Abstract
Simple Summary
Stress Granules (SGs) were discovered in 1999 and while the first decade of research has focused on some fundamental questions, the field is now investigating their role in human pathogenesis. Since then, evidences of a link between SGs and cancerology are accumulating in vitro and in vivo. In this work we summarized the role of SGs proteins in cancer development and their prognostic values. We find that level of expression of protein involved in SGs formation (and not mRNA level) could serve a prognostic marker in cancer. With this review we strongly suggest that SGs (proteins) could be targets of choice to block cancer development and counteract resistance to improve patients care.
Abstract
Cancer treatments are constantly evolving with new approaches to improve patient outcomes. Despite progresses, too many patients remain refractory to treatment due to either the development of resistance to therapeutic drugs and/or metastasis occurrence. Growing evidence suggests that these two barriers are due to transient survival mechanisms that are similar to those observed during stress response. We review the literature and current available open databases to study the potential role of stress response and, most particularly, the involvement of Stress Granules (proteins) in cancer. We propose that Stress Granule proteins may have prognostic value for patients.
Keywords: stress granules, G3BP1, G3BP2, CAPRIN-1, USP10, TIA-1, TIAR, cancer prognosis, biomarker, metastasis, resistance, cell death, pro-survival properties
1. Generalities
According to the World Health Organization, cancer is the second leading cause of death worldwide. In most cases, mortality is not due to the primary tumor itself, but to the occurrence of metastasis. At diagnosis, non-metastatic diseases are treated with first line systemic adjuvant (post-operative) and/or neoadjuvant (pre-operative) treatments in order to avoid tumor relapse by eradicating potential residual or micro-metastatic foci. In case of metastatic disease, the aim of systemic treatment is to achieve clinical remissions as durable as possible, which remains a palliative situation for most of solid cancers. In both situations, if the therapy does not eliminate all malignant cells, residual cancer cells may acquire migratory properties and develop drug resistance mechanisms, further contributing to difficult-to-treat metastasis [1].
While the number of drugs approved for cancer treatment increases, the number of complete remissions after first line treatment remains too low and metastases keep occurring because primary or secondary resistance develops. Different strategies are being developed in order to improve the results of systemic treatments. One of them is precision medicine which aims to overcome this situation by increasing the specificity of the administered treatment: DNA and RNA of patients’ metastases are now sequenced to identify genome alterations and determine the best therapeutic option after first line therapy based on actionable gene mutations and genome alterations. We are now entering the “one tumor at one time-point for one therapeutic strategy” era. However, this remains difficult in practice and the benefit remains to be proven on a large scale. Tumors are heterogeneous and metastases are the sum of years of genomic instability and individual environmental history. We might not have the ability to decipher this degree of complexity yet. However, in the meantime studies can identify new resistance pathways to increase efficiency of systemic treatments and improve patients’ remission. Growing evidence indicates that the malignant cell can respond to exposure to anti-cancer drugs through fast, “acute”, and “ready-to-use” biological components or processes [2]. For all these reasons, elements of the stress response could be an interesting path to explore in this context.
The stress response is an ancestral evolutionary mechanism acquired by the earliest cellular organisms to protect them from sudden environmental or intracellular changes [3]. At the cellular level, any sudden change in the environment that diverges from its optimal growth condition is considered as an insult that triggers a stress response. The stress response includes the activation of stress-responsive genes expression, such as those coding for heat shock proteins (hours to days’ time scale). Stress response also includes some really fast mechanisms (minutes to hours), such as the formation of “Stress Granules” (SGs) [4,5]. The fate of a stressed cell is ultimately determined by its ability to recover from the stress [6,7,8]. Actually, cancer cells face a diverse set of stress conditions during oncogenesis, such as hypoxia, nutrient deprivation, oxidative stress, and even anti-cancer drugs. They have all been reported to induce the formation of SGs in vitro [4,9,10,11,12,13,14,15,16]. It is surmised that SGs are transient triage centers that are designed to help cells to quickly and transiently modify translation to overcome stress and enhance survival, both of which could be used to escape cancer therapies [17,18,19].
In this review, we will focus on stress granules (SGs) as actors of cancer development. We will first start by presenting what SGs are and how they form, discuss their pro-survival effect and how they could be involved in cancer progression. Second, we will analyze the prognostic value of the major proteins that are involved in SGs formation. Finally, we will discuss the involvement and contribution of cellular stress responses to cancer evolution.
2. SGs Are Composed of Proteins Involved in the Regulation of mRNA Translation
Stress granules, first discovered in 1999, are membrane-less cytoplasmic condensates, visible by conventional and electron microscopy [4,20,21,22]. More recently, SGs have been show to handle like hydrogel structures in the cytoplasm and form what is called a liquid-liquid phase separation [23,24]. They have been observed in plants, yeast, worms, insects, and mammalian cells [4,25,26,27,28,29]. This high degree of conservation across species highlights their importance for cell survival and the maintenance of cell integrity [30]. These foci are composed of mRNAs, RNA-binding proteins, and 40S ribosomes [4,31]. It is generally admitted that the absence of a membrane ensures the rapid execution of SGs formation and the extreme lability and adaptability of their components (SGs’ proteins generally are present in cells in homeostatic conditions and only switch their localization and functions between basal and stress conditions). However, this has hindered the purification of these structures and the precise identification of SGs components by global analysis. Currently, even if methods have been reported to purify SGs markers [32,33], candidate approaches have been for a long time the only way to identify specific components. Most studies have used immunofluorescence and Florescent In Situ Hybridization (FISH) to identify proteins and mRNAs that are included in these structures. In 2015, an inventory of the literature indicated more than a hundred proteins recruited to SGs [31]. These proteins form an eclectic mix involved in various signaling pathways. Even if there is still no consensus to predict the recruitment of specific proteins to SGs, most of them interact with RNA or are involved in the metabolism of RNA. SGs are also composed of components that are involved in translation initiation, such as Eukaryotic Initiation Factor (EIF) 3 and EIF4 complex proteins or PABP (PolyA Binding Protein) [31]. The presence of these components is the consequence of a general translation inhibition that precedes SGs formation.
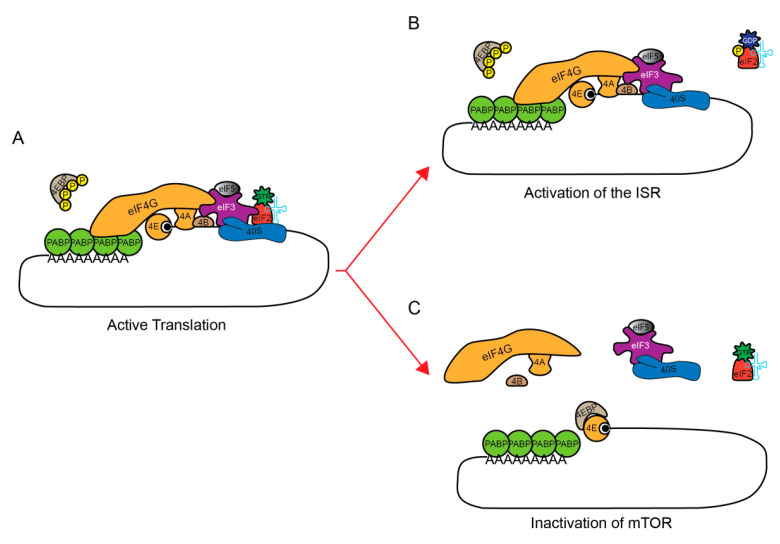
In homeostatic conditions, active translation is facilitated by the formation of a closed-loop mRNA during specific steps of translation [34] (Figure 1A). This is a situation where the 5′ and 3′ ends of an mRNA are brought in close proximity. The 5′ mRNA cap is bound by EIF4E and the 3′ poly (A) tail is bound by PABP. These two proteins are bridged by the large scaffolding protein EIF4G. To initiate translation, the ternary complex, composed of EIF2:tRNAiMet:GTP facilitates the decoding of the start codon, which results in GTP-to-GDP hydrolysis. In response to stress, translation is rapidly inhibited at this initiation step, which, in most cases, results in the induction of SGs formation and the storage of EIFs. The inhibition of two translation pathways can induce the formation of SGs [20]: the phosphorylation of a subunit of EIF2, EIF2α (or EIF2S1) [9], prevents the formation of the translation initiation complex EIF2α-tRNAmet-GTP (Figure 1A,B), and the dephosphorylation of 4EBP that inhibits mRNA circularization, so as to impair re-initiation at the start codon (Figure 1C) [35,36]. These two pathways are not mutually exclusive. In the current state of the literature, most of the investigated stressors induce the formation of SGs via the phosphorylation of EIF2α. However, one or both pathways could be activated, depending on the type of stress [9,37]. While it is intuitive to think that cells shut down translation to preserve energy, one can wonder what would be the other consequences of SGs formation in response to environmental stress.
Figure 1.
