Abstract
The Amazonian red side-necked turtle Rhynemis rufipes is an endemic Amazonian Chelidae species that occurs in small streams throughout Colombia and Brazil river basins. Little is known about various biological aspects of this species, including its sex determination strategies. Among chelids, the greatest karyotype diversity is found in the Neotropical species, with several 2n configurations, including cases of triploidy. Here, we investigate the karyotype of Rhinemys rufipes by applying combined conventional and molecular cytogenetic procedures. This allowed us to discover a genetic sex-determining mechanism that shares an ancestral micro XY sex chromosome system. This ancient micro XY system recruited distinct repeat motifs before it diverged from several South America and Australasian species. We propose that such a system dates back to the earliest lineages of the chelid species before the split of South America and Australasian lineages.
Keywords: genetic sex determination, comparative genome hybridization, Chelidae, Neotropical region, karyotype
1. Introduction
The side-necked turtles of the family Chelidae comprise over 60 valid species currently restricted to the southern hemisphere and can be found throughout Australasian and South American regions [1,2,3] Chelids stand out as the most diverse group of Pleurodira regarding their karyotype configurations, i.e., difference in ploidy numbers, varying numbers of Mac (macrochromosomes) and mic (microchromosomes), as well as the presence of sex chromosomes involving Mac and mic [4,5,6]. Despite such variability, some level of karyotype conservation is also evident. For example, except for one species, Peltocephalus dumerilianus, which has 2n = 26 [7], the Amazonian podocnemidids analyzed so far have 2n = 28 chromosomes, predominantly Mac type [8]. Even in chelids, which are the most speciose among Pleurodires, an evolutionarily conserved trend in karyotypes is visible. All Australasian species possess 2n = 50 or 2n = 54 chromosomes, for instance [4,6,9,10]. However, karyotypes of the South American lineages remain a puzzle, with several diploid numbers being described even in closely related species, as reported in Mesoclemmys species, which exhibit 2n = 40, 42, 52, 58, and 60 chromosomes [4,7,11].
The Amazonian red side-necked turtle (Rhinemys rufipes) is an exclusively Amazonian species that has a nocturnal lifestyle and occurs in small streams of close canopy forests, extending from the tributaries of the Apaporís river in Colombia throughout the upper Negro and Amazon river basins in Brazil [12,13,14,15,16,17,18,19,20]. However, little is known about many aspects of this species; for instance, the nesting sites of Rhinemys have yet to be discovered. Likewise, its sex determination strategies are still unknown, which, like the vast majority of Chelidae species across Australasia and South America lineages, may also share a genetic sex-determination (GSD) mechanism [9,10,21,22]. The stability of sex ratios for hatchlings in some Neotropical Chelidae species suggests GSD as a sex-determining mechanism for South American lineages [21,22,23,24]. For Rhinemys, however, it is not known whether the GSD mechanism is present or whether this species has an environmental sex determination (ESD). Rhinemys has a breeding season directly related to seasonal rain and precipitation patterns of the Amazon rainforest, which vary widely over the extent of its distribution [14,15,17,20]. This species is likely to use subterranean formations and galleries for nesting [19] at different seasons along the year [14,17,19], probably reflecting the non-stability of environmental variables (e.g., luminosity, temperature, and humidity). Overall, this suggests that these environmental features are not primary factors in the determination of the sex proportion of hatchlings in this species. The diploid number (2n) for Chelidae species ranges from 40 to 64 chromosomes, with the presence of well-differentiated Mac and mic sex chromosomes in some species [5,7,9,10,25,26]. However, the greatest karyotype diversity is found in the Neotropical species, with several 2n configurations, including cases of triploidy in the Neotropical twist-neck turtle (Platemys platycephala, 3n = 96) [4,5,7,25,26,27]. To date, only a single and contradictory reference exists concerning chromosomal data for R. rufipes: McBee et al. [5] reported data of 2n = 58 chromosomes and fundamental number NF = 64. However, this same study focused on the karyotypic composition of the Acanthochelys species, and no metaphase of Rhinemys was presented. Moreover, details regarding origin, sex, and number of individuals analyzed were not mentioned either. Therefore, reinvestigation of fresh material, with known gender and known origin individuals of R. rufipes, is required to corroborate these previous findings.
In this study, we aim to provide a full description of the karyotype composition of R. rufipes from the Amazon rainforest. For that, we applied multiple conventional (Giemsa staining and C-banding) and molecular cytogenetic tools, as comparative genomic hybridization (CGH), FISH mapping of telomeric (TTAAGG)n, 18S rDNA, and simple short repeats (SSRs) sequences. Additionally, we identified sex chromosomes and modes of sex determination in this species, and are thus able to discuss the evolution of sex chromosomes in the broader context of turtle sex chromosomes.
2. Material and Methods
2.1. Sampling, Mitotic Chromosomes Preparation, and C-Banding
Turtles were sampled from natural populations from the central Amazon region (Lower Rio Negro) under permission granted by Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) number 45275. We analyzed 5 males and 4 females of the red side-necked turtle R. rufipes. Chromosomal preparations were cultured from small blood samples at 29 °C, following Viana et al. [7]. C-positive heterochromatin staining was performed according to Sumner et al. [28]. It is highlighted that no animal needed to be euthanized in this study.
2.2. Probes for Chromosome Hybridization
18S rDNA and telomeric (TTAGGG)n probes were isolated according to Gross et al. [29] and Ijdo et al. [30], respectively. Both probes were labeled with aminoallyl-dUTPATTO-550 (red) by Nick-translation means (Jena Bioscience, Jena, Germany). Simple short repeats (SSRs), (GT)15, (AG)15, (AAC)10, (GGAT)8, (ACGC)8, (AATC)8, (ATCC)8, (AATG)8, (GACA)8, and (GATA)8 were used, directly labeled with Cy-3, during the synthesis [31].
2.3. Fluorescence In Situ Hybridization (FISH) for Repetitive DNA Mapping
FISH experiments followed the protocol described by Pinkel et al. [32], with modifications detailed in Viana et al. [33]. Briefly, the chromosome slides were denatured in 70% formamide/2xSSC at 70 °C; spreads were dehydrated in ethanol (100%). Then, 20 µL of the hybridization mixture (100 ng of each probe, 50% deionized formamide, and 10% dextran sulfate) was dropped on the slides, and the hybridization was carried out for 24 h at 37 °C in a moist chamber containing distilled water. The posthybridization washes were performed once in 2× SSC (44 °C, 5 min) and a final wash in 4× SSC/0.1% Tween (5 min, room temperature). The chromosomes were counterstained with DAPI (4′,6-Diamidino-2-Phenylindole, 1.2 µg/mL) and mounted in antifade solution (Vector, Burlingame, CA, USA).
2.4. Preparation of Probes for Comparative Genomic Hybridization (CGH)
The gDNA of males and females of R. rufipes was extracted from blood using the Wizard Genomic Purification Kit (Promega), following the manufacturer’s recommendations. Female-derived gDNA was labeled with aminoallyl-dUTPATTO-550 (red) and males’ gDNA with aminoallyl-dUTPATTO-488 (green) using a Nick-translation Labeling Kit (Jena Bioscience, Jena, Germany). The final hybridization mixture for each slide (20 μL) was composed of male- and female-derived gDNA (500 ng each), 20 μg of male-derived Cot-1 DNA (i.e., the fraction of genomic DNA enriched for highly repetitive sequences), prepared according to Zwick et al. [34], and the hybridization buffer containing 50% formamide, 2× SSC, 10% SDS, 10% dextran sulfate, and Denhardt’s buffer, pH 7.0. The probes were ethanol-precipitated, and the dried pellets were resuspended in hybridization buffer, as mentioned above.
