Abstract
The cardioprotective properties of extracellular vesicles (EVs) derived from mesenchymal stromal cells (MSCs) are currently being investigated in preclinical studies. Although microRNAs (miRNAs) encapsulated in EVs have been identified as one component responsible for the cardioprotective effect of MSCs, their potential off-target effects have not been sufficiently characterized. In the present study, we aimed to investigate the miRNA profile of EVs isolated from MSCs that were derived from cord blood (CB) and adipose tissue (AT). The identified miRNAs were then compared to known targets from the literature to discover possible adverse effects prior to clinical use. Our data show that while many cardioprotective miRNAs such as miR-22-3p, miR-26a-5p, miR-29c-3p, and miR-125b-5p were present in CB- and AT-MSC-derived EVs, a large number of known oncogenic and tumor suppressor miRNAs such as miR-16-5p, miR-23a-3p, and miR-191-5p were also detected. These findings highlight the importance of quality assessment for therapeutically applied EV preparations.
Keywords: mesenchymal stromal cells, extracellular vesicles, microRNA, oncomiR, tumor suppressor, cardioprotection, adipose tissue, cord blood
1. Introduction
Mesenchymal stromal cells (MSCs) have been extensively studied in preclinical and clinical trials over the past few decades for their promising capabilities in regenerative medicine [1]. There is consensus that MSCs cannot regenerate damaged human heart tissue. However, preclinical studies showed that MSCs may provide cardioprotective effects after myocardial damage by modulating the immune response, promoting neoangiogenesis, and reducing fibrosis in the myocardial scar [2]. The therapeutic efficacy of MSCs is mainly attributed to their paracrine secretion of various growth factors, chemokines, cytokines, and extracellular vehicles (EVs) [3]. Studies in rodents and pigs showed a reduction in scar size after a single injection of MSCs after myocardial injury [4,5]. In clinical trials, the results regarding the therapeutic effect of MSCs after single treatments in patients with myocardial infarction are more inconsistent [6]. Potential issues associated with the use of MSCs include:
-
(i)
the difficulty in generating a consistent source of cells with a stable phenotype,
-
(ii)
a significant first-pass effect due to entrapment of large cells in the lung and liver microvasculature, and
-
(iii)
patient-specific comorbidities in autologous applications [7].
In addition, less than 2% of the injected human cells remain at the target site after 60 min [8]. In a porcine model of acute myocardial ischemia, intramyocardial injections resulted in a retention rate of just over 10% after 60 min [9]. Furthermore, the same study showed that less than 1% of the engrafted cells were still present four weeks after transplantation. This, in turn, means that the release time of the cardioprotective MSC secretome at the site of injury is significantly shorter than the overall process of myocardial remodeling, which prompted scientists to further investigate the secretome of MSCs, specifically MSC-derived EVs. In general, EVs are membranous nanoparticles produced by cells that are divided into three categories based on their biosynthesis: apoptotic bodies, microvesicles, and exosomes [10]. All of them are considered intercellular messengers that, when stimulated, can transmit biological signals through the blood and lymphatic system to neighboring cells and distant tissues. Proteins, messengerRNAs (mRNAs), and microRNAs (miRNAs) partially encapsulated and protected by the lipid membrane of EVs act as the biological mediators between cells. In fact, gain-of-function and loss-of-function assays have demonstrated that miRNAs transported by EVs are primarily responsible for the cardioprotective effect of MSCs [11]. MiRNAs are short nucleotide sequences of 18–22 base pairs that can bind to the 3′ untranslated region of their target mRNAs, either to interfere with their transport to the ribosome or to prevent their translation at the ribosomal site [12]. Because of their short length, miRNAs usually target more than one mRNA, making specific target prediction difficult. To date, more than 150 miRNAs have been identified in MSC-derived EVs [13]. Although there are some differences in the miRNA profile depending on the source of MSCs, a number of cardioprotective miRNAs have been identified that are commonly transported by EVs from various MSC tissue origins [14]. MiRNAs encapsulated in EVs have several functions including regulation of cell physiology, proliferation, cell differentiation, and apoptosis. For example, they can regulate the expression of members of the hypoxia-inducible factor family, which are important for the modulation of vascular sprouting in the setting of hypoxia, via the RNA interference pathway [15]. Furthermore, miRNAs can also target mRNAs that regulate fibrosis and fibroblast activation, such as tissue growth factor-beta (TGF-beta) and members of the SMAD family [16].
Since it was shown that EVs isolated from MSCs can recapitulate the cardioprotective effects of their parent cells, it was hypothesized that the use of EVs may offer significant advantages over their cellular counterparts due to a higher safety profile, lower immunogenicity, and the inability to directly induce tumors [17]. However, whereas many preclinical studies use multiple direct myocardial injections to deliver EVs, this strategy may not be optimal for many patients in clinical practice. Direct access to the heart (i.e., intracoronary or intramyocardial) is achieved either through catheter-based techniques or by cardiovascular surgery, and both methods are associated with a risk of complications. In turn, a single intramyocardial injection may not be sufficient to improve tissue remodeling after a myocardial injury due to the short half-life of EVs and patient-associated comorbidities that can reduce the intrinsic wound healing capacity seen in healthy animal subjects. As a result, several groups are currently investigating methods for intravenous application of EVs that would allow for sufficient titers of therapeutic EVs in myocardial tissues [18,19]. Despite their small size, EVs, like other lipid-based nanoparticles, undergo a significant first-pass effect with accumulation in the liver and lung tissue [19]. While several teams are currently working on targeted delivery strategies for EVs, another pharmacological component must also be considered: application of EVs over long periods translates into the systemic application of a considerable amount of miRNAs, despite their short half-life of less than 24 h [20]. In the field of cancer biology, a multitude of studies describe the role of miRNAs in cancer progression, transformation, and metastasis. In this context, miRNAs are divided into three classes:
-
(i)
oncogenic miRNAs,
-
(ii)
tumor suppressor miRNAs, and
-
(iii)
miRNAs with a dual role in cancer progression.
However, to the best of our knowledge, likely due to the limited number of preclinical trials with systemic EV applications, their miRNA cargo was not analyzed in connection with possible off-target effects. In particular, the presence or absence of pro-oncogenic miRNAs in EV preparations has not been conclusively proven. These potential risks need to be assessed for the clinical use of EVs, especially when treating patients with undetected tumors or predispositions to tumor development. The aim of the present study was therefore to characterize the miRNA cargo of EVs isolated from two clinically relevant MSC sources (i.e., cord blood (CB) and adipose tissue (AT)) and then to compare the EV miRNA cargo to well-known miRNAs involved in cancer biology.
2. Materials and Methods
2.1. Cell Isolation and Cell Culture
Human AT-derived MSCs were isolated from patients undergoing liposuction, as described previously [21]. Four donors (three female, one male, mean age 41.8 ± 9.3 years) were included in this study. None of the lipoaspirate donors were obese (body mass index was below 25 for all donors) and none of the donors reported any medical conditions at the time of liposuction. CB-derived MSCs were isolated from CB of four healthy newborns (two female, two male) at the Charité University Hospital Berlin, as described elsewhere [22]. Neither mother nor infant suffered from any medical conditions at the time of donation. All procedures were approved by the local medical ethics committees (Charité University Hospital Ethics Committee, registration number EA2/178/13; Cantonal Ethics Committee Zurich, registration number KEK-ZH 2010-0476/0) and written consent was obtained from patients or relatives. All MSCs were cultured in MesenPRO RS medium (Life Technologies, Grand Island, NY, USA, catalog no. 12747-010) containing 10% fetal bovine serum (FBS; Life Technologies, Carlsbad, CA, USA, catalog no. 10270106), 1% penicillin/streptomycin (P/S; Merck Millipore, Burlington, MA, USA, catalog no. A2213), and 2 ng/mL recombinant human fibroblast growth factor-basic (FGF-b; PeproTech, Hamburg, Germany, catalog no. 100-18C) in a humidified atmosphere of 5% carbon dioxide at 37 °C.
2.2. EV Isolation
EVs were isolated from MSC-conditioned medium using (i) sequential ultracentrifugation (UC) or (ii) the exoEasy Maxi Kit (Qiagen, Hilden, Germany, catalog no. 76064) according to the manufacturer’s instructions. Briefly, MSCs were expanded to a confluence of about 80% and washed once with Dulbecco’s phosphate-buffered saline (DPBS, Dulbecco’s phosphate-buffered saline; Life Technologies, Bleiswijk, The Netherlands, catalog no. 14190-144). The cells were switched to Dulbecco’s modified eagle medium (DMEM 1X)-GlutaMAX (Life Technologies, Paisley, United Kingdom, catalog no. 21885-025) containing 10% exosome-depleted FBS (Life Technologies, Bleiswijk, The Netherlands, catalog no. A2720803), 1% P/S, and 2 ng/mL FGF-b for 48 h, followed by a transfer to starvation medium (DMEM 1X-GlutaMAX supplemented with 1% P/S and 2 ng/mL FGF-b) for 24 h. For the isolation of EVs using sequential UC, the supernatant of approximately 3 × 107 cells at early passages (passages 5–7) was processed according to the protocol of Beez et al. [23]. For the isolation of EVs using the Qiagen kit, an MSC-conditioned medium of approximately 3 × 106 cells at early passages was collected and centrifuged at 2000× g for 15 min at 4 °C (Allegra X-15R Centrifuge, Beckman Coulter, Indianapolis, IN, USA). The supernatant was decanted and filtered using a 0.2 μm syringe filter (Sartorius, Hanover, Germany, catalog no. 16534) to remove any remaining cell debris and large aggregates. Thereafter, 8 mL of the filtered solution were mixed with 8 mL XBP buffer by gently inverting the tube. The mixture was transferred to the exoEasy spin column, centrifuged at 500× g for 1 min at room temperature (R.T) and the flow-through was discarded. Then, the bound EVs were washed with 10 mL XWP buffer and centrifuged at 5000× g for 5 min to remove residual buffer from the column. To elute EVs, 0.5 mL XE buffer was added and the column was centrifuged at 500× g for 5 min to collect the eluate, which was re-applied to the same column and centrifuged at 5000× g for 5 min. Final EV preparations were transferred to low-binding tubes (Sarstedt, Numbrecht, Germany, catalog no. 72.706.600) and stored at −80 °C until further use.
2.3. Nanoparticle Tracking Analysis (NTA) and Total Protein Analysis
Particle concentration and size distribution of EV preparations were examined using the ZetaView instrument (Particle Metrix, Inning, Germany). Particles were automatically tracked and sized based on Brownian motion and the diffusion coefficient. The NTA measurement conditions were as follows: temperature = 26.6 ± 2.2 °C, viscosity = 0.87 ± 0.04 cP, frames per second = 30, and measurement time = 75 s. Sample videos were analyzed using NTA software (ZetaView, Particle Metrix, Inning, Germany, version 8.04.02).
Total protein content of EV preparations was determined using the commercially available Bicinchoninic Acid (BCA) Protein Assay Kit with bovine serum albumin as a standard (Thermo Scientific, catalog no. 23227). Briefly, 20 µL of samples or standards were mixed with 200 µL of freshly made BCA working reagent and incubated for 30 min at 50 °C. Absorbance was measured at 560 nm with a Mithras LB940 plate reader (Berthold Technologies, Pforzheim, Germany) and analyzed with MikroWin 2000 software (Mikrotek Laborsysteme, Overath, Germany, version 4.41).
2.4. Transmission Electron Microscopy (TEM)
Isolated EV preparations were stained according to the protocol of Théry et al. [24] and morphologically evaluated at the electron microscopy (EM,) facility of the Charité—Universitätsmedizin Berlin. Briefly, 20 µL of MSC-derived EVs were first placed on formvar carbon-coated copper EM grids (Plano, Wetzlar, Germany, catalog no. G2430N) for 20 min. Then, the samples were incubated for 20 min in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA, catalog no. 15714), followed by 5 min in 1% glutaraldehyde (Serva, Heidelberg, Germany, catalog no. 23114). After several washing steps with water, the samples were stained for 10 min in a freshly prepared solution of 4% uranyl acetate (Serva, Heidelberg, Germany, catalog no. 77870) and 2% methylcellulose (Sigma-Aldrich, St. Louis, MO, USA, catalog no. M-6385). Imaging was performed using the Leo 906 microscope (Carl Zeiss, Oberkochen, Germany), equipped with ImageSP Viewer software (SYS-PROG, Minsk, Belarus, version 1.2.7.11).
2.5. Immunofluorescence Staining and Flow Cytometry
Expression of surface molecules was measured as described before [23]. Briefly, 2 µg of MSC-derived EV protein were incubated with 15 µL of 4 μm aldehyde/sulfate latex beads (Thermo Fisher, catalog no. A37304) for 15 min at R.T. The sample volume was filled up to 1 mL with DPBS and incubated for 1 h at R.T with gentle shaking. Thereafter, samples were centrifuged for 10 min at 300× g, and after discarding the supernatant, samples were washed once with 1% fetal calf serum in DPBS (flow cytometry buffer). Next, the beads loaded with EVs were incubated with the following fluorescence-conjugated antibodies: anti-CD9/FITC (BioLegend, San Diego, CA, USA, catalog no. 312104), anti-CD63/PE (BioLegend, San Diego, CA, USA, catalog no. 353004), anti-CD73/APC (BioLegend, San Diego, CA, USA, catalog no. 344006), anti-CD81/FITC (BioLegend, San Diego, CA, USA, catalog no. 349504), anti-HLA-ABC/PE (BioLegend, catalog no. 311405), or anti-HLA-DR/APC (BioLegend, San Diego, CA, USA, catalog no. 307610), each at a dilution of 1:25 in flow cytometry buffer. After 30 min at 4 °C, the beads were washed twice with flow cytometry buffer, fixed with flow cytometry buffer supplemented with 0.5% PFA, and stored at 4 °C until measurement using a MACSQuant VYB flow cytometer (Miltenyi Biotec, Bergisch Gladbach, Germany). Beads incubated with antibodies but no EVs served as negative controls, respectively. Analysis was performed using FlowJo software (Tree Star, Ashland, OR, USA, version 10.6.1).
2.6. MiRNA Analysis
MiRNA was extracted from 200 µL of isolated EVs using the miRNeasy Mini Kit (Qiagen, Hilden, Germany, catalog no. 74104) according to the manufacturer’s instructions. The RNA quantity and purity were assessed with the Agilent 2100 Bioanalyzer system (Agilent Technologies, Waldbroon, Germany). Reverse transcription (RT) was performed using the miRCURY LNA Universal cDNA Synthesis Kit II (Exiqon-Qiagen, Hilden, Germany, catalog no. 203301). RT thermocycling parameters were as follows: 42 °C for 60 min and 95 °C for 5 min. Quantitative polymerase chain reaction (qPCR) was performed using the miRCURY LNA Universal RT microRNA PCR system (Exiqon-Qiagen, catalog no. 339340) with 752 known human miRNAs and 3 interplate calibrators and 1 spike-in miRNA as an internal control. All primer/probe sets for miRNAs were custom designed by the supplier. Three extraction controls and two cDNA synthesis controls were additionally used as indicated by the provider. Two real-time qPCR amplifications were performed for each RT reaction. Reactions were performed according to the manufacturers’ instructions using a LightCycler 480 II system (Roche, Rotkreuz, Switzerland). QPCR thermocycling conditions were as follows: 95 °C for 10 min, followed by 40 cycles at 95 °C for 10 s and 60 °C for 1 min. Melt curve analysis was performed between 60 and 95 °C at a ramp rate of 0.11 °C/s. After interpolation calibration, the examined miRNAs were classified into three categories:
-
(i)
miRNAs with mean corrected CT (CTcorr) values below 30.00 were considered as detected with certainty,
-
(ii)
miRNAs with mean CTcorr values between 30.00 and 32.99 were considered as detected with uncertainty, and
-
(iii)
miRNAs with mean CTcorr values equal or greater than 33.00 were considered as not detected.
All analyzed miRNAs and their expression values are listed in Supplementary Materials Table S1. The obtained CT values of miRNAs were normalized using the geNorm method, which calculates a normalization factor based on multiple reference miRNAs [25]. In brief, the arithmetic mean of the CT values of miRNAs that were stably expressed across all samples, namely hsa-miR-1260a, hsa-miR-125b-5p, hsa-miR-21-5p, hsa-miR-23a-3p, hsa-miR-24-3p, hsa-miR-221-3p, hsa-let-7i-5p, hsa-miR-199a-3p, and hsa-miR-100-5p, were subtracted from CTcorr values to calculate delta CT (dCT) values for every sample. In order to plot miRNA expression on heatmaps, Z-scores were determined from logarithmically transformed dCT values for each miRNA. The Z-scores were calculated as a numerical measurement of the mean value group with z = (x − μ)/σ, where x is the raw score, μ is the population mean, and σ is the population standard deviation. Finally, heatmaps of miRNAs were created with the gplots package of RStudio (version 1.3.959).
2.7. Literature Search for miRNAs
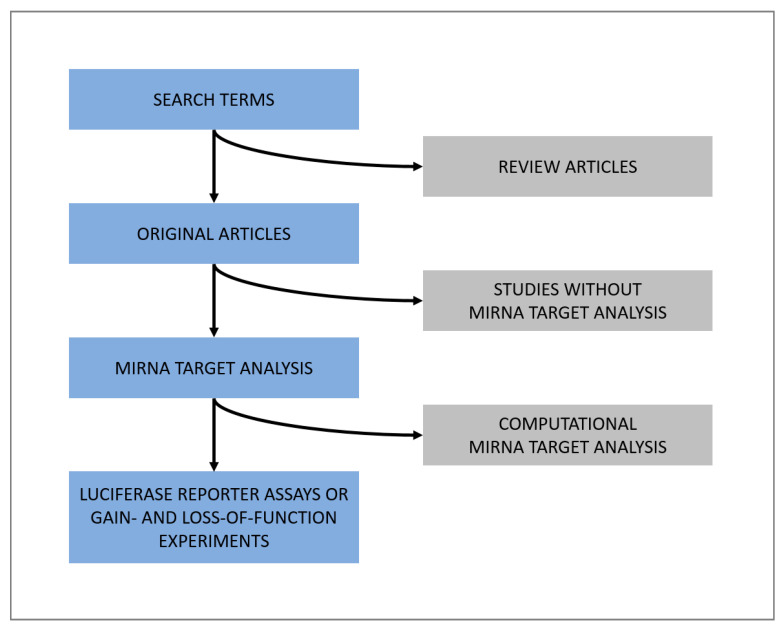
A systematic literature search was conducted for all miRNAs with a low mean CTcorr value (≤29.99) in both CB- and AT-MSC-derived EVs. Pubmed, Medline, and Scopus were used as search engines with the following search terms: “name of miRNA”, “name of miRNA” AND “heart”, “name of miRNA” AND “fibrosis”, “name of miRNA” AND “cancer”, “name of miRNA” AND “fibroblasts”, “name of miRNA” AND “endothelial cells”, “name of miRNA” AND “angiogenesis”, “name of miRNA” AND “immunomodulation”, “name of miRNA” AND “macrophages”, “name of miRNA” AND “t-cells”, and “name of miRNA” AND “immune cells”. For published miRNA targets, only studies were considered that confirmed miRNA targets by luciferase reporter assays or gain- and loss-of-function experiments. The findings are summarized in Appendix A Table A1, Table A2 and Table A3.
2.8. Statistical Analysis
GraphPad Prism (GraphPad Software, San Diego, CA, USA, versions 6.0 and 8.3.0) was used for performing data analysis and generating graphs. The statistical significance of differences in EV particle concentration, total protein amount, and surface marker expression was determined by the Mann–Whitney test; a p-value of less than 0.05 was considered significant. All miRNA data are shown as median with interquartile range, if not indicated otherwise. Data were tested with Shapiro–Wilk test for normal distribution. Statistical differences between two groups with only one variable in paired observations were determined either with the Wilcoxon matched-pairs signed rank test for non-parametric samples or with the unpaired t-test for parametric samples. Results were considered significant with * p < 0.05, ** p < 0.01, and *** p < 0.001.
