Abstract
Simple Summary
Many bilateral breast cancer patients with increased hereditary susceptibility to breast cancer result negative for BRCA1 or BRCA2 pathogenic variants and, thus, need a further genetic testing through a broader gene panel. Some patients with negative test result for BRCA1/2 pathogenic variants may harbor pathogenic variants in other breast cancer susceptibility genes, including ATM, CHEK2, PALB2, PTEN, TP53. Of course, the use of a multi-gene panel provides clinicians more information through a single test. Therefore, we focused on potential clinical impact of a NGS-based multi-gene panel testing in bilateral breast cancer patients, in order to evaluate the utility of perform a most comprehensive genetic analysis in these subjects, regardless the criteria concerning personal and family history of cancer established by the current guidelines. Our study revealed that the use of a NGS-based multiple-gene panel testing could increase the detection rates of germline alterations in bilateral breast cancer patients.
Abstract
Patients with unilateral breast cancer (UBC) have an increased risk of developing bilateral breast cancer (BBC). The annual risk of contralateral BC is about 0.5%, but increases by up to 3% in BRCA1 or BRCA2 pathogenic variant (PV) carriers. Our study was aimed to evaluate whether all BBC patients should be offered multi-gene panel testing, regardless their cancer family history and age at diagnosis. We retrospectively collected all clinical information of 139 BBC patients genetically tested for germline PVs in different cancer susceptibility genes by NGS-based multi-gene panel testing. Our investigation revealed that 52 (37.4%) out of 139 BBC patients harbored germline PVs in high- and intermediate-penetrance breast cancer (BC) susceptibility genes including BRCA1, BRCA2, PTEN, PALB2, CHEK2, ATM, RAD51C. Nineteen out of 53 positively tested patients harbored a PV in a known BC susceptibility gene (no-BRCA). Interestingly, in the absence of an analysis performed via multi-gene panel, a significant proportion (14.4%) of PVs would have been lost. Therefore, offering a NGS-based multi-gene panel testing to all BBC patients may significantly increase the detection rates of germline PVs in other cancer susceptibility genes beyond BRCA1/2, avoiding underestimation of the number of individuals affected by a hereditary tumor syndrome.
Keywords: ATM, BRCA1, BRCA2, breast cancer, bilateral breast cancer, CHECK2, germline pathogenic variants, multi-gene panel testing, PALB2, PTEN
1. Introduction
Breast cancer (BC) is the most commonly diagnosed female malignancies worldwide [1]. Women with unilateral breast cancer (UBC) have an increased risk of developing bilateral breast cancer (BBC) [2]. The cumulative incidence rate of developing contralateral BC at 10 years is about 3.4% for UBC patients [1,3]. In recent years, the increasing BC incidence rates observed through screening programs, improved treatment, and growing life expectancy have resulted in the early detection of increasing incidence of developing BBC [1,4]. Multiple criteria are associated with increased risk of second primary BC, including an early age of onset, hormone receptor-positivity of the initial tumor, race and ethnicity, as well as presence of known pathogenic variants (PVs) in hereditary cancer-associated genes [5]. Although BC is frequently a sporadic tumor (75–80%), approximately 15–20% are familial type and about 5–10% of cases are hereditary, caused by germline PVs in specific breast cancer-associated genes [5,6,7,8]. The two genes mainly involved in the BC genetic predisposition are BRCA1, located on chromosome 17 [9] and BRCA2, located on chromosome 13 [10]. Generally, the cumulative risk of developing BC by age 80 years has been observed to be 72% for germline BRCA1 PV carriers and 69% for germline BRCA2 PV carriers, respectively [11]. Specifically, the annual risk of contralateral BC is about 0.5%, but increases by up to 3% in BRCA1 or BRCA2 PV carriers, increasing 10-year cumulative risk up to 13–40% [1,12,13]. In a recent study, Kuchenbaecker et al. [11] have reported that the 20-year cumulative risk for contralateral BC, after a first BC diagnosis, was 40% for germline BRCA1 PV carriers and 26% for germline BRCA2 PV carriers. Moreover, germline BRCA1 PV carriers showed a 15–45% risk of developing ovarian cancer (OC) or tubal carcinoma, whereas germline BRCA2 PV carriers exhibited a 10–20% risk of developing OC [14,15,16].
Currently, both the National Comprehensive Cancer Network (NCCN) guidelines and European Society for Medical Oncology (ESMO) guidelines recommend genetic testing for BRCA1 and BRCA2 for women with multiple primary breast cancers, if first diagnosis was ≤50 years old [17,18]. Additionally, the Italian guidelines, established by the Italian Medical Oncology Association (AIOM), provide the same recommendation, also suggesting to perform the genetic tests on patients with a personal history of BC diagnosed before age 50 years with a family history of one or more first-degree relatives with BBC. With the increase in the acquisition of multi-gene panels, genetic testing used for detecting alterations in other genes beyond BRCA1 and BRCA2 has become more prevalent [5,7]. Moreover, the role of genetic testing in BC is rapidly changing, becoming an interesting area of research. In fact, although BRCA1 and BRCA2 are the two high-penetrance genes mainly correlated with increased risk of hereditary BC, several studies have identified many other susceptibility genes for BC [5]. Recently, Tebaldi et al. [19] found high rates of PVs in another BC susceptibility gene, named PALB2, in individuals with BBC, also observing an overall higher rate of BBC in patients with non-BRCA PVs than those that were BRCA1- or BRCA2-positive. Additionally, a recent study has defined high-risk BC genes as those with a BC odds ratio >5.0 (BRCA1, BRCA2, PALB2, STK11, TP53, PTEN, and CDH1) [20]. In particular, PALB2 and PTEN are considered high-risk genes with lifetime risk above 40%. Low/moderate-risk BC genes have been reported as those with a BC odds ratio between 2.0 and 5.0 (ATM, CHEK2, and NBN) [20]. The identification of germline PVs in high-risk individuals allowed an increase in surveillance and earlier implementation of the risk-reduction strategies [5,21,22]. However, germline PVs detected in low/moderate-risk BC genes are also interesting and may have a significant clinical impact [23]. It was observed that approximately 1% of women of European origin affected by BC harbor germline CHEK2 PVs, including the most commonly detected founder mutation named 1100delC [24]. The probability of carrying a CHEK2 PV is high in the Netherlands [24], where the risk of a contralateral BC was estimated to be increased by up to 6.5-fold for women with a CHEK2 1100delC mutation [25].
Based on a Breast Cancer BRCA System database retrospectively collected at University Hospital Policlinico “P. Giaccone” of Palermo, the aim of this work was to describe the typology and gene location of germline PVs detected both in BRCA1/2 and other susceptibility genes, in order to investigate the prevalence of different inherited genetic variants in BBC patients and evaluate the utility of performing a NGS-based multi-gene panel testing in these individuals. This information could be useful and interesting in order to decide if the use of a multi-gene panel testing is recommendable for BBC patients, regardless their cancer family history and age at diagnosis.
