Abstract
The contribution of genetic ancestry on chronic obstructive pulmonary disease (COPD) predisposition remains unclear. To explore this relationship, we analyzed the associations between 754,159 single nucleotide polymorphisms (SNPs) and risk of COPD (n = 214 cases, 193 healthy controls) in Talca, Chile, considering the genetic ancestry and established risk factors. The proportion of Mapuche ancestry (PMA) was based on a panel of 45 Mapuche reference individuals. Five PRDM15 SNPs and two PPP1R12B SNPs were associate with COPD risk (p = 0.05 to 5 × 10−4) in those individuals with lower PMA. Based on linkage disequilibrium and sliding window analyses, an adjacent PRDM15 SNPs were associated with COPD risk in the lower PMA group (p = 10−3 to 3.77 × 10−8). Our study is the first to report an association between PPP1R12B and COPD risk, as well as effect modification between ethnicity and PRDM15 SNPs in determining COPD risk. Our results are biologically plausible given that PPP1R12B and PRDM15 are involved in immune dysfunction and autoimmunity, providing mechanistic evidence for COPD pathogenesis and highlighting the importance to conduct more genome wide association studies (GWAS) in admixed populations with Amerindian descent.
Keywords: chronic obstructive pulmonary disease (COPD), ancestry, autoimmunity, genome wide association studies (GWAS), personalized medicine, Hispanic paradox, immune dysfunction
1. Introduction
Chronic obstructive pulmonary disease (COPD) is a common disorder characterized by a persistent and progressive airflow limitation that is associated with an enhanced chronic inflammatory response in the airways and lungs to noxious particles or gases, particularly those attributed to cigarette and biomass smoke [1,2]. COPD is the fourth leading cause of death worldwide and it is projected to become the third by 2020 [1,3]. According to the World Health Organization, approximately 3 million people in the world die as a consequence of COPD every year, and the disease burden is increasing in Latin America [4]. Latin American countries experience a COPD prevalence of 13.4%, with in-hospital mortality rate ranging from 6.7% to 29.5% [5]. The prevalence of COPD increases steeply with age, with the highest prevalence among those people over 60 years [6]. Considering the high rates of tobacco use and biomass burning in several Latin American countries [7,8], COPD may become an even greater health problem in Latin America than previously hypothesized. In Chile, for example, respiratory diseases are the third most common cause of death, with COPD accounting for 22% of the deaths and being the second cause of decease [9].
In general, Latin Americans have been considered homogenous in most genome-wide association studies (GWAS), ignoring the degree of admixture variation between Latin American countries according to the major ancestry population component. However, both Latin Americans and Hispanics/Latinos in other parts of the world are characterized by broad ethnic diversity [10], with different admixture proportions of Amerindian, European, and African ancestry between and within countries, attributed to distinct populations of origin and historic characteristics of European settlement and slave-trading [11,12,13,14]. In Chile, for example, Amerindian ancestry proportions are higher in the north and the south, whereas European ancestry proportions are highest in central regions [15,16]. Conversely, the contribution of African ancestry proportions decreases from north to south [15,16].
Most genetic association studies of COPD have been in Caucasian, African, and Asian populations, often with controversial results depending on the population studied [17], suggesting that COPD susceptibility may depend on diverse ethnic gene–environment interactions. To date, over 97 independent genetic loci have been associated with the lung parameters defining COPD—forced expiratory volume in one second (FEV1), forced vital capacity (FVC), or the FEV1/FVC ratios—and with COPD risk [18,19]. Notably, single nucleotide polymorphisms (SNPs) near or in HHIP, FAM13A, ADAM19, and CHRNA3/5 have been frequently associated with COPD risk [18,20,21,22,23]. Regarding lung function or COPD-related phenotypes, only four GWAS have been focused on Hispanic/Latino populations, reporting novel loci (in or near the genes KLHL7/NUPL2, DLG2, PDZD2, and PRDM15), as well as others previously identified in non-Hispanic populations [24,25,26,27]. Therefore, further genetic studies on non-Caucasian populations are required in order to understand whether ancestry plays a key role in the pathogenesis of COPD [28,29].
The present study, conducted in El Maule, a rural region of Chile, explored the risk of COPD associated with established risk factors and identified susceptibility variants both independently and in conjunction with genetic ancestry.
2. Materials and Methods
2.1. Study Population
The El Maule Region, located about 250 km south of Santiago, is one of the most rural counties in Chile. El Maule experiences some of the highest levels of air pollution attributed to biomass burning in Chile. Moreover, according to the Chilean Ministry of Health, El Maule shows high mortality rates attributed to COPD, pneumonia, and respiratory system malignancies (Supplementary Figure S1). For this study, we recruited 214 Maulean patients with COPD at the respiratory service of the Hospital Regional de Talca, where they attended to undergo diagnostic tests after suspected COPD or for COPD monitoring visits. Control subjects (n = 193) with no record of any specific illness were enrolled in parallel at the same Hospital through a volunteer recruitment program. The Ethics Committees of the Maulean Health Service and Universidad Autónoma de Chile approved the study and all subjects provided written informed consent (approval code: 063-15).
