Abstract
Rheumatoid arthritis (RA) is a chronic systemic autoimmune disease. New evidence suggested that linc02381 suppressed colorectal cancer progression by regulating PI3 K signaling pathway, but the role of linc02381 in other diseases, such as RA, remains unclear. This study aimed to reveal the mechanism of linc02381 in RA progression. In vivo and in vitro, we found that linc02381 was upregulated in RA synovial tissues or RA fibroblast-like synoviocytes (RA-FLSs, P < 0.01), which were detected by quantitative real-time polymerase chain reaction. Cell Counting Kit-8, EDU, and Transwell assays revealed that linc02381 overexpression enhanced cell proliferation and invasion, and linc02381 knockdown inhibited cell proliferation and invasion in FLSs. Moreover, the results of bioinformatics analysis, luciferase reporter gene assay, and pull-down assay verified that linc02381 could directly bind with miR-590-5p. MiR-590-5p was downregulated in RA-FLSs, and overexpression of linc02381 suppressed expression of miR-590-5p that post-transcriptionally suppressed the expression of mitogen-activated protein kinase kinase 3 (MAP2K3), and overexpression of miR-590-5p reversed the effect of linc02381 overexpression on MAP2K3 expression. MiR-590-5p inhibitor reversed the inhibition effect of linc02381 knockdown on proliferation and invasion of FLSs, which enhanced expression of MAP2K3, and activation of p38 and AP-1 in the MAPK signaling pathway. In summary, linc02381 was upregulated in RA synovial tissues and RA-FLSs, and it exacerbated RA by adsorbing miR-590-5p to activate the MAPK signaling pathway.
Keywords: linc02381, rheumatoid arthritis, miR-590-5p, RNA–RNA interaction, the MAPK signaling pathway
Introduction
Rheumatoid arthritis (RA) is a chronic systemic autoimmune disease, typically characterized by synovial hyperplasia and joint destruction, leading to irreversible joint damage. Pathophysiology suggests that RA is a clinical syndrome that spans multiple disease subtypes with typical symptoms of persistent synovial inflammation and associated damage to articular cartilage and underlying bone1,2. The major local cell populations in joints affected by RA are synovial and chondrocytes. It is reported that 50%–60% of the risk of RA can be attributed to genetic factors, and smoking is the main environmental risk3,4. RA causes progressive joint destruction, increased disability and mortality, and it is difficult to cure and requires lifelong treatment once it is fully developed. Therefore, it is necessary to diagnose RA accurately and early5.
Long noncoding RNAs (lncRNAs) are a functional RNA of more than 200 nucleotides in length. Numerous evidences suggested that lncRNAs are involved in the development and progression of tumors and a variety of diseases and played a variety of roles in regulating gene expression at the transcriptional, post-transcriptional, and epigenetic levels. It is reported that lncRNA MEG3 inhibits the proliferation of RA fibroblast-like synoviocytes (RA-FLSs) by acting on miR-141 and protein kinase B (AKT)/mammalian target of rapamycin6. In addition, downregulation of lncRNA DILC in the plasma was detected in RA-FLSs7, while lncRNA PVT1 enhances proliferation and invasion of the RA-FLSs8. Linc02381 is a novel lncRNA, which is located at 12q13.13 region. Recently, it has been reported that linc02381 (NR_026656.1) functioned as a tumor suppressor to inhibit the colorectal carcinoma cell proliferation and invasion9. Bioinformatics analysis reveals that linc02381 may be involved in activation of several inflammation pathways closely related to autoimmune diseases.
MiRNAs are a small class of noncoding RNA molecules, which are related to many human diseases. It was reported that miR-590-5p regulated tumorigenesis of colorectal cancer10, the growth of gastric cancer cells11, and lymph node metastasis12. A most recent study revealed that miR-590-5p affects the mineralization of and apoptosis of subchondral osteoblasts in osteoarthritis13, suggesting that miR-590-5p may play important roles in inflammation-related orthopedic diseases. However, miR-590-5p is rarely reported to regulate RA, so it is necessary to explore the role of miR-590-5p in RA. Furthermore, Clash-seq analysis showed that linc02381 could potentially bind with multiple miRNAs, such as miR-21-5p, miR-18-5p, miR-96-5p, miR-27a-3p, miR-590-5p/3p, etc9.
Mitogen-activated protein kinase (MAPK) is a kind of serine/threonine protein kinase in all eukaryotic cells. After phosphorylation of MAPK, upstream signals can be transmitted downstream14. MAPKs coordinately regulate cell proliferation, differentiation, motility, and survival15. At present, MAPK1, MAPK3, MAPK6, MAPK14, MAPK12, and MAPK10 are the main researches of MAPK-activated protein kinase16. MAP3 K, MAP2 K, and MAPK kinases are located at the highest and lowest positions, respectively, in the canonical three-layer MAPK signaling cascade17. There have been a few reports on MAPK signaling pathway in the development of RA18,19. For example, resveratrol reduces RA by inhibiting the MAPK signaling20. In addition, activation of MAPK signaling pathway by bispecific tyrosine regulated kinase 1A promotes proliferation, migration, and invasion of rheumatoid synovial fibroblasts21. Therefore, deactivating MAPK has been regarded as an effective way to inhibit RA progression.
In our study, we demonstrated that linc02381 was upregulated in RA tissues and RA-FLSs. In addition, we confirmed that linc02381 could enhance the proliferation and invasion of RA-FLSs. In all, our results indicated that linc02381 inhibits RA through adsorbing miR-590-5p and inactivation of MAP2K3/p38 signaling pathway.
The purpose of this study is to explore the role of linc02381 in RA progression. We found that overexpression of linc02381 promoted the proliferation and invasion of RA-FLSs. In addition, our results indicated that linc02381 promotes RA deterioration through adsorbing miR-590-5p and activating the MAP2K3/p38 signaling pathway, which suggested that linc02381 may be a potential therapy target for RA.
Materials and Methods
Patients and Samples Collection
In this study, a total of 24 male patients (46.8 ± 4.5 years) with RA diagnosed in Huaihe Hospital of Henan University (Kaifeng, China) from June 2017 to February 2019 were included. And basic information of the patients was shown in supplemental Table 1. According to the American Society of Rheumatology (revised classification standard in 1987), patients can be diagnosed with RA if they are excluded other arthritis and have more than or equal to four of the following indexes: (1) morning stiffness for at least 1 h (≥6 weeks); (2) involvement of three or more joints (≥6 weeks); (3) hand joints (wrist, metacarpophalangeal, or proximal interphalangeal joints) (≥6 weeks); (4) symmetric arthritis (≥6 weeks); (5) rheumatoid subcutaneous nodules; (6) X-ray changes; and (7) serum rheumatoid factor positive. RA synovial tissues were collected from the patients’ wrist during surgical treatment in Huaihe Hospital of Henan University, and normal human synovium tissues were donated by 12 male volunteers (32.6 ± 6.7 years), who underwent cut-off surgery due to severe trauma. Informed consent was obtained from all of the participants. This study was approved by the Ethical Review Board for Research in the Huaihe Hospital of Henan University. The collected tissue samples were placed in liquid nitrogen and stored at −80°C for later use.