2. Results
2.1. Clinical Features of Bilateral Breast Cancer Patients
In total, 139 BBC patients (137 of which females and two males) were recruited and studied over a period ranging from October 2015 to June 2020 at the “Regional Center for the prevention, diagnosis and treatment of rare and heredo-familial tumors of adults” of the Section of Medical Oncology of the University Hospital Policlinico “P. Giaccone” of Palermo.
Ninety-three patients had metachronous BC, and 46 had synchronous BC. The average age of the first malignancy diagnosis was 45 years (range: 21–77 years). The average age of the second tumor was 51 years (range: 21–80 years). The average interval between the first and second BC diagnosis was 4 ± 6.08 years. In our study, 88 out of 139 patients had a second contralateral tumor within 5 years of the first tumor diagnosis, 27 subjects between 6 and 10 years, while only 24 individuals developed second tumor 10 years after the first diagnosis.
Considering the general clinical features and family history of the study population, 53 women (38.1%) were postmenopausal, five women (3.6%) had previous ovarian cancer, 82 patients (59%) had a BC family history, 44 (31.6%) of which with one relative and 38 (27.4%) with two or more family members affected by BC. Moreover, 11 patients (7.9%) had a family history of ovarian cancer, eight (5.7%) of which with one relative and three (2.2%) with two or more family members affected by ovarian cancer (Table 1).
Table 1.
Clinical characteristics of bilateral breast cancer patients.
| BBC (n = 139) | No. Patients (%) |
|---|---|
| Tumor description | |
| Metachronous | 93 (66.9%) |
| Synchronous | 46 (33.1%) |
| Sex | |
| Female | 137 |
| Male | 2 |
| Age at diagnosis (years) | |
| Primary tumor | |
| Median (range) | 45 (21–77) |
| ≤40 | 41 (29.5%) |
| 41–50 | 53 (38.12%) |
| 51–60 | 29 (20.9%) |
| ≥60 | 16 (11.5%) |
| Secondary tumor | |
| Median (range) | 51 (21–80) |
| ≤40 | 21 (15.1%) |
| 41–50 | 48 (34.5%) |
| 51–60 | 40 (28.8%) |
| ≥60 | 30 (21.6%) |
| Time between 1st and 2nd tumors (years) | |
| Median ± DS | 4 ± 6.08 |
| ≤5 | 88 (63.3%) |
| 6–10 | 27 (19.4%) |
| >10 | 24 (17.3%) |
| Histology 1st breast cancer | |
| Ductal in situ | 96 (69%) |
| Ductal invasive | 13 (9.4%) |
| Lobular invasive | 12 (8.6%) |
| Others | 18 (13%) |
| Histology 2nd breast cancer | |
| Ductal in situ | 16 (11.5%) |
| Ductal invasive | 92 (66.2%) |
| Lobular invasive | 15 (10.8%) |
| Others | 16 (11.5%) |
| Molecular phenotype 1st breast cancer | |
| Luminal A | 28 (20.1%) |
| Luminal B/HER2 - | 49 (35.3%) |
| Luminal B/HER2 + | 7 (5%) |
| HER2 + (non-luminal) | 6 (4.3%) |
| Triple negative | 12 (8.6%) |
| Unknown | 37 (26.7%) |
| Molecular phenotype 2nd breast cancer | |
| Luminal A | 35 (25.2%) |
| Luminal B/HER2 - | 35 (25.2%) |
| Luminal B/HER2 + | 10 (7.2%) |
| HER2 + (non-luminal) | 5 (3.6%) |
| Triple negative | 17 (12.2%) |
| Unknown | 37 (26.6%) |
| Genetic testing results | |
| Pathogenic variant BRCA1 | 13 (9.3%) |
| Pathogenic variant BRCA2 | 19 (13.7%) |
| Pathogenic variant BRCA1/BRCA2 | 1 (0.7%) |
| Pathogenic variant CHEK2 | 5 (3.6%) |
| Pathogenic variant PALB2 | 8 (5.8 %) |
| Pathogenic variant ATM | 3 (2.2%) |
| Pathogenic variant PTEN | 1 (0.7%) |
| Pathogenic variant RAD51C | 2 (1.4%) |
| Pathogenic variant MUTHY | 1 (0.7%) |
| Variant of uncertain significance | 27 (19.4%) |
| Negative | 59 (42.5%) |
| Type of surgery 1st breast cancer | |
| Mastectomy | 50 (36.2%) |
| Breast conserving therapy | 74 (53.2%) |
| Unknown | 15 (10.9%) |
| Type of surgery 2nd breast cancer | |
| Mastectomy | 48 (34.5%) |
| Breast conserving therapy | 55 (39.9%) |
| Unknown | 35 (25.3%) |
As regards the tumor histology of first BC, 96 (69%) out of 139 patients had ductal carcinoma in situ (DCIS), 13 (9.4%) invasive ductal carcinoma (IDC), 12 (8.6%) invasive lobular carcinoma (ILC), and finally 18 patients showed other types of BC. Considering the histology of the second tumor, 16 patients (11.5%) had DCIS, 92 (66.2%) IDC, 15 (10.8%) ILC, 16 patients (11.5%) other types of BC. Finally, both for the first and second diagnosis, the most representative molecular phenotypes of BC were luminal B/HER2- (35.3% and 25.2%, respectively) and luminal A (20.1% and 25.1%, respectively) (Table 1).
2.2. Detection of Germline Pathogenic Variants in Cancer Susceptibility Genes by Multi-Gene Panel Testing
All 139 probands were genetically tested for germline PVs in different cancer susceptibility genes, including BRCA1 and BRCA2, by NGS-based multi-gene panel testing. The mutational screening showed that 52 (37.4%) out of 139 genetically tested BBC patients harbored germline PVs (class V) in several high- and intermediate-penetrance BC susceptibility genes including BRCA1 and BRCA2, one patient showed a heterozygous PV in MUTYH gene, 27 (19.4%) patients were carriers of variants of uncertain significance (VUS; class III), six of which in BRCA1/2, whereas 59 (42.5%) showed no genetic variants (classes III, IV, and V) (Table 1).
Our analysis revealed that 33 (62.3%) out of 53 PV-positive BBC patients have been shown to harbor germline BRCA1/2 PVs, 13 (24.5%) of which in BRCA1 and 19 (35.8%) in BRCA2, and only one (1.9%) in both genes (double heterozygosity for BRCA1 and BRCA2 PVs) (Table 2). Nineteen (35.8%) out of 53 individuals positively tested by multi-gene panel have been shown to carry a PV in a known BC susceptibility gene (no-BRCA). In particular, five (9.4%) out of 53 PV-positive BBC patients have been shown to harbor CHEK2 PVs, eight patients (15.1%) showed PALB2 PVs, three individuals (5.7%) carried ATM PVs, only one patient (1.9%) exhibited a PTEN PV, while PVs in RAD51C were detected in two (3.8%) triple-negative BC patients (Table 3). No PVs were detected in MMR genes (MLH1, MSH2, MSH6, PMS2, and EPCAM).
Table 2.