3. Results
3.1. Characterization of EVs
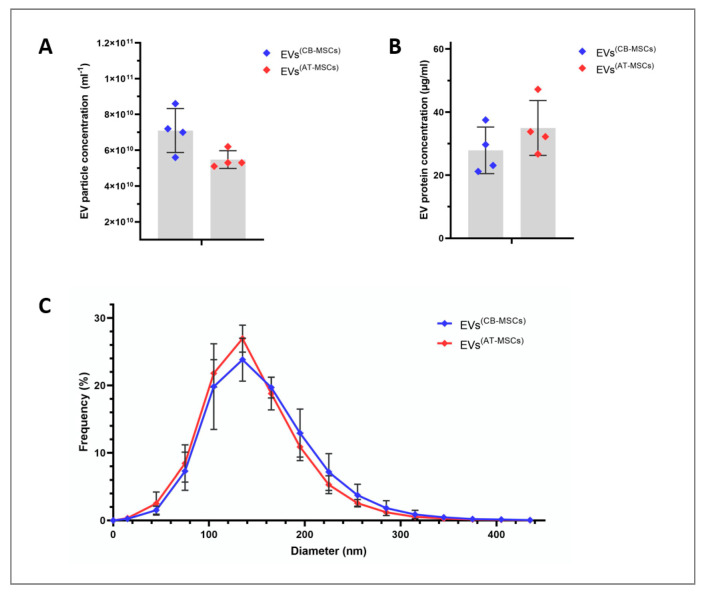
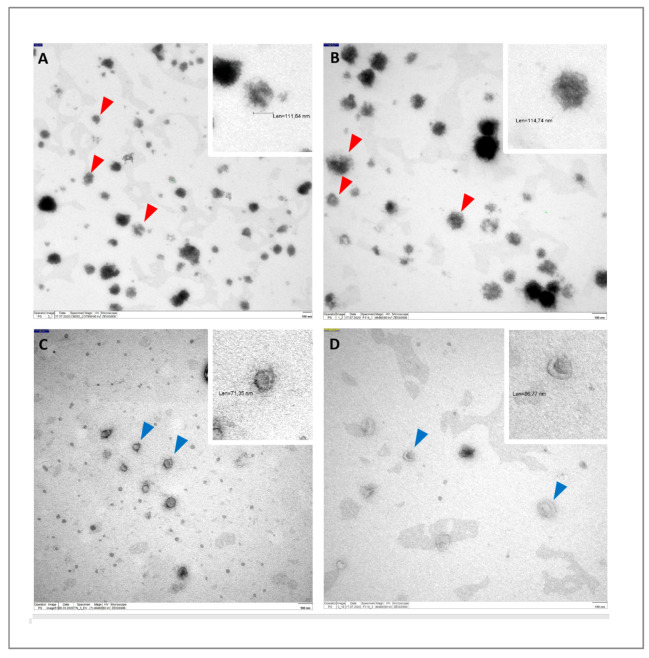
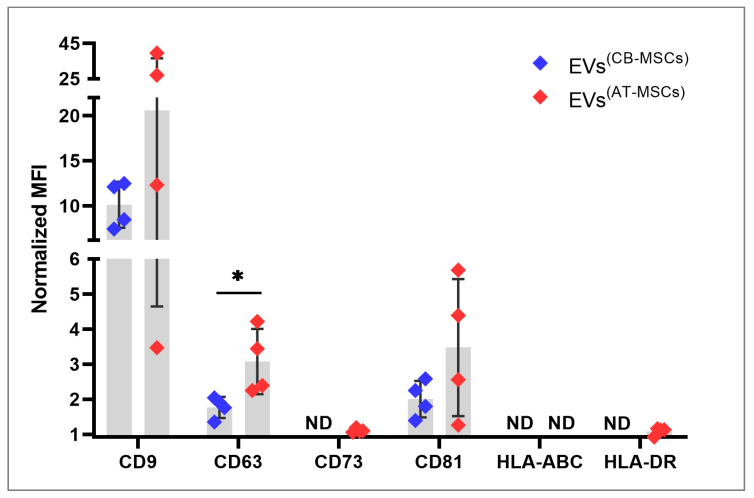
All EVs were harvested from the supernatants of in vitro-cultured CB- and AT-MSCs, which were derived from tissues of four healthy subjects each. Although isolated from different sources, both MSC lines showed a typical spindle-shaped cell morphology under EV biogenesis conditions (Figure 1). The mean number of EV particles obtained was 7.1 ± 1.2 × 1010 per mL for CB-MSC-derived EVs and 5.5 ± 0.5 × 1010 per mL for AT-MSC-derived EVs (Figure 2A), but this difference was not significant (p = 0.057). Similarly, protein concentrations between EVs from CB- and AT-MSCs were not statistically significant (p = 0.343), with mean values of 27.9 ± 7.4 and 35.0 ± 8.7 µg/mL protein (Figure 2B). Quantitative analysis of EV diameters demonstrated an asymmetrical distribution, with a mean diameter of 132.7 ± 12.1 nm for EVs from CB-MSCs and a mean diameter of 123.9 ± 6.6 nm for EVs from AT-MSCs (Figure 2C), indicating the presence of exosomes, which are typically 40 to 150 nm in diameter [26]. Furthermore, both EV variants, which were isolated with the Qiagen kit, exhibited typical cup-like shapes as observed by TEM (Figure 3A,B). In comparison, EVs isolated by sequential UC showed a similar shape (Figure 3C,D). However, in contrast to the EVs isolated by UC, the EVs isolated by Qiagen membrane affinity columns were covered by a corona that bound larger amounts of uranyl acetate (Figure 3A,B, red triangles). EVs isolated by sequential UC have not been further examined because this manuscript focuses on EVs isolated by the Qiagen exoEasy Maxi Kit due to its excellent scalability, which is needed for the production of large EV amounts for clinical application. Next, we analyzed the isolated EV preparations for selected membrane proteins that have been associated with EVs in the past. Regardless of the cell source, it was possible to detect on all EV preparations CD9, CD63, and CD81, with CD9 exhibiting the highest normalized mean fluorescence intensities (MFIs) (Figure 4). Interestingly, all of the aforementioned markers tended to have higher values in AT-MSC-derived EVs than in CB-MSC-derived EVs, while only CD63 levels were significantly higher (p = 0.029). Figure 4 also shows that CD73 was only detected in EVs from AT-MSCs, but not from CB-MSCs. Since it was hypothesized that MSC-derived EVs do not carry human leukocyte antigens (HLAs) and are therefore less immunogenic [23], we also included HLA-ABC and HLA-DR in the flow cytometry analysis. Our data indicate that EVs from CB-MSCs did not exhibit a signal for HLA-ABC and HLA-DR (Figure 4). For EVs from AT-MSCs, HLA-ABC was also not present, while HLA-DR was detected in small amounts (Figure 4). In sum, these results indicated that the isolated EVs contained exosomes.
Figure 1.
Cord blood (CB)- and adipose tissue mesenchymal stromal cells (AT-MSCs) maintain their spindle-shaped morphology under extracellular vesicles (EV) biogenesis conditions. MSCs were expanded to a confluence of about 80%, washed with Dulbecco’s phosphate-buffered saline and cultivated for 48 h in exosome-depleted medium. Then, the cells were switched to starvation medium for 24 h to derive the conditioned medium for EV isolation. Representative bright-field images of cell morphology of CB-MSCs (A) and AT-MSCs (B) were taken by phase-contrast microscopy at the time of EV isolation. Bars, 200 µm.
Figure 2.
Particle number, protein amount and size distribution of EVs isolated from CB- and AT-MSCs. Particle concentration (A) and size distribution (C) of EV preparations were measured by nanoparticle tracking analysis. Protein content (B) was determined by the bicinchoninic acid assay. In (A–C), the results are mean values ± standard deviation (SD) obtained from four different donors per cell type.
Figure 3.
Identification of EV-like structures via transmission electron microscopy. CB- and AT-MSC-derived EVs shown in (A,B) were isolated using the Qiagen exoEasy Maxi Kit, and CB- and AT-MSC-derived EVs shown in (C,D) were isolated using sequential ultracentrifugation. All EVs exhibit the expected cup-like shape, an artefact of the fixation method. In (A,B), EVs are covered with phosphate-rich matter, and red triangles indicate structures in which covered EVs were detected. In (C,D), exemplary EVs are indicated by blue triangles. In (A–D), enlarged regions of selected EVs are shown on that top right.
Figure 4.
CB- and AT-MSC-derived EVs display a distinct surface marker profile. Detection of the surface marker proteins CD9, CD63, CD73, CD81, HLA-ABC, and HLA-DR using flow cytometry on EV preparations. The data are presented as means ± SD of normalized mean fluorescence intensities (MFIs), which were calculated as the ratio of the geometric MFI of EV samples (beads + EVs + antibodies) to control samples (beads + antibodies). Statistical analysis was performed by the Mann-Whitney test with * p < 0.05. ND indicates not detected. EVs from four different donors per cell type were included.
3.2. MiRNA Profile of CB- and AT-MSC-Derived EVs
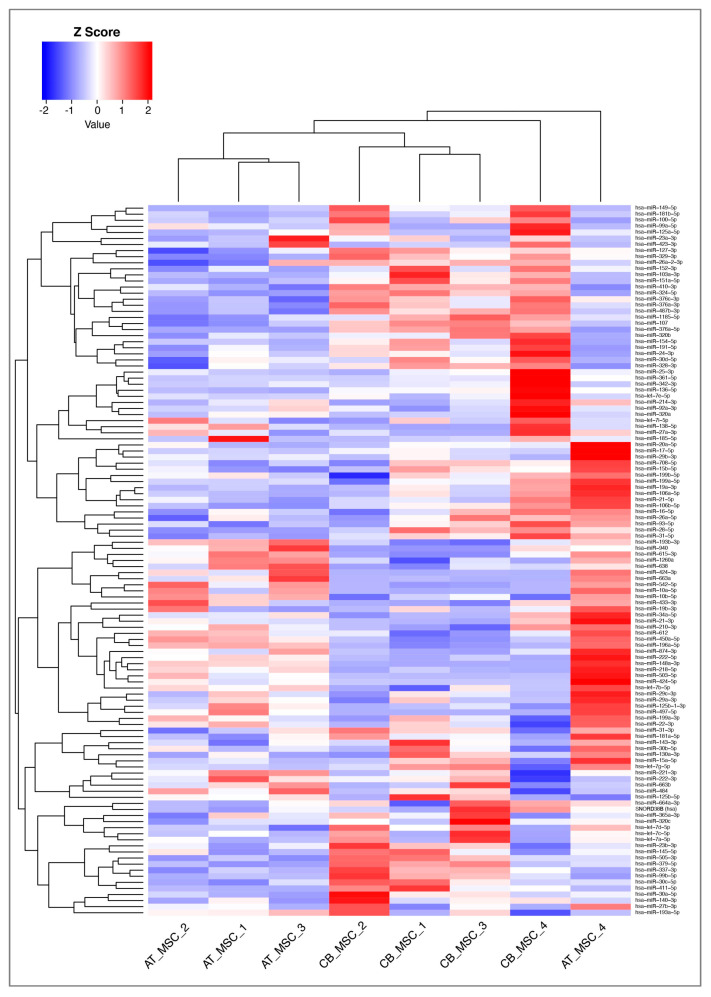
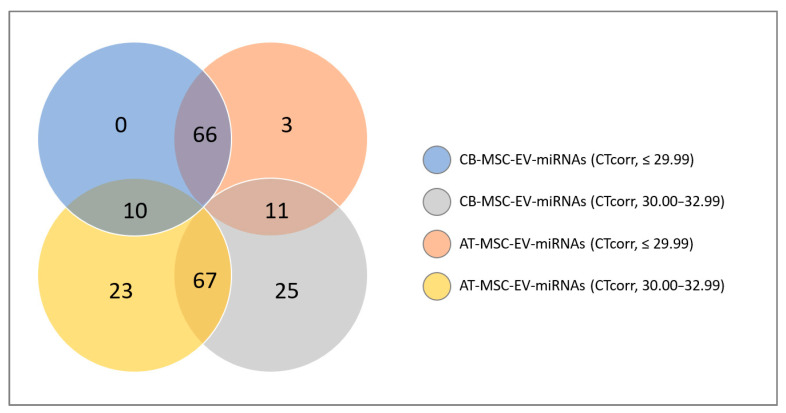
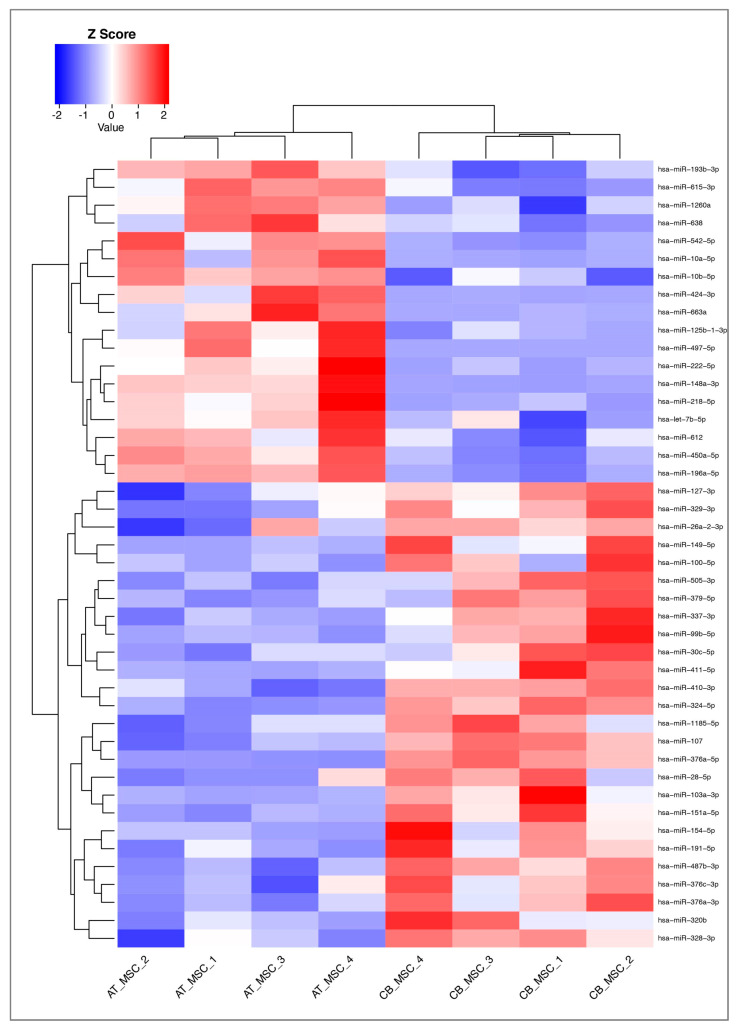
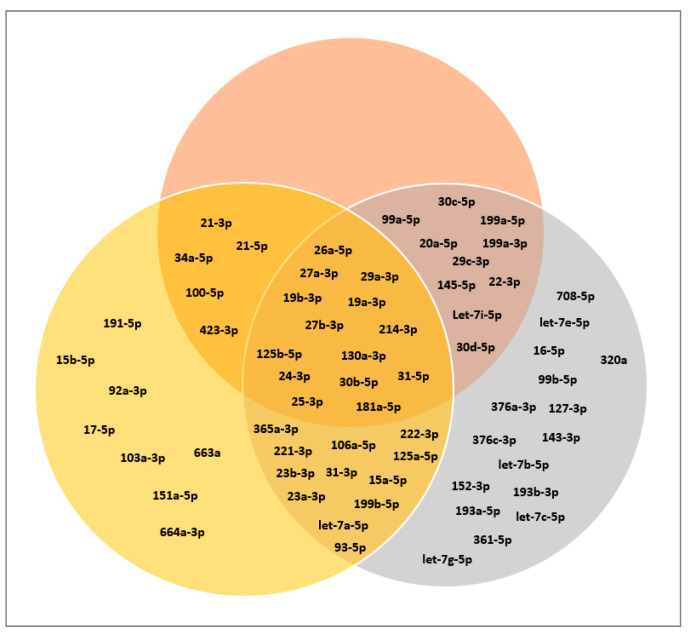
Of the 752 miRNAs examined in this study, 117 were detected with certainty according to the guidelines of the Qiagen-Exiqon miRCURY LNA Universal RT microRNA PCR system. Based on these miRNAs, a heatmap was created (Figure 5). The grouping of donors shows a consistent clustering with only one outlier per group (CB_MSC_4 and AT-MSC_4). Interestingly, the expression profile of EV surface markers for these donors also differed from the other donors in the same group. For further analysis, all miRNAs with mean CTcorr values below 33.00 in at least one group were included. Following this, 205 miRNAs were detected in EV samples, while the majority of miRNAs (547) were not detected (Figure 6). From our analysis, 76 miRNAs were highly expressed in CB-MSC-derived EVs and 80 miRNAs were strongly expressed in AT-MSC-derived EVs with mean CTcorr values of less than 30.00. Intriguingly, among them, 66 miRNAs were found in EVs from both MSC sources. Only 10 were uniquely highly expressed in CB-MSC-derived EVs, namely let-7d-5p, miR-30a-5p, miR-106b-5p, miR-107, miR-136-5p, miR-140-3p, miR-181b-5p, miR-320b, and miR-320c, and miR-342-3p, and 14 were uniquely highly expressed in AT-MSC-derived EVs, namely miR-10b-5p, miR-29b-3p, miR-138-5p, miR-148a-3p, miR-185-5p, miR-210-3p, miR-424-3p, miR-424-5p, miR-433-3p, miR-484, miR-503-5p, miR-663b, miR-874-3p, and miR-940. Furthermore, 100 and 103 miRNAs in CB-MSC-derived EVs and AT-MSC-derived EVs, respectively, which showed mean CTcorr values of 30.00 to 32.99, were considered to be low expressed. To visualize differential miRNA expression profiles, a heatmap of all miRNAs that were significantly different in expression between CB- and AT-MSC-derived EVs was created, showing a clear clustering of CB-MSC-EV-miRNAs and AT-MSC-EV-miRNAs (Figure 7, 44 miRNAs). Overall, the differences in expression after normalization did not exceed a two-fold increase or decrease for almost all miRNAs, except for miR-10b-5p (8.23-fold higher in AT-MSC-derived EVs), miR-103a-3p (3.35-fold higher in CB-MSC-derived EVs), miR-222-5p (8.28-fold higher in AT-MSC-derived EVs), miR-376a-3p (2.45-fold higher in CB-MSC-derived EVs), miR-663a (7.68-fold higher in AT-MSC-derived EVs), and miR-1260a (2.87-fold higher in AT-MSC-derived EVs). Three miRNAs were only found to be highly expressed in AT-MSC-derived EVs, but were absent in CB-MSC-derived EVs, namely miR-148a-3p, miR-424-3p, miR-503-5p. In sum, CB- and AT-MSC-derived EVs are similar in their miRNA composition, with the exception of a small number of miRNAs.
Figure 5.
Heatmap and dendrograms of all microRNAs (miRNAs) detected with certainty according to the guidelines of the Qiagen-Exiqon miRCURY LNA Universal RT microRNA PCR system. Sample IDs are shown on the x-axis. Samples with similar miRNA expression are clustered together. The heatmap was generated by RStudio and 2dCT was used for data input. Z-scores of more than zero indicate a higher expression of miRNAs in one sample compared to the others; Z-scores of less than zero indicate the opposite.
Figure 6.
Venn diagram of miRNAs found in CB- and AT-MSC-derived EVs. In total, 752 miRNAs were analyzed and categorized according to mean CTcorr values. High miRNA expression means CTcorr value ≤ 29.99; low miRNA expression means CTcorr value = 30.00–32.99. Five hundred and forty-seven miRNAs were not detected in EVs from CB-MSCs or in EVs from AT-MSCs (mean CTcorr value ≥ 33.00).
Figure 7.
Heatmap and dendrograms of miRNAs that were significantly changed in AT-MSC-derived EVs compared to CB-MSC-derived EVs. Sample IDs are shown on the x-axis. Samples with similar miRNA expression are clustered together. The heatmap was generated by RStudio and 2dCT was used for data input. Z-scores of more than zero indicate a higher expression of miRNAs in one sample compared to the others; Z-scores of less than zero indicate the opposite.
3.3. Classification of miRNAs: Tumor Suppressor miRNAs, Oncogenic miRNAs, and Cardioprotective miRNAs
We then conducted a literature research (Figure 8) to group all 66 miRNAs found at high levels in both CB- and AT-MSC-derived EVs based on their function. As indicated in Figure 9, the majority of identified miRNAs have a well-known role as tumor suppressor. We also found many miRNAs, such as miR-103a-3p, miR-151a-5p, and miR-191-5p, which are known oncogenic miRNAs (oncomiRs). Interestingly, we also identified a large number of miRNAs (26) known to act both as oncomiRs and as tumor suppressor. The EV samples examined in this study also showed positive hits for well-known cardioprotective miRNAs, such as miR-21-3p, miR-22-3p, miR-26a-5p, and miR-125b-5p. While having cardioprotective properties, most of them are also associated with oncogenic and tumor suppressor properties. In summary, these data indicate that both CB- and AT-MSC-derived EVs not only transfer a certain set of miRNAs that are involved in one particular mechanism, but rather a multitude of miRNAs that are linked to several biochemical processes, including tumor suppression, tumorigenesis, and cardioprotection.
Figure 8.
Diagram of literature search rules applied for all miRNAs with a low mean CTcorr value (≤ 29.99) in both CB- and AT-MSC-derived EVs. Search terms were “name of miRNA”, “name of miRNA” AND “heart”, “name of miRNA” AND “cancer”, “name of miRNA” AND “fibrosis”, “name of miRNA” AND “endothelial cells”, “name of miRNA” AND “angiogenesis”, “name of miRNA” AND “immunomodulation”, “name of miRNA” AND “macrophages”, “name of miRNA” AND “t-cells”, and “name of miRNA” AND “immune cells”.
Figure 9.
Venn diagram of selected miRNAs based on their function. Gray, tumor suppressor miRNAs; yellow, oncogenic miRNAs; red, cardioprotective miRNAs. With the exception of miR-1260a, all miRNAs with a low mean CTcorr value (≤29.99) in both CB- and AT-MSC-derived EVs were included. MiR-1260a could not be included, as no targets were described in the literature so far. Further details on these miRNAs are given in Table A1, Table A2 and Table A3.