Cell Culture and Treatment
Human Normal-FLSs (Cat.#408K-05a) were purchased from Cell Applications Inc. (San Diego, CA, USA), and human RA-FLSs (MH7A) (Cat.#C0878) were purchased from Shanghai Guandao Bioengineering Co. Ltd. (Shanghai, China). Normal-FLSs were cultured in minimal essential medium, and RA-FLSs were cultured in Dulbecco’s modified Eagle’s medium (DMEM). They were both supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA). Cells were cultured in a humidified incubator with 5% CO2 at 37°C.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
Total RNA of cells and tissues was extracted by using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. We used the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, USA) to reverse transcribe linc02381 into cDNA. Next, the commercial kit SYBR Premix Dimer Eraser kit (Takara, Dalian, China) was used to identify gene expression according to the manufacturer’s instructions with U6 as a control. Quantitative real-time polymerase chain reaction (RT-qPCR) was performed by fluorescence quantitative PCR (ABI 7900 HT, Thermo Fisher Scientific, Waltham, MA, USA). Twenty microliters of reaction system was used as follows: SYBR Premix Ex Taq II (2×) 10.0 μl, forward primer (10 μmol/l) 0.8 μl, reverse primer (10 μmol/l) 0.8 μl, cDNA template 2.0 μl, and dH2O (RNase free) 6.4 μl. PCR program was performed as follows: 95°C 1 min, 1 cycle, 95°C 20 s, 56°C 1 s, 72°C 15 s, 35 cycles. Specificity of the obtained PCR products was confirmed by melting curve. The relative gene expression of linc02381 was quantified by using the 2-ΔΔCt method. Primer sequences are as follows: linc02381 forward: 5′-CCC TGC CCA TAA GCT ACT CA-3′, reverse: 5′-AAC TTT GAC CCC CAA ATG CC-3′; miR-590-5p forward: 5′-TCC ATT GAA ACG CCT AGG AGA ATT TGC-3′, reverse: 5′-GCA AAT TCT CCT AGG CGT TTC AAT GGA-3′; U6 forward: 5′-CTC GCT TCG GCA GCA CA-3′, reverse: 5′-AAC GCT TCA CGA ATT TGC GT-3′; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward: 5′-TTG GTA TCG TGG AAG GAC TCA-3′, reverse: 5′-TGT CAT CAT ATT TGG CAG GT-3′.
Construction and Transfection of Overexpression Plasmid Vector and Small Interfering RNAs
Full-length linc02381 gene was cloned to pcDNA3.1 vector for overexpression plasmid vector (pcDNA3.1-linc02381), which constructed and purchased from Geneseed Biotech (Guangzhou, China). Linc02381 small interfering RNA (siRNA; 20 and 50 nM), MAP2K3 siRNA, and their negative control (NC siRNA) were synthesized and purchased from Geneseed Biotech. Cells were inoculated in a six-well plate at a density of 1.2 × 105/cell about 70% confluence. 20 and 50 nM linc02381 siRNA, MAP2K3 siRNA, and 1 μg/ml pcDNA3.1-linc02381 were transfected into RA-FLSs using Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions.
Dual Luciferase Reporter Gene Assay
We used StarBase ((http://starbase.sysu.edu.cn/index.php) to predict potential binding sites between linc02381 and miR-590-5p. To further identify potential relationships, we performed a luciferase reporter assay. First, we cloned the wild-type and mutant 3’-UTR sequences of linc02381 into the pGL3 promoter vector containing the luciferase reporter gene, respectively. RA-FLSs were seeded into 24-well plates and cotransfected with luciferase plasmid and wild-type linc02381 or mutant linc02381 using Lipofectamine 3000 (Invitrogen) when grown to approximately 70% confluence. Twenty-four hours after transfection, cells were harvested and cells were analyzed using a dual luciferase reporter kit (Promega, Madison, WI, USA) to normalize luciferase reporter activity to Renilla luciferase activity.
RNA Pull-Down Assay
To further verify the relationship between linc02381 and miR-590-5p, biotin-labeled miR-590-5p was synthesized and transfected into HEK293 T cells. Bio-miR-590-5p probe was transcribed and purified by GenePharma Company (Shanghai, China) via the AmpliScribe T7-Flash Biotin-RNA Transcription Kit (Epicenter, Madison, WI, USA). After 48 h, the cells were washed and lysed, then the extracts were incubated with the streptavidin coated magnetic beads at 4°C for 3 h. Then, the beads were washed twice with ice-cold buffer, three times with low salt buffer, and once with high salt buffer. Finally, the RNA–RNA complex was eluted, and the RNA pull-down product was applied in the following RT-qPCR.
Western Blotting
The cells were lysed with radioimmunoprecipitation assay buffer at 4°C, and then separated by high speed centrifugation (14,000 rpm) at 4°C for 10 min. Next, the supernatant was quantified using the BCA Protein Assay Kit (Shanghai Biotech Biotechnology Research Institute). The protein sample was then loaded onto 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis at 70 V for 30 min then 120 V for 90 min and transferred to a nitrocellulose membrane (Millipore, Boston, MA, USA) at 300 mA for 2 h. After blocking with 5% skim milk, the blot was probed with a primary antibody. After washing with phosphate buffered saline, the horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G secondary antibodies (ab150077, 1:1,000, Abcam, Cambridge, UK) were added. The primary antibodies used in this study include rabbit anti-MAP2K3 antibody (ab195037, 1:500, Abcam), rabbit anti-AP-1 antibody (ab21981, 1:200, Abcam), and rabbit anti-p38 antibody (ab31828, 1:300, Abcam). We then uniformly add the chemiluminescence reagent to the membrane and develop with the developer. The expression level of the protein was analyzed by chemiluminescence and quantified by Image J software (Version 1. 46; National Institutes of Health, Bethesda, MD, USA). We used GAPDH as an endogenous control to normalize protein expression.
Cell Viability Assay
The cells were seeded into 96-well plates (Corning, New York, NY, USA) and cultured in 5% CO2 at 37°C, and then examined for cell proliferation activity using the Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories, Kumamoto, Japan). After 24 or 72 h of incubation, 10 µl of CCK-8 was added to each well and then incubated for an additional hour at 37°C. The absorbance optical delnsity (OD) value was read at 450 nm using a microplate reader.