BRCA1/2 pathogenic variants detected in patients with bilateral breast cancers.
| Gene | Variant Type | HGVS Nomenclature | Protein Change | No. Patients | Allele Frequency (ExAC */GnomAD **) | Allele Frequency (Study Cohort) |
|---|---|---|---|---|---|---|
| BRCA1 | fs | c.4964_4982del | p.Ser1655fs | 4 (7.4%) | ExAC 0.000008 | 0.0.14 |
| BRCA1 | fs | c.3226_3227AG [1] | p.Gly1077fs | 2 (3.8%) | GnomAD 0.000004 | 0.0072 |
| BRCA1 | NS | c.3904G>T | p.Glu1302Ter | 2 (3.8%) | / | 0.0072 |
| BRCA1 | fs | c.1531del | p.Gly512Terfs | 1 (1.9%) | / | 0.0036 |
| BRCA1 | fs | c.514del | p.Gln172fs | 1 (1.9%) | ExAC 0.000008 GnomAD 0.000004 |
0.0036 |
| BRCA1 | fs | c.5266dupC | p.Gln1756Profs | 1 (1.9%) | ExAC 0.000156 GnomAD 0.00018 |
0.0036 |
| BRCA1 | fs | c.3266del | p.Leu1089fs | 1 (1.9%) | / | 0.0036 |
| BRCA1 | NS | c.4327C > T | p.Arg1443Ter | 1 (1.9%) | GnomAD 0.000024 | 0.0036 |
| BRCA2 | fs | c.1238del | p.Leu413fs | 4 (7.4%) | GnomAD 0.000004 | 0.0.14 |
| BRCA2 | fs | c.9026_9030del | p.Tyr3009fs | 2 (3.8%) | GnomAD 0.000004 | 0.0072 |
| BRCA2 | fs | c.9253dupA | p.Thr3085Asnfs | 2 (3.8%) | / | 0.0072 |
| BRCA2 | fs | c.6082_6086del | p.Glu2028fs | 2 (3.8%) | ExAC 0.000008 GnomAD 0.000004 | 0.0072 |
| BRCA2 | fs | c.9098_9099insA | p.Gln3034fs | 1 (1.9%) | / | 0.0036 |
| BRCA2 | NS | c.8594T > A | p.Leu2865Ter | 1 (1.9%) | / | 0.0036 |
| BRCA2 | M | c.631G > A | p.Val211Ile | 1 (1.9%) | / | 0.0036 |
| BRCA2 | fs | c.5851_5854del | p.Ser1951fs | 1 (1.9%) | / | 0.0036 |
| BRCA2 | fs | c.2808_2811del | p.Ala938Profs | 1 (1.9%) | ExAC 0.000017 GnomAD 0.000008 | 0.0036 |
| BRCA2 | IVS | c.7008-2A > T | / | 1 (1.9%) | / | 0.0036 |
| BRCA2 | IVS | c.476-2A > G | / | 1 (1.9%) | / | 0.0036 |
| BRCA2 | M | c.7007G > A | p.Arg2336His | 1 (1.9%) | / | 0.0036 |
| BRCA2 | NS | c.93G > A | p.Trp31Ter | 1 (1.9%) | GnomAD 0.000004 | 0.0036 |
| BRCA2/ BRCA1 | IVS/M | c.8331 + 2T > C/c.181T > G | /p.Cys61Gly | 1 (1.9%) | /-ExAC 0.000067 GnomAD 0.000032 |
0.0036 |
Abbreviations: fs, frameshift; IVS, intronic variants; M, missense; NS, non-sense. * Dataset ExAC v1.0; ** Dataset GnomAD v2.1.1.
Table 3.
Pathogenic variants detected in no-BRCA genes of patients with bilateral breast cancers positively tested by multi-gene panel.
| Gene | Variant Type | HGVS Nomenclature | Protein Change | No. Patients | Allele Frequency (ExAC */GnomAD **) | Allele Frequency (Study Cohort) |
|---|---|---|---|---|---|---|
| CHEK2 | fs | c.1100del | p.Thr367fs | 5 (9.2%) | gnomAD 0.00204 ExAC 0.00182 |
0.018 |
| RAD51C | NS | c.224dup | p.Tyr75Ter | 1 (1.9%) | gnomAD 0.00001 | 0.0036 |
| ATM | NS | c.8818_8821dup | p.Ser2941Ter | 1 (1.9%) | / | 0.0036 |
| PALB2 | fs | c.758dup | p.Ser254fs | 2 (3.8%) | gnomAD 0.00002 ExAC 0.00003 |
0.0072 |
| PALB2 | NS | c.2566C > T | p.Gln856Ter | 2 (3.8%) | gnomAD 0.00000 ExAC 0.00001 |
0.0072 |
| PALB2 | fs | c.1050_1053del | p.Thr351fs | 2 (3.8%) | / | 0.0072 |
| PALB2 | NS | c.2257C > T | p.Arg753Ter | 2 (3.8%) | gnomAD 0.00002 ExAC 0.00003 |
0.0072 |
| MUTYH | M | c.1103G > A | p.Gly368Asp | 1 (1.9%) | gnomAD 0.00303 ExAC 0.00280 |
0.0036 |
| PTEN | M | c.284C > A | p.Pro95Gln | 1 (1.9%) | / | 0.0036 |
| ATM | M | c.8147T > C | p.Val2716Ala | 1 (1.9%) | gnomAD 0.00003 ExAC 0.00004 |
0.0036 |
| RAD51C | IVS | c.1026 + 5_1026 + 7del | / | 1 (1.9%) | / | 0.0036 |
| ATM | LGR | Exon 57–61del | / | 1 (1.9%) | / | 0.0036 |
Abbreviations: fs, frameshift; IVS, intronic variants; LGR, large genomic rearrangement; M, missense; NS, non-sense. *Dataset ExAC v1.0; ** Dataset GnomAD v2.1.1.
Additionally, based on the classification criteria developed by the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) consortium and according to IARC recommendations [26], 28 different genetic variants detected in 27 BBC patients were classified as VUS (class III). The VUS analysis showed that these variants were mainly located on 13 genes, including ATM, BRCA1, BRCA2, RAD50, CHEK2, APC, BARD1, TP53, etc. (Table 4). Most VUS were found on the ATM gene in a heterozygous state (seven different variants, 25%), but to a lesser degree also on BRCA2 (14%), APC (11%), and BRCA1 (7%) (Figure 1). Finally, our analysis also showed that some patients were carriers of more than one VUS on different genes.
Table 4.