The Global Initiative for Chronic Obstructive Lung Disease (GOLD) criteria [30] were used for the diagnostic evaluation of subjects. Standard clinical information and medical history was collected. Pulmonary function—including measurements of forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC), and carbon monoxide diffusing capacity of the lung (DLCO)—was assessed in all subjects using standard procedures [31] and equipment (Masterlab; Jaeger, Würzburg, Germany). Oxygen saturation was also measured by pulse-oximetry (Ohmeda TuffSat, Soma Technology, Bloomfield, Connecticut, USA). Body mass index (BMI) was calculated using the current weight and height of each study participant. Dyspnea was determined using the modified Medical Research Council scale (mMRC) and exercise capacity was determined with the distance walked in 6 minutes test (6MWT). Health-related quality of life and symptom burden in patients with COPD was quantified using the COPD assessment test (CAT). A Body mass, airflow Obstruction, Dyspnea, and Exercise (BODE) score was assigned to each COPD patient. Cigarette smoking history was measured by pack-years and cumulative exposure to biomass smoke (hour-years) was calculated as previously described [32].
Finally, patients with asthma–COPD overlap syndrome were identified and excluded if they had a history of asthma, rhinitis, or any extra-pulmonary disease affecting lung function, and with positive bronchodilator test, FEV1 increasing by ≥12% and 200 mL. Participants with a COPD exacerbation or hospitalization record during the previous two months were also excluded, to ensure that COPD patients were stable.
2.2. Genotyping, Imputation and Ancestry Estimation
When visiting the respiratory service, 5 mL of blood was collected from each participant and stored in plastic vacutainer tubes containing ethylenediaminetetraacetic acid (EDTA). DNA from peripheral blood cells was extracted using the GeneJET Genomic DNA purification kit #K0722 (Thermo Fisher Scientific, Waltham, Massachusetts, USA), following the manufacturer’s instructions. Samples were stored at −80 °C until genotyped using the Illumina Global Screening Array [27].
We investigated 89 SNPs (Supplementary Table S1), that were previously associated with COPD risk at genome-wide significance in large European GWAS [21,33,34,35]. Since 46 of these SNPs were not included in the array, we imputed them using the IMPUTE2 software version 2.3.2 with version 3 of the 1000 Genomes Project data as the reference set, as previously described [36,37]. We used the ADMIXTURE software, version 1.3, for supervised estimation of individual European, African, Mapuche, and Aymara ancestry components [38]. The 1000 Genome project contributed individual surrogates of European and African ancestry: 99 Utah residents with northern and western European ancestry (CEU), 107 individuals from Iberian populations in Spain (IBS), and 108 Yorubans in Ibadan, Nigeria (YRI) [37]. Lorenzo et al. had identified nine Mapuche and nine Aymara in the Chilean study population of the Consortium for the Analysis of the Diversity and Evolution of Latin America [39]. The Mapuche reference individuals were complemented by four Huilliche from Reich et al. and 32 Huilliche–Pehuenche from Lindo et al., and the Aymara reference individuals were complemented by 22 Aymara and 40 Quechua from Reich et al. [40,41]. Finally, we assessed the distribution of all the SNPs previously evaluated in a part of this Chilean cohort [27] considering the genetic ancestry.
To explore linkage disequilibrium (LD) pattern in the PRDM15 gene, we accessed the 1000 Genomes genetic data of 629 individuals from different ethnic backgrounds via FTP download (available at ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20100804/ALL.2of4intersection.20100804.genotypes.vcf.gz), including 261 Europeans (EUR), 177 East Asians (EAS), 169 Africans (AFR), and 22 admixed Americans (AMR).
2.3. Statistical Analyses
SNPs that met the quality control (QC) criteria of a minor allele frequency (MAF) > 0.01, missing call rate < 0.2, and/or Hardy–Weinberg equilibrium (HWE) p > 0.001 were considered for inclusion in the association analyses. Of the 89 SNPs, 80 passed QC and were included in the analysis (Supplementary Table S1). When we analyzed all the SNPs previously evaluated (n = 455,564) [27], allele frequencies were compared between COPD patients and controls by χ2 test, and odds ratios (OR) with 95% confidence intervals (95% CI) were calculated using PLINK software (v1.07) [42]. Sliding window analyses, assessing the frequency of composite genotypes of a fixed number of contiguous SNPs, were tested for association analyses by χ2 test with PLINK software [42]. Haplotype analysis was performed with Haploview version 4.1 software [43], using the CI method to perform LD assessment and define haplotype blocks. Associations between identified COPD susceptibility genetic markers and potential interactions with genetic ancestry were tested using logistic regression, considering COPD diagnosis as response variable. An additive model was assumed for individual genotypes and ancestry proportions were included as continuous covariates. The SNPAssoc package of R was used to investigate the associations between COPD risk and the combined effects of SNPs associated with COPD risk, established COPD risk factors and individual ancestry proportions, using multiple logistic regression models [44].