Inhibition of translation pathways to induce the formation of SGs. (A) Active translation initiation complex (basal condition). Two pathways could be activated to induce translation inhibition upstream SGs formation. Under basal conditions, EIF2α is not phosphorylated and could allow the formation of the EIF2: tRNAiMet: GTP ternary complex of translation initiation. Additionally, MTOR is active and constitutively phosphorylates EIF4E-Binding Protein (4EBP). (B) The phosphorylation of a subunit of EIF2, EIF2α (or EIF2S1), by one (or more) kinase(s), notably HRI/EIF2AK1, PKR/EIF2AK2, PERK/EIF2AK3 and/or GCN2/EIF2AK4, prevents the hydrolyzed GDP from leaving the ternary complex EIF2α-tRNAmet-GTP by blocking the formation of an active complex with ATP necessary for translation initiation. (C) Hyper-phosphorylated 4EBP cannot interact with EIF4E, the mRNA cap-binding protein. However, induction of a stress response inactivates MTOR leading to a rapid de-phosphorylation of 4EBP, thereby allowing it to interact with EIF4E. The EIF4E: 4EBP interaction prevents EIF4E: EIF4G complex formation.
3. Stress Granules Are Pro-Survival Entities at the Cytoplasmic Level
Stressors triggering the formation of SGs can be as diverse as extreme temperatures (hot or cold), oxidative stress, osmotic stress, endoplasmic reticulum (ER) stress, mitochondrial stress, or UV irradiation (previously reviewed [30,31]). Several lines of evidence point toward pro-survival benefits of SGs formation, possibly explaining the evolutionary conservation of this process. Indeed, the mutation or complete knock-out of specific proteins that are involved in SGs formation, or treatments that impair SGs formation, results in rapid cell death after stress exposure [38,39,40,41,42,43,44]. This pro-survival effect of SGs formation could be explained by several independent mechanisms:
First, many pro-apoptotic signaling molecules are sequestered in SGs and it has been proposed that it prevents them from activating the pro-apoptotic cascade. This is the case for RACK1 (Receptor of Activated Protein C Kinase 1), TRAF2 (TNF receptor-associated factor 2), and RSK2 (Ribosomal S6 kinase 2) [45,46,47].
Second, while not fully characterized, SGs protect cells from oxidative insults by reducing the level of cellular reactive oxygen species (ROS) [39,41,48]. Indeed, when the expression level of a major SGs regulator G3BP1 (RAS GTPase-activating Protein-Binding Protein 1) drops, ROS generation after exposure to oxidative insult increases. Moreover, the overexpression of G3BP1 reduces the level of ROS as compared to wild type cells. Cells expressing a truncated form of the protein that abrogates SGs formation have increased ROS production. Similar results have been obtained with USP10 (Ubiquitin carboxyl-terminal hydrolase 10), another SGs regulating protein [39].
Third, SGs formation reduces the cellular energetic needs during stress by restricting the process of translation, which consumes a lot of ATP. By protecting mRNAs from stress-induced degradation, this allows cells to restart translation as soon as the stress is resolved without having to synthetize de novo RNAs [49]. Additionally, SGs sequester untranslated mRNAs concomitantly with the global inhibition of translation [17]. Some mRNAs, such as chaperone mRNAs, are excluded from the SGs structures, so that they can be preferentially translated during the time of the stress and participate in the proper protein folding and avoid functional defects [17,19,50]. By these actions, SGs are described as triage centers for translation of mRNAs during stress exposure [17]. One leading hypothesis is that SGs are able to reshape translational patterns under stress exposure [19,51].
4. Stress Granules Are Involved in Cancer Progression
In some cases, SGs pro-survival role might not be beneficial for the host, such as in the context of cancer. Cancer cells are frequently, if not systematically, exposed to stresses, such as hypoxia and nutrient deprivation, two stressors able to induce the formation of SGs [10] and promote resistance to therapies, which suggest a pro-survival role of SGs in this context [18]. Chemotherapies (CT) can also induce SGs formation [11,12,14,15,16,52]. The cancer cell capacity to form SGs in response to CT is correlated with cell survival in vitro [11,12,14,15,16,52]. Blocking the induction of these chemotherapy-induced granules by interfering with the phosphorylation of EIF2α increases the efficiency of CT [16,53]. Along the same lines, some molecules can prevent SGs formation and restore sensitivity to CT. This was shown in a study using hypoxia to induce chemo-resistance in HeLa cells. A screen of small molecules revealed that β-estradiol, progesterone and stanolone prevent SGs formation and restore the sensitivity to CT in HeLa cells [18]. The same molecules used in the MCF7 breast cancer cell line did not block SGs formation, or the chemoresistance induction, suggesting a cell-specific effect of these molecules on SGs [16,18]. The ability to block SGs formation seems to vary from patient-to-patient, just as treatment response. Overall, this study suggests that blocking SGs formation in cancer may be an interesting option against cancer cells, provided that a compound targeting SGs and able to overcome the patient/tissue heterogeneity could be found [18].
During cancer development and progression, tumor cells acquire driver mutations that are responsible for cell transformation, then progression, and aggressiveness of the disease. Malignant transformation is a complex and multifactorial mechanism, involving major changes in the genome, and in transcription and translation programs of the cell. For example, Epithelial-to-Mesenchymal Transition (EMT), acquisition of stemness features, or the acquisition of drug resistance involve specific modifications of translation programs. A growing tumor is an extremely dynamic environment where stressors, such as mechanical constriction, hypoxia and/or starvation (nutrient and/or glucose) play a role at multiple levels. SGs could play a critical role in integrating these stressors into changes in translation that leads to cancer progression [18,51]. Recent studies have demonstrated that such changes occur after exposure to hypoxia stress [18,54,55,56].
5. SGs Regulators and Their Role in Cancer
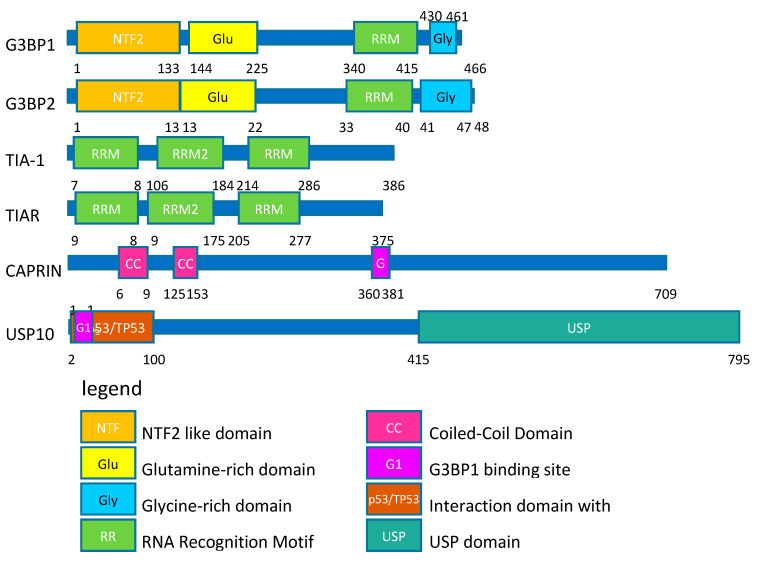
SGs are composed of numerous proteins [31] and the number of regulators increases with the expansion of the field. As a proof of concept, here we focus here on the proteins that are the most used as SGs markers: TIA-1 (T-cell-restricted intracellular antigen-1), TIAR (TIA-1-related protein), G3BP1 (RAS GTPase-activating protein-binding protein 1), and G3BP2 (RAS GTPase-activating protein-binding protein 2) [31]. We also looked at two robust regulators of SGs aggregation: the SGs formation enhancer CAPRIN-1 (Cell Cycle associated protein 1) and the inhibitor of SGs formation USP10 (Ubiquitin carboxyl-terminal hydrolase 10) [57] (Figure 2).
Figure 2.
Structure of stress granules (SGs) related proteins. SGs related proteins are represented according to domains described in the Uniprot database.
TIA-1 and TIAR have documented roles in immunity, RNA splicing, and translation. Structurally, they bind RNA through RNA Recognition Motifs (RRM) (Figure 2). They are the historical markers for SGs [4,31]. Their overexpression induces the spontaneous formation of SGs [4], and their individual knock-out reduces the cell ability to SGs formation in MEF (mouse embryonic fibroblast) in response to oxidative and thermal stress [58]. Surprisingly, the depletion of TIAR using a doxycyclin-inducible system triggers stress by activation of PKR and SGs induction in 50% of the HEK293 cells [59]. This discrepancy could be explained by the difference in the cell line used and the way to inhibit expression. The knockout is a stable and permanent protein depletion, whereas the doxycyclin system induces a sudden change in the cell environment that could be perceived as a stress for the cell. The role of TIA-1 and TIAR in cancer is still debated, because, in some studies, their depletion accelerates mitotic entry and proliferation [60,61], whereas, in others, they contribute to angiogenesis, tumor growth, and chemoresistance [62]. This discrepancy could be explained by post-translational modifications. For example, in basal conditions, expression of TIA-1 enhances SGs formation, but TIA-1 oxidation results in the opposite effect [41].