Variant of uncertain significance detected in patients with bilateral breast cancers.
| Gene | Variant Type | HGVS Nomenclature | Protein Change | Variant Interpretation | No. Patients | Allele Frequency (ExAC a/GnomAD b) | Allele Frequency (Cohort Study) |
|---|---|---|---|---|---|---|---|
| BRCA1 | M | c.4054G > A | p.Glu1352Lys | VUS | 1 (3.70%) | gnomAD 0.00002 ExAC 0.00004 |
0.0036 |
| BRCA1 | IVS | c.670 + 31A > C | / | VUS | 1 (3.70%) | / | 0.0036 |
| BRCA2 | M | c.5267T > A | p.Val1756Glu | VUS | 1 (3.70%) | gnomAD 0.00000 ExAC 0.00001 |
0.0036 |
| BRCA2 | M | c.8299C > T | p.Pro2767Ser | VUS | 1 (3.70%) | / | 0.0036 |
| BRCA2 | M | c.1769T > G | p.Phe590Cys | CIP | 1 (3.70%) | gnomAD 0.00001 ExAC 0.00002 |
0.0036 |
| BRCA2 | M | c.9581C > A | p.Pro3194Gln | CIP | 1 (3.70%) | gnomAD 0.00001 ExAC 0.00001 |
0.0036 |
| TP53 | M | c.446C > T * | p.Ser149Phe | VUS | 1 (3.70%) | / | 0.0036 |
| ATM | M | c.1229T > C * | p.Val410Ala | CIP | gnomAD 0.00222 ExAC 0.00217 |
0.0036 | |
| EPCAM | M | c.334G > A | p.Gly112Ser | VUS | 1 (3.70%) | / | 0.0036 |
| MUTYH | M | c.1378 C > T ** | p.Arg460Cys | VUS | 1 (3.70%) | gnomAD 0.00005 ExAC 0.00005 |
0.0036 |
| NBN | M | c.283G > A ** | p.Asp95Asn | CIP | gnomAD 0.00173 ExAC 0.00186 |
0.0036 | |
| ATM | M | c.6407G > C | p.Arg2136Thr | VUS | 1 (3.70%) | / | 0.0036 |
| MUTYH | M | c.202T > C | p.Ser68Pro | VUS | 1 (3.70%) | gnomAD 0.00001 ExAC 0.00001 |
0.0036 |
| CHEK2 | M | c.1388G > A | p.Cys463Tyr | VUS | 2 (7.4%) | gnomAD 0.00000 | 0.0072 |
| MSH2 | M | c.1111G > C | p.Glu371Gln | VUS | 1 (3.70%) | / | 0.0036 |
| APC | M | c.3920T > A | p.Ile1307Lys | CIP, Risk Factor | 1 (3.70%) | gnomAD 0.00201 ExAC 0.00169 |
0.0036 |
| ATM | M | c.4060C > A *** | p.Pro1354Thr | CIP | 1 (3.70%) | gnomAD 0.00019 ExAC 0.00021 |
0.0036 |
| CHEK2 | M | c.1441 G > T *** | p.Asp481Tyr | CIP | 2 (7.4%) | gnomAD 0.00039 ExAC 0.00028 |
0.0072 |
| ATM | M | c.6983C > T | p.Pro2328Leu | VUS | 1 (3.70%) | gnomAD 0.00000 | 0.0036 |
| APC | M | c.3949G > C **** | p.Glu1317Gln | CIP | 1 (3.70%) | gnomAD 0.00438 ExAC 0.00413 |
0.0036 |
| ATM | M | c.4258C > T **** | p.Leu1420Phe | CIP | gnomAD 0.01104 ExAC 0.01271 |
0.0036 | |
| BARD1 | M | c.1793C > T | p.Thr598Ile | VUS | 1 (3.70%) | gnomAD 0.000008 | 0.0036 |
| ATM | M | c.6067G > A | p.Gly2023Arg | CIP | 1 (3.70%) | gnomAD 0.00143 ExAC 0.00157 |
0.0036 |
| MSH6 | M | c.663A > C | p.Glu221Asp | CIP | 2 (7.4%) | gnomAD 0.00071 ExAC 0.00063 |
0.0072 |
| ATM | M | c.6293T > C | p.Leu2098Pro | VUS | 1 (3.70%) | gnomAD 0.00000 ExAC 0.00001 |
0.0036 |
| RAD50 | M | c.1277A > G | p.Gln426Arg | CIP | 1 (3.70%) | gnomAD 0.00014 ExAC 0.00015 |
0.0036 |
| APC | M | c.5338C > T | p.Pro1780Ser | VUS | 1 (3.70%) | / | 0.0036 |
| RAD50 | M | c.1094G > A | p.Arg365Gln | CIP | 1 (3.70%) | gnomAD 0.00046 ExAC 0.00040 |
0.0036 |
Abbreviations: CIP, conflicting interpretations of pathogenicity; M, missense; VUS, variant of uncertain significance; a Dataset ExAC v1.0; b Dataset GnomAD v2.1.1 * These variants in ATM and TP53 genes were observed in the same patient. ** These variants in MUTYH and NBN genes were observed in the same patient. *** These variants in ATM and CHEK2 genes were observed in the same patient. **** These variants in ATM and APC genes were observed in the same patient.
Figure 1.
Distribution and frequency of variants of uncertain significance (VUS) identified in different genes of bilateral breast cancer patients. The OncoPrint, showing the identified VUS by heatmap, was obtained by the informatic tool Mutation Mapper-cBioPortal for Cancer Genomics.
2.3. Type and Gene Location of Genetic Variants in Bilateral Breast Cancers
The most frequently detected PVs in most BRCA-positive carriers were c.4964_4982del (p.Ser1655fs) and c.3226_3227AG [1] (p.Gly1077fs) for BRCA1, and c.1238del (p.Leu413fs) for BRCA2. The most common Sicilian founder variant named c.4964_4982del [27] resulted as among the most recurrent BRCA1 PVs in BBC patients, since this variant was detected in four families, whereas the other most frequent BRCA1 PV named c.3226_3227AG [1] was reported as a founder mutation both in Italy (Tuscany) and Norway [28].
Additionally, the BRCA2 PV named c.1238del, frequently detected in the luminal B molecular subtypes observed in BBCs, could be a good candidate for becoming a potential variant with founder effect in Sicily, but further in-depth studies are needed.
Interestingly, other two BRCA2 PVs named c.9098_9099insA (p.Gln3034fs) and c.8594T > A (p.Leu2865Ter), albeit found in only one BBC proband, respectively, were detected in 18 and 10 family members, respectively.
As regards the analysis of other moderate-risk susceptibility genes, the c.1100delC variant, as already previously demonstrated by other authors [29,30], has been shown to be the most recurrent CHEK2 PV in our study population, since it was detected in five BBC patients. Interestingly, we found that this variant mainly correlates with a luminal A/B phenotype, estrogen receptor positivity >60%, and progesterone receptor positivity between 20% and 60%.
Although PALB2 has been shown to be the most frequently altered gene in BBC patients, besides BRCA1 and BRCA2, however four different variants were identified. Among no-BRCA genes, also heterozygous ATM resulted as altered in BBC individuals, showing three different genetic variants, including a large genomic rearrangement (LRG). The ATM PVs harbored by all three BBC patients were mainly observed in the luminal B tumor phenotype. Finally, a monoallelic PV in MUTYH gene, whose homozygous alterations are usually correlated with MUTYH-associated colon polyposis (MAP) syndrome and colorectal cancer [31], was unusually observed in one BBC patient.
Our study also aimed to evaluate the typology and gene location of germline PVs in BBCs, in order to investigate potential associations between specific PVs and bilaterality in BC.