4. Discussion
4.1. EV Phenotype
Overall, the EVs analyzed in our study showed the expected proteins to be present in both CB- and AT-MSCs, such as the tetraspanins CD9, CD63, and CD81. The latter was present in significantly lower amounts in EVs from CB-MSCs than in EVs from AT-MSCs, an observation that was not made in other comparative studies before. The phenomenon that EVs from MSCs have only little or no HLAs present on their surface and therefore have a low immunogenicity [23] was confirmed in our study, since HLA-ABC was not found in both CB- and AT-MSC-derived EVs. Furthermore, HLA-DR was not detected in CB-MSC-derived EVs and it was only slightly above the detection level for the flow cytometry assay in AT-MSC-derived EVs. Consequently, the phenotype of the EVs might reflect the low expression of HLA molecules of the parent CB- and AT-MSCs.
It is known that the isolation method can significantly influence the composition of miRNAs in EV preparations [26,27]. To date, there is a multitude of different EVs isolation protocols available [28], and an ideal isolation method for clinical use remains to be determined. In this study, EVs were isolated using a commercially available EV isolation kit from Qiagen. In contrast to protocols using sequential UC to isolate EVs, this kit is more appropriate for scaling up the production of EVs. Initially, we performed side-to-side comparisons for the isolation of EVs using sequential UC and Qiagen membrane affinity columns. A similar comparison reported by the group of Streanska et al. [29] demonstrated that both methods lead to EVs with encapsulated miRNAs. However, they found differences in EV size and surface protein expression depending on the isolation method. While in their study, they were not able to detect the tetraspanins CD63 and CD81 using the Qiagen kit for EV isolation, we were able to detect tetraspanins such as CD9, CD63 and CD81, considered as typical EV markers. It should be noted, however, that we performed flow cytometry analysis, whereas the others used the Western blot. Furthermore, TEM analysis revealed that EVs isolated by the Qiagen kit were coated with either proteins or nucleic acids. For this experiment, EVs were incubated with uranyl acetate to stain phosphate groups of the lipid membrane. However, the presence of phosphate-rich proteins or nucleic acids in the so-called EV corona can also result in strong staining. We therefore hypothesize that the structures surrounding the EVs are most likely a mixture of proteins and nucleic acids. In line with this, the group of Varga et al. [30] has recently shown that EVs in vivo are also surrounded by a variety of different proteins that are not integrated in their own membrane. Furthermore, Jeppesen et al. [26] were able to separate a protein fraction from a pure vesicle fraction and they demonstrated that different EV isolation methods impact the EV-miRNA composition. Our data suggest that the Qiagen membrane affinity method produces EVs with an intact corona, indicating that miRNAs may also be bound to proteins in the corona. However, it cannot be conclusively determined whether the analyzed miRNAs were encapsulated, bound to co-isolated proteins, or bound within the EV protein corona. Studies that have so far investigated the therapeutic potential of EVs did not purify the EVs in their in vivo models prior to injection. Therefore, regardless of the isolation method, co-purified miRNAs will be injected together with the EV fraction. However, when EVs are used clinically, it is expected that additionally administered miRNAs could also play a role in the cardioprotective mechanism. It is therefore of importance to validate the miRNA profiles for each isolation method before conducting downstream experiments or even clinical studies. A more in depth analysis of the isolated EVs might have answered this question, but would be beyond the scope of this project.
4.2. MiRNA Profile
As mentioned above, miRNA analysis of EVs derived from CB- and AT-MSCs showed that a large number of detected miRNAs play an important role in tumor biology. Due to the multiple targets a miRNA can have, it is difficult to predict all possible targets of each miRNA. In this study, we therefore only reviewed targets that were confirmed already by other groups through in vitro assays. Since the PCR array used in our experiment focused on cellular miRNAs, which play a well-known role in cancer biology, it is not surprising that most miRNAs (547 out of 752) were not detected in the EV samples. Our data show a biological variability that is expected from human-derived samples [31]: EV samples derived from both CB- and AT-MSCs contain one outlier in terms of their surface marker configuration and their miRNA profiles (Figure 4 and Figure 7). Due to the small number of donors examined in this study, the effect of donor-specific confounding factors (e.g., gender, age, or race) on the miRNA profiles cannot be determined. Our literature search revealed that most studies focused on the therapeutic aspect of miRNAs of MSC-derived EVs. Only a few studies made the data of their miRNA arrays publicly available [32,33,34]. Additionally, the role of MSC-derived miRNAs in cancer biology has been discussed and investigated by other groups. Even here, however, only few groups made all collected data available for secondary analysis. In the case of AT-MSCs, one group has investigated the role of AT-MSC-derived EVs in the development and treatment of osteoarthritis [32,33]. In both publications, the raw data of the miRNA array were made available by the authors. A side-to-side comparison revealed that 71.0% and 73.3% of the 65 highest expressed miRNAs in both data sets were identical to the miRNAs found in our EV samples. The discrepancy could be explained by the difference in treatment of AT-MSCs at time of isolation and the isolation method itself.
4.2.1. Anti-fibrotic Signaling via Suppression of the TGF-Beta Pathway
MiRNAs were initially examined in the context of cancer biology. Target search was therefore biased and provided a greater number of miRNAs related to cancer than, for instance, to cardioprotection. However, some miRNAs with cardioprotective properties often interfere with proteins that are also regulated in cancer cells. For instance, miRNAs that advantageously modulate fibrosis and activation of fibroblasts usually target either the mRNA of proteins in the TGF-beta/SMAD-axis or promoter and receptor mRNAs that modulate cell cycle activation. Typically, miRNA-mediated suppression of TGF-beta signaling leads to decreased fibrosis in different tissues [35]. Both CB-and AT-MSC-derived EVs contain sets of miRNAs that target TGF-beta receptors directly or downstream signaling proteins such as SMAD proteins. In the context of TGF-beta signaling, SMAD2, 3, and 4 are the downstream promoters that can activate pro-fibrotic gene expression in multiple tissues including the heart [36]. MiR-16-5p (−1.03-fold change, p = 0.84), miR-23a-3p (−1.14-fold change, p = 0.99), and miR-130a-3p (−1.11-fold change, p = 0.75), which showed no difference in relative amounts for the comparison of CB-MSC-derived EVs to AT-MSC-derived EVs, all target the SMAD mRNA directly and exhibit an anti-fibrotic, and in most cancers, a tumor suppressor effect [37,38,39]. At the same time, miR-130a-3p can also act as an oncomiR in esophageal cancer by inhibiting the expression of SMAD4 [40], which incidentally leads to a tumor suppressor effect in hepatoma cells [38]. This dual role of miRNAs in cancer biology is well known and shows the complexity of gene expression regulation via RNA interference [41]. Similarly, while miR-130a-3p suppresses fibrosis in hepatic steatosis by suppressing the TGF-beta receptors 1 and 2 [37], the suppression of TGF-beta receptor 3 by miR-23b-3p and miR-27b-3p in atrial fibroblasts leads to increased fibrosis in the context of atrial fibrillation [42]. This underlines that, similar to the effect of miRNAs in cancer, a dual role of miRNAs and thus potential off-target effects can be hypothesized. It also highlights that adverse effects, such as increased fibrosis, may depend on the presence of miRNA clusters. For the EV samples investigated in the present study, both miR-23b-3p and miR-27b-3p were found with mean CTcorr values of 25.2 ± 1.4 and 25.5 ± 1.1 versus 27.4 ± 0.9 and 27.2 ± 1.1 in CB- and AT-MSC-derived EVs, respectively.
4.2.2. Role of miRNA-Mediated Mammalian Target of Rapamycin (mTOR) Suppression
The miRNA target analysis also revealed that some miRNAs found in CB- and AT-MSC-derived EVs target mTOR or mTOR-associated proteins, including miR-99b/a, miR-100-5p, miR-143-3p, miR-199a-5p/3p, and miR-199b-5p. MTOR is a protein kinase that regulates cell growth, autophagy, and cell survival [43]. Since activation of mTOR plays a crucial role in maintaining growth and inducing metastasis in many cancers, it has been intensively studied as a potential target for cancer therapy [44]. For all of the miRNAs mentioned, overexpression in cancer cell lines led to the induction of apoptosis and autophagy. Interestingly, miR-100-5p can also suppress angiogenesis by preventing cell proliferation in vascular smooth muscle cells, an effect that could counteract a potential cardioprotective effect [45]. Similarly, both miR-143-3p and miR-199a-3p can increase apoptosis during hypoxic or inflammatory injury in kidney and synovial cells, respectively [46,47]. One could therefore postulate that miRNAs that inhibit mTOR signaling are unproblematic in the context of promoting preexisting tumors at the time of EV therapy. However, further studies are needed to elucidate whether MSC-derived EVs suppress mTOR signaling and how this affects the injured heart. There is some evidence that mTOR plays an important role in the activation of cell autophagy in myocardial injuries, which can prevent cell apoptosis and necrosis in the myocardial scar [48]. In the EV samples examined in this work, at least six miRNAs were found that can target mTOR or mTOR signaling related protein mRNAs (Table A1, Table A2 and Table A3). A prolonged exposure to EVs containing these miRNAs may therefore either aggravate myocardial injury by increasing apoptosis in the early stages of myocardial infarction or improve wound healing and remodeling via autophagy.
4.2.3. OncomiRs in MSC-Derived EVs
At least six MSC-EV-miRNAs found in the present study are known oncomiRs, namely miR-24-3p, miR-92a-3p, miR-103a-3p, 151a-5p, miR-191-5p, and miR-423-3p. Remarkably, miR-24-3p and miR-423-3p were also associated with cardioprotective properties. Most of these miRNAs target proteins of the Wnt signaling pathway and/or the phosphatase and tensin homolog deleted from chromosome ten (PTEN) protein (Table A1, Table A2 and Table A3). PTEN is an intracellular membrane-bound phosphatase that hydrolyzes phosphatidylinositol (3,4,5)-trisphosphate to phosphatidylinositol (4,5)-bisphosphate and therefore reduces phosphoinositide-dependent kinase-1- and AKT-mediated activation of cell cycle progression and anti-apoptotic signaling [49]. It is a well-described tumor suppressor and often affected by mutations in various cancers. MiR-103a, for example, targets PTEN in endothelial cells and promotes proliferation and thus angiogenesis [50]. At the same time, miRNA-103a acts as an inhibitor of Wnt signaling in squamous cell carcinoma and promotes cell proliferation [51]. Similarly, the inhibition of Wnt signaling is also promoted by miR-92a and miR-221-3p, which in turn also inhibits PTEN expression in esophageal, gastric, and pancreatic cancer [52,53,54]. While Wnt signaling inhibition and PTEN inhibition are desirable targets for miR-10b-5p, miR-27b-3p, and miR-103a-3p in the context of cardioprotection [50,55,56], this may also promote progression of undetected tumors in recipients of EVs containing miRNAs.
5. Conclusions
The administration of MSC-derived EVs containing miRNAs offers a promising therapeutic approach for cardiovascular disease due to their proposed cardioprotective effects. In the present work, we have isolated EVs from two clinically relevant MSC sources, i.e., CB and AT, using membrane affinity columns and analyzed their miRNA cargo by qRT-PCR. Our data show that EVs from CB- and AT-MSCs are similar in their miRNA composition. Although a large number of miRNAs found in EVs from both MSC sources have been associated with cardioprotective properties, our literature research for known miRNA targets has revealed that they may also play a critical role in the tumor biology of various cancers. Given that EVs and miRNAs have a half-life of less than 24 h, a single administration of EVs may not be sufficient to improve tissue remodeling after a myocardial injury and multiple EV administrations would be required. However, this procedure, in turn, could lead to the accumulation of miRNAs in patients with early-stage cancers that may not have been recognized prior to treatment. Therefore, careful screening of patients for preexisting neoplasms prior to EV administration is important to reduce the risk of potential side effects that could facilitate or even worsen existing tumors. Further reports and functional studies are needed to evaluate both the therapeutic and adverse effects of EVs and their transported miRNAs, depending on the dose and duration of treatment.
Acknowledgments
We thank the Core Facility for Electron Microscopy of the Charité—Universitätsmedizin Berlin for support in acquisition and analysis of the data.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/10/9/1353/s1, Table S1: Analyzed miRNAs and their expression values.
Appendix A
Table A1.
MiRNAs that are known tumor suppressors (TS). Selected miRNAs are also involved in cardioprotection (CP). Targets are given for each miRNA, with no claim to completeness. Pubmed IDs (PMIDs) are given as references when no DOI numbers are available.
| MiRNA Function | MiRNA Name | CB-MSC-EV [dCT ± SD] |
AT-MSC-EV [dCT ± SD] |
Fold Difference | p-Value | Confirmed Target GeneGLOBE ID | Cell/Tissue/Cancer Type | MiRNA Cluster | Biological Effect | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| TS | miR-127-3p | 4.05 ± 0.24 | 4.98 ± 0.72 | –1.9 | 0.03 | BCL6 | fibroblasts | — | proliferation inhibition in senescent fibroblasts | doi:10.1371/journal.pone.0080266 |
| KMT5a | chondrocytes | — | proliferation inhibition in osteoarthritis | doi:10.1016/j.bbrc.2018.06.104 | ||||||
| MMP13 | chondrocytes | — | enhances proliferation of chondrocytes in osteoarthritis | doi:10.1111/jcmm.14400 | ||||||
| ITGA6 | osteosarcoma | — | tumor suppressor (cell growth, invasion) | doi:10.1002/iub.1710 | ||||||
| KIF3B | squamous cell carcinoma | — | tumor suppressor (cell growth) | doi:10.26355/eurrev_201901_16877 | ||||||
| KIF3B | pancreatic beta cells | — | proliferation inhibition, diabetes | doi:10.18632/aging.101835 | ||||||
| TS, CP | miR-30c-5p | 4.01 ± 0.5 | 4.84 ± 0.38 | –1.7 | 0.04 | PAI1 | breast cancer | — | suppression of vasculogenesis | doi:10.1172/JCI123106 |
| CTGF | cardiac fibroblasts | miR-133 | cardioprotection (anti-fibrotic) | doi:10.1161/CIRCRESAHA.108.182535 | ||||||
| TGFB1, TGFBR2 | cardiac fibroblasts | — | suppression of fibrosis | doi:10.1111/jcmm.13548 | ||||||
| CTGF | fibroblasts | — | suppression of cardiac and renal fibrosis | doi:10.1016/j.jdiacomp.2015.12.011 | ||||||
| ADAM19 | colorectal carcinoma | — | tumor suppressor (cell proliferation) | doi:10.1371/journal.pone.0120698 | ||||||
| SNAI1 | squamous cell carcinoma | — | tumor suppressor (cell proliferation) | doi:10.1016/j.biopha.2017.12.095 | ||||||
| BCL9 | prostate cancer | — | tumor suppressor (Wnt signaling suppression, cell proliferation) | doi:10.3892/ol.2016.4161 | ||||||
| TS | miR-99b-5p | 4.45 ± 0.46 | 5.45 ± 0.18 | –2.01 | 0.02 | mTOR, AKT, IGF1 | hepatocytes | — | promotes hepatitis B virus replication | doi:10.1111/cmi.12709 |
| mTOR, AKT, IGF1 | gastric cancer | — | tumor suppressor (cell autophagy) | doi:10.3892/ol.2018.9269 | ||||||
| IGF1 | keratinocytes | — | cell proliferation | doi:10.1016/j.biopha.2015.07.013 | ||||||
| PI2K, AKT7, mTOR | cervical cancer | — | tumor suppressor (cell proliferation) | doi:10.1002/jcp.27645 | ||||||
| TS | miR-376a-3p | 3.76 ± 0.47 | 5.05 ± 0.51 | –2.45 | 0.01 | c-MYC | non-small cell lung carcinoma | — | tumor suppressor (cell proliferation, invasion) | doi:10.1002/cbin.10828 |
| COA1, PDIA6 | giant cell tumor | miR-127-3p | tumor suppressor (cell proliferation, invasion) | doi:10.1016/j.canlet.2017.08.029 | ||||||
| NRP1 | breast cancer | — | tumor suppressor (tumor progression) | doi:10.2147/OTT.S173416 | ||||||
| COA1, GLE1, PDIA6 | giant cell tumor | miR-127-3p | tumor suppressor (tumor progression) | doi:10.3390/cancers11122019 | ||||||
| TS | mir-376c-3p | 3.98 ± 0.35 | 4.85 ± 0.57 | –1.81 | 0.03 | HOXB7 | squamous cell carcinoma | — | tumor suppressor (cell proliferation) | doi:10.1016/j.biopha.2017.04.050 |
| BCL2, SYF2 | gastric cancer | — | tumor suppressor (cell proliferation) | doi:10.1155/2016/9604257 | ||||||
| CKD1 | neuroblastoma cells | — | tumor suppressor (cell proliferation) | doi:10.3892/ol.2018.9431 | ||||||
| HB-EGF | medullary thyroid carcinoma | — | tumor suppressor (cell proliferation) | doi:10.5114/aoms.2019.85244 | ||||||
| TS | let-7b-5p | 3.03 ± 0.8 | 1.98 ± 0.36 | 2.07 | 0.04 | FAS | macrophages | — | inhibits clearance of mycobacterium tuberculosis | doi:10.1093/femsle/fny040 |
| KIAA1377 | squamous cell carcinoma | — | tumor suppressor (cell proliferation, invasion) | doi:10.1002/cbin.11136 | ||||||
| IGF1R | multiple melanoma | — | tumor suppressor (cell proliferation, enhances apoptosis) | doi:10.1093/abbs/gmu089 | ||||||
| CDC25B, CDK1 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation, metastasis) | doi:10.1002/jcb.29477 | ||||||
| TS | miR-193b-3p | 3.16 ± 0.45 | 2.21 ± 0.16 | 1.93 | 0.003 | MORC4 | breast cancer | — | tumor suppressor (cell proliferation, enhances apoptosis) | doi:10.1002/jcb.27751 |
| p21-AK2 | ovarian carcinoma | — | tumor suppressor (cell autophagy) | doi:10.1016/j.biopha.2017.11.086 | ||||||
| HDAC3 | brain | — | suppression of NFkB signaling, reduction of inflammation in brain injury | doi:10.1186/s12974-020-01745-0 | ||||||
| CKD1, AJUBA, HEG1 | lung cancer | — | tumor suppressor (cell proliferation, metastasis) | doi:10.1042/BSR20190634 | ||||||
| TGFB1 | liver | — | decreases fibrosis | doi:10.1111/jcmm.14210 | ||||||
| TS | mir-143-3p | 3.28 ± 0.49 | 3.48 ± 0.51 | –1.15 | 0.6 | LIMK1 | breast cancer | — | tumor suppressor (tumor progression) | PMID: 28559978 |
| FOSL2 | osteosarcoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1038/s41598-017-18739-3 | ||||||
| BMPR2 | bone marrow-derived MSCs | — | promotes cartilage differentiation | doi:10.26355/eurrev_201812_16649 | ||||||
| IGF1R, IGFBP5 | synovial cells | — | promotes inflammation and increases apoptosis in RA | doi:10.3892/etm.2018.5907 | ||||||
| BCL2, IGF1R | squamous cell carcinoma | — | tumor suppressor (tumor progression) | doi:10.1016/j.bbrc.2019.08.075 | ||||||
| TS, CP | miR-199a-3p | 1.92 ± 0.96 | 1.1 ± 0.45 | 1.82 | 0.16 | ITGB8 | ovarian carcinoma | — | tumor suppressor (chemoresistance) | doi:10.3892/or.2018.6259 |
| GRP78 | non-small cell lung carcinoma | miR-495 (low detection) |
tumor suppressor (tumor progression) | doi:10.1016/j.gene.2017.03.032 | ||||||
| DDIT4, ING4 | cardiomyocytes | miR-214 | cardioprotective (inhibit cardiomyocyte apoptosis during injury) | doi:10.1152/ajpheart.00807.2015 | ||||||
| mTOR | kidney | — | induces injury induced apoptosis | doi:10.1002/jcb.29030 | ||||||
| AXL | osteosarcoma | — | tumor suppressor (tumor progression) | PMID: 25520864 | ||||||
| mTOR | endometrial endometrioid adenocarcinoma | — | tumor suppressor (cell autophagy) | PMID: 31966798 | ||||||
| KL | kidney | — | activation of NFkB signaling in lupus nephritis | doi:10.1016/j.molimm.2018.10.003 | ||||||
| SMAD1 | prostate cancer | — | tumor suppressor (cell proliferation, invasion) | doi:10.18632/oncotarget.17191 | ||||||
| mTOR | hepatocellular carcinoma | — | tumor suppressor (chemosensitivity) | doi:10.1186/s13046-019-1512-5 | ||||||
| AGAP2 | glioma cells | — | tumor suppressor (tumor progression) | doi:10.18632/aging.102092 | ||||||
| PTGIS | endothelial cells | miR-199a-5p | nitrovasodilatator resistance | doi:10.1161/CIRCULATIONAHA.117.029206 | ||||||
| CD44 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation) | doi:10.1016/j.bbrc.2010.10.130 | ||||||
