Abstract
Head and neck squamous cell carcinoma (HNSCC) is one of the most common cancers worldwide. Long noncoding RNAs were proved to be associated with the development and progression in HNSCC. However, the mechanism of LINC00460 in HNSCC needs to be further investigated. The study used quantitative real-time polymerase chain reaction assay to detect the expression of LINC00460 in cancer tissues and cell lines. Gain and loss of function experiments were conducted to analyze the effects of LINC00460 and miR-4443 on cell proliferation, invasion, and apoptosis of HNSCC cells in vitro. The interactions among miR-4443 and LINC00460 were detected by dual-luciferase reporter assay. Here, the study showed that LINC00460 was highly expressed in HNSCC tissues and cell lines. Functionally, knockdown of LINC00460 inhibited HNSCC cell proliferation and migration in vitro. Besides, LINC00460 promoted cell progression by sponging miR-4443, and miR-4443 inhibitor could reverse the effects of si-LINC00460 on cell proliferation and migration. In summary, LINC00460 could potentially promote cell progression and epithelial mesenchymal transition by sponging miR-4443 in HNSCC. LINC00460 could be used as a potential therapeutic target for HNSCCs.
Keywords: LINC00460, head and neck squamous cell carcinoma, miR-4443, long noncoding RNA
Introduction
Head and neck squamous cell carcinoma (HNSCC) is one of the most common cancers worldwide1. In recent years, the prevalence rate of HNSCC ranks the sixth worldwide2. And 600,000 patients3 suffer from recurrence and metastasis every year even the diagnosis and treatment of HNSCC has largely processed in recent years. The 5-year survival rate of HNSCC is still 60%. Therefore, more efforts should be made to explore a novel therapeutic target of HNSCC, which is of clinical significance to early diagnosis and treatment.
Noncoding RNA (ncRNA), a molecule that is lack of protein-coding ability, includes microRNA and long noncoding RNAs (lncRNAs)4,5. lncRNAs are a novel group of RNAs identified as transcripts longer than 200 nucleotides in length without potential protein-coding ability6. In recent years, accumulating study reveals that lncRNAs are associated with the development and progression of cancers. High expression of HOTAIR may lead to alteration of histone H3 lysine 27 methylation and increase cancer invasiveness and metastasis depending on PRC27–9. The dysregulated expression of lncRNAs can participate in carcinoma’s recurrence and metastasis, through regulating mechanisms and transcription of genes in three levels, including epigenetic modification, transcription, protein function regulation, and post-transcriptional regulation, and can be involved in genomic imprinting and chromatin modification10–13. Although the function and mechanism of lncRNAs are still unclear, more and more studies reveal that lncRNAs might function as oncogenes.
LINC00460 has been reported to act as an oncogenic RNA in several human cancers, such as lung cancer14, colorectal cancer15, nasopharyngeal carcinoma13, as well as HNSCC16. In HNSCC, high expression of LINC00460 promoted epithelial mesenchymal transition (EMT) by promoting peroxiredoxin-1 into the nucleus16. Through inhibiting the expression of miR-612, LINC00460 could contribute to cell progression and invasion by targeting AKT2 in HNSCC cell lines17. These findings suggest that LINC00460 is associated with the progression of HNSCC.
MicroRNAs play a crucial role in the development of cancer and can be potential treatment targets. Some studies report that lncRNAs can function as competitive endogenous RNAs (ceRNAs) or miRNA sponges to achieve their functions18. Hsa-miR-4443 is involved in a variety of tumors19,20, and many studies have confirmed the relationship between lncRNA and miR-4443. It is suggested that lncRNA FEZF1-AS1 could contribute to hepatocellular carcinoma progression by regulating miR-444321. Also, lncRNA MNX1-AS1 was reported to promote the progression of glioblastoma by inhibiting miR-444322. Up to the present, there is still no study about its role in HNSCC.
Our study aims to investigate whether lncRNA LINC00460 could regulate miR-4443 mediating proliferation and migration of HNSCC, thus providing evidences for potential clinical application. Our study provides insights into how LINC00460 promotes HNSCC cell proliferation, migration, and EMT, but inhibits cell apoptosis by sponging miR-4443.
Materials and Methods
Collection of Tissue Specimens
The study collected a total of 15 fresh HNSCC tissues and paired adjacent normal tissues from patients undergoing surgery between 2013 and 2014 at the Department of Oral and Maxillofacial Surgery, Affiliated Stomatology Hospital of Nanjing Medical University, China. Histopathological diagnosis was confirmed by two experienced pathologists. All samples were immediately placed on ice, snap-frozen in liquid nitrogen, and stored at −80°C until the extraction of total RNA. Patients who had been treated with molecular targeted therapy, chemotherapy, or radiotherapy before surgery were excluded. All patients provided written informed consent before sample collection, and the study protocol was approved by the Ethics Committee of Nanjing Medical University.
Cell Culture
Three HNSCC cell lines (CAL-27, WSU-HN4, and WSU-HN6) were obtained from the Cell Bank of the Chinese Academy of Sciences, Shanghai, China. WSU-HN4 and WSU-HN6 were cultured and maintained in Dulbecco’s modified eagle’s medium (DMEM)/F12 (1:1) medium (Gibco, Grand Island, NY, USA), and CAL-27 was cultured in DMEM (Gibco). The media were supplemented with 10% fetal bovine serum (FBS), 100 mg/ml penicillin, and 100 mg/ml streptomycin (Invitrogen, Carlsbad, CA, USA). All the cells were cultured at 37°C in an incubator with a humidified 5% CO2 atmosphere. All experiments were repeated in triplicate.
