Abstract
RMND1 (required for meiotic nuclear division 1 homolog) pathogenic variants are known to cause combined oxidative phosphorylation deficiency (COXPD11), a severe multisystem disorder. In one patient, a homozygous RMND1 pathogenic variant, with an established role in COXPD11, was associated with a Perrault-like syndrome. We performed a thorough clinical investigation and applied a targeted multigene hearing loss panel to reveal the cause of hearing loss, ovarian dysfunction (two cardinal features of Perrault syndrome) and chronic kidney disease in two adult female siblings. Two compound heterozygous missense variants, c.583G>A (p.Gly195Arg) and c.818A>C (p.Tyr273Ser), not previously associated with disease, were identified in RMND1 in both patients, and their segregation with disease was confirmed in family members. The patients have no neurological or intellectual impairment, and nephrological evaluation predicts a benign course of kidney disease. Our study presents the mildest, so far reported, RMND1-related phenotype and delivers the first independent confirmation that RMND1 is causally involved in the development of Perrault syndrome with renal involvement. This highlights the importance of including RMND1 to the list of Perrault syndrome causative factors and provides new insight into the clinical manifestation of RMND1 deficiency.
Keywords: RMND1 (required for meiotic nuclear division 1 homolog), Perrault syndrome, renal disease, hearing loss, ovarian dysfunction, COXPD11 (combined oxidative phosphorylation deficiency), mitochondria
1. Introduction
RMND1 (required for meiotic nuclear division 1 homolog) is a nuclear gene that encodes a protein needed for proper functioning of mitochondria. Although the data on its exact role are still limited, it has been shown that the RMND1 protein belongs to a large mitochondrial inner membrane complex that supports translation of the mtDNA-encoded polypeptides [1,2], all of which represent essential structural components of the oxidative phosphorylation (OXPHOS) complexes. It has been proposed that RMND1 tethers mitochondrial ribosomes close to the sites where the primary mRNAs are matured, spatially coupling mitochondrial transcription with translation [3]. In line with this, RMND1 pathogenic variants cause a generalized mitochondrial translation defect and are detected in patients with combined oxidative phosphorylation deficiency (COXPD11; MIM #614922), a severe recessive condition characterized by the presence of lactic acidosis, deafness, renal and liver dysfunction, central nervous system and muscle involvement with an onset at birth or early infancy [4,5].
In 2018, a different clinical presentation consistent with a diagnosis of Perrault syndrome (PRLTS) was associated with a known RMND1 (c.713A>G; p.Asn238Ser) homozygous variant. The individual reported by Demain et al. suffered from sensorineural hearing loss (HL) and primary ovarian insufficiency (POI), defining clinical features of PRLTS, in addition to renal dysfunction and short stature. The phenotype was delineated based on exome sequencing data from a single patient [6]. Considering the absence of another reported disease-causing variant, a doubt may arise as to whether RMND1 is related to PRLTS development. Here, we present novel data delivering the first independent confirmation that RMND1 is causally involved in the development of PRLTS with chronic kidney disease.
2. Materials and Methods
2.1. Study Subjects
Two affected sisters from a nonconsanguineous Polish family, together with their parents and two other unaffected sisters, participated in the study (Figure 1A). The proband was born at term after an uneventful pregnancy, and her development in the first years of life was considered normal until the age of four, when bilateral HL was diagnosed. She received hearing aids at the age of six; the degree of HL progressed gradually and was accompanied by tinnitus and vertigo from the age of 31. No ear malformations were observed on temporal bone CT scans. Cochlear implantation was performed for the right ear at the age of 34 and for the left ear at the age of 36 with a good outcome. From the age of 17, she was under gynecological care due to irregular and scanty menstruation (menarche at age 14). Hypergonadotropic hypogonadism and small ovaries and uterus were recognized. Infertility was diagnosed, and hormone replacement therapy was introduced at the age of 28. Hypertension was diagnosed at the age of 31. At the age of 33, her left adrenal gland was removed because of lymphangioma, and chronic kidney disease (CKD) was diagnosed.
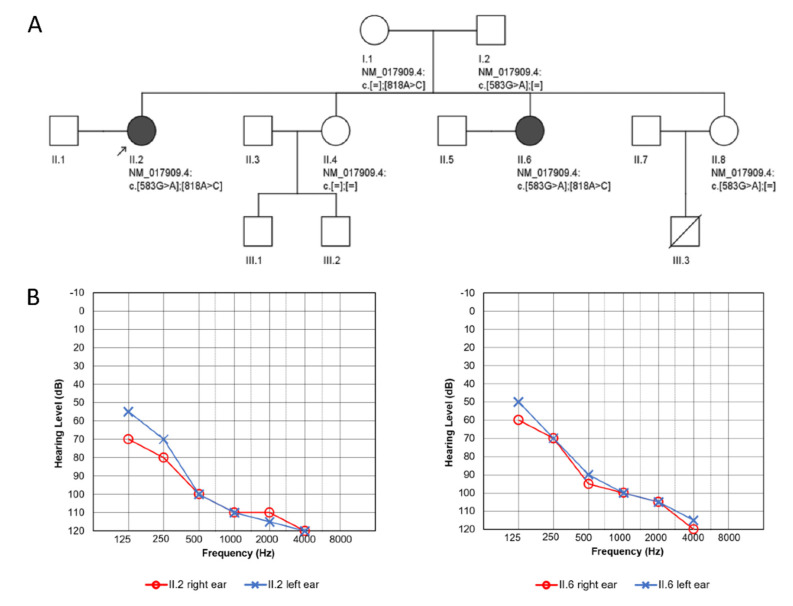
Figure 1.