3. Results
3.1. Demographic and Clinical Findings
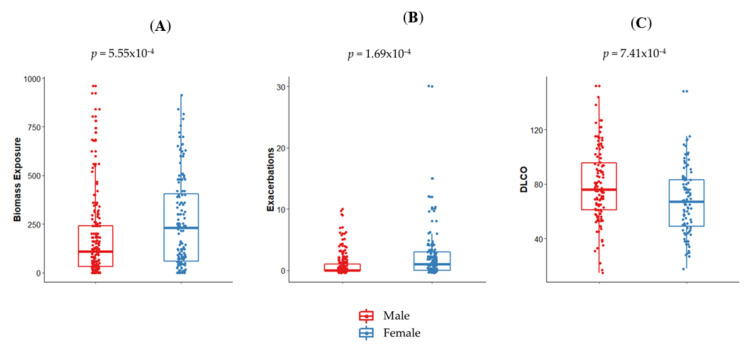
In our population, COPD patients and controls were of similar age (p = 0.69) but differed in sex proportions (p = 7.3 × 10−6), and controls showed a significantly higher average of schooling years (p = 2.2 × 10−15; Table 1). Interestingly, BMI was lower in COPD patients (p = 1.79 × 10−5), although both groups were generally overweight (Table 1). COPD patients exhibited higher amounts of cigarette smoking and cumulative exposure to biomass smoke than controls (p = 2.45 × 10−12 and p = 3.98 × 10−4, respectively). COPD patients also had reduced FEV1, FEV1/FVC, DLCO, oxygen saturation, and 6MWT compared to controls (p = 2.2 × 10−15 to p = 2.15 × 10−7). Most COPD patients were GOLD stage 2 (FEV1 between 50% and 79% of predicted). Female patients showed a higher exposure to biomass smoke and an increased number of exacerbations in the previous year (p = 5.5 × 10−4 and p = 1.69 × 10−4, respectively; Figure 1A,B). In contrast, male patients exhibited an increased DLCO (p = 7.41 × 10−4; Figure 1C).
Table 1.
Demographic, clinical, and genetic ancestry data of the study population.
| - | Control Subjects n = 193 |
COPD Patients n = 214 |
|---|---|---|
| Sex, Male/Female | 60/133 | 121/93 |
| Age, years | 68.66 ± 3.25 | 70.97 ± 4.69 |
| Smoking history, pack-years | 7.75 ± 3.25 | 30.47 ± 14.82 * |
| Current Smoker | 31 (16.06%) | 33 (15.42%) |
| Former smoker | 71 (36.79%) | 145 (67.76%) |
| Never smoker | 91 (47.15%) | 36 (16.82%) |
| Biomass exposure, hour-years | 96.87 ± 32.57 | 225.62 ± 54.28 * |
| Scholarship, years | 14.33 ± 2.57 | 7.21 ± 3.98 * |
| BMI, kg/m2 | 29.45 ± 5.02 | 26.96 ± 5.02 * |
| Exacerbations in the previous year | –– | 1.37 ± 1.50 |
| FEV1, % predicted | 108.84 ± 18.40 | 61.47 ± 24.56 * |
| FEV1/FVC, % | 83.00 ± 6.27 | 58.25 ± 10.48 * |
| DLCO, % predicted | 87.43 ± 24.48 | 72.33 ± 25.13 * |
| Oxygen Saturation, % | 96.14 ± 2.34 | 92.36 ± 4.76 * |
| 6MW, meters | 462.95 ± 87.82 | 351.50 ± 155.61 * |
| mMRC | –– | 2.28 ± 1.39 |
| CAT | –– | 14.94 ± 8.46 |
| BODE | –– | 3.18 ± 2.74 |
| European ancestry, % | 55.93 ± 6.89 | 56.32 ± 10.27 |
| African ancestry, % | 1.73 ± 1.00 | 1.73 ± 1.12 |
| Mapuche ancestry, % | 35.39 ± 6.71 | 35.11 ± 8.54 |
| Aymara ancestry, % | 6.96 ±.45 | 6.73 ± 3.52 |
Data presented as mean ± standard deviation, unless otherwise indicated. Definition of abbreviations: BMI: body mass index; FEV1: forced expiratory volume in 1 second; FVC: forced vital capacity; DLCO: carbon monoxide diffusing capacity; 6MW: 6 minutes walking test; mMRC: modified Medical Research Council scale; CAT: Chronic obstructive pulmonary disease (COPD) assessment test; BODE: Body mass, airflow Obstruction, Dyspnea, and Exercise index. * Different from control subjects (p < 0.05).
Figure 1.
Sex differences in clinical and exposure parameters in chronic obstructive pulmonary disease (COPD) patients. Female COPD patients showed an increased exposure to in-home biomass smoke (A) and exacerbations in the previous year (B), whereas males exhibited a better carbon monoxide diffusing capacity of the lung (DLCO) (C).
3.2. Analysis of Population Structure
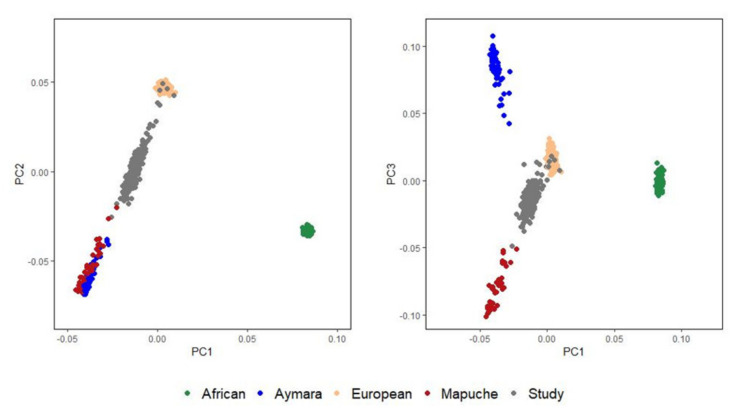
We conducted genetic principal component analyses using the EIGENSTRAT function available at popgen.dk/software/index.php/Rscripts [45]. The first principal component (PC1) distinguished Africans from non-Africans and the second principal component (PC2) separated European and Native American ancestry components. The third principal component (PC3) separated the Mapuche and Aymara Native American subcomponents. Major influences of the European and Mapuche ancestries in the study population were revealed (Figure 2). In our cohort, the average Aymara proportion was 6.48%, whereas the average Mapuche percentage was 35.24%. The average European and African proportions were 55.61% and 1.59%, respectively. COPD patients and controls were similar with respect to ancestry proportions (Table 1).