2.5. Comparative Genomic Hybridization (CGH)
CGH experiments were performed according to our previous studies [35,36,37,38]. The slides were incubated at 37 °C in a dark, humid chamber for 72 h. Posthybridization washes were performed once in 50% formamide/2× SSC, pH 7.0 (44 °C, 5 min), once in 2× SSC (44 °C, 5 min), and a final wash in 4× SSC/0.1% Tween (3 min, room temperature). The chromosomes were counterstained with DAPI (1.2 µg/mL) and mounted in an antifade solution (Vector, Burlingame, CA, USA).
2.6. Microscopic Analyses
For the 5 males and 4 females, at least 10 metaphase spreads were analyzed for each in order to confirm the karyotype structure and FISH results. Images were captured using an Olympus BX51 microscope (Olympus Corporation, Ishikawa, Japan) with CoolSNAP. Chromosomes were classified as macrochromosomes (Mac) and microchromosomes (mic) or as metacentric (m), submetacentric (sm), subtelocentric (st), and acrocentric (a), according to [39].
3. Results
3.1. Karyotype and C-Positive Heterochromatin
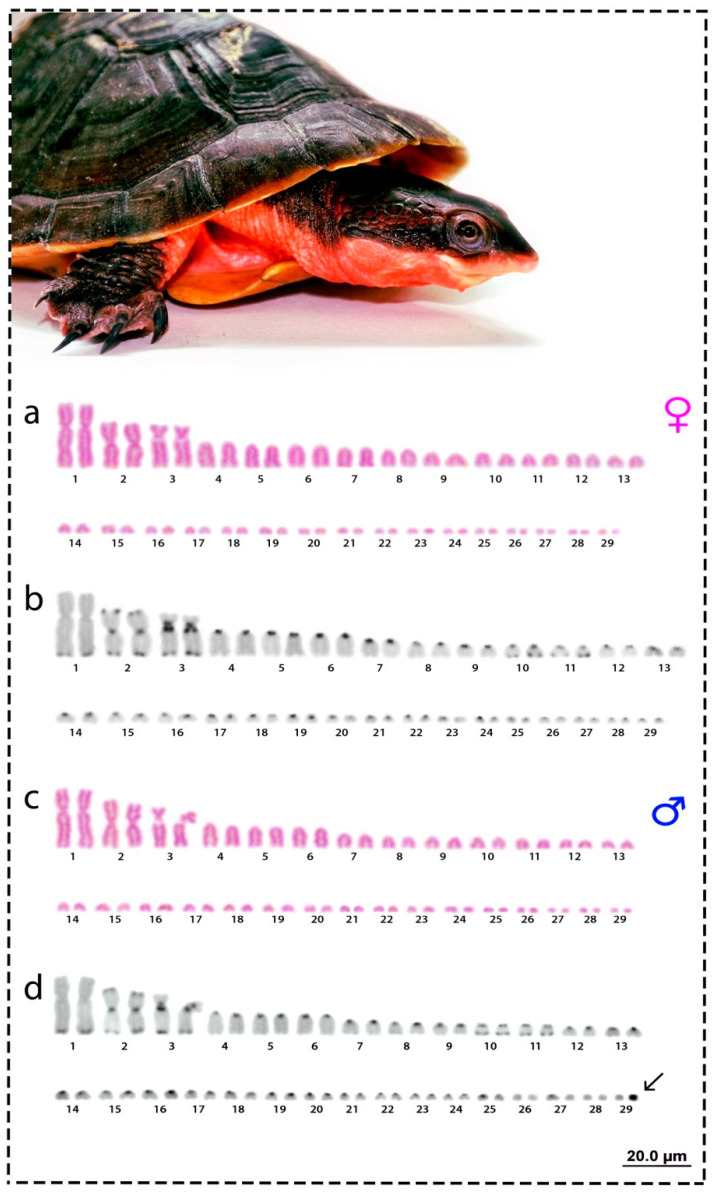
The red side-necked turtle has 2n = 58 chromosomes, with 26 Mac and 32 mic. The karyotype is composed of 2m + 2sm + 2st + 20a and 32 mic (mic predominantly acrocentrics), with a fundamental number (NF) equal to 64 for males and females (Figure 1). The C-positive heterochromatin was found in all chromosomes, with a preferential localization in the centromeric regions (pairs 2 to 29) and in telomeric regions for some Mac pairs (1, 2, 3, 10, 11) in both males and females (Figure 1). However, one of the smallest mic in all males was completely heterochromatic and was not observed in any females. This identifies R. rufipes as having a XX/XY sex chromosome system (Figure 1).
Figure 1.
Karyotypes of females (a,b) and males (c,d) of R. rufipes in Giemsa-staining (a,c) and C-banding (b,d). Arrow indicates the tiny heterochromatic microchromosome, which corresponds to the Y sex chromosome. Bar = 20 µm.
3.2. Comparative Genomic Hybridization (CGH)
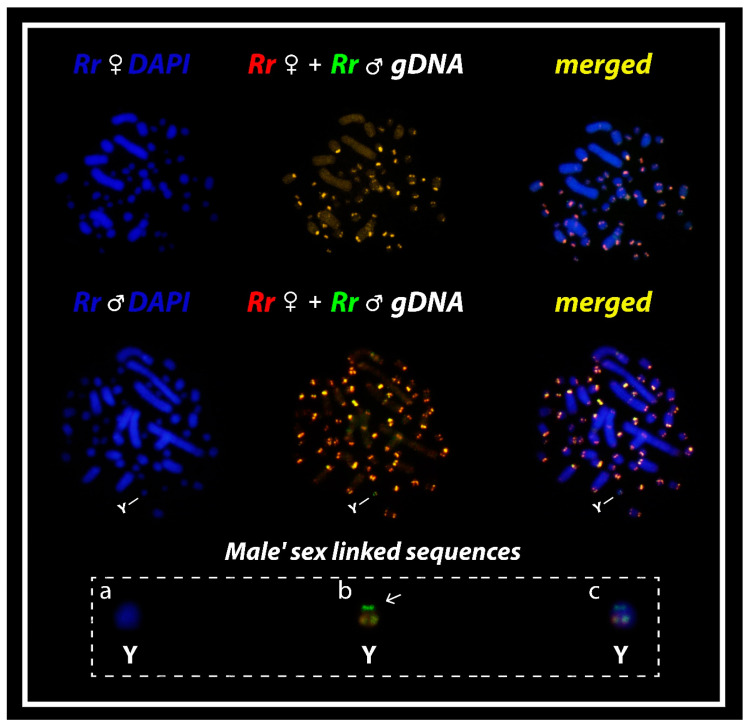
Our intraspecific comparison between males and females of R. rufipes produced intense hybridization signals in both Mac and mic, mostly collocated with heterochromatic portions (Figure 2). The merged images identified male-specific sequences accumulated in a small region of a tiny mic of the karyotype, the micro Y sex chromosome (boxed). This identifies R. rufipes as a GSD species with a micro XY sex chromosome system. However, we are not able to properly identify the X chromosome due to the morphological similarity with many other mics and the absence of chromosomal-specific signal patterns (Figure 2).