Pedigree and audiological data of the investigated family. (A) Pedigree showing affected family members (proband II.2, proband’s sister II.6) and the identified RMND1 variants. (B) Pure tone audiometry results of the proband (left panel) and her sister (right panel) at the age of 32.
The proband’s younger sister was also born at term without any complications, and her development was normal. Bilateral HL was diagnosed at the age of 3. She does not have tinnitus or vertigo, and her HL is stable. She is using hearing aids sporadically and prefers to communicate with sign language. Evaluation for primary amenorrhea and delayed pubertal development at the age of 18 revealed gonadal dysgenesis with a normal female karyotype 46,XX. Vitamin B12 deficiency anemia and osteoporosis were diagnosed at the age of 26. At the age of 32, hypertension and CKD were recognized. Due to hypertension, enarenal and indapamide were implemented in the proband and amlodipine and torasemide in her sister. Both receive oestradiol and dydrogesterone supplementation to reduce the complications of POI.
Written informed consent was obtained from all participants. The study was approved by the ethics committee at the Institute of Physiology and Pathology of Hearing (KB.IFPS.25/2017) and performed according to the Declaration of Helsinki.
2.2. Nephrological and Neurological Examinations
The proband and her sister underwent thorough nephrological evaluation including whole blood count, electrolytes, venous blood gases, serum creatinine, calcium/phosphate balance, uric acid, lipid profile, urinalysis as well as urine albumin to creatinine ratio (UACR), urinary tract ultrasound with kidney size, and cortical thickness evaluation. To estimate glomerular filtration rate (eGFR), CKD-EPI (CKD Epidemiology Collaboration) creatinine equation was used [7]. A detailed neurological examination was performed. To assess the occurrence of neurological signs, the scale for assessment rating of ataxia—5th version (SARA) and Inventory of Non-Ataxia symptoms—6th version (INAS) were used [8,9].
2.3. Targeted HL Gene Panel, Data Analysis and Interpretation
Genomic DNA was extracted using a standard salting out procedure. Libraries were prepared with a custom HL 237-gene panel (SeqCap EZ Choice, Roche, Switzerland), containing genes related to PRLTS, i.e., HSD17B4, HARS2, LARS2, TWNK, ERAL1, CLPP, and RMND1 and sequenced on a MiSeq Illumina platform. The quality control of raw FASTQ reads was performed, followed by adapter trimming and low quality reads removal with Trimmomatic [10]. Burrows–Wheeler Aligner [11] was used to map reads on hg38, followed by sorting and duplication removal using Samblaster [12]. Variant identification was done using multiple algorithms: HaplotypeCaller from GATK (Genome Analysis Toolkit) [13], Freebayes [14], DeepVariant [15], and MuTect2 [16]. Identified variants were annotated using Ensembl VEP [17] as well as multiple databases, including dbSNP [18], dbNSFP [18], GnomAD [19], ClinVar [20], and HGMD [21]. Inhouse databases of previously identified variants were used for annotation, to identify sequencing artifacts as well as variants common in the Polish population. The pathogenicity of identified variants was predicted based on the biochemical properties of the codon change and degree of evolutionary conservation using PolyPhen-2 [22], SIFT [23], Mutation Taster [24], LRT [25], and CADD [26]. Pathogenicity of the identified single nucleotide (SNV) and INDEL variants was evaluated by analyzing allele frequency, in silico predictions, annotations from public variant databases, matches in the inhouse variants database, and related medical literature. Evolutionary conservation was evaluated using GERP++ score [27]. Multiple protein sequence alignment was performed using COBALT [28], and variant localization across evolutionary diverse species was visualized with Jalview v2.11.1.0 software [29]. Detected variants were assigned according to standards and guidelines for the interpretation of sequence variants [30,31]. Selected probably causative variants were confirmed using direct Sanger sequencing and reported based on the RMND1 NM_017909.4 and NP_060379.2 reference sequences.
3. Results
3.1. Clinical Presentation
The major clinical features of the proband, a 44-year old female, and her sister, a 36-year old female, were severe-to-profound bilateral sensorineural HL (Figure 1B) and ovarian dysfunction accompanied by CKD that developed in the fourth decade of life. Both had a normal stature. Laboratory findings on renal involvement, blood lactate concentration and core parameters of venous acid-base balance are given in Table 1, as shown by eGFR and UACR both patients were in stage G3, A1 of CKD [32]. The proband’s calculated one-year eGFR decline was −0.45 mL/min and in her sister −0.66 mL/min. On repeated ultrasound evaluations, the size and cortex thickness of the kidneys was slightly diminished, but generally, the kidneys’ dimensions did not change within a twelve-year follow up. The proband’s sister had a more complex nephrological profile. Although on ultrasound, both kidneys and their cortex were of normal size, on scintigraphy at the age of 32, substantial asymmetry of ERPL (effective renal plasma flow; 64% left, 36% right kidney) with uneven radiotracer accumulation in the right organ was found. It was interpreted as post-inflammatory scars even though the patient denied urinary tract infections.
Table 1.