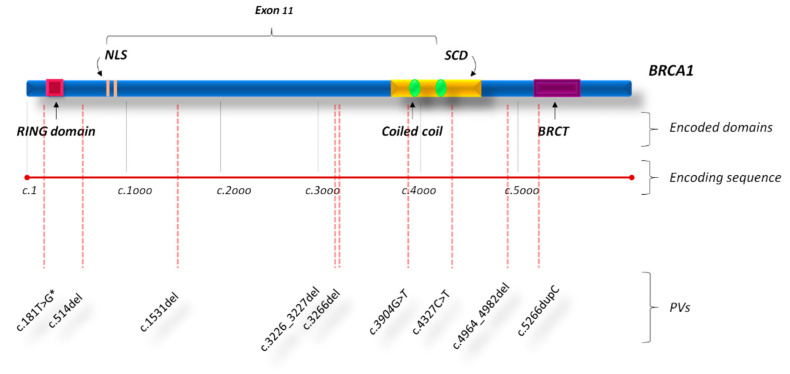
As regards the gene location of BRCA1 PVs detected in BBC patients of our study cohort, most of observed BRCA1 PVs (6/9) were frameshift mutations and were equally distributed along the entire gene sequence, in particular inside three hypothetical cluster regions present in the BRCA1 protein structure which includes the RING domain at the N-terminus, a region encoded by exon 11, and BRCT domain near the C-terminus. Five out of nine BRCA1 PVs were detected in the exon 11 of BBC patients (nucleotides: 1650–4446; codons: 512–1143), whereas two in the sequence corresponding to the BRCT repeats (nucleotides: 5083–5382; codons: 1655–1756) and the other two in the RING domain (nucleotides: 300–633; codons: 61–172) (Figure 2).
Figure 2.
BRCA1 functional domains and gene location of BRCA1 PVs in bilateral breast cancer patients. Abbreviations: BRCT, BRCA1 C-terminus domain; NLS, nuclear localization sequence; PVs, pathogenic variants; SCD, serine cluster domain. * This PV is present together with the BRCA2 PV named c.8331+2T>C in one patient showing double heterozygosity for BRCA1 and BRCA2 PVs.
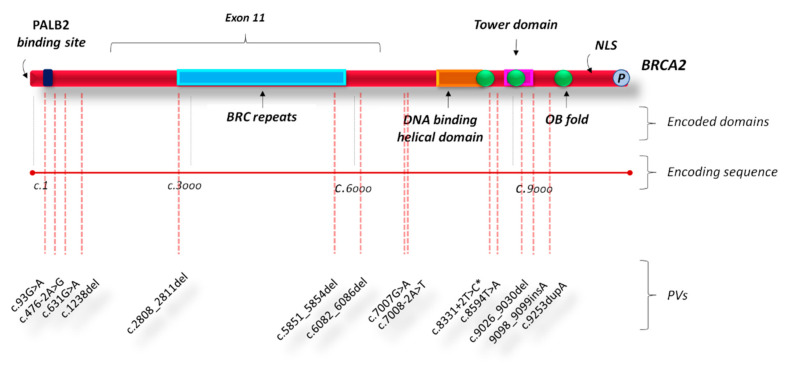
Conversely, most of observed BRCA2 PVs were mainly localized inside three other putative cluster regions present in the BRCA2 protein structure which includes the PALB2 binding site at the N-terminus, BRC repeats (located within the exon 11), and DNA binding helical domain near the C-terminus. Four BRCA2 PVs were located at the N-terminus, near the PALB2 binding site (nucleotides: 321–1466; codons: 31–413), whereas three inside the exon 11 (nucleotides: 3036–6310; codons: 938–2028), two of which within the BRC repeats, and, finally, five variants were detected at the C-terminus, near DNA binding helical domain (nucleotides: 8559–9481; codons: 2865–3085) (Figure 3). Half of the BRCA2 PVs found in BBC patients were frameshift mutations, three were intronic variants (IVS), while two were missense and two nonsense.
Figure 3.
BRCA2 functional domains and gene location of BRCA2 PVs in bilateral breast cancer patients. Abbreviations: NLS, nuclear localization sequence; OB, oligonucleotide binding; PVs, pathogenic variants. * This PV is present together with the BRCA1 PV named c.181T>G in one patient showing double heterozygosity for BRCA1 and BRCA2 PVs.
3. Discussion
The incidence of BBC is slightly higher, 3%, than all BCs. More precisely, synchronous tumors (contemporary bilaterality) represent 0.6%, while those metachronous constitute 2.2% [32,33].
Some of these BBCs have been shown to be related to the Hereditary Breast and Ovarian Cancer (HBOC) syndrome, since the annual risk of contralateral BC significantly increases in BRCA1 or BRCA2 PV carriers [13]. However, many BBC patients with increased hereditary susceptibility to BC result negative for BRCA1 or BRCA2 PVs and, thus, need further genetic testing through a broader gene panel. Some patients with negative test result for BRCA1/2 PVs may harbor a still undiscovered PV in these genes, probably due to the sequencing limitations, or PVs in other cancer susceptibility genes, including ATM, CHEK2, PALB2, PTEN, TP53, and others, in 3–4% of cases [34,35]. For this reason, today, thanks also to the progress obtained by next-generation sequencing (NGS) technology which revolutionized the clinical approach to genetic testing, the use of multi-gene panels containing several susceptibility genes beyond BRCA1/2 is becoming increasingly frequent [36,37]. Recent studies showed that, because of overlapping phenotypes, a significant number of PVs, which would be missed if individually tested for HBOC, Cowden syndrome, Li–Fraumeni syndrome, Lynch syndrome, or other hereditary cancer syndromes, could be more easily detected by multi-gene panel testing. Therefore, single gene panels covering all these syndromes rather than individual syndrome-specific panels were generated [38]. Of course, the use of a multi-gene panel provides clinicians more information about one or more syndromes in a single test [39]. The clinical significance of comprehensive multi-gene panels useful in the clinical management of hereditary breast and ovarian tumors was recently investigated in numerous studies [36,40]. However, today, the debate about the choice of genes to include for each syndrome and identification of specific groups of high-risk patients who should be offered multi-gene panel testing still remains open. Indeed, to date, no well-defined guidelines have been written to identify criteria for selecting the most suitable patients to perform multi-gene panel testing on.
In this work, we retrospectively collected and analyzed all clinical information of 139 BBC patients who have been genetically tested for germline PVs in different cancer susceptibility genes, including BRCA1 and BRCA2, by NGS-based multi-gene panel testing. This study aimed to evaluate whether all BBC patients should be offered multi-gene panel testing, regardless of their cancer family history and age at diagnosis.
Our investigation revealed that 52 (37.4%) out of 139 genetically tested BBC patients were shown to carry germline PVs in high- and intermediate-penetrance BC susceptibility genes such as BRCA1, BRCA2, PTEN, PALB2, CHEK2, ATM, RAD51C. Interestingly, a noteworthy correlation between PVs in PALB2 or CHEK2 and BBCs was observed. In addition, our study showed that CHEK2 PVs correlate with a luminal A/B phenotype and ATM PVs with a luminal B subtype. Lower percentages of PVs were found also in PTEN and RAD51C genes. Furthermore, 19.4% of patients with BBC (27/139) were shown to be carriers of VUS in several cancer susceptibility genes.