Figure 2.
Genetic principal component analyses of study individuals and European, African, Mapuche, and Aymara reference individuals.
3.3. Influence of Genetic Ancestry on the Association of SNPs and COPD
Four of our 80 a priori SNPs were associated with COPD risk: rs626750 (OR 0.57, 95% CI 0.37–0.87, p = 0.01), rs9599114 (OR 0.75, 95% CI 0.57–0.99, p = 0.05), rs7181486 (OR 1.40, 95% CI 1.01–1.93, p = 0.04), and rs8034191 (OR 1.44, 95% CI 1.04–1.99, p = 0.03). Only rs207675 showed suggestive association after accounting for covariates (OR 0.50, 95% CI 0.28–0.90, p = 0.02; Table 2).
Table 2.
Common variants associated with COPD susceptibility in Caucasian and Chilean population (only SNPs with p ≤ 0.05 are shown).
| CAUCASIAN | CHILEAN | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | SNP Risk Allele |
Risk Allele Frequency |
OR | A1 | MAF Controls |
MAF COPD Cases |
OR1 | p | *OR2 | 95% CI | p |
| rs626750 | T | 0.17 | 0.74 | T | 0.15 | 0.09 | 0.57 | 0.01 | 0.50 | 0.28–0.90 | 0.02 |
| rs8034191 | C | 0.33 | 1.40 | C | 0.21 | 0.27 | 1.44 | 0.03 | 1.33 | 0.86–2.07 | 0.20 |
| rs7181486 | C | 0.39 | 1.32 | T | 0.21 | 0.27 | 1.40 | 0.04 | 1.39 | 0.90–2.15 | 0.14 |
| rs9599114 | C | 0.42 | 0.11 | C | 0.47 | 0.40 | 0.75 | 0.05 | 0.77 | 0.53–1.13 | 0.18 |
SNP: single nucleotide polymorphism; A1: minor allele nucleotide; MAF: minor allele frequency; OR: odds ratio; p: p-value; CI: confidence interval. *OR adjusted for sex, smoking status, biomass exposure, scholarship, and body mass index.
When exploring all variants of the Illumina Global Screening Array, rs12741415 (PPP1R12B gene), rs116062217 (PPP1R12B gene), rs1054761 (PRDM15 gene), rs4075967 (PRDM15 gene), and rs4862451 (intergenic) were associated to COPD risk (p < 10−5; Supplementary Table S2 and Supplementary Figure S2). The top four hits map to PPP1R12B and PRDM15, on chromosomes 1 and 21, respectively. In order to explore the influence of the proportion of Mapuche ancestry (PMA) on the association between the SNPs markers and COPD, we stratified the analysis by PMA levels expressed in two categories (low vs. high PMA) based on the median value of this variable (35%). The association between the PRDM15 variants and COPD was present in those individuals within the lower PMA group (Table 3). In addition, PRDM15 rs1054761 (OR = 0.23, p = 0.02), rs2236696 (OR = 10.41, p = 1.00 × 10−3), rs4075967 (OR = 0.22, p = 0.05), rs4075970 (OR = 6.76, p = 8.25 × 10−4) and rs28360603 (OR = 7.55, p = 5.41 × 10−4), were associated with risk in those with lower PMA after adjusting for age, sex, smoking status, biomass exposure, scholarship, and body mass index (Table 3).
Table 3.
Association between PRMD15 SNPs and COPD risk in participants with low and high PMA (proportion of Mapuche ancestry, <35% vs. ≥35%).
| Low PMA | |||||||
| SNP | A1 | OR1 | 95%CI | p | *OR2 | 95%CI | p |
| rs1054761 | T | 0.31 | 0.18–0.51 | 7.24 × 10–7 | 0.23 | 0.09–0.89 | 0.02 |
| rs2236696 | T | 2.45 | 1.5–3.99 | 1.89 × 10–4 | 10.41 | 1.99–54.59 | 1.00 × 10−3 |
| rs8184900 | G | 0.4 | 0.26–0.62 | 9.99 × 10–6 | 0.40 | 0.11–1.49 | 0.16 |
| rs4075967 | A | 0.27 | 0.16–0.46 | 9.46 × 10–8 | 0.22 | 0.04–1.12 | 0.05 |
| rs4075970 | A | 2.43 | 1.57–3.78 | 3.32 × 10–5 | 6.76 | 1.84–24.84 | 8.25 × 10−4 |
| rs28360603 | A | 2.26 | 1.46–3.48 | 1.24 × 10–4 | 7.55 | 2–28.54 | 5.41 × 10−4 |
| rs7275618 | C | 1.79 | 1.18–2.71 | 4.75 × 10–3 | 2.04 | 0.57–7.30 | 0.25 |
| rs35109371 | C | 1.87 | 1.24–2.82 | 2.16 × 10–3 | 2.02 | 0.56–7.26 | 0.27 |
| High PMA | |||||||
| SNP | A1 | OR1 | 95%CI | p | *OR2 | 95%CI | p |
| rs1054761 | T | 0.63 | 0.41–0.96 | 0.03 | 0.41 | 0.13–1.25 | 0.10 |
| rs2236696 | T | 1.02 | 0.7–1.5 | 0.92 | 2.34 | 0.80–6.92 | 0.11 |
| rs8184900 | G | 0.73 | 0.5–1.08 | 0.12 | 0.58 | 0.29–1.16 | 0.12 |
| rs4075967 | A | 0.73 | 0.47–1.13 | 0.15 | 0.45 | 0.15–1.40 | 0.15 |
| rs4075970 | A | 0.98 | 0.68–1.43 | 0.93 | 2.08 | 0.77–5.64 | 0.13 |
| rs28360603 | A | 1.22 | 0.82–1.79 | 0.32 | 1.04 | 0.34–3.17 | 0.94 |
| rs7275618 | C | 1.18 | 0.77–1.83 | 0.44 | 0.81 | 0.22–3.01 | 0.75 |
| rs35109371 | C | 1.32 | 0.88–1.96 | 0.17 | 1.08 | 0.31–3.71 | 0.90 |
SNP: single nucleotide polymorphism; A1: minor allele nucleotide; OR: odds ratio; p: p-value; CI: confidence interval; PMA: proportion of Mapuche ancestry.*OR adjusted for sex, smoking status, biomass exposure, scholarship, and body mass index.