The G3BP1 protein also contains an RRM (Figure 2) and has been reported to have helicase and RNAse activity under normal conditions [22,58]. G3BP1 is closely related to another protein, G3BP2, with which it shares 98% identity. G3BP1 and G3BP2 are currently considered as the master regulators of SGs. Their overexpression also induces spontaneous formation of SGs [22,63]. Individual knockout partially inhibits or delays the formation of SGs in response to oxidative, ER or mitochondrial stress [49,57], but the double knock out completely abolishes the formation of SGs in response to oxidative, ER, mitochondrial stress, as well as EIF2α-independent stress [57]. G3BP1/2 have been related to tumor initiation [64], proliferation, [65,66,67], migration [66,67,68], and invasion [66,67] of tumor cells. Those functions are attributed to the RRM and the NTF2 protein domains [65,66], which are also essential for SGs formation [22]. G3BP1 and G3BP2 have been linked to chemo- and radio-resistance [69,70], as their depletions in cell lines increase treatment efficiency. Ultimately, two studies investigated the role of G3BP1 in cancer development in in vivo experiments. In the first one, hepatocellular carcinoma cells were injected into the tail vein to mimic metastatic dissemination by lung colonization [68]. The downregulation of G3BP1 decreased lung colonization, whereas the overexpression of G3BP1 increased lung colonization [68]. In the second study, orthotopic tumor xenografts from ACHN renal carcinoma cell were used and the primary tumor site formation as well as metastasis in the lung and liver were monitored. G3BP1 depletion strongly inhibited both the primary site and the metastasis formation [67].
Finally, USP10 and CAPRIN-1 are two interactors of G3BP1 (Figure 2) that compete with each other to interact with their target [57]. Both bind G3BP1 on a short linear motif Phe-Gly-Asp-Phe (FGDF-motif), and they have opposing effects on SGs formation: CAPRIN-1:G3BP1/2 interaction favors SGs formation, whereas USP10:G3BP1/2 interaction inhibits their formation [57]. CAPRIN-1 overexpression is also reported to induce spontaneous formation of SGs and its inhibition decreased SGs formation in response to oxidative and ER stress as well as in response to EIF2α-independent stress [57]. In contrast, the overexpression of USP10 inhibits SGs formation in response to oxidative, mitochondrial, and EIF2α-independent stress. USP10 depletion does not significantly impact SGs formation in response to oxidative, mitochondrial and EIF2α-independent stress [57]. Those two proteins are not the sole regulators of G3BP1 aggregation, because the removal of the FGDF-motif does not influence the formation of SGs. CAPRIN-1 and USP10 not only have opposite effect on SGs regulation, but also have opposed incidence on cell proliferation and tumor development. CAPRIN-1 enhances proliferation [71,72,73], whereas USP10 inhibits tumor progression and invasion [74,75].
6. SGs Proteins as Prognostic Markers: At the mRNA or Protein Level?
SGs proteins regulator expression level can be used to estimate the capacity of cells composing a tissue to form SGs [22,57,58,76]. In line with this, we analyzed publicly available gene expression data of breast [77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111], colon [112,113,114,115,116,117,118,119,120,121], and pancreatic [122,123,124,125,126,127,128,129,130,131,132,133,134,135] cancers as a proof of concept. mRNA levels for G3BP1, TIA-1, TIAR, and CAPRIN-1 are mostly upregulated (p ≤ 1.07 × 10−4) in primary tumors when compared to healthy tissues (Figures S1–S3). This correlates with observations made in several patient cohorts, where SGs proteins were upregulated in tumor as compared to healthy tissues [64,65,68,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152]. Even if other possibilities exist to explain this phenomenon, we can hypothesize that tumor cells divert and exacerbate a pro-survival mechanism potentially based on SGs, in order to facilitate their survival in response to the numerous stresses encountered. This is true for most primary solid tumors. CAPRIN-1 and G3PB1 mRNA show the most noticeable/prominent upregulation in primary tumors (Figures S1–S3).
The difference in expression between healthy tissues and metastases was not as pronounced as between healthy and primary tumors (Figures S1–S3). For breast and colon cancer, mRNAs encoding SGs proteins level between metastases and primary tumors stay steady or decrease. This might be explained by the fact that tumor cells have already adapted their growth to major stresses or because it involves other regulatory mechanisms for SGs formation in breast an in colon cancer. From the three cancers studied here, pancreatic cancer is the most aggressive and the one with lower survival rate (9% five-year survival, as compared to 91% for breast cancer and 63% for colon cancer according to the American Society of Clinical Oncology). It is also the only one where mRNAs encoding SGs proteins are still increasing between primary tumors and metastatic sites, suggesting that cancer cells are still evolving/adapting toward a more aggressive disease.
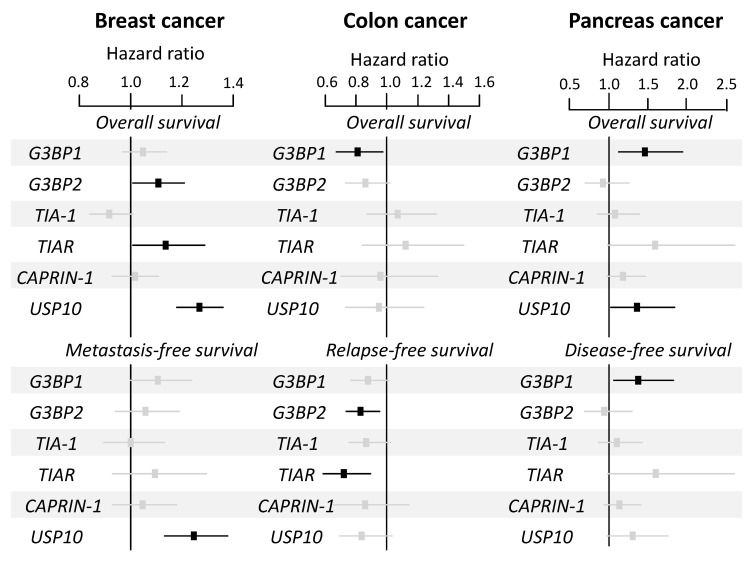
Despite the fact that an increased expression of mRNA encoding for SGs proteins was a marker of tumor transformation (Figure S1–S3), hazard ratio analyses revealed few significant correlations with patients’ survival (Figure 3). This was not consistent with data from the literature showing that SGs proteins upregulation and poor prognosis are often correlated, in multiple cancer types. Indeed, G3BP1/2high proteins expression is linked to poor prognosis in various tumors, including sarcomas [138], breast [136,137], lung [139], stomach [140,141], liver [68], and prostate cancers [142]. High TIA-1 protein levels correlate with poor prognosis in patients with colorectal cancer [143], lymphoma [144], and hepatocellular carcinoma [145]. A high expression of CAPRIN-1 protein correlates with poor prognosis for patients with osteosarcoma [146], and hepatocellular carcinoma [147,148]. Consistent with an inhibitory effect of USP10 on SGs formation, high protein expression correlates with better prognosis in patients with ovarian [150], lung [151], small intestine [152], prostate [142], and stomach carcinoma [149] (Table 1).
Figure 3.
Forest plot showing the hazard ratio for survival events of the mRNA expression of SGs genes according to overall and metastasis-free survival in breast cancer, overall and relapse-free survival in colon cancer, and overall and disease-free survival in pancreatic cancer patients. A ratio greater than one indicates a poor prognosis and a ratio lower than one indicates good prognosis. The black squares correspond to significant genes and the grey ones to non-significant genes. The analysis was performed according to supplementary material 1 and Table S1.
Table 1.
Kaplan–Maier analysis on SGs protein (high vs. low expression). Summary of Kaplan–Meier survival analysis found in the literature [64,65,68,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152]. Data are presented for patients with high expression of the protein compared with patient with low expression of the protein. «Poor» means that patient with high protein expression have a worse prognosis than patients with a lower expression. «Good» means that patients with high protein expression have a better prognosis than patients with a lower expression.
| Cancer Type | G3BP1 | G3BP2 | TIA-1 | CAPRIN-1 | USP10 |
|---|---|---|---|---|---|
| Breast | Poor [136] | Poor [137] | |||
| Colon/Colo-Rectal | Poor [143] | ||||
| Sarcoma | Poor [138] | ||||
| Stomach | Poor [140,141] | Good [149] | |||
| Lung | Poor [139] | Good [151] | |||
| Liver | Poor [68] | Poor [145] | Poor [147,148] | ||
| Prostate | Poor [142] | Good [142] | |||
| Intestinal | Good [152] | ||||
| Ovarian | Good [150] | ||||
| Osteosarcoma | Poor [146] | ||||
| Lymphoma | Poor [144] |
7. Conclusions
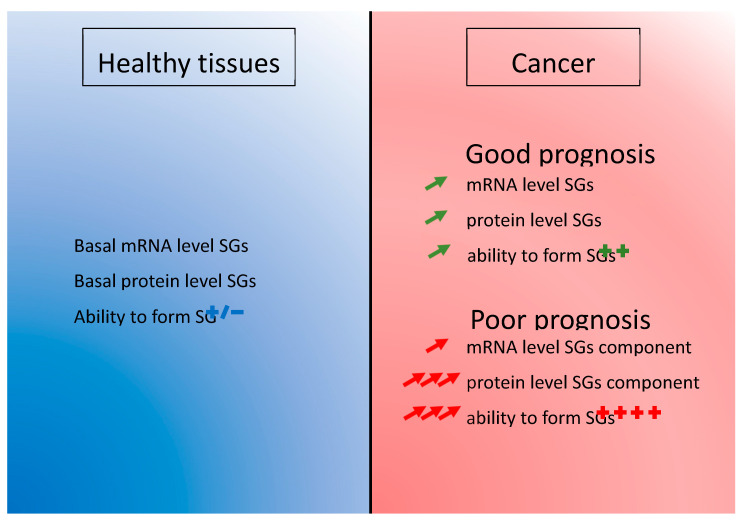
Taken together, the discordant results between mRNA and proteins expression levels regarding the prognostic value suggest (Figure 3, Table 1):
An early increased of mRNA level in the pathology achieved by upregulation of transcription or decrease of RNA degradation. This phenomenon could insure an efficient level of the proteins that are involved in SGs function. This basal level is potentially the result of cancer cells subversion of the SGs mechanism to ensure their survival and it is present in the majority of cancer patients (Figure 4).