Total RNA Extraction, Reverse Transcription, and Real-Time Quantitative Polymerase Chain Reaction
Total RNA was isolated from cells or frozen tissues in accordance with the TRIZOL reagent (Invitrogen) manufacturer’s protocol. Subsequently, 1 µg of total RNA was used to reverse transcribed into cDNA by using the PrimerScript reagent Kit (TaKaRa, Shiga, Japan). The quantitative real-time polymerase chain reaction (qRT-PCR) experiments were performed with the use of the SYBR Premix Ex Taq II (TaKaRa) on the ABI PRISM 7300 Real-Time PCR system. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and 5 S were selected as the internal reference. The PCR primer sequences are as follows: GAPDH 5′-GAACGGGAAGCTCACTGG-3′- (sense) and 5′-GCCTGCTTCACCACCTTCT-3′- (antisense), U6 5′-CTCGCTTCGGCAGCACA-3′- (sense) and 5′-AACGCTTCACGAATTTGCGT-3′- (antisense), and LINC00460 5′-ATGCACACTTCTCGGCTAAG-3′- (sense) and 5′-GGTCGTAACCTTCGTTCTCATC-3′- (antisense). All Bulge-Loop miRNA primers were designed by RiboBio (Guangzhou, China). Mature miR-4689, miR-4443, miR-4665-5p, and U6 were transcribed by Bulge-Loop miRNA qRT-PCR Starter Kit (RiboBio). Relative expression of genes was calculated by 2−ΔΔCt. All experiments were repeated in triplicate.
Cell Transfection
The short interfering RNA (siRNA), negative control (NC), miR-4443 mimic, miR-4443 inhibitor, and pEX-3 vector containing LINC00460 sequence or empty plasmid used in our study were synthesized from Gene Pharma (Shanghai, China). Besides, GP-miRGLO plasmids encoding wild-type (WT) LINC00460 or mutant (MUT) LINC00460 were constructed by Gene Pharma. Transfection was conducted with Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instruction, and after cells were seeded on plates for 24 h later. The sequences are summarized in Table S1.
Cell Viability Assay
A Cell Counting Kit-8 (APE×BIO Technology, Houston, TX, USA) was used to assess cell proliferation. Twenty-four hours after transfected with si-LINC00460 or NC, approximately 2 × 103 cells per well were seeded into 96-well plates with a volume of 100 µl medium, and then incubated at 37°C later. At 0, 24, 48, 72, 96, and 120 h after gene transfection, 10 µl of CCK-8 reagent was added to each well. After incubation at 37°C for 2 h, the absorbance levels were measured at a wavelength of 450 nm using a microplate reader. All experiments were repeated in triplicate.
Transwell Migration Assays
For transwell migration assay, CAL-27 or WSU-HN4 cells (6 × 104) after transfected 24 h were plated in a total volume of 200 µl serum-free medium on the upper transwell chamber (Corning, Corning, NY, USA) with 8-µm porosity polycarbonate filters. The lower chambers contain 600 µl DMEM supplemented with 10% FBS, which function as a chemoattractant. Then, the chambers were put into 24-well plates, and cells were incubated for 24 or 36 h at 37°C. Subsequently, cells migrated from the membrane were fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet (Beyotime, Shanghai, China) for 30 min. The migrated cells were calculated in five random microscopic fields with the use of a microscope. All migrated cells were counted using Image-J(USA) software. The experiments were repeated in triplicate.
Flow Cytometry
Cell apoptosis were detected by Annexin V-FITC/PI double staining (BD Biosciences, Franklin Lakes, NJ, USA). CAL-27 or WSU-HN4 cells were harvested 48 h after transfected with siRNA or overexpression plasmids. Then, cells were washed by phosphate-buffered saline for two times and suspending cells with 400 µl 1 ×Binding Buffer at a concentration of 1 × 106 cells/ml. For analysis, cells were incubated for 15 min after the addition of 5 µl Annexin V-FITC, and then 10 µl PI was added into the solution for 15 min incubation in the dark. All samples were detected by the use of flow cytometry (BD Biosciences).
Western Blot Assay and Antibodies
Cells were lysed after transfection for 24 h on ice with the aid of radioimmunoprecipitation assay buffer containing phenylmethane sulfonyl fluoride (Beyotime). Then the solution was centrifuged for 15 min, at 12,000 rpm, 4°C after incubation for 15 min on ice. The equal protein (20 μg) was separated on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and then separated gel electrophoresis was transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Boston, MA, USA) in a wet manner. After blocking using 5% of the skimmed milk for 2 h, the PVDF membrane was washed in Tris-buffered saline Tween (TBST) for three times. Then, the membranes were marked with primary antibodies at 4°C over night, recognized as E-cadherin, N-cadherin, and Vimentin (1:1,000 dilution; Cell Signaling Technology, Danvers, MA, USA), and GAPDH (1:1,000 dilution; Cell Signaling Technology) antibody was used as control. The next day, the membranes were washed three times with TBST. Protein bands were incubated with the combination of secondary antibody conjugated horseradish peroxidase for 1 h. The protein strips were visualized by the Image QuantLAS 4000 mini imaging system under the exposure to enhanced chemiluminescent reagent.