| SOCS7, STAT3 | kidney | — | suppress renal fibrosis | doi:10.1038/srep43409 | ||||||
| TS, CP | miR-199a-5p | 2.63 ± 0.92 | 2.35 ± 0.65 | 1.22 | 0.68831 | AA1B | monocytes | — | inhibits differentiation | doi:10.1189/jlb.1A0514-240R |
| MAP3K11 | non-small cell lung carcinoma | — | tumor suppressor (tumor progression) | doi:10.7150/jca.29426 | ||||||
| SNAI1 | papillary thyroid carcinoma | — | tumor suppressor (tumor progression) | doi:10.1016/j.bbrc.2018.02.051 | ||||||
| HIF1A | hemangioma cells | — | tumor suppressor (cell proliferation, autophagy) | doi:10.1177/0394632017749357 | ||||||
| CCR7 | bladder cancer | — | tumor suppressor (metastasis) | doi:10.1186/s12894-016-0181-3 | ||||||
| ETS1 | breast cancer | — | tumor suppressor (cell invasion) | doi:10.1111/cas.12952 | ||||||
| CLTC | hepatocellular carcinoma | — | tumor suppressor (tumorigenesis) | doi:10.1002/cbf.3252 | ||||||
| PIAS3 | cervical cancer | — | tumor suppressor (metastasis, suppresses epithelial–mesenchymal transition) | doi:10.1002/jcb.28631 | ||||||
| ROCK1 | colorectal carcinoma | — | tumor suppressor (cell proliferation, metastasis) | doi:10.1177/1533034618775509 | ||||||
| ECE1 | spinal cord nerves | — | inhibition of ischemia-reperfusion injury | doi:10.1007/s10571-018-0597-2 | ||||||
| TET2 | osteoblasts | — | promote differentiation | doi:10.1016/j.gene.2019.144193 | ||||||
| DDR1 | brain | — | protect against ischemia-reperfusion injury | doi:10.1016/j.wneu.2019.07.203 | ||||||
| DRAM1 | acute myeloid leukemia | — | tumor suppressor (chemosensitivity) | doi:10.1155/2019/5613417 | ||||||
| CDH1 | squamous cell carcinoma | — | tumor suppressor (cell invasion) | doi:10.3892/ol.2016.4602 | ||||||
| MAP4K3 | hepatocellular carcinoma | let-7c | tumor suppressor (invasion, metastasis) | doi:10.18632/oncotarget.14623 | ||||||
| ATF6, GRP78 | cardiomyocytes | — | downregulation in myocardial hypoxic preconditioning | doi:10.1007/s13105-018-0657-6 | ||||||
| MAP3K11 | esophageal cancer | — | tumor suppressor (cell proliferation) | doi:10.18632/oncotarget.6752 | ||||||
| ZEB1 | ovarian ectopic endometrial stromal cell | — | inhibition of epithelial–mesenchymal transition | doi:10.1007/s43032-019-00016-5 | ||||||
| HIF1A, OSGIN2 | sarcoma | — | tumor suppressor (tumor progression) | doi:10.3892/ol.2016.5320 | ||||||
| PHLPP1 | colorectal carcinoma | — | tumor suppressor (chemosensitivity) | doi:10.1517/14728222.2015.1057569 | ||||||
| BIP | kidney | — | protect against ischemia-reperfusion injury | doi:10.1096/fj.201801821R | ||||||
| WNT2 | urothelial cells | — | inhibiting smooth muscle cell proliferation | doi:10.1074/jbc.M114.618694 | ||||||
| MAGT1 | gliomal cells | — | tumor suppressor (tumor progression) | doi:10.1002/jcb.28791 | ||||||
| CD44, SIRT1 | squamous cell carcinoma | — | tumor suppressor (repress stemness) | doi:10.1080/15384101.2019.1689482 | ||||||
| KL | gastric cancer | — | oncomiR (promotes tumor progression) | doi:10.1186/1471-2407-14-218 | ||||||
| Mrz 08 | gliomal cells | — | tumor suppressor (tumor progression) | doi:10.26355/eurrev_201909_18858 | ||||||
| JunB | cardiomyocytes | — | promotes apoptosis in the failing heart) | doi:10.1038/s41598-018-24932-9 | ||||||
| CAV1 | lung | — | promotes lung fibrosis | doi:10.1371/journal.pgen.1003291 | ||||||
| NFKB | ovarian carcinoma | — | tumor suppressor (cell proliferation, invasion) | doi:10.3892/ol.2018.9170 | ||||||
| SIRT1, ENOS | endothelial cells | — | promotes migration and tube formation | doi:10.1007/s00705-013-1744-1 | ||||||
| HIF1A | prostate adeno-carcinoma | — | tumor suppressor (tumor progression) | doi:10.18632/oncotarget.18315 | ||||||
| TS, CP | miR-99a-5p | 4.11 ± 0.87 | 4.32 ± 0.33 | –1.15 | 0.49 | mTOR | urothelial carcinoma | — | tumor suppressor (cell autophagy) | doi:10.2147/OTT.S114276 |
| HOXA1 | smooth muscle cells | — | cardioprotective (inhibits smooth muscle cell proliferation and atherosclerosis) | doi:10.1016/j.lfs.2019.116664 | ||||||
| mTOR | bladder cancer | — | tumor suppressor (cell proliferation) | doi:10.1002/jcb.27318 | ||||||
| NOX4 | oral cancer | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.4149/neo_2017_503 | ||||||
| CDC25A | breast cancer | — | tumor suppressor (tumor progression) | doi:10.3390/genes11040369 | ||||||
| TS | miR-16-5p | 1.76 ± 0.43 | 1.81 ± 0.33 | –1.03 | 0.84 | AKT3 | breast cancer | — | tumor suppressor (tumor progression) | doi:10.1042/BSR20191611 |
| SMAD3 | chordoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1038/s41419-018-0738-z | ||||||
| PIK3R1 | fibroblasts | — | inhibits proliferation | doi:10.3390/ijms20051036 | ||||||
| ANXA11 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation, metastasis) | doi:10.1186/s13046-019-1188-x | ||||||
| MYCN | neuroblastoma cells | miR-15a-5p, miR-15b-5p |
tumor suppressor (tumor progression) | doi:10.1002/1878-0261.12588 | ||||||
| IGA2 | colorectal carcinoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1002/jcp.28747 | ||||||
| SESN1 | myoblasts | — | myoblast differentiation and proliferation | doi:10.1038/s41419-018-0403-6 | ||||||
| BACH2 | gingival epithelial cells | miR-145-5p | induce apoptosis | PMID: 32509061 | ||||||
| SMAD3 | chondrocytes | — | promotes osteoarthritis | doi:10.2174/1381612821666150909094712 | ||||||
| CARM | cervical cancer | — | tumor suppressor (promotes radiosensitivty) | doi:10.1111/pin.12867 | ||||||
| VEGFA | MSCs | — | suppresses osteogenic potential of MSCs | doi:10.18632/aging.103223 | ||||||
| VEGFA | breast cancer | — | tumor suppressor (cell proliferation, invasion, autophagy) | doi:10.18632/oncotarget.20398 | ||||||
| VEGFA | colorectal carcinoma | — | tumor suppressor (cell proliferation, invasion, autophagy) | doi:10.1016/j.omtn.2020.03.006 | ||||||
| EPT1 | preadipocytes | — | promotes differentiation | doi:10.1016/j.bbrc.2019.04.179 | ||||||
| TS, CP | miR-22-3p | 2.52 ± 0.7 | 1.83 ± 0.31 | 1.62 | 0.09 | HMGB1 | arterial smooth muscle cells | — | inhibits atherosclerosis | doi:10.1159/000480212 |
| MAPK14 | brain | — | prevents Alzheimer’s disease | doi:10.2174/1567202616666191111124516 | ||||||
| WRNIP1 | small cell lung cancer | — | tumor suppressor (radiosensitivity) | doi:10.1002/jcb.29032 | ||||||
| AE1 | retinoblastoma | — | tumor suppressor (cell proliferation) | doi:10.1016/j.biopha.2018.06.038 | ||||||
| EIF4EBP3 | cervical cancer | — | oncomiR (tumuorogenesis | doi:10.7150/ijms.21645 | ||||||
| PTEN | kidney | — | suppresses sepsis-induced kidney injury | doi:10.1042/BSR20200527 | ||||||
| AKT3 | Wilm’s tumor | — | tumor suppressor (cell growth) | doi:10.26355/eurrev_202006_21493 | ||||||
| SP1 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | PMID: 27904693 | ||||||
| SIRT1 | peridontal stem cells | — | increases proliferation and differentiation | doi:10.1002/cbin.11271 | ||||||
| PTAFR | cardiac fibroblasts | — | cardioprotective (reduces activation of cardiac fibroblasts) | doi:10.26355/eurrev_202004_20869 | ||||||
| YAP1 | non-small cell lung carcinoma | — | tumor suppressor (tumor progression) | doi:10.1111/1759-7714.13280 | ||||||
| DDIT4 | glioblastoma | — | tumor suppressor (cell proliferation) | doi:10.1016/j.neulet.2020.134896 | ||||||
| SIRT1 | ectopic endometrial cells | — | enhances proliferation and invasion | doi:10.26355/eurrev_202001_20033 | ||||||
| FTO | MSCs | — | promotes osteogenic differentiation | doi:10.1186/s13287-020-01707-6 | ||||||
| NFIB | gastric cancer | — | tumor suppressor (tumor progression) | doi:10.4149/neo_2020_190418N350 | ||||||
| TS | miR-152-3p | 4.42 ± 0.66 | 5.16 ± 0.62 | –1.9 | 0.09 | SOS1 | glioblastoma | — | tumor suppressor (chemosensitivity) | doi:10.2147/OTT.S210732 |
| p27 | chronic myeloid leukemia | — | oncomiR (tumorigenesis) | doi:10.26355/eurrev_201812_16646 | ||||||
| KLF4 | prostate cancer | — | tumor suppressor (tumor progression) | doi:10.1002/jcb.28984 | ||||||
| FOXF1 | fibroblasts | — | promotes cell proliferation, invasion and extracellular matrix production | doi:10.1016/j.lfs.2019.116779 | ||||||
| CDK8 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation) | doi:10.1016/j.prp.2019.03.034 | ||||||
| TMEM97 | prostate cancer | — | tumor suppressor (tumor progression) | doi:10.1186/s13148-018-0475-2 | ||||||
| SPIN1 | breast cancer | miR-148 | tumor suppressor (chemosensitivity) | doi:10.1186/s13046-018-0748-9 | ||||||
| PIK3CA | breast cancer | — | tumor suppressor (tumor progression) | doi:10.3727/096504017x14878536973557 | ||||||
| TS, CP | miR-145-5p | 1.55 ± 0.65 | 1.90 ± 0.37 | –1.28 | 0.3 | FLT1 | trophoblast | — | promote cell proliferation, invasion | doi:10.1016/j.lfs.2019.117008 |
| KLF4 | lung | — | promotes chronic obstructive pulmonary disease | doi:10.1016/j.cbi.2019.01.011 | ||||||
| CD40 | cardiomyocytes | — | cardioprotection (in ischemia-reperfusion injury) | doi:10.1007/s11010-017-2982-4 | ||||||
| KLF5 | gastric cancer | — | tumor suppressor (tumor progression) | doi:10.1002/jcp.27525 | ||||||
| TAGLN2 | bladder cancer | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.3892/ol.2018.9436 | ||||||
| SOX2 | breast cancer | — | tumor suppressor (tumor progression) | doi:10.1016/j.jss.2018.11.030 | ||||||
| RHBDD1 | colorectal carcinoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1016/j.biocel.2019.105641 | ||||||
| FSCN1 | squamous cell carcinoma | — | tumor suppressor (tumor progression) | doi:10.1016/j.ymthe.2018.09.018 | ||||||
| TPT1 | prolactinoma | — | tumor suppressor (chemosensitivity) | doi:10.1007/s40618-018-0963-4 | ||||||
| TGFB1 | vascular smooth muscle cells | — | inhibits proliferation | doi:10.12659/MSM.910986 | ||||||
| TLR4 | melanoma | — | tumor suppressor (cell autophagy) | doi:10.1002/jcb.28388 | ||||||
| SEMA3A | AT-MSCs | — | suppresses osteogenic potential of MSCs | doi:10.1007/s11626-019-00318-7 | ||||||
| AKAP12 | prostate cancer | — | tumor suppressor (chemosensitivity) | doi:10.1111/jcmm.13604 | ||||||
| SMAD2/3 | hepatocellular carcinoma | — | reduces extracellular matrix production | doi:10.1016/j.bbrc.2019.11.040 | ||||||
| MTDH | squamous cell carcinoma | — | tumor suppressor (tumor progression) | doi:10.1177/1533033819850189 | ||||||
| NRAS | melanoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1002/cam4.1030 | ||||||
| MTDH | non-small cell lung carcinoma | — | tumor suppressor (tumor progression) | doi:10.1096/fj.201701237RR | ||||||
| TS | miR-193a-5p | 4.52 ± 0.67 | 4.35 ± 0.23 | 1.13 | 0.82 | CDK8 | leukemia | — | tumor suppressor (cell proliferation, apoptosis) | doi:10.3892/ijmm.2020.4671 |
| COL1A1 | colorectal carcinoma | — | tumor suppressor (inhibits epithelial–mesenchymal transition) | doi:10.2147/OTT.S255485 | ||||||
| COL1A1 | colorectal carcinoma | — | tumor suppressor (inhibits epithelial–mesenchymal transition) | doi:10.3389/fonc.2020.00850 | ||||||
| HOXA1 | breast cancer | — | tumor suppressor (tumor progression) | doi:10.18632/aging.103123 | ||||||
| HOXA7 | ovarian carcinoma | — | tumor suppressor (cell proliferation, apoptosis) | doi:10.4149/neo_2020_190730N687 | ||||||
| CCNE1 | esophageal cancer | — | tumor suppressor (tumor progression) | doi:10.1007/s13402-019-00493-5 | ||||||
| ERBB2 | colorectal carcinoma | — | tumor suppressor (tumor progression) | doi:10.2147/CMAR.S234620 | ||||||
| SRSF6 | pancreatic cancer | — | oncomiR (metastasis) | PMID: 32064152 | ||||||
| DPEP1 | hepatoblastoma | — | tumor suppressor (tumor progression) | doi:10.1038/s41419-019-1943-0 | ||||||
| TS, CP | miR-20a-5p | 4.83 ± 0.27 | 4.72 ± 0.93 | 1.08 | 0.58 | ABCA1 | artery smooth muscle cells | — | promotes cell proliferation and migration | doi:10.1002/jbt.22589 |
| PTEN | endothelial cells | — | pro-angiogenic, inhibits autophagy and apoptosis | doi:10.1038/s41419-020-02745-x | ||||||
| ERBB2 | hepatocellular carcinoma | miR-17-5p | tumor suppressor (metastasis) | doi:10.7150/thno.41365 | ||||||
| TGFBR2 | liver | — | anti-fibrotic | doi:10.3389/fonc.2020.00107 | ||||||
| STAT3 | endometrial carcinoma | — | tumor suppressor (inhibits epithelial–mesenchymal transition, invasion) | PMID: 31949657 | ||||||
| STAT3 | bronchial epithelial cells | — | suppresses apoptosis | doi:10.1016/j.mcp.2019.101499 | ||||||
| SRCIN1 | osteoclasts | — | promote proliferation and differentiation | doi:10.1002/cam4.2454 | ||||||
| TGFB1 | endothelial cells | — | anti-angiogenic | doi:10.1002/jcp.29111 | ||||||
| TS, CP | miR-29c-3p | 3.27 ± 0.92 | 2.48 ± 0.99 | 1.73 | 0.28 | STAT3 | cardiac fibroblasts | — | cardioprotection (inhibits cell proliferation) | doi:10.23736/S0031-0808.20.03975-0 |
| TNFAIP1 | neuroblastoma cells | — | oncomiR (inhibits apoptosis) | doi:10.1007/s11064-020-03096-x | ||||||
| FOS | lens epithelial cells | — | inhibits epithelial–mesenchymal transition | doi:10.1016/j.biopha.2020.110290 | ||||||
| VEGFA | colorectal carcinoma | — | tumor suppressor (inhibit angiogenesis) | doi:10.1186/s13046-020-01594-y | ||||||
| TFAP2C | T-cell acute lymphoblastic leukemia | miR-29b-3p | tumor suppressor (cell proliferation) | doi:10.1016/j.bbrc.2020.03.170. Epub 2020 | ||||||
| NFAT | brain | — | inhibit inflammation in Parkinson’s disease | doi:10.1111/gtc.12764 | ||||||
| CCNA2 | esophageal cancer | — | tumor suppressor (cell proliferation, migration, and invasion) | doi:10.3389/fbioe.2020.00075 | ||||||
| TRIM31 | hepatocellular carcinoma | — | tumor suppressor (tumor progression) | doi:10.3892/or.2020.7469 | ||||||
| FOXP1 | ovarian carcinoma | — | tumor suppressor (chemosensitivity) | doi:10.1080/15384101.2019 | ||||||
| TS, CP | miR-30d-5p | 4.70 ± 0.44 | 5.43 ± 0.63 | 1.66 | 0.09 | SIRT1 | cardiomyocytes | — | cardioprotection (inhibits hypoxia induced apoptosis) | PMID: 32098921 |
| SMAD2 | ovarian granulosa cells | — | promotes apoptosis | doi:10.3892/etm.2019.8184 | ||||||
| NT5E | prostate cancer | — | tumor suppressor (cell proliferation, migration) | doi:10.1089/cbr.2018.2457 | ||||||
| RUNX2 | colon cancer | — | tumor suppressor (tumor progression) | PMID: 29552759 | ||||||
| TS | miR-320a | 2.51 ± 0.58 | 2.64 ± 0.20 | –1.09 | 0.60 | CXCL9 | synovial cells | — | suppress cell proliferation | doi:10.3389/fphys.2020.00441 |
| SIRT4 | ovaries | — | prevent premature ovarian insufficiency | doi:10.1016/j.omtn.2020.05.013 | ||||||
| HIF1A | endometrial carcinoma | — | tumor suppressor (anti-angiogenic) | doi:10.1016/j.yexcr.2020.112113 | ||||||
| SMAD5 | bone marrow-derived MSCs | — | promote osteogenic differentiation | doi:10.26355/eurrev_202003_20648 | ||||||
| ANRIL | papillary thyroid carcinoma | — | tumor suppressor (tumorigenesis) | doi:10.1016/j.prp.2020.152856 | ||||||
| LOX1 | endothelial cells | — | inhibit apoptosis upon low-density lipoprotein exposure | doi:10.1007/s11010-020-03688-9 | ||||||
| CPEB1 | osteosarcoma | — | tumor suppressor (invasion, migration) | doi:10.1002/cam4.2919 | ||||||
| TXNRD1 | osteosarcoma | — | tumor suppressor (cell proliferation, migration) | doi:10.1080/15384047.2019.1702405 | ||||||
| FOXM1 | hepatocellular carcinoma | — | tumor suppressor (inhibits epithelial–mesenchymal transition, tumor progression) | doi:10.3390/biom10010020 | ||||||
| PBX3 | gastric cancer | — | tumor suppressor (tumor progression) | doi:10.4251/wjgo.v11.i10.842 | ||||||
| PKCG | cancer | — | tumor suppressor (cell invasion) | doi:10.1038/s41419-019-1921-6 | ||||||
| MAFB | retina | — | promotes diabetic retinopathy | doi:10.18632/aging.101962 | ||||||
| MAPK | synovial cells | — | promote apoptosis, inhibit proliferation | doi:10.26355/eurrev_201903_17228 | ||||||
| IGFR1 | endometrial carcinoma | — | tumor suppressor (tumor progression) | doi:10.3892/ijmm.2019.4051 | ||||||
| TS | miR-361-5p | 4.82 ± 0.92 | 5.33 ± 0.31 | –1.43 | 0.34 | ITGB1 | cervical cancer | — | tumor suppressor (cell proliferation) | doi:10.1007/s43032-019-00008-5 |
| FOXO1 | chondrocytes | — | promotes apoptosis and inhibits cell proliferation | doi:10.1186/s12920-019-0649-6 | ||||||
| SDCBP | gastric cancer | — | tumor suppressor (tumor progression) | doi:10.1097/CAD.0000000000000846 | ||||||
| WT1 | hepatocellular carcinoma | — | tumor suppressor (tumorigenesis) | doi:10.26355/eurrev_201910_19277 | ||||||
| ABCA1 | vascular smooth muscle cells | — | inhibits proliferation | PMID: 31312370 | ||||||
| CLDN8 | retinoblastoma | — | tumor suppressor (cell proliferation, promotes apoptosis) | doi:10.1007/s00381-019-04199-9 | ||||||
| VEGFA | hemangioma cells | — | tumor suppressor (anti-angiogenic) | doi:10.1016/j.bbrc.2019.03.084 | ||||||
| FOXM1 | cervical cancer | — | tumor suppressor (tumor progression) | doi:10.1080/21691401.2019.1577883 | ||||||
| FOXM1 | osteosarcoma | — | tumor suppressor (tumorigenesis) | doi:10.1002/jcp.28026 | ||||||
| SIRT1 | liver | — | promotes hepatosteatosis | doi:10.1016/j.metabol.2018.08.007 | ||||||
| SND1 | glioma cells | — | tumor suppressor (invasion, migration) | doi:10.2147/OTT.S171539 | ||||||
| MMP3, MMP9, VEGF | gastric cancer | — | tumor suppressor (inhibits epithelial–mesenchymal transition, tumor progression) | doi:10.1016/j.gene.2018.06.095 | ||||||
| RQCD1 | breast cancer | — | tumor suppressor (invasion, migration) | doi:10.17305/bjbms.2018.3399 | ||||||
| ROCK1 | papillary thyroid carcinoma | — | tumor suppressor (tumor progression) | doi:10.1016/j.biopha.2018.03.122 | ||||||
| RPL22L1 | ovarian carcinoma | — | tumor suppressor (tumorigenesis) | PMID: 31938372 | ||||||
| FOXM1 | gastric cancer | — | tumor suppressor (chemoresistance) | doi:10.18632/oncotarget.23513 | ||||||
| FGFR1, MMP1 | breast cancer | — | tumor suppressor (cell proliferation, metastasis, metabolism) | doi:10.1186/s13046-017-0630-1 | ||||||
| FOXM1 | lung cancer | — | tumor suppressor (tumor progression) | doi:10.4149/neo_2017_406 | ||||||
| TWIST1 | glioma cells | — | tumor suppressor (inhibits epithelial–mesenchymal transition) | doi:10.3892/or.2017.5406 | ||||||
| TS | miR-708-5p | 5.32 ± 0.62 | 5.41 ± 0.72 | –1.06 | 0.94 | PGE2 | lung cancer | — | tumor suppressor (tumorigenesis) | doi:10.18632/oncotarget.27614 |
| CTNNB1 | colon cancer | — | tumor suppressor (tumor progression) | doi:10.1016/j.biopha.2020.110292 | ||||||
| TLR4 | macrophages | — | immunomodulation of controlling inflammatory factors | doi:10.26355/eurrev_201909_19019 | ||||||
| ZEB1 | osteosarcoma | — | tumor suppressor (cell proliferation, invasion) | doi:10.3892/mmr.2019.10013 | ||||||
| URGCP | pancreatic ductal adenocarcinoma | — | tumor suppressor (tumor progression) | doi:10.1016/j.prp.2019.01.026 | ||||||
| TS | let-7c-5p | 3.67 ± 1.01 | 3.94 ± 0.38 | –1.21 | 0.43 | TGFBR1 | kidney | — | chronic kidney disease | doi:10.1155/2020/6960941 |
| PBX3 | squamous cell carcinoma | — | tumor suppressor (tumor progression) | doi:10.1186/s12943-020-01215-4 | ||||||
| CMYC | hepatocellular carcinoma | — | tumor suppressor (cell proliferation) | doi:10.1016/j.bbrc.2019.09.091 | ||||||
| HMGA2 | dental pulp stem cells | — | promotes osteogenic differentiation | doi:10.1111/1440-1681.13059 | ||||||
| DMP1MNF | dental pulp stem cells | — | inhibits inflammation | doi:10.12659/MSM.909093 | ||||||
| NAP1L1 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation, migration) | doi:10.1016/j.canlet.2018.08.024 | ||||||
| TS | let-7e-5p | 4.48 ± 1.39 | 5.18 ± 0.15 | –1.63 | 0.28 | CCR7 | squamous cell carcinoma | — | tumor suppressor (cell proliferation, metastasis) | doi:10.7150/jca.29536 |
| FASLG | endothelial progenitors | — | prevents deep vein thrombosis | doi:10.1016/j.thromres.2015.12.020 | ||||||
| RCN1 | nasopharyngeal carcinoma | — | tumor suppressor (cell autophagy) | doi:10.1152/ajpcell.00352.2019 | ||||||
| TS | let-7g-5p | 4.69 ± 1.15 | 4.60 ± 0.52 | 1.07 | 0.84 | HMGA2 | glioblastoma | — | tumor suppressor (tumor progression) | doi:10.1111/jcmm.14884 |
| IGF1R | nasopharyngeal carcinoma | — | tumor suppressor (cell migration, invasion) | doi:10.12659/MSM.914555 | ||||||
| PRKCA | mammary cells | — | regulates differentiation | doi:10.1002/jcp.27676 | ||||||
| VSIG4 | glioblastoma | — | tumor suppressor (inhibits epithelial–mesenchymal transition) | doi:10.3892/or.2016.5098 | ||||||
| TS, CP | let-7i-5p | 1.14 ± 0.89 | 1.39 ± 0.93 | –1.19 | 0.72 | GALE | glioblastoma | — | tumor suppressor (cell proliferation, metastasis) | doi:10.2147/CMAR.S221585 |
| HMGA1 | bladder cancer | — | tumor suppressor (cell proliferation, metastasis) | doi:10.1186/s12894-019-0485-1 | ||||||
| CCND2, E2F2 | cardiomyocytes | — | cardioprotection (promotes proliferation after injury) | doi:10.1042/CS20181002 | ||||||
| KLK6 | colon cancer | — | tumor suppressor (cell proliferation, metastasis) | doi:10.3892/or.2018.6577 |
Table A2.