A post-transcriptional regulation impacting the final level of proteins. Increasing the translation rate of the mRNA encoding a SGs protein would be more efficient than producing more mRNAs. This could be done by specific signal in the non-translated region of the mRNA such as AU-rich element (ARE) [153] or to the length of the mRNA as it was already described [154]. Favoring translation over transcription is perhaps also a mechanism allowing an economy of energy in cancer cells (Figure 4).
Or a post-translational modification enhancing the half-life of the protein. The degradation of the protein is delayed and it enhances the overall level of protein (Figure 4).
Figure 4.
The two steps regulation of SGs component in cancer. During oncogenesis, there is a global increased expression of mRNA encoding SGs-related proteins. Protein, but not mRNA, expression levels are prognostic for survival, suggesting different layers of regulation for SGs in cancer.
8. Perspectives
The cancer field is still facing the challenge of metastasis and chemoresistance during the disease evolution, despite global survival improvement of cancer patients over the years. Of course, the search for molecular alterations have been successful in the past, contributing to improved tumor classification and the development of efficient targeted therapies. However, some patients do not respond or become refractory to given therapies, meaning that finding treatments that are able to counter drug-induced mechanisms of resistance is the challenge to address. One reason that has limited progress in this field so far is the fact that mechanisms of resistance might only be present during the course of the treatment, and may recess once therapy is stopped. The reversibility of the phenomenon, also termed “plasticity”, makes it difficult to apprehend, since it may not be identifiable in the resected specimen [2]. The mechanisms involved are most probably fast, “acute” and involve “ready-to-use” components, like those of the stress pathways. In this line, the cancer field is accumulating evidence for a role for SGs proteins in the adaptation and survival of cancer cells during tumor growth [64,65,136] and chemoresistance [41,42,43,44,45,46]. All of the SGs proteins that are described in this review have been previously reported to have a role in cancer development and/or cell cycle regulation [60,61,62,64,65,66,67,68,71,72,73,74,75]. These studies collectively suggest that tumor cells have an increased ability to form SGs when compared to non-pathological cells based on their increased expression of SG-related proteins. In addition, prognostic analyses correlate this SGs protein expression with poor survival in patients (Figure 4). The exact mechanism for this increased protein level without transcriptional change is not known yet and could be the result of increased specific translation or decreased protein degradation. Currently, we have no way to distinguish between the roles of these proteins by themselves or as part of the SGs. However, we noticed that all SGs nucleators (TIA-1, TIAR, G3BP1, G3BP2, and CAPRIN-1) have an “enhancing” effect on cancer development properties [62,64,65,66,67,68,71,72,73], whereas USP10, a SGs inhibitor, has anti-cancer properties [74,75] (Figure 5). There are some interesting correlation between other SGs markers and development, such as eIF3s and eIF4s subunits and complexes, UBAP2L or YB1, for example [138,155,156,157,158,159,160]. All of those proteins have, in common, to be recruited to SGs or to have role in their regulation [31,138,161,162]. This collection of evidences points toward a role of SGs in cancer. Meanwhile in vivo experimental evidences supporting a role for SGs in cancer development are accumulating. Limiting the formation of SGs through genetic [67,68] or chemical inhibition [53,163] showed promising results on the inhibition of primary and metastasis formation. Of course, we cannot exclude that SGs proteins have additional and complementary roles to SGs in tumorigenesis. SGs have already been observed in vivo [26,51,164] and the presence of SG-like foci has been reported in in vivo mouse xenografted tumors [138]. Recently, the field is starting to observe SGs in clinical samples from patients with pancreatic adenocarcinoma [165]. While promising, SGs observations in patients are still at the leading edge and need to be reproduced in other kinds of cancer. Overall, SGs might thus turn out to be important actors of tumor cells plasticity in response to the various stresses encountered, which will make them a target of choice in the fight against tumor development, progression, and prevention of chemoresistance. Further studies are warranted in order to understand SGs the mechanism of action in the development of aggressive and invasive cancers and how to block them to improve patient care, especially for refractory patients or in the prevention of chemoresistance.
Figure 5.
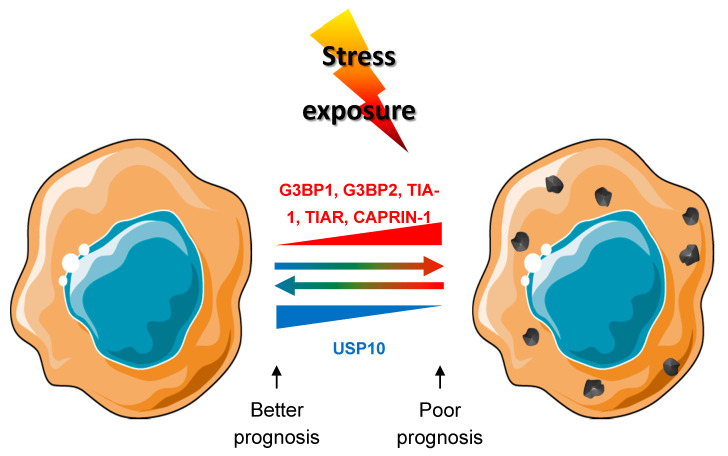
Link between SGs proteins and prognosis in patients. Overexpression of G3BP1, G3BP2, Table 1. TIAR and CAPRIN1 are linked to increased ability to form SG. Whereas increased expression of USP10 decreases the cell ability to form SGs in response to stimuli. Increased expression G3BP1, G3BP2, TIA-1, TIAR and CAPRIN1 proteins are reported as linked to poor prognosis, whereas increased expression of USP10 is a good prognosis for patients.
Acknowledgments
We thank Christine Vande Velde for reviewing the manuscript and providing helpful feedback. We also want to thank the preprint platform for hosting the pre-peer reviewed versions of the paper (https://www.preprints.org/manuscript/202001.0180/v1).
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/9/2470/s1, Figure S1: mRNA expression of SGs genes in breast cancer. SGs genes mRNA expression level (log2) reported as box plots according to normal breast, primary breast cancer, and breast metastatic samples. The p-value reported for each comparison was defined by Tukey’s range test. Figure S2: mRNA expression of SGs genes in colon cancer. SGs genes mRNA expression level (log2) reported as box plots according to normal colon, primary breast cancer, and breast metastatic samples. The p-value reported for each comparison was defined by Tukey’s range test. Figure S3: mRNA expression of SGs genes in pancreatic cancer. SGs genes mRNA expression level (log2) reported as box plots according to normal pancreas, primary breast cancer, and breast metastatic samples. The p-value reported for each comparison was defined by Tukey’s range test, Table S1: List of data sets included in the study.
Author Contributions
A.A. and E.M. had the original idea. P.F. and F.B. performed the bioinformatics analysis of the open data bases. A.A., P.F. and S.M.L. did the figures. A.A. did the literature overview. A.A., S.M.L., F.B., D.B., C.A. and E.M. wrote or corrected the review. All authors have read and agreed to the published version of the manuscript.