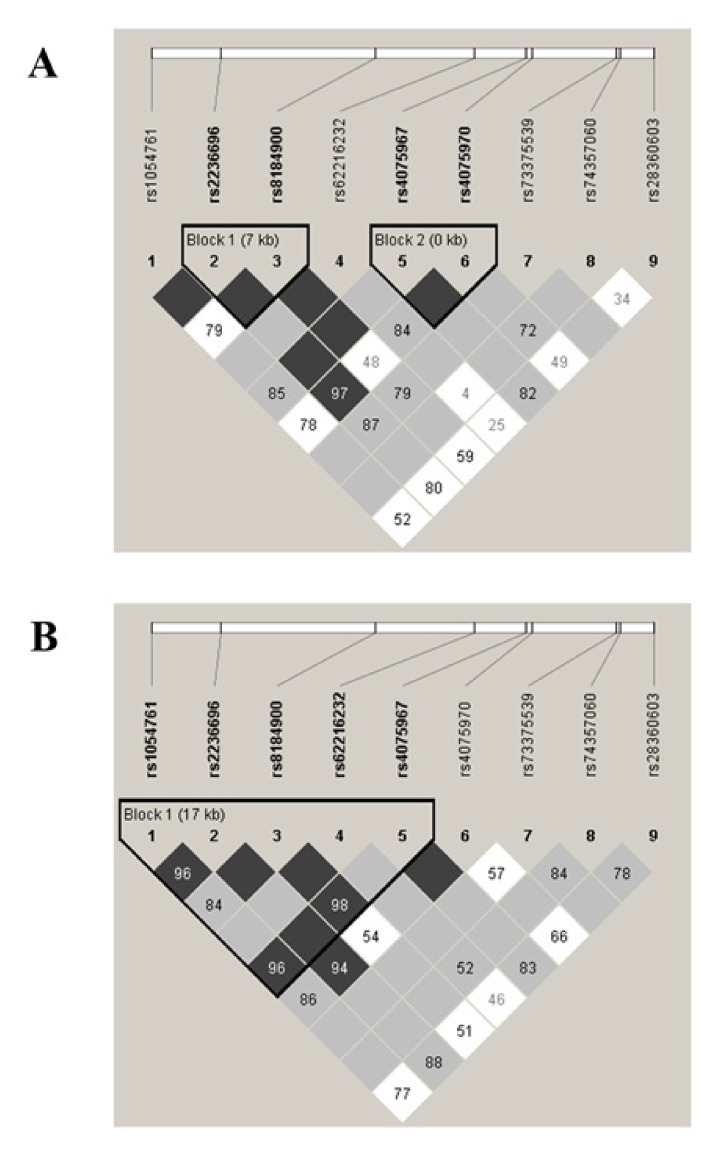
The sliding window omnibus test revealed several SNP blocks associated with COPD in the lower PMA group (Table 4). Hence, the p-values obtained for the sliding window test of a region comprising PRDM15 gene (Chr21: 43.229.099 to 43.256.172) were lower than 10−6. We also performed case–control regression analysis based on haplotype block reconstruction, and detailed haplotype block information and the LD plot around the PRDM15 gene are shown in Figure 3. In the lower PMA group, PRDM15 was divided into two LD blocks, both with two SNPs (Figure 3A), showing association with COPD (positions 43,221,826 through 43,229,099, CG (p = 7.40 × 10−6, p-value using 10,000 permutations <10−3); and positions 43,236,176 through 43,236,481, AG (p = 1.01 × 10−7, p-value using 10,000 permutations <10−4]). Differences were also found in the LD pattern among the higher PMA group (Figure 3B). No differences were observed around PRDM15 gene. When data across the four superpopulations (EUR, EAS, AMR, and AFR) were compared, high inter-ethnic differences were found in the LD patterns and haplotype blocks compared to our control population (Supplementary Figure S3).
Table 4.