Laboratory results of the proband and her sister.
| RBC (T/L) | Hb (g/dL) | Creatinine (mg/dL) | eGFR CKD EPI (mL/min) | Acid Base Venous Balance | Blood Lipids (mg/dL) | Calcium (mg/dL) | Phosphates (mg/dL) | PTH (pg/mL) | UACR (mg/g) | |
|---|---|---|---|---|---|---|---|---|---|---|
| proband | 4.49 (4.2–6.3) |
13.3 (12–16) |
1.53 (0.6–1.3) |
41 (>90) |
pH 7.31 (7.35–7.45) HCO3− 20.7 (22–26 mmol/L) BE −1.7 (−2 to +2mmol/l) Anion gap 12 (12 ± 4 mEq/L) Cl− 105 (98–106 mmol/L) Lactic acid 1.2 (0.5–1.6 mmol/L) K+ 5.4 (3.4–4.5 mEq/L) Na+ 137 (136–146 mEq/L) |
T chol 159 (<190) HDL 71 (35–65) LDL 72 (<115) TG 76 (<150) |
9.2 (8.5–10.1) |
3.6 (2.5–4.9) |
72.6 (12–68.3) |
2.9 (<30) |
|
proband’s
sister |
4.08 (4.2–6.3) |
12.4 (12–16) |
1.38 (0.6–1.3) |
49 (>90) |
pH 7.35 (7.35–7.45) HCO3− 22 (22–26 mmol/L) BE −0.6 (−2 to +2mmol/L) Anion gap 8.9 (12 ± 4 mEq/L) Cl− 105 (98–106 mmol/L) Lactic acid 1.7 (0.5–1.6 mmol/L) K+ 5.2 (3.4–4.5 mEq/L) Na+ 138 (136–146 mEq/L) |
T chol 213 (<190) HDL 68 (35–65)LDL 145 (<115) TG 57 (<150) |
9.4 (8.5–10.1) |
3.6 (2.5–4.9) |
148.8 (12–68.3) |
6.5 (<30) |
Abnormal values are given in bold; reference values are in parentheses.
Neurological assessment did not reveal any features of cerebellar, pyramidal or extrapyramidal syndromes either in the proband or her affected sister. They presented normal muscle tone and strength as well as reflexes in the upper and lower limbs. Both have completed higher education.
3.2. Identification of Pathogenic Variants
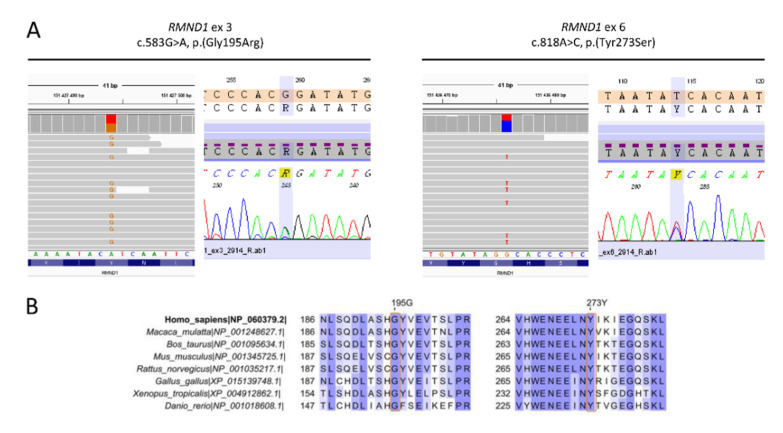
After performing next-generation sequencing (NGS), two heterozygous variants, c.583G>A and c.818A>C in RMND1, corresponding to missense changes p.Gly195Arg and p.Tyr273Ser, respectively, were identified in the proband (Figure 2A). The vast majority of computational algorithms predicted a probably pathogenic character of detected variants, and they were identified only in heterozygous, individual cases in the gnomAD population database (Table 2). Conservation analyses showed 100% identity of the analyzed regions among all tested species (Figure 2B), with GERP++ scores of 4.57 and 5.95. Based on the applicable standards and guidelines, we have classified the identified RMND1 variants as likely pathogenic. No other pathogenic variants related to isolated or syndromic hereditary HL, in particular to PRLTS, were found. The same RMND1 variant constellation was identified in her affected younger sister. Both parents and another healthy sister were heterozygous carriers of one of the RMND1 variants. In the third sister, none of the RMND1 variants was identified (Figure 1A).
Figure 2.
Genetic data of the investigated family. (A) Results of next-generation sequencing (NGS) and Sanger sequencing showing c.583G>A transition (p.Gly195Arg) and c.818A>C transversion (p.Tyr273Ser) in the RMND1 gene. (B) Multiple protein sequence alignment of selected RMND1 regions among different species.
Table 2.
Characteristics of RMND1 variants detected in this study.
| Variant cDNA Level | Variant Protein Level | Reference SNP ID | Population Frequencies | Pathogenicity Predictions | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| gnomAD | 1000 Genomes | ESP 6500 | SIFT | PolyPhen-2 | Mutation Taster | LRT | CADD | ACMG Classification * | |||
| c.583G>A | p.(Gly195Arg) | rs776083030 | 0.00002388 (6/251308) | 0 | 0 | D (0.011) | PD (0.997) | D (1) | D (0) | D (29.7) | LP (PM2, PP1_M, PP3, PP4) |
| c.818A>C | p.(Tyr273Ser) | rs766739125 | 0.00000399 (1/250612) | 0 | 0 | D (0) | PD (1) | D (0.99) | N (0.001742) | D (26.4) | LP (PM2, PP1_M, PP3, PP4) |
Abbreviations: D, damaging; PD, probably damaging; N, neutral; * ACMG classification criteria legend: LP, likely pathogenic; PM, moderate pathogenicity evidence; PP, supporting pathogenicity evidence; _M, moderate.