MiRNAs that are known oncomiRs (O). Selected miRNAs are also involved in cardioprotection (CP). Targets are given for each miRNA, with no claim to completeness. Pubmed IDs (PMIDs) are given as references when no doi numbers are available.
| MiRNA Function | MiRNA Name | CB-MSC-EV [dCT ± SD] | AT-MSC-EV [dCT ± SD] | Fold Difference | p-Value | Confirmed Target GeneGLOBE ID | Cell/Tissue/Cancer Type | MiRNA Cluster | Biological Effect | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| O, CP | miR-100-5p | 1.11 ± 0.67 | 2.05 ± 0.28 | –1.9 | 0.05 | ANGPT2 | hepatocellular carcinoma | — | suppression of angiogenesis | doi:10.1002/path.4804 |
| p53 | pancreatic ductal adenocarcinoma | — | oncomiR (promotes cell growth) | doi:10.1038/s41467-018-03962-x | ||||||
| mTOR | endometrial carcinoma | miR-199a-3p, miR-199b-5p | tumor suppressor (cell autophagy) | PMID: 31966798 | ||||||
| mTOR | breast cancer | — | tumor suppressor (anti-angiogenic) | doi:10.1007/s13402-017-0335-7 | ||||||
| mTOR | osteosarcoma | — | tumor suppressor (cell autophagy) | doi:10.26355/eurrev_201809_15913 | ||||||
| mTOR | vascular smooth muscle cells | — | suppression of angiogenesis | doi:10.1161/CIRCULATIONAHA.110.000323 | ||||||
| O | miR-151a-5p | 4.0 ± 0.38 | 5.09 ± 0.2 | –2.14 | 0.007 | CDH1 | non-small cell lung carcinoma | — | oncomiR (promotes epithelial–mesenchymal transition, proliferation, invasion) | doi:10.1038/oncsis.2017.66 |
| p53 | nasopharyngeal carcinoma | — | oncomiR (promotes cell proliferation, invasion) | doi:10.1042/BSR20191357 | ||||||
| O | miR-103a-3p | 2.49 ± 0.64 | 4.24 ± 0.18 | –3.35 | 0.02 | APC/APC2 | colorectal carcinoma | miR-1872 (not tested) |
oncomiR (activator of Wnt signaling, cell proliferation) | doi:10.1002/jcb.26357 |
| CDK5 | bladder cancer | miR-107 | oncomiR (promotes cell proliferation, invasion) | doi:10.1038/emm.2015.39 | ||||||
| CDK6 | AT-MSCs | — | inhibit proliferation | doi:10.1038/srep30919 | ||||||
| GPRC5A | prostate cancer | — | oncomiR, tumor suppressor (depending on cancer type) | doi:10.1261/rna.045757.114 | ||||||
| CDH11, NR3C1 | squamous cell carcinoma | — | oncomiR (promotes cell proliferation) | doi:10.26355/eurrev_202006_21505 | ||||||
| PTEN | endothelial progenitor cells | — | promotes migration and angiogenesis | doi:10.1016/j.avsg.2019.10.048 | ||||||
| SNRK | glomerular endothelial cells | — | promotes NFkB/p65 activation, renal inflammation and fibrosis | doi:10.1038/s41467-019-11515-z | ||||||
| O | miR-191-5p | 3.25 ± 0.42 | 4.26 ± 0.42 | –2.01 | 0.02 | SOX4 | breast cancer | — | oncomiR (promotes cell proliferation) | doi:10.1261/rna.060657.117 |
| EGR1, UBE2D3 | hepatocellular carcinoma | — | oncomiR (promotes cell proliferation) | PMID: 31933962 | ||||||
| EGR1 | osteosarcoma | — | oncomiR (activates PI3K/AKT pathway, proliferation, invasion) | doi:10.26355/eurrev_201905_17783 | ||||||
| ENOS, MMP1, MMP9 | endothelial cells | — | antiangiogenic | doi:10.1096/fj.201601263R | ||||||
| O | miR-92a-3p | 3.05 ± 0.75 | 3.12 ± 0.26 | –1.05 | 0.67 | WNT5A | chondrocytes | — | enhance chondrogenesis | doi:10.1186/s13287-018-1004-0 |
| PTEN | squamous cell carcinoma | — | oncomiR (promotes cell proliferation, metastasis) | doi:10.3892/ijmm.2019.4258 | ||||||
| CDH1 | glioma cells | — | oncomiR (promotes tumor progression) | doi:10.3390/ijms17111799 | ||||||
| PTEN | pancreatic cancer | — | oncomiR (promotes cell proliferation, invasion) | doi:10.11817/j.issn.1672-7347.2020.180459 | ||||||
| O, CP | miR-423-3p | 4.49 ± 0.12 | 4.47 ± 0.14 | 1.01 | 0.83 | RAP2C | cardiomyocytes | — | cardioprotection (in ischemic postconditioning secreted by cardiac fibroblast-EVs) | doi:10.1093/cvr/cvy231 |
| p21CIP1, WAF1 | colorectal carcinoma | — | oncomiR (promotes cell growth) | doi:10.1159/000430230 | ||||||
| PANX2 | glioma cells | — | oncomiR (promotes tumor progression) | PMID: 29928399 | ||||||
| ADIPOR2 | laryngeal cancer | — | oncomiR (promotes tumor progression) | PMID: 25337209 | ||||||
| O, CP | miR-21-5p | -0.90 ± 0.52 | -0.78 ± 0.82 | –1.09 | 0.98 | FASLG | hepatocellular carcinoma | — | oncomiR (chemoresitance) | doi:10.1089/dna.2018.4529 |
| CCR7 | chondrosarcoma | — | tumor suppressor (tumor progression) | doi:10.1080/03008207.2019.1702650 | ||||||
| TIAM1 | colon cancer | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1159/000493457 | ||||||
| PDCD4 | breast cancer | — | oncomiR (chemoresitance) | doi:10.4149/neo_2018_181207N930 | ||||||
| BCL2, TLR4 | macrophages | — | regulates mycobacterial survival | doi:10.1002/1873-3468.13438 | ||||||
| SET, TAF-IA | lung adenocarcinoma | — | oncomiR (promotes tumor progression) | doi:10.1016/j.lfs.2019.06.014 | ||||||
| RAB11A | neurons | — | neuroprotection during traumatic brain injury | doi:10.12659/MSM.915727 | ||||||
| PTEN, PDCD4 | lung | — | anti-apoptotic during ischemia-reperfusion injury | doi:10.1016/j.ejphar.2019.01.022 | ||||||
| SOX7 | non-small cell lung carcinoma | — | oncomiR (chemoresitance) | doi:10.2147/OTT.S146423 | ||||||
| TGFB1 | non-small cell lung carcinoma | — | oncomiR (promotes cell proliferation) | doi:10.3892/etm.2018.6752 | ||||||
| CHL1 | colon adenocarcinoma | — | oncomiR (promotes cell proliferation, invasion) | doi:10.1186/s10020-018-0034-5 | ||||||
| PTEN, PDCD4 | lung cancer | — | oncomiR (promotes cell proliferation, metastasis m2 polarization) | doi:10.1186/s13046-019-1027-0 | ||||||
| SMAD7 | non-small cell lung carcinoma | — | oncomiR (promotes tumor progression) | doi:10.2147/OTT.S172393 | ||||||
| PI3K | cardiomyocytes | — | cardioprotection (improves contractility) | doi:10.1161/CIRCRESAHA.118.312420 | ||||||
| SPRY1 | joints | — | suppresses angiogenesis and matrix degeneration | doi:10.1186/s13075-020-2145-y | ||||||
| PDCD4 | squamous cell carcinoma | — | oncomiR (anti-apoptotic) | doi:10.3892/etm.2019.7970 | ||||||
| MASPIN | endothelial cells | — | suppresses angiogenesis and proliferation | doi:10.1080/09168451.2018.1459179 | ||||||
| CDKN2C | melanoma | — | oncomiR (promotes cell proliferation) | doi:10.1002/2211-5463.12819 | ||||||
| CCL1, TIMP3 | neurons | — | inhibits neuropathic pain development | doi:10.1002/jcb.28920 | ||||||
| SMAD7 | fibroblasts | — | promote fibrosis in tendon injury | doi:10.1016/j.omtn.2018.11.006 | ||||||
| FASLG | cardiomyocytes | — | cardioprotection (in ischemia-reperfusion injury) | doi:10.1042/BSR20190597 | ||||||
| WWC2 | lung adenocarcinoma | — | oncomiR (promotes cell proliferation, metastasis) | doi:10.3233/CBM-201489 | ||||||
| PTEN, mTOR | brain | — | protects against seizure damage | doi:10.1016/j.eplepsyres.2018.05.001 | ||||||
| SMAD7 | fibroblasts | — | activation of spinal fibrosis | doi:10.7150/ijbs.24074 | ||||||
| PDCD4 | osteosarcoma | — | oncomiR (promotes cell proliferation, metastasis) | doi:10.3892/ijo.2017.4127 | ||||||
| HMSH2 | non-small cell lung carcinoma | — | oncomiR (chemoresitance) | doi:10.1159/000481839 | ||||||
| PTEN | smooth muscle cells | — | promotes proliferation and remodeling | doi:10.3390/ijms20040875 | ||||||
| MAPK10 | breast cancer | — | oncomiR (promotes tumor progression) | doi:10.1042/BSR20181000 | ||||||
| PTEN | fibroblasts | — | prevents radiation-induced autophagy | doi:10.1038/s41374-019-0323-9 | ||||||
| CADM1 | tongue cancer | — | oncomiR (chemoresitance) | doi:10.1007/s00109-016-1417-0 | ||||||
| O, CP | miR-34a-5p | 3.49 ± 1.45 | 2.09 ± 0.84 | 2.65 | 0.23 | NOTCH1 | cardiomyocytes | — | cardiotoxic | doi:10.31083/j.rcm.2019.03.545 |
| BCL2 | endothelial cells | — | hypoxia induced autophagy | doi:10.1002/jcb.29207 | ||||||
| ZEB1 | cardiomyocytes | — | aggravates hypoxia induced apoptosis | doi:10.1515/hsz-2018-0195 | ||||||
| ACSL1 | hepatocytes | — | increases hepatic triglyceride and cholesterol levels | doi:10.3390/ijms20184420 | ||||||
| SIRT1 | kidney | — | promotes injury induced fibrosis | doi:10.1038/s41419-018-0527-8 | ||||||
| SIRT1 | kidney | — | promotes injury induced fibrosis | doi:10.1016/j.bbrc.2017.12.048 | ||||||
| DLL1 | osteosarcoma | — | oncomiR (chemoresitance) | doi:10.1038/srep44218 | ||||||
| AGTR1 | osteosarcoma | — | oncomiR (chemoresitance) | doi:10.1186/s12885-016-3002-x | ||||||
| CD117 | osteosarcoma | — | oncomiR (chemoresitance) | doi:10.18632/oncotarget.8546 | ||||||
| PD-L1 | ovarian carcinoma | — | oncomiR (chemoresitance) | doi:10.4149/neo_2019_190202N106 | ||||||
| BCL2 | ovarian carcinoma | — | oncomiR (promotes cell proliferation) | doi:10.2147/OTT.S142446 | ||||||
| O | miR-15b-5p | 4.41 ± 0.46 | 4.88 ± 1.04 | –1.38 | 0.69 | AKT3 | arteries | — | inhibits ateriogenesis, angiogenesis | doi:10.1161/ATVBAHA.116.308905 |
| PAQR3 | gastric cancer | — | oncomiR (promotes metastasis) | doi:10.3892/or.2017.5673 | ||||||
| AXIN2 | hepatocellular carcinoma | — | oncomiR (promotes cell proliferation, invasion) | doi:10.3892/ol.2019.11056 | ||||||
| RECK | prostate cancer | — | oncomiR (tumorigenesis) | doi:10.3892/ol.2019.11056 | ||||||
| SEMA3A | podocytes | — | repressing apoptosis and inflammation in high glucose injury | doi:10.1002/jcp.28691 | ||||||
| PDK4 | osteosarcoma | — | oncomiR (promotes cell proliferation) | doi:10.1016/j.bbrc.2018.08.035 | ||||||
| BMPR1A | cardiomyocytes | — | promotes doxorubicin induced injury | doi:10.1007/s12012-018-9495-6 | ||||||
| HPSE2 | breast cancer | — | oncomiR (promotes cell proliferation, metastasis) | doi:10.3389/fonc.2020.00108 | ||||||
| O | miR-17-5p | 5.57 ± 0.15 | 5.34 ± 0.62 | 1.10 | 0.57 | BAMBI | nasopharyngeal carcinoma | — | oncomiR (promotes angiogenesis) | doi:10.7150/jca.30757 |
| ETV1 | breast cancer | — | tumor suppressor (cell proliferation) | doi:10.1186/s12885-017-3674-x | ||||||
| RBL2, E2F4 | pancreatic cancer | — | oncomiR (promotes cell proliferation) | doi:10.1016/j.canlet.2017.09.044 | ||||||
| ANKH | fibroblasts | — | increased ostegenesis | doi:10.1016/j.omtn.2019.10.003 | ||||||
| BRCC2 | osteosarcoma | — | oncomiR (promotes cell growth) | doi:10.3892/or.2016.4542 | ||||||
| NTN4 | breast cancer | — | oncomiR (promotes metastasis, invasion) | PMID: 31933983 | ||||||
| SKSI1 | osteosarcoma | — | oncomiR (promotes epithelial–mesenchymal transition) | doi:10.1002/jcb.27832 | ||||||
| SMAD7 | fibroblasts | — | promotes liver fibrosis | doi:10.1111/jcmm.14432 | ||||||
| TGFB2 | cervical cancer | — | oncomiR (promotes cell proliferation) | doi:10.26355/eurrev_201804_14712 | ||||||
| E2F1 | granulosa cells | — | promotes cell proliferation | doi:10.1111/rda.13551 | ||||||
| SMAD5 | myoblasts | miR-106b-5p | promotes osteogenic differentiation | doi:10.1016/j.yexcr.2016.07.010 | ||||||
| MFN2 | satellite cells | — | modulates mitochondrial function | PMID: 31198013 | ||||||
| P21 | nasopharyngeal carcinoma | — | oncomiR (promotes cell proliferation) | doi:10.1002/cam4.863 | ||||||
| HOXB13 | prostate cancer | — | oncomiR (promotes tumor progression) | doi:10.1186/s12935-019-0994-8 | ||||||
| P21 | astrocytes | — | inhibits apoptosis during hypoxia | doi:10.1186/s12935-019-0994-8 | ||||||
| CMYC | hepatocellular carcinoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1007/s13277-015-4355-5 | ||||||
| VEGFA | endothelial cells | — | mitigates endometriosis | doi:10.1007/s12038-020-00049-y | ||||||
| TMOD1 | gastric cancer | — | oncomiR (tumorigenesis) | doi:10.26355/eurrev_201907_18430 | ||||||
| SMAD7 | osteoblasts | — | promotes osteogenic differentiation | doi:10.1038/emm.2014.43 | ||||||
| RUNX3 | gastric cancer | — | oncomiR (promotes cell proliferation, metastasis) | doi:10.1016/j.biopha.2020.110246 | ||||||
| SOCS6 | gastric cancer | — | oncomiR (promotes cell proliferation) | doi:10.1016/j.febslet.2014.04.036 | ||||||
| PTEN | thyroid cancer | — | oncomiR (promotes cell proliferation) | doi:10.4149/neo_2019_190110N29 | ||||||
| PTEN, GAINT7 | hepatocellular carcinoma | — | oncomiR (tumorigenesis) | doi:10.1242/jcs.122895 | ||||||
| PIK3R1 | squamous cell carcinoma | — | tumor suppressor (cell autophagy) | doi:10.1007/s10620-012-2400-4 | ||||||
| P21/PTEN | smooth muscle cells | — | promotes hypoxia induced proliferation | doi:10.1186/s12931-018-0902-0 | ||||||
| SMAD7 | nasal epithelial cells | — | aggravates inflammatory response | doi:10.1186/s12860-018-0152-5 | ||||||
| TGFB2 | gastric cancer | — | oncomiR (promotes cell proliferation) | doi:10.18632/oncotarget.8946 | ||||||
| HBP1 | breast cancer | — | oncomiR (promotes metastasis, invasion) | doi:10.1007/s10549-010-0954-4 | ||||||
| SMAD7 | hepatic stellate cells | — | activates stellate cells | doi:10.1038/labinvest.2015.58 | ||||||
| O, CP | miR-21-3p | 5.28 ± 1.27 | 3.89 ± 1.01 | 2.26 | 0.26 | SPRY1 | fibroblasts | — | promotes wound healing | doi:10.18632/aging.103610 |
| MAT2B | brain | — | attenuate ischemia-reperfusion injury | doi:10.3325/cmj.2019.60.439 | ||||||
| PTEN | vascular smooth muscle cells | — | promote migration and proliferation (pro-atherogenic) | doi:10.7150/thno.37357 | ||||||
| VEGFA | granulosa cells | — | inhibits autophagy | doi:10.1530/REP-19-0285 | ||||||
| TGS4 | retinal pigment epithelial cells | — | modulates apoptosis and inflammation | doi:10.1111/1440-1681.13142 | ||||||
| AKT, CDK2 | kidney | — | regulates metabolic alterations in acute kidney injury | doi:10.1155/2019/2821731 | ||||||
| P53 | multiple cancers | — | oncomiR (inhibit apoptosis) | doi:10.1016/j.abb.2019.05.026 | ||||||
| PTEN | liver cancer | — | oncomiR (inhibit apoptosis) | doi:10.2147/CMAR.S183328 | ||||||
| HDAC1 | epithelium | — | inhibits influenca virus replication | doi:10.3389/fcimb.2018.00175 | ||||||
| SORBS2 | cardiomyocytes | — | promoted myocardial dysfunction in sepsis | doi:10.1016/j.yjmcc.2016.03.014 | ||||||
| HDAC1 | cardiomyocytes | — | cardioprotection (suppression of myocardial hypertrophy) | doi:10.1093/cvr/cvu254 | ||||||
| O | miR-663a | 4.43 ± 0.38 | 1.49 ± 1.06 | 7.68 | 0.02 | MYL9 | osteosarcoma | — | oncomiR (tumorigenesis) | doi:10.1177/0960327120937330 |
| TGFB1 | liver | — | reduces hepatic stellar cell activation | doi:10.1155/2020/3156267 | ||||||
| ZBTB7A | osteosarcoma | — | oncomiR (inhibits apoptosis) | doi:10.1016/j.canlet.2019.01.046 | ||||||
| TGFB1 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation, invasion) | doi:10.1186/s12885-018-5016-z | ||||||
| NFIX | spermatogonial stem cells | — | promote proliferation and inhibit apoptosis | doi:10.1016/j.omtn.2018.05.015 | ||||||
| EMP3 | gallbladder cancer | — | oncomiR (tumor progression) | doi:10.1016/j.canlet.2018.05.022 | ||||||
| O | miR-664a-3p | 4.67 ± 1.55 | 4.77 ± 0.54 | –1.07 | 0.44 | FHL1 | lung | — | Progression of chronic obstructive pulmonary disease | doi:10.2147/COPD.S224763 |
| FOXP3 | gastric cancer | — | oncomiR (tumorigenesis) | doi:10.1111/cpr.12567 |
Table A3.