Finally, we observed that most of BBC-related germline BRCA1/2 PVs were frameshift mutations mainly located within the exon 11 for BRCA1, and near the PALB2 binding site (at the N-terminus) and the DNA binding helical domain (at the C-terminus) for BRCA2.
Our work showed that a deeper genetic analysis through NGS-based multi-gene panel testing may help us to identify PVs in high- and moderate-risk susceptibility genes different from most common BRCA1 and BRCA2 genes, allowing to stratify a considerable portion of hereditary BBC patients who may benefit from the screening programs, active surveillance strategies, or risk-reducing surgery interventions, where necessary. In addition, this information could allow to identify family members with a greater risk of developing BC (or other tumors) and implement prevention and surveillance programs for these subjects. However, larger study cohorts are needed in order to determine more precise rates of PVs in BBC patients with previously negative BRCA genetic testing. Finally, our study has shown been to be compatible with the recommendations of the current NCCN guidelines, providing a more cost-effective cancer risk evaluation compared with a gene-by-gene approach.
4. Patients and Methods
4.1. Study Population
A retrospective cohort study was performed at the “Sicilian Regional Center for the Prevention, Diagnosis and Treatment of Rare and Heredo-Familial Tumors” of the Section of Medical Oncology of University Hospital Policlinico “P. Giaccone” of Palermo. All patients diagnosed with BBC, enrolled from October 2015 to June 2020 at a single institution, underwent germline genetic testing by multi-gene panel including: (i) high-risk BC susceptibility genes such as BRCA1, BRCA2, CDH1, PALB2, PTEN, STK11, and TP53; (ii) moderate-risk BC susceptibility genes such as ATM, BARD1, BRIP1, CHEK2, NBN, RAD50, RAD51C, and RAD51D; (iii) cancer predisposition genes related to other hereditary tumor syndromes such as MLH1, MSH2, MSH6, PMS2, EPCAM, APC, MUTYH. The only criterion used for performing genetic testing was the presence of a BBC in patients above age 18 years, regardless of criteria concerning the cancer family history and age at diagnosis established by the guidelines.
Genetic counselling was carried out by a multidisciplinary team mainly consisting of an oncologist, a geneticist, and a psychologist. The information concerning the personal and familial history of tumor, family geographical origin, age of cancer diagnosis, disease stages (I–IV), histological tumor subtype, molecular phenotype, risk factors were anonymously recorded for all patients who previously provided a written informed consent. The estrogen-receptor (ER), progesterone-receptor (PgR), HER2-receptor status (HER2), Ki67 status, and histological grade (Grades I, II, and III) of the primary and secondary tumors were extrapolated from histological report of diagnostic core biopsies or tumor resections for clinical use.
When a pathogenic or likely pathogenic variant was identified in a patient, the genetic test result was considered informative. Conversely, genetic test result was defined not informative, when no pathogenic or likely pathogenic variant was detected, but its presence could not be excluded, or a VUS to which it was not possible to attribute a risk value was detected.
Patients carrying a germline PV in any of analyzed genes were addressed to enhanced screening programs and/or risk-reducing surgical strategies by an oncologist with expertise in cancer genetics. Targeted genetic testing was proposed and extended to the first-degree family members of patients harboring a mutation, after providing informed consent.
4.2. Sample Collection and Next-Generation Sequencing Analysis by Multi-Gene Panel
Peripheral blood samples were collected from BBC patients. Genomic DNA was isolated from the peripheral blood using the DNeasy® Blood Kit (QIAGEN), quantified by Qubit® 3.0 fluorometer (Thermofisher Scientific, Waltham, MA, USA) and its quality was assessed by using 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). We used 4 ng of DNA to prepare the barcoded library using a Multi-Gene Hereditary Cancer Panel Testing named HEVA SCREEN (4bases SA) that has allowed to perform a mutational screening on 22 genes involved in risk of hereditary breast, ovarian and colorectal cancer, and other inherited tumor syndromes (ATM, APC, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD50, RAD51C, RAD51D, STK11, and TP53). The kit consists of three multiplex PCR primer pools. We used 20 ng of DNA per primer pool for multiplex PCR amplification, followed by barcode ligation and purification with Agentcourt AMPureXP reagent (Beckman Coulter, Beverly, MA, USA). Quantity and quality of prepared libraries were assessed by Qubit® 3.0 fluorometer (Thermofisher Scientific) and Agilent 2100 Bioanalyzer on-chip electrophoresis (Agilent Technologies), respectively, as previously described [41]. Subsequently, libraries were mixed in an equimolar ratio and emulsion PCR was performed with the Ion OneTouch OT2 System (Thermofisher Scientific) using Ion 540 Kit-OT2 (Thermofisher Scientific). Finally, sequencing was performed with Ion 540 Chip (Thermofisher Scientific) using Ion Torrent S5 (Thermofisher Scientific) instrument. The sequencing data was analyzed with Amplicon Suite (SmartSeq s.r.l.) and Ion Reporter Software v.5.12 (Thermofisher Scientific).
4.3. Sanger Sequencing
Genetic variants detected in the analyzed genes were validated by Sanger sequencing using a BigDye Therminator 3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA) and read through the 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), according to manufacturer’s protocols.
4.4. Genetic Variant Classification
Based on the classification criteria developed by the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) consortium (https://enigmaconsortium.org/) and according to IARC recommendations [26], the detected genetic variants were divided into five classes: benign (class I), likely benign (class II), variant of uncertain significance (VUS, class III), likely pathogenic (class IV), and pathogenic (class V). For the identification and classification of genetic variants, several databases, such as ClinVar, BRCA Exchange, LOVD, were used. The localization of the variants on the different genetically tested genes was described and graphically represented using the informatic tool Mutation Mapper-cBioPortal for Cancer Genomics [42,43].
The variants identified in the different genes were named according to the recommendations for the description of sequence variants provided by the Human Genome Variation Society (HGVS). HGVS nomenclature was approved by the HGVS, Human Variome Project (HVP), and the Human Genome Organization (HUGO) [44].
5. Conclusions
In this work, we focused on potential clinical impact of a NGS-based multi-gene panel testing in BBC individuals, in order to assess the utility of performing a most comprehensive genetic analysis in these patients, regardless the criteria concerning personal and family history of cancer established by the current guidelines.
Our study revealed that the use of a NGS-based multiple-gene panel testing could improve the detection rates of germline deleterious alterations in patients affected by BBC, since we observed that, in the absence of an analysis performed via multi-gene panel, a significant proportion (14.4%) of PVs would have been lost.
Acknowledgments
All authors thank Chiara Drago for the language English revision.