Figure 2.
Mitotic chromosome spreads of R. rufipes after comparative genomic hybridization (CGH) procedures using male- and female-derived genomic probes. The common genomic regions are highlighted in yellow. The Y chromosome after DAPI (a), probed with male- and female-derived genomic probes (b) and merged image (hybridization signals + DAPI) (c), is highlighted in boxes. Please note the male-specific Y-linked sequences (b) in green.
3.3. Mapping of 18S rDNA, Telomeric Repeats and SSRs Motifs
All microsatellite repeat motifs used, namely, (GT)15, (AG)15, (AAC)10, (GGAT)8, (ACGC)8, (AATC)8, (ATCC)8, (AATG)8, (GACA)8 and (GATA)8, showed hybridization signals on chromosomes of R. rufipes (Figure 3 and Figure 4). However, a Y-specific pattern of accumulation for some SSRs was observed (Figure 4b). The (GT)15, (AG)15, (ATCC)8, (GACA)8 and (GGAT)8 motifs showed hybridization signals on both Mac and mic of males and females, but with a particular amplification of such SSRs on the Y sex chromosome, nevertheless (Figure 3 and Figure 4). On the other hand, the motifs (AATC)8, (AATG)8 and (GATA)8 were found to be male sex-specific, with exclusive Y-linked amplification (Figure 4a,b). On the contrary, of all other SSRs used here, (ACGC)8 was the sole repeat motif with no differential pattern between males and females (Figure 3).
Figure 3.
Mapping of several simple short repeats (SSRs) on the chromosomes of males and females of R. rufipes, with highlights to the particular accumulation for some of them on the micro Y sex chromosome (arrows). Bar = 20 µm.
Figure 4.
Mapping of several SSRs, telomeric (TTAGGG)n, and 18S rDNA on the chromosomes of males and females of R. rufipes (a). (b) Different patterns of SSR motif recruitment to enlarged forms of micro Y sex chromosome. Bar = 20 µm.
The mapping of the 18S rDNA sequences showed simple markings on the secondary constriction of the submetacentric pair (Figure 4). The telomeric motifs were evidenced on all terminal portions of all Mac and mic; however, an interstitial telomeric sequence (ITS) is evident on the centromeric region of the sole metacentric pair of the karyotype (pair 2; Figure 4).
4. Discussion
Although reptiles have been extensively studied from the cytogenetic point of view, the lack of karyotype data for many turtle species impairs a comprehensive analysis of evolutionary trends and their chromosomal relationships. In this sense, this study is the first to provide classical and molecular cytogenetic data for one representative of Chelidae species, the Amazonian red side-necked turtle R. rufipes. Our study demonstrates that this species has GSD with an XY sex microchromosome system, where the mic Y sex chromosome can easily be identified by the accumulation of SSRs. Moreover, our CGH experiments identified the presence of male-specific sequences concentrated on a small portion of the Y sex chromosome.
The side-necked turtles of the Chelidae family are phylogenetically related to Pelomedusidae (Africa) and Podocnemididae (Amazonia and Madagascar) turtle lineages, forming a monophyletic group (Pleurodira), predominantly widespread in the southern hemisphere [2,40,41,42,43]. Most Podocnemididae species (7 out of 8) exhibit 28 Mac, whereas Pelomedusidae species (for those with known karyotypes) present 34 and 36 chromosomes with the presence of Mac and mic [4,44,45,46]. Interestingly, the sole existing reference in the literature regarding the karyotype for R. rufipes [5] also reports 2n = 58 and NF = 64, very similar to our findings (Figure 1) [5]. This same karyotype configuration is also observed in some Mesoclemmys and Phrynops species [4,47], and although R. rufipes is an eccentric species among Neotropical chelids and easily recognized, given the paucity of additional information (e.g., locality of original samples or even the source of the samples), we cannot rule out the possibility that McBee and colleagues [5] have analyzed a Mesoclemmys or even Phrynops species, the latter inclusive, a genus in which Rhinemys was first placed. Nonetheless, Phrynops are older lineages than R. rufipes, whose origins date back ~34 mya, at the end of Oligocene (Figure 5), and much closer to M. tuberculata, M. perplexa, and M. vanderhaegei. However, despite presenting a GSD system [24,48], the configuration of sex chromosomes in the Mesoclemmys species needs further investigation to infer whether Mac or mic sex chromosomes frequently occur and whether transitions between Mac and mic are governing the karyotype diversity present in this group.
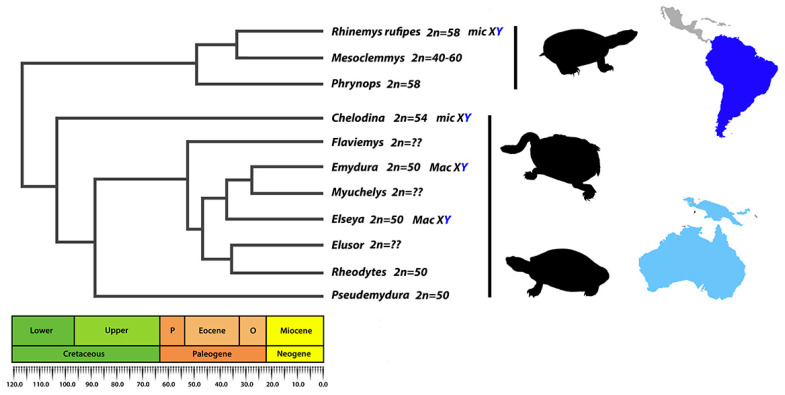
Figure 5.
Chronogram for some chelid species, adapted from Pereira et al. [42]. P = Paleocene and O = Oligocene.
Sex chromosomes in turtles evolved independently and multiple times along its evolution [49], and several intra/interchromosomal rearrangements, sometimes involving a candidate for master sex-determining genes, are likely to represent the main factor that drove the landscape of sex chromosomes in turtles [50,51,52]. In Australasian chelids, for instance, translocations/fusion events orchestrated the origin of Mac and mic sex chromosomes in the ancestor of the species, with 2n = 50 and 2n = 54 [6,9,10,51,53].
The ancestor state for sex chromosomes in Australasia Chelidae is still under debate, namely, whether a Mac or mic ancestor sex chromosome was present before the split (~105mya) between the clade composed of species 2n = 50/Mac sex chromosomes and the clade with 2n = 54 and mic sex chromosomes (Chelodina spp.) (Figure 5) [4,6,9]. A mic sex chromosome ancestor state likely represents the most realistic scenario, which fused with a macro, resulting in the species with 2n = 50 and Mac sex chromosomes [53]. However, the reverse scenario is also plausible, suggesting that Mac ancestor could equally represent the ancestor condition, assuming Chelodina as the sister group to Emydura [6].
Australasian chelids comprise two major lineages, basically, a lineage with 2n = 50 and a lineage with 2n = 54. The Emydura species is closer to Myuchelys and Elseya, which forms a sister group to the lineages composed of Elusor and Rheodytes, jointly with Flaviemys and Pseudoemydura, species with Mac sex chromosomes and 2n = 50. All species with Mac sex chromosomes and 2n = 50 comprise a sister group to the Chelodina species that possesses 2n = 54 and mic sex chromosomes, representing the oldest lineages of Australasian chelids (Figure 5).