5-Ethynyl-2’- deoxyuridine (EdU) analysis is also used to detect cell proliferation. The logarithmic growth stage cells were cultured in 96-well plates with 4 × 103 cells per well. Diluting the EdU solution with a ratio of 1:1,000 in the cell culture medium to prepare a proper amount of 10 μM EdU culture medium. Then cell culture medium was replaced with EdU culture medium. Cells were incubated for 2 hours, and then cells were harvested, fixed with 4% formaldehyde and stained with Apollo. Finally, cells were analyed with fluorescence microscope (WMF-3580, Shanghai Yuguang Instrument Co., Ltd, Shanghai, China).
Transwell Invasion Assay
We used 8.0 µm chamber plates to determine cell invasiveness. The upper surface of the Transwell filter we used was coated with Matrigel (BD, Franklin Lake, New Jersey, USA). Firstly, cells were planted into the 8.0 µm chamber plates and then 300 µl of serum-free DMEM medium was added to the upper compartment of the chamber, and 500 µl of DMEM medium supplemented with 10% FBS was added to the lower chamber for 48 h incubations. Then, the noninvasive cells on the upper side of the chamber were suspended with a cotton swab, and then the invasive cells were fixed in 4% paraformaldehyde and stained with a crystal violet solution. We stained infiltrating cells using an Olympus IX70 inverted microscope (Olympus Corp., Tokyo, Japan) and randomly selected the best six fields of view, and each experiment was repeated three times.
Cell Apoptosis
Flow cytometry was used to detect apoptosis. After the cells were harvested, the cells were washed with phosphate buffered saline three times, then 5 µl annexin V-fluorescein isothiocyanate and 5 µl propidium iodide were used for 20 min in the dark at room temperature, and apoptosis was detected by flow cytometry (BD Biosciences, USA).
Enzyme-Linked Immunosorbent Assay
After transfection, the cells were harvested and centrifuged, and IL-12, IL-16, IL-1β, and tumor necrosis factor α (TNF-α) levels in the cell supernatant were determined by an enzyme-linked immunosorbent assay kits (Thermo Fisher Scientific) following the manufacturer’s instructions.
Statistical Analysis
Data are represented as the mean ± standard deviation (standard error of the mean) and processed with SPSS version 22.0 software (IBM Corp., Armonk, NY, USA). We used t-test or one-way analysis of variance to determine statistical significance. P-values <0.05 were considered to be statistically significant. All experiments were guaranteed to be repeated three times.
Results
Linc02381 was Upregulated in RA Synovial Tissues and RA-FLSs
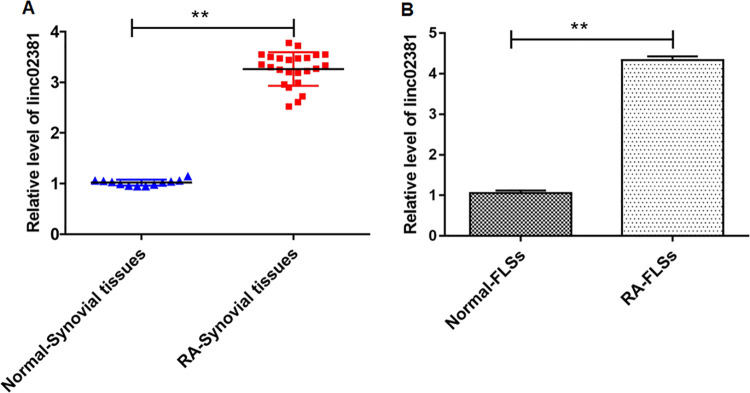
The relative level of linc02381 in RA synovial tissues and RA-FLSs was determined by RT-qPCR. As shown in Fig. 1A, B, the expression of linc02381 was upregulated in RA synovial tissues and RA-FLSs (P < 0.01). The result suggested that linc02381 might be used as a marker of RA diagnosis.
Figure 1.
Linc02381 was upregulated in RA synovial tissues and RA-FLSs. (A, B) The expression of linc02381 in RA synovial tissues and RA-FLSs was determined by RT-qPCR. **P < 0.01. Independent samples t-test was used to analyze the statistical significance. Three independent experiments were carried out, respectively. RA: rheumatoid arthritis; RA-FLSs: RA fibroblast-like synoviocytes; RT-qPCR: quantitative real-time polymerase chain reaction.
Overexpression of Linc02381 Enhanced FLS Proliferation and Invasion, and Knockdown of Linc02381 Inhibited Cell Proliferation and Invasion
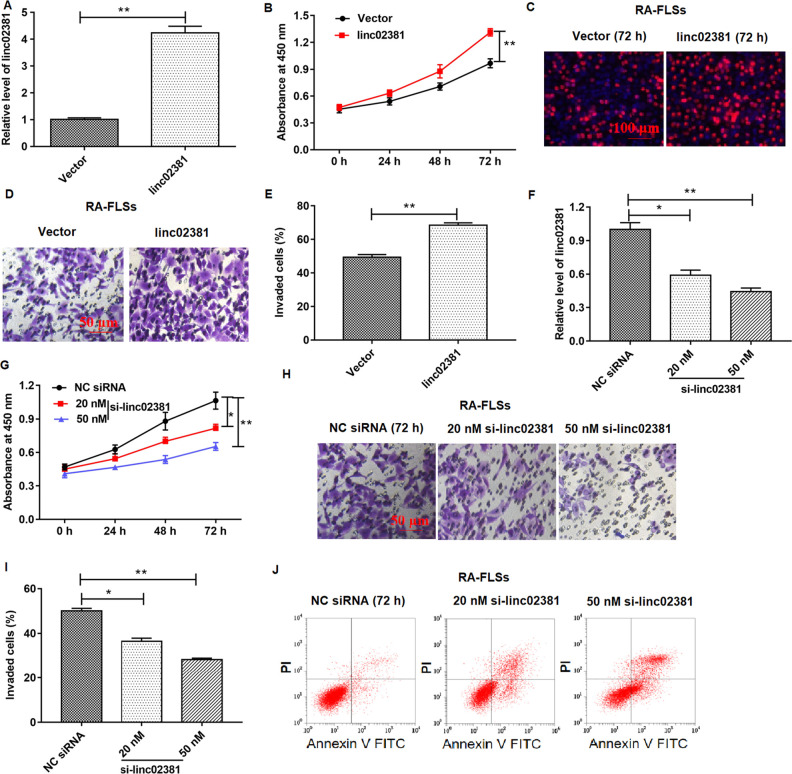
We studied the effects of linc02381 on proliferation and invasion of RA-related cells in vitr o. Blank and pcDNA3.1-linc02381 vectors were transfected into FLSs. As shown in Fig. 2A, linc02381 was successfully overexpressed in RA-FLSs (P < 0.01), and overexpression of linc02381 enhanced the cell proliferation and invasion in FLSs in 72 h, which was detected by CCK-8 assay, EdU assay, and Transwell assay (P < 0.01; Fig. 2B–E). In addition, we transfected FLSs with 50 or 20 nM linc02381 siRNA, respectively. The results showed that 20 nM linc02381 siRNA transfection suppressed cell proliferation and invasion, and 50 nM further suppressed cell proliferation and invasion, which were determined by CCK-8 assay and Transwell assay (Fig. 2F-I). In addition, flow cytometry assay revealed that 20 nM siRNA and 50 nM siRNA both enhanced FLS apoptosis (Fig. 2J). In all, linc02381 plays an important role in enhancing FLS proliferation and invasion.