4. Discussion
Our clinical and genetic investigation shows that a combination of HL, ovarian dysfunction, and CKD constitutes a milder end of the RMND1-related phenotypic spectrum. Presence of the three clinical features can be defined as Perrault-like syndrome [6], PRLTS with renal involvement or just PRLTS with a respective consecutive number, according to the nomenclature used by OMIM (Online Mendelian Inheritance in Man, https://omim.org/), where subsequent numbers are assigned to a syndrome in order to distinguish the causative gene. PRLTS is characterized by the presence of sensorineural HL in both males and females and ovarian dysfunction ranging from gonadal dysgenesis to POI in females. These are the two PRLTS cardinal features; however, in some individuals additional, usually neurological conditions (e.g., developmental delay, cognitive impairment, ataxia or sensory axonal neuropathy) have been also reported (Table 3) [33]. Taking into account the heterogeneity of PRLTS phenotypic manifestations, in our opinion, it seems justified to recognize RMND1 as the seventh PRLTS gene, where renal involvement represents an additional characteristic finding and no neurological signs or symptoms are found (neither in the patient reported by Demain [6] nor in our patients).
Table 3.
Genes causally involved in the development of PRLTS and PRLTS-like features.
| Gene (Locus) | Protein | Subcellular Localization | Function | Additional Clinical Features * | Inheritance Mode | Ref. |
|---|---|---|---|---|---|---|
|
CLPP (19p13.3) |
caseinolytic mitochondrial matrix peptidase proteolytic subunit | mitochondrial | mitochondrial protein degradation (component of a proteolytic complex) |
|
AR | [33,38,39,40,41,42,43] |
|
ERAL1 (17q11.2) |
Era like 12S mitochondrial rRNA chaperone 1 | mitochondrial | mitochondrial protein translation (assembly of mitochondrial ribosomal subunit) |
|
AR | [44] |
|
GGPS1 (1q42.3) |
geranylgeranyl diphosphate synthase 1 | cytoplasmic | acts on peroxisomal products, part of mevalonate pathway |
|
AR | [33,45] |
|
HARS2 (5q31.3) |
histidyl-tRNA synthetase 2 | mitochondrial | mitochondrial protein translation (synthesis of histidyl-transfer RNA) |
|
AR | [39,46,47] |
|
HSD17B4 (17q21.2) |
hydroxysteroid 17-β dehydrogenase 4 | peroxisomal | β-oxidation pathway for fatty acids in peroxisomes |
|
AR | [39,41,48,49,50,51] |
|
LARS2 (3p21.31) |
leucyl-tRNA synthetase | mitochondrial | mitochondrial protein translation (synthesis of leucyl-transfer RNA) |
|
AR | [33,39,41,52,53,54,55,56,57,58,59,60] |
|
PEX6 (6p21.1) |
peroxisomal biogenesis factor 6 | peroxisomal | peroxisomal protein import (ATPase activity) |
|
AR | [33] |
|
RMND1 (6q25.1) |
required for meiotic nuclear division 1 homolog | mitochondrial | mitochondrial protein translation |
|
AR | [6] Present study |
|
TFAM (10q21.1) |
transcription factor A, mitochondrial | mitochondrial | key mitochondrial transcription factor |
|
AR | [33] |
|
TWNK (10q24.31) |
twinkle mtDNA helicase | mitochondrial | mitochondrial DNA replication and transcription (unwinds double-stranded DNA) |
|
AR | [39,41,61,62,63,64,65] |
* Clinical features additional to hearing loss (HL) and ovarian dysfunction observed in some patients. AR, autosomal recessive.
Kidney function is frequently affected in patients with RMND1 deficiency. Analyzing a large group of patients with COXPD11 due to RMND1 pathogenic variants, Ng et al. found that renal involvement was present in more than two thirds of patients [4]. It was manifested by cystic dysplasia, renal tubular acidosis (persistent hyponatremia and hyperkalemia), end stage renal failure with subsequent kidney transplantation, anemia, proteinuria or CKD at different stages. The single, so far described, patient with PRLTS and RMND1 homozygous pathogenic variant [6] had distal renal tubular acidosis with hyperchloremic metabolic acidosis, a normal anion gap, mildly elevated uric acid, low urine citrate levels, normal calcium levels, and a normal renal ultrasound. CKD was mentioned, but exact kidney function has not been given. In our patients, we found CKD of mild to moderate severity. The proband was affected by metabolic acidosis with normal fasting lactic acid concentration, hyperkalemia, normal chloride, and a normal anion gap. In her sister, we did not find metabolic acidosis, although the fasting lactic acid concentration was slightly above normal values. Both sisters presented with hypertension that may be secondary to CKD, and applied antihypertensive medications might have had an influence on electrolyte abnormalities. The calculated yearly filtration losses that we assessed in the patients were similar to the value of eGFR slope (−0.48 mL/min) found in women aged 35 to 49 years and renal stage IIIa (45–59 mL/min) [34]. This, together with a low-grade UACR of our patients, predicts a benign kidney disease course and makes reaching kidney failure and a requirement of renal replacement therapy less likely [32].