Association analyses of sliding windows of 2–10 single nucleotide polymorphisms each, using chi-square statistics within PLINK software (only p-values <10−6 are shown).
| SNP Markers | p-Value |
|---|---|
| rs4075967|rs4075970 | 1.01 × 10−7 |
| rs1054761|rs2236696|rs8184900 | 9.53 × 10−7 |
| rs2236696|rs8184900|rs62216232 | 3.77 × 10−8 |
| rs62216232|rs4075967|rs4075970 | 1.43 × 10−7 |
| rs2236696|rs8184900|rs62216232|rs4075967 | 1.01 × 10−7 |
| rs8184900|rs62216232|rs4075967|rs4075970 | 1.16 × 10−7 |
| rs62216232|rs4075967|rs4075970|rs73375539 | 1.44 × 10−7 |
| rs4075967|rs4075970|rs73375539|rs74357060 | 1.42 × 10−7 |
| rs2236696|rs8184900|rs62216232|rs4075967|rs4075970 | 9.48 × 10−8 |
| rs8184900|rs62216232|rs4075967|rs4075970|rs73375539 | 8.36 × 10−8 |
| rs62216232|rs4075967|rs4075970|rs73375539|rs74357060 | 1.09 × 10−7 |
| rs4075967|rs4075970|rs73375539|rs74357060|rs28360603 | 4.38 × 10−7 |
| rs2236696|rs8184900|rs62216232|rs4075967|rs4075970|rs73375539 | 9.52 × 10−8 |
| rs8184900|rs62216232|rs4075967|rs4075970|rs73375539|rs74357060 | 5.81 × 10−8 |
| rs62216232|rs4075967|rs4075970|rs73375539|rs74357060|rs28360603 | 5.80 × 10−7 |
| rs2236696|rs8184900|rs62216232|rs4075967|rs4075970|rs73375539|rs74357060 | 1.61 × 10−7 |
| rs8184900|rs62216232|rs4075967|rs4075970|rs73375539|rs74357060|rs28360603 | 4.78 × 10−7 |
| rs62216232|rs4075967|rs4075970|rs73375539|rs74357060|rs28360603|rs28708536 | 4.29 × 10−7 |
| rs2236696|rs8184900|rs62216232|rs4075967|rs4075970|rs73375539|rs74357060| rs28360603 |
6.39 × 10−7 |
| rs8184900|rs62216232|rs4075967|rs4075970|rs73375539|rs74357060|rs28360603| rs28708536 |
3.76 × 10−7 |
| rs62216232|rs4075967|rs4075970|rs73375539|rs74357060|rs28360603|rs28708536| rs76291974 |
2.28 × 10−7 |
| rs8184900|rs62216232|rs4075967|rs4075970|rs73375539|rs74357060|rs28360603| rs28708536|rs76291974 |
1.79 × 10−7 |
| rs62216232|rs4075967|rs4075970|rs73375539|rs74357060|rs28360603|rs28708536| rs76291974|rs62214694 |
2.86 × 10−7 |
| rs8184900|rs62216232|rs4075967|rs4075970|rs73375539|rs74357060|rs28360603| rs28708536|rs76291974|rs62214694 |
2.17 × 10−7 |
| rs62216232|rs4075967|rs4075970|rs73375539|rs74357060|rs28360603|rs28708536| rs76291974|rs62214694|rs17766525 |
2.66 × 10−7 |
Figure 3.
Linkage disequilibrium plots around the PRDM15 gene in the lower (A) and higher (B) proportion of Mapuche ancestry (PMA) groups. Squares represent the linkage disequilibrium (D’) value between every two single nucleotide polymorphisms: the darker the square, the higher D’ value. The five triangular shapes surrounding these markers indicate the haplotype blocks that were defined using the confidence interval method.
4. Discussion
The present study is the first to report the contribution of established risk factors, genetic ancestry, and SNPs associated to COPD risk in a Latin American population, characterized by a large admixture of Amerindian ancestry.
Our analysis highlighted the unique demographic and clinical characteristics of our study population. Almost a fifth of all COPD patients were never-smokers, whereas most experienced a high exposure to biomass smoke. These exposure scenarios, coupled with the low educational level, reflect the rural nature of our population. Maulean COPD patients were also overweight, coinciding with studies in Chile and elsewhere [46,47]. Interestingly, females were more exposed to indoor biomass smoke than males, as have been reported in other studies performed in middle-income or developing countries [2]. Considering the advanced age of the participants, this probably reflects a sociocultural effect, since decades ago Chilean women followed a different lifestyle than men, remaining at unventilated homes during long periods, cooking, or being near woodstoves. The higher exposure to biomass smoke could also explain the fact that female COPD participants showed a decreased DLCO than men. Indeed, it has been reported that biomass smoke-related COPD is associated with milder emphysema and DLCO reduction than tobacco-related COPD [48]. Our results are similar to recent reports of higher COPD exacerbation frequency among women [49,50], which may reflect the higher hyperresponsiveness prevalence reported in females [51]. Further, exposure to household biomass smoke pollutants, such as PM2.5 and NO2, have been associated with increased risk of COPD exacerbations [52].
