Abstract
The application of nanotechnology for antimicrobial delivery has capacity to improve antibacterial efficacy. Currently, the usage of various inorganic and organic carriers, such as metal ions, nano-silicon and surfactants, might increase the potential toxicity of nanoparticles and make their clinical transformation more difficult. Herein, a nano-delivery system was constructed by direct self-assembly of antibacterial phytochemicals (berberine and rhein) originated from traditional Chinese medicine Coptis chinensis Franch. and Rheum palmatum L., respectively. Combining X-ray single crystal diffraction, nuclear magnetic resonance and other spectra characterizations, the stacked structure of nanoparticles was profoundly demonstrated. Briefly, rhein acted as the layered backbone and berberine embedded in it. In vitro bacteriostasis experiment showed the minimum bactericidal concentration of nanoparticles was 0.1 μmol/mL, which was lower than that of berberine and rhein. The results of confocal laser scanning microscope, biofilm quantitive assay and scanning electron microscopy indicated that nanoparticles had strong inhibitory effects on Staphylococcus aureus biofilm. More importantly, transmission electron microscopy and mass spectra indicated the further bacteriostatic mechanism of nanoparticles. Meanwhile, the nanoparticles had well biocompatibility and safety. Current study will open up new prospect that the design of self-assemblies between active phytochemicals can be originated from traditional Chinese medicine combination.
Keywords: Natural phytochemicals, Self-assembly, Antibacterial, Synergistic effect, Staphylococcus aureus, Traditional Chinese medicine
Graphical abstract
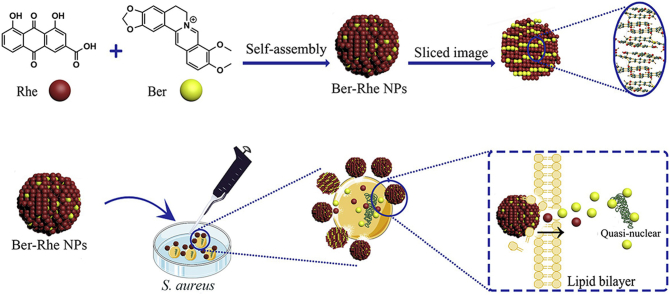
Ber-Rhe nanoparticles (NPs) were formed by direct self-assembly of Ber and Rhe. Combining X-ray single crystal diffraction and spectral analyses, the self-assembly mechanism of NPs had been demonstrated. Meanwhile, Ber–Rhe NPs demonstrated the inhibitory effect against Staphylococcus aureus by destroying the cell wall & membrane and cleaning the biofilm.
1. Introduction
Bacterial infection is a major factor that restricts the quality of human life, which leads to various diseases and even death1. According to the World Health Organization, bacterial infection was likely to bright about an increasing proportion of diarrhea-associated death, and was responsible for killing approximately half a million children every year. In recent years, in the field of nanotechnology, more and more nano dosage forms on antibacterial drugs have been explored. Nano-drug carriers have many advantages, such as small size, large specific surface area, controlled release of matrix, targeted drug delivery, etc. Metal ion nanoparticles2, antibacterial hydrogel3, 4, 5, and cationic polymers6, 7, 8, have been widely explored for their applications particularly in the field of bacteriostasis. Among above nanocarriers, one of their common characteristics is the antibiotics prepared into/onto nano dosage forms to achieve drug delivery by pharmacy means.
Although many nano-drug delivery systems were reported9, 10, 11, only a few had been transformed into clinical applications, like the amphotericin B12. Inevitably, some inorganic adjuvants could not be eliminated by human metabolism, such as silica, metal ions or skeletons, and magnetic nanoparticles13,14. At the same time, nanoemulsion or nanosphere prepared by polymer materials might accumulate in the viscera and produce toxicity15,16, such as anaphylases, teratogenic or carcinogenic reactions17,18. These characteristics were described as poor biocompatibility. Therefore, exploring antibiotics with simple synthesis, carrier-free and excellent biocompatibility would be of great significance in the clinical antibacterial treatment.
In recent years, more and more researchers have been focusing on the active ingredients in traditional herbal medicine19,20. Medicinal plants recorded by ancient Greek, Roman medical books and traditional Chinese medicine (TCM) have broad prospects for research at this stage. In particular, the combination spirit of TCM has been widely accepted in clinic. Many studies have shown that plant-derived natural small molecules had prominent performance in bacterial inhibition, such as berberine21 (Ber), rhein22 (Rhe), matrine23, curcumin24, gallic acid25, glycyrrhizic acid26, etc. Our previous research indicated that Ber could be self-assembled into nanoparticles and nanofibers with baicalin and wogonoside respectively, which showed different antibacterial effects27. Notably, berberine hydrochloride tablet prepared by Ber was famous in clinic to treat bacterial diarrhea28,29. Besides, because of the unique anthracene skeleton, Rhe was prone to form a stacked structure in solution due to hydrogen bonding, π−π interaction and hydrophobic effect according to the recent publications30,31. Inspired by the combination of TCM, we successfully gained self-assembled nanoparticles (Ber-Rhe NPs) of Rhe and Ber, which were originated from the classic herb–drug combination of Rheum palmatum L. and Coptis chinensis Franch28,32. A series of characterization methods, especially nuclear magnetic resonance (NMR) and X-ray diffraction, demonstrated the self-assembly mechanism of the Ber-Rhe NPs. Briefly, Rhe and Ber formed the basic units by π−π interaction and electrostatic interaction. Subsequently, the hydrogen bonds between Rhe molecules derived the basic units to further construct a layered skeleton material in the aqueous phase. In a word, nanoparticles with Rhe as the basic framework and Ber embedded in the layer gap were formed. Meanwhile, the antimicrobial activity of Ber-Rhe NPs against Staphylococcus aureus was improved. Furtherly, a series of assays were carried to evaluate the NPs’ antibacterial mechanism. Confocal laser scanning microscope (CLSM), 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) test and scanning electron microscopy (SEM) demonstrated that Ber-Rhe NPs had strong inhibitory effects on S. aureus biofilm. SEM, transmission electron microscopy (TEM), drug release test and aggregation quantity of Ber around S. aureus further indicated the deep bacteriostatic mechanism of nanoparticles. Schematic representation of the self-assembly and antibacterial mechanism of the Ber-Rhe NPs were shown in Fig. 1A. Driven by weak bonds, self-assembly of the effective constitutions would exhibit greater advantages than nanocarriers, including the enhancement of drug-loading efficiency, ability to sidestep the metabolic difficulties and toxicities associated with nanocarriers33, 34, 35. At the same time, natural small molecules did not require modification, harsh reaction conditions or complicated synthesis process, made it more economical and conducive to the conversion into clinical drugs. Furthermore, it was reported that self-assembly of small natural phytochemicals was still a severe challenge31. Because this relied heavily on molecular configurations, intermolecular forces and so on, the development largely depended on contingency. Current study will provide a promising mode to design and prepare self-assembling nanostructures without nano adjuvants from the enlightenment of TCM combination.
Figure 1.