Ovarian dysfunction (ovarian atrophy and hypergonadotropic hypogonadism) as a consequence of RMND1 pathogenic variants has been previously reported only once in a patient described by Demain et al. [6] and in none of the approximately 40 patients with OXPHOS deficiency. This could be explained by the early, prepubertal age at which the majority of children were investigated [4,5,35,36]. In only two patients examined at the age of 14 and 17, no reference was made to their sexual development [36]. Thus, at the moment, it is not clear how frequently ovaries are affected in patients with RMND1 deficiency.
RMND1 is a nuclear-encoded protein involved in mitochondrial translation. Disruption of this process is a well-known mechanism leading to PRLTS development (Table 3). In this study, we have identified two likely pathogenic RMND1 variants not previously associated with disease. Presence of the detected RMND1 variants in a trans configuration is consistent with the autosomal recessive mode of inheritance. Our study provides an independent confirmation on the causative role of RMND1 in Perrault syndrome with renal involvement. Hearing loss and renal dysfunction are typical for of RMND1-related disorders. These two clinical features accompanied by ovarian dysfunction were present in our patients and they are consistent with the phenotype reported in the original study by Demain et al. [6]. The identification of two ultra-rare RMND1 variants that are in a trans configuration, co-occur in two affected family members (having an almost identical phenotype) and do not co-occur in two other healthy siblings, strongly supports their pathogenic potential.
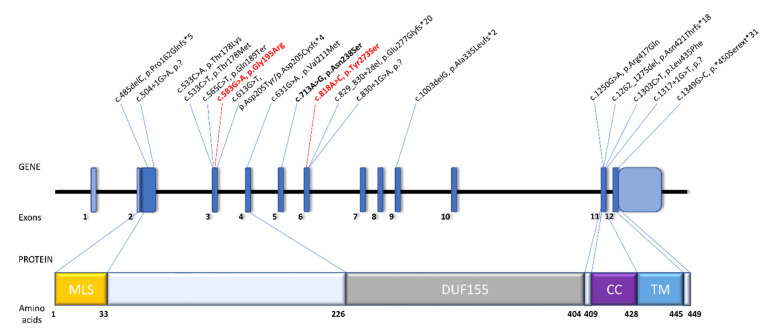
One of the identified variants (p.Gly195Arg) localizes close to the DUF155 domain at the protein N-terminus and the second one (p.Tyr273Ser) within the DUF155 domain (Figure 3). Considering that RMND1 has three protein-encoding transcripts all of which contain the DUF155 domain [1], one may assume that all three proteins arising from the p.(Tyr273Ser)-carrying allele will be dysfunctional. It is not applicable for the second RMND1 variant, that will affect two out of three alternative transcripts, leaving some functional RMND1 protein in the cells. Although the tissue-specific ratio of RMND1 transcripts remains unknown, this observation may provide a possible explanation for the milder phenotype in our patients. It could also be owed to some other yet unidentified modifying factors. It is still a conundrum why the single patient with a homozygous p.(Asn238Ser) variant, localizing within the DUF155 domain, presented a relatively mild phenotype resembling PRLTS [6], in contrast to the, currently, four other patients with the same causative variant and a more severe infantile-onset multisystem disorder [4,5,37].
Figure 3.
Schematic representation of RMND1 gene and protein organization. Gene and protein structure is depicted based on the canonical transcript NM_017909.4 and reference protein sequence NP_060379.2. Previously reported RMND1 pathogenic variants involved in development of combined oxidative phosphorylation deficiency (COXPD11) are written in black. Variants identified in this study are shown in red. Bolded are variants involved in the development of Perrault syndrome (PRLTS) with renal involvement. Abbreviations: MLS, mitochondrial localization sequence; DUF155, domain of unknown function; CC, coiled-coil; TM, transmembrane.
5. Conclusions
In summary, we report two novel RMND1 likely pathogenic variants leading to the mildest, so far reported, RMND1-related phenotype that corresponds to PRLTS with renal involvement. It was identified in two adult siblings with a very similar clinical presentation. Our study highlights the importance of including RMND1 to the list of PRLTS causative factors and directs attention to ovaries as yet another organ affected by RMND1 deficiency. Future functional studies could be helpful to clarify the molecular mechanisms underlying the differences in phenotype severity of RMND1-related disorders.
Acknowledgments
We are grateful to the patients and their family for participation in this study.