MiRNAs that are known for their tumor suppressor and oncogenic potential (TS/O). Selected miRNAs are also involved in cardioprotection (CP). Targets are given for each miRNA, with no claim to completeness. Pubmed IDs (PMIDs) are given as references when no doi numbers are available.
| MiRNA Function | MiRNA Name | CB-MSC-EV [dCT ± SD] | AT-MSC-EV [dCT ± SD] | Fold Difference | p-Value | Confirmed Targets GeneGLOBE ID | Cell/Tissue/Cancer Type | MiRNA Cluster | Biological Effect | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| TS/O | miR-31-3p | 4.93 ± 0.56 | 5.08 ± 0.49 | –1.11 | 0.64 | RASA1 | colorectal carcinoma | — | oncomiR (promotes cell proliferation, tumor progression) | doi:10.1074/jbc.M112.367763 |
| SEMA4C | cervical cancer | — | tumor suppressor (chemoresistance) | doi:10.1038/s41598-019-54177-z | ||||||
| TIAM1 | colorectal carcinoma | miR-21 | oncomiR (promotes epithelial–mesenchymal transition, invasion) | doi:10.1074/jbc.M110.160069 | ||||||
| TS/O | miR-199b-5p | 4.39 ± 1.5 | 3.73 ± 0.43 | 1.58 | 0.53 | HER2 | osteosarcoma | — | oncomiR (promotes tumor progression) | PMID: 30610808 |
| STON2 | papillary thyroid carcinoma | — | tumor suppressor (metastasis, suppresses epithelial–mesenchymal transition) | doi:10.1002/iub.1889 | ||||||
| DYRK1A, NOTCH1, JAG1 | — | — | promotes pathological myocardial remodeling | doi:10.1016/j.ncrna.2016.12.002 | ||||||
| KLK10 | cervical cancer | — | oncomiR (promotes cell proliferation, metastasis) | doi:10.1016/j.bbrc.2018.05.165 | ||||||
| mTOR | endometrial endometrial adenocarcinoma | miR-100-5p, miR-199a-3p | tumor suppressor (cell autophagy) | PMID: 31966798 | ||||||
| GSK3B | monocytes | — | inhibition of NFkB signaling, anti-inflammatory | doi:10.1007/s10753-018-0799-2 | ||||||
| JAG1 | ligamentum flavum cells | — | inhibition of osteogenic differentiation | doi:10.1111/jcmm.13047 | ||||||
| CAV1 | non-small cell lung carcinoma | — | oncomiR (promotes cell proliferation) | doi:10.1038/s41419-019-1740-9 | ||||||
| ALK1 | breast cancer | — | tumor suppressor (angiogenesis) | doi:10.3389/fgene.2019.01397 | ||||||
| DDR1 | breast cancer | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.3892/ol.2018.9255 | ||||||
| BICC1 | oral cancer | miR-101-3p (not detected) |
tumor suppressor (cell autophagy) | doi:10.1016/j.mcp.2020.101567 | ||||||
| MLK | pancreatic beta cells | — | increases cell proliferation | doi:10.2174/2211536605666160607082214 | ||||||
| JAG1, DDR1 | colorectal carcinoma | — | tumor suppressor (cell proliferation, invasion) | doi:10.1002/path.5238 | ||||||
| PODXL, DDR1 | acute myeloid leukemia | — | tumor suppressor (cell proliferation) | doi:10.1002/ajh.23129 | ||||||
| HES1 | medulloblastoma | — | tumor suppressor (impairs cancer stem cell function) | doi:10.1371/journal.pone.0004998 | ||||||
| ITGA3 | squamous cell carcinoma | miR-199a-3p/5p | tumor suppressor (cell proliferation) | doi:10.1111/cas.13298 | ||||||
| TS/O | miR-221-3p | 1.25 ± 0.67 | 0.65 ± 0.25 | 1.51 | 0.11 | AXIN2 | - | miR-15b-5p | oncomiR (promotes cell proliferation, invasion) | doi:10.3892/ol.2019.11056 |
| THBS2 | squamous cell carcinoma | — | oncomiR (promotes angiogenesis) | doi:10.1007/s10456-019-09665-1 | ||||||
| SDF1 | cartilage | — | prevent cartilage degradation in osteoarthritis | doi:10.1007/s00109-017-1516-6 | ||||||
| VASH1 | squamous cell carcinoma | — | oncomiR (promotes metastasis) | doi:10.1038/s41388-018-0511-x | ||||||
| THBS1 | trophoblast | — | promotes invasion and proliferation | doi:10.1016/j.biopha.2018.10.009 | ||||||
| JAK3 | macrophages | — | regulates M1 to M2 transition | doi:10.3389/fimmu.2019.03087 | ||||||
| ARF4 | epithelial ovarian cancer | — | tumor suppressor (cell proliferation, metastasis) | doi:10.1016/j.bbrc.2017.01.002 | ||||||
| THBS2 | squamous cell carcinoma | — | oncomiR (promotes metastasis) | doi:10.1038/s41419-017-0077-5 | ||||||
| MMP22 | macrophages | — | prevent low-density lipoprotein-induced oxidative stress | doi:10.1002/jcb.27917 | ||||||
| EIF5A2 | medulloblastoma | — | tumor suppressor (cell proliferation, enhances apoptosis) | doi:10.1080/09168451.2018.1553604 | ||||||
| PTEN | gastric cancer | — | oncomiR (promotes tumor progression) | doi:10.3727/096504016 × 14756282819385 | ||||||
| TIMP3 | retina | — | promotes microvascular dysfunction | doi:10.1007/s00424-020-02432-y | ||||||
| PARP1 | breast cancer | — | tumor suppressor (tumor progression) | doi:10.18632/oncotarget.21561 | ||||||
| RB1 | pancreatic cancer | — | oncomiR (chemoresistance) | doi:10.1007/s13277-016-5445-8 | ||||||
| JNK1, TGFBR1, ETS-1 | cardiac fibroblasts | — | cardioprotective (inhibits fibroblast activation) | doi:10.1161/HYPERTENSIONAHA.117.10094 | ||||||
| TS/O, CP | miR-25-3p | 4.52 ± 0.53 | 4.8 ± 0.13 | –1.21 | 0.33 | BTG2 | breast cancer | — | tumor suppressor (cell proliferation) | doi:10.1186/s12943-017-0754-0 |
| PTEN | retinoblastoma | — | oncomiR (promotes tumor progression) | doi:10.1016/j.biopha.2019.109111 | ||||||
| FBXW7, DKK3 | glioma cells | — | oncomiR (promotes cell proliferation, metastasis) | doi:10.3892/etm.2019.7583 | ||||||
| BTG2 | breast cancer | — | oncomiR (promotes cell proliferation, metastasis) | doi:10.1155/2019/7024675 | ||||||
| SEMA4C | cervical cancer | — | tumor suppressor (suppresses EMT) | doi:10.1111/cas.13104 | ||||||
| ADAM10 | endothelial cells | — | inhibit NFkB Signaling and reduces inflammation | doi:10.3389/fimmu.2019.02205 | ||||||
| EZH2 | cardiomyocytes | — | cardioprotective (inhibit cardiomyocyte apoptosis during injury) | doi:10.1080/0886022X.2020.1745236 | ||||||
| TS/O | miR-23b-3p | 0.73 ± 0.69 | 1.02 ± 0.26 | –1.23 | 0.307825 | SIRT1 | lens epithelial cells | — | reduces apoptosis in oxidative stress | doi:10.1002/jcb.29270 |
| TGFBR3 | atrial fibroblasts | miR-27b-3p | promote atrial fibrosis in atrial fibrillation | doi:10.1111/jcmm.14211 | ||||||
| CB1R | gastric cancer | miR-130a-5p (not detected) |
tumor suppressor (cell proliferation) | doi:10.2147/OTT.S181706 | ||||||
| ANXA2 | pancreatic ductal adenocarcinoma | — | tumor suppressor (cell proliferation) | doi:10.1159/000494468 | ||||||
| ETS1 | hepatocytes | — | downregulate Apo(a) expression | doi:10.1002/cbin.10896 | ||||||
| PGC1A | osteosarcoma | — | oncomiR (promotes cell proliferation) | doi:10.1038/s41419-019-1614-1 | ||||||
| EBF3 | squamous cell carcinoma | — | oncomiR (promotes cell proliferation, metastasis) | doi:10.1093/abbs/gmy049 | ||||||
| ZEB1 | hepatocellular carcinoma | — | tumor suppressor (suppresses epithelial–mesenchymal transition) | doi:10.1016/j.gene.2018.05.061 | ||||||
| CMET | cervical cancer | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1038/s41598-020-60143-x | ||||||
| ATG12, HMGB2 | gastric cancer | — | tumor suppressor (chemosensitivity) | doi:10.1038/cddis.2015.123 | ||||||
| HS6ST2 | chondrocytes | — | enhances matrix degradation in osteoarthritis | doi:10.1038/s41419-018-0729-0 | ||||||
| TGIF1 | keratinocytes | — | regulation of keratinocyte differentiation | doi:10.1111/exd.13119 | ||||||
| PTEN | renal cancer | — | oncomiR (promotes cell proliferation) | doi:10.1371/journal.pone.0050203 | ||||||
| TS/O, CP | miR-27b-3p | 2.97 ± 0.79 | 2.77 ± 0.5 | 1.15 | 0.84 | TGFBR3 | atrial fibroblasts | miR-27b-3p | promote atrial fibrosis in atrial fibrillation | doi:10.1111/jcmm.14211 |
| HOXA10 | colorectal carcinoma | — | oncomiR (promotes cell invasion, metastasis) | doi:10.1042/BSR20191087 | ||||||
| CBLB, GRB2 | breast cancer | — | tumor suppressor (cell proliferation, chemoresistance) | doi:10.1038/s41419-017-0211-4 | ||||||
| WNT3A | atrial fibroblasts | — | cardioprotection (reduces atrial fibrosis during atrial fibrillation) | doi:10.1155/2019/5703764 | ||||||
| MARCH7 | endometrial carcinoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1093/abbs/gmz030 | ||||||
| SMAD7 | endothelial cells | — | suppresses endothelial cell proliferation and migration in Kawasaki disease | doi:10.1159/000492354 | ||||||
| HIPK2 | chondrocytes | — | inhibits apoptosis in rheumatoid arthritis | doi:10.1080/21691401.2019.1607362 | ||||||
| PPARG | thyroid cancer | — | oncomiR (chemoresistance) | doi:10.1111/bcpt.13076 | ||||||
| FZD7 | lung cancer | — | tumor suppressor (tumor progression) | PMID: 29028088 | ||||||
| SP7 | maxillary sinus membrane stem cells | — | suppress osteogenic differentiation | doi:10.1097/ID.0000000000000637 | ||||||
| LIMK1 | colorectal carcinoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | PMID: 31966797 | ||||||
| GSPT1 | gastric cancer | — | tumor suppressor (tumor progression) | doi:10.1016/j.biopha.2019.109417 | ||||||
| YAP1 | glioma cells | — | tumor suppressor (tumorigenesis) | doi:10.1139/bcb-2019-0300 | ||||||
| ROR1 | gastric cancer | — | tumor suppressor (cell proliferation) | doi:10.1186/s13046-015-0253-3 | ||||||
| PPARG | oocytes | — | maturation | doi:10.1016/j.bbrc.2016.09.046 | ||||||
| GSPT1 | non-small cell lung carcinoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.2147/OTT.S196865 | ||||||
| NRF2 | squamous cell carcinoma | — | tumor suppressor (tumor progression) | doi:10.1007/s13577-020-00329-7 | ||||||
| TRAF3 | chondrocytes | — | inhibits IL1B-induced injury | doi:10.1016/j.intimp.2019.106052 | ||||||
| NR5A2, CREB1 | breast cancer | — | tumor suppressor (chemosensitivity) | doi:10.1038/cddis.2016.361 | ||||||
| TS/O, CP | miR-24-3p | 0.43 ± 0.62 | 1.48 ± 0.56 | –2.05 | 0.091 | KEAP1 | cardiomyocytes | — | cardioprotection (in ischemia-reperfusion injury) | doi:10.1155/2018/7042105 |
| FGF11 | T-cells | — | oncomiR (immune evasion) | doi:10.1002/path.4781 | ||||||
| SOX7 | lung cancer | — | oncomiR (promotes metastasis, invasion) | doi:10.1002/jcb.26553 | ||||||
| SOCS6 | prostate cancer | — | oncomiR (promotes metastasis, invasion, proliferation) | PMID: 31938287 | ||||||
| p27KIP1 | papillary thyroid carcinoma | — | oncomiR (promotes metastasis, invasion, proliferation) | doi:10.26355/eurrev_201907_18327 | ||||||
| BIM | breast cancer | — | oncomiR (chemoresistance) | doi:10.1002/jcb.28568 | ||||||
| RIPK1 | cardiomyocytes | — | cardioprotection (in ischemia-reperfusion injury) | doi:10.1159/000495161 | ||||||
| DEDD | bladder cancer | — | oncomiR (promotes tumor progression) | doi:10.3892/or.2016.5326 | ||||||
| PRKCH | Lacrimal adenoid cystic carcinoma | — | tumor suppressor (tumor progression) | doi:10.1371/journal.pone.0158433 | ||||||
| MTT1 | hepatocellular carcinoma | — | oncomiR (promotes cell proliferation) | doi:10.1002/cbf.3213 | ||||||
| SMAD5 | peridontal stem cells | — | inhibit osteogenic differentiation | doi:10.1002/jcp.27499 | ||||||
| JAB1/CSN5 | nasopharyngeal carcinoma | — | tumor suppressor (radiosensitivity) | doi:10.1038/onc.2016.147 | ||||||
| ATG4A | small cell lung cancer | — | tumor suppressor (chemosensitivity) | doi:10.18632/oncotarget.2787 | ||||||
| IGFBP5 | intervertebrate discs | — | induces disc degeneration | doi:10.1016/j.lfs.2020.117288 | ||||||
| NOTCH1, DLL1 | endothelial cells | — | inhibit angiogenesis after myocardial infarction | doi:10.3390/ijms21051733 | ||||||
| LAMB3 | pancreatic ductal adenocarcinoma | — | tumor suppressor (tumor progression) | doi:10.3389/fonc.2019.01499 | ||||||
| CHD5 | squamous cell carcinoma | — | oncomiR (promotes cell proliferation, chemoresistance) | doi:10.2217/fon-2016-0179 | ||||||
| FGF11 | fibroblasts | — | activation of fibrosis and proliferation in renal fibrosis | doi:10.1002/jcp.29329 | ||||||
| TS/O | miR-23a-3p | -0.14 ± 0.54 | 0.06 ± 0.95 | –1.14 | 0.99 | PNRC2 | renal cell carcinoma | — | oncomiR (promotes tumor progression) | doi:10.1016/j.biopha.2018.11.065 |
| KLF3 | melanoma | — | oncomiR (promotes tumor progression) | doi:10.1186/s12935-019-0927-6 | ||||||
| FGF2 | squamous cell carcinoma | — | tumor suppressor (cell proliferation) | doi:10.1016/j.prp.2018.12.021 | ||||||
| CHD17 | hepatocellular carcinoma | — | oncomiR (promotes cell proliferation) | doi:10.1007/s13105-020-00726-4 | ||||||
| SMAD3 | chondrocytes | — | promotes osteoarthritis | doi:10.1016/j.bbrc.2016.06.071 | ||||||
| PTEN | gliomal cells | — | oncomiR (promotes cell proliferation) | doi:10.1002/ar.24410 | ||||||
| TS/O, CP | miR-130a-3p | 4.86 ± 0.83 | 5.01 ± 0.74 | –1.11 | 0.75 | PDE4D | cardiomyocytes | — | cardioprotection (improves cardiac cell proliferation after myocardial infarction) | doi:10.1002/jcp.26327 |
| SMAD4 | esophageal cancer | — | oncomiR (promotes epithelial–mesenchymal transition) | doi:10.1002/cam4.1981 | ||||||
| RAB5B | breast cancer | — | tumor suppressor ( invasion, metastasis) | doi:10.1016/j.bbrc.2018.05.018 | ||||||
| SOX4 | non-small cell lung carcinoma | — | tumor suppressor (chemosensitivity) | doi:10.1080/15384047.2017.1385679 | ||||||
| SMAD4 | hepatoma cells | — | tumor suppressor (invasion, metastasis) | doi:10.1186/s13046-016-0296-0 | ||||||
| SNON | kidney | — | inhibition of renal fibrosis | doi:10.1016/j.yexmp.2019.104358 | ||||||
| TGFBR1/2 | hepatic stellate cells | — | decreases hepatic fibrosis | doi:10.1038/cddis.2017.10 | ||||||
| BACH2 | nasopharyngeal carcinoma | — | tumor suppressor (cell autophagy) | doi:10.1042/BSR20160576 | ||||||
| TS/O | miR-15a-5p | 5.52 ± 2.85 | 4.42 ± 1.07 | 2.14 | 0.73 | VEGFA | chondrocytes | — | aggravates osteoarthritis | doi:10.5582/bst.2016.01187 |
| VEGFA | peritoneal mesothelial cells | — | suppresses inflammation and fibrosis | doi:10.1002/jcp.27660 | ||||||
| WNT3A | endometrial carcinoma | — | tumor suppressor (cell growth) | PMID: 29164582 | ||||||
| CXCL10 | chronic myeloid leukemia | — | tumor suppressor (cell autophagy), metastasis | PMID: 28979704 | ||||||
| MYCN | neuroblastoma cells | miR-15b-5p, miR-16-5p | tumor suppressor (tumor progression) | doi:10.1002/1878-0261.12588 | ||||||
| PTHrP | chondrocytes | — | promotes osteoarthritis | doi:10.1155/2019/3904923 | ||||||
| FASN | arteries | — | alleviates atherosclerosis and vascular inflammation | doi:10.1042/BSR20181852 | ||||||
| PHLPP2 | gastric cancer | — | oncomiR (chemoresistance) | doi:10.4149/neo_2020_190904N861 | ||||||
| TP53INP1 | cervical cancer | — | oncomiR (anti-apoptotic) | doi:10.26355/eurrev_201910_19129 | ||||||
| BDNF | hepatocellular carcinoma | — | tumor suppressor (cell proliferation) | doi:10.1007/s13277-015-4427-6 | ||||||
| TGFB3, VEGF | retinal endothelial cells | — | promote endothelial cell tight junction formation | doi:10.1016/j.visres.2017.07.007 | ||||||
| VEGFA | endometrial mesenchymal stem cells | — | promote endometriosis | PMID: 27608888 | ||||||
| HOXA3 | thyroid cancer | — | tumor suppressor (tumor progression) | doi:10.1089/hum.2018.109 | ||||||
| TS/O, CP | miR-181a-5p | 5.5 ± 0.68 | 6.54 ± 0.7 | –1.09 | 0.9 | PBX1 | ligaments | — | promotes osteogenesis | doi:10.7150/thno.44309 |
| CBLB | esophageal cancer | — | tumor suppressor (chemosensitivity) | doi:10.2147/CMAR.S251264 | ||||||
| E2F7 | non-small cell lung carcinoma | — | oncomiR (tumor progression) | doi:10.2147/CMAR.S240964 | ||||||
| ESM1 | retina | — | anti-angiogenesis | doi:10.1002/jcp.29733 | ||||||
| SIRT1 | cardiomyocytes | — | promotes apoptosis in hypoxic injury | doi:10.1080/09168451.2020.1750943 | ||||||
| KLF17 | prostate cancer | — | oncomiR (promotes epithelial–mesenchymal transition) | PMID: 32195032 | ||||||
| PDGFRA | endothelial cells | — | anti-angiogenesis | doi:10.1002/cbf.3472 | ||||||
| AKT3 | gastric adenocarcinoma | — | tumor suppressor (cell proliferation, apoptosis) | doi:10.1098/rsob.190095 | ||||||
| p53 | cardiomyocytes | — | cardioprotection (reduces high glucose induced apoptosis) | doi:10.1538/expanim.19-0058 | ||||||
| ATG7 | hepatocellular carcinoma | — | oncomiR (inhibits autophagy) | doi:10.1002/jcb.29064 | ||||||
| TS/O | miR-106a-5p | 5.19 ± 0.76 | 5.42 ± 1.07 | –1.17 | 0.97 | HK2 | squamous cell carcinoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1007/s11010-020-03840-5 |
| STAT3 | endothelial cells | — | allelviates atherosclerosis and vascular inflammation | doi:10.3892/mmr.2020.11147 | ||||||
| RBM24 | prostate cancer | — | oncomiR (tumor progression) | doi:10.2147/OTT.S246274 | ||||||
| TGFBR2 | colorectal carcinoma | — | oncomiR (chemoresistance) | PMID: 31949649 | ||||||
| ARHGAP24 | ovarian carcinoma | — | oncomiR (cell proliferation, invasion) | doi:10.1016/j.lfs.2020.117296 | ||||||
| TGFBR2 | palate | — | promotes cleft palate formation | doi:10.1016/j.yexcr.2019 | ||||||
| TS/O | miR-125a-5p | 2.3 ± 0.54 | 2.64 ± 0.22 | –1.27 | 0.27 | FUT4 | osteosarcoma | — | tumor suppressor (tumor progression) | doi:10.3389/fgene.2020.00672 |
| LIN28B | ovarian carcinoma | — | tumor suppressor (cell proliferation, metastasis) | doi:10.3892/mmr.2020.11223 | ||||||