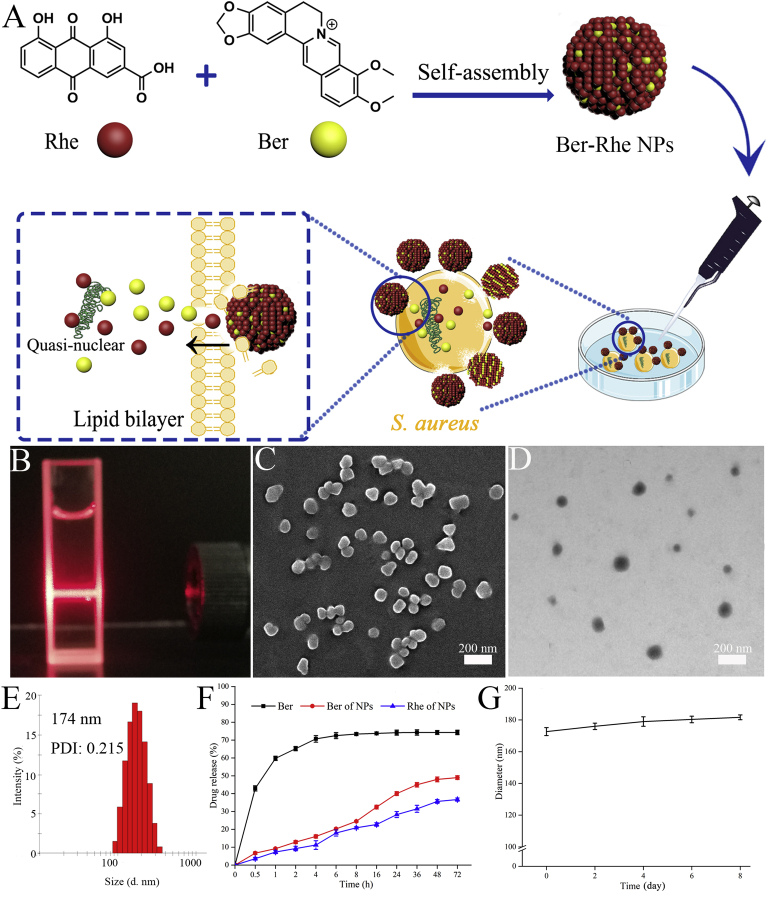
(A) Schematic representation of the self-assembly and antibacterial mechanism of the Ber-Rhe NPs. (B) Tyndall effect of nanoparticles. (C) SEM image of nanoparticles. (D) TEM image of nanoparticles. (E) The hydrodynamic diameters and distributions of nanoparticles. (F) The release of Ber and Rhe from Ber-Rhe NPs at PBS (pH = 7.4). (G) Stability of nanoparticles measured by DLS.
2. Materials and methods
2.1. Preparation of Ber-Rhe NPs
Ber-Rhe NPs were prepared by a one-step self-assembly process. Briefly, 10 mmol/L Rhe dimethyl sulfoxide (DMSO) solution and 1 mmol/L Ber methanol solution were mixed in a round-bottomed flask (molar ratio, 1:1). Then, 5 g/L sodium hydroxide solution was added in above miscible liquids under vigorous stirring for 10 min. Subsequently, the mixture of Ber and Rhe was gradually added to PBS at 60 °C with stirring. After a bath sonication for 1 h, the solution in dialysis bag (MWCO = 2.5 kDa) was dialyzed against ultrapure water for 12 h. Finally, the self-assembled Ber-Rhe NPs were obtained.
2.2. Crystallography
Recrystallization was used in this section36. 20 mg Ber-Rhe NPs was fully dissolved in 5 mL methanol solution. The beaker was not sealed, placed it in the crystal incubator and set the temperature to 25 °C. After stewing for 3 days, the sheet crystal was placed on the copper target and analyzed by single crystal X-ray diffractometer (Bruker, Karlsruhe, Germany). The diffraction data were collected and analyzed using a series of softwares, including Smart, Shelxtl, Platon, Diamond, etc. Cambridge Crystallographic Data Centre (CCDC, Cambridge, UK) deposition numbers 1842747 contain crystallographic data for this article. These data could be acquired free of charge by http://www.ccdc.cam.ac.uk/conts/retrieving.html or from the CCDC, 12 Union Road, Cambridge CB21EZ, UK.
2.3. Drug released test
For studying the release behavior of Ber-Rhe NPs, PBS solution of Ber-Rhe NPs (1 μmol/mL, 2 mL) in dialysis bag (MW = 3500) was immersed into PBS (20 mL) at 37 °C. At desired time points (0.5, 1, 2, 4, 6, 8, 16, 24, 36, 48 and 72 h), dialysis solution (0.5 mL) was collected and analyzed with high performance liquid chromatography (HPLC). The chromatographic parameters were similar to those of the bacterial uptake test (Supporting Information Section 2) except that the gradient solution was modified as follows: 0–10 min, 29% B; 11–17 min, 65% B; 18–20 min, 65%–80% B. Ber and Rhe solution, which were equal to Ber-Rhe NPs, were the control group, each experiment was repeated three times.
2.4. Determination of minimum inhibitory concentration (MIC) and minimal bactericidal concentration (MBC)
The culture method of S. aureus was recorded in Supporting Information Section 3. The MIC and MBC of Ber-Rhe NPs were evaluated by microporous dilution method37. Briefly, 100 μL samples (Ber, Rhe and Ber-Rhe NPs) were injected to 96-well microtiter plates using broth micro dilution methods, then 100 μL bacterial suspension (2 × 106 CFU/mL) was added to each well with an ultimate volume of 200 μL. And the gradient concentrations of Ber, Rhe and Ber-Rhe NPs were 0.2, 0.1, 0.05, 0.025 and 0.0125 μmol/mL, respectively. After fully mixing, the 96-well microtiter plates were placed in an incubator at 37 °C for 16 h. The bacterial reproduction was obtained through measuring the optical density at 600 nm (OD600) via microplate reader. After that, the medium of the above 96-well plate was applied to the nutrient agar mediums with the inoculation ring. After being cultured for 18 h in a 37 °C incubator, the nutrient agar mediums were observed. This experiment was conducted in triplicate. As proof of concept, MIC is the minimum concentration for samples with transparent wells. And, MBC is the minimum concentration that maintained less than 5 colonies on the nutrient agar mediums.
2.5. Determination of biofilm clearance effect, CLSM and SEM observation
S. aureus biofilms were prepared as described in Supporting Information Section 4. The quantification of anti-biofilm ability of Ber, Rhe and Ber-Rhe NPs was carried out in 96-well plates by XTT method. After incubation with samples at 37 °C for 24 h, biofilms were swilled by PBS for 3 times. Menadione was dissolved in acetone to obtain an ultimate concentration of 1.72 mg/mL. XTT was dissolved in PBS and the final concentration was 0.5 mg/mL. Before the determination of S. aureus biofilm formation, XTT solution was mixed with menadione at a ratio of 1000:1 and 200 μL of it was added to the micropores. After incubation with XTT-menadione solution at 37 °C in the dark for 2 h, the absorbance of the biofilms at 490 nm was recorded with microplate reader. With normal saline as the control group, each sample was repeated 3 times in micropores and the mean value was recorded. Before the CLSM observation, biofilms on the bottom of the petri dishes were stained with live/dead BacLight bacterial viability kit (Thermo Fisher Scientific, Waltham, MA, USA) for 0.5 h in the dark. Setting the excitation wavelength to 488 and 561 nm, CLSM was used to image S. aureus biofilms. We used Imaris software (Bitplane Imaris 7.4.2) to process the images and obtained the 3D renderings, which made the bacteriostatic effect of different samples more obvious. The SEM analysis of biofilms was based on previous studies38. S. aureus in logarithmic growth phase was inoculated into a 12-well plate with silicon wafers on the bottom for 24 h. Then, the medium was removed completely; fresh medium containing different samples was added to the 12-well plate. After incubation for 24 h at 37 °C, the biofilms were washed by PBS for 3 times. Then, we fixed the biofilms using 2.5% glutaraldehyde for 4 h. Graded ethanol solutions were used to dehydrate the cells. After dehydrated with acetone, biofilms were observed by SEM.
