Abstract
Generalized modules for membrane antigens (GMMA) represent a technology particularly attractive for designing affordable vaccines against Gram-negative bacteria. We explored such technology for the development of O-antigen-based vaccines against Shigella and nontyphoidal Salmonella. Adsorption of GMMA on Alhydrogel was required for abrogation of pyrogenicity in rabbits, and Shigella sonnei GMMA on Alhydrogel was well tolerated and immunogenic in humans. Quantification of key antigens in formulated vaccines was fundamental for release and to check stability overtime. Traditionally, the direct quantification of antigens adsorbed on aluminum salts has been challenging, and the quantification of each active ingredient in multicomponent formulated vaccines has been even more complicated. To directly quantify each active ingredient and unbound drug substances in formulated vaccines, we developed the Formulated Alhydrogel competitive ELISA (FAcE) and the competitive ELISA method, respectively. The methods were both fully characterized, assessing specificity, repeatability, intermediate precision, and accuracy, for S. sonnei OAg quantification, both in a single component or multicomponent GMMA formulation also containing S. flexneri GMMA. The developed immunological methods allowed us to fully characterize Shigella GMMA drug products, supporting their preclinical and clinical development. The same methods, already extended to GMMA from nontyphoidal Salmonella and Neisseria meningitidis, could be potentially extended to any antigen formulated on Alhydrogel.
Keywords: GMMA, vaccines, Shigella, Alhydrogel formulations, assay characterization, FAcE, cELISA
1. Introduction
Generalized modules for membrane antigens (GMMA) are outer membrane exosomes shed from Gram-negative bacteria that are genetically modified to enhance the level of particle production [1]. Similar to native outer membrane vesicles, GMMA contain outer membrane lipids, outer membrane proteins, and soluble periplasmic components. They are self-adjuvanted particles, containing Toll-like receptor agonists, and present multiple bacterial antigens in their natural conformation as they faithfully resemble the composition of bacterial outer membrane. GMMA represent an attractive technology to develop high-yield and highly immunogenic particles, ideal for using as vaccines, especially to fight neglected diseases for which the balance between price and effectiveness is essential [2,3]. We have used GMMA technology to develop vaccines against enteric diseases, for example, Shigella and nontyphoidal Salmonella [3,4,5]. In both cases the O-antigen (OAg) portion of lipopolysaccharide (LPS) molecules was the key target for protective immunity. GMMA producing strains have been further manipulated to reduce the risk of inducing systemic reactogenicity, by playing with the lipid A acylation pattern [6,7]. The genetic manipulation of lipid A resulted in a reduction of in vitro pro-inflammatory response of over 500-fold as compared with wild-type lipid A [7]. Furthermore, the adsorption of GMMA with Alhydrogel allowed the abrogation of pyrogenicity in rabbits [5]. S. sonnei GMMA have been tested on Alhydrogel in humans, and the results have shown that they are well tolerated, immunogenic [8,9], and able to induce a strong anamnestic response upon boosting [10].
A panel of analytical techniques needs to be put in place to control each step of drug substance and drug product production. In particular it is essential to determine, with high specificity, the quality and quantity of each active ingredient present in the final formulation, as well as to follow over time their stability, as required by regulatory authorities. Traditionally, the direct quantification of antigens adsorbed on aluminium salts has been challenging, and antigens needed to be extracted using laborious and often ineffective desorption procedures [11,12,13,14,15]. Even more complicated is the quantification of each active ingredient of multicomponent formulated vaccines. To directly quantify the OAg in GMMA-based Alhydrogel-formulated vaccines, we have developed the Formulated Alhydrogel competive ELISA (FAcE) method [16]. Similarly, for the direct quantification of unbound drug substances in formulates, we developed a competitive ELISA (cELISA) assay.
Here, we present full development and characterization of the two methods in terms of accuracy, repeatability, reproducibility, specificity, and quantification range. In particular, we characterize cELISA and FAcE for S. sonnei OAg quantification, both when GMMA are in monovalent or multicomponent drug product formulations. FAcE assay on drug products and cELISA on drug product supernatants allow us to fully characterize the Alhydrogel-formulated vaccines, including lot-to-lot variations and the possibility of following their stability over time. The methods also work properly for multicomponent formulations and can be virtually extended to quantification of any Alhydrogel-formulated antigen.
2. Materials and Methods
2.1. GMMA Preparation (Drug Substance, DS)
Shigella sonnei 1790-GMMA [5], as well as GMMA from three different strains of S. flexneri, were produced by fermentation in a chemically defined media, purified by two tangential flow filtrations, and fully characterized using a panel of analytical methods, as previously reported [5].
Single component GMMA DS were diluted in PBS supplemented with Tween-20 (Sigma, St. Louis, MO, USA) 0.05% and 0.1% BSA (Sigma) at a nominal final concentration of 15 μg/mL (OAg based) prior to performing the assay.
A multicomponent GMMA DS mixture was obtained by mixing three different S. flexneri GMMA samples with S. sonnei GMMA in PBS Tween-20 0.05% and 0.1% BSA with a final nominal concentration of each sugar target of ~0.3 μg/mL (for S. sonnei OAg) and ~1 μg/mL (for each S. flexneri OAg).
2.2. Formulation of GMMA (Drug Product, DP)
S. sonnei 1790-GMMA were formulated at ~15.5 μg/mL or ~4 μg/mL nominal OAg content (used, respectively, for characterization of assay for single component and multicomponent DP) by mixing them for 2 h, at room temperature, with Alhydrogel 2% (Brenntag Biosector, Frederikssund, Denmark) (final concentration of 0.7 mg Al3+/mL) in Tris buffer (final concentration of 10 mM Tris, 9 g/L NaCl and pH 7.4).
GMMA purified from three different S. flexneri strains were also formulated (either as single component or as mixture of the three components) by mixing them for 2 h at room temperature (at ~26 μg/mL or ~15 μg/mL nominal OAg content of each active ingredient, respectively, for characterization of assay for single component as negative controls, and multicomponent DP) with Alhydrogel 2% (Brenntag Biosector, Denmark) (final concentration of 0.7 mg Al3+/mL) in water, and then conditioned with phosphate buffer (final concentration of 20 mM phosphate buffer, pH 6.5) and NaCl (final concentration 9 g/L).
The multicomponent formulation was obtained by bedside mixing the tri-component S. flexneri GMMA formulation (resulting in 15 μg/mL of each S. flexneri OAg) and S. sonnei single component drug product (resulting in ~4 μg/mL of S. sonnei OAg).
All formulations were characterized in terms of pH, osmolarity, and by visual inspection. Total proteins in DS used for formulations were quantified using the Lowry assay method [17] (in case of S. sonnei) or microBCA method (for S. flexneri), and non-adsorbed proteins in DP evaluated by SDS-PAGE Silver Staining analysis of the supernatant, obtained after centrifuging the formulation. GMMA adsorption on Alhydrogel was confirmed to be ≥95% for all single component and for the multicomponent formulations used here.