If Elusor, Flaviemys, and, especially, Myuchelis (sister group to Emydura spp. 2n = 50 and Mac sex chromosomes) somehow presented mic sex chromosomes, this scenario would require the occurrence of one turnover considering a Mac as the ancestral state and two turnovers considering a mic as an ancestor. Further analyses in Myuchelys, Elusor, and Flaviemys will uncover if these species also possess 2n = 50 and Mac sex chromosomes, as seen in their sister groups with known karyotype, and clarify the turnovers involving Mac and mic sex chromosomes in Australasian lineages.
Chelids, undoubtedly, possess South American origins, with fossil records dating to lower Cretaceous [2,54,55]; several studies also show that Chelidae (both South America and Australasia lineages) started to diversify before the split of Australasia from Gondwana [42,56,57,58]. This implies that besides vicariant events, they also experienced dispersal events, resulting in a widespread distribution at the beginning of the Gondwana fragmentation [2].
The mic sex chromosomes of Rhinemys date back to the early Oligocene, before the diversification of many Australasian species [42]. This suggests the possibility that mic sex chromosomes could already be present in the common ancestor of Australasian and South American chelids. In this scenario, an interesting question emerges as a new piece of the puzzle. If the mic sex chromosomes of R. rufipes somehow share homologies with the Australasia chelid sex chromosomes (Mac in the 2n = 50 species or mic in 2n = 54), this mic sex chromosome highlights ancestry dating back the Upper Jurassic, ~160 myr. This implies a much older turnover event involving mic and Mac sex chromosomes than the turnovers that occurred before the split of Chelodina and Emydura lineages [6,53]. Indeed, translocations involving a sex-determining locus via mic to Mac seems to have been the evolutionary pathway chosen by the chelids. However, further studies in other South American chelids are necessary and indispensable to investigate this hypothesis and decipher the real ancestor state for the sex chromosomes of chelids, as well as likely transition events involving macro and micro sex chromosomes along their evolutionary trajectories.
As seen in other chelids with poorly or highly differentiated sex chromosomes [6,53,59], Rhinemys rufipes also recruited some specific Y-linked SSR motifs (Figure 4b). These SSRs are commonly found on sex chromosomes across a range of Squamata reptile species [35,59] and are inherently thought to display a key role in chromosome sex differentiation [60,61,62,63]. Indeed, Squamata reptiles exhibit a dynamic genome regarding the rich content of SSRs [64,65,66,67]; however, turtles are relatively SSR-poor [67,68,69] even though sharing the same repeat motifs present in sex chromosomes of several Squamata lineages [59].
Regardless, Rhinemys rufipes and Australasia chelids share several SSRs on their sex chromosomes [6,59] despite a long divergence time, dating back to the split of South America and Australasia lineages in the Late Jurassic [42,70]. This could indeed represent a remnant of a common evolutionary history and origin of their sex chromosome. Chromosomics [71,72] is required to deeply infer the homology of sex chromosomes spanning ~115 myr of independent evolution among Chelidae species, especially because SSRs are polymorphic, showing high variability and elevated rates of mutation reflected in divergent microsatellites landscapes and rapid shifts even in closely related species [67]. In this sense, the presence of the recurrent recruitment of the same simple short repeats on the sex chromosomes of many reptiles, including Rhinemys and other chelid turtles, could simply be the product of co-optation and convergent accumulation of such repeats due to evolutionary advantages facilitated by the dynamic behavior of SSRs (e.g., recombination, control of genic expression, chromosomal organization).
SSR landscapes and their genome-wide distribution may inherently impact on chromosomal architecture; for instance, they are one of the major promoters of the expansion and/or contraction of repetitive sequences with which they are associated [73,74], for instance, transposable elements and rDNAs [63,75,76]. Here, we found at least three SSRs (GT, AG, AAC) bearing 18S rDNA of the 3rd pair, with minute differences in size, sometimes in both males and females (not shown in the figures), suggesting that these small repeats (di- and trinucleotides) are likely involved in regulatory processes, reflective of uneven crossing and ectopic recombination mediated by the association of such SSRs and the activity of the ribosomal sites. Interestingly, the other SSRs (the tetranucleotides: AATC, ACGC, GGAT, GACA), similarly present on both autosomes and sex chromosomes, did not present any association with the 18S rDNA sites, but had prominent signals on some small acrocentric chromosomes, perhaps displaying structural roles in the chromosomal architecture since they are mostly colocated with heterochromatic segments in both males and females (Figure 3 and Figure 4a).
Likewise, interstitial telomeric sequences (ITSs) can be found to be associated with rDNAs, several times attributed to represent hotspots prone to chromosomal rearrangements [77,78]. However, they are relatively rare in some turtle lineages [79]. Except for the Y-linked accumulation of telomeric motifs in Elseya novaeguinae [6], hitherto, TTAGGGn repeats on the centromeric position of the sole metacentric chromosome pair of R. rufipes (Figure 4) seem to be the first evidence of ITSs in Chelidae species. On the contrary, such repeats, as the remnant of chromosomal fusion events, is a widespread feature in its sister group, the Podocnemididae family [8]. In many vertebrate groups, ITS repeat has constantly been attributed to relics of ancient chromosomal rearrangements [33,80,81]; however, ITSs exhibit a random amplification across lineages [80,82], and, due to their dynamic functioning [78], sometimes do not represent artifacts or ghosts of past intra/interchromosomal rearrangements. Since the most of closely related species to R. rufipes also exhibits the 2n = 58, with very similar karyotype structure [5,7,42,47], the ITS present on the centromeric position of the sole metacentric pair could indeed be the result of past intrachromosomal rearrangements, but combined with the fact that these interstitial telomeric motifs are rarely found in some turtle lineages [79] and taking into account their dynamic behavior [78], it is also plausible that such ITSs may have arisen from other mechanisms, such as telomerase activity adding these motifs to nonterminal regions, duplication events, or even association with mobile elements and/or other repetitive sequences [83,84,85,86,87,88]. The karyotype of other South American chelids is also required to infer whether this ITS present in R. rufipes is a classical exception to the rule [82] or ITS sites are frequent and are shaping the karyotype diversity evident in Neotropical chelids.
The mic Y sex chromosome of R. rufipes can be easily identified by the accumulation of at least nine SSRs (Figure 4b); however, as with other chelid species possessing mic sex chromosomes, it was not possible to identify the mic X [6,59]. Undifferentiated sex chromosomes are notably frequent across the vertebrate phylogeny and are more common than previously thought [89,90,91]. Although the levels of degeneration are not primarily related to the age of the sex chromosomes and the lineages where they arose [92,93,94,95], they are more frequent in ancient lineages that did not acquire sex-specific signatures [37,96,97]. Interestingly, as revealed here in our study, even with ancient origin, they can also be found practically intact in recent lineages, where the X and Y of R. rufipes are undistinguished from each other, with the Y sex chromosome being identified solely by comparative genomic hybridization and recruitment of several SSR motifs. The sex chromosomes may remain undifferentiated for many reasons, for example, transitions and turnovers involving XY and ZW, evolutionary advantages promoted by the recombination between XY and/or Z and W, as well as shifts involving autosomes [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94]. In this pathway, the biased male recruitment of repetitive sequences for the Y mic sex chromosome and the maintenance of the chromosomal morphology and similarity of the X and Y in R. rufipes seem to be another classical scape from the evolutionary trap of sex chromosomes [92,93,98].