Isolation of Cytoplasmic and Nuclear RNA
Isolation of nuclear and cytoplasmic RNA was conducted using the PARIS kit (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol. All samples were purified and treated by DNase I. The separated RNA from nuclear and cytoplasmic fractions was subsequently used for reverse transcription, and real-time PCR reactions (TaKaRa) were applied to every sample. The U6 was described as controls for the nucleus, while GAPDH was treated as internal controls for the cytoplasm.
Dual-Luciferase Reporter Assay
For dual-luciferase reporter assay, the binding sites of LINC00460 and miR-4443 were predicted by online websites. In the study, the WT sequences of LINC00460 and the MUT fragments were synthesized by Gene Pharma, then inserted into the pEX-3 vector, respectively. Two distinctively constructed plasmids, namely pEX-3-LINC00460-WT and pEX-3-LINC00460-MUT, were obtained. CAL-27 or WSU-HN4 cells were seeded into 24-well plates and incubated for 24 h. The reagent Lipofectamine 2000 (Invitrogen) was used to cotransfect 50 nmol miR-4443 mimic or mimics NC with 2 µg pEX-3-LINC00460-WT or pEX-3-LINC00460-MUT. Next, cells were harvested and lysed for luciferase assay 48 h after transfection. The luciferase activities were tested using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) while the Renilla luciferase function as an internal control.
Statistical Analysis
Statistical analyses were conducted using SPSS software (version 22.0, SPSS Inc., Chicago, IL, USA). Student’s t-test or analysis of variance (ANOVA, with Sidak post hoc test) was performed to analyze the significance of differences between groups. P-values <0.05 were considered statistically significant.
Results
The Expression of LINC00460 is Upregulated in HNSCC Tissue Samples and is Correlated With Poor Prognosis in HNSCC
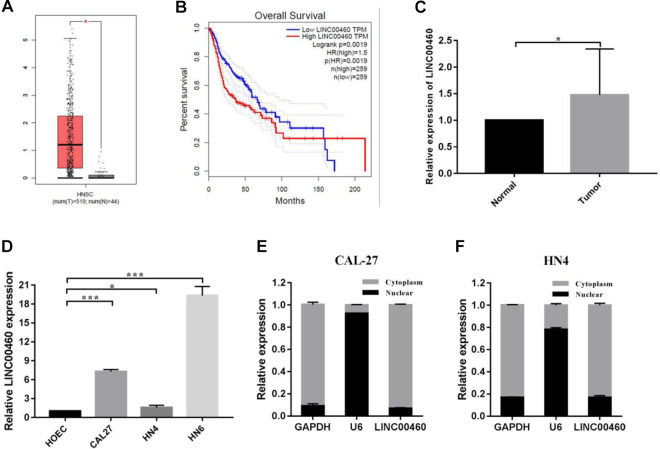
In the GEPIA database, we found that the expression of LINC00460 was significantly high in HNSCC patients (Fig. 1A) and always indicated poor prognosis (Fig. 1B). To investigate the clinical significance of LINC00460 with HNSCC, we detect the expression level of LINC00460 in 15 pairs of HNSCC tissues and adjacent normal tissues by RT-qPCR. The results indicated that the expression of LINC00460 was significantly upregulated in HNSCC tissues compared with adjacent normal tissues (Fig. 1C). Furthermore, the expression of LINC00460 is higher in three HNSCC cell lines (CAL-27, WSU-HN4, and WSU-HN6) compared with HOEC, a normal human oral mucosa cell line (Fig. 1D). By isolation of cytoplasmic and nuclear RNA in CAL-27 and WSU-HN4 cell lines, we observed that LINC00460 was predominantly distributed in the cytoplasm by using RT-qPCR (Fig. 1E, F). In summary, these findings suggested that the upregulated LINC00460 might serve as a vital regulator in the HNSCC development.
Fig. 1.
The expression of LINC00460 was upregulated in HNSCC tissue samples and was correlated with poor prognosis in HNSCC. (A) Relative LINC00460 expression in HNSCC from TCGA database. In all, 519 HNSCC tumor tissues and 44 normal tissues, respectively. (B) Kaplan–Meier survival analysis of overall survival according to LINC00460 expression in HNSCC from TCGA database. (C) LINC00460 expression in 15 pair samples of HNSCC and adjacent normal tissues. (D) Relative levels of LINC00460 in HOEC, a normal human oral mucosa cell line and three types of HNSCC cells, CAL-27, WSU-HN4, and WSU-HN6, were determined by qRT-PCR. (E, F) Subcellular location of LINC00460 in CAL-27, and WSU-HN4 cells. U6 and GAPDH acted as nucleus and cytoplasm marker, respectively. Data are presented as the mean ± SD, analyzed using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. GAPDH: glyceraldehyde 3-phosphate dehydrogenase; HNSCC: head and neck squamous cell carcinoma; qRT-PCR: quantitative real-time polymerase chain reaction.