2.6. Slice preparation for SEM and TEM observation
When preparing SEM photographs of S. aureus, S. aureus cells treated by Ber, Rhe, Ber-Rhe NPs and PBS were collected via a centrifuge (Heraeus Pico 17, Thermo Fisher Scientific) at 3000 rpm for 10 min. In particular, undiluted and immobilized bacteria were photographed with nanoparticles attached to them. In another handling method, S. aureus treated by different samples were swilled by PBS for 3 times and then fixed with 2.5% glutaraldehyde for 4 h. Afterwards, we fixed them using 0.1% osmic acid for 1 h, swilled them through PBS for 3 times, dehydrated the S. aureus cells using graded ethanol solutions with 30%, 50%, 70%, 80%, 90% and 100% (v/v, in water), ultimately dehydrated the S. aureus cells through acetone twice for 10 min. To this step, some cell samples were imaged by SEM. The remaining cell samples were embedded in aseptic 2% AGAR for 48 h. Ultimately, ultrathin sections (60–70 nm) were stained through 2% uranyl acetate and 2% lead citrate solution, and then observed by TEM39.
2.7. Hemolysis assay of Ber-Rhe NPs
Hemolysis test of Ber-Rhe NPs was carried out using mice erythrocytes40. Erythrocytes were obtained and washed with normal saline for three times and prepared into 20% RBC solution. Ber-Rhe NPs were diluted with normal saline into different concentrations (2, 4, 8, and 16 MIC). Then, we mixed Ber-Rhe NPs dispersion (1 mL) with 20% RBC solution (100 μL) to obtain the administration of blood cell suspension. The ultrapure water was positive control and normal saline was the negative control. After incubated for 2 h at 37 °C, the RBC suspension was recorded at 540 nm with a microplate reader. The hemolysis ratio of Ber-Rhe NPs was calculated by Eq. (1):
| (1) |
where An was the absorption at different concentration of the Ber-Rhe NPs, A0 was the absorption of negative control and A1 was the absorption of positive control.
2.8. Cell viability
The cytotoxicity of Ber-Rhe NPs against MDCK cells was evaluated by MTT assay. MDCK cells suspension was added into 96-well plates at about 5 × 103 cells per well and then incubated at 37 °C for 24 h. Afterwards the medium was removed and added fresh culture medium containing serial concentration of Ber-Rhe NPs, Ber and Rhe. After incubated at 37 °C for 24 and 48 h, 96-well plates were added 10 μL 5% MTT solution and further incubated for 4 h. Then the medium containing samples and unreacted dye was removed and the formazan crystals were dissolved by 150 μL DMSO. A microplate reader was used to record the absorbance at a wavelength of 490 nm.
2.9. Assessment of toxicological response in zebrafish
One-day-old zebrafish were used to evaluate the potential toxicity of Ber, Rhe and Ber-Rhe NPs in this assay. Malformation or mortality of zebrafish reflected the toxicity of the samples. Healthy zebrafish were placed in a 12-well plate (20 zebrafish per well), incubated with different concentrations of Ber, Rhe and Ber-Rhe NPs in Holtfreter's solution at 27 °C. The morphology and survival status of zebrafish were observed at different time intervals (24, 48 and 72 h), and ultimately reflected the safety of different samples. The experimental procedures with zebrafish used in this study were reviewed and approved by the Animal Care Committee of Beijing University of Chinese Medicine, Beijing, China.
2.10. Statistical analysis
The obtained data were analyzed and exhibited as mean ± standard deviations (SD) of n ≥ 3. Statistical analyses of various parameters were carried out by t-test. Differences were determined statistically significant for *P < 0.05, **P < 0.01 and ***P < 0.001, respectively.
3. Results and discussion
3.1. Synthesis and morphological characterization of Ber-Rhe NPs
Natural phytochemicals with certain pharmacological effects were extracted from medicinal herbs. They were often recognized as lead compounds to enhance their medical value by performing a series of structural modifications. Coincidentally, we found Ber and Rhe could self-assemble into nanoparticles without any involvement of carriers and excipients. This discovery just solved the above possible shortcomings. Meanwhile, Rhe was also demonstrated as antibacterial ingredient, which might result in synergistic effect with Ber. Hence, a carrier-free, biocompatible, biodegradable, low-cost and harmfulness nano self-assembly was constructed in this section (Fig. 1A).
Tyndall effect of nanoparticles was significant (Fig. 1B). The morphology and size of Ber-Rhe NPs were characterized by dynamic light scattering (DLS), SEM and TEM, respectively. The DLS image in Fig. 1E showed that the average size of nanoparticles was approximately 174 nm, which was consistent with that characterized by SEM and TEM (Fig. 1C and D). Meanwhile, the morphology of Ber-Rhe NPs changed obviously, compared with the parent phytochemicals (Supporting Information Fig. S1). At the same time, we found that the mixture of the individuals also had weak Tyndall phenomenon. The results of DLS and SEM jointly substantiated that the simple mixture contained particles of different morphologies and sizes (Supporting Information Fig. S2). It was nanotechnology that helped us prepare Rhe and Ber into a uniform nano-platform. In terms of NPs' release and stability characteristics, after 72 h of dialysis in PBS (37 °C), about 50% of the Ber and 36% of the Rhe in nanoparticles were released, indicating that the stacking structure could sustainably release in physiological environment (Fig. 1F). What's more, the stability of Ber-Rhe NPs was measured by DLS. As shown in Fig. 1G, the nanoparticles exhibited excellent stability for 10 days (at 25 °C in dark). This result was associated with the further antibacterial mechanism analysis.
3.2. Self-assembly process and characteristics of Ber-Rhe NPs
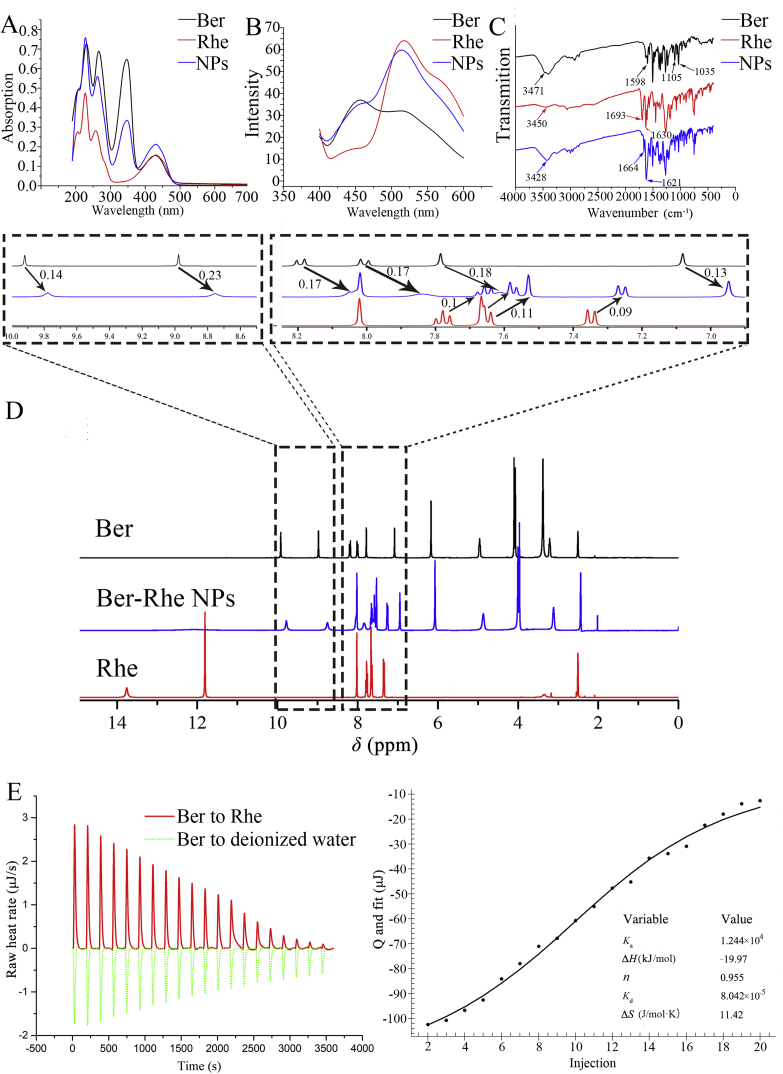
The composition analysis of Ber-Rhe NPs was based on a series of spectroscopy characterizations. As shown in Fig. 2A, the UV–Vis spectrum of Ber in deionized water exhibited 4 significant absorption peaks. The characteristic absorption peaks at 350 and 428 nm were produced by a conjugated system of isoquinoline rings. The UV–Vis spectrum of Rhe in deionized water showed 4 absorption peaks, the same absorption peaks with Ber were at 204, 230, 265 and 428 nm, respectively. The UV–Vis spectra of nanoparticles had the same of both Ber and Rhe, indicating the successful construction of binary self-assembly. Similarly, the combined characteristics of fluorescence emission spectrum of the nanoparticles were same as the UV–Vis analysis results, when excited at 350 nm (Fig. 2B). The fourier transform infrared spectra (FT-IR) of nanoparticles provided more information for the interaction between Ber and Rhe. As shown in Fig. 2C, Ber exhibited vibration bands 1598, 1105 and 1035 cm−1, which were ascribed to a C–N stretching band and alicyclic ether, respectively. Rhe showed typical C O groups in 1693 and 1630 cm−1 as well as associated O–H stretching band. After forming Ber-Rhe NPs, C O groups of Rhe exhibited blue shift from 1693 and 1630 to 1664 and 1621 cm−1, indicating the formation of hydrogen bonding or π−π stacking between Ber and Rhe. In a word, the above spectral analyses had proved that the nanoparticles were binary aggregates composed of Ber and Rhe.