The karyotype of R. rufipes surely brings important novelties to the knowledge of chromosome evolution and the evolutionary history of South America and Australasia Chelidae species. The likely ancestry for sex chromosomes also highlights that recruitment of particular SSR motifs on sex chromosomes could already have shaped the pathways that have resulted in the current configurations of Mac and mic sex chromosomes in chelids, long prior its diversification. Future analyses in other South America and Australasia chelids will uncover the events of rearrangements that the sex chromosomes underwent and decipher what scenario corresponds to the oldest sex chromosome configuration in Chelidae, namely, whether a Mac or a mic represents the ancestral state.
Acknowledgments
This study is in memory of Jabson Franco da Costa, who dedicated his life to studying Amazonian turtles. We are also grateful to Milena and Breno Almeida from the Amazonian Center for Herpetology (Centro Amazônico de Herpetologia) for all the valuable support provided.
Author Contributions
Conceptualization, designed the study, and initial structure, P.F.V.; methodology, P.F.V., E.F., M.B.C., V.T.d.C., S.M., R.C.V., T.L., and T.E.; validation, P.F.V., E.F., M.B.C., and T.E.; formal analysis and investigation, P.F.V., E.F., M.B.C., V.T.d.C., S.M., R.C.V., T.L., and T.E. All authors analyzed and interpreted the data. P.V. wrote the manuscript with contributions from all coauthors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Fundação de Amparo à Pesquisa do Amazonas (FAPEAM), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES; Grant Number: 88881.190036/2018-01), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; Grant numbers: 302449/2018-3 and 302449/2018-3), Center for Studies on Adaptations of Aquatic Biota of the Amazon (ADAPTA), Projects (Grant Number: Pronex/FAPEAM/CNPq 003/2009), and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; Grant Number 2018/220033-1). V.T.d.C. is grateful to the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—CAPES (PNPD).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ernst C.H.E., Barbour R.W. Turtles of the World. 1st ed. Smithsonian Institution Press; Washington, DC, USA: 1989. p. 313. [Google Scholar]
- 2.Ferreira G.S., Bronzati M., Langer M.C., Sterli J. Phylogeny, biogeography and diversification patterns of side-necked turtles (Testudines: Pleurodira) R. Soc. Open Sci. 2018;5:171773. doi: 10.1098/rsos.171773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Reptile Database. [(accessed on 22 May 2020)]; Available online: http://www.reptile-database.org.
- 4.Bull J.J., Legler J.M. Karyotypes of side-necked turtles (Testudines: Pleurodira) Can. J. Zool. 2018;58:828–841. doi: 10.1139/z80-115. [DOI] [Google Scholar]
- 5.McBee K., Bickham J.W., Rhodin A.G.J., Mittermeier R.A. Karyotipic variation in the genus Platemys (Testudines: Pleurodira) Copeia. 1985;2:445–449. doi: 10.2307/1444856. [DOI] [Google Scholar]
- 6.Mazzoleni S., Augstenová B., Clemente L., Auer M., Fritz U., Praschag P., Protiva T., Velenský P., Kratochvíl L., Rovatsos M. Sex is determined by XX/XY sex chromosomes in Australasian side-necked turtles (Testudines: Chelidae) Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-61116-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viana P.F., Ribeiro L.B., Lima T., de Carvalho V.T., Vogt R.C., Gross M.C., Feldberg E. An optimized protocol for obtaining mitotic chromosomes from cultured reptilian lymphocytes. Nucleus. 2016;59:191–195. doi: 10.1007/s13237-016-0174-3. [DOI] [Google Scholar]
- 8.Cavalcante M.G., Bastos C.E.M.C., Nagamachi C.Y., Pieczarka J.C., Vicari M.R., Noronha R.C.R. Physical mapping of repetitive DNA suggests 2n reduction in Amazon turtles Podocnemis (Testudines: Podocnemididae) PLoS ONE. 2018;13:e0197536. doi: 10.1371/journal.pone.0197536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ezaz T., Valenzuela N., Grützner F., Miura I., Georges A., Burke R.L., Graves J.A.M. An XX/XY sex microchromosome system in a freshwater turtle, Chelodina longicollis (Testudines: Chelidae) with genetic sex determination. Chromosome Res. 2006;14:139–150. doi: 10.1007/s10577-006-1029-6. [DOI] [PubMed] [Google Scholar]
- 10.Martinez P.A., Ezaz T., Valenzuela N., Georges A., Graves J.A.M. An XX/XY heteromorphic sex chromosome system in the Australian chelid turtle Emydura macquarii: A new piece in the puzzle of sex chromosome evolution in turtles. Chromosome Res. 2008;16:815–825. doi: 10.1007/s10577-008-1228-4. [DOI] [PubMed] [Google Scholar]
- 11.Viana P.F. The puzzle of karyotype evolution in Neotropical chelids. Status (manuscript in preparation)
- 12.Magnusson W.E., Lima A.C., Costa V.L., Vogt R.C. Home range of the turtle, Phrynops rufipes, in an isolated reserve in central Amazônia, Brazil. Chelonian Conserv. Biol. 1997;2:494–499. [Google Scholar]
- 13.Magnusson W.E., Lima A.C., Costa V.L., Lima O.P. Growth of the turtle, Phrynops rufipes, in central Amazônia, Brazil. Chelonian Conserv. Biol. 1997;2:576–581. [Google Scholar]
- 14.Lamar W.W., Mendem F. Notes on the chelid turtle Phrynops rufipes in Colombia. Salamandra. 1984;18:305–321. [Google Scholar]
- 15.Rueda-Almonacid J.V., Carr J.L., Mittermeier R.A., Rodríguez-Mahecha J.V., Mast R.B., Vogt R.C., Rhodin A.G.J., De La Ossa-Velásquez J., Rueda J.N., Mittermeier C.G. Las Tortugas y los Cocodrilianos de los Países Andinos del Trópico. 1st ed. Conservación Internacional; Bogotá, Colombia: 2007. p. 538. [Google Scholar]
- 16.Páez V.P., Morales-Betancourt M.A., Lasso C.A., Castaño-Mora O.V., Bock B.C. Biología y Conservación de las Tortugas Continentales de Colombia. 1st ed. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt; Bogotá, Colombia: 2012. p. 528. [Google Scholar]
- 17.Alvarenga C.C.E. Master’s Thesis. Universidade Federal do Amazonas; Manaus, Brazil: 2006. Aspectos de Biologia Reprodutiva de Rhinemys rufipes (Spix, 1824) (Chelidae, Testudines) na Reserva Florestal Adolpho Ducke, Amazonas, Brasil. [Google Scholar]
- 18.Sanchez D.E.A. Master’s Thesis. Universidade Federal do Amazonas; Manaus, Brazil: 2008. Home range of the turtle, Phrynops rufipes, in an isolated reserve in central Amazônia, Brazil. [Google Scholar]
- 19.Vogt R.C. Tartarugas da Amazônia. 1st ed. Instituto Nacional de Pesquisas da Amazônia; Manaus, Brazil: 2008. p. 104. [Google Scholar]
- 20.Ferrara C.R., Fagundes C.K., Morcatty T.Q., Vogt R.C. Quelônios Amazônicos: Guia de Identificação e Distribuição. 1st ed. Wildlife Conservation Society; Manaus, Brazil: 2017. p. 182. [Google Scholar]