Funding
A.A. is supported by a postdoctoral fellowship from the Fondation ARC (n°PDF20180507565) and the Fondation de France (n°00107936). This work has been supported by Inserm, Institut Paoli-Calmettes (SIRIC INCa-DGOS-Inserm 6038) and grants from the Ligue Nationale Contre Le Cancer (EL2016.LNCC/DaB–EL2019.LNCC/FB), Association Ruban Rose, and Fondation Groupe EDF (Project Nano-S) all to EM. This work was also funded by grants from the United States NIH (n°GM124458) to SML.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- 1.Norouzi S., Gorgi Valokala M., Mosaffa F., Zirak M.R., Zamani P., Behravan J. Crosstalk in cancer resistance and metastasis. Crit. Rev. Oncol. Hematol. 2018;132:145–153. doi: 10.1016/j.critrevonc.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 2.Echeverria G.V., Ge Z., Seth S., Zhang X., Jeter-Jones S., Zhou X., Cai S., Tu Y., McCoy A., Peoples M., et al. Resistance to neoadjuvant chemotherapy in triple-negative breast cancer mediated by a reversible drug-tolerant state. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aav0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J.Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kedersha N., Ivanov P., Anderson P. Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hetz C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 7.Adams C.J., Kopp M.C., Larburu N., Nowak P.R., Ali M.M.U. Structure and Molecular Mechanism of ER Stress Signaling by the Unfolded Protein Response Signal Activator IRE1. Front. Mol. Biosci. 2019;6:11. doi: 10.3389/fmolb.2019.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulda S., Gorman A.M., Hori O., Samali A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010;2010:214074. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aulas A., Fay M.M., Lyons S.M., Achorn C.A., Kedersha N., Anderson P., Ivanov P. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 2017 doi: 10.1242/jcs.199240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moeller B.J., Cao Y., Li C.Y., Dewhirst M.W. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–441. doi: 10.1016/S1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 11.Kaehler C., Isensee J., Hucho T., Lehrach H., Krobitsch S. 5-Fluorouracil affects assembly of stress granules based on RNA incorporation. Nucl. Acids Res. 2014;42:6436–6447. doi: 10.1093/nar/gku264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimura K., Sasaki A.T., Anderson P. Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucl. Acids Res. 2012;40:8099–8110. doi: 10.1093/nar/gks566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adjibade P., St-Sauveur V.G., Quevillon Huberdeau M., Fournier M.J., Savard A., Coudert L., Khandjian E.W., Mazroui R. Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells. Oncotarget. 2015;6:43927–43943. doi: 10.18632/oncotarget.5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier M.J., Gareau C., Mazroui R. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int. 2010;10:12. doi: 10.1186/1475-2867-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szaflarski W., Fay M.M., Kedersha N., Zabel M., Anderson P., Ivanov P. Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget. 2016;7:30307–30322. doi: 10.18632/oncotarget.8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilas-Boas Fde A., da Silva A.M., de Sousa L.P., Lima K.M., Vago J.P., Bittencourt L.F., Dantas A.E., Gomes D.A., Vilela M.C., Teixeira M.M., et al. Impairment of stress granule assembly via inhibition of the eIF2alpha phosphorylation sensitizes glioma cells to chemotherapeutic agents. J. Neurooncol. 2016;127:253–260. doi: 10.1007/s11060-015-2043-3. [DOI] [PubMed] [Google Scholar]
- 17.Kedersha N., Anderson P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 18.Timalsina S., Arimoto-Matsuzaki K., Kitamura M., Xu X., Wenzhe Q., Ishigami-Yuasa M., Kagechika H., Hata Y. Chemical compounds that suppress hypoxia-induced stress granule formation enhance cancer drug sensitivity of human cervical cancer HeLa cells. J. Biochem. 2018;164:381–391. doi: 10.1093/jb/mvy062. [DOI] [PubMed] [Google Scholar]
- 19.Somasekharan S.P., Zhang F., Saxena N., Huang J.N., Kuo I.C., Low C., Bell R., Adomat H., Stoynov N., Foster L., et al. G3BP1-linked mRNA partitioning supports selective protein synthesis in response to oxidative stress. Nucl. Acids Res. 2020 doi: 10.1093/nar/gkaa376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tourriere H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song M.S., Grabocka E. Stress Granules in Cancer. Rev. Physiol. Biochem. Pharmacol. 2020 doi: 10.1007/112_2020_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nover L., Scharf K.D., Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol. Cell. Biol. 1983;3:1648–1655. doi: 10.1128/MCB.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangiardi D.A., McLaughlin-Williamson K., May K.E., Messana E.P., Mountain D.C., Cotanche D.A. Progression of hair cell ejection and molecular markers of apoptosis in the avian cochlea following gentamicin treatment. J. Comp. Neurol. 2004;475:1–18. doi: 10.1002/cne.20129. [DOI] [PubMed] [Google Scholar]
- 27.Anderson E.N., Gochenaur L., Singh A., Grant R., Patel K., Watkins S., Wu J.Y., Pandey U.B. Traumatic injury induces Stress Granule Formation and enhances Motor Dysfunctions in ALS/FTD Models. Hum. Mol. Genet. 2018 doi: 10.1093/hmg/ddy047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scharf K.D., Heider H., Hohfeld I., Lyck R., Schmidt E., Nover L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell Biol. 1998;18:2240–2251. doi: 10.1128/MCB.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., Kwon S.H., Su Y., Dong Z. Stress granules are formed in renal proximal tubular cells during metabolic stress and ischemic injury for cell survival. Am. J. Physiol. Ren. Physiol. 2019;317:F116–F123. doi: 10.1152/ajprenal.00139.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souquere S., Mollet S., Kress M., Dautry F., Pierron G., Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 2009;122:3619–3626. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- 31.Aulas A., Vande Velde C. Alterations in stress granule dynamics driven by TDP-43 and FUS: A link to pathological inclusions in ALS? Front. Cell Neurosci. 2015;9:423. doi: 10.3389/fncel.2015.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheeler J.R., Jain S., Khong A., Parker R. Isolation of yeast and mammalian stress granule cores. Methods. 2017;126:12–17. doi: 10.1016/j.ymeth.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.C., Krach F., Yang D., Sen A., et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell. 2018;172:590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adivarahan S., Livingston N., Nicholson B., Rahman S., Wu B., Rissland O.S., Zenklusen D. Spatial Organization of Single mRNPs at Different Stages of the Gene Expression Pathway. Mol. Cell. 2018;72:727–738.e5. doi: 10.1016/j.molcel.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazroui R., Sukarieh R., Bordeleau M.E., Kaufman R.J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.e06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang Y., Kedersha N., Low W.K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J.O. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 37.Aulas A., Lyons S.M., Fay M.M., Anderson P., Ivanov P. Nitric oxide triggers the assembly of “type II” stress granules linked to decreased cell viability. Cell Death Dis. 2018;9:1129. doi: 10.1038/s41419-018-1173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald K.K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G.A., Vande Velde C. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 2011;20:1400–1410. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi M., Higuchi M., Matsuki H., Yoshita M., Ohsawa T., Oie M., Fujii M. Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol. Cell. Biol. 2013;33:815–829. doi: 10.1128/MCB.00763-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maharjan N., Kunzli C., Buthey K., Saxena S. C9ORF72 Regulates Stress Granule Formation and Its Deficiency Impairs Stress Granule Assembly, Hypersensitizing Cells to Stress. Mol. Neurobiol. 2017;54:3062–3077. doi: 10.1007/s12035-016-9850-1. [DOI] [PubMed] [Google Scholar]
- 41.Arimoto-Matsuzaki K., Saito H., Takekawa M. TIA1 oxidation inhibits stress granule assembly and sensitizes cells to stress-induced apoptosis. Nat. Commun. 2016;7:10252. doi: 10.1038/ncomms10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orru S., Coni P., Floris A., Littera R., Carcassi C., Sogos V., Brancia C. Reduced stress granule formation and cell death in fibroblasts with the A382T mutation of TARDBP gene: Evidence for loss of TDP-43 nuclear function. Hum. Mol. Genet. 2016;25:4473–4483. doi: 10.1093/hmg/ddw276. [DOI] [PubMed] [Google Scholar]