Author Contributions
Conceptualization, M.O. and D.O.; methodology, D.O., J.P., I.S., and H.S.; formal analysis, D.O., J.P., and I.S.; investigation, D.O., J.P., and I.S.; resources, M.O. and H.S.; writing—original draft preparation, M.O. and D.O.; writing—review and editing, J.P., I.S., and H.S.; visualization, D.O. and J.P.; supervision, M.O.; project administration, M.O.; funding acquisition, M.O. and H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Science Centre Grant No. 2016/22/E/NZ5/00470 and the Institute of Physiology and Pathology of Hearing. The APC was funded by the Institute of Physiology and Pathology of Hearing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Garcia-Diaz B., Barros M.H., Sanna-Cherchi S., Emmanuele V., Akman H.O., Ferreiro-Barros C.C., Horvath R., Tadesse S., El Gharaby N., DiMauro S., et al. Infantile encephaloneuromyopathy and defective mitochondrial translation are due to a homozygous RMND1 mutation. Am. J. Hum. Genet. 2012;91:729–736. doi: 10.1016/j.ajhg.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janer A., Antonicka H., Lalonde E., Nishimura T., Sasarman F., Brown G.K., Brown R.M., Majewski J., Shoubridge E.A. An RMND1 Mutation causes encephalopathy associated with multiple oxidative phosphorylation complex deficiencies and a mitochondrial translation defect. Am. J. Hum. Genet. 2012;91:737–743. doi: 10.1016/j.ajhg.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janer A., van Karnebeek C.D., Sasarman F., Antonicka H., Al Ghamdi M., Shyr C., Dunbar M., Stockler-Ispiroglu S., Ross C.J., Vallance H., et al. RMND1 deficiency associated with neonatal lactic acidosis, infantile onset renal failure, deafness, and multiorgan involvement. Eur. J. Hum. Genet. 2015;23:1301–1307. doi: 10.1038/ejhg.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng Y.S., Alston C.L., Diodato D., Morris A.A., Ulrick N., Kmoch S., Houstek J., Martinelli D., Haghighi A., Atiq M., et al. The clinical, biochemical and genetic features associated with RMND1-related mitochondrial disease. J. Med. Genet. 2016;53:768–775. doi: 10.1136/jmedgenet-2016-103910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shayota B.J., Le N.T., Bekheirnia N., Rosenfeld J.A., Goldstein A.C., Moritz M., Bartholomew D.W., Pastore M.T., Xia F., Eng C., et al. Characterization of the renal phenotype in RMND1-related mitochondrial disease. Mol. Genet. Genomic Med. 2019;7:e973. doi: 10.1002/mgg3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demain L.A.M., Antunes D., O’Sullivan J., Bhaskhar S.S., O’Keefe R.T., Newman W.G. A known pathogenic variant in the essential mitochondrial translation gene RMND1 causes a Perrault-like syndrome with renal defects. Clin. Genet. 2018;94:276–277. doi: 10.1111/cge.13255. [DOI] [PubMed] [Google Scholar]
- 7.Levey A.S., Stevens L.A. Estimating GFR using the CKD Epidemiology Collaboration (CKD-EPI) creatinine equation: More accurate GFR estimates, lower CKD prevalence estimates, and better risk predictions. Am. J. Kidney Dis. 2010;55:622–627. doi: 10.1053/j.ajkd.2010.02.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz-Hubsch T., du Montcel S.T., Baliko L., Berciano J., Boesch S., Depondt C., Giunti P., Globas C., Infante J., Kang J.S., et al. Scale for the assessment and rating of ataxia: Development of a new clinical scale. Neurology. 2006;66:1717–1720. doi: 10.1212/01.wnl.0000219042.60538.92. [DOI] [PubMed] [Google Scholar]
- 9.Schmitz-Hubsch T., Coudert M., Bauer P., Giunti P., Globas C., Baliko L., Filla A., Mariotti C., Rakowicz M., Charles P., et al. Spinocerebellar ataxia types 1, 2, 3, and 6: Disease severity and nonataxia symptoms. Neurology. 2008;71:982–989. doi: 10.1212/01.wnl.0000325057.33666.72. [DOI] [PubMed] [Google Scholar]
- 10.Lohse M., Bolger A.M., Nagel A., Fernie A.R., Lunn J.E., Stitt M., Usadel B. RobiNA: A user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faust G.G., Hall I.M. SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv Prepr. 20121207.3907 [Google Scholar]
- 15.Poplin R., Chang P.C., Alexander D., Schwartz S., Colthurst T., Ku A., Newburger D., Dijamco J., Nguyen N., Afshar P.T., et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 16.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. bioRxiv. 2020;10:531210. doi: 10.1101/531210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 22.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;76:7–20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 25.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papadopoulos J.S., Agarwala R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 29.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oza A.M., DiStefano M.T., Hemphill S.E., Cushman B.J., Grant A.R., Siegert R.K., Shen J., Chapin A., Boczek N.J., Schimmenti L.A., et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat. 2018;39:1593–1613. doi: 10.1002/humu.23630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin A., Stevens P.E., Bilous R.W., Coresh J., De Francisco A.L.M., De Jong P.E., Griffith K.E., Hemmelgarn B.R., Iseki K., Lamb E.J., et al. Kidney disease: Improving global outcomes (KDIGO) CKD work group. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. Suppl. 2013;3:150. doi: 10.1038/kisup.2012.73. [DOI] [PubMed] [Google Scholar]