Figure 2.
Effect of linc02381 overexpression and linc02381 knockdown on RA-FLS proliferation and invasion. Blank vector, pcDNA-linc02381 (linc02381, 1 μg/ml), NC siRNA, or linc02381 siRNA (20 or 50 nM) vector was transfected into FLSs. (A) The overexpression efficiency of pcDNA-linc02381 expression in FLSs was confirmed by RT-qPCR. (B–E) Cell proliferation was detected by CCK-8 assay or EdU assay, and cell invasion was detected by Transwell assay after FLSs were transfected with pcDNA3.1-linc02381. (F) The expression of linc02381 was examined by RT-qPCR in FLSs transfected with 20 or 50 nM linc02381 siRNA. (G–I) Cell proliferation and invasion of FLSs following transfection with 20 nM linc02381 siRNA or 50 nM linc02381 siRNA. (J) Cell apoptosis was detected by flow cytometry assay in FLSs transfected with 20 or 50 nM linc02381 siRNA. *P < 0.05 or **P < 0.01. Three independent experiments were carried out, respectively. CCK-8: Cell Counting Kit-8; NC: negative control; RA: rheumatoid arthritis; RA-FLSs: RA fibroblast-like synoviocytes; RT-qPCR: quantitative real-time polymerase chain reaction; siRNA: small interfering RNA.
Linc02381 Directly Interacted With miR-590-5p, and They Negatively Regulated Expression of Each Other
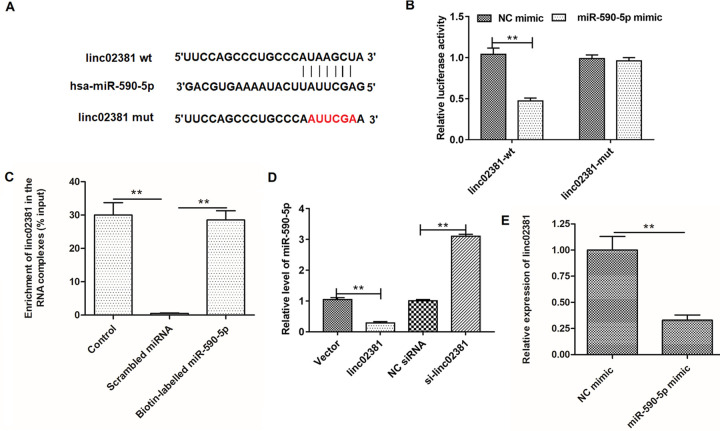
Using the online bioinformatics tool StarBase (http://starbase.sysu.edu.cn/index.php), we found that linc02381 potentially interacted with multiple miRNAs. Among them, miR-590-5p was a most downregulated miRNAs and associated with inflammation of articulation. As shown in Fig. 3A, linc02381 binds with miR-590-5p with its seed sequence in linc02381 sequence (chr12: 54526177-54526193[+]). We demonstrated the adsorption of miR-590-5p by wild-type linc02381 instead of mutant linc02381 by using a double-luciferase reporter gene assay (Fig. 3B). RNA pull-down assay also revealed that abundant linc02381 was detected in biotin-labeled miR-590-5p but not in the biotin-labeled scrambled miRNA (P < 0.01; Fig. 3C). We then transfected RA-FLSs with empty vector, pcDNA3.1-linc02381 expression vector, NC siRNA, and linc02381 siRNA and measured the relative level of miR-590-5p in cell lysates. As shown in Fig. 3D, overexpression or knockdown of linc02381 significantly inhibited or enhanced the relative level of miR-590-5p, while overexpression of miR-590-5p suppressed the relative level of linc02381 compared with NC mimic, which was detected by RT-qPCR (P < 0.01; Fig. 3E).
Figure 3.
Prediction and confirmation of interaction between linc02381 and miR-590-5p. (A) Details of the potential binding site of linc02381 and miR-590-5p in wild-type linc02381 and the mutant linc02381 sequences. (B) Verification of the relationship between linc02381 and miR-590-5p by the luciferase gene reporter assay. (C) Verification of the relationship between linc02381 and miR-590-5p by RNA pull-down assay based on the biotinylated miR-590-5p. (D) The effect of linc02381 overexpression and knockdown on the expression of miR-590-5p, respectively. (E) NC mimic or miR-590-5p mimic was transfected into RA-FLSs, and the expression of linc02381 was detected by RT-qPCR. **P < 0.01. Three independent experiments were carried out, respectively. NC: negative control; RA: rheumatoid arthritis; RA-FLSs: RA fibroblast-like synoviocytes; RT-qPCR: quantitative real-time polymerase chain reaction.