Figure 2.
(A) UV–Vis spectra of Ber, Rhe and Ber-Rhe NPs. (B) Fluorescence spectra of Ber, Rhe and Ber-Rhe NPs. (C) FT-IR spectra of Ber, Rhe and Ber-Rhe NPs. (D) 1H NMR spectrum of Ber, Rhe and Ber-Rhe NPs. (E) Calorimetric titration of Ber/Rhe and heating curve of Rhe solution titrated by Ber solution.
To further confirm the weak interaction types and positions between Ber and Rhe, 1H NMR of nanoparticles was measured in dimethylsulfoxide-d6, and then compared with the monomer Ber and Rhe under the identical conditions. Fig. 2D showed representative 1H NMR spectra, the nanoparticles had the H signals of Ber and Rhe, illustrating the successful synthesis of the conjugated Ber-Rhe NPs. The attribution of H signal was recorded in Supporting Information Section 5. It was obvious that the chemical shifts of the H signals in Ber moved up-field systematically from 9.92, 8.98, 8.19, 8.01, 7.79 and 7.08 ppm to 9.78, 8.75, 8.02, 7.84, 7.61 and 6.95 ppm after Ber and Rhe formed nanoparticles (Supporting Information Table S1). After the formation of Ber-Rhe NPs, the chemical shifts of four C–H peaks in the Rhe's anthraquinone ring also changed apparently from 7.78, 7.66, 7.64 and 7.35 ppm to 7.66, 7.57, 7.53 and 7.26 ppm, respectively (Table S1). The H signals on isoquinoline ring which were involved in conjugation of Ber had obvious shift from down-field to up-field. In particular, the H-11 and H-12 signals in Ber changed from the original double peaks to single peak after complexation. Similarly, the H signals on anthracene ring in Rhe also showed obvious shift from down-field to up-field. The fact was that the anthracene ring in Rhe and the isoquinoline ring in Ber became a structure of π−π stacking, which enhanced the delocalization of electrons and made the electronic fraction more extensive. At the same time, it proved that Ber and Rhe complexed with an equimolar in hydrofacies.
Isothermal titration calorimetry (ITC) analysis curves of the solutions of Ber and Rhe were shown in Fig. 2E. Two experiments were carried out in this part, including the titration of Ber solution (2.5 mmol/L) into deionized water and Ber solution (2.5 mmol/L) into Rhe solution (0.5 mmol/L). The energy changes data of the two interactions was shown in Supporting Information Table S2. The binding heat of deionized water titrated by Ber was set as benchmark, and the experimental group was the titration process of Ber into Rhe. The heating curve of the titration process was fitted with Nano Analyze software (NanoAnalyze v3.10.0). The titration of Ber solution into deionized water was an endothermic dilution process; however, the titration of Ber into Rhe released a lot of heat. By fitting titration curve, we obtained the binding thermodynamic parameters (ΔS, ΔH, ΔG, n, Ka and Kd) of Ber and Rhe, summarized in Supporting Information Table S3. Ka = 1.244 × 104, ΔG = −23.37 kJ/mol, indicating the spontaneity of the reaction between Ber and Rhe. The fitted stoichiometric ratio (n) was 0.955, indicating the binding ratio of Ber and Rhe was 1:1, which conformed to the result of 1H NMR of nanoparticles. This was direct evidence that Ber and Rhe were self-assembled. ΔH = −19.97 kJ/mol and −TΔS = 3.40 kJ/mol, which demonstrated that enthalpy was most of the binding energy, because the entropy change (ΔS) of binding contributed adversely. At the same time, this also demonstrated that the aggregation process was a chemical reaction, such as the formation of electrostatic interaction, π−π stacking or hydrogen bonding27, rather than a physical combination. These conclusions were consistent with the results of the UV–Vis, fluorescence, FT-IR spectrums and the 1H NMR spectrum.
3.3. Single crystal analysis disclosed the noncovalent bonding and self-assembly mechanisms
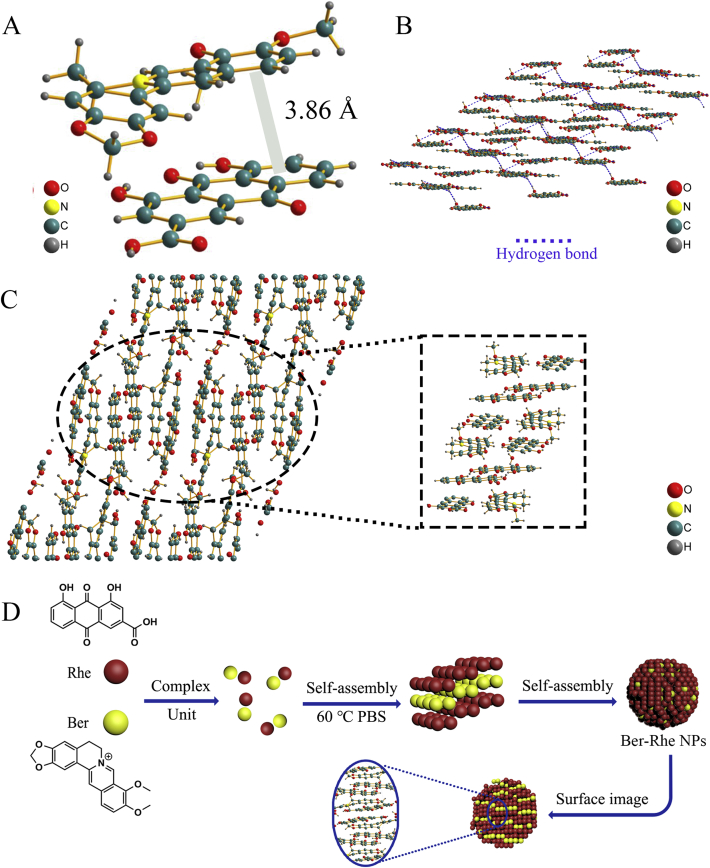
To prove the above analyses and disclose the detailed formation mechanism of nanoparticle, X-ray single crystal diffraction was operated to explore the π−π stacking and hydrogen bonds. Coincidentally, we also observed the Ber-Rhe complex units in the crystal. What's more, the interaction force was also clearly displayed. As shown in Fig. 3A, the crystal structure contained basic units of Rhe and Ber induced by π−π interaction (the ring/ring distance ∼3.86 Å). The electrostatic interaction between the unit cells also made the whole assembly structure more reasonable. In stereoscopic view, Rhe formed a layered stacking structure by hydrogen bonding (Fig. 3B). Subsequently, Ber was embedded in the layered structure formed by Rhe (Fig. 3C). Finally, nanoparticles with Rhe as the basic framework and Ber embedded in the layer gap were obtained. In this way, Rhe played the role of skeleton structure, in which Ber was embedded to make the system more stable (Fig. 3D). Other crystal diagrams including skeleton diagram and stacking diagram at different angles were shown in Supporting Information Fig. S4, which also proved that the above explanations were reasonable. Above analyses confirmed that the nanoparticles were formed by self-assembly of the Ber-Rhe complex units, which were consistent with the analyses results of 1H NMR and ITC data. At the same time, the hydrogen bonds between Rhe also confirmed the correctness of FT-IR analysis. In a word, Rhe molecules were layered by hydrogen bonding, and Ber were pulled into the layers by π−π stacking and electrostatic interaction; then Ber-Rhe NPs were formed eventually.