2.3. Working Principle of Assays
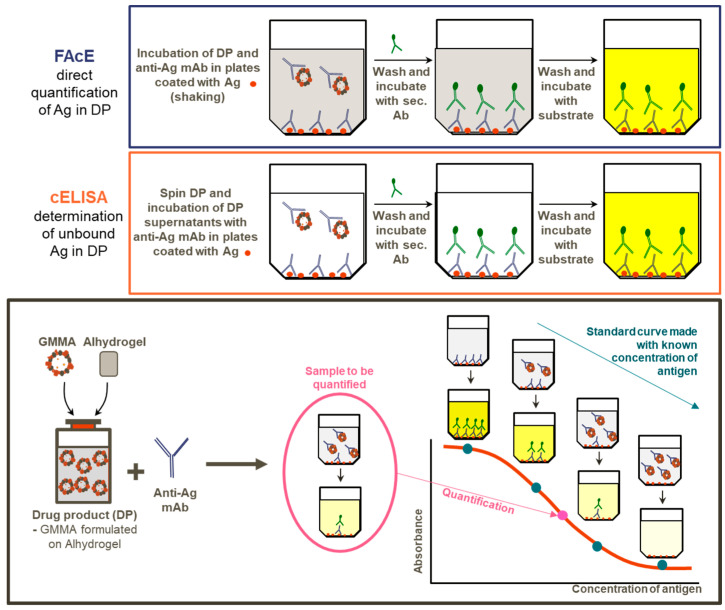
Both cELISA for the direct quantification of S. sonnei OAg in DP supernatant and FAcE assay for the direct quantification of S. sonnei OAg in DP are based on the competition of an anti-S. sonnei LPS monoclonal antibody (mAb) for binding to the S. sonnei LPS antigen coated on the ELISA plate or the LPS of the GMMA suspension present in the ELISA wells. The OAg part of the LPS is the active ingredient of Shigella GMMA-based vaccines, and therefore we refer to this across the text. The more antigen that is present in the sample to be measured, the more mAb will bind to it, and less will be available to bind to the coated antigen. Consequently, the lowest signal is obtained with the highest quantity of measured antigen in the solution, and vice versa (Figure 1).
Figure 1.
Visual descriptions of methods. Generalized modules for membrane antigens (GMMA) are formulated in Alhydrogel and their OAg content (active ingredient) can be directly quantified by Formulated Alhydrogel competitive ELISA (FAcE), whereas the quantity of unbound material can be determined by competititve ELISA (cELISA) performed on drug product supernatant. Both assays are quantitative and rely on standard curves made with a known quantity of antigen assayed in parallel to samples with unknown quantity. Samples and standard curve points compete with coated antigen for a limited amount of anti-antigen (anti-Ag) specific monoclonal antibodies (mAb) in a similar way.
2.4. cELISA Protocol for Determination of Non-Absorbed S. sonnei GMMA in DP Supernatants
Our cELISA method was based on a reference standard curve built by 10 sequential dilutions (2.3-fold apart) of a freshly prepared S. sonnei GMMA solution, starting from a known concentration in terms of OAg of about 15 µg/mL, quantified through a combination of high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) and NMR analysis that determine sugar content of the core region and OAg repeats to core ratio, respectively [5]. Two blank wells were also added containing dilution buffer only (PBS Tween 0.05% and 0.1% BSA). The test samples were GMMA solutions diluted in Tris buffer (final concentration of 10 mM Tris, 9 g/L NaCl, and pH 7.4) in the case of single components, or GMMA diluted in Tris buffer mixed 1:1 with phosphate NaCl buffer (20 mM phosphate buffer, pH 6.5, and NaCl 9 g/L) for multicomponent GMMA solutions; such samples containing GMMA in DP alum-free buffer mimic the potentially unbound GMMA in DP supernatants.
Five identical samples (A–E) were independently prepared by diluting the same solution containing GMMA (either for single component or for multicomponent assay characterization) (Figure S1A) in DP alum-free buffer. Each sample was assayed at 3 independent dilutions, each dispensed in 4 replicates within the same plate. The three independent dilutions were selected to fit within the most linear and central part of the standard curve, and were 0.25, 0.125, and 0.0625 μg/mL respectively. Samples not containing the S. sonnei OAg (samples N1, N2, and N3 constituted by 3 different S. flexneri GMMA solutions) were also assayed as negative controls to determine specificity of the assay at 0.25 μg/mL (the highest concentration assayed for samples containing S. sonnei OAg), in 4 replicates within the same plate.
The assay was run in independent triplicate in 96-well plates (Nunc round bottom Maxisorp ELISA plates), with the standard curve in duplicate on each plate. Plates were coated overnight at 4 °C with 100 μL/well of Shigella sonnei LPS at 0.5 µg/mL in PBS and blocked with 200 µL/well of 5% fat-free milk dissolved in PBS buffer at 25 °C for 1 h. Then, plates were washed three times with 250 µL of washing buffer (PBS with 0.05% Tween-20 (PBS-T)). Then, 800 μL of each independent dilution of test samples (and of positive and negative controls), as well as 400 μL of each dilution point of the standard curves, were spiked with an equal volume of anti-LPS mAb (Inbios International, Inc., Seattle, WA, USA, INB3192 purified mouse anti-S. sonnei IgG1) finally diluted 1:10,000 in PBS with 0.1% BSA and 0.05% Tween-20 in 2 mL Eppendorf tubes prior to be added into the test ELISA plates and immediately incubated for 2 h at 25 °C. Then, plates were washed three times with PBS-T. Next, 100 µL of secondary anti-mouse IgG conjugated to alkaline phosphatase (Sigma-Aldrich, A3438) were added to the plates and incubated for 1 h at 25 °C. After three more washes in PBS-T, 100 µL of p-nitrophenyl phosphate substrate solution (Sigma, N2770) were added and plates were incubated for 1 h at 25 °C. Absorbances at 405 and 490 nm were read and the difference between them (OD405nm–OD490nm) was calculated.
2.5. FAcE Protocol for Determination of S. sonnei OAg in DP
The standard curves were prepared by 10 sequential dilutions (2.3-fold apart) of Alhydrogel freshly formulated S. sonnei GMMA, starting from a known concentration in terms of OAg µg/mL (about 15.5 µg/mL), and two blanks (Alhydrogel diluent only). A standard curve was run in duplicate on each plate. The same freshly formulated sample used to build the standard curve was used as internal positive control (and therefore assayed as the other samples) to validate the whole plate within the assay. A plate was considered valid if the measured OAg concentration in the positive control was within a 70–130% accuracy range as compared with its nominal value. Test samples were represented by formulated GMMA (either as single component or as a multicomponent mixture). For each test sample, either five identical samples (A–E) in case of FAcE assay characterization for S. sonnei single component formulation, or four identical samples (sample A–D) in the case of FAcE characterization for quantification of S. sonnei OAg in Shigella multicomponent GMMA formulation, were independently prepared by diluting them in DP buffer (Figure S1B). Each sample was assayed at 3 independent dilutions, each dispensed in 4 replicates within the same plate. The three independent dilutions were selected to fit within the most linear (and central) part of the standard curve, and were 0.25, 0.125, and 0.0625 μg/mL, respectively. Samples not containing the S. sonnei OAg (samples N1, N2, and N3 constituted by 3 different S. flexneri single component formulates) were also assayed as negative controls to determine specificity of the assay at 0.25 μg/mL (the highest concentration assayed for samples containing S. sonnei OAg) in 4 replicates within the same plate.