MiR-590-5p was Downregulated in RA-FLSs, and Its Expression was Negatively Regulated by linc02381
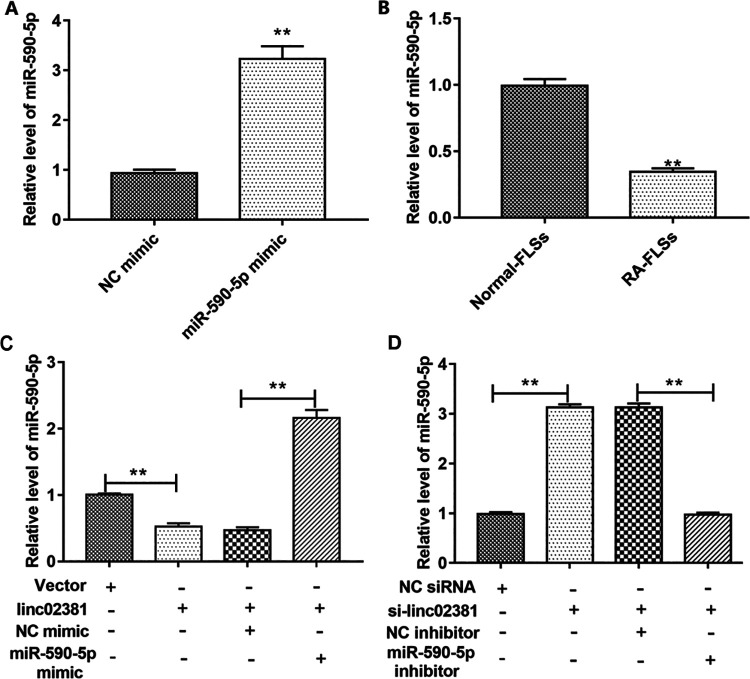
Empty vector or pcDNA3.1-linc02381 vector was transfected into FLSs individually or together with NC mimic or miR-590-5p mimic. NC siRNA or linc02381 siRNA was also transfected into FLSs alone, or together with NC inhibitor or miR-590-5p inhibitor. The overexpression efficiency of the miR-590-5p mimic was confirmed by qPCR (Fig. 4A). Since the negative regulation between miR-590-5p and linc02381, it’s not hard to speculate that miR-590-5p must displayed the opposite expression to linc02381. As expected, miR-590-5p was downregulated in RA-FLSs compared with normal FLSs (Fig. 4B). Then, our results showed that linc02381 overexpression inhibited the expression of miR-590-5p, and overexpression of miR-590-5p reversed the inhibition effect of linc02381 on miR-590-5p expression (Fig. 4C). Correspondingly, we also found that knockdown of linc02381 enhanced the expression of miR-590-5p and miR-590-5p inhibitor reversed the facilitation of linc02381 knockdown on miR-590-5p expression (Fig. 4D).
Figure 4.
Overexpression of miR-590-5p could reverse the effect of linc02381 overexpression in FLSs. (A) The expression of miR-590-5p was detected by RT-qPCR in RA-FLSs transfected with NC mimic or miR-590-5p mimic. (B) The expression of miR-590-5p in RA synovial tissues and RA-FLSs was determined by RT-qPCR. (C) Expression of miR-590-5p in FLSs was detected after transfected with pcDNA3.1-linc02381 alone or together with miR-590-5p mimic. (D) Expression of miR-590-5p in FLSs was detected after transfected with linc02381 siRNA alone or together with miR-590-5p inhibitor. **P < 0.01. Three independent experiments were carried out, respectively. NC: negative control; RA: rheumatoid arthritis; RA-FLSs: RA fibroblast-like synoviocytes; RT-qPCR: quantitative real-time polymerase chain reaction.
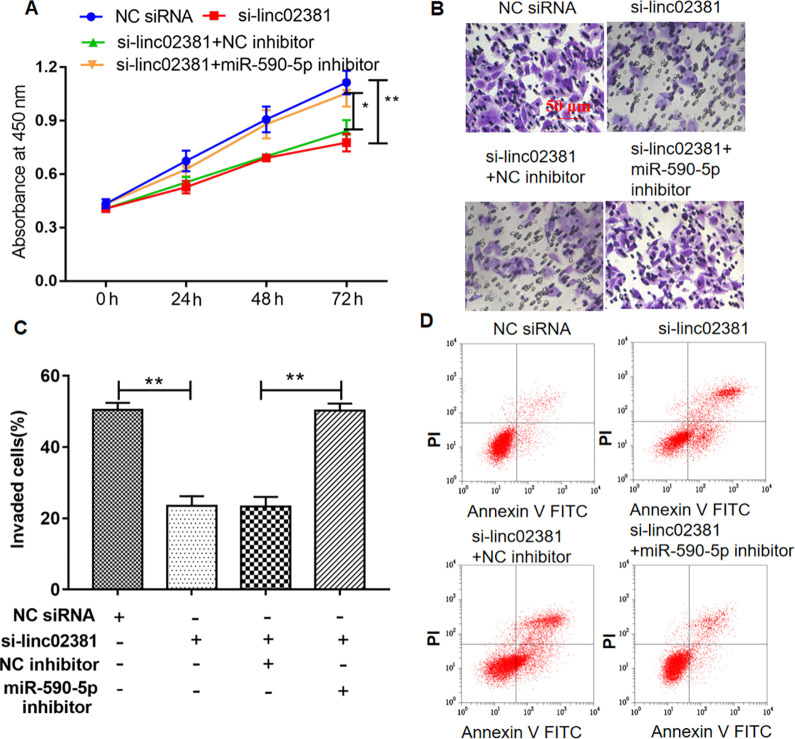
MiR-590-5p Inhibitor Reversed the Inhibition of linc02381 Knockdown on Proliferation and Invasion of FLSs
Cells were treated as Fig. 4D. As shown in Fig. 5A–C, miR-590-5p inhibitor rescued the inhibition of linc02381 knockdown on proliferation in 72 h and invasion in 24 h of FLSs, which were determined by CCK-8 assay and Transwell assay. However, the level of cell apoptosis was just opposite to proliferation (Fig. 5D).
Figure 5.
MiR-590-5p inhibitor reversed the effect of linc02381 overexpression on FLS proliferation and invasion. (A–C) FLS proliferation and invasion were investigated after transfected with linc02381 siRNA alone or together with miR-590-5p inhibitor, which were detected by CCK-8 assay and Transwell assay. (D) FLSs were treated as same as (A), and cell apoptosis was detected by flow cytometry assay. *P < 0.05. Three independent experiments were carried out, respectively. CCK-8: Cell Counting Kit-8; FLSs: fibroblast-like synoviocytes; siRNA: small interfering RNA.