Figure 3.
(A) Basic unit of Ber-Rhe NPs crystal under the action of π−π stacking; (B) the layered framework structure formed by Rhe under hydrogen bonding; (C) the accumulation structure of Ber-Rhe NPs crystals; (D) self-assembly process diagram of Ber and Rhe.
3.4. Antibacterial activity of Ber-Rhe NPs
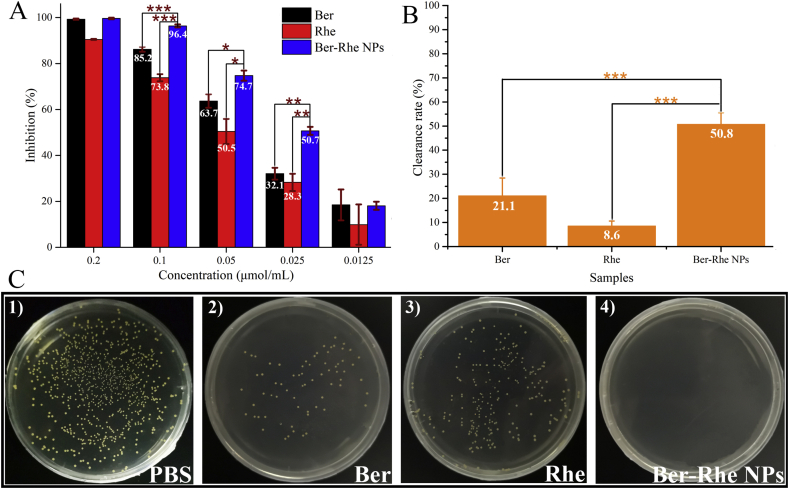
The antibacterial activity of Ber-Rhe NPs was explored by growth inhibition against S. aureus. After incubated for 16 h, the S. aureus suspension was recorded at 600 nm (OD600) by microplate reader. The MIC of NPs toward S. aureus was 0.05 μmol/mL by turbidimetry. 0.1 μmol/mL was the MBC of nano-assembly toward S. aureus, which was lower than that of Ber (MBC, 0.2 μmol/mL) and Rhe (MBC, >0.2 μmol/mL). As shown in Fig. 4A, the inhibition rate of 0.1 μmol/mL (MBC) Ber-Rhe NPs against S. aureus reached 96%, while Ber and Rhe at the same concentration were 85.2% and 73.8%. To confirm the synergistic effects between Ber and Rhe, plate scribing method was applied in this section. The bacteriostatic effect of the three samples at 0.1 μmol/mL was shown in Fig. 4C. The antibacterial effect of Ber was obviously enhanced by self-assembling into nanoparticles with Rhe.
Figure 4.
(A) The inhibitory rates of different concentrations of Ber, Rhe and Ber-Rhe NPs against S. aureus. (B) Effect of Ber-Rhe NPs on S. aureus biofilm formation. (C) Colony forming units (CFU) for S. aureus treated with PBS, Ber, Rhe and Ber-Rhe NPs at 0.1 μmol/mL (106 times diluted).
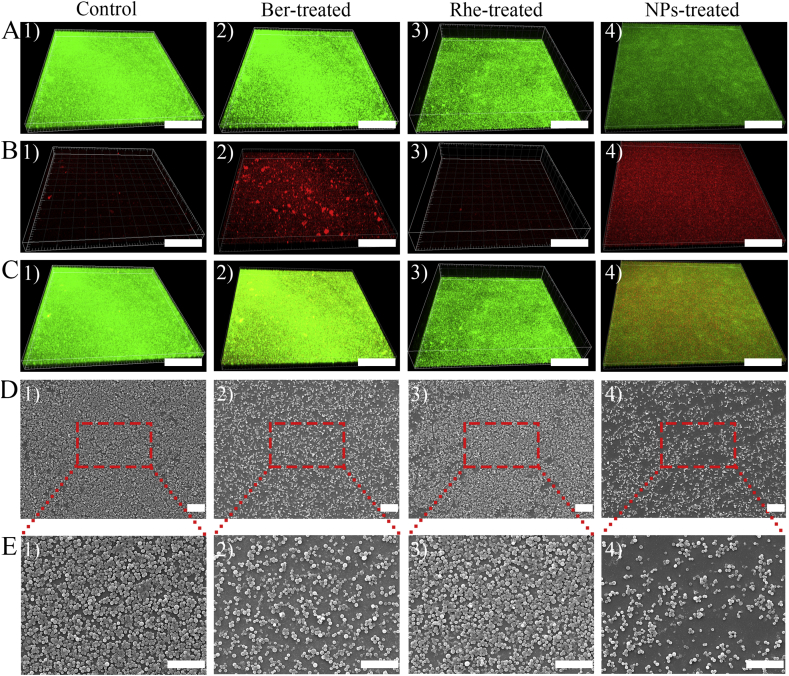
Moreover, biofilm plays a significant role in the growth of S. aureus. It acts as a barrier against foreign interference, protecting bacteria from extermination. In quantitative analysis (Fig. 4B), the concentration of three samples was 0.1 μmol/mL (2 MIC of NPs). At this concentration, Rhe had no obvious scavenging effect on biofilm. The S. aureus biofilm treated with Ber had fallen by 21.1%; whereas, Ber-Rhe NPs cleaned S. aureus biofilm up to 50.8% when compared to control group. The biofilm clearance of Ber-Rhe NPs was much higher than Ber and Rhe (P < 0.001). Before the observation with CLSM, biofilms were stained with 100 μL live/dead stain for 0.5 h in the dark. The green-fluorescent stain labeled live bacterial cells (Fig. 5A). In contrast, the red-fluorescent labeled dead bacterial cells (Fig. 5B). Fluorescence stacking diagram of dead bacteria and live bacteria were shown in Fig. 5C. S. aureus produced numerous microbial colonies covering the whole surface of the horizon. In the culture with 0.1 μmol/mL Ber, the number of dead colonies was substantially increased. The inhibitory effect of Rhe on biofilm was weaker than that of Ber, which was consistent with the above inhibition results. A further increased in dead colonies occurred in the culture with 0.1 μmol/mL Ber-Rhe NPs, and the number of live colonies was substantially decreased under the identical imaging settings. Obviously, Ber-Rhe NPs demonstrated a stronger inhibitory effect on S. aureus biofilms than Ber. Consistently, the same biofilm scavenging ability of the samples was observed in SEM images (Fig. 5D and E).
Figure 5.
CLSM and SEM analysis of S. aureus biofilms; (A) living bacteria in biofilms treated by PBS, Ber, Rhe and Ber-Rhe NPs; (B) dead bacteria in biofilms treated by PBS, Ber, Rhe and Ber-Rhe NPs; (C) fluorescence stacking diagram of dead bacteria and living bacteria (scale bar = 200 μm); (D) SEM images of biofilms incubated with PBS, Ber, Rhe and Ber-Rhe NPs (scale bar = 10 μm); (E) SEM images of partial biofilms (scale bar = 10 μm).