Knockdown of LINC00460 on HNSCC Cells Inhibits Growth and Induces Apoptosis In Vitro
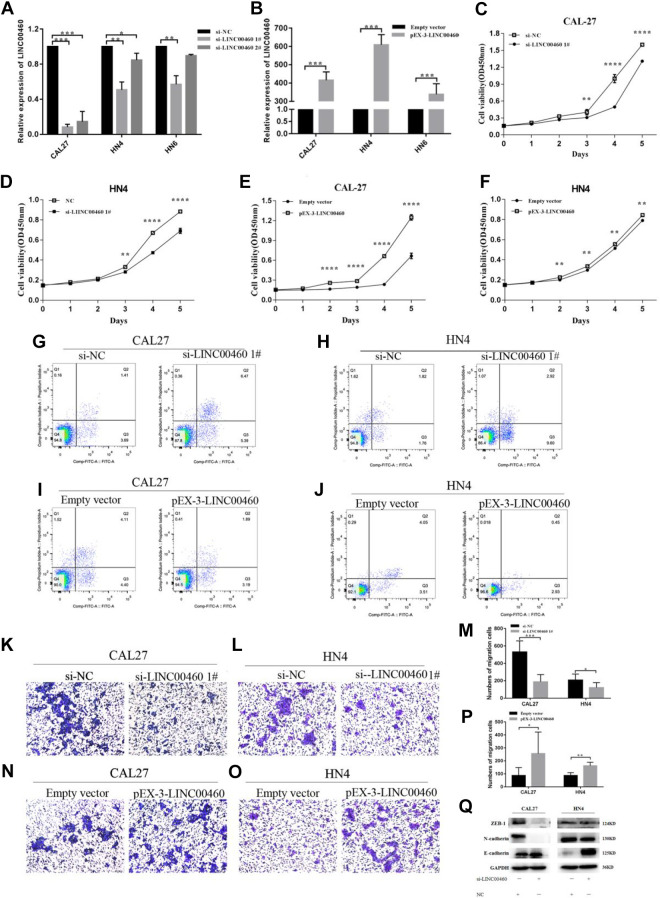
To determine the effects of LINC00460 on cell proliferation and apoptosis, small interfering RNAs (siRNAs) were transfected to knockdown LINC00460 in CAL-27, WSU-HN4, and WSU-HN6 cell lines, and plasmid-mediated overexpression was used for exogenously controlling the expression of LINC0460 in CAL-27 and WSU-HN4 (Fig. 2A, B).
Fig. 2.
Silence of LINC00460 inhibited the migration of HNSCC cells. (A, B) Validation of siRNA knockdown and overexpression vector efficiency in CAL-27 and WSU-HN4 cells as determined by qRT-PCR. (C, D) CCK-8 proliferation assays in CAL-27 and WSU-HN4 cells after transfection with si-LINC00460. (E, F) CCK-8 proliferation assays in CAL-27 and WSU-HN4 cells after transfection with pEX-3-LINC00460 or empty vector. (G, H) Flow cytometry was used to evaluate the effect of apoptosis after transfection with si-LINC00460 or negative control. (I, J) Flow cytometry was used to evaluate the effect of apoptosis after transfection with pEX-3-LINC00460 or empty vector. (K–M) Transwell migration assay in si-LINC00460 or negative control transfected CAL-27 and WSU-HN4 cells. Magnification: ×100. (N–P) Transwell migration assay in pEX-3-LINC00460 or empty vector transfected CAL-27 and WSU-HN4 cells. Magnification: ×100. (Q) Expression of E-cadherin, ZAB1, N-cadherin, and GAPDH protein in CAL-27 and WSU-HN4 cells after transfection with si-LINC00460 or negative control. Data are presented as the mean ± SD, analyzed using Student’s t-test or analysis of variance. *P < 0.05, **P < 0.01, ***P < 0.001. GAPDH: glyceraldehyde 3-phosphate dehydrogenase; HNSCC: head and neck squamous cell carcinoma; qRT-PCR: quantitative real-time polymerase chain reaction; SD: standard deviation.
The effects of LINC00460 on HNSCC cell growth were detected by CCK8. And the results demonstrated that knockdown of LINC00460 significantly decreased cell proliferation in CAL-27 and WSU-HN4 compared to the NC group (Fig. 2C, D). Besides, after transfected by pEX-3-LINC00460, cell proliferation was largely increased in CAL-27 and WSU-HN4 cells compared to the group transfected with an empty vector (Fig. 2E, F). Next, flow cytometry analysis was applied to verify whether the inhibition of cell growth was induced by apoptosis. Then, we found that the percentage of apoptotic cells was remarkably increased in the si-LINC00460 transfected CAL-27 and WU-HN4 cells group compared to the NC group, which proved that HNSCC cell proliferation was induced by apoptosis (Fig. 2G, H). And the percentage of apoptotic cells was decreased in the group transfected with pEX-3-LINC00460 compared to the group transfected with an empty vector (Fig. 2I, J).
Silence of LINC00460 Inhibits the Migration of HNSCC Cells
In the study, the ability of migration of HNSCC cells was detected by transwell assay. As shown in Fig. 2K–M after the silence of LINC00460, the migration abilities were significantly decreased both in CAL-27 and WSU-HN4 cells compared to the NC group. And the migration abilities were enhanced in the group transfected with pEX-3-LINC00460 compared to the group transfected with an empty vector (Fig. 2N–P). Moreover, western blot analysis was implemented to confirm better the mechanisms of LINC00460 regulated metastasis in HNSCC. Those findings revealed that the expression level of E-cadherin was increased while N-cadherin, ZEB-1, was decreased compared with the NC cells when knockdown of LINC00460 in CAL-27 and WSU-HN4 cells (Fig. 2Q). The results convinced us that knockdown of LINC00460 could inhibit EMT ability.