- 43.Aulas A., Stabile S., Vande Velde C. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol. Neurodegener. 2012;7:54. doi: 10.1186/1750-1326-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aulas A., Stabile S., Vande Velde C. Erratum to: Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol. Neurodegener. 2015;10:45. doi: 10.1186/s13024-015-0041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eisinger-Mathason T.S., Andrade J., Groehler A.L., Clark D.E., Muratore-Schroeder T.L., Pasic L., Smith J.A., Shabanowitz J., Hunt D.F., Macara I.G., et al. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell. 2008;31:722–736. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 47.Kim W.J., Back S.H., Kim V., Ryu I., Jang S.K. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cellul. Biol. 2005;25:2450–2462. doi: 10.1128/MCB.25.6.2450-2462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thedieck K., Holzwarth B., Prentzell M.T., Boehlke C., Klasener K., Ruf S., Sonntag A.G., Maerz L., Grellscheid S.N., Kremmer E., et al. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell. 2013;154:859–874. doi: 10.1016/j.cell.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 49.Aulas A., Caron G., Gkogkas C.G., Mohamed N.V., Destroismaisons L., Sonenberg N., Leclerc N., Parker J.A., Vande Velde C. G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J. Cell Biol. 2015;209:73–84. doi: 10.1083/jcb.201408092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gareau C., Fournier M.J., Filion C., Coudert L., Martel D., Labelle Y., Mazroui R. p21(WAF1/CIP1) upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. PLoS ONE. 2011;6:e20254. doi: 10.1371/journal.pone.0020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goncalves A.C., Towers E.R., Haq N., Porco J.A., Jr., Pelletier J., Dawson S.J., Gale J.E. Drug-induced Stress Granule Formation Protects Sensory Hair Cells in Mouse Cochlear Explants During Ototoxicity. Sci. Rep. 2019;9:12501. doi: 10.1038/s41598-019-48393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adjibade P., Mazroui R. Control of mRNA turnover: Implication of cytoplasmic RNA granules. Semin. Cell Dev. Biol. 2014 doi: 10.1016/j.semcdb.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen H.G., Conn C.S., Kye Y., Xue L., Forester C.M., Cowan J.E., Hsieh A.C., Cunningham J.T., Truillet C., Tameire F., et al. Development of a stress response therapy targeting aggressive prostate cancer. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aar2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quail D.F., Taylor M.J., Walsh L.A., Dieters-Castator D., Das P., Jewer M., Zhang G., Postovit L.M. Low oxygen levels induce the expression of the embryonic morphogen Nodal. Mol. Biol. Cell. 2011;22:4809–4821. doi: 10.1091/mbc.e11-03-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conley S.J., Gheordunescu E., Kakarala P., Newman B., Korkaya H., Heath A.N., Clouthier S.G., Wicha M.S. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc. Natl. Acad Sci. USA. 2012;109:2784–2789. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jewer M.L., Zhang G., Liu J., Findlay S., Vincent K., Tandoc K., Dieters-Castator D., Quail D., Dutta I., Coatham M., et al. Translational control of breast cancer plasticity. BioRxviv. 2019 doi: 10.1038/s41467-020-16352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G.M., Ivanov P., et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.e04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer C., Garzia A., Mazzola M., Gerstberger S., Molina H., Tuschl T. The TIA1 RNA-Binding Protein Family Regulates EIF2AK2-Mediated Stress Response and Cell Cycle Progression. Mol. Cell. 2018;69:622–635.e6. doi: 10.1016/j.molcel.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez-Jimenez C., Ludena M.D., Izquierdo J.M. T-cell intracellular antigens function as tumor suppressor genes. Cell Death Dis. 2015;6:e1669. doi: 10.1038/cddis.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lafarga V., Sung H.M., Haneke K., Roessig L., Pauleau A.L., Bruer M., Rodriguez-Acebes S., Lopez-Contreras A.J., Gruss O.J., Erhardt S., et al. TIAR marks nuclear G2/M transition granules and restricts CDK1 activity under replication stress. EMBO Rep. 2019;20 doi: 10.15252/embr.201846224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamdollah Zadeh M.A., Amin E.M., Hoareau-Aveilla C., Domingo E., Symonds K.E., Ye X., Heesom K.J., Salmon A., D’Silva O., Betteridge K.B., et al. Alternative splicing of TIA-1 in human colon cancer regulates VEGF isoform expression, angiogenesis, tumour growth and bevacizumab resistance. Mol. Oncol. 2015;9:167–178. doi: 10.1016/j.molonc.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsuki H., Takahashi M., Higuchi M., Makokha G.N., Oie M., Fujii M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells. 2013;18:135–146. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- 64.Gupta N., Badeaux M., Liu Y., Naxerova K., Sgroi D., Munn L.L., Jain R.K., Garkavtsev I. Stress granule-associated protein G3BP2 regulates breast tumor initiation. Proc. Natl. Acad. Sci. USA. 2017;114:1033–1038. doi: 10.1073/pnas.1525387114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guitard E., Parker F., Millon R., Abecassis J., Tocque B. G3BP is overexpressed in human tumors and promotes S phase entry. Cancer Lett. 2001;162:213–221. doi: 10.1016/S0304-3835(00)00638-8. [DOI] [PubMed] [Google Scholar]
- 66.Taniuchi K., Nishimori I., Hollingsworth M.A. The N-terminal domain of G3BP enhances cell motility and invasion by posttranscriptional regulation of BART. Mol. Cancer Res. MCR. 2011;9:856–866. doi: 10.1158/1541-7786.MCR-10-0574. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y., Fu D., Chen Y., Su J., Wang Y., Li X., Zhai W., Niu Y., Yue D., Geng H. G3BP1 promotes tumor progression and metastasis through IL-6/G3BP1/STAT3 signaling axis in renal cell carcinomas. Cell Death Dis. 2018;9:501. doi: 10.1038/s41419-018-0504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dou N., Chen J., Yu S., Gao Y., Li Y. G3BP1 contributes to tumor metastasis via upregulation of Slug expression in hepatocellular carcinoma. Am. J. Cancer Res. 2016;6:2641–2650. [PMC free article] [PubMed] [Google Scholar]
- 69.Cho E., Than T.T., Kim S.H., Park E.R., Kim M.Y., Lee K.H., Shin H.J. G3BP1 Depletion Increases Radiosensitisation by Inducing Oxidative Stress in Response to DNA Damage. Anticancer. Res. 2019;39:6087–6095. doi: 10.21873/anticanres.13816. [DOI] [PubMed] [Google Scholar]
- 70.Heijink A.M., Everts M., Honeywell M.E., Richards R., Kok Y.P., de Vries E.G.E., Lee M.J., van Vugt M. Modeling of Cisplatin-Induced Signaling Dynamics in Triple-Negative Breast Cancer Cells Reveals Mediators of Sensitivity. Cell Rep. 2019;28:2345–2357.e5. doi: 10.1016/j.celrep.2019.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang B., David M.D., Schrader J.W. Absence of caprin-1 results in defects in cellular proliferation. J. Immunol. 2005;175:4274–4282. doi: 10.4049/jimmunol.175.7.4274. [DOI] [PubMed] [Google Scholar]
- 72.Gong B., Hu H., Chen J., Cao S., Yu J., Xue J., Chen F., Cai Y., He H., Zhang L. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed. Pharmacother. 2013;67:629–636. doi: 10.1016/j.biopha.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 73.Yang Z.S., Qing H., Gui H., Luo J., Dai L.J., Wang B. Role of caprin-1 in carcinogenesis. Oncol. Lett. 2019;18:15–21. doi: 10.3892/ol.2019.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun J., Li T., Zhao Y., Huang L., Sun H., Wu H., Jiang X. USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Mol. Cell Biochem. 2018;441:1–7. doi: 10.1007/s11010-017-3170-2. [DOI] [PubMed] [Google Scholar]
- 75.Lu C., Ning Z., Wang A., Chen D., Liu X., Xia T., Tekcham D.S., Wang W., Li T., Liu X., et al. USP10 suppresses tumor progression by inhibiting mTOR activation in hepatocellular carcinoma. Cancer lett. 2018;436:139–148. doi: 10.1016/j.canlet.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 76.Lee A.K., Klein J., Fon Tacer K., Lord T., Oatley M.J., Oatley J.M., Porter S.N., Pruett-Miller S.M., Tikhonova E.B., Karamyshev A.L., et al. Translational Repression of G3BP in Cancer and Germ Cells Suppresses Stress Granules and Enhances Stress Tolerance. Mol. Cell. 2020 doi: 10.1016/j.molcel.2020.06.037. [DOI] [PubMed] [Google Scholar]
- 77.van de Vijver M.J., He Y.D., van’t Veer L.J., Dai H., Hart A.A., Voskuil D.W., Schreiber G.J., Peterse J.L., Roberts C., Marton M.J., et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 78.van’t Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 79.Farmer P., Bonnefoi H., Becette V., Tubiana-Hulin M., Fumoleau P., Larsimont D., Macgrogan G., Bergh J., Cameron D., Goldstein D., et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 80.Minn A.J., Gupta G.P., Siegel P.M., Bos P.D., Shu W., Giri D.D., Viale A., Olshen A.B., Gerald W.L., Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y., Klijn J.G., Zhang Y., Sieuwerts A.M., Look M.P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M.E., Yu J., et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 82.Hess K.R., Anderson K., Symmans W.F., Valero V., Ibrahim N., Mejia J.A., Booser D., Theriault R.L., Buzdar A.U., Dempsey P.J., et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J. Clin. Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 83.Ivshina A.V., George J., Senko O., Mow B., Putti T.C., Smeds J., Lindahl T., Pawitan Y., Hall P., Nordgren H., et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 84.Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B., et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 85.Bonnefoi H., Potti A., Delorenzi M., Mauriac L., Campone M., Tubiana-Hulin M., Petit T., Rouanet P., Jassem J., Blot E., et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: A substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8:1071–1078. doi: 10.1016/S1470-2045(07)70345-5. [DOI] [PubMed] [Google Scholar]
- 86.Desmedt C., Piette F., Loi S., Wang Y., Lallemand F., Haibe-Kains B., Viale G., Delorenzi M., Zhang Y., d’Assignies M.S., et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 87.Miller W.R., Larionov A. Changes in expression of oestrogen regulated and proliferation genes with neoadjuvant treatment highlight heterogeneity of clinical resistance to the aromatase inhibitor, letrozole. Breast Cancer Res. 2010;12:R52. doi: 10.1186/bcr2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klein A., Wessel R., Graessmann M., Jurgens M., Petersen I., Schmutzler R., Niederacher D., Arnold N., Meindl A., Scherneck S., et al. Comparison of gene expression data from human and mouse breast cancers: Identification of a conserved breast tumor gene set. Int. J. Cancer. 2007;121:683–688. doi: 10.1002/ijc.22630. [DOI] [PubMed] [Google Scholar]