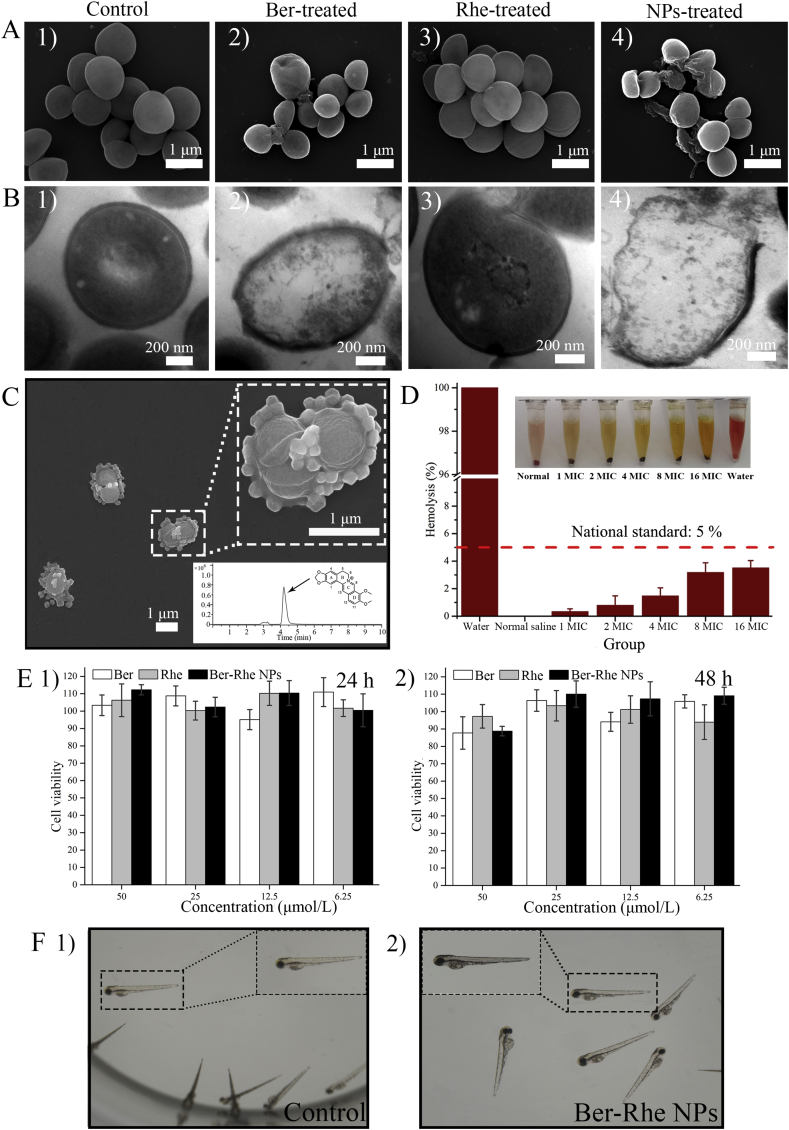
Finally, in order to exhibit the bacteriostatic difference between Ber-Rhe NPs and individuals intuitively, SEM and TEM were operated to observe the morphological change of S. aureus cells which were treated by Ber, Rhe and Ber-Rhe NPs at the concentration of 0.1 μmol/mL for 12 h. The bacteria treated by PBS were imaged as control. In SEM images (Fig. 6A), the control group had clear edges and surface integrity with regular ellipsoid. When incubated with Rhe, the morphology of bacteria remained basically unchanged. However, the membrane of bacteria treated with Ber was pitted; morphological changes were also observed. Obviously, the membrane morphology of bacterial exhibited shrunk and fusion with massive cytoplasmic leakage after incubation with Ber-Rhe NPs. As shown in TEM images (Fig. 6B), untreated bacteria exhibited a smooth surface; the nucleoid of the cell center was intact. In contrast, the bacterial membrane became fracture after incubated with Ber-Rhe NPs. Moreover, the morphology of the bacteria was severely altered with numerous cytoplasmic releasing.
Figure 6.
(A) SEM images of S. aureus treated by (1) PBS, (2) Ber, (3) Rhe and (4) Ber-Rhe NPs; (B) TEM images of S. aureus incubated with (1) PBS, (2) Ber, (3) Rhe and (4) Ber-Rhe NPs; (C) nanoparticles attached to the S. aureus; (D) hemolytic activities of Ber-Rhe NPs vary from 1 to 16 MIC. (E) Cell viability after incubated with Ber, Rhe and Ber-Rhe NPs for (1) 24 and (2) 48 h. (F) Image of zebrafish exposed to Holtfreter's solution and 100 μmol/L Ber-Rhe NPs for 72 h.
It is reported that the cytoderm of gram-positive bacteria is the main acting site of emodin41, 42, 43. Analysis showed that anthraquinone ring had strong affinity to membrane. This was similar to the antibacterial mechanism of vancomycin (Supporting Information Fig. S3C), which inhibited cytoderm formation by forming hydrogen bond complexes due to the amide and carboxyl groups at the peptidoglycan terminal on the cytomembrane of the bacteria44. Coincidentally, Rhe also has multiple alternating phenolic hydroxyl, carbonyls and carboxyl groups (Fig. S3B), which can form hydrogen bonds with amide and carboxyl groups on the peptidoglycan, providing a possible explanation for the adhesion of nanoparticles on the bacterial surface. Rhe from self-assembled nanoparticles could be regarded as a functional antibacterial skeleton material as well as a bacteriostatic active ingredient. As shown in Fig. 6C, plenty of nanoparticles were adhered to the surface of the bacteria, local high concentration of Ber and Rhe would damage the membrane ion channel protein and lyse the bacteria2. Simultaneously, HPLC−MS/MS disclosed that S. aureus was accessed or adhered by Ber (Supporting Information Fig. S5). As clearly shown in Fig. 6B 4, floccules out of bacteria were also observed. Considering the above images of TEM and drug release test, we inferred that Ber-Rhe NPs disintegrated on the S. aureus membrane, and then Ber was released into the cell. After releasing and entering the cell, Ber and Rhe could further affect the physiological function and structural integrity of the bacteria and lead to its death2,45.
In summary, Ber-Rhe NPs inhibited the formation of S. aureus biofilm and made it lose barrier function. Due to the part of Rhe which distributed on the nanoparticles, hydrogen bonding between in Rhe molecules increased the affinity of the nanoparticles to the surface of bacteria. After Ber-Rhe NPs adhered to the membrane, Ber and Rhe were released to lyse the membrane. Ber and Rhe affected the physiological function of bacteria. The synergistic antibacterial effect of Ber and Rhe made the nanoparticles more effective.
3.5. Biocompatibility analysis
The biocompatibility of Ber-Rhe NPs was evaluated by hemolysis test, cytotoxicity test and zebrafish. The hemolysis test was performed as follows, 2% red blood cells solution was incubated with Ber-Rhe NPs (0.05, 0.1, 0.2, 0.4, and 0.8 μmol/mL) for 4 h. As shown in Fig. 6D, Ber-Rhe NPs exhibited low hemolytic activities (<5%), even at the highest concentration of 0.8 μmol/mL (16 MIC). Cytotoxicity of Ber-Rhe NPs was investigated using MDCK cells. As shown in Fig. 6E, cell viability of Ber-Rhe NPs was over 80% even at concentration of 50 μmol/L. Simultaneously, to further evaluate in vivo biocompatibility, zebrafish was used to examine the biosafety of the Ber, Rhe and Ber-Rhe NPs at the concentrations ranging from 6.25 to 100 μmol/L. Zebrafish as a feasible in vivo biosafety model have attracted much attention in recent years46. The deformity and death of zebrafish were considered as important indicators to evaluate toxicity. As shown in Fig. 6F, after incubation with Ber-Rhe NPs for 72 h, zebrafish were all kept survival, which indicated the low toxicity of the Ber-Rhe NPs. These results demonstrated that Ber-Rhe NPs was highly biocompatible and had greater potential for applications in clinic.