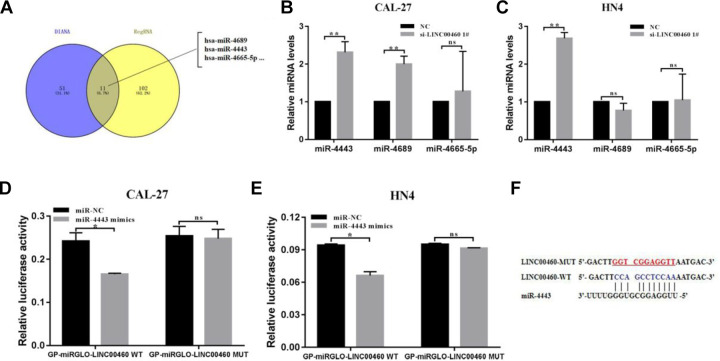
LINC00460 Regulates miR-4443 in HNSCC Cells
Recently, accumulating study indicated that lncRNAs might target and modulate miRNA to function as an oncogene. In our study, the bioinformatics analysis was applied to investigate the mechanism of how LINC00460 regulates HNSCC cell growth and metastasis. The online bioinformatics databases such as DIANA TOOLS (http://diana.imis.athena-innovation.gr) and RegRNA 2.0 (http://regrna2.mbc.nctu.edu.tw) were used to predict the potential miRNA that could target LINC00460. According to the results, we selected three miRNAs (miR-4689, miR-4443, and miR-4665-5p) for subsequent experiments (Fig. 3A). The previous study had confirmed that abnormal expression of those miRNA could contribute to tumorigenesis in several cancers. Then, we calculated the expression of miR-4689, miR-4443, and miR-4665-5p by qRT-PCR; the results showed that only miR-4443 was highly expressed in LINC00460 knockdown groups compared with the NC group both in CAL-27 and WSU-HN4 cells, while the level of miR-4689 and miR-4665-5p exhibited no significant change (Fig. 3B, C). Those results reveal that miR-4443 might be negatively modulated by LINC00460.
Fig. 3.
LINC00460 targeted and regulated miR-4443 in HNSCC cells. (A) The potential miRNA that could target LINC00460 by the bioinformatics analysis. (B, C) Relative miR-4689, miR-4443, and miR-4665-5p expression in CAL-27 and WSU-HN4 cells following knockdown of LINC00460. (D, E) miR-4443 mimics or mimics NC and GP-miRGLO-LINC00460-WT or GP-miRGLO-LINC00460-MUT were cotransfected into CAL-27 and WSU-HN4 cells. Luciferase activity was detected 24 h after transfection. (F) The predicted binding sites of miR-4443 to the LINC00460 sequence. Data are presented as the mean ± SD, analyzed using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. HNSCC: head and neck squamous cell carcinoma; NC: negative control; SD: standard deviation.
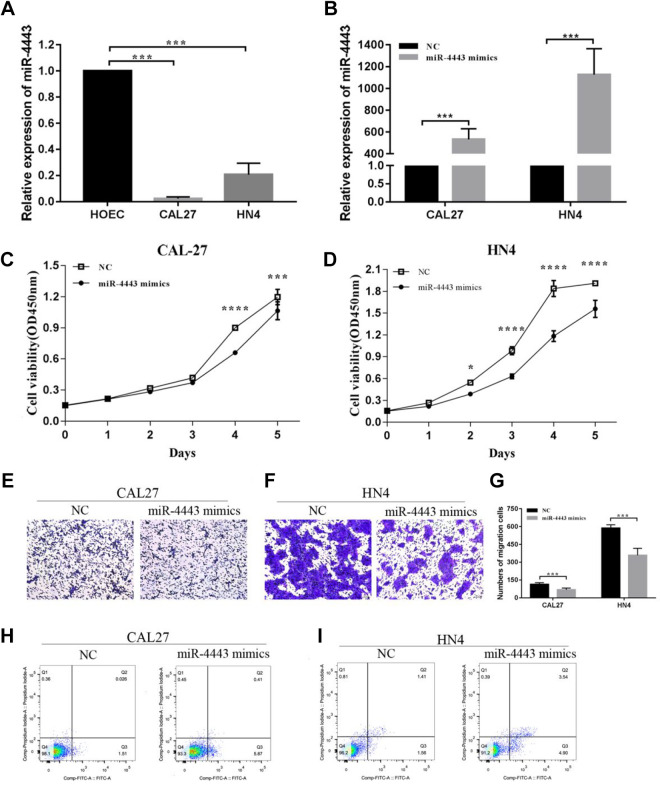
Overexpression of miR-4443 Inhibits Cell Growth, Migration, and Induces Apoptosis In Vitro
Because the role of miR-4443 in HNSCC is unknown, we performed the following experiments to verify the effect of miR-4443 on the biological function of HNSCC. First, we analyze the expression level of miR-4443 in HOEC, CAL-27, and WSU-HN4 cells by RT-qPCR. The results indicated that miR-4443 expression was significantly decreased in CAL-27 and WSU-HN4 cells (Fig. 4A). Subsequently, miR-4443 mimics were transfected to CAL-27 and WSU-HN4 cells to increase the expression of miR-4443 (Fig. 4B). We found that overexpression of miR-4443 significantly inhibited the proliferation of CAL-27 and WSU-HN4 cells detected by CCK8 (Fig. 4C, D). In contrast, overexpression of miR-4443 increased the apoptotic cells both in CAL-27 and WU-HN4 compared to the NC group (Fig. 4H, I). And the migration abilities were significantly decreased in miR-4443 mimics groups both in CAL-27 and WU-HN4, compared to the NC group (Fig. 4E–G). Compared to the si-LINC00460 group, those results demonstrated that the opposite effects on proliferation, migration, and apoptosis of CAL-27 and WU-HN4 cells were observed in miR-4443 mimics group.
Fig. 4.