- 21.Ewert M.A., Etcheberger C.R., Nelson C.E. Turtle sex-determination modes and TSD patterns, and some TSD patterns correlates. In: Valenzuela N., Lance V.A., editors. Temperature-Dependent Sex Determination in Vertebrates. 1st ed. Volume 1. Smithsonian Books; Washington, DC, USA: 2004. pp. 21–32. [Google Scholar]
- 22.Ferreira-Júnior P.D. Aspectos Ecológicos da Determinação Sexual em Tartarugas. Acta Amaz. 2009;39:139–154. doi: 10.1590/S0044-59672009000100014. [DOI] [Google Scholar]
- 23.Vogt R.C. Ecologia reprodutiva de uma comunidade de quelônios da Amazônia; Proceedings of the Congresso Latino-Americano de Zoologia; Belém, Brazil. 26–31 July 1992; p. 131. [Google Scholar]
- 24.Cunha F.A., Fernandes T., Franco J., Vogt R.C. Reproductive Biology and Hatchling Morphology of the Amazon Toad-headed Turtle (Mesoclemmys raniceps) (Testudines: Chelidae), with Notes on Species Morphology and Taxonomy of the Mesoclemmys Group. Chelonian Conserv. Biol. 2019;18:195–209. doi: 10.2744/CCB-1271.1. [DOI] [Google Scholar]
- 25.Bickham J.W., Hanks B.G. Diploid-triploid mosaicism and tissue ploidy diversity within Platemys platycephala from Suriname. Cytogenet Genome. Res. 2009;127:280–286. doi: 10.1159/000297716. [DOI] [PubMed] [Google Scholar]
- 26.Barros R.M., Sampaio M.M., Assis M.F., Ayres M., Cunha O.R. General considerations on the karyotypic evolution of Chelonia from the Amazon region of Brazil. Cytologia. 1976;41:559–565. doi: 10.1508/cytologia.41.559. [DOI] [Google Scholar]
- 27.Bickham J.W., Tucker P.K., Legler J.M. Diploid-triploid mosaicism: An unusual phenomenon in side-necked turtles (Platemys platycephala) Science. 1985;227:1591–1593. doi: 10.1126/science.227.4694.1591. [DOI] [PubMed] [Google Scholar]
- 28.Sumner A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972;75:204–206. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- 29.Gross M.C., Schneider C.H., Valente G.T., Porto J.I.R., Martins C., Feldberg E. Variability of 18S rDNA locus among Symphysodon fishes: Chromosomal rearrangements. J. Fish. Biol. 2009;76:1117–1127. doi: 10.1111/j.1095-8649.2010.02550.x. [DOI] [PubMed] [Google Scholar]
- 30.Ijdo J.W., Wells R.A., Baldini A., Reeders S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991;19:4780. doi: 10.1093/nar/19.17.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubat Z., Hobza R., Vyskot B., Kejnovsky E. Microsatellite accumulation in the Y chromosome of Silene latifolia. Genome. 2008;51:350–356. doi: 10.1139/G08-024. [DOI] [PubMed] [Google Scholar]
- 32.Pinkel D., Straume T., Gray J.W. Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viana P.F., Ribeiro L.B., Souza G.M., Chalkidis H.M., Gross M.C., Feldberg E. Is the Karyotype of Neotropical Boid Snakes Really Conserved? Cytotaxonomy, Chromosomal Rearrangements and Karyotype Organization in the Boidae Family. PLoS ONE. 2016;11:e0160274. doi: 10.1371/journal.pone.0160274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwick M.S., Hanson R.E., Mcknight T.D., Islam-Faridi M.H., Stelly D.M., Wing R.A., Price H.J. A rapid procedure for the isolation of C0t-1 DNA from plants. Genome. 1997;40:138–142. doi: 10.1139/g97-020. [DOI] [PubMed] [Google Scholar]
- 35.Viana P.F., Ezaz T., Cioffi M.B., Almeida B.J., Feldberg E. Evolutionary Insights of the ZW Sex Chromosomes in Snakes: A New Chapter Added by the Amazonian Puffing Snakes of the Genus Spilotes. Genes. 2019;10:288. doi: 10.3390/genes10040288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cioffi M.B., Ráb P., Ezaz T., Bertollo L.A.C., Lavoué S., de Oliveira E.A., Sember A., Molina W.F., Souza F.H.S., Majtánová Z., et al. Deciphering the Evolutionary History of Arowana Fishes (Teleostei, Osteoglossiformes, Osteoglossidae): Insight from Comparative Cytogenomics. Int. J. Mol. Sci. 2019;20:4296. doi: 10.3390/ijms20174296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viana P.F., Ezaz T., Cioffi M.B., Liehr T., Al-Rikabi A., Goll L.G., Rocha A.M., Feldberg E. Landscape of snake’ sex chromosomes evolution spanning 85 MYR reveals ancestry of sequences despite distinct evolutionary trajectories. Sci. Rep. 2020;10:1–14. doi: 10.1038/s41598-020-69349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sassi F.D., Hatanaka T., Moraes R.L.R.D., Toma G.A., Oliveira E.A.D., Liehr T., Rab P., Bertollo L.A.C., Viana P.F., Feldberg E., et al. An Insight into the Chromosomal Evolution of Lebiasinidae (Teleostei, Characiformes) Genes. 2020;11:365. doi: 10.3390/genes11040365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levan A., Fredga K., Sandberg A.A. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–220. doi: 10.1111/j.1601-5223.1964.tb01953.x. [DOI] [Google Scholar]
- 40.Guillon J.-M., Guéry L., Hulin V., Girondot M. A large phylogeny of turtles (Testudines) using molecular data. Contrib. Zool. 2012;81:147–158. doi: 10.1163/18759866-08103002. [DOI] [Google Scholar]
- 41.Crawford N.G., Parham J.F., Sellas A.B., Faircloth B.C., Glenn T.C., Papenfuss T.J., Henderson J.B., Hansen M.H., Simison W.B. A phylogenomic analysis of turtles. Mol. Phylogenet. Evol. 2015;83:250–257. doi: 10.1016/j.ympev.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 42.Pereira A.G., Sterli J., Moreira F.R., Schrago C.G. Multilocus phylogeny and statistical biogeography clarify the evolutionary history of major lineages of turtles. Mol. Phylogenet. Evol. 2017;113:59–66. doi: 10.1016/j.ympev.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 43.Colston T.J., Kulkarni P., Jetz W., Pyron R.A. Phylogenetic and Spatial Distribution of Evolutionary Isolation and Threat in Turtles and Crocodilians (Non-Avian Archosauromorphs) BMC Evol. Biol. 2019:20. doi: 10.1186/s12862-020-01642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ayres M., Sampaio M.M., Barros R.M.S., Dias L.B., Cunha O.R. A karyological study of turtles from the Brazilian Amazon region. Cytogenetics. 1969;8:401–409. doi: 10.1159/000130051. [DOI] [PubMed] [Google Scholar]
- 45.Killebrew F.C. Mitotic chromosomes of turtles: I. the Pelomedusidae. J. Herpetol. 1975;9:281–285. doi: 10.2307/1563192. [DOI] [Google Scholar]
- 46.De Smet W.H. The chromosomes of 11 species of Chelonia. (REPTILIA) Acta Zool. Pathol. Antverp. 1978;70:15–34. [Google Scholar]
- 47.Mittermeier R.A., Fedullo L.P. Cytogenetic analysis of the pleurodine turtle Phrynops hogei and its taxonomic implications. Amphibia-Reptilia. 1991;12:203–212. [Google Scholar]