- 33.Tucker E.J., Rius R., Jaillard S., Bell K., Lamont P.J., Travessa A., Dupont J., Sampaio L., Dulon J., Vuillaumier-Barrot S., et al. Genomic sequencing highlights the diverse molecular causes of Perrault syndrome: A peroxisomal disorder (PEX6), metabolic disorders (CLPP, GGPS1), and mtDNA maintenance/translation disorders (LARS2, TFAM) Hum. Genet. 2020 doi: 10.1007/s00439-020-02176-w. [DOI] [PubMed] [Google Scholar]
- 34.Baba M., Shimbo T., Horio M., Ando M., Yasuda Y., Komatsu Y., Masuda K., Matsuo S., Maruyama S. Longitudinal Study of the Decline in Renal Function in Healthy Subjects. PLoS ONE. 2015;10:e0129036. doi: 10.1371/journal.pone.0129036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broenen E., Ranchin B., Besmond C., Freychet C., Fouilhoux A., Perouse de Montclos T., Ville D., Bacchetta J. RMND1 mutations in two siblings: Severe renal hypoplasia but different levels of extrarenal abnormality severity: The ethics of decision making. Arch. Pediatr. 2019;26:377–380. doi: 10.1016/j.arcped.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Ravn K., Neland M., Wibrand F., Duno M., Ostergaard E. Hearing impairment and renal failure associated with RMND1 mutations. Am. J. Med. Genet. Part A. 2016;170:142–147. doi: 10.1002/ajmg.a.37399. [DOI] [PubMed] [Google Scholar]
- 37.Parikh S., Karaa A., Goldstein A., Ng Y.S., Gorman G., Feigenbaum A., Christodoulou J., Haas R., Tarnopolsky M., Cohen B.K., et al. Solid organ transplantation in primary mitochondrial disease: Proceed with caution. Mol. Genet. Metab. 2016;118:178–184. doi: 10.1016/j.ymgme.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Theunissen T.E., Szklarczyk R., Gerards M., Hellebrekers D.M., Mulder-Den Hartog E.N., Vanoevelen J., Kamps R., de Koning B., Rutledge S.L., Schmitt-Mechelke T., et al. Specific MRI Abnormalities Reveal Severe Perrault Syndrome due to CLPP Defects. Front. Neurol. 2016;7:203. doi: 10.3389/fneur.2016.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerat J., Jonard L., Loundon N., Christin-Maitre S., Lacombe D., Goizet C., Rouzier C., Van Maldergem L., Gherbi S., Garabedian E.N., et al. An Application of NGS for Molecular Investigations in Perrault Syndrome: Study of 14 Families and Review of the Literature. Hum. Mutat. 2016;37:1354–1362. doi: 10.1002/humu.23120. [DOI] [PubMed] [Google Scholar]
- 40.Dursun F., Mohamoud H.S., Karim N., Naeem M., Jelani M., Kirmizibekmez H. A Novel Missense Mutation in the CLPP Gene Causing Perrault Syndrome Type 3 in a Turkish Family. J. Clin. Res. Pediatr. Endocrinol. 2016;8:472–477. doi: 10.4274/jcrpe.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demain L.A., Urquhart J.E., O’Sullivan J., Williams S.G., Bhaskar S.S., Jenkinson E.M., Lourenco C.M., Heiberg A., Pearce S.H., Shalev S.A., et al. Expanding the Genotypic Spectrum of Perrault syndrome. Clin. Genet. 2017;91:302–312. doi: 10.1111/cge.12776. [DOI] [PubMed] [Google Scholar]
- 42.Ahmed S., Jelani M., Alrayes N., Mohamoud H.S., Almramhi M.M., Anshasi W., Ahmed N.A., Wang J., Nasir J., Al-Aama J.Y. Exome analysis identified a novel missense mutation in the CLPP gene in a consanguineous Saudi family expanding the clinical spectrum of Perrault Syndrome type-3. J. Neurol. Sci. 2015;353:149–154. doi: 10.1016/j.jns.2015.04.038. [DOI] [PubMed] [Google Scholar]
- 43.Jenkinson E.M., Rehman A.U., Walsh T., Clayton-Smith J., Lee K., Morell R.J., Drummond M.C., Khan S.N., Naeem M.A., Rauf B., et al. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am. J. Hum. Genet. 2013;92:605–613. doi: 10.1016/j.ajhg.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatzispyrou I.A., Alders M., Guerrero-Castillo S., Zapata Perez R., Haagmans M.A., Mouchiroud L., Koster J., Ofman R., Baas F., Waterham H.R., et al. A homozygous missense mutation in ERAL1, encoding a mitochondrial rRNA chaperone, causes Perrault syndrome. Hum. Mol. Genet. 2017;26:2541–2550. doi: 10.1093/hmg/ddx152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foley A.R., Zou Y., Dunford J.E., Rooney J., Chandra G., Xiong H., Straub V., Voit T., Romero N., Donkervoort S., et al. GGPS1 Mutations Cause Muscular Dystrophy/Hearing Loss/Ovarian Insufficiency Syndrome. Ann. Neurol. 2020 doi: 10.1002/ana.25772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karstensen H.G., Rendtorff N.D., Hindbaek L.S., Colombo R., Stein A., Birkebaek N.H., Hartmann-Petersen R., Lindorff-Larsen K., Hojland A.T., Petersen M.B., et al. Novel HARS2 missense variants identified in individuals with sensorineural hearing impairment and Perrault syndrome. Eur. J. Med. Genet. 2020;63:103733. doi: 10.1016/j.ejmg.2019.103733. [DOI] [PubMed] [Google Scholar]