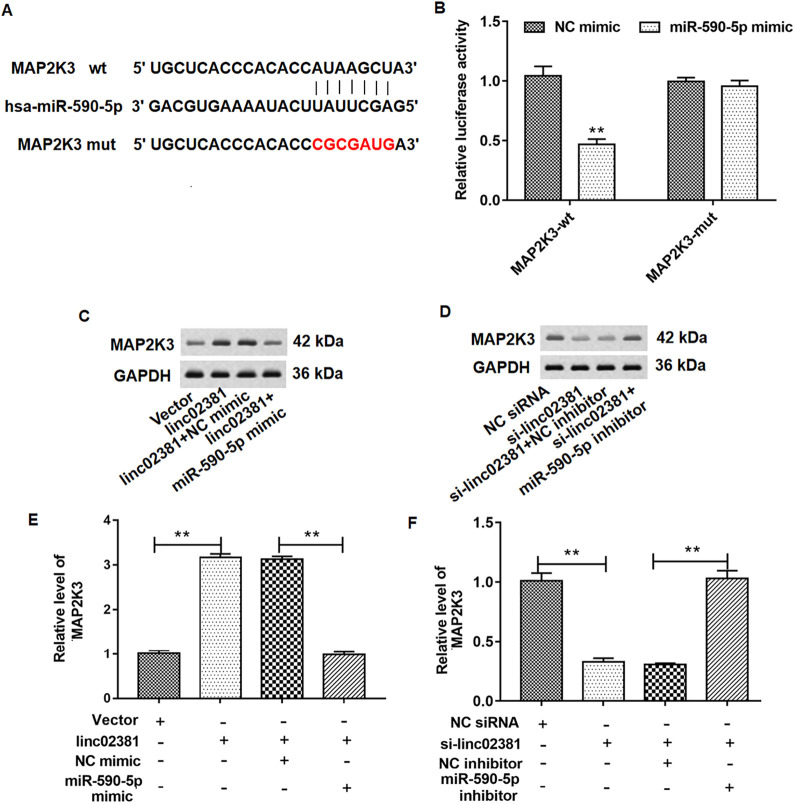
MiR-590-5p Bound to the 3’-UTR of Wild-type MAP2K3, and Overexpression of Linc02381 Promoted the Expression of MAP2K3 Protein
Cell were treated as Fig. 4C and D. Firstly, we used StarBase (http://starbase.sysu.edu.cn/index.php) to predict the binding the relationship between MAP2K3 gene and miR-590-5p. Next, the results of Double-luciferase reporter gene assay vertified that miR-590-5p bound to the 3'UTR of wild-type MAP2K3 (Fig. 6A and Fig. 6B). We found that overexpression of linc02381 enhanced MAP2K3 protein level, which was detected by Western blotting (Fig. 6C and Fig. 6E). Overexpression of miR-590-5p reversed the facilitation of linc02381 overexpression on MAP2K3 protein level (Fig. 6C and Fig. 6E). On the contrary, We found that knockdown of linc02381 inhibited MAP2K3 protein level, which was also determined by Western blotting (Fig. 6D and Fig. 6F). MiR-590-5p inhibitor reversed the inhibition of linc02381 knockdown on expression of MAP2K3 protein (Fig. 6D and Fig. 6F).
Figure 6.
MiR-590-5p targets MAP2K3, and overexpression of miR-590-5p rescued the effect of linc02381 overexpression on MAP2K3 expression. (A) Binding details of MAP2K3 and miR-590-5p. (B) Relationship between miR-590-5p and MAP2K3 was verified by dual luciferase gene reporter assay. (C, E) MAP2K3 protein levels were determined in FLSs by Western blotting after transfection with pcDNA3.1-linc02381 alone or together with miR-590-5p mimic. (D, F) MAP2K3 protein levels were determined in FLSs by Western blotting after transfected with linc02381 siRNA alone or together with miR-590-5p inhibitor. **P < 0.01. Three independent experiments were carried out, respectively. FLSs: fibroblast-like synoviocytes; MAP2K3: mitogen-activated protein kinase kinase 3; siRNA: small interfering RNA.
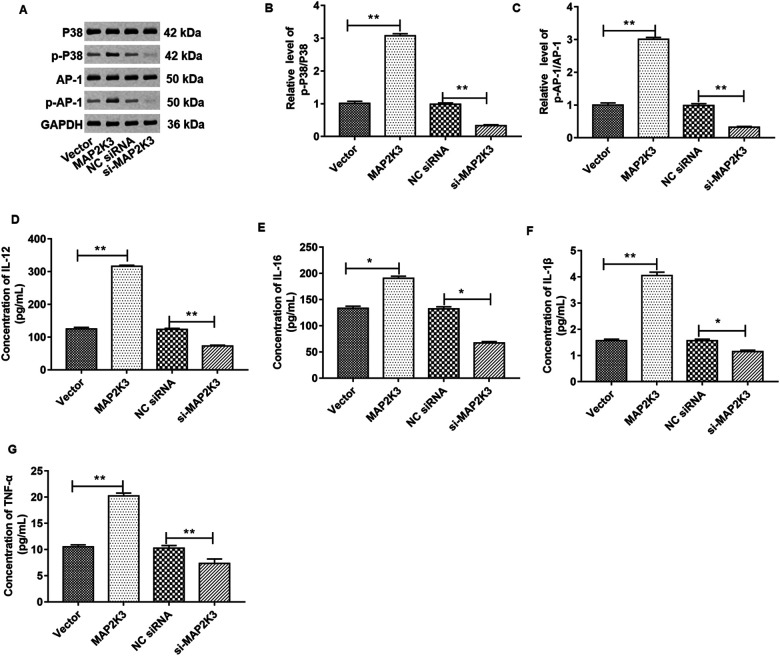
MAP2K3 Positively Regulated the MAPK Signaling Pathway and MAPK-Induced Production of Proinflammatory Cytokines
We transfected the empty vector, pcDNA-MAP2K3 expression vector, NC siRNA, and MAP2K3 siRNA into FLSs and studied their effects on the MAPK signaling pathway. As shown in Fig. 7A, Western blotting was used to detect the relative level of total p38, phosphorylated p38, total AP-1, and phosphorylated AP-1 proteins. Fig. 7B–D are the quantification of Fig. 7A. The results manifested that overexpression or inhibition of MAP2K3 enhances or inhibits phosphorylation levels of p38 and AP-1. As expected, overexpression of MAP2K3 increased the secretion of IL-12, IL-16, IL-1β, and TNF-α in the supernatant (Fig. 7D–G).
Figure 7.
Overexpression of MAP2K3 activated the MAPK signaling pathway, and knockdown of MAP2K3 inactivated the MAPK signaling pathway. And overexpression of MAP2K3 induced secretion of proinflammatory cytokines. (A-C) The expression of total p38, phosphorylated p38, AP-1, and phosphorylated AP-1 proteins in FLSs was determined by Western blotting after transfection with pcDNA3.1-MAP2K3 or MAP2K3 siRNA. (D-G) ELISA was used to detect the levels of cytokines IL-12, IL-16, IL-1β, and TNF-α in the supernatant of the treated FLSs. *P < 0.01. Three independent experiments were carried out, respectively. ELISA: enzyme-linked immunosorbent assay; FLSs: fibroblast-like synoviocytes; MAP2K3: mitogen-activated protein kinase kinase 3; siRNA: small interfering RNA; TNF-α: tumor necrosis factor α.
Discussion
In our study, we confirmed that linc02381 was upregulated in RA tissues and RA cell lines, which enhanced cell proliferation and invasion in FLSs. We predicted and proved that wild-type linc02381 binds with and negatively regulates miR-590-5p. In addition, miR-590-5p could reverse the promotion of linc02381 proliferation and invasion of FLSs. Finally, we also demonstrated that linc02381 promotes the proliferation and invasion of FLS by promoting MAPK signaling via upregulation of MAP2K3.