Overexpression of miR-4443 inhibit cell growth, migration, and induced apoptosis in vitro. (A) Relative levels of miR-4443 on HOEC, a normal human oral mucosa cell line and two types of HNSCC cells, CAL-27 and WSU-HN4, were determined by qRT-PCR. (B) Relative miR-4443 expression in CAL-27 and WSU-HN4 cells after transfection with miR-4443 mimics. (C, D) CCK-8 proliferation assays in CAL-27 and WSU-HN4 cells after transfection with miR-4443 mimics or mi-NC. (E, F, G) Transwell migration assay in CAL-27 and WSU-HN4 cells after transfection with miR-4443 mimics or mi-NC. Magnification: ×100. (H, I) Apoptosis rate in CAL-27 and WSU-HN4 cells after transfection with miR-4443 mimics or mi-NC by flow cytometry. Data are presented as the mean ± SD, analyzed using Student’s t-test or analysis of variance. *P < 0.05, **P < 0.01, ***P < 0.001. HNSCC: head and neck squamous cell carcinoma; qRT-PCR: quantitative real-time polymerase chain reaction; SD: standard deviation.
MiR-4443 is a Direct Target of LINC00460
To verify the hypothesis that miR-4443 might a target of LINC00460, we carried out a dual-luciferase reporter system to confirm whether LINC00460 could bind to miR-4443. Then we found that the cotransfection of miR-4443 mimic and LINC00460-WT significantly suppressed the luciferase activity both in CAL-27 and WSU-HN4 cells, while the cotransfection of miR-4443 mimic and LINC00460 mutant type (LINC00460-MT) in CAL-27 and WSU-HN4 cells did not affect the luciferase activity. These results directly demonstrated that LINC00460 could be the target of miR-4443 (Fig. 3D–F).
LINC00460 Regulates miR-4443 Expressions in CAL-27 and WSU-HN4 Cells in the Rescue Experiment
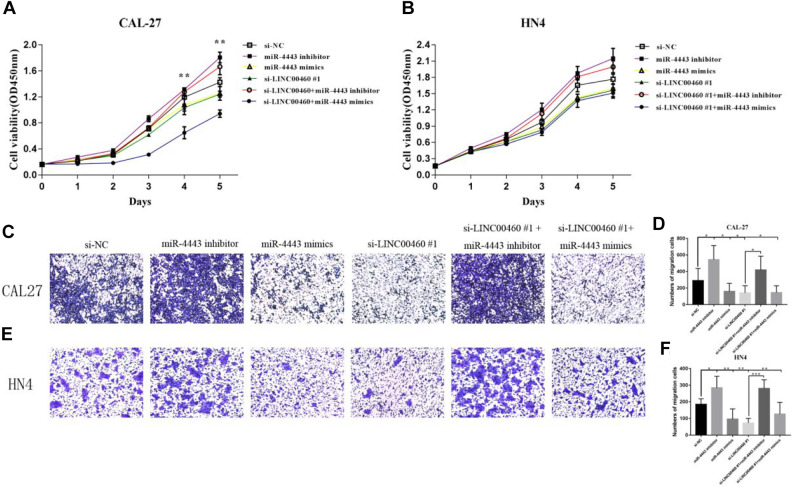
To confirm whether LINC00460 exerted its function through miR-4443 in CAL-27 and WU-HN4 cells, we cotransfected si-LINC00460 and miR-4443 inhibitor or miR-4443 mimics into the CAL-27 and WU-HN4 cells. As shown in Fig. 5A–F, the proliferation and migration abilities of si-LINC00460 or miR-4443 mimics transfected cells were decreased, while those abilities were increased in miR-4443 inhibitor transfected group. In the rescue experiment, the suppression of cell proliferation induced by si-LINC00460 was reversed by miR-4443 inhibitor after cotransfection of si-LINC00460 and miR-4443 inhibitor in HNSCC cell lines. Transwell assays showed that inhibition of migration abilities in CAL-27 and WSU-HN4 cells induced by si-LINC00460 were reversed by miR-4443 inhibitor. These results confirmed that LINC00460 functions as an oncogene in HNSCC cells relying on miR-4443.
Fig. 5.
miR-4443 inhibitor could partially reverse the effects of si-LINC00460 on cell proliferation and migration. (A, B) CCK-8 proliferation assay. (C–F) Transwell migration assay. Magnification: ×100. Data are presented as the mean ± SD, analyzed using Student’s t-test or analysis of variance. *P < 0.05, **P < 0.01, ***P < 0.001. SD: standard deviation.
Discussion
HNSCC is a non-skin malignancy cancer with high mortality rate23. Multiple studies have demonstrated that lncRNAs modulate biological processes in human cancers by regulating protein-coding, or affecting gene expression at the transcriptional levels or post-transcriptional levels, even through chromatin modification24. They can function as oncogenes to promote proliferation or metastasis in multiple cancers.
LINC00460 is a lncRNA of 935 bp in length and encoded on chromosome13q3314. It was reported that upregulation of LINC00460 could promote cell progression in HNSCC, and LINC00460 could inhibit autophagy of HNSCC cell lines by negatively regulating miR-20623. In our study, LINC00460 was highly upregulated in 15 HNSCC tumor tissues compared to the adjacent normal tissues. It has been demonstrated that LINC00460 could promote cell proliferation, migration, and invasion in HNSCC cell lines16. In our study, further investigation also showed that knockdown of LINC00460 could significantly inhibit cell proliferation, migration, and facilitate apoptosis in HNSCC cell lines.