- 89.Bos P.D., Zhang X.H., Nadal C., Shu W., Gomis R.R., Nguyen D.X., Minn A.J., van de Vijver M.J., Gerald W.L., Foekens J.A., et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoeflich K.P., O’Brien C., Boyd Z., Cavet G., Guerrero S., Jung K., Januario T., Savage H., Punnoose E., Truong T., et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin. Cancer Res. 2009;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 91.Marty B., Maire V., Gravier E., Rigaill G., Vincent-Salomon A., Kappler M., Lebigot I., Djelti F., Tourdes A., Gestraud P., et al. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res. 2008;10:R101. doi: 10.1186/bcr2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Merritt W.M., Lin Y.G., Han L.Y., Kamat A.A., Spannuth W.A., Schmandt R., Urbauer D., Pennacchio L.A., Cheng J.F., Nick A.M., et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmidt M., Bohm D., von Torne C., Steiner E., Puhl A., Pilch H., Lehr H.A., Hengstler J.G., Kolbl H., Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 94.Yu K., Ganesan K., Tan L.K., Laban M., Wu J., Zhao X.D., Li H., Leung C.H., Zhu Y., Wei C.L., et al. A precisely regulated gene expression cassette potently modulates metastasis and survival in multiple solid cancers. PLoS Genet. 2008;4:e1000129. doi: 10.1371/journal.pgen.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barry W.T., Kernagis D.N., Dressman H.K., Griffis R.J., Hunter J.D., Olson J.A., Marks J.R., Ginsburg G.S., Marcom P.K., Nevins J.R., et al. Intratumor heterogeneity and precision of microarray-based predictors of breast cancer biology and clinical outcome. J. Clin. Oncol. 2010;28:2198–2206. doi: 10.1200/JCO.2009.26.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iwamoto T., Bianchini G., Booser D., Qi Y., Coutant C., Shiang C.Y., Santarpia L., Matsuoka J., Hortobagyi G.N., Symmans W.F., et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J. Natl. Cancer Inst. 2011;103:264–272. doi: 10.1093/jnci/djq524. [DOI] [PubMed] [Google Scholar]
- 97.Korde L.A., Lusa L., McShane L., Lebowitz P.F., Lukes L., Camphausen K., Parker J.S., Swain S.M., Hunter K., Zujewski J.A. Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer. Breast Cancer Res. Treat. 2010;119:685–699. doi: 10.1007/s10549-009-0651-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Prat A., Parker J.S., Karginova O., Fan C., Livasy C., Herschkowitz J.I., He X., Perou C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Silver D.P., Richardson A.L., Eklund A.C., Wang Z.C., Szallasi Z., Li Q., Juul N., Leong C.O., Calogrias D., Buraimoh A., et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J. Clin. Oncol. 2010;28:1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tabchy A., Valero V., Vidaurre T., Lluch A., Gomez H., Martin M., Qi Y., Barajas-Figueroa L.J., Souchon E., Coutant C., et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clin. Cancer Res. 2010;16:5351–5361. doi: 10.1158/1078-0432.CCR-10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jonsson S., Olsson B., Jacobsson S., Palmqvist L., Ricksten A., Ekeland-Sjoberg K., Wadenvik H. BCR-ABL1 transcript levels increase in peripheral blood but not in granulocytes after physical exercise in patients with chronic myeloid leukemia. Scand. J. Clin. Lab. Investig. 2011;71:7–11. doi: 10.3109/00365513.2010.521981. [DOI] [PubMed] [Google Scholar]
- 102.Desmedt C., Di Leo A., de Azambuja E., Larsimont D., Haibe-Kains B., Selleslags J., Delaloge S., Duhem C., Kains J.P., Carly B., et al. Multifactorial approach to predicting resistance to anthracyclines. J. Clin. Oncol. 2011;29:1578–1586. doi: 10.1200/JCO.2010.31.2231. [DOI] [PubMed] [Google Scholar]
- 103.Guedj M., Marisa L., de Reynies A., Orsetti B., Schiappa R., Bibeau F., MacGrogan G., Lerebours F., Finetti P., Longy M., et al. A refined molecular taxonomy of breast cancer. Oncogene. 2012;31:1196–1206. doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hatzis C., Pusztai L., Valero V., Booser D.J., Esserman L., Lluch A., Vidaurre T., Holmes F., Souchon E., Wang H., et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Popovici V., Chen W., Gallas B.G., Hatzis C., Shi W., Samuelson F.W., Nikolsky Y., Tsyganova M., Ishkin A., Nikolskaya T., et al. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010;12:R5. doi: 10.1186/bcr2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ellis M.J., Ding L., Shen D., Luo J., Suman V.J., Wallis J.W., Van Tine B.A., Hoog J., Goiffon R.J., Goldstein T.C., et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sabatier R., Finetti P., Adelaide J., Guille A., Borg J.P., Chaffanet M., Lane L., Birnbaum D., Bertucci F. Down-regulation of ECRG4, a candidate tumor suppressor gene, in human breast cancer. PLoS ONE. 2011;6:e27656. doi: 10.1371/journal.pone.0027656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang Y., Sieuwerts A.M., McGreevy M., Casey G., Cufer T., Paradiso A., Harbeck N., Span P.N., Hicks D.G., Crowe J., et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res. Treat. 2009;116:303–309. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]
- 110.Chen D.T., Nasir A., Culhane A., Venkataramu C., Fulp W., Rubio R., Wang T., Agrawal D., McCarthy S.M., Gruidl M., et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res. Treat. 2010;119:335–346. doi: 10.1007/s10549-009-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jorissen R.N., Gibbs P., Christie M., Prakash S., Lipton L., Desai J., Kerr D., Aaltonen L.A., Arango D., Kruhoffer M., et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin. Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sheffer M., Bacolod M.D., Zuk O., Giardina S.F., Pincas H., Barany F., Paty P.B., Gerald W.L., Notterman D.A., Domany E. Association of survival and disease progression with chromosomal instability: A genomic exploration of colorectal cancer. Proc. Natl. Acad. Sci. USA. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Staub E., Groene J., Heinze M., Mennerich D., Roepcke S., Klaman I., Hinzmann B., Castanos-Velez E., Pilarsky C., Mann B., et al. An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types. J. Mol. Med. 2009;87:633–644. doi: 10.1007/s00109-009-0467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Smith J.J., Deane N.G., Wu F., Merchant N.B., Zhang B., Jiang A., Lu P., Johnson J.C., Schmidt C., Bailey C.E., et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kennedy R.D., Bylesjo M., Kerr P., Davison T., Black J.M., Kay E.W., Holt R.J., Proutski V., Ahdesmaki M., Farztdinov V., et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J. Clin. Oncol. 2011;29:4620–4626. doi: 10.1200/JCO.2011.35.4498. [DOI] [PubMed] [Google Scholar]
- 117.Sveen A., Agesen T.H., Nesbakken A., Rognum T.O., Lothe R.A., Skotheim R.I. Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival. Genome Med. 2011;3:32. doi: 10.1186/gm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Laibe S., Lagarde A., Ferrari A., Monges G., Birnbaum D., Olschwang S., Project C.O.L. A seven-gene signature aggregates a subgroup of stage II colon cancers with stage III. OMICS. 2012;16:560–565. doi: 10.1089/omi.2012.0039. [DOI] [PubMed] [Google Scholar]
- 119.Marisa L., de Reynies A., Duval A., Selves J., Gaub M.P., Vescovo L., Etienne-Grimaldi M.C., Schiappa R., Guenot D., Ayadi M., et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Sousa E.M.F., Colak S., Buikhuisen J., Koster J., Cameron K., de Jong J.H., Tuynman J.B., Prasetyanti P.R., Fessler E., van den Bergh S.P., et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9:476–485. doi: 10.1016/j.stem.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 121.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Badea R., Socaciu M., Lupsor M., Mosteanu O., Pop T. Evaluating the liver tumors using three-dimensional ultrasonography. A pictorial essay. J. Gastrointestin Liver Dis. 2007;16:85–92. [PubMed] [Google Scholar]
- 123.Van den Broeck A., Vankelecom H., Van Eijsden R., Govaere O., Topal B. Molecular markers associated with outcome and metastasis in human pancreatic cancer. J. Exp. Clin. Cancer Res. 2012;31:68. doi: 10.1186/1756-9966-31-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lunardi S., Jamieson N.B., Lim S.Y., Griffiths K.L., Carvalho-Gaspar M., Al-Assar O., Yameen S., Carter R.C., McKay C.J., Spoletini G., et al. IP-10/CXCL10 induction in human pancreatic cancer stroma influences lymphocytes recruitment and correlates with poor survival. Oncotarget. 2014;5:11064–11080. doi: 10.18632/oncotarget.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park M., Kim M., Hwang D., Park M., Kim W.K., Kim S.K., Shin J., Park E.S., Kang C.M., Paik Y.K., et al. Characterization of gene expression and activated signaling pathways in solid-pseudopapillary neoplasm of pancreas. Mod. Pathol. 2014;27:580–593. doi: 10.1038/modpathol.2013.154. [DOI] [PubMed] [Google Scholar]
- 126.Grutzmann R., Saeger H.D., Luttges J., Schackert H.K., Kalthoff H., Kloppel G., Pilarsky C. Microarray-based gene expression profiling in pancreatic ductal carcinoma: Status quo and perspectives. Int. J. Colorectal Dis. 2004;19:401–413. doi: 10.1007/s00384-003-0563-3. [DOI] [PubMed] [Google Scholar]
- 127.Monzon F.A., Lyons-Weiler M., Buturovic L.J., Rigl C.T., Henner W.D., Sciulli C., Dumur C.I., Medeiros F., Anderson G.G. Multicenter validation of a 1,550-gene expression profile for identification of tumor tissue of origin. J. Clin. Oncol. 2009;27:2503–2508. doi: 10.1200/JCO.2008.17.9762. [DOI] [PubMed] [Google Scholar]