MiRNAs are a small class of noncoding RNA molecules that are involved in many human diseases. There are many studies on miR-590-5p, and it was found that miR-590-5p inhibits tumorigenesis of colorectal cancer by targeting YAP1 and breast cancer stem cells and metastasis by targeting SOX2. MiR-590-5p also regulates the growth of gastric cancer cells via the AKT/ERK pathway11. In vulvar squamous cell carcinoma studies, upregulation of miR-590-5p promotes lymph node metastasis. Studies have found that upregulation of miRNA-590 in RA promotes osteoblast apoptosis via transforming growth factor-β1/phosphoinositol 3-kinase/Akt signaling, and the study found that miRNA-590-5p was downregulated in RA, which is similar to our results in RA-FLSs. Studies have found that upregulation of miRNA-590 promotes osteoblast apoptosis via transforming growth factor-β1/phosphoinositol 3-kinase/Akt signaling22, while it is largely unknown that the role of miRNA-590 in RA. Our study found that miRNA-590-5p was downregulated in RA-FLSs. Therefore, further study can explore the effect of miRNA-590-5p on the activity and behavior of RA-FLSs.
MAP2K3 is considered to be a mitogen-activated protein kinase expressed in all eukaryotic cells and it is also a dual-specific kinase that is activated in vivo by cytokines and environmental stress, catalyzing the concomitant phosphorylation of threonine and tyrosine residues in MAP kinase p38. MAP2K3 has been found in human breast epithelial cells to promote cell senescence and has been identified as MAP2K3, a protein that promotes senescence in these cells23. There is evidence to confirm that the gene MAP2K3 is knocked down in human and mouse hearts, saving the fibrotic/hypertrophic phenotype that activates transcription factor 3 knockout cells24. The study found that targeted inhibition of MAP2K3 blocking MAPK signaling pathway can prevent the prevention of blood-brain barrier disruption and is beneficial for the treatment of cerebral ischemia25. However, MAP2K3 has been less studied in RA. This study found that inhibition of MAP2K3 inactivates MAPK pathway, thereby inhibiting the occurrence and progression of RA26. Certainly, the MKK3/p38 axis has been increasingly studied in cancer. Studies have shown that the MKK3/p38 axis regulates cell proliferation, apoptosis, and cell cycle progression in hepatoma cells27. Furthermore, we have demonstrated that linc02381 was upregulated in RA tissues and RA cell line, and linc02381 activated MAPK signal pathway by absorbing miR-590-5p to promote RA-FLS proliferation and invasion.
P38 pathway regulates the decomposition of inflammatory response by increasing the expression of anti-inflammatory cytokines, thus promoting wound healing and homeostasis. It has been confirmed that MAP2K3 (or MKK3) and MAP2K6 (or MKK6) are the main regulators of p38 phosphorylation and activation, both of which can form stable signal complexes with p38 and phosphorylate the downstream substrate28. However, it has also been reported that MKK3 and MKK6 activate p38 pathway, but they regulate different proinflammatory cytokine subsets. MKK6 mainly promotes the colocalization of p38 and MAPKAPK2 (MK2), rather than phosphorylation of p3829. It has been reported that the activation of p38 MAPKs by MKK3 or MKK6 has the opposite effect on the activity of MMP-1 promoter. The activation of p38 by MKK6 may enhance the activity of MMP-1 promoter and the activation of ERK1,2 pathway. Activation of the downstream signal cascade of MKK3/MKK3b seems to inhibit ERK1,2-mediated MMP-1 gene transcription induction30. Therefore, the mechanism of MKK6 activating p38 pathway remains to be explored. The lack of MKK6 and MKK3 reduced the severity of arthritis, while the arthritis MKK-6–/– mice had the least joint damage, indicating that MKK6 was the key regulatory factor of collagen arthritis joint damage31. P38 MAPK pathway plays an important role in regulating the expression of cytokines and metalloproteinases in RA and other diseases, so it has become a potential pathway29.
In summary, linc02381 was upregulated in RA synovial tissues and RA-FLSs, which was involved in the regulation of proliferation and invasion of FLSs. In addition, our results demonstrated that linc02381 could adsorb miR-590-5p and upregulate MAP2K3 protein to activate the MAPK signaling pathway and promote FLS cell proliferation and invasion.
Supplemental Material
Supplementary_table_1 for Linc02381 Exacerbates Rheumatoid Arthritis Through Adsorbing miR-590-5p and Activating the Mitogen-Activated Protein Kinase Signaling Pathway in Rheumatoid arthritis-fibroblast-like synoviocytes by Jing Wang and Qing Zhao in Cell Transplantation
Footnotes
Ethical Approval: This study was approved by the Ethics Committee at Huaihe Hospital of Henan University (Kaifeng, China).
Statement of Human and Animal Rights: All procedures in this study were conducted in accordance with Huaihe Hospital of Henan University of Ethics Committee’s or Institutional Review Board’s (Approval Number: 00009) approved protocols.
Statement of Informed Consent: Written informed consent was obtained from the patients for their anonymized information to be published in this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Qing Zhao  https://orcid.org/0000-0003-4248-0754
https://orcid.org/0000-0003-4248-0754
Supplemental Material: Supplemental material for this article is available online.