We found that Amerindian ancestry-specific alleles in PRDM15 may confer a protection against COPD. Our results are biologically plausible given that PRDM15 has been shown to play a key role in regulating WNT and MAPK-ERK signaling [53] through regulation of the MAPK-ERK signaling cascade. In addition, rs12741415 and rs116062217, located in the PPP1R12B, were associated with COPD susceptibility in our population. The protein encoded by the gene PPP1R12B, a protein phosphatase 1 regulatory subunit 12B, is a component of myosin phosphatase complex, which regulates contractile processes in muscular and other cells [54]. Interestingly, Freĭdin and coworkers postulated PPP1R12B as a candidate gene for childhood asthma [55], but these data have not been replicated in other populations. It is also noteworthy that both PRDM15 and PPP1R12B have been related to immune dysfunction and/or autoimmunity, mechanisms that are thought to play a key role in COPD pathogenesis [56]. Hence, PRDM15 is overexpressed in B-cell dysregulation [57,58], a characteristic feature of COPD [59], whereas PPP1R12B and its “M20” transcript are higher in patients with celiac disease autoimmunity [60]. Although further research is needed to deeper understand the potential contribution of PRDM15 and PPP1R12B to COPD, resequencing PRDM15 and PPP1R12B may clarify the nature of the genetic variation and develop a functional study of their protein products in this complex disease.
The results of the present study suggest that the risk allele frequencies of some SNP may vary in Europeans and Chileans. Given our findings, and that allele frequencies of disease-associated SNPs may vary by ethnic group due to genetic drift or selection [61], it is important to account for ancestry to avoid false associations. The results of the present study showed little replication of previously reported genetic associations with COPD, with only 10.1% of known COPD loci from GWAS in populations of European origin showing a suggestive association in our Latin American cohort (p < 0.1). For example, our population exhibited as smaller allele frequency of rs207675-C (our control population = 0.48) relative to European populations (https://www.internationalgenome.org/; 1000 genomes project (EUR) = 0.64). Assuming our findings are replicated in another population, they will have implications for the implementation of personalized medicine which most often focuses on specific biological, genetic, and clinical characteristics of an individual. Our results suggest that there are other factors that provide additional complexity that need to be considered, such as ethnicity, life-style differences, and environmental factors. In this context, there is a relevant gap to overcome between European and non-European target populations, which is necessary for the implementation of the personalized medicine in all populations [62].
While these results are intriguing, we must acknowledge that our study is limited by the sample size and lack of replication. However, due to the heterogeneity of its genetic structure, getting ideal ancestral populations in Latin Americans is more difficult than in other admixed populations, which confers relevance to what is found in our population [63]. Moreover, since approximately 79% of all GWAS participants are Caucasians, and the fraction of non-European individuals in GWAS has stagnated or declined since late 2014 [64], our preliminary results give a valuable idea of the heterogeneity in genetic susceptibility to COPD in diverse ethnic groups.
In summary, the present study shows that PRDM15 and PPP1R12B are associated with COPD risk in a Latin American population. Moreover, the association of PRDM15 variants conferring genetic protection to COPD is higher in patients with more Amerindian ancestry. Although the relative low sample size and the lack of replication limit the implications of the present study, our results stress the need to conduct more GWAS in admixed populations, such as those with Amerindian descent.
Acknowledgments
The authors would like to thank Cristian Vergara, Alejandra Muñoz, Fabiola Lastra, Carolina Lara, Cintia Muñoz, Hanuxa Celedón, and Victor Smolski for their technical assistance. They also thank Marta Chmielewska for linguistic advice.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4426/10/3/93/s1: Supplementary Figure S1: Analysis of mortality rate in Chile; Supplementary Table S1: Eighty-nine common single nucleotide polymorphisms (SNPs) included in the study from previous genome wide association studies (GWAS) in populations of diverse ethnic background; Supplementary Figure S2: Single-marker allelic association results; Supplementary Table S2: Single-SNP allelic association analysis (p < 10−5); Supplementary Figure S3: Linkage disequilibrium plots around the PRDM15 gene.
Author Contributions
Study concept and design: J.O., R.D.-P., R.S.S., and J.L.B.; Data acquisition: R.S.S., S.J., and J.O.; Data analysis: R.D.-P., F.B., J.L.B, and J.O.; Data interpretation: R.D.-P., H.D.H.III, A.A., M.M., F.B., J.L.B., and J.O.; Funding acquisition: J.O.; Investigation: J.O. and R.D.-P.; Methodology: J.O., R.D.-P., F.B., and J.L.B.; Supervision: J.O.; Writing – original draft: J.O. and R.D.-P.; Writing – review and editing: J.O., R.D.-P., H.D.H.III, R.S.S., S.J., M.M., A.A., F.B., and J.L.B. All authors have read and agreed to the published version of the manuscript.
Funding
Funding support for this study was provided by the Chilean National Science and Technology Fund (CONICYT), FONDECYT Project N° 11150022.
Conflicts of Interest
Marc Miravitlles has received speaker fees from AstraZeneca, Boehringer Ingelheim, Chiesi, Cipla, Menarini, Rovi, Bial, Sandoz, Zambon, CSL Behring, Grifols and Novartis; consulting fees from AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Bial, Gebro Pharma, CSL Behring, Laboratorios Esteve, Ferrer, Mereo Biopharma, Verona Pharma, TEVA, pH Pharma, Novartis and Grifols; and research grants from GlaxoSmithKline and Grifols; all outside the submitted work. The rest of the authors declare no conflicts of interest.