The assay was run in independent triplicate 96-well plates (Nunc round bottom Maxisorp ELISA plates), with the standard curve in duplicate for each plate. Plates were coated overnight at 4 °C with 100 μL/well of S. sonnei LPS at 0.5 µg/mL and blocked with 200 µL/well of 5% fat-free milk dissolved in PBS buffer at 25 °C, for 1 h. Then, plates were washed three times with 250 µL of washing buffer (PBS with 0.05% Tween-20 (PBS-T)). Then, 800 μL of each independent dilution of test samples (and of positive and negative controls) and 400 μL of each dilution point of the standard curves were spiked with an equal volume of anti-LPS mAb (Inbios International, Inc. INB3192 purified mouse anti-S. sonnei IgG1), finally diluted 1:10,000 in PBS with 1% BSA and 0.05% Tween-20 in 2 mL Eppendorf tubes prior to be added in ELISA test plates, and immediately incubated for 2 h, at room temperature, shaking at 600 rpm using a MixMate (Eppendorf). Then, plates were washed three times with PBS-T and 100 µL of secondary anti-mouse IgG conjugated to alkaline phosphatase (Sigma-Aldrich, A3438) were added to the plates and incubated for 1 h, at 25 °C. After three more washes in PBS-T, 100 µL of p-nitrophenyl phosphate substrate solution (Sigma, N2770) were added, and plates were incubated for 1 h, at 25 °C. Absorbances at 405 and 490 nm were read and the difference between them (OD405nm−OD490nm) was calculated.
2.6. Calculations
Four parameter nonlinear (4PL) regression was applied to the standard curve and the amount of test samples were calculated from the equation Y = ((b/(a − Log10(X)))^(d) − 1)/c, where a, b, c, and d represents values of the curve parameters (respectively, being a = minimum asymptote, b = Hill’s slope, c = inflection point, and d = maximum asymptote), Y the amount determined, and X the absorbance reading. For the standard curve, the back calculation was performed by converting the observed readings of the standards to concentrations of the antigens using the 4PL equation described above. The OD values on which the 4PL curve was linear were automatically calculated and represented the quantification ranges of the standard curves.
Each plate was considered valid if the fitting of the standard curve presented R2 > 0.9, and if the OAg concentrations calculated from the internal positive control sample fell within 70–130% of its nominal concentration (in the case of FAcE assay). For each sample (test samples and positive control), OAg concentration was expressed as the average of all the values entering in the quantification range of the standard curve for each of the three plates assayed.
For precision determination, analysis was performed on three different days by two operators, each time in triplicate. Two different sets of experiments (for each assay) were performed to characterize the assays for monocomponent and multicomponent drug products.
Intermediate precision and repeatability were calculated using MiniTab software v.18 with Gage R & R protocol, considering as factors day and operator. For each experimental value obtained, the accuracy was also determined with the following formula: (experimental value/nominal value) * 100. To assess the overall accuracy for each assay, the average obtained from the differences of each experimental value from the nominal value was calculated with its confidence interval at 95% (CI 95%-indicated as percentage of nominal value).
3. Results
For both cELISA and FAcE, the following parameters were evaluated to characterize the assays: quantification range, precision, accuracy, and specificity. The parameters were evaluated both for single component and multicomponent preparations. Quantification of S. sonnei OAg was used for this purpose as the model antigen, because it was the active ingredient in the most advanced GMMA-based vaccine [5].
3.1. Quantification Range
The quantification range represents the range between the lowest and the highest amount of analyte in a sample which can be quantitatively determined with suitable precision and accuracy. Quantification range was calculated as the range between the minimum and maximum accepted OD (the linear part of the 4PL standard curve fitted to ODs). Results for each plate on each day and by each operator obtained for quantitation of single component formulations were considered to calculate the mean of the minimum and maximum OD and of the resulting minimum and maximum OAg (μg/mL)/well and corresponding standard deviations (Table 1). Both FAcE and cELISA showed a dynamic range ≥2 OD, resulting in a broad range of OAg (µg/mL)/well that could be directly determined within assays. Those ranges were 0.035–1.39 µg/mL and 0.015–1.12 µg/mL for FAcE and cELISA, respectively.
Table 1.
Quantification range.
| Quantification Range | |||
|---|---|---|---|
| Mean All Repeats | Standard Deviation | ||
| FAcE | Maximum OD | 2.7 | 0.20 |
| Min. OAg (µg/mL)/well | 0.04 | 0.01 | |
| Minimum OD | 0.67 | 0.09 | |
| Max. OAg (µg/mL)/well | 1.39 | 0.23 | |
| cELISA | Maximum OD | 2.69 | 0.13 |
| Min. OAg (µg/mL)/well | 0.01 | 0.01 | |
| Minimum OD | 0.46 | 0.04 | |
| Max. OAg (µg/mL)/well | 1.12 | 0.24 | |
3.2. Accuracy
Accuracy is the measure of the closeness agreement between the nominal value of analyte measured (in our case the overall mean of the obtained values was considered to be the nominal value) and the values obtained within the assay. The average value of S. sonnei OAg measured by FAcE in DP was 17.47 and 4.95 µg/mL, respectively, for single component and multicomponent formulations, whereas the average of S. sonnei OAg measured in Alhydrogel-free DP buffer by cELISA were 1.67 and 0.32 µg/mL, respectively, for single component and multicomponent formulations. Accuracy and precision were not evaluated at multiple concentration levels, as the assay was performed by analyzing the sample at different dilutions and by averaging all the results fitting within the quantification range of the standard curves (the minimum and maximum accepted OD for each plate) obtained in each of the three plates. Table 2 and Table 3 (for cELISA and FAcE, respectively) report accuracy %, standard deviation, coefficient of variation %, standard error, and standard error % for each single OAg measurement performed (for test samples and controls, either when quantitating S. sonnei OAg in single or multicomponent drug products or drug product supernatants).