4. Conclusions
In conclusion, we have successfully constructed a natural phytochemicals-based self-assemble nano-platform against S. aureus. All experiments, especially NMR and X-ray single crystal diffraction analyses demonstrated that Rhe acted as layered backbone under hydrogen bonding and Ber embedded in it by π−π interaction and electrostatic interaction. Thereafter, nanoparticles formed a stable three-dimensional configuration of layered deposits in aqueous phase.
Due to the synergistic bacteriostasis of Ber and Rhe which were commonly used as antibacterial agents in clinic, the antimicrobial activity of Ber-Rhe NPs was enhanced. Under the leadership of Rhe, the novel nanoparticles could adhere to the surface of the bacteria and increase the individuals’ concentration around S. aureus. Cell integrity was severely compromised, which leaded to its death. Notably, instead of violent reaction conditions, the self-assembly system was directly owing to the noncovalent bonding between Ber and Rhe, indicating that this was a green strategy of simple process and efficient performance. The easy obtained, multifunctional and biocompatible self-assemble system was supposed to provide new opportunities for the construction of new agents from enlightenment of TCM. Moreover, this study maybe come up with a new angle that self-assembly between natural phytochemicals could be originated from TCM.
Acknowledgments
This research was funded by the Beijing Municipal Natural Science Foundation (No. 7202116, China), National Natural Science Foundation of China (No. 81603256), project of China Association of Chinese Medicine (CACM-2018-QNRC2-B08), the Fundamental Research Funds for the Central Universities (BUCM-2019-JCRC002, BUCM-2018-2020 and 2019-JYB-TD005, China), Beijing Key Laboratory for Basic and Development Research on Chinese Medicine (Beijing, China). We thank institute of automation, Chinese academy of sciences for providing the technical assistance of SEM.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.12.014.
Contributor Information
Penglong Wang, Email: wpl581@126.com.
Haimin Lei, Email: hm_lei@126.com.
Author contributions
Penglong Wang and Haimin Lei designed the research, obtained the funding and superintend the whole study; Penglong Wang, Bing Xu, Tao Ma, Feifei Li and Cong Yan directed the conduct of the experiments; Xuehao Tian, Tong Li and Xuemei Huang performed the all experiments; Xuehao Tian, Yuqin Yang, Wenbo Guo, Mengmeng Yan, Desheng Cai, Hao Zhang and Xiaohui Jia analyzed all data from experiments; Xuehao Tian and Penglong Wang wrote and revised the manuscript. All authors read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Zhai Z., Boquete J.P., Lemaitre B. Cell-specific Imd-NF-κB responses enable simultaneous antibacterial immunity and intestinal epithelial cell shedding upon bacterial infection. Immunity. 2018;48:897–910. doi: 10.1016/j.immuni.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Mei L., Lu Z., Zhang X., Li C., Jia Y. Polymer-Ag nanocomposites with enhanced antimicrobial activity against bacterial infection. ACS Appl Mater Interfaces. 2014;6:15813–15821. doi: 10.1021/am502886m. [DOI] [PubMed] [Google Scholar]
- 3.Ding F., Nie Z., Deng H., Xiao L., Du Y., Shi X. Antibacterial hydrogel coating by electrophoretic co-deposition of chitosan/alkynyl chitosan. Carbohydr Polym. 2013;98:1547–1552. doi: 10.1016/j.carbpol.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 4.Yang W., Tao X., Zhao T., Weng L., Kang E., Wang L. Antifouling and antibacterial hydrogel coatings with self-healing properties based on a dynamic disulfide exchange reaction. Polym Chem. 2015;6:7027–7035. [Google Scholar]
- 5.Fullenkamp D.E., Rivera J.G., Gong Y.K., Lau K.A., He L., Varshney R., et al. Mussel-inspired silver-releasing antibacterial hydrogels. Biomaterials. 2012;33:3783–3791. doi: 10.1016/j.biomaterials.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strassburg A., Kracke F., Wenners J., Jemeljanova A., Kuepper J., Petersen H., et al. Nontoxic, hydrophilic cationic polymers—identified as class of antimicrobial polymers. Macromol Biosci. 2015;15:1710–1723. doi: 10.1002/mabi.201500207. [DOI] [PubMed] [Google Scholar]
- 7.Guo J., Qin J., Ren Y., Wang B., Cui H., Ding Y., et al. Antibacterial activity of cationic polymers: side-chain or main-chain type?. Polym Chem. 2018;9:4611–4616. [Google Scholar]
- 8.Yang Y., Cai Z., Huang Z., Tang X., Zhang X. Antimicrobial cationic polymers: from structural design to functional control. Polym J. 2018;50:33–44. [Google Scholar]
- 9.Zhou Y., Quan G., Wu Q., Zhang X., Niu B., Wu B., et al. Mesoporous silica nanoparticles for drug and gene delivery. Acta Pharm Sin B. 2018;8:165–177. doi: 10.1016/j.apsb.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R., He Y., Zhang S., Qin J., Wang J. Cell membrane-based nanoparticles: a new biomimetic platform for tumor diagnosis and treatment. Acta Pharm Sin B. 2018;8:14–22. doi: 10.1016/j.apsb.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamble R.N., Mehta P.P., Kumar A. Efavirenz self-nano-emulsifying drug delivery system: in vitro and in vivo evaluation. AAPS PharmSciTech. 2016;17:1240–1247. doi: 10.1208/s12249-015-0446-2. [DOI] [PubMed] [Google Scholar]
- 12.Thompson G.R., Barker B.M., Wiederhold N.P. Large-scale evaluation of in vitro amphotericin B, triazole, and echinocandin activity against Coccidioides species from US institutions. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02634-16. e02634-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Napierska D., Thomassen L.C., Lison D., Martens J.A., Hoet P.H. The nanosilica hazard: another variable entity. Part Fibre Toxicol. 2010;7:39. doi: 10.1186/1743-8977-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y.S., Church J.S., Woodhead A.L., Moussa F. Preparation and characterization of silica coated iron oxide magnetic nano-particles. Spectrochim Acta, Part A. 2010;76:484–489. doi: 10.1016/j.saa.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Liu J., Zeng F., Allen C. In vivo fate of unimers and micelles of a poly(ethylene glycol)-block-poly(caprolactone) copolymer in mice following intravenous administration. Eur J Pharm Biopharm. 2007;65:309–319. doi: 10.1016/j.ejpb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Elsabahy M., Wooley K.L. Data mining as a guide for the construction of cross-linked nanoparticles with low immunotoxicity via control of polymer chemistry and supramolecular assembly. Acc Chem Res. 2015;48:1620–1630. doi: 10.1021/acs.accounts.5b00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potter T.M., Neun B.W., Dobrovolskaia M.A. In vitro and in vivo methods for analysis of nanoparticle potential to induce delayed-type hypersensitivity reactions. Methods Mol Biol. 2018;1682:197–210. doi: 10.1007/978-1-4939-7352-1_17. [DOI] [PubMed] [Google Scholar]
- 18.Xia G., Liu T., Wang Z., Hou Y., Dong L., Zhu J., et al. The effect of silver nanoparticles on zebrafish embryonic development and toxicology. Artif Cells Nanomed Biotechnol. 2016;44:1116–1121. doi: 10.3109/21691401.2015.1011803. [DOI] [PubMed] [Google Scholar]
- 19.Normile D. The new face of traditional Chinese medicine. Science. 2003;299:188–190. doi: 10.1126/science.299.5604.188. [DOI] [PubMed] [Google Scholar]