References
- 1. McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365(23):2205–2219. [DOI] [PubMed] [Google Scholar]
- 2. Lee DM, Weinblatt ME. Rheumatoid arthritis. Lancet. 2001;358(9285):903–911. [DOI] [PubMed] [Google Scholar]
- 3. Scott DL, Wolfe F, Huizinga TWJ. Rheumatoid arthritis. Lancet. 2010;376(9746):1094–1108. [DOI] [PubMed] [Google Scholar]
- 4. Neumann E, Frommer K, Diller M, Müller-Ladner U. Rheumatoide arthritis [in German]. Z Rheumatol. 2018;77(9):769–775. [DOI] [PubMed] [Google Scholar]
- 5. Finckh A, Alpizar-Rodriguez D, Roux-Lombard P. Value of biomarkers in the prevention of rheumatoid arthritis. Clin Pharmacol Ther. 2017;102(4):585–587. [DOI] [PubMed] [Google Scholar]
- 6. Li G, Liu Y, Meng F, Xia Z, Wu X, Fang Y, Zhang C, Zhang Y, Liu D. LncRNA MEG3 inhibits rheumatoid arthritis through miR-141 and inactivation of AKT/mTOR signalling pathway. J Cell Mol Med. 2019;23(10):7116–7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang G, Tang L, Zhang X, Li Y. LncRNA DILC participates in rheumatoid arthritis by inducing apoptosis of fibroblast-like synoviocytes and down-regulating IL-6. Biosci Rep. 2019;39(5):BSR20182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang T, Liu HW, Chen JQ, Wang SH, Hao LQ, Liu M, Wang B. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed Pharmacother. 2017;88:302–308. [DOI] [PubMed] [Google Scholar]
- 9. Jafarzadeh M, Soltani BM, Soleimani M, Hosseinkhani S. Epigenetically silenced LINC02381 functions as a tumor suppressor by regulating PI3K-Akt signaling pathway. Biochimie. 2020;171–172:63–71. [DOI] [PubMed] [Google Scholar]
- 10. Ou C, Sun Z, Li X, Li X, Ren W, Qin Z, Zhang X, Yuan W, Wang J, Yu W, Zhang S, et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53–63. [DOI] [PubMed] [Google Scholar]
- 11. Shen B, Yu S, Zhang Y, Yuan Y, Li X, Zhong J, Feng J. miR-590-5p regulates gastric cancer cell growth and chemosensitivity through RECK and the AKT/ERK pathway. Onco Targets Ther. 2016;9:6009–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang X, Wu X. miRNA expression profile of vulvar squamous cell carcinoma and identification of the oncogenic role of miR-590-5p. Oncol Rep. 2016;35(1):398–408. [DOI] [PubMed] [Google Scholar]
- 13. Prasadam I, Batra J, Perry S, Gu W, Crawford R, Xiao Y. Systematic identification, characterization and target gene analysis of microRNAs involved in osteoarthritis subchondral bone pathogenesis. Calcif Tissue Int. 2016;99(1):43–55. [DOI] [PubMed] [Google Scholar]
- 14. Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6(5):322–327. [DOI] [PubMed] [Google Scholar]
- 15. Jiang Y, Yin X, Wu L, Qin Q, Xu J. MAPK/P53-mediated FASN expression in bone tumors. Oncol Lett. 2017;13(6):4035–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010;1802(4):396–405.20079433 [Google Scholar]
- 17. Pan W, Wei N, Xu W, Wang G, Gong F, Li N. MicroRNA-124 alleviates the lung injury in mice with septic shock through inhibiting the activation of the MAPK signaling pathway by downregulating MAPK14. Int Immunopharmacol. 2019;76:105835. [DOI] [PubMed] [Google Scholar]
- 18. Zhang L, Luo J, Wen H, Zhang T, Zuo X, Li X. MDM2 promotes rheumatoid arthritis via activation of MAPK and NF-kappaB. Int Immunopharmacol. 2016;30:69–73. [DOI] [PubMed] [Google Scholar]
- 19. Zhai KF, Duan H, Cui CY, Cao YY, Si JL, Yang HJ, Wang YC, Cao WG, Gao GZ, Wei ZJ. Liquiritin from glycyrrhiza uralensis attenuating rheumatoid arthritis via reducing inflammation, suppressing angiogenesis, and inhibiting MAPK signaling pathway. J Agric Food Chem. 2019;67(10):2856–2864. [DOI] [PubMed] [Google Scholar]
- 20. Yang G, Chang CC, Yang Y, Yuan L, Xu L, Ho CT, Li S. Resveratrol alleviates rheumatoid arthritis via reducing ROS and inflammation, inhibiting MAPK signaling pathways, and suppressing angiogenesis. J Agric Food Chem. 2018;66(49):12953–12960. [DOI] [PubMed] [Google Scholar]
- 21. Guo X, Zhang D, Zhang X, Jiang J, Xue P, Wu C, Zhang J, Jin G, Huang Z, Yang J, Zhu X, et al. Dyrk1A promotes the proliferation, migration and invasion of fibroblast-like synoviocytes in rheumatoid arthritis via down-regulating Spry2 and activating the ERK MAPK pathway. Tissue Cell. 2018;55:63–70. [DOI] [PubMed] [Google Scholar]
- 22. Yang J, Zhuang Y, Liu J. Upregulation of microRNA-590 in rheumatoid arthritis promotes apoptosis of bone cells through transforming growth factor-β1/phosphoinositide 3-kinase/Akt signaling. Int J Mol Med. 2019;43(5):2212–2220. [DOI] [PubMed] [Google Scholar]
- 23. Jia M, Souchelnytskyi N, Hellman U, O’Hare M, Jat PS, Souchelnytskyi S. Proteome profiling of immortalization-to-senescence transition of human breast epithelial cells identified MAP2K3 as a senescence-promoting protein which is downregulated in human breast cancer. Proteomics Clin Appl. 2010;4(10-11):816–828. [DOI] [PubMed] [Google Scholar]
- 24. Li Y, Li Z, Zhang C, Li P, Wu Y, Wang C, Bond Lau W, Ma XL, Du J. Cardiac fibroblast-specific activating transcription factor 3 protects against heart failure by suppressing MAP2K3-p38 signaling. Circulation. 2017;135(21):2041–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He F, Xiao Z, Yao H, Li S, Feng M, Wang W, Liu Z, Liu Z, Wu J. The protective role of microRNA-21 against coxsackievirus B3 infection through targeting the MAP2K3/P38 MAPK signaling pathway. J Transl Med. 2019;17(1):335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie X, Song J, Li G. MiR-21a-5p suppresses bisphenol A-induced pre-adipocyte differentiation by targeting map2k3 through MKK3/p38/MAPK. Biochem Biophys Res Commun. 2016;473(1):140–146. [DOI] [PubMed] [Google Scholar]
- 27. Luo S, Ren B, Zou G, Liu J, Chen W, Huang Y, Chen X, Fu Y. SPAG9/MKK3/p38 axis is a novel therapeutic target for liver cancer. Oncol Rep. 2019;41(4):2329–2336. [DOI] [PubMed] [Google Scholar]
- 28. Chabaud-Riou M, Firestein GS. Expression and activation of mitogen-activated protein kinase kinases-3 and -6 in rheumatoid arthritis. Am J Pathol. 2004;164(1):177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoshizawa T, Hammaker D, Boyle DL, Corr M, Flavell R, Davis R, Schett G, Firestein GS. Role of MAPK kinase 6 in arthritis: distinct mechanism of action in inflammation and cytokine expression. J Immunol. 2009;183(2):1360–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Westermarck J, Li SP, Kallunki T, Han J, Kahari VM. p38 mitogen-activated protein kinase-dependent activation of protein phosphatases 1 and 2A inhibits MEK1 and MEK2 activity and collagenase 1 (MMP-1) gene expression. Mol Cell Biol. 2001;21(7):2373–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hammaker D, Topolewski K, Edgar M, Yoshizawa T, Fukushima A, Boyle DL, Burak EC, Sah RL, Firestein GS. Decreased collagen-induced arthritis severity and adaptive immunity in MKK-6-deficient mice. Arthritis Rheum. 2012;64(3):678–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_table_1 for Linc02381 Exacerbates Rheumatoid Arthritis Through Adsorbing miR-590-5p and Activating the Mitogen-Activated Protein Kinase Signaling Pathway in Rheumatoid arthritis-fibroblast-like synoviocytes by Jing Wang and Qing Zhao in Cell Transplantation