Table 2.
cELISA results for each single test performed. n.d., not detectable; St. Dev, standard deviation; SE, standard error; CV, coefficient of variation.
| Operator | Day | OAg (µg/mL) | St. Dev (µg/mL) | CV % | SE | SE % | Accuracy % | |
|---|---|---|---|---|---|---|---|---|
| Determination of S. sonnei OAg in single component DP supernatant | A | 1 | 1.60 | 0.19 | 11.67 | 0.031 | 1.945 | 95.97 |
| A | 1 | 1.75 | 0.18 | 10.08 | 0.029 | 1.680 | 104.64 | |
| A | 1 | 1.76 | 0.22 | 12.46 | 0.037 | 2.077 | 105.35 | |
| A | 1 | 1.71 | 0.32 | 18.51 | 0.053 | 3.085 | 102.61 | |
| A | 1 | 1.66 | 0.31 | 18.66 | 0.052 | 3.111 | 99.49 | |
| A | 2 | 1.69 | 0.26 | 15.17 | 0.043 | 2.528 | 101.23 | |
| A | 2 | 1.74 | 0.12 | 7.03 | 0.020 | 1.172 | 104.22 | |
| A | 2 | 1.74 | 0.14 | 8.28 | 0.024 | 1.381 | 103.97 | |
| A | 2 | 1.82 | 0.36 | 19.66 | 0.060 | 3.277 | 108.76 | |
| A | 2 | 1.80 | 0.35 | 19.55 | 0.059 | 3.259 | 108.01 | |
| A | 3 | 1.58 | 0.18 | 11.63 | 0.031 | 1.939 | 94.63 | |
| A | 3 | 1.65 | 0.19 | 11.37 | 0.031 | 1.896 | 99.08 | |
| A | 3 | 1.72 | 0.15 | 8.47 | 0.024 | 1.412 | 103.31 | |
| A | 3 | 1.74 | 0.14 | 7.87 | 0.023 | 1.343 | 104.22 | |
| A | 3 | 1.64 | 0.30 | 18.10 | 0.050 | 3.016 | 98.48 | |
| B | 1 | 1.58 | 0.16 | 10.09 | 0.027 | 1.682 | 94.89 | |
| B | 1 | 1.67 | 0.17 | 10.36 | 0.029 | 1.726 | 100.11 | |
| B | 1 | 1.73 | 0.14 | 8.19 | 0.024 | 1.365 | 103.57 | |
| B | 1 | 1.73 | 0.31 | 18.02 | 0.052 | 3.003 | 103.91 | |
| B | 1 | 1.73 | 0.31 | 17.75 | 0.051 | 2.959 | 103.68 | |
| B | 2 | 1.44 | 0.22 | 15.03 | 0.036 | 2.506 | 86.26 | |
| B | 2 | 1.60 | 0.18 | 11.22 | 0.030 | 1.871 | 95.80 | |
| B | 2 | 1.64 | 0.18 | 11.18 | 0.031 | 1.863 | 98.19 | |
| B | 2 | 1.71 | 0.33 | 19.19 | 0.055 | 3.198 | 102.22 | |
| B | 2 | 1.71 | 0.33 | 19.32 | 0.055 | 3.219 | 102.36 | |
| B | 3 | 1.62 | 0.23 | 14.39 | 0.039 | 2.399 | 96.82 | |
| B | 3 | 1.61 | 0.13 | 8.09 | 0.022 | 1.348 | 96.41 | |
| B | 3 | 1.58 | 0.15 | 9.35 | 0.025 | 1.558 | 94.88 | |
| B | 3 | 1.57 | 0.30 | 19.15 | 0.050 | 3.192 | 94.03 | |
| B | 3 | 1.55 | 0.30 | 19.64 | 0.051 | 3.274 | 92.91 | |
| Determination of S. sonnei OAg on multicomponent DP supernatant | A | 1 | 0.35 | 0.05 | 14.29 | 0.008 | 2.382 | 107.84 |
| A | 1 | 0.35 | 0.04 | 10.89 | 0.006 | 1.815 | 108.80 | |
| A | 1 | 0.35 | 0.03 | 7.28 | 0.004 | 1.248 | 109.72 | |
| A | 1 | 0.34 | 0.03 | 8.47 | 0.005 | 1.474 | 106.41 | |
| A | 1 | 0.34 | 0.04 | 12.37 | 0.008 | 2.337 | 104.84 | |
| A | 2 | 0.35 | 0.02 | 6.49 | 0.004 | 1.082 | 108.61 | |
| A | 2 | 0.36 | 0.03 | 7.06 | 0.004 | 1.176 | 112.31 | |
| A | 2 | 0.35 | 0.03 | 7.49 | 0.004 | 1.285 | 108.89 | |
| A | 2 | 0.35 | 0.02 | 6.36 | 0.004 | 1.107 | 108.55 | |
| A | 2 | 0.34 | 0.02 | 6.00 | 0.004 | 1.134 | 106.06 | |
| A | 3 | 0.30 | 0.03 | 10.61 | 0.005 | 1.768 | 93.29 | |
| A | 3 | 0.31 | 0.02 | 7.05 | 0.004 | 1.176 | 95.22 | |
| A | 3 | 0.30 | 0.03 | 9.73 | 0.005 | 1.669 | 94.26 | |
| A | 3 | 0.30 | 0.03 | 10.97 | 0.006 | 1.910 | 93.36 | |
| A | 3 | 0.30 | 0.03 | 8.71 | 0.005 | 1.646 | 94.89 | |
| B | 1 | 0.30 | 0.03 | 11.11 | 0.006 | 1.851 | 94.59 | |
| B | 1 | 0.31 | 0.04 | 13.52 | 0.007 | 2.253 | 97.28 | |
| B | 1 | 0.31 | 0.04 | 12.16 | 0.006 | 2.085 | 96.93 | |
| B | 1 | 0.31 | 0.10 | 30.77 | 0.017 | 5.356 | 97.69 | |
| B | 1 | 0.31 | 0.03 | 8.94 | 0.005 | 1.690 | 96.16 | |
| B | 2 | 0.31 | 0.02 | 5.92 | 0.003 | 0.986 | 97.23 | |
| B | 2 | 0.33 | 0.02 | 6.38 | 0.003 | 1.063 | 101.45 | |
| B | 2 | 0.32 | 0.02 | 7.52 | 0.004 | 1.289 | 100.11 | |
| B | 2 | 0.32 | 0.09 | 28.60 | 0.016 | 4.979 | 98.95 | |
| B | 2 | 0.32 | 0.03 | 8.95 | 0.005 | 1.692 | 99.90 | |
| B | 3 | 0.34 | 0.05 | 14.19 | 0.008 | 2.365 | 105.10 | |
| B | 3 | 0.28 | 0.03 | 10.03 | 0.005 | 1.672 | 88.59 | |
| B | 3 | 0.29 | 0.05 | 18.89 | 0.009 | 3.240 | 89.02 | |
| B | 3 | 0.30 | 0.09 | 31.41 | 0.016 | 5.468 | 93.48 | |
| B | 3 | 0.29 | 0.04 | 14.58 | 0.008 | 2.756 | 90.47 | |
| N1 | A | 1 | n.d | --- | --- | --- | --- | --- |
| A | 2 | n.d | --- | --- | --- | --- | --- | |
| A | 3 | n.d | --- | --- | --- | --- | --- | |
| B | 1 | n.d | --- | --- | --- | --- | --- | |
| B | 2 | n.d | --- | --- | --- | --- | --- | |
| B | 3 | n.d | --- | --- | --- | --- | --- | |
| N2 | A | 1 | n.d | --- | --- | --- | --- | --- |
| A | 2 | n.d | --- | --- | --- | --- | --- | |
| A | 3 | n.d | --- | --- | --- | --- | --- | |
| B | 1 | n.d | --- | --- | --- | --- | --- | |
| B | 2 | n.d | --- | --- | --- | --- | --- | |
| B | 3 | n.d | --- | --- | --- | --- | --- | |
| N3 | A | 1 | n.d | --- | --- | --- | --- | --- |
| A | 2 | n.d | --- | --- | --- | --- | --- | |
| A | 3 | n.d | --- | --- | --- | --- | --- | |
| B | 1 | n.d | --- | --- | --- | --- | --- | |
| B | 2 | n.d | --- | --- | --- | --- | --- | |
| B | 3 | n.d | --- | --- | --- | --- | --- |
Table 3.