EMT plays a vital role in cell migration and invasion25. In HNSCC, LINC00460 was found to induce the activation of the EMT signaling pathway16. In our study, the epithelial marker (E-cadherin) was increased while the mesenchymal phenotypes (N-cadherin and ZEB-1) were inhibited after silencing LINC00460 in CAL-27 and WU-HN4 cell lines, which reminded that tumor progression and cell migration were hindered, consistent with the previous study. The results of GEPIA database indicated that patients with high expression of LINC00460 had worse prognosis in HNSCC patients. Therefore, LINC00460 may play an important role in the progression of HNSCC.
To explore the mechanism by which LINC00460 accelerates the procession of HNSCC, we examined the expression of LINC00460 both in the cytoplasm and nuclear. LINC00460 is mainly distributed in the cytoplasm by RT-qPCR. It has been demonstrated that LncRNA located in cytoplasm may function as ceRNA for specific miRNA16. So we speculate that LINC00460 may serve as a ceRNA for specific miRNA in HNSCC. Previous studies demonstrated that LINC00460 could function as a ceRNA or a target of miRNA to modulate cancer development. It is shown to promote cell proliferation by sponging miR-149-5p in the cytoplasm to upregulate the expression of CUL4A in colorectal cancer26. And LINC00460 promoted the expression of MMP-9 to accelerate the proliferation and metastasis of meningioma by targeting miR-53927. Also, LINC00460 has been reported to function as an oncogene in HNSCC. It is demonstrated that LINC00460 could inhibit autophagy and increase the expression of STC2 by sponging miR-206 in HNSCC23. And LINC00460 could promote cell migration and proliferation by sponging miR-612 to increase the expression of AKT2. However, the relationship between LINC00460 and miR-4443 has not been reported. Our study predicted that miR-4689, miR-4443, and miR-4665-5p might be the potential molecule in HNSCC by bioinformatics tools. By RT-qPCR, the expression of miR-4443 was significantly upregulated in cells transfected with si-LINC00460 compared to the NC group. So we speculate miR-4443 might be the potential miRNA that could be downregulated by LINC00460. According to the previous studies, overexpression of miR-4443 suppressed the proliferation and invasion in glioblastoma, and the progression of ovarian cancer was inhibited by miR-444321,22. But the function of miR-4443 in HNSCC remains to be explored. Through the functional assay, we confirmed that miR-4443 is a tumor suppressor in HNSCC. The function of miR-4443 on proliferation, migration, and apoptosis is opposite to that of LINC00460 in HNSCC cells. In the rescue experiment, the silence of LINC00460 suppresses the proliferation and migration of HNSCC cells, whereas miR-4443 inhibitor could reverse those effect after the cotransfection of miR-4443 inhibitor and si-LINC00460 in HNSCC cell lines. Dual-luciferase reporter assays confirmed that miR-4443 is a direct target of LINC00460. It is suggested that LINC00460 functions as an oncogene by sponging miR-4443 in HNSCC.
In conclusion, it is confirmed that LINC00460 was highly expressed in HNSCC tissues and cell lines. The expression of LINC00460 is positively related with poor prognosis. The silence of LINC00460 could inhibit cell growth, migration, and EMT ability in HNSCC cells. It is indicated that LINC00460 exerts its oncogenic abilities in HNSCC by binding to miR-4443. However, the work has some limitations. The tissue sample size needs to be expanded to analyze the relationship between the expression level of LINC00460 and clinicopathological features in HNSCC. And the correlations between LINC00460 and miR-4443 expression in HNSCC tissues should also be explored. In addition, the molecular mechanism of how LINC00460 regulates HNSCC progression by sponging miR-4443 needs to be further studied. Overall, LINC00460 may be a novel therapeutic biomarker in HNSCC.
Supplemental Material
Supplementary_Table_1 for Long Noncoding RNA LINC00460 Promotes Cell Progression by Sponging miR-4443 in Head and Neck Squamous Cell Carcinoma by Meng Li, Xiaomin Zhang, Xu Ding, Yang Zheng, Hongming Du, Huaiqi Li, Huan Ji, Zeyu Wang, Pengfei Jiao, Xiaomeng Song, Yi Zhong and HeMing Wu in Cell Transplantation
Acknowledgments
Thank all the authors since everyone contributed to the project.
Footnotes
Ethical Approval: The study was approved by the Ethics Committee of Nanjing Medical University, Jiangsu Province, China.
Statement of Human and Animal Rights: All procedures in the study involving human subjects were conducted in accordance with the human ethics committee of Affiliated Stomatology Hospital of Nanjing Medical University, Jiangsu Province, China. This study does not contain any studies with animal.
Statement of Informed Consent: We confirm that guidelines on patient consent have been met and any details of informed consent obtained are indicated within the text of the submitted manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (No. 81600908), the Nature Science Foundation of Jiangsu Province (Grant No. BK2018040793), and the National Nature Science Foundation of China (No. 81772887).