Author Contributions
Conceptualization, D.F., L.I., A.R. and V.B.; genetic counselling D.F., V.C., C.F.; sample collection and gene testing, M.B., A.F. and N.B.; data curation and analysis, L.I., D.F., L.R.C., C.B., and G.B.; writing D.F., L.I. and C.F.; supervision, A.R., G.B. and V.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pan B., Xu Y., Zhou Y.D., Yao R., Wu H.W., Zhu Q.L., Wang C.J., Mao F., Lin Y., Shen S.J., et al. The Prognostic Comparison Among Unilateral, Bilateral, Synchronous Bilateral, and Metachronous Bilateral Breast Cancer: A Meta-Analysis of Studies from Recent Decade (2008–2018) Cancer Med. 2019 doi: 10.1002/cam4.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sim Y., Tan V.K.M., Sidek N.A.B., Chia D.K.A., Tan B.K.T., Madhukumar P., Yong W.S., Wong C.Y., Ong K.W. Bilateral Breast Cancers in An Asian Population, and A Comparison Between Synchronous and Metachronous Tumours. ANZ J. Surg. 2018;88:982–987. doi: 10.1111/ans.14773. [DOI] [PubMed] [Google Scholar]
- 3.Lu W., Schaapveld M., Jansen L., Bagherzadegan E., Sahinovic M.M., Baas P.C., Hanssen L.M.H.C., van Der Mijle H.C.J., Brandenburg J.D., Wiggers T., et al. The Value of Surveillance Mammography of the Contralateral Breast in Patients with a History of Breast Cancer. Eur. J. Cancer. 2009;45:3000–3007. doi: 10.1016/j.ejca.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Hartman M., Czene K., Reilly M., Adolfsson J., Bergh J., Adami H.-O., Dickman P.W., Hall P. Incidence and Prognosis of Synchronous and Metachronous Bilateral Breast Cancer. J. Clin. Oncol. 2007;25:4210–4216. doi: 10.1200/JCO.2006.10.5056. [DOI] [PubMed] [Google Scholar]
- 5.Corredor J., Woodson A.H., Gutierrez Barrera A., Arun B. Multigene Panel Testing Results in Patients with Multiple Breast Cancer Primaries. Breast J. 2020 doi: 10.1111/tbj.13762. [DOI] [PubMed] [Google Scholar]
- 6.Apostolou P., Fostira F. Hereditary Breast Cancer: The Era of New Susceptibility Genes. Biomed Res. Int. 2013;2013:1–11. doi: 10.1155/2013/747318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valencia O.M., Samuel S.E., Viscusi R.K., Riall T.S., Neumayer L.A., Aziz H. The Role of Genetic Testing in Patients with Breast Cancer. JAMA Surg. 2017;152:589. doi: 10.1001/jamasurg.2017.0552. [DOI] [PubMed] [Google Scholar]
- 8.Kotsopoulos J., Huzarski T., Gronwald J., Singer C.F., Moller P., Lynch H.T., Armel S., Karlan B., Foulkes W.D., Neuhausen S.L., et al. Bilateral Oophorectomy and Breast Cancer Risk Inbrca1andbrca2mutation Carriers. JNCI J. Natl. Cancer Inst. 2017;109 doi: 10.1093/jnci/djw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L., Ding W., et al. A Strong Candidate for the Breast and Ovarian Cancer Susceptibility Gene Brca1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 10.Wooster R., Bignell G., Lancaster J., Swift S., Seal S., Mangion J., Collins N., Gregory S., Gumbs C., Micklem G., et al. Identification of the Breast Cancer Susceptibility Gene Brca2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 11.Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.-A., Mooij T.M., Roos-Blom M.-J., Jervis S., van Leeuwen F.E., Milne R.L., Andrieu N., et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for Brca1 and Brca2 Mutation Carriers. JAMA. 2017;317:2402. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 12.Metcalfe K., Gershman S., Lynch H.T., Ghadirian P., Tung N., Kim-Sing C., Olopade O.I., Domchek S., Mclennan J., Eisen A., et al. Predictors of Contralateral Breast Cancer in Brca1 and Brca2 Mutation Carriers. Br. J. Cancer. 2011;104:1384–1392. doi: 10.1038/bjc.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graeser M.K., Engel C., Rhiem K., Gadzicki D., Bick U., Kast K., Froster U.G., Schlehe B., Bechtold A., Arnold N., et al. Contralateral Breast Cancer Risk in Brca1 and Brca2 Mutation Carriers. J. Clin. Oncol. 2009;27:5887–5892. doi: 10.1200/JCO.2008.19.9430. [DOI] [PubMed] [Google Scholar]
- 14.Incorvaia L., Fanale D., Badalamenti G., Bono M., Calò V., Cancelliere D., Castiglia M., Fiorino A., Pivetti A., Barraco N., et al. Hereditary Breast and Ovarian Cancer in Families from Southern Italy (Sicily)—Prevalence and Geographic Distribution of Pathogenic Variants in Brca1/2 Genes. Cancers. 2020;12:1158. doi: 10.3390/cancers12051158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson D. Cancer Incidence in Brca1 Mutation Carriers. J. Natl. Cancer Inst. 2002;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 16.Breast Cancer Linkage Consortium Cancer Risks in Brca2 Mutation Carriers. J. Natl. Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 17.Bevers T.B., Helvie M., Bonaccio E., Calhoun K.E., Daly M.B., Farrar W.B., Garber J.E., Gray R., Greenberg C.C., Greenup R., et al. Breast Cancer Screening and Diagnosis, Version 3.2018, Nccn Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018;16:1362–1389. doi: 10.6004/jnccn.2018.0083. [DOI] [PubMed] [Google Scholar]
- 18.Senkus E., Kyriakides S., Ohno S., Penault-Llorca F., Poortmans P., Rutgers E., Zackrisson S., Cardoso F. Primary Breast Cancer: Esmo Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2015;26:8–30. doi: 10.1093/annonc/mdv298. [DOI] [Google Scholar]
- 19.Tedaldi G., Tebaldi M., Zampiga V., Danesi R., Arcangeli V., Ravegnani M., Cangini I., Pirini F., Petracci E., Rocca A., et al. Multiple-Gene Panel Analysis in A Case Series of 255 Women with Hereditary Breast and Ovarian Cancer. Oncotarget. 2017;8:47064–47075. doi: 10.18632/oncotarget.16791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slavin T.P., Maxwell K.N., Lilyquist J., Vijai J., Neuhausen S.L., Hart S.N., Ravichandran V., Thomas T., Maria A., Villano D., et al. The Contribution of Pathogenic Variants in Breast Cancer Susceptibility Genes to Familial Breast Cancer Risk. NPJ Breast Cancer. 2017;3 doi: 10.1038/S41523-017-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Economopoulou P., Dimitriadis G., Psyrri A. Beyond Brca: New Hereditary Breast Cancer Susceptibility Genes. Cancer Treat. Rev. 2015;41:1–8. doi: 10.1016/j.ctrv.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Singer C.F., Balmaña J., Bürki N., Delaloge S., Filieri M.E., Gerdes A.-M., Grindedal E.M., Han S., Johansson O., Kaufman B., et al. Genetic Counselling and Testing of Susceptibility Genes for Therapeutic Decision-Making in Breast Cancer—An European Consensus Statement and Expert Recommendations. Eur. J. Cancer. 2019;106:54–60. doi: 10.1016/j.ejca.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Narod S.A. Bilateral Breast Cancers. Nat. Rev. Clin. Oncol. 2014;11:157–166. doi: 10.1038/nrclinonc.2014.3. [DOI] [PubMed] [Google Scholar]