| MACC1 | hepatocellular carcinoma | miR-34a | tumor suppressor (cell proliferation, metastasis) | doi:10.4149/neo_2020_191019N1062 | ||||||
| FNDC3B | colorectal carcinoma | miR-217 | oncomiR (cell proliferation, invasion) | doi:10.2147/OTT.S226520 | ||||||
| HK2 | lung | — | inhibits glycolysis and improved pulmonary arterial hypertension | doi:10.18632/aging.103163 | ||||||
| TRAF6 | macrophages | — | promotes M2 polarization | doi:10.1007/s10753-020-01231-y | ||||||
| GALNT7 | cervical cancer | — | tumor suppressor (cell proliferation, invasion) | doi:10.1186/s12935-020-01209-8 | ||||||
| VEGFA | trophoblast | — | suppresses migration and proliferation | doi:10.1016/j.bbrc.2020.02.137 | ||||||
| GAB2 | breast cancer | — | tumor suppressor (cell proliferation, invasion) | doi:10.3934/mbe.2019347 | ||||||
| SIRT7 | non-small cell lung carcinoma | — | tumor suppressor (radioresistance) | doi:10.3233/CBM-190381 | ||||||
| TAZ | ovarian carcinoma | — | tumor suppressor (inhibits epithelial–mesenchymal transition) | doi:10.3233/CBM-190381 | ||||||
| TS/O, CP | miR-125b-5p | −1.36 ± 0.51 | −1.21 ± 0.36 | –1.11 | 0.58 | p53, BAK1 | cardiomyocytes | — | cardioprotection (inhibits apoptosis in ischemia-reperfusion injury) | doi:10.7150/thno.28021 |
| p53, BNIP3 | cardiomyocytes | — | cardioprotection (inhibits apoptosis in ischemia-reperfusion injury) | doi:10.1161/CIRCRESAHA.118.312758 | ||||||
| SMAD7 | cardiomyocytes | — | cardiotoxic (increase hypoxia induced injury signaling) | doi:10.3892/ijmm.2018.3496 | ||||||
| BAK1, KLF13 | cardiomyocytes | — | cardioprotection (inhibits apoptosis in ischemia-reperfusion injury) | doi:10.1016/j.yjmcc.2017.11.003 | ||||||
| EIF5A2 | melanoma | — | tumor suppressor (cell proliferation, metastasis) | doi:10.1186/s13046-020-01599-7 | ||||||
| BTG2 | lung adenocarcinoma | — | oncomiR (cell proliferation, migration and promotes epithelial–mesenchymal transition) | doi:10.26355/eurrev_202004_20841 | ||||||
| PAK3 | prenatal follicles | — | inhibits steroidogenesis | doi:10.1016/j.metabol.2020.154241 | ||||||
| BACE1 | neurons | — | attenuate neurotoxicity | doi:10.1016/j.jns.2020.116793 | ||||||
| TRIB2 | squamous cell carcinoma | — | tumor suppressor (tumor progression) | doi:10.1042/BSR20193172 | ||||||
| PDK1 | cervical cancer | — | tumor suppressor (tumorigenesis) | doi:10.1155/2020/4351671 | ||||||
| NLRC5 | cardiomyocytes | — | cardioprotection (inhibits apoptosis in ischemia-reperfusion injury) | doi:10.3892/etm.2019.8309 | ||||||
| STAT3 | embryonic stem cells | — | tumor suppressor (tumorigenesis) | doi:10.7150/jca.33696 | ||||||
| TRAF6 | skeletal muscle | — | relieves skeletal muscle atrophy | doi:10.21037/atm.2019.08.39 | ||||||
| HK2 | bladder cancer | — | tumor suppressor (tumor progression) | doi:10.1007/s13577-019-00285-x | ||||||
| AKT3 | keratinocytes | miR-181b-5p (not tested) |
inhibit proliferation | doi:10.1016/j.ejphar.2019.172659 | ||||||
| LIMK1 | brain | — | neuroprotection | doi:10.2174/1567202616666190906145936 | ||||||
| TXNRD1 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation, invasion, metastasis) | doi:10.1186/s12935-019-0919-6 | ||||||
| TRAF6 | chondrocytes | — | anti-inflammatory in the setting of osteoarthritis | doi:10.1038/s41598-019-42601-3 | ||||||
| TS/O, CP | miR-19a-3p | 3.07 ± 0.62 | 3.29 ± 1.04 | –1.16 | 0.99 | PTEN | brain | — | alleviates ischemia-reperfusion injury-induced apoptosis | doi:10.1016/j.neuroscience.2020.04.020 |
| IGFBP3 | brain | — | alleviates ischemia-reperfusion injury | doi:10.1186/s40659-020-00280-9 | ||||||
| FAS | rectal cancer | — | tumor suppressor (induces apoptosis) | doi:10.1177/1533033820917978 | ||||||
| PIK3IP1 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation) | doi:10.7150/jca.37748 | ||||||
| FOXF2 | colorectal carcinoma | — | tumor suppressor (inhibits epithelial–mesenchymal transition) | doi:10.3748/wjg.v26.i6.627 | ||||||
| IGFBP3 | ovarian carcinoma | — | oncomiR (tumor progression) | doi:10.1002/mc.23113 | ||||||
| SOCS3 | synovial cells | — | promote cell proliferation | 10.1002/jcb.28442 | ||||||
| PTEN | osteosarcoma | — | oncomiR (chemoresistance) | doi:10.3892/ol.2018.9592 | ||||||
| PFN1 | hepatocellular carcinoma | — | oncomiR (tumor progression) | doi:10.1016/j.prp.2018.12.012 | ||||||
| PTEN | hepatocellular carcinoma | — | oncomiR (chemoresistance, metastasis) | doi:10.1016/j.biopha.2018.06.097 | ||||||
| PITX1 | gastric cancer | — | oncomiR (tumor progression) | doi:10.1159/000489590 | ||||||
| TSC1 | osteoblasts | — | mediates dexamethasone resistance | doi:10.18632/oncotarget.23326 | ||||||
| SMAD2/4 | prostate cancer | — | tumor suppressor (invasion, metastasis) | doi:10.3892/or.2017.6096 | ||||||
| TGFBR2 | cardiac fibroblasts | miR-19b-3p | cardioprotection: anti-fibrotic | doi:10.1038/srep24747 | ||||||
| TS/O, CP | miR-19b-3p | 3.29 ± 0.35 | 3.07 ± 0.81 | 1.16 | 0.47 | NRP1 | gastric cancer | — | tumor suppressor (tumor progression) | doi:10.1186/s12935-020-01257-0 |
| CCDC6 | cholangiosarcoma | — | oncomiR (promotes proliferation, epithelial–mesenchymal transition) | doi:10.1016/j.abb.2020.108367 | ||||||
| HIF1A | endothelial cells | — | anti-angiogenic after hypoxia | doi:10.1096/fj.201902434R | ||||||
| BACE1 | brain | miR-16-5p | prevent amyloid beta induced apoptosis | doi:10.1097/WNR.0000000000001379 | ||||||
| TNFAIP3 | endothelial cells | — | pro-inflammatory in the setting of meningitis | doi:10.3390/pathogens8040268 | ||||||
| HOXA9 | non-small cell lung carcinoma | — | oncomiR (promotes proliferation, migration, invasion) | doi:10.2147/OTT.S216320 | ||||||
| PTEN | pancreatic cancer | — | oncomiR (cell proliferation) | doi:10.21037/atm.2019.04.61 | ||||||
| GRK6 | chondrocytes | — | reduces inflammation and matrix degradation | doi:10.1007/s11010-019-03563-2 | ||||||
| PTEN | muscle cells | — | osteogenic differentiation | doi:10.1002/cbin.11133 | ||||||
| TS/O CP | miR-214-3p | 2.19 ± 1.04 | 1.96 ± 0.59 | 1.17 | 0.99 | PTEN | cardiomyocytes | — | cardioprotection: inhibiting autophagy in sepsis | doi:10.1155/2020/1409038 |
| ATM | lung | — | reduce radiation induced pulmonary injury | doi:10.1089/ars.2019.7965 | ||||||
| CENPM | hepatocellular carcinoma | — | tumor supressor (tumor progression) | doi:10.1093/jb/mvaa073 | ||||||
| PLAGL2 | colorectal carcinoma | — | tumor suppressor (cell proliferation) | doi:10.18632/aging.103233 | ||||||
| LIVIN | colorectal carcinoma | — | tumor supressor (tumor progression) | doi:10.1080/21655979.2020 | ||||||
| IL17 | myocardium | — | cardioprotective (anti-fibrotic) | doi:10.3389/fcell.2020.00243 | ||||||
| WNT23 | vascular smooth muscle cells | — | inhibits cell proliferation | doi:10.26355/eurrev_202003_20696 | ||||||
| ABCB1, XIAP | retinoblastoma | — | oncomiR (chemoresistance) | doi:10.2147/OTT.S235862 | ||||||
| PSMD10 | papillary thyroid carcinoma | — | tumor suppressor (tumor progression) | doi:10.1002/jcp.29557 | ||||||
| TWIST1 | endometrial carcinoma | — | tumor suppressor (inhibit epithelial–mesenchymal transition) | doi:10.2147/OTT.S181037 | ||||||
| LHX6 | ovarian carcinoma | — | oncomiR (tumorigenesis) | doi:10.3390/cancers11121917 | ||||||
| ST6GAL1 | breast cancer | — | oncomiR (cell proliferation, inhibit apoptosis) | doi:10.1007/s10616-019-00352-z | ||||||
| FOXP3 | breast cancer | — | oncomiR (cell proliferation) | doi:10.26355/eurrev_201910_19156 | ||||||
| HDGF | pancreatic cancer | — | tumor suppressor (chemosensitivity) | doi:10.2147/OTT.S222703 | ||||||
| BIRC5 | breast cancer | — | tumor suppressor (cell proliferation) | doi:10.26355/eurrev_201909_18856 | ||||||
| NLRC5 | myocardium | — | cardioprotection: anti-fibrotic | doi:10.1042/CS20190203 | ||||||
| CTNNB1 | preadipocytes | — | promote differentiation | doi:10.3390/ijms20081816 | ||||||
| TS/O | miR-222-3p | 2.34 ± 1.60 | 1.61 ± 0.57 | 1.66 | 0.50 | PUMA | non-small cell lung carcinoma | — | oncomiR (cell proliferation, inhibit apoptosis) | doi:10.1177/1533033820922558 |
| PDCD10 | ovarian carcinoma | — | tumor suppressor (inhibit epithelial–mesenchymal transition) | doi:10.7150/thno.43198 | ||||||
| GILZ | airway epithelial cells | — | ameliorates glucocorticoid induced inhibition of cell repair | doi:10.1080/10799893.2020.1742739 | ||||||
| IGF1 | bone marrow-derived MSCs | — | promote osteogenic differentiation | doi:10.1016/j.diabres.2020.108121 | ||||||
| TMP2 | renal clear cell carcinoma | — | oncomiR (tumor progression) | doi:10.3233/CBM-190264 | ||||||
| IRF2, INPP4B | acute myeloid leukemia | — | tumor suppressor (cell proliferation) | doi:10.1016/j.mcp.2020.101513 | ||||||
| CDKN1B | squamous cell carcinoma | — | oncomiR (tumorigenesis) | doi:10.1111/jop.12986 | ||||||
| PPP2R2A | large B-cell lymphoma | — | oncomiR (cell proliferation, inhibit apoptosis) | doi:10.1177/1533033819892256 | ||||||
| GAS5, PTEN | colorectal carcinoma | — | oncomiR (promotes cell proliferation, migration, invasion) | doi:10.1016/j.omtn.2019.06.009 | ||||||
| PDE3A | endothelial cells | miR-27a-3p | promote vascular integrity | doi:10.1007/s12035-018-1446-5 | ||||||
| TIMP3 | osteosarcoma | — | oncomiR (promote metastasis and invasion) | doi:10.2147/OTT.S175745 | ||||||
| PTEN | papillary thyroid carcinoma | — | oncomiR (inhibit apoptosis) | doi:10.18632/oncotarget.23336 | ||||||
| TS/O, CP | miR-26a-5p | 3.75 ± 0.54 | 4.06 ± 0.72 | –1.24 | 0.53 | RANBP9 | brain | — | inhibit injury induced apoptosis | doi:10.1016/j.acthis.2020.151571 |
| TLR4 | kidney | — | protect against diabetic nephropathy | doi:10.1074/jbc.RA120.012522 | ||||||
| HMGA2 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation, promote apoptosis) | doi:10.2147/CMAR.S237752 | ||||||
| CTGF | macrophages | — | modulates TLR signaling upon activation | doi:10.1042/BSR20192598 | ||||||
| CREB1 | renal cell carcinoma | miR-27a-3p, miR-221-3p | tumor suppressor (cell proliferation, promote apoptosis) | doi:10.1038/s41598-020-63403-y | ||||||
| DYRK1A | brain | — | inhibit development Alzheimer’s disease | doi:10.2174/1567202617666200414142637 | ||||||
| WNT5A | gastric cancer | — | tumor suppressor (cell proliferation) | doi:10.2147/OTT.S241199 | ||||||
| ADAM17 | cardiomyocytes | — | cardioprotection (inhibit apoptosis) | doi:10.1007/s10863-020-09829-5 | ||||||
| COL10A1 | gastric cancer | — | tumor suppressor (cell proliferation, migration, and invasion) | doi:10.26355/eurrev_202002_20170 | ||||||
| HOXA5 | osteosarcoma | — | oncomiR (promotes cell proliferation, migration) | doi:10.2147/OTT.S232100 | ||||||
| PTEN | myocardium | — | cardioprotection (inhibits apoptosis in ischemia-reperfusion injury) | doi:10.1590/1414-431 × 20199106 | ||||||
| PTGS2 | joints | — | alleviate osteoarthritis | doi:10.1016/j.intimp.2019.105946 | ||||||
| AURKA | hepatocellular carcinoma | — | tumor suppressor (chemosensitivity) | doi:10.1177/1533033819851833 | ||||||
| WNT5A | papillary thyroid carcinoma | — | tumor suppressor (cell proliferation, migration, and invasion) | doi:10.2147/OTT.S205994 | ||||||
| PTEN | myocardium | — | cardioprotection (inhibits apoptosis in ischemia-reperfusion injury) | doi:10.26355/eurrev_201908_18661 | ||||||
| PTEN | synovial cells | — | promote cell proliferation and inhibit apoptosis | doi:10.1042/BSR20182192 | ||||||
| TS/O, CP | miR-27a-3p | 1.87 ± 0.98 | 1.80 ± 0.88 | 1.05 | 0.96 | SLIT2 | endothelial cells | — | promotes apoptosis, autophagy during inflammation | doi:10.1016/j.jss.2020.05.102 |
| SP7 | preosteoblasts | — | promotes differentiation | doi:10.3892/mmr.2020.11246 | ||||||
| TAB3 | kidney | — | promotes apoptosis during kidney injury | doi:10.1080/09168451.2020.1792760 | ||||||
| PDL1 | macrophages | — | oncomiR (promotes immune evasion of breast cancer) | doi:10.1111/jcmm.15367 | ||||||
| BNIP3 | pancreatic cancer | — | oncomiR (inhibits apoptosis) | doi:10.3892/ijmm.2020.4632 | ||||||
| SMURF2 | lung | — | anti-fibrotic after bleomycin exposure | PMID: 32538751 | ||||||
| TGFBR1 | cardiomyocytes | — | cardioprotection (inhibits apoptosis in ischemia-reperfusion injury) | doi:10.1155/2020/2016259 | ||||||
| FBXW7 | cervical cancer | — | oncomiR (tumor progression) | doi:10.2147/CMAR.S234897 | ||||||
| ICOS | lung adenocarcinoma | — | tumor suppressor (promotes antitumor immunity) | doi:10.1111/1759-7714.13411 | ||||||
| NOVA | gastric cancer | — | oncomiR (promotes epithelial–mesenchymal transition) | doi:10.3892/mmr.2020.10949 | ||||||
| BNIP3 | cardiomyocytes | — | cardioprotection (inhibits apoptosis in ischemia-reperfusion injury) | doi:10.1016/j.omtn.2019.11.017 | ||||||
| TS/O, CP | miR-29a-3p | 2.50 ± 0.87 | 1.64 ± 0.66 | 1.82 | 0.19 | E2F1 | ovarian carcinoma | — | oncomiR (promotes epithelial–mesenchymal transition) | doi:10.18632/aging.103388 |
| PTEN | aorta | — | promotes development of aortic aneurysms | doi:10.1002/jcp.29746 | ||||||
| DRP1 | myocardium | — | cardioprotection (prevent myocardial hypertrophy) | doi:10.2174/0929866527666200416144459 | ||||||
| COL4A2 | hepatocellular carcinoma | — | tumor suppressor (cell proliferation, migration, and invasion) | doi:10.1039/c9mt00266a | ||||||
| COL5A1 | breast cancer | — | tumor suppressor (cell proliferation, migration) | doi:10.1016/j.lfs.2019.117179 | ||||||
| TNFR1 | endothelial cells | — | reduces TNF-alpha injury response | doi:10.1016/j.omtn.2019.10.014 | ||||||
| TS/O, CP | miR-30b-5p | 5.04 ± 1.10 | 4.56 ± 0.62 | 1.40 | 0.62 | KIF18A | prostate cancer | — | oncomiR (radioresistance) | doi:10.1089/cbr.2019.3538. |
| MYBL2 | medulloblastoma | — | tumor suppressor (cell proliferation, promotes apoptosis) | doi:10.1136/jim-2020-001354 | ||||||
| CAMK2D | dermal papilla cells | — | inhibits proliferation | doi:10.1186/s12864-020-06799-1 | ||||||
| ASPP2 | breast cancer | — | oncomiR (cell proliferation, migration, and invasion) | doi:10.1155/2020/7907269 | ||||||
| PTAFR | myocardium | — | cardioprotection: anti-fibrotic | doi:10.26355/eurrev_202004_20869 | ||||||
| PPARGC1A | Huh-7 cells | — | regulate lipid metabolism | doi:10.1186/s12944-020-01261-3 | ||||||
| CTNNB1 | cardiomyocytes | — | cardiotoxic (increased apoptosis during myocardial injury) | doi:10.23736/S00264806.20.06565-9 | ||||||
| AVEN | cardiomyocytes | — | cardiotoxic (increased apoptosis during myocardial injury) | doi:10.1186/s11658-019-0187-4 | ||||||
| TS/O, CP | miR-31-5p | 1.42 ± 0.46 | 2.24 ± 0.74 | –1.76 | 0.15 | YAP | colorectal carcinoma | — | tumor suppressor (cell proliferation, metastasis, chemosensitivity) | doi:10.1016/j.yexcr.2020.112176 |
| FLOT1 | renal clear cell carcinoma | — | tumor suppressor (cell proliferation, promote apoptosis) | doi:10.2147/OTT.S254634 | ||||||
| HOXA7 | trophoblast | — | inhibit proliferation | doi:10.1111/jog.14344 | ||||||
| PEX5 | hepatocellular carcinoma | — | oncomiR (radioresistance) | doi:10.7150/thno.42371 | ||||||
| TNS1 | colon adenocarcinoma | — | oncomiR (tumor progression) | doi:10.18632/aging.103096 | ||||||
| PKCG | cardiomyocytes | — | cardioprotective, inhibit cardiomyocyte hypertrophy | doi:10.26355/eurrev_202002_20351 | ||||||
| PAN3 | cardiomyocytes | — | cardioprotective: attenuates doxorubicin induced cardiotoxicity | doi:10.1016/j.yjmcc.2020.02.009 | ||||||
| ETBR, VEGFA | endothelial cells | — | anti-angiogenic | doi:10.1016/j.lfs.2020.117306 | ||||||
| MEGEA3 | hepatocellular carcinoma | — | oncomiR (chemoresistance, cell proliferation) | doi:10.1016/j.omtn.2019.10.035 | ||||||
| LATS2 | colorectal carcinoma | — | oncomiR (chemoresistance) | doi:10.3390/cancers11101576 | ||||||
| MLH1 | renal cell carcinoma | — | oncomiR (chemoresistance) | doi:10.1002/ijc.32543 | ||||||
| VEGFA | gliomal cells | — | tumor suppressor (anti-angiogenic) | doi:10.1002/ijc.32483 | ||||||
| TS/O | miR-365a-3p | 3.99 ± 0.84 | 4.34 ± 0.67 | –1.27 | 0.43 | ABCC4 | gastric cancer | — | tumor suppressor (tumor progression) | doi:10.2147/OTT.S245557 |
| ADAM10 | colorectal carcinoma | — | tumor suppressor (cell proliferation, migration) | doi:10.7150/jca.42731 | ||||||
| CREL | pancreatic cancer | — | tumor suppressor (tumor progression) | doi:10.1016/j.canlet.2019.03.025 | ||||||
| TET1 | hepatocellular carcinoma | — | tumor suppressor (tumor progression, invasion) | doi:10.4149/neo_2018_171119N752 | ||||||
| USP33 | lung cancer | — | oncomiR (tumorigenesis) | doi:10.1186/s12935-018-0563-6 | ||||||
| TS/O | miR-93-5p | 5.04 ± 0.59 | 5.43 ± 0.62 | –1.31 | 0.39 | MAP3K2 | hepatocellular carcinoma | — | oncomiR (tumor progression) | doi:10.1038/s41388-020-01401-0 |
| RGMB | squamous cell carcinoma | — | oncomiR (migration and invasion) | doi:10.7150/jca.43854 | ||||||
| FOXA1 | colorectal carcinoma | — | oncomiR (radioresistance) | doi:10.1186/s13046-019-1507-2 | ||||||