ORCID iDs: Meng Li  https://orcid.org/0000-0001-5937-0709
https://orcid.org/0000-0001-5937-0709
Hongming Du  https://orcid.org/0000-0002-0909-0773
https://orcid.org/0000-0002-0909-0773
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Xu C, Jiang C, Wu Q, Liu L, Yan X, Shi R. A feed-forward regulatory loop between HUR and the long noncoding RNA hotair promotes head and neck squamous cell carcinoma progression and metastasis. Cell Physiol Biochem. 2016;40(5):1039–1051. [DOI] [PubMed] [Google Scholar]
- 2. Zou AE, Ku J, Honda TK, Yu V, Kuo SZ, Zheng H, Xuan Y, Saad MA, Hinton A, Brumund KT, Lin JH, et al. Transcriptome sequencing uncovers novel long noncoding and small nucleolar RNAs dysregulated in head and neck squamous cell carcinoma. RNA. 2015;21(6):1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hedberg ML, Goh G, Chiosea SI, Bauman JE, Freilino ML, Zeng Y, Wang L, Diergaarde BB, Gooding WE, Lui VW, Herbst RS, et al. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J Clin Invest. 2016;126(1):169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun W, Califano JA. Sequencing the head and neck cancer genome: implications for therapy. Ann N Y Acad Sci. 2014;1333:33–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji T, Chen WT, Zou X. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. [DOI] [PubMed] [Google Scholar]
- 7. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zornig M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feng J, Zhou Q, Yi H, Ma S, Li D, Xu Y, Wang J, Yin S. A novel lncRNA n384546 promotes thyroid papillary cancer progression and metastasis by acting as a competing endogenous RNA of miR-145-5p to regulate AKT3. Cell Death Dis. 2019;10(6):433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M, Hung T, Argani P, Rinn JL, Wang Y, et al. Long non-coding RNA hotair reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang R, Ma Z, Feng L, Yang Y, Tan C, Shi Q, Lian M, He S, Ma H, Fang J. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol Cancer. 2018;17(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Avgeris M, Tsilimantou A, Levis PK, Tokas T, Sideris DC, Stravodimos K, Ardavanis A, Scorilas A. Loss of GAS5 tumour suppressor lncRNA: an independent molecular cancer biomarker for short-term relapse and progression in bladder cancer patients. Br J Cancer. 2018;119(12):1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jin S, Yang X, Li J, Yang W, Ma H, Zhang Z. p53-targeted lincRNA-p21 acts as a tumor suppressor by inhibiting JAK2/STAT3 signaling pathways in head and neck squamous cell carcinoma. Mol Cancer. 2019;18(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kong Y, Cui M, Chen S, Xu Y, Xu Y, Tao Z. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. [DOI] [PubMed] [Google Scholar]
- 14. Yue QY, Zhang Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(4):1003–1010. [DOI] [PubMed] [Google Scholar]
- 15. Wang X, Mo F, Bo H, Xiao L, Chen G, Zeng P, Huang Y, Lei Z, Yuan W, Chen Z. Upregulated expression of long non-coding RNA, LINC00460, suppresses proliferation of colorectal cancer. Cancer. 2018;9(16):2834–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang Y, Cao W, Wu K, Qin X, Wang X, Li Y, Yu B, Zhang Z, Wang X, Yan M, Xu Q, et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J Exp Clin Cancer Res. 2019;38(1):365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie X, Xiong G, Wang Q, Ge Y, Cui X. Long non-coding RNA LINC00460 promotes head and neck squamous cell carcinoma cell progression by sponging miR-612 to up-regulate AKT2. Am J Transl Res. 2019;11(10):6326–6340. [PMC free article] [PubMed] [Google Scholar]
- 18. Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J, Rao X, Li M, Sun M, Jiang M, Xu Y, et al. Long noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 2019;156(3):676–6691. [DOI] [PubMed] [Google Scholar]
- 19. Chen X, Zhong S, Lu P, Wang D, Zhou S, Yang S, Shen H, Zhang L, Zhang X, Zhao J, Tang JH. miR-4443 participates in the malignancy of breast cancer. PLoS One. 2016;11(8):e0160780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ebrahimi SO, Reiisi S. Downregulation of miR-4443 and miR-5195-3p in ovarian cancer tissue contributes to metastasis and tumorigenesis. Arch Gynecol Obstet. 2019;299(5):1453–1458. [DOI] [PubMed] [Google Scholar]
- 21. Gong J, Wang J, Liu T, Hu J, Zheng J. lncRNA FEZF1AS1 contributes to cell proliferation, migration and invasion by sponging miR4443 in hepatocellular carcinoma. Mol Med Rep. 2018;18(6):5614–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao Y, Xu Y, Wang J, Yang X, Wen L, Feng J. . lncRNA MNX1-AS1 promotes glioblastoma progression through inhibition of miR-4443. Oncol Res. 2019;27(3):341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xue K, Li J, Nan S, Zhao X, Xu C. Downregulation of LINC00460 decreases STC2 and promotes autophagy of head and neck squamous cell carcinoma by up-regulating microRNA-206. Life Sci. 2019;231:116459. [DOI] [PubMed] [Google Scholar]
- 24. Sun Q, Hao Q, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34(2):142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. [DOI] [PubMed] [Google Scholar]
- 26. Lian Y, Yan C, Xu H, Yang J, Yu Y, Zhou J, Shi Y, Ren J, Ji G, Wang K. A novel lncrna, linc00460, affects cell proliferation and apoptosis by regulating KLF2 and CUL4A expression in colorectal cancer. Mol Ther Nucleic Acids. 2018;12:684–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xing H, Wang S, Li Q, Ma Y, Sun P. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed Pharmacother. 2018;105:677–682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Table_1 for Long Noncoding RNA LINC00460 Promotes Cell Progression by Sponging miR-4443 in Head and Neck Squamous Cell Carcinoma by Meng Li, Xiaomin Zhang, Xu Ding, Yang Zheng, Hongming Du, Huaiqi Li, Huan Ji, Zeyu Wang, Pengfei Jiao, Xiaomeng Song, Yi Zhong and HeMing Wu in Cell Transplantation