References
- 1.Global Initiative for Chronic Obstructive Lung Diesease (GOLD) 2020 Global Strategy for Prevention, Diagnosis and Management of COPD. Global Initiative for Chronic Obstructive Lung Diesease (GOLD); Fontana, WI, USA: 2020. [Google Scholar]
- 2.Silva R., Oyarzun M., Olloquequi J. Pathogenic mechanisms in chronic obstructive pulmonary disease due to biomass smoke exposure. Arch. Bronconeumol. 2015;51:285–292. doi: 10.1016/j.arbres.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Han M.K., Hanania N.A., Martinez F.J. Confronting the Challenge of COPD: What Is New in the Approaches to Diagnosis, Treatment, and Patient Outcomes. Chest. 2018;154:984–985. doi: 10.1016/j.chest.2018.08.1024. [DOI] [PubMed] [Google Scholar]
- 4.Vestbo J., Hurd S.S., Agusti A.G., Jones P.W., Vogelmeier C., Anzueto A., Barnes P.J., Fabbri L.M., Martinez F.J., Nishimura M., et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 5.Ciapponi A., Alison L., Agustina M., Demian G., Silvana C., Edgardo S. The epidemiology and burden of COPD in Latin America and the Caribbean: Systematic review and meta-analysis. COPD. 2014;11:339–350. doi: 10.3109/15412555.2013.836479. [DOI] [PubMed] [Google Scholar]
- 6.Menezes A.M., Perez-Padilla R., Jardim J.R., Muino A., Lopez M.V., Valdivia G., Montes de Oca M., Talamo C., Hallal P.C., Victora C.G. Chronic obstructive pulmonary disease in five Latin American cities (the PLATINO study): A prevalence study. Lancet. 2005;366:1875–1881. doi: 10.1016/S0140-6736(05)67632-5. [DOI] [PubMed] [Google Scholar]
- 7.Palloni A., Novak B., Pinto-Aguirre G. The enduring effects of smoking in Latin America. Am. J. Public Health. 2015;105:1246–1253. doi: 10.2105/AJPH.2014.302420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres-Duque C., Maldonado D., Pérez-Padilla R., Ezzati M., Viegi G. Biomass fuels and respiratory diseases: A review of the evidence. Proc. Am. Thorac. Soc. 2008;5:577–590. doi: 10.1513/pats.200707-100RP. [DOI] [PubMed] [Google Scholar]
- 9.SZOT M. J. Mortalidad por enfermedades respiratorias en Chile durante 1999. Rev. Chil. Enf. Respir. 2003;19:8–14. doi: 10.4067/S0717-73482003000100002. [DOI] [Google Scholar]
- 10.Wade P. Race and Ethnicity in Latin America. Pluto Press; London, UK: 2010. [Google Scholar]
- 11.Klimentidis Y.C., Miller G.F., Shriver M.D. Genetic admixture, self-reported ethnicity, self-estimated admixture, and skin pigmentation among Hispanics and Native Americans. Am. J. Phys. Anthropol. 2009;138:375–383. doi: 10.1002/ajpa.20945. [DOI] [PubMed] [Google Scholar]
- 12.Sans M. Admixture studies in Latin America: From the 20th to the 21st century. Hum. Biol. 2000;72:155–177. [PubMed] [Google Scholar]
- 13.Ruiz-Linares A., Adhikari K., Acuña-Alonzo V., Quinto-Sanchez M., Jaramillo C., Arias W., Fuentes M., Pizarro M., Everardo P., de Avila F., et al. Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals. PLoS Genet. 2014;10:e1004572. doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S., Ray N., Rojas W., Parra M.V., Bedoya G., Gallo C., Poletti G., Mazzotti G., Hill K., Hurtado A.M., et al. Geographic Patterns of Genome Admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuentes M., Pulgar I., Gallo C., Bortolini M.-C., Canizales-Quinteros S., Bedoya G., González-José R., Ruiz-Linares A., Rothhammer F. Geografía génica de Chile: Distribución regional de los aportes genéticos americanos, europeos y africanos. Rev. Méd. de Chile. 2014;142:281–289. doi: 10.4067/S0034-98872014000300001. [DOI] [PubMed] [Google Scholar]
- 16.Eyheramendy S., Martinez F.I., Manevy F., Vial C., Repetto G.M. Genetic structure characterization of Chileans reflects historical immigration patterns. Nat. Commun. 2015;6:6472. doi: 10.1038/ncomms7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smolonska J., Wijmenga C., Postma D.S., Boezen H.M. Meta-analyses on suspected chronic obstructive pulmonary disease genes: A summary of 20 years’ research. Am. J. Respir. Crit. Care Med. 2009;180:618–631. doi: 10.1164/rccm.200905-0722OC. [DOI] [PubMed] [Google Scholar]
- 18.Wain L.V., Shrine N., Artigas M.S., Erzurumluoglu A.M., Noyvert B., Bossini-Castillo L., Obeidat M., Henry P.A., Portelli M.A., Hall R.J., et al. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat. Genet. 2017;49:416–425. doi: 10.1038/ng.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho M., Tantisira K. ¡HOLA! The Influence of Being Hispanic on Lung Ancestry. Am. J. Respir. Crit. Care Med. 2018;198:148–149. doi: 10.1164/rccm.201802-0306ED. [DOI] [PubMed] [Google Scholar]
- 20.Kheirallah A.K., Miller S., Hall I.P., Sayers I. Translating Lung Function Genome-Wide Association Study (GWAS) Findings: New Insights for Lung Biology. Adv. Genet. 2016;93:57–145. doi: 10.1016/bs.adgen.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Lutz S.M., Cho M.H., Young K., Hersh C.P., Castaldi P.J., McDonald M.L., Regan E., Mattheisen M., DeMeo D