- 24.Zhang S., Phelan C.M., Zhang P., Rousseau F., Ghadirian P., Robidoux A., Foulkes W., Hamel N., Mccready D., Trudeau M., et al. Frequency of the Chek2 1100delc Mutation Among Women with Breast Cancer: An international Study. Cancer Res. 2008;68:2154–2157. doi: 10.1158/0008-5472.CAN-07-5187. [DOI] [PubMed] [Google Scholar]
- 25.Broeks A., De Witte L., Nooijen A., Huseinovic A., Klijn J.G.M., van Leeuwen F.E., Russell N.S., van’t Veer L.J. Excess Risk for Contralateral Breast Cancer in Chek2*1100delc Germline Mutation Carriers. Breast Cancer Res. Treat. 2004;83:91–93. doi: 10.1023/B:BREA.0000010697.49896.03. [DOI] [PubMed] [Google Scholar]
- 26.Plon S.E., Eccles D.M., Easton D., Foulkes W.D., Genuardi M., Greenblatt M.S., Hogervorst F.B.L., Hoogerbrugge N., Spurdle A.B., Tavtigian S.V. Sequence Variant Classification and Reporting: Recommendations for Improving the Interpretation of Cancer Susceptibility Genetic Test Results. Hum. Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo A., Calò V., Bruno L., Schirò V., Agnese V., Cascio S., Foddai E., Fanale D., Rizzo S., Di Gaudio F., et al. Is Brca1-5083del19, Identified in Breast Cancer Patients of Sicilian Origin, a Calabrian Founder Mutation? Breast Cancer Res. Treat. 2008;113:67–70. doi: 10.1007/s10549-008-9906-7. [DOI] [PubMed] [Google Scholar]
- 28.Janavičius R. Founder Brca1/2 Mutations in the Europe: Implications for Hereditary Breast-Ovarian Cancer Prevention and Control. EPMA J. 2010;1:397–412. doi: 10.1007/s13167-010-0037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding D., Zhang Y., He X., Meng W., Ma W., Zheng W. Frequency of the Chek2 1100delc Mutation among Women with Early-Onset and Bilateral Breast Cancer. Breast Cancer Res. 2012;14 doi: 10.1186/bcr3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weischer M., Nordestgaard B.G., Pharoah P., Bolla M.K., Nevanlinna H., van’t Veer L.J., Garcia-Closas M., Hopper J.L., Hall P., Andrulis I.L., et al. Chek2*1100delc Heterozygosity in Women with Breast Cancer Associated with Early Death, Breast Cancer–Specific Death, and Increased Risk of a Second Breast Cancer. J. Clin. Oncol. 2012;30:4308–4316. doi: 10.1200/JCO.2012.42.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stjepanovic N., Moreira L., Carneiro F., Balaguer F., Cervantes A., Balmaña J., Martinelli E. Hereditary Gastrointestinal Cancers: Esmo Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2019;30:1558–1571. doi: 10.1093/annonc/mdz233. [DOI] [PubMed] [Google Scholar]
- 32.Padmanabhan N. Synchronous Bilateral Breast Cancers. J. Clin. Diagn. Res. 2015 doi: 10.7860/JCDR/2015/14880.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huber A., Seidler S.J., Huber D.E. Clinicopathological Characteristics, Treatment and Outcome of 123 Patients with Synchronous or Metachronous Bilateral Breast Cancer in a Swiss Institutional Retrospective Series. Eur. J. Breast Health. 2020;16:129–136. doi: 10.5152/ejbh.2020.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desmond A., Kurian A.W., Gabree M., Mills M.A., Anderson M.J., Kobayashi Y., Horick N., Yang S., Shannon K.M., Tung N., et al. Clinical Actionability of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Risk Assessment. JAMA Oncol. 2015;1:943. doi: 10.1001/jamaoncol.2015.2690. [DOI] [PubMed] [Google Scholar]
- 35.Tung N., Battelli C., Allen B., Kaldate R., Bhatnagar S., Bowles K., Timms K., Garber J.E., Herold C., Ellisen L., et al. Frequency of Mutations in Individuals with Breast Cancer Referred Forbrca1 and brca2 testing Using Next-Generation Sequencing with a 25-Gene Panel. Cancer. 2015;121:25–33. doi: 10.1002/cncr.29010. [DOI] [PubMed] [Google Scholar]
- 36.Easton D.F., Pharoah P.D.P., Antoniou A.C., Tischkowitz M., Tavtigian S.V., Nathanson K.L., Devilee P., Meindl A., Couch F.J., Southey M., et al. Gene-Panel Sequencing and the Prediction of Breast-Cancer Risk. N. Engl. J. Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin H.-C., Lee H.-B., Yoo T.-K., Lee E.-S., Kim R.N., Park B., Yoon K.-A., Park C., Lee E.S., Moon H.-G., et al. Detection of Germline Mutations in Breast Cancer Patients with Clinical Features of Hereditary Cancer Syndrome Using A Multi-Gene Panel Test. Cancer Res. Treat. 2020;52:697–713. doi: 10.4143/crt.2019.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crawford B., Adams S.B., Sittler T., van den Akker J., Chan S., Leitner O., Ryan L., Gil E., van ’t Veer L. Multi-Gene Panel Testing for Hereditary Cancer Predisposition in Unsolved High-Risk Breast and Ovarian Cancer Patients. Breast Cancer Res. Treat. 2017;163:383–390. doi: 10.1007/s10549-017-4181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurian A.W., Hare E.E., Mills M.A., Kingham K.E., Mcpherson L., Whittemore A.S., Mcguire V., Ladabaum U., Kobayashi Y., Lincoln S.E., et al. Clinical Evaluation of a Multiple-Gene Sequencing Panel for Hereditary Cancer Risk Assessment. J. Clin. Oncol. 2014;32:2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laduca H., Stuenkel A.J., Dolinsky J.S., Keiles S., Tandy S., Pesaran T., Chen E., Gau C.-L., Palmaer E., Shoaepour K., et al. Utilization of Multigene Panels in Hereditary Cancer Predisposition Testing: Analysis of More Than 2000 Patients. Genet. Med. 2014;16:830–837. doi: 10.1038/gim.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoheisel J.D., Simbolo M., Gottardi M., Corbo V., Fassan M., Mafficini A., Malpeli G., Lawlor R.T., Scarpa A. DNA Qualification Workflow for Next Generation Sequencing of Histopathological Samples. PLoS ONE. 2013;8:e62692. doi: 10.1371/Journal.Pone.0062692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci. Signal. 2013;6:Pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., Mcgowan-Jordan J., Roux A.-F., Smith T., Antonarakis S.E., Taschner P.E.M. Hgvs Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]