- 48.Valenzuela N., Adams D.C., Janzen F.J. Pattern does not equal process: Exactly when is sex environmentally determined? Am. Nat. 2003;161:676–683. doi: 10.1086/368292. [DOI] [PubMed] [Google Scholar]
- 49.Valenzuela N., Adams D.C. Chromosome number and sex determination coevolve in turtles. Evolution. 2011;65:1808–1813. doi: 10.1111/j.1558-5646.2011.01258.x. [DOI] [PubMed] [Google Scholar]
- 50.Montiel E.E., Badenhorst D., Tamplin J., Burke R.L., Valenzuela N. Discovery of the youngest sex chromosomes reveals first case of convergent co-option of ancestral autosomes in turtles. Chromosoma. 2017;126:105–113. doi: 10.1007/s00412-016-0576-7. [DOI] [PubMed] [Google Scholar]
- 51.Lee L., Montiel E.E., Valenzuela N. Discovery of putative XX/XY male heterogamety in Emydura subglobosa turtles exposes a novel trajectory of sex chromosome evolution in Emydura. Cytogenet Genome Res. 2019;158:160–169. doi: 10.1159/000501891. [DOI] [PubMed] [Google Scholar]
- 52.Bista B., Valenzuela N. Turtle Insights into the Evolution of the Reptilian Karyotype and the Genomic Architecture of Sex Determination. Genes. 2020;11:416. doi: 10.3390/genes11040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee L., Montiel E.E., Navarro-Domínguez B.M., Valenzuela N. Chromosomal rearrangements during turtle evolution altered the synteny of genes involved in vertebrate sex determination. Cytogenet Genome Res. 2019;157:77–88. doi: 10.1159/000497302. [DOI] [PubMed] [Google Scholar]
- 54.Ferreira G.S., Langer M.C. A pelomedusoid (Testudines, Pleurodira) plastron from the lower Cretaceous of Alagoas, Brazil. Cretac. Res. 2013;46:267–271. doi: 10.1016/j.cretres.2013.10.001. [DOI] [Google Scholar]
- 55.Romano P.S., Gallo V., Ramos R.R., Antonioli L. Atolchelys lepida, a new side-necked turtle from the Early Cretaceous of Brazil and the age of crown Pleurodira. Biol. Lett. 2014;10:20140290. doi: 10.1098/rsbl.2014.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de la Fuente M.S., Umazano A.M., Sterli J., Carballido J.L. New chelid turtles of the lower section of the Cerro Barcino formation (Aptian-Albian?), Patagonia, Argentina. Cretac. Res. 2011;32:527–537. doi: 10.1016/j.cretres.2011.03.007. [DOI] [Google Scholar]
- 57.Joyce W.G. Origin, Evolution and Biogeographic History of South American Turtles. Ameghiniana. 2014;51:81. doi: 10.5710/AMGH.v51i1.1. [DOI] [Google Scholar]
- 58.Shaffer H.B., McCartney-Melstad E., Near T.J., Mount G.G., Spinks P.Q. Phylogenomic analyses of 539 highly informative loci dates a fully resolved time tree for the major clades of living turtles (Testudines) Mol. Phylogenet. Evol. 2017;115:7–15. doi: 10.1016/j.ympev.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 59.Matsubara K., O’Meally D., Azad B., Georges A., Sarre S.D., Graves J.A.M., Matsuda Y., Ezaz T. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma. 2016;125:111–123. doi: 10.1007/s00412-015-0531-z. [DOI] [PubMed] [Google Scholar]
- 60.Richard G.F., Pâques F. Mini- and microsatellite expansions: The recombination connection. EMBO Rep. 2000;1:122–126. doi: 10.1093/embo-reports/kvd031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin P., Makepeace K., Hill S.A., Hood D.W., Moxon E.R. Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl. Acad. Sci. USA. 2005:3800–3804. doi: 10.1073/pnas.0406805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balaresque P., King T.E., Parkin E.J., Heyer E., Carvalho-Silva D., Kraaijenbrink T., de Knijff P., Tyler-Smith C., Jobling M.A. Gene conversion violates the stepwise mutation model for microsatellites in Y-chromosomal palindromic repeats. Hum. Mutat. 2014;35:609–617. doi: 10.1002/humu.22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Figliuolo V.S.P., Goll L., Viana P.F., Feldberg E., Gross M.C. First Record on Sex Chromosomes in a Species of the Family Cynodontidae: Cynodon gibbus (Agassiz, 1829) Cytogenet Genome Res. 2020;160:29–37. doi: 10.1159/000505889. [DOI] [PubMed] [Google Scholar]
- 64.Alföldi J., Di Palma F., Grabherr M., Williams C., Kong L., Mauceli E., Russell P., Lowe C.B., Glor R.E., Jaffe J.D., et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castoe T.A., de Koning A.J., Hall K.T., Yokoyama K.D., Gu W., Smith E.N., Feschotte C., Uetz P., Ray D.A., Dobry J., et al. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol. 2011;12:406. doi: 10.1186/gb-2011-12-7-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castoe T.A., Hall K.T., Mboulas M.L.G., Gu W., de Koning A.J., Fox S.E., Poole W.E., Vemulapalli V., Daza J.M., Mockler T., et al. Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol. Evol. 2011;3:641–653. doi: 10.1093/gbe/evr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams R.H., Blackmon H., Reyes-Velasco J., Schield D.R., Card D.C., Andrew A.L., Waynewood N., Castoe T.A. Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome. 2016;59:295–310. doi: 10.1139/gen-2015-0124. [DOI] [PubMed] [Google Scholar]
- 68.Shedlock A.M., Botka C.W., Zhao S., Shetty J., Zhang T., Liu J.S., Deschavanne P.J., Edwards S.V. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc. Natl. Acad. Sci. USA. 2007;104:2767–2772. doi: 10.1073/pnas.0606204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Card D.C., Schield D.R., Reyes-Velasco J., Fujita M.K., Andrew A.L., Oyler-McCance S.J., FIke J.A., Tomback D.F., Ruggiero R.P., Castoe T.A. Two low coverage bird genomes and a comparison of reference-guided versus de novo genome assemblies. PLoS ONE. 2014;9:e106649. doi: 10.1371/journal.pone.0106649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joyce W.G., Parham J.F., Lyson T.R., Warnock R.C.M., Donoghue P.C.J. A divergence dating analysis of turtles using fossil calibrations: An example of best practices. J. Paleontol. 2013;87:612–634. doi: 10.1666/12-149. [DOI] [Google Scholar]
- 71.Deakin J.E., Ezaz T. Understanding the Evolution of Reptile Chromosomes through Applications of Combined Cytogenetics and Genomics Approaches. Cytogenet. Genome Res. 2019;157:7–20. doi: 10.1159/000495974. [DOI] [PubMed] [Google Scholar]
