitive detection of Xanthomonas fragariae in strawberry. PLoS ONE. 2016;11:e0147122. doi: 10.1371/journal.pone.0147122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kebede M., Timilsina S., Ayalew A., Admassu B., Potnis N., Minsavage G.V., Jones J.B. Molecular characterization of Xanthomonas strains responsible for bacterial spot of tomato in Ethiopia. Eur. J. Plant Pathol. 2014;140:677–688. doi: 10.1007/s10658-014-0497-3. [DOI] [Google Scholar]
- 35.Leite R.P., Minsavage G.V., Bonas U., Stall R.E. Detection and identification of phytopathogenic Xanthomonas strains by amplification of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 1994;60:1068–1077. doi: 10.1128/AEM.60.4.1068-1077.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keremane M.L., Ramadugu C., Rodriguez E., Kubota R., Shibata S., Hall R.F. A rapid field detection system for citrus huanglongbing associated‘Candidatus Liberibacter asiaticus’ from the psyllid vector, Diaphorina citri Kuwayama and its implications in disease management. Crop. Prot. 2015;68:41–48. doi: 10.1016/j.cropro.2014.10.026. [DOI] [Google Scholar]
- 37.Golmohammadi M., Llop P., Scuderi G., Gell I., Graham J.H., Cubero J. mRNA from selected genes is useful for specific detection and quantification of viable Xanthomonas citri subsp. citri. Plant Pathol. 2012;61:479–488. doi: 10.1111/j.1365-3059.2011.02526.x. [DOI] [Google Scholar]
- 38.Simões T.H., Gonçalves E.R., Rosato Y.B., Mehta A. Differentiation of Xanthomonas species by PCR-RFLP of rpfB and atpD genes. FEMS Microbiol. Lett. 2007;271:33–39. doi: 10.1111/j.1574-6968.2007.00691.x. [DOI] [PubMed] [Google Scholar]