- 47.Pierce S.B., Chisholm K.M., Lynch E.D., Lee M.K., Walsh T., Opitz J.M., Li W., Klevit R.E., King M.C. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc. Natl. Acad. Sci. USA. 2011;108:6543–6548. doi: 10.1073/pnas.1103471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim M.J., Kim S.J., Kim J., Chae H., Kim M., Kim Y. Genotype and phenotype heterogeneity in perrault syndrome. J. Pediatr. Adolesc. Gynecol. 2013;26:e25–e27. doi: 10.1016/j.jpag.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Pierce S.B., Walsh T., Chisholm K.M., Lee M.K., Thornton A.M., Fiumara A., Opitz J.M., Levy-Lahad E., Klevit R.E., King M.C. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome. Am. J. Hum. Genet. 2010;87:282–288. doi: 10.1016/j.ajhg.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lieber D.S., Hershman S.G., Slate N.G., Calvo S.E., Sims K.B., Schmahmann J.D., Mootha V.K. Next generation sequencing with copy number variant detection expands the phenotypic spectrum of HSD17B4-deficiency. BMC Med. Genet. 2014;15:30. doi: 10.1186/1471-2350-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen K., Yang K., Luo S.S., Chen C., Wang Y., Wang Y.X., Li D.K., Yang Y.J., Tang Y.L., Liu F.T., et al. A homozygous missense variant in HSD17B4 identified in a consanguineous Chinese Han family with type II Perrault syndrome. BMC Med. Genet. 2017;18:91. doi: 10.1186/s12881-017-0453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pierce S.B., Gersak K., Michaelson-Cohen R., Walsh T., Lee M.K., Malach D., Klevit R.E., King M.C., Levy-Lahad E. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am. J. Hum. Genet. 2013;92:614–620. doi: 10.1016/j.ajhg.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Solda G., Caccia S., Robusto M., Chiereghin C., Castorina P., Ambrosetti U., Duga S., Asselta R. First independent replication of the involvement of LARS2 in Perrault syndrome by whole-exome sequencing of an Italian family. J. Hum. Genet. 2016;61:295–300. doi: 10.1038/jhg.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kosaki R., Horikawa R., Fujii E., Kosaki K. Biallelic mutations in LARS2 can cause Perrault syndrome type 2 with neurologic symptoms. Am. J. Med. Genet. A. 2018;176:404–408. doi: 10.1002/ajmg.a.38552. [DOI] [PubMed] [Google Scholar]
- 55.Carminho-Rodrigues M.T., Klee P., Laurent S., Guipponi M., Abramowicz M., Cao-van H., Guinand N., Paoloni-Giacobino A. LARS2-Perrault syndrome: A new case report and literature review. BMC Med. Genet. 2020;21:109. doi: 10.1186/s12881-020-01028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Jaroudi D., Enabi S., AlThagafi M.S. Perrault syndrome with amenorrhea, infertility, Tarlov cyst, and degenerative disc. Gynecol. Endocrinol. 2019;35:1037–1039. doi: 10.1080/09513590.2019.1637407. [DOI] [PubMed] [Google Scholar]
- 57.Riley L.G., Rudinger-Thirion J., Frugier M., Wilson M., Luig M., Alahakoon T.I., Nixon C.Y., Kirk E.P., Roscioli T., Lunke S., et al. The expanding LARS2 phenotypic spectrum: HLASA, Perrault syndrome with leukodystrophy, and mitochondrial myopathy. Hum. Mutat. 2020;41:1425–1434. doi: 10.1002/humu.24050. [DOI] [PubMed] [Google Scholar]
- 58.Pan Z., Xu H., Tian Y., Liu D., Liu H., Li R., Dou Q., Zuo B., Zhai R., Tang W., et al. Perrault syndrome: Clinical report and retrospective analysis. Mol. Genet. Genomic Med. 2020:e1445. doi: 10.1002/mgg3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Knaap M.S., Bugiani M., Mendes M.I., Riley L.G., Smith D.E.C., Rudinger-Thirion J., Frugier M., Breur M., Crawford J., van Gaalen J., et al. Biallelic variants in LARS2 and KARS cause deafness and (ovario)leukodystrophy. Neurology. 2019;92:e1225–e1237. doi: 10.1212/WNL.0000000000007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cherot E., Keren B., Dubourg C., Carre W., Fradin M., Lavillaureix A., Afenjar A., Burglen L., Whalen S., Charles P., et al. Using medical exome sequencing to identify the causes of neurodevelopmental disorders: Experience of 2 clinical units and 216 patients. Clin. Genet. 2018;93:567–576. doi: 10.1111/cge.13102. [DOI] [PubMed] [Google Scholar]
- 61.Gotta F., Lamp M., Geroldi A., Trevisan L., Origone P., Fugazza G., Fabbri S., Nesti C., Rubegni A., Morani F., et al. A novel mutation of Twinkle in Perrault syndrome: A not rare diagnosis? Ann. Hum. Genet. 2020;84:417–422. doi: 10.1111/ahg.12384. [DOI] [PubMed] [Google Scholar]
- 62.Oldak M., Ozieblo D., Pollak A., Stepniak I., Lazniewski M., Lechowicz U., Kochanek K., Furmanek M., Tacikowska G., Plewczynski D., et al. Novel neuro-audiological findings and further evidence for TWNK involvement in Perrault syndrome. J. Transl. Med. 2017;15:25. doi: 10.1186/s12967-017-1129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dominguez-Ruiz M., Garcia-Martinez A., Corral-Juan M., Perez-Alvarez A.I., Plasencia A.M., Villamar M., Moreno-Pelayo M.A., Matilla-Duenas A., Menendez-Gonzalez M., Del Castillo I. Perrault syndrome with neurological features in a compound heterozygote for two TWNK mutations: Overlap of TWNK-related recessive disorders. J. Transl. Med. 2019;17:290. doi: 10.1186/s12967-019-2041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fekete B., Pentelenyi K., Rudas G., Gal A., Grosz Z., Illes A., Idris J., Csukly G., Domonkos A., Molnar M.J. Broadening the phenotype of the TWNK gene associated Perrault syndrome. BMC Med. Genet. 2019;20:198. doi: 10.1186/s12881-019-0934-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morino H., Pierce S.B., Matsuda Y., Walsh T., Ohsawa R., Newby M., Hiraki-Kamon K., Kuramochi M., Lee M.K., Klevit R.E., et al. Mutations in Twinkle primase-helicase cause Perrault syndrome with neurologic features. Neurology. 2014;83:2054–2061. doi: 10.1212/WNL.0000000000001036. [DOI] [PMC free article] [PubMed] [Google Scholar]