- 20.Stone R. Lifting the veil on traditional Chinese medicine. Science. 2008;319:709–710. doi: 10.1126/science.319.5864.709. [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa K., Takahashi K., Hosoi S., Takeda H., Yoshida H., Wakana D., et al. Antimicrobial agent isolated from Coptidis rhizome extract incubated with Rhodococcus sp. strain BD7100. J Antibiot (Tokyo) 2019;72:71–78. doi: 10.1038/s41429-018-0114-3. [DOI] [PubMed] [Google Scholar]
- 22.Gong R., Lee D., Lee J., Choi D., Kim G., Lee S., et al. Potentiating activity of rhein in targeting of resistance genes in methicillin-resistant Staphylococcus aureus. Asian Pac J Trop Med. 2019;12:14–18. [Google Scholar]
- 23.Lin B., Liu X., Wu S., Zheng H., Huo K., Qi S., et al. Phytochemicals content, antioxidant and antibacterial activities of Sophora viciifolia. Chem Biodivers. 2019;16 doi: 10.1002/cbdv.201900080. [DOI] [PubMed] [Google Scholar]
- 24.Itzia Azucena R.C., José Roberto C.L., Martin Z.R., Rafael C.Z., Leonardo H.H., Gabriela T.P., et al. Drug susceptibility testing and synergistic antibacterial activity of curcumin with antibiotics against enterotoxigenic Escherichia coli. Antibiotics. 2019;8 doi: 10.3390/antibiotics8020043. pii: E43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Pu C., Tang W., Wang S., Sun Q. Gallic acid liposomes decorated with lactoferrin: characterization, in vitro digestion and antibacterial activity. Food Chem. 2019;293:315–322. doi: 10.1016/j.foodchem.2019.04.116. [DOI] [PubMed] [Google Scholar]
- 26.Wang L., Yang R., Yuan B., Liu Y., Liu C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm Sin B. 2015;5:310–315. doi: 10.1016/j.apsb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T., Wang P., Guo W., Huang X., Tian X., Wu G., et al. Natural berberine-based Chinese herb medicine assembled nanostructures with modified antibacterial application. ACS Nano. 2019;13:6770–6781. doi: 10.1021/acsnano.9b01346. [DOI] [PubMed] [Google Scholar]
- 28.Zhou F., Yang W., Wang L. Study on in vitro antibacterial effect of huanglian jiedu decoction on 100 clinical multi-drug resistant bacteria. Internet J Lab Med. 2018;39:3061–3065. [Google Scholar]
- 29.Ling X., Xiang Y., Chen F., Tang Q., Zhang W., Tan X. Intestinal absorption differences of major bioactive compounds of Gegenqinlian decoction between normal and bacterial diarrheal mini-pigs in vitro and in situ. J Chromatogr B Analyt Technol Biomed Life Sci. 2018;1083:93–101. doi: 10.1016/j.jchromb.2018.02.045. [DOI] [PubMed] [Google Scholar]
- 30.Bandyopadhyay S., Patra P.H., Mahanti A., Mondal D.K., Dandapat P., Bandyopadhyay S., et al. Potential antibacterial activity of berberine against multi drug resistant enterovirulent Escherichia coli isolated from yaks (Poephagus grunniens) with haemorrhagic diarrhoea. Asian Pac J Trop Med. 2013;6:315–319. doi: 10.1016/S1995-7645(13)60063-2. [DOI] [PubMed] [Google Scholar]
- 31.Zheng J., Fan R., Wu H., Yao H., Yan Y., Liu J., et al. Directed self-assembly of herbal small molecules into sustained release hydrogels for treating neural inflammation. Nat Commun. 2019;10:1604. doi: 10.1038/s41467-019-09601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu D., Lv Y., Wang J., Yang M., Kong L. Deciphering the mechanism of Huang-Lian-Jie-Du-Decoction on the treatment of sepsis by formula decomposition and metabolomics: enhancement of cholinergic pathways and inhibition of HMGB-1/TLR4/NF-κB signaling. Pharmacol Res. 2017;121:94–113. doi: 10.1016/j.phrs.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 33.DeLoach A.S., Conrad B.R., Einstein T.L., Dougherty D.B. Coverage dependent molecular assembly of anthraquinone on Au(111) J Chem Phys. 2017;147:184701. doi: 10.1063/1.4999623. [DOI] [PubMed] [Google Scholar]
- 34.Wang D., Yu C., Li X., Shi L., Tong G., Wu J., et al. Nucleoside analogue-based supramolecular nanodrugs driven by molecular recognition for synergistic cancer therapy. J Am Chem Soc. 2018;140:8797–8806. doi: 10.1021/jacs.8b04556. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y., Chen F., Pan Y., Li Z., Xue X., Okeke C.I., et al. Nanodrug formed by coassembly of dual anticancer drugs to inhibit cancer cell drug resistance. ACS Appl Mater Interfaces. 2015;7:19295–19305. doi: 10.1021/acsami.5b05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ujihara T., Hayashi N. Association of catechin molecules in water: quantitative binding study and complex structure analysis. J Nat Prod. 2015;79:66–73. doi: 10.1021/acs.jnatprod.5b00658. [DOI] [PubMed] [Google Scholar]
- 37.Li Q., Wu Y., Lu H., Wu X., Chen S., Song N., et al. Construction of supramolecular nanoassembly for responsive bacterial elimination and effective bacterial detection. ACS Appl Mater Interfaces. 2017;9:10180–10189. doi: 10.1021/acsami.7b00873. [DOI] [PubMed] [Google Scholar]
- 38.Singh P., Pandit S., Garnæs J., Tunjic S., Mokkapati V.R., Sultan A., et al. Green synthesis of gold and silver nanoparticles from Cannabis sativa (industrial hemp) and their capacity for biofilm inhibition. Int J Nanomed. 2018;13:3571–3591. doi: 10.2147/IJN.S157958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y., Tian Y., Cui Y., Liu W., Ma W., Jiang X. Small molecule-capped gold nanoparticles as potent antibacterial agents that target gram-negative bacteria. J Am Chem Soc. 2010;132:12349–12356. doi: 10.1021/ja1028843. [DOI] [PubMed] [Google Scholar]
- 40.Hong W., Gao X., Qiu P., Yang J., Qiao M., Shi H., et al. Synthesis, construction, and evaluation of self-assembled nano-bacitracin A as an efficient antibacterial agent in vitro and in vivo. Int J Nanomed. 2017;12:4691–4708. doi: 10.2147/IJN.S136998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M. Army Medical University; Chongqing: 2015. Investigation on its anti-MRSA effects of emodin in vitro and in vivo, and its mechanisms [dissertation] [Google Scholar]
- 42.Liu M., Peng W., Qin R., Yan Z., Cen Y., Zheng X., et al. The direct anti-MRSA effect of emodin via damaging cell membrane. Appl Microbiol Biotechnol. 2015;99:7699–7709. doi: 10.1007/s00253-015-6657-3. [DOI] [PubMed] [Google Scholar]
- 43.Cao F., Peng W., Li X., Liu M., Li B., Qin R., et al. Emodin is identified as the active component of ether extracts from Rhizoma Polygoni Cuspidati, for anti-MRSA activity. Can J Physiol Pharmacol. 2015;93:485–493. doi: 10.1139/cjpp-2014-0465. [DOI] [PubMed] [Google Scholar]
- 44.Wei L., Li G. Advances in the study of vancomycin resistance in Staphylococcus aureus. Med Recapitulate. 2019;25:119–123. [Google Scholar]
- 45.Lu B., Lu F., Ran L., Yu K., Xiao Y., Li Z., et al. Self-assembly of natural protein and imidazole molecules on gold nanoparticles: applications in wound healing against multi-drug resistant bacteria. Int J Biol Macromol. 2018;119:505–516. doi: 10.1016/j.ijbiomac.2018.07.167. [DOI] [PubMed] [Google Scholar]
- 46.Pitt J.A., Kozal J.S., Jayasundara N., Massarsky A., Trevisan R., Geitner N., et al. Uptake, tissue distribution, and toxicity of polystyrene nanoparticles in developing zebrafish (Danio rerio) Aquat Toxicol. 2018;194:185–194. doi: 10.1016/j.aquatox.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.