| AHNAK | gastric cancer | — | oncomiR (promotes epithelial–mesenchymal transition) | doi:10.1186/s12935-019-1092-7 | ||||||
| PD-L1 | colorectal carcinoma | — | tumor suppressor (tumor progression) | doi:10.1002/cbin.11323 | ||||||
| FOXK2 | cervical cancer | — | oncomiR (tumor progression) | doi:10.1007/s43032-020-00140-7 | ||||||
| CASC2 | chondrocytes | — | inhibits apoptosis in osteoarthritis | doi:10.1186/s12891-019-3025-y | ||||||
| MMP2 | gliomal cells | — | tumor suppressor (cell proliferation, migration) | doi:10.26355/eurrev_201911_19446 | ||||||
| TS/O | let-7a-5p | 2.46 ± 0.96 | 2.79 ± 0.46 | –1.26 | 0.40 | SMAD2 | chondrocytes | — | promotes hypertrophic differentiation | doi:10.1152/ajpcell.00039.2020 |
| SAMD2 | lens epithelial cells | — | inhibits proliferation, migration and invasion | PMID: 32345785 | ||||||
| DUSP7 | breast cancer | — | tumor suppressor (chemoresistance) | doi:10.2147/CMAR.S238513 | ||||||
| BCLXL | lung cancer | — | tumor suppressor (cell autophagy) | doi:10.1016/j.omto.2019.08.010 | ||||||
| BCL2L1 | lung cancer | — | tumor suppressor (induce apoptosis) | doi:10.3389/fonc.2019.00808 | ||||||
| EGFR | breast cancer | — | oncomiR (chemoresistance) | doi:10.1002/iub.2075 | ||||||
| HMGA2 | kidney | — | promotes diabetic nephropathy | doi:10.3892/mmr.2019.10057 |
Author Contributions
T.Z.N.-S.: conceptualization, investigation, formal analysis, writing—original draft, writing—review and editing, supervision. S.N.: investigation, formal analysis, writing—original draft, writing—review & editing. A.G.D.: investigation, formal analysis, writing—original draft. V.E.: investigation, formal analysis. H.M.: investigation. K.K.: investigation. C.M.B.: investigation, formal Analysis. P.W.: investigation. J.B.: investigation. R.B.: investigation. M.S.: writing—review and editing. V.F.: writing—review and editing, funding acquisition. M.Y.E.: conceptualization, writing—review and editing, supervision, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by institutional funds. Nazari-Shafti is a scholar in the BIH Charité Clinician Scientist Program funded by the Charité—Universitätsmedizin Berlin and the Berlin Institute of Health. Neuber was funded by the German Centre for Cardiovascular Research (FKZ 81Z0100302).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Han Y., Li X., Zhang Y., Han Y., Chang F., Ding J. Mesenchymal stem cells for regenerative medicine. Cells. 2019;8:886. doi: 10.3390/cells8080886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karantalis V., Hare J.M. Use of mesenchymal stem cells for therapy of cardiac disease. Circ. Res. 2015;116:1413–1430. doi: 10.1161/CIRCRESAHA.116.303614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spees J.L., Lee R.H., Gregory C.A. Mechanisms of mesenchymal stem/stromal cell function. Curr. Stem Cell Res. 2016;7:1–3. doi: 10.1186/s13287-016-0363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Spoel T.I., Jansen of Lorkeers S.J., Agostoni P., van Belle E., Gyöngyösi M., Sluijter J.P., Cramer M.J., Doevendans P.A., Chamuleau S.A. Human relevance of pre-clinical studies in stem cell therapy: Systematic review and meta-analysis of large animal models of ischaemic heart disease. Cardiovasc. Res. 2011;91:649–658. doi: 10.1093/cvr/cvr113. [DOI] [PubMed] [Google Scholar]
- 5.Jansen of Lorkeers S.J., Eding J.E., Vesterinen H.M., van der Spoel T.I., Sena E.S., Duckers H.J., Doevendans P.A., Macleod M.R., Chamuleau S.A. Similar effect of autologous and allogeneic cell therapy for ischemic heart disease: Systematic review and meta-analysis of large animal studies. Circ. Res. 2015;116:80–86. doi: 10.1161/CIRCRESAHA.116.304872. [DOI] [PubMed] [Google Scholar]
- 6.Cambria E., Pasqualini F.S., Wolint P., Günter J., Steiger J., Bopp A., Hoerstrup S.P., Emmert M.Y. Translational cardiac stem cell therapy: Advancing from first-generation to next-generation cell types. NPJ Regen. Med. 2017;2:1–10. doi: 10.1038/s41536-017-0024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendt M., Rezvani K., Shpall E. Mesenchymal stem cell-derived exosomes for clinical use. Bone Marrow Transplant. 2019;54:789–792. doi: 10.1038/s41409-019-0616-z. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann M., Wollert K.C., Meyer G.P., Menke A., Arseniev L., Hertenstein B., Ganser A., Knapp W.H., Drexler H. Monitoring of bone marrow cell homing into the infarcted human myocardium. Circulation. 2005;111:2198–2202. doi: 10.1161/01.CIR.0000163546.27639.AA. [DOI] [PubMed] [Google Scholar]
- 9.Zeng L., Hu Q., Wang X., Mansoor A., Lee J., Feygin J., Zhang G., Suntharalingam P., Boozer S., Mhashilkar A., et al. Bioenergetic and functional consequences of bone marrow-derived multipotent progenitor cell transplantation in hearts with postinfarction left ventricular remodeling. Circulation. 2007;115:1866–1875. doi: 10.1161/CIRCULATIONAHA.106.659730. [DOI] [PubMed] [Google Scholar]
- 10.Doyle L.M., Wang M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8:727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moghaddam A.S., Afshari J.T., Esmaeili S.A., Saburi E., Joneidi Z., Momtazi-Borojeni A.A. Cardioprotective microRNAs: Lessons from stem cell-derived exosomal microRNAs to treat cardiovascular disease. Atherosclerosis. 2019;285:1–9. doi: 10.1016/j.atherosclerosis.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Felekkis K., Touvana E., Stefanou C.H., Deltas C. microRNAs: A newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236. [PMC free article] [PubMed] [Google Scholar]
- 13.Latysheva N.S., Babu M.M. Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res. 2016;44:4487–4503. doi: 10.1093/nar/gkw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nazari-Shafti T.Z., Exarchos V., Biefer H.R., Cesarovic N., Meyborg H., Falk V., Emmert M.Y. MicroRNA Mediated Cardioprotection–Is There a Path to Clinical Translation? Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serocki M., Bartoszewska S., Janaszak-Jasiecka A., Ochocka R.J., Collawn J.F., Bartoszewski R. miRNAs regulate the HIF switch during hypoxia: A novel therapeutic target. Angiogenesis. 2018;21:183–202. doi: 10.1007/s10456-018-9600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng X.M., Nikolic-Paterson D.J., Lan H.Y. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 17.Gowen A., Shahjin F., Chand S., Odegaard K.E., Yelamanchili S.V. Mesenchymal Stem Cell-Derived Extracellular Vesicles: Challenges in Clinical Applications. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandergriff A., Huang K.E., Shen D., Hu S., Hensley M.T., Caranasos T.G., Qian L., Cheng K. Targeting regenerative exosomes to myocardial infarction using cardiac homing peptide. Theranostics. 2018;8:1869. doi: 10.7150/thno.20524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Chen Y., Zhao Z., Meng Q., Yu Y., Sun J., Yang Z., Chen Y., Li J., Ma T., et al. Engineered Exosomes with Ischemic Myocardium-Targeting Peptide for Targeted Therapy in Myocardial Infarction. J. Am. Heart Assoc. 2018;7:e008737. doi: 10.1161/JAHA.118.008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marzi M.J., Ghini F., Cerruti B., De Pretis S., Bonetti P., Giacomelli C., Gorski M.M., Kress T., Pelizzola M., Muller H., et al. Degradation dynamics of microRNAs revealed by a novel pulse-chase approach. Genome Res. 2016;26:554–565. doi: 10.1101/gr.198788.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuk P.A., Zhu M.I., Mizuno H., Huang J., Futrell J.W., Katz A.J., Benhaim P., Lorenz H.P., Hedrick M.H. Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 22.Bieback K., Netsch P. Isolation, Culture, and Characterization of Human Umbilical Cord Blood-Derived Mesenchymal Stromal Cells in Methods in Molecular Biology. Vol. 1416. Humana Press Inc.; Totowa, NJ, USA: 2016. pp. 245–258. [DOI] [PubMed] [Google Scholar]
- 23.Beez C.M., Haag M., Klein O., Van Linthout S., Sittinger M., Seifert M. Extracellular vesicles from regenerative human cardiac cells act as potent immune modulators by priming monocytes. J. Nanobiotechnol. 2019;17:1–8. doi: 10.1186/s12951-019-0504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;30:3–22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 25.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., et al. Reassessment of exosome composition. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buschmann D., Kirchner B., Hermann S., Märte M., Wurmser C., Brandes F., Kotschote S., Bonin M., Steinlein O.K., Pfaffl M.W., et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles. 2018;7:1481321. doi: 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell A.E., Sneider A., Witwer K.W., Bergese P., Bhattacharyya S.N., Cocks A., Cocucci E., Erdbrügger U., Falcon-Perez J.M., Freeman D.W., et al. Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: An ISEV position paper arising from the ISEV membranes and EVs workshop. J. Extracell. Vesicles. 2019;8:1684862. doi: 10.1080/20013078.2019.1684862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stranska R., Gysbrechts L., Wouters J., Vermeersch P., Bloch K., Dierickx D., Andrei G., Snoeck R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018;16:1–9. doi: 10.1186/s12967-017-1374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga Z., Fehér B., Kitka D., Wacha A., Bóta A., Berényi S., Pipich V., Fraikin J.L. Size Measurement of Extracellular Vesicles and Synthetic Liposomes: The Impact of the Hydration Shell and the Protein Corona. Colloids Surf. B Biointerfaces. 2020;19:111053. doi: 10.1016/j.colsurfb.2020.111053. [DOI] [PubMed] [Google Scholar]
- 31.Wiklander O.P., Bostancioglu R.B., Welsh J.A., Zickler A.M., Murke F., Corso G., Felldin U., Hagey D.W., Evertsson B., Liang X.M., et al. Systematic methodological evaluation of a multiplex bead-based flow cytometry assay for detection of extracellular vesicle surface signatures. Front. Immunol. 2018;9:1326. doi: 10.3389/fimmu.2018.01326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ragni E., Orfei C.P., De Luca P., Lugano G., Viganò M., Colombini A., Valli F., Zacchetti D., Bollati V., De Girolamo L. Interaction with hyaluronan matrix and miRNA cargo as contributors for in vitro potential of mesenchymal stem cell-derived extracellular vesicles in a model of human osteoarthritic synoviocytes. Stem Cell Res. Therapy. 2019;10:109. doi: 10.1186/s13287-019-1215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragni E., Perucca O.C., De Luca P., Colombini A., Viganò M., de Girolamo L. Secreted Factors and EV-miRNAs Orchestrate the Healing Capacity of Adipose Mesenchymal Stem Cells for the Treatment of Knee Osteoarthritis. Int. J. Mol. Sci. 2020;21:1582. doi: 10.3390/ijms21051582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reis M., Mavin E., Nicholson L., Green K., Dickinson A.M., Wang X.N. Mesenchymal stromal cell-derived extracellular vesicles attenuate dendritic cell maturation and function. Front. Immunol. 2018:2538. doi: 10.3389/fimmu.2018.02538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel V., Noureddine L. MicroRNAs and fibrosis. Curr. Opin. Nephrol. Hypertens. 2012;21:410. doi: 10.1097/MNH.0b013e328354e559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khalil H., Kanisicak O., Prasad V., Correll R.N., Fu X., Schips T., Vagnozzi R.J., Liu R., Huynh T., Lee S.J., et al. Fibroblast-specific TGF-β–Smad2/3 signaling underlies cardiac fibrosis. J. Clin. Investig. 2017;127:3770–3783. doi: 10.1172/JCI94753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Du J., Niu X., Fu N., Wang R., Zhang Y., Zhao S., Sun D., Nan Y. MiR-130a-3p attenuates activation and induces apoptosis of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis by directly targeting TGFBR1 and TGFBR2. Cell Death Dis. 2017;8:e2792. doi: 10.1038/cddis.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Li Y., Wang R., Qin S., Liu J., Su F., Yang Y., Zhao F., Wang Z., Wu Q. MiR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J. Exp. Clin. Cancer Res. 2016;35:1. doi: 10.1186/s13046-016-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H., Yang K., Ren T., Huang Y., Tang X., Guo W. miR-16-5p inhibits chordoma cell proliferation, invasion and metastasis by targeting Smad3. Cell Death Dis. 2018;9:1–3. doi: 10.1038/s41419-018-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian X., Fei Q., Du M., Zhu H., Ye J., Qian L., Lu Z., Zhang W., Wang Y., Peng F., et al. miR-130a-3p regulated TGF-β1-induced epithelial-mesenchymal transition depends on SMAD4 in EC-1 cells. Cancer Med. 2019;8:1197–1208. doi: 10.1002/cam4.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svoronos A.A., Engelman D.M., Slack F.J. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z., Xiao Z., Guo H., Fang X., Liang J., Zhu J., Yang J., Li H., Pan R., Yuan S., et al. Novel role of the clustered miR-23b-3p and miR-27b-3p in enhanced expression of fibrosis-associated genes by targeting TGFBR3 in atrial fibroblasts. J. Cell Mol. Med. 2019;23:3246–3256. doi: 10.1111/jcmm.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hua H., Kong Q., Zhang H., Wang J., Luo T., Jiang Y. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019;12:71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Easton J.B., Houghton P.J. mTOR and cancer therapy. Oncogene. 2006;25:6436–6446. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]
- 45.Grundmann S., Hans F.P., Kinniry S., Heinke J., Helbing T., Bluhm F., Sluijter J.P., Hoefer I., Pasterkamp G., Bode C., et al. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation. 2011;123:999–1009. doi: 10.1161/CIRCULATIONAHA.110.000323. [DOI] [PubMed] [Google Scholar]
- 46.Yang A., Liu F., Guan B., Luo Z., Lin J., Fang W., Liu L., Zuo W. p53 induces miR-199a-3p to suppress mechanistic target of rapamycin activation in cisplatin-induced acute kidney injury. J. Cell. Biochem. 2019;120:17625–17634. doi: 10.1002/jcb.29030. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z., Wang J., Pan Z., Zhang Y. miR-143-3p regulates cell proliferation and apoptosis by targeting IGF1R and IGFBP5 and regulating the Ras/p38 MAPK signaling pathway in rheumatoid arthritis. Exp. Ther. Med. 2018;15:3781–3790. doi: 10.3892/etm.2018.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi B., Ma M., Zheng Y., Pan Y., Lin X. mTOR and Beclin1: Two key autophagy-related molecules and their roles in myocardial ischemia/reperfusion injury. J. Cell. Physiol. 2019;234:12562–12568. doi: 10.1002/jcp.28125. [DOI] [PubMed] [Google Scholar]
- 49.Chalhoub N., Baker S.J. PTEN and the PI3-kinase pathway in cancer. Annual Review of Pathology. Mech. Dis. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang P., Zhao Q., Gong K., Long Y., Zhang J., Li Y., Guo X. Downregulation of miR-103a-3p Contributes to Endothelial Progenitor Cell Dysfunction in Deep Vein Thrombosis Through PTEN Targeting. Ann. Vasc. Surg. 2020;64:339–346. doi: 10.1016/j.avsg.2019.10.048. [DOI] [PubMed] [Google Scholar]
- 51.Gao D.C., Hou B., Zhou D., Liu Q.X., Zhang K., Lu X., Zhang J., Zheng H., Dai J.G. Tumor-derived exosomal miR-103a-2-5p facilitates esophageal squamous cell carcinoma cell proliferation and migration. Eur. Rev. Med. Pharmacol. Sci. 2020;24:6097–6110. doi: 10.26355/eurrev_202006_21505. [DOI] [PubMed] [Google Scholar]
- 52.Shi J., Zhang Y., Jin N., Li Y., Wu S., Xu L. MicroRNA-221-3p plays an oncogenic role in gastric carcinoma by inhibiting PTEN expression. Oncol. Res. 2017;25:523–536. doi: 10.3727/096504016X14756282819385. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Li X., Guo S., Min L., Guo Q., Zhang S. miR-92a-3p promotes the proliferation, migration and invasion of esophageal squamous cell cancer by regulating PTEN. Int. J. Mol. Med. 2019;44:973–981. doi: 10.3892/ijmm.2019.4258. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Liu Y., Hu Q., Ao J., Li H., Li M. Role of miR-92a-3p/PTEN Axis in Regulation of Pancreatic Cancer Cell Proliferation and Metastasis. J. Cent. South Univ. Med. Sci. 2020;45:280–289. doi: 10.11817/j.issn.1672-7347.2020.180459. [DOI] [PubMed] [Google Scholar]
- 55.Wu L., Chen Y., Chen Y., Yang W., Han Y., Lu L., Yang K., Cao J. Effect of HIF-1α/miR-10b-5p/PTEN on Hypoxia-Induced Cardiomyocyte Apoptosis. J. Am. Heart Assoc. 2019;8:e011948. doi: 10.1161/JAHA.119.011948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lv X., Li J., Hu Y., Wang S., Yang C., Li C., Zhong G. Overexpression of miR-27b-3p targeting Wnt3a regulates the signaling pathway of Wnt/β-catenin and attenuates atrial fibrosis in rats with atrial fibrillation. Oxid. Med. Cell. Longev. 2019;2019:5703764. doi: 10.1155/2019/5703764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.