- 72.Deakin J.E., Potter S., O’Neill R., Ruiz-Herrera A., Cioffi M.B., Eldridge M.D., Fukui K., Marshall Graves J.A.M., Griffin D., Grutzner F., et al. Chromosomics: Bridging the gap between genomes and chromosomes. Genes. 2019;10:627. doi: 10.3390/genes10080627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Payseur B.A., Nachman M.W. Microsatellite variation and recombination rate in the human genome. Genetics. 2000;156:1285–1298. doi: 10.1093/genetics/156.3.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y.C., Korol A.B., Fahima T., Beiles A., Nevo E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol. Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294X.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 75.Ramsay L., Macaulay M., Cardle L., Morgante M., Ivanissevich S.D., Maestri E., Powell W., Waugh R. Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant. J. 1999;17:415–425. doi: 10.1046/j.1365-313X.1999.00392.x. [DOI] [PubMed] [Google Scholar]
- 76.Prizon A.C., Bruschi D.P., Gazolla C.B., Borin-Carvalho L.A., Portela-Castro A.L.D.B. Chromosome Spreading of the Retrotransposable Rex-3 Element and Microsatellite Repeats in Karyotypes of the Ancistrus Populations. Zebrafish. 2018;15:504–514. doi: 10.1089/zeb.2018.1620. [DOI] [PubMed] [Google Scholar]
- 77.Barros A.V., Wolski M.A.V., Nogaroto V., Almeida M.C., Moreira-Filho O., Vicari M.R. Fragile sites, dysfunctional telomere and chromosome fusions: What is 5S rDNA role? Gene. 2017;608:20–27. doi: 10.1016/j.gene.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 78.Aksenova A.Y., Mirkin S.M., Clemente L., Mazzoleni S., Pensabene-Bellavia E., Augstenová B., Auer M., Praschag P., Protiva M., Velenský P., et al. At the beginning of the end and in the middle of the beginning: Structure and maintenance of telomeric DNA repeats and interstitial telomeric sequences. Genes. 2019;10:118. doi: 10.3390/genes10020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clemente L., Mazzoleni S., Pensabene-Bellavia E., Augstenová B., Auer M., Praschag P., Protiva T., Velenský P., Wagner P., Fritz U., et al. Interstitial Telomeric Repeats Are Rare in Turtles. Genes. 2020;11:657. doi: 10.3390/genes11060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ocalewicz K. Telomeres in fishes. Cytogenet. Genome Res. 2013;141:114–125. doi: 10.1159/000354278. [DOI] [PubMed] [Google Scholar]
- 81.Montiel E.E., Badenhorst D., Lee L.S., Literman R., Trifonov V., Valenzuela N. Cytogenetic insights into the evolution of chromosomes and sex determination reveal striking homology of turtle sex chromosomes to amphibian autosomes. Cytogenet. Genome Res. 2016;148:292–304. doi: 10.1159/000447478. [DOI] [PubMed] [Google Scholar]
- 82.Rovatsos M., Kratochvíl L., Altmanova M., Pokorna M.J. Interstitial telomeric motifs in squamate reptiles: When the exceptions outnumber the rule. PLoS ONE. 2015;10:e0134985. doi: 10.1371/journal.pone.0134985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meyne J., Baker R.J., Hobart H.H., Hsu T.C., Ryder O.A., Ward O.G., Wiley J.E., Wurster-Hill D.H., Yates T.L., Moyzis R.K. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma. 1990;99:3–10. doi: 10.1007/BF01737283. [DOI] [PubMed] [Google Scholar]
- 84.Garrido-Ramos M.A., Herrán R., Ruiz-Rejón C., Ruiz-Rejón M. A satellite DNA of the Sparidae family (Pisces, Perciformes) associated with telomeric sequences. Cytogenet. Cell Genet. 1998;83:3–9. doi: 10.1159/000015151. [DOI] [PubMed] [Google Scholar]
- 85.Nergadze S., Santagostino M., Salzano A., Mondello C., Giulotto E. Contribution of telomerase RNA retrotranscription to DNA double-strand break repair during mammalian genome evolution. Genome Biol. 2007;8:260. doi: 10.1186/gb-2007-8-12-r260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Flint J., Craddock C.F., Villegas A., Bentley D.P., Williams H.J., Galanello R., Cao A., Wood W.G., Ayyub H., Higgs D.R. Healing of broken human chromosomes by the addition of telomeric repeats. Am. J. Hum. Genet. 1994;55:505–512. [PMC free article] [PubMed] [Google Scholar]
- 87.Azzalin C.M., Nergadze S.G., Giulotto E. Human intrachromosomal telomeric-like repeats: Sequence organization and mechanisms of origin. Chromosoma. 2001;110:75–82. doi: 10.1007/s004120100135. [DOI] [PubMed] [Google Scholar]
- 88.Viana P.F., Ezaz T., Marajó L., Ferreira M., Zuanon J., Cioffi M.B., Bertollo L., Gross M.C., Feldberg E. Genomic Organization of Repetitive DNAs and Differentiation of an XX/XY Sex Chromosome System in the Amazonian Puffer Fish, Colomesus asellus (Tetraodontiformes) Cytogenet. Genome Res. 2017;153:96–104. doi: 10.1159/000484423. [DOI] [PubMed] [Google Scholar]
- 89.Bachtrog D., Mank J.E., Peichel C.L., Kirkpatrick M., Otto S.P., Ashman T.L., Hahn M.W., Kitano J., Mayrose I., Ming R., et al. Tree of Sex Consortium. Sex determination: Why so many ways of doing it? PLoS Biol. 2014;12:e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tree of Sex Consortium Tree of Sex: A database of sexual systems. Sci. Data. 2014;24:140015. doi: 10.1038/sdata.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pennell M.W., Mank J.E., Peichel C.L. Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 2018;27:3950–3963. doi: 10.1111/mec.14540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Smet W.H.O. Chromosomes of 23 species of snakes. Acta Zool. Pathol. Antverp. 1978;70:85–118. [Google Scholar]
- 93.Baker R.J., Mengden G.A., Bull J.J. Karyotypic studies of thirty-eight species of North American snakes. Copeia. 1972:257–265. doi: 10.2307/1442486. [DOI] [Google Scholar]
- 94.Wright A.E., Dean R., Zimmer F., Mank J.E. How to make a sex chromosome. Nat. Commun. 2016;7:12087. doi: 10.1038/ncomms12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Darolti I., Wright A.E., Sandkam B.A., Morris J., Bloch N.I., Farré M., Fuller R.C., Bourne G.R., Larkin D.M., Breden F., et al. Extreme heterogeneity in sex chromosome differentiation and dosage compensation in livebearers. Proc. Natl. Acad. Sci. USA. 2019;116:19031–19036. doi: 10.1073/pnas.1905298116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stock M., Horn A., Grossen C., Lindtke D., Sermier R., Betto-Colliard C., Dufresnes C., Bonjour E., Dumas Z., Luquet E., et al. Ever-young sex chromosomes in European tree frogs. PLoS Biol. 2011;9:e1001062. doi: 10.1371/journal.pbio.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ogata M., Lambert M., Ezaz T., Miura I. Reconstruction of female heterogamety from admixture of XX-XY and ZZ-ZW sex-chromosome systems within a frog species. Mol. Ecol. 2018;27:4078–4089. doi: 10.1111/mec.14831. [DOI] [PubMed] [Google Scholar]
- 98.Nielsen S.V., Guzmán-Méndez I.A., Gamble T., Blumer M., Pinto B.J., Kratochvíl L., Rovatsos M. Escaping the evolutionary trap? Sex chromosome turnover in basilisks and related lizards (Corytophanidae: Squamata) Biol. Lett. 2019;15:20190498. doi: 10.1098/rsbl.2019.0498. [DOI] [PMC free article] [PubMed] [Google Scholar]