FAcE assay results for each single test performed. n.d., not detectable; St. Dev, standard deviation; SE, standard error; CV, coefficient of variation.
| Operator | Day | OAg (µg/mL] | St. Dev (µg/mL) | CV % | SE | SE % | Accuracy % | |
|---|---|---|---|---|---|---|---|---|
| Determination of S. sonnei OAg in single component DP supernatant | A | 1 | 17.73 | 2.98 | 16.82 | 0.497 | 2.803 | 101.44 |
| A | 1 | 14.77 | 1.84 | 12.48 | 0.307 | 2.079 | 84.50 | |
| A | 1 | 15.71 | 1.88 | 11.94 | 0.322 | 2.047 | 89.91 | |
| A | 1 | 17.88 | 2.08 | 11.61 | 0.361 | 2.021 | 102.30 | |
| A | 1 | 15.77 | 3.96 | 25.10 | 0.748 | 4.743 | 90.23 | |
| A | 2 | 16.83 | 1.40 | 8.30 | 0.233 | 1.383 | 96.32 | |
| A | 2 | 18.23 | 4.12 | 22.59 | 0.740 | 4.058 | 104.32 | |
| A | 2 | 19.04 | 1.93 | 10.15 | 0.322 | 1.691 | 108.95 | |
| A | 2 | 21.36 | 3.14 | 14.70 | 0.523 | 2.449 | 122.24 | |
| A | 2 | 18.56 | 2.89 | 15.54 | 0.481 | 2.591 | 106.22 | |
| A | 3 | 17.59 | 2.05 | 11.64 | 0.362 | 2.059 | 100.68 | |
| A | 3 | 17.74 | 3.22 | 18.14 | 0.569 | 3.206 | 101.52 | |
| A | 3 | 19.03 | 1.91 | 10.06 | 0.338 | 1.778 | 108.88 | |
| A | 3 | 18.47 | 2.69 | 14.58 | 0.476 | 2.578 | 105.69 | |
| A | 3 | 18.74 | 2.16 | 11.54 | 0.382 | 2.040 | 107.27 | |
| B | 1 | 15.46 | 2.54 | 16.43 | 0.423 | 2.738 | 88.46 | |
| B | 1 | 17.09 | 1.62 | 9.50 | 0.271 | 1.584 | 97.78 | |
| B | 1 | 17.85 | 2.34 | 13.13 | 0.390 | 2.188 | 102.12 | |
| B | 1 | 17.25 | 2.30 | 13.32 | 0.388 | 2.251 | 98.72 | |
| B | 1 | 18.47 | 3.13 | 16.94 | 0.521 | 2.823 | 105.68 | |
| B | 2 | 17.25 | 3.49 | 20.21 | 0.581 | 3.369 | 98.69 | |
| B | 2 | 19.90 | 3.00 | 15.06 | 0.499 | 2.509 | 113.89 | |
| B | 2 | 19.09 | 3.25 | 17.03 | 0.542 | 2.838 | 109.24 | |
| B | 2 | 18.67 | 2.46 | 13.15 | 0.415 | 2.223 | 106.81 | |
| B | 2 | 18.02 | 1.71 | 9.50 | 0.289 | 1.605 | 103.11 | |
| B | 3 | 17.02 | 3.87 | 22.76 | 0.732 | 4.302 | 97.39 | |
| B | 3 | 15.05 | 2.33 | 15.48 | 0.440 | 2.925 | 86.15 | |
| B | 3 | 15.47 | 2.14 | 13.80 | 0.404 | 2.608 | 88.55 | |
| B | 3 | 15.40 | 2.44 | 15.85 | 0.470 | 3.051 | 88.15 | |
| B | 3 | 14.82 | 1.72 | 11.62 | 0.331 | 2.236 | 84.79 | |
| Determination of S. sonnei OAg on multicomponent DP supernatant | A | 1 | 4.62 | 0.54 | 11.74 | 0.090 | 1.957 | 93.39 |
| A | 1 | 4.55 | 0.54 | 11.80 | 0.092 | 2.023 | 91.89 | |
| A | 1 | 4.70 | 0.36 | 7.55 | 0.062 | 1.315 | 95.05 | |
| A | 1 | 4.61 | 0.51 | 11.07 | 0.096 | 2.091 | 93.28 | |
| A | 1 | 4.57 | 0.51 | 11.09 | 0.084 | 1.848 | 92.29 | |
| A | 2 | 4.77 | 0.73 | 15.24 | 0.125 | 2.613 | 96.51 | |
| A | 2 | 4.62 | 0.65 | 14.12 | 0.114 | 2.457 | 93.47 | |
| A | 2 | 4.70 | 0.61 | 13.05 | 0.116 | 2.466 | 94.97 | |
| A | 2 | 4.73 | 0.58 | 12.29 | 0.097 | 2.048 | 95.56 | |
| A | 2 | 4.75 | 0.48 | 10.03 | 0.082 | 1.721 | 95.99 | |
| A | 3 | 4.85 | 0.70 | 14.42 | 0.122 | 2.509 | 98.06 | |
| A | 3 | 5.05 | 0.73 | 14.40 | 0.138 | 2.721 | 102.19 | |
| A | 3 | 5.47 | 0.49 | 8.87 | 0.081 | 1.478 | 110.67 | |
| A | 3 | 5.46 | 0.61 | 11.16 | 0.105 | 1.914 | 110.47 | |
| A | 3 | 5.46 | 0.73 | 13.28 | 0.126 | 2.312 | 110.42 | |
| B | 1 | 5.25 | 0.65 | 12.30 | 0.122 | 2.325 | 106.04 | |
| B | 1 | 5.46 | 0.67 | 12.29 | 0.112 | 2.049 | 110.45 | |
| B | 1 | 5.41 | 0.81 | 15.04 | 0.140 | 2.579 | 109.43 | |
| B | 1 | 5.22 | 0.78 | 14.94 | 0.136 | 2.600 | 105.52 | |
| B | 1 | 5.15 | 0.77 | 15.04 | 0.146 | 2.842 | 104.14 | |
| B | 2 | 4.84 | 0.33 | 6.83 | 0.055 | 1.138 | 97.75 | |
| B | 2 | 4.98 | 0.47 | 9.50 | 0.081 | 1.630 | 100.67 | |
| B | 2 | 4.69 | 0.53 | 11.24 | 0.092 | 1.956 | 94.72 | |
| B | 2 | 4.80 | 0.52 | 10.82 | 0.098 | 2.045 | 97.09 | |
| B | 2 | 12.51 | 1.16 | 9.27 | 0.193 | 1.544 | 108.52 | |
| B | 3 | 11.50 | 1.02 | 8.88 | 0.170 | 1.480 | 99.79 | |
| B | 3 | 10.60 | 1.48 | 14.00 | 0.247 | 2.333 | 91.98 | |
| B | 3 | 12.4 | 1.18 | 9.49 | 0.196 | 1.582 | 107.60 | |
| B | 3 | 11.16 | 1.86 | 16.66 | 0.310 | 2.777 | 96.84 | |
| B | 3 | 10.98 | 1.07 | 9.73 | 0.178 | 1.622 | 95.28 | |
| Positive control (single component formulations) | A | 1 | 12.51 | 1.16 | 9,.27 | 0.193 | 1.54 | 108.52 |
| A | 2 | 11.5 | 1.02 | 8.88 | 0.170 | 1.48 | 99.79 | |
| A | 3 | 10.6 | 1.48 | 14.00 | 0.247 | 2.33 | 91.98 | |
| B | 1 | 12.4 | 1.18 | 9.49 | 0.196 | 1.58 | 107.60 | |
| B | 2 | 11.16 | 1.86 | 16.66 | 0.310 | 2.77 | 96.84 | |
| B | 3 | 10.98 | 1.07 | 9.73 | 0.178 | 1.62 | 95.28 | |
| N1 | A | 1 | n.d | --- | --- | --- | --- | --- |
| A | 2 | n.d | --- | --- | --- | --- | --- | |
| A | 3 | n.d | --- | --- | --- | --- | --- | |
| B | 1 | n.d | --- | --- | --- | --- | --- | |
| B | 2 | n.d | --- | --- | --- | --- | --- | |
| B | 3 | n.d | --- | --- | --- | --- | --- | |
| N2 | A | 1 | n.d | --- | --- | --- | --- | --- |
| A | 2 | n.d | --- | --- | --- | --- | --- | |
| A | 3 | n.d | --- | --- | --- | --- | --- | |
| B | 1 | n.d | --- | --- | --- | --- | --- | |
| B | 2 | n.d | --- | --- | --- | --- | --- | |
| B | 3 | n.d | --- | --- | --- | --- | --- | |
| N3 | A | 1 | n.d | --- | --- | --- | --- | --- |
| A | 2 | n.d | --- | --- | --- | --- | --- | |
| A | 3 | n.d | --- | --- | --- | --- | --- | |
| B | 1 | n.d | --- | --- | --- | --- | --- | |
| B | 2 | n.d | --- | --- | --- | --- | --- | |
| B | 3 | n.d | --- | --- | --- | --- | --- |
The results for both assays were accurate, either when OAg content was determined in single component or multicomponent drug products and drug product supernatants, respectively, for FAcE and cELISA. All measurements showed an accuracy with a maximum percentage deviation from the nominal value of 11.05% (considering a confidence interval at 95%) (Table 4).
Table 4.
Evaluation of accuracy and precision for FAcE and cELISA. CI, confidence interval; CV, coefficient of variation.
| CI 95% (Deviation from the Nominal Value %) | CV% IP (Intermediate Precision) | CV% R (Repeatability) | CV% Operator | CV% Day | ||
|---|---|---|---|---|---|---|
| FAcE | Single component | 6.52–11.05 | 9.83 | 7.88 | 2.03 | 5.52 |
| Multicomponent | 5.18–7.62 | 8.11 | 4.52 | 6.63 | 1.14 | |
| cELISA | Single component | 3.31–6.16 | 5.53 | 4.31 | 3 | 1.71 |
| Multicomponent | 5.49–7.88 | 8.23 | 3.87 | 4.9 | 5.37 | |
3.3. Precision, Repeatability, and Reproducibility
Precision of the method expresses the closeness of agreement among multiple analyses of the same homogeneous representative sample tested under the prescribed conditions. Precision was considered at two levels, i.e., repeatability (intra-assay variation) and reproducibility (inter-assay variation). Repeatability measures the precision of the method under the same working conditions (same sample tested several times by the same operator in the same day, on the same lot of plates and using the same lot of critical reagents), and is due to those factors that have not been considered in the analysis (factors other than operator and day of analysis). Reproducibility is a measure of variability within laboratory variations. Results of repeatability and reproducibility are summarized in Table 4. Intermediate precision (CV% IP) estimates the assay reproducibility, hence, the variability in the same laboratory by performing the analysis in different days, by different operators, whereas repeatability (CV% R) measures the precision of the method under the same working conditions.
The results of both assays were reproducible (CV% IP < 10%) and repeatable (CV% R < 8%), with the majority of variance generally attributed to factors other than operator and day. In particular, cELISA for quantification of unbound material in drug product supernatants showed a good intermediate precision (CV% IP = 5.5% for single component and 8.2% for multicomponent) and good repeatability (CV% R = 4.3% for single component and 3.9% for multicomponent). In the case of assay performed on single components, the majority of variance was attributable to factors not considered in the analysis (% of variance component for repeatability = 60.86%) and, between the factors considered, only the operator was significant (p value = 0.008). In contrast, both day and operator were significant (p = 0.0001, the operator contributed to 35.42% of the variance and day contributed to 42.51% of the variance) in the case of quantification of S. sonnei OAg in multicomponent samples, and, for 22.7%, the variance was attributed to repeatability.
Similar to the observations for cELISA, in the case of the FAcE assay for direct quantification of S. sonnei OAg in monocomponent drug products, the majority of variance was also attributed to factors not considered in the analysis (% of variance component for repeatability = 64.21%) and between operator and day, only day was significant (p value = 0.008). For S. sonnei OAg quantification in multicomponent drug products, instead, the majority of variance was attributed to factors considered in the analysis (operator with 66.92% and day with 1.99% of contribution to variance components) with a contribution of repeatability equal to 31.09%; in this case contribution of operator was significant (p value = 0.0001).
3.4. Specificity
Another critical parameter of the assay is the ability to assess unequivocally the analyte in the presence of components which are expected to be present. In our case, other than the active ingredient represented by the S. sonnei OAg, the other GMMA components present in the formulations are lipids, proteins, lipoproteins, etc. Therefore, specificity was evaluated by assaying in FAcE and cELISA assays all the other single component drug products composing the multicomponent vaccines. The inability to determine the OAg concentration in all the wells assayed for those samples indicated the specificity of both FAcE and cELISA assays (Table 2 and Table 3). Moreover, the specificity of both assays was confirmed as the precision of both cELISA and FAcE remained good, also when other non-S. sonnei sugar components were present in the mixture (i.e., in assays characterized with multicomponent GMMA-based vaccines).
4. Discussion
GMMA is a promising technology for the development of affordable vaccines to fight neglected bacterial diseases causing millions of deaths every year. The most advanced GMMA-based vaccine, 1790-GAHB (S. sonnei GMMA formulated on Alhydrogel), has been tested in multiple clinical trials, and was well tolerated and immunogenic in European [8] and Kenyan [9] adults, as well as able to induce a strong anamnestic response after boosting [10]. Results with 1790-GAHB have pushed toward the development of a multicomponent GMMA-based vaccine, in which the S. sonnei component is formulated in Alhydrogel with GMMA from various S. flexneri strains, to cover the majority of the Shigella serotypes epidemiologically relevant worldwide. Similarly, a GMMA-based approach is under development and is entering clinical trials with the objective of producing a vaccine to fight invasive nontyphoidal Salmonella disease [3,4,6].
The direct and specific quantification of antigen content in Alhydrogel formulates, as well as the detection of unbound active ingredients in drug products, are essential for the full characterization of vaccines and represent a critical parameter to follow their stability over time. Using the FAcE method, the antigen/s of interest in Alhydrogel-formulated vaccines have been directly quantified [16] without the need to be extracted, and thus preventing possible antigen loss and alterations of critical epitopes. Similarly, by using the cELISA assay, the direct quantification of unbound drug substances in formulates have been effectively evaluated. This is critical to ensure that the quantity of unbound GMMA is below critical levels, as the presence of unbound GMMA over 10% in 1790GAHB, at the same dose used in humans, has been shown to induce pyrogenicity in rabbits [5]; thus, the possibility of quantifying with high sensitivity unbound GMMA in DP supernant is central to confirm that the drug product maintains a profile with low risk of inducing systemic reactogenicity.
In the present work, a full characterization of both methods has been described. S. sonnei OAg was selected as the model active ingredient and quantified both in single component and multicomponent DP formulations.
Our results demonstrate that FAcE and cELISA are extremely sensitive methods which are able to quantify up to 37 and 15 ng/mL of active ingredient, respectively, in a single component formulation, and up to 34 and 20 ng/mL of active ingredient, respectively, in a multicomponent formulation. They also showed good repeatability and high intermediate precision with CV% < 8% and 10%, respectively, and very good accuracy, represented by a maximum deviation of 11.05% from the expected value with a 95% CI. Importantly, good accuracy was maintained in both assays when multicomponent formulations were analyzed, demonstrating the lack of interference in more complex mixtures. Furthermore, both FAcE and cELISA results were highly specific, because they were unable to detect and quantify unrelated GMMA components from different Shigella serotypes. As further optimization and characterization steps for the cELISA method, spike and recovery assay could be considered to enforce accuracy. In contrast, for the FacE assay, the addition of a spike formulated on Alhydrogel could cause changes in the original formulation, therefore rendering a spike/recovery assay technically difficult.
The methods presented here support S. sonnei GMMA-based vaccine drug product release and should be critical for following its stability over time. Applications of the methods cover assessment of different quality aspects of vaccine characterization, such as efficiency of different formulation conditions, stability of formulated drug product, and lot-to-lot variations. Moreover, the possibility of characterizing specifically active ingredients in multicomponent vaccines is a crucial attribute in the modern vaccinology, as modern vaccines often contain multiple antigens in the same formulation, either to increase the broadness of vaccines to protect against multiple serotypes of the same pathogen, or to cover multiple diseases in one shot. For a direct quantification, FAcE is preferable to the O-phthalaldehyde assay (OPA) [15], direct Alhydrogel formulation immunoassay (DAFIA) [18], and flow cytometry [19] in terms of range of quantification, sensitivity, and specificity, with only the Luminex-based assay [20] showing a higher sensitivity, but a lower detection range. MicroBCA is suitable for direct quantification of Alhydrogel formulations, but it is not able to quantify specifically each active ingredient in complex vaccines mixtures.
cELISA is superior in terms of sensitivity and specificity as compared with other methods developed to obtain GMMA quantification in drug product supernatant such as micro-BCA or SDS-PAGE followed by silver staining. By using this assay, it is possible to quantify directly and specifically the active ingredients (e.g., S. sonnei OAg), whereas, for the other methods, the quantification is indirect. When tested in our lab, SDS-PAGE was characterized by lack of reproducibility and both micro-BCA and SDS-PAGE analysis had a sensitivity at least 10 times less than cELISA.
The same methods characterized here for S. sonnei GMMA candidate vaccine can be extended to GMMA from different Shigella serotypes (either as single component or in multicomponent formulations), as well as to other pathogens, as already verified in our lab for drug products containing Salmonella and Neisseria meningitidis GMMA. Furthermore, both assays have been verified to be applicable for detection of active ingredients of non-GMMA analytes formulated in Alhydrogel (i.e., protein antigens or classical polysaccharide-protein conjugates). The broad applicability of the methods makes them key analytical techniques to directly quantify active ingredients in complex Alhydrogel-formulated vaccines, supporting their preclinical and clinical development.
Acknowledgments
We thank Allan Saul and Laura B. Martin for project leadership and for critically reviewing the manuscript. We thank Emilia Cappelletti and Ivan Pisoni for technical support on preparation of the DS and DP used in the assays.
Supplementary Materials
The following are available online at https://www.mdpi.com/2409-9279/3/3/62/s1, Figure S1: Plates layout. (A) Plates layout for cELISA (to characterize assay both for single and multicomponent) and for FAcE characterization (single components); (B) Plates layout for FAcE (to characterize assay for multiple components).
Author Contributions
Conceived and designed the experiments, O.R., M.G.A., F.M. (Francesca Micoli), and F.N.; Performed the experiments, O.R., M.G.A., A.A., and F.M. (Francesca Mancini); Analyzed the data, O.R., M.G.A., F.M. (Francesca Micoli), and F.N.; Contributed to the writing of the manuscript, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was sponsored by the GlaxoSmithKline Biologicals SA and funded in part by a grant from the Bill & Melinda Gates Foundation (OPP1133860). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors have the following conflict of interest to declare: all authors are currently employee of the GSK Vaccines Institute for Global Health (Srl) (GVGH), an affiliate of GlaxoSmithKline Biologicals SA. This does not alter the authors’ adherence to all Journal policies on data and material sharing.
References
- 1.Berlanda Scorza F., Colucci A.M., Maggiore L., Sanzone S., Rossi O., Ferlenghi I., Pesce I., Caboni M., Norais N., Di Cioccio V., et al. High yield production process for Shigella outer membrane particles. PLoS ONE. 2012;7:e35616. doi: 10.1371/journal.pone.0035616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kis Z., Shattock R., Shah N., Kontoravdi C. Emerging technologies for low-cost, rapid vaccine manufacture. Biotechnol. J. 2019;14:1–2. doi: 10.1002/biot.201970055. [DOI] [PubMed] [Google Scholar]
- 3.Micoli F., Rondini S., Alfini R., Lanzilao L., Necchi F., Negrea A., Rossi O., Brandt C., Clare S., Mastroeni P., et al. Comparative immunogenicity and efficacy of equivalent outer membrane vesicle and glycoconjugate vaccines against nontyphoidal Salmonella. Proc. Natl. Acad. Sci. USA. 2018;115:10428–10433. doi: 10.1073/pnas.1807655115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Benedetto G., Alfini R., Cescutti P., Caboni M., Lanzilao L., Necchi F., Saul A., MacLennan C.A., Rondini S., Micoli F. Characterization of O-antigen delivered by Generalized Modules for Membrane Antigens (GMMA) vaccine candidates against nontyphoidal Salmonella. Vaccine. 2017;35:419–426. doi: 10.1016/j.vaccine.2016.11.089. [DOI] [PubMed] [Google Scholar]
- 5.Gerke C., Colucci A.M., Giannelli C., Sanzone S., Vitali C.G., Sollai L., Rossi O., Martin L.B., Auerbach J., Di Cioccio V., et al. Production of a Shigella sonnei vaccine based on Generalized Modules for Membrane Antigens (GMMA), 1790GAHB. PLoS ONE. 2015;10:e0134478. doi: 10.1371/journal.pone.0134478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi O., Caboni M., Negrea A., Necchi F., Alfini R., Micoli F., Saul A., MacLennan C.A., Rondini S., Gerke C. Toll-Like Receptor Activation by Generalized Modules for Membrane Antigens from Lipid A Mutants of Salmonella enterica Serovars Typhimurium and Enteritidis. Clin. Vaccine Immunol. 2016;23:304–314. doi: 10.1128/CVI.00023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi O., Pesce I., Giannelli C., Aprea S., Caboni M., Citiulo F., Valentini S., Ferlenghi I., MacLennan C.A., D′Oro U., et al. Modulation of endotoxicity of Shigella generalized modules for membrane antigens (GMMA) by genetic lipid A modifications: Relative activation of TLR4 and TLR2 pathways in different mutants. J. Biol. Chem. 2014;289:24922–24935. doi: 10.1074/jbc.M114.566570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Launay O., Lewis D.J.M., Anemona A., Loulergue P., Leahy J., Scire A.S., Maugard A., Marchetti E., Zancan S., Huo Z., et al. Safety Profile and Immunologic Responses of a Novel Vaccine Against Shigella sonnei Administered Intramuscularly, Intradermally and Intranasally: Results From Two Parallel Randomized Phase 1 Clinical Studies in Healthy Adult Volunteers in Europe. EBioMedicine. 2017;22:164–172. doi: 10.1016/j.ebiom.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obiero C.W., Ndiaye A.G.W., Scire A.S., Kaunyangi B.M., Marchetti E., Gone A.M., Schutte L.D., Riccucci D., Auerbach J., Saul A., et al. A Phase 2a Randomized Study to Evaluate the Safety and Immunogenicity of the 1790GAHB Generalized Modules for Membrane Antigen Vaccine against Shigella sonnei Administered Intramuscularly to Adults from a Shigellosis-Endemic Country. Front. Immunol. 2017;8:1884. doi: 10.3389/fimmu.2017.01884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Launay O., Ndiaye A.G.W., Conti V., Loulergue P., Scire A.S., Landre A.M., Ferruzzi P., Nedjaai N., Schutte L.D., Auerbach J., et al. Booster Vaccination With GVGH Shigella sonnei 1790GAHB GMMA Vaccine Compared to Single Vaccination in Unvaccinated Healthy European Adults: Results From a Phase 1 Clinical Trial. Front. Immunol. 2019;10:335. doi: 10.3389/fimmu.2019.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burrell L.S., White J.L., Hem S.L. Stability of aluminium-containing adjuvants during aging at room temperature. Vaccine. 2000;18:2188–2192. doi: 10.1016/s0264-410x(00)00031-1. [DOI] [PubMed] [Google Scholar]
- 12.Rinella J.V., White J.L., Hem S.L. Effect of pH on the Elution of Model Antigens from Aluminum-Containing Adjuvants. J. Colloid Interface Sci. 1998;205:161–165. doi: 10.1006/jcis.1998.5648. [DOI] [PubMed] [Google Scholar]
- 13.Rinella J.V., Workman R.F., Hermodson M.A., White J.L., Hem S.L. Elutability of Proteins from Aluminum-Containing Vaccine Adjuvants by Treatment with Surfactants. J. Colloid Interface Sci. 1998;197:48–56. doi: 10.1006/jcis.1997.5230. [DOI] [PubMed] [Google Scholar]
- 14.Zhu D., Huang S., McClellan H., Dai W., Syed N.R., Gebregeorgis E., Rausch K.M., Mullen G.E., Long C., Martin L.B., et al. Efficient extraction of vaccines formulated in aluminum hydroxide gel by including surfactants in the extraction buffer. Vaccine. 2012;30:189–194. doi: 10.1016/j.vaccine.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu D., Saul A., Huang S., Martin L.B., Miller L.H., Rausch K.M. Use of o-phthalaldehyde assay to determine protein contents of Alhydrogel-based vaccines. Vaccine. 2009;27:6054–6059. doi: 10.1016/j.vaccine.2009.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Necchi F., Carducci M., Pisoni I., Rossi O., Saul A., Rondini S. Development of FAcE (Formulated Alhydrogel competitive ELISA) method for direct quantification of OAg present in Shigella sonnei GMMA-based vaccine and its optimization using Design of Experiments approach. J. Immunol. Methods. 2019;471:11–17. doi: 10.1016/j.jim.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Rossi O., Maggiore L., Necchi F., Koeberling O., MacLennan C.A., Saul A., Gerke C. Comparison of colorimetric assays with quantitative amino acid analysis for protein quantification of Generalized Modules for Membrane Antigens (GMMA) Mol. Biotechnol. 2015;57:84–93. doi: 10.1007/s12033-014-9804-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu D., Huang S., Gebregeorgis E., McClellan H., Dai W., Miller L., Saul A. Development of a Direct Alhydrogel Formulation Immunoassay (DAFIA) J. Immunol. Methods. 2009;344:73–78. doi: 10.1016/j.jim.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ugozzoli M., Laera D., Nuti S., Skibinski D.A., Bufali S., Sammicheli C., Tavarini S., Singh M., O′Hagan D.T. Flow cytometry: An alternative method for direct quantification of antigens adsorbed to aluminum hydroxide adjuvant. Anal. Biochem. 2011;418:224–230. doi: 10.1016/j.ab.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Agnolon V., Bruno C., Galletti B., Mori E., Ugozzoli M., Pergola C., O′Hagan D.T., Baudner B.C. Multiplex immunoassay for in vitro characterization of acellular pertussis antigens in combination vaccines. Vaccine. 2016;34:1040–1046. doi: 10.1016/j.vaccine.2016.01.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.