- 128.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., Miller D.K., Christ A.N., Bruxner T.J., Quinn M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 129.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., Cooc J., Weinkle J., Kim G.E., Jakkula L., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kirby M.K., Ramaker R.C., Gertz J., Davis N.S., Johnston B.E., Oliver P.G., Sexton K.C., Greeno E.W., Christein J.D., Heslin M.J., et al. RNA sequencing of pancreatic adenocarcinoma tumors yields novel expression patterns associated with long-term survival and reveals a role for ANGPTL4. Mol. Oncol. 2016;10:1169–1182. doi: 10.1016/j.molonc.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang G., Schetter A., He P., Funamizu N., Gaedcke J., Ghadimi B.M., Ried T., Hassan R., Yfantis H.G., Lee D.H., et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS ONE. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Winter C., Kristiansen G., Kersting S., Roy J., Aust D., Knosel T., Rummele P., Jahnke B., Hentrich V., Ruckert F., et al. Google goes cancer: Improving outcome prediction for cancer patients by network-based ranking of marker genes. PLoS Comput. Biol. 2012;8:e1002511. doi: 10.1371/journal.pcbi.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cancer Genome Atlas Research Network Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stratford J.K., Bentrem D.J., Anderson J.M., Fan C., Volmar K.A., Marron J.S., Routh E.D., Caskey L.S., Samuel J.C., Der C.J., et al. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 2010;7:e1000307. doi: 10.1371/journal.pmed.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen D.T., Davis-Yadley A.H., Huang P.Y., Husain K., Centeno B.A., Permuth-Wey J., Pimiento J.M., Malafa M. Prognostic Fifteen-Gene Signature for Early Stage Pancreatic Ductal Adenocarcinoma. PLoS ONE. 2015;10:e0133562. doi: 10.1371/journal.pone.0133562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.French J., Stirling R., Walsh M., Kennedy H.D. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem. J. 2002;34:223–231. doi: 10.1023/A:1021737413055. [DOI] [PubMed] [Google Scholar]
- 137.Wei S.C., Fattet L., Tsai J.H., Guo Y., Pai V.H., Majeski H.E., Chen A.C., Sah R.L., Taylor S.S., Engler A.J., et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 2015;17:678–688. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Somasekharan S.P., El-Naggar A., Leprivier G., Cheng H., Hajee S., Grunewald T.G., Zhang F., Ng T., Delattre O., Evdokimova V., et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J. Cell Biol. 2015;208:913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng H., Zhan Y., Zhang Y., Liu S., Lu J., Yang Y., Wen Q., Fan S. Elevated expression of G3BP1 associates with YB1 and p-AKT and predicts poor prognosis in nonsmall cell lung cancer patients after surgical resection. Cancer Med. 2019;8:6894–6903. doi: 10.1002/cam4.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Min L., Ruan Y., Shen Z., Jia D., Wang X., Zhao J., Sun Y., Gu J. Overexpression of Ras-GTPase-activating protein SH3 domain-binding protein 1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2015;67:677–688. doi: 10.1111/his.12695. [DOI] [PubMed] [Google Scholar]
- 141.Xiong R., Gao J.L., Yin T. G3BP1 activates the TGF-beta/Smad signaling pathway to promote gastric cancer. Onco Targets Ther. 2019;12:7149–7156. doi: 10.2147/OTT.S213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Takayama K.I., Suzuki T., Fujimura T., Takahashi S., Inoue S. Association of USP10 with G3BP2 Inhibits p53 Signaling and Contributes to Poor Outcome in Prostate Cancer. Mol. Cancer Res. MCR. 2018;16:846–856. doi: 10.1158/1541-7786.MCR-17-0471. [DOI] [PubMed] [Google Scholar]
- 143.Zlobec I., Karamitopoulou E., Terracciano L., Piscuoglio S., Iezzi G., Muraro M.G., Spagnoli G., Baker K., Tzankov A., Lugli A. TIA-1 cytotoxic granule-associated RNA binding protein improves the prognostic performance of CD8 in mismatch repair-proficient colorectal cancer. PLoS ONE. 2010;5:e14282. doi: 10.1371/journal.pone.0014282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hasselblom S., Sigurdadottir M., Hansson U., Nilsson-Ehle H., Ridell B., Andersson P.O. The number of tumour-infiltrating TIA-1+ cytotoxic T cells but not FOXP3+ regulatory T cells predicts outcome in diffuse large B-cell lymphoma. Br. J. Haematol. 2007;137:364–373. doi: 10.1111/j.1365-2141.2007.06593.x. [DOI] [PubMed] [Google Scholar]
- 145.Tak H., Kang H., Ji E., Hong Y., Kim W., Lee E.K. Potential use of TIA-1, MFF, microRNA-200a-3p, and microRNA-27 as a novel marker for hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2018;497:1117–1122. doi: 10.1016/j.bbrc.2018.02.189. [DOI] [PubMed] [Google Scholar]
- 146.Sabile A.A., Arlt M.J., Muff R., Husmann K., Hess D., Bertz J., Langsam B., Aemisegger C., Ziegler U., Born W., et al. Caprin-1, a novel Cyr61-interacting protein, promotes osteosarcoma tumor growth and lung metastasis in mice. Biochim. Biophys. Acta. 2013;1832:1173–1182. doi: 10.1016/j.bbadis.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 147.Tan N., Dai L., Liu X., Pan G., Chen H., Huang J., Xu Q. Upregulation of caprin1 expression is associated with poor prognosis in hepatocellular carcinoma. Pathol. Res. Pract. 2017;213:1563–1567. doi: 10.1016/j.prp.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 148.Zhang Y., You W., Zhou H., Chen Z., Han G., Zuo X., Zhang L., Wu J., Wang X. Downregulated miR-621 promotes cell proliferation via targeting CAPRIN1 in hepatocellular carcinoma. Am. J. Cancer Res. 2018;8:2116–2129. [PMC free article] [PubMed] [Google Scholar]
- 149.Zeng Z., Wu H.X., Zhan N., Huang Y.B., Wang Z.S., Yang G.F., Wang P., Fu G.H. Prognostic significance of USP10 as a tumor-associated marker in gastric carcinoma. Tumour Biol. 2014;35:3845–3853. doi: 10.1007/s13277-013-1509-1. [DOI] [PubMed] [Google Scholar]
- 150.Han G.H., Chay D.B., Yi J.M., Cho H., Chung J.Y., Kim J.H. Loss of Both USP10 and p14ARF Protein Expression Is an Independent Prognostic Biomarker for Poor Prognosis in Patients With Epithelial Ovarian Cancer. Cancer Genom. Proteom. 2019;16:553–562. doi: 10.21873/cgp.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zeng Z., Li D., Yu T., Huang Y., Yan H., Gu L., Yuan J. Association and clinical implication of the USP10 and MSH2 proteins in non-small cell lung cancer. Oncol. Lett. 2019;17:1128–1138. doi: 10.3892/ol.2018.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Song J.S., Yi J.M., Cho H., Choi C.H., Park Y., Chung E.J., Song J., Chung J.Y., Hong S.M. Dual loss of USP10 and p14ARF protein expression is associated with poor prognosis in patients with small intestinal adenocarcinoma. Tumour Biol. 2018;40:1010428318808678. doi: 10.1177/1010428318808678. [DOI] [PubMed] [Google Scholar]
- 153.Otsuka H., Fukao A., Funakami Y., Duncan K.E., Fujiwara T. Emerging Evidence of Translational Control by AU-Rich Element-Binding Proteins. Front. Genet. 2019;10:332. doi: 10.3389/fgene.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Khong A., Matheny T., Jain S., Mitchell S.F., Wheeler J.R., Parker R. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell. 2017;68:808–820.e5. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dai L., Lin Z., Cao Y., Chen Y., Xu Z., Qin Z. Targeting EIF4F complex in non-small cell lung cancer cells. Oncotarget. 2017;8:55731–55735. doi: 10.18632/oncotarget.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chan K., Robert F., Oertlin C., Kapeller-Libermann D., Avizonis D., Gutierrez J., Handly-Santana A., Doubrovin M., Park J., Schoepfer C., et al. eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat. Commun. 2019;10:5151. doi: 10.1038/s41467-019-13086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mamane Y., Petroulakis E., Rong L., Yoshida K., Ler L.W., Sonenberg N. eIF4E--from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 158.Hershey J.W. The role of eIF3 and its individual subunits in cancer. Biochim. Biophys. Acta. 2015;1849:792–800. doi: 10.1016/j.bbagrm.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 159.Yin Y., Long J., Sun Y., Li H., Jiang E., Zeng C., Zhu W. The function and clinical significance of eIF3 in cancer. Gene. 2018;673:130–133. doi: 10.1016/j.gene.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 160.Yoshida K., Kajiyama H., Inami E., Tamauchi S., Ikeda Y., Yoshikawa N., Nishino K., Utsumi F., Niimi K., Suzuki S., et al. Clinical Significance of Ubiquitin-associated Protein 2-like in Patients With Uterine Cervical Cancer. In Vivo. 2020;34:109–116. doi: 10.21873/invivo.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Frydryskova K., Masek T., Borcin K., Mrvova S., Venturi V., Pospisek M. Distinct recruitment of human eIF4E isoforms to processing bodies and stress granules. BMC Mol. Biol. 2016;17:21. doi: 10.1186/s12867-016-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cirillo L., Cieren A., Barbieri S., Khong A., Schwager F., Parker R., Gotta M. UBAP2L Forms Distinct Cores that Act in Nucleating Stress Granules Upstream of G3BP1. Curr. Biol. 2020;30:698–707.e6. doi: 10.1016/j.cub.2019.12.020. [DOI] [PubMed] [Google Scholar]
- 163.Palam L.R., Gore J., Craven K.E., Wilson J.L., Korc M. Integrated stress response is critical for gemcitabine resistance in pancreatic ductal adenocarcinoma. Cell Death Dis. 2015;6:e1913. doi: 10.1038/cddis.2015.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sun Y., Yang P., Zhang Y., Bao X., Li J., Hou W., Yao X., Han J., Zhang H. A genome-wide RNAi screen identifies genes regulating the formation of P bodies in C. elegans and their functions in NMD and RNAi. Protein Cell. 2011;2:918–939. doi: 10.1007/s13238-011-1119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Grabocka E., Bar-Sagi D. Mutant KRAS Enhances Tumor Cell Fitness by Upregulating Stress Granules. Cell. 2016;167:1803–1813.e12. doi: 10.1016/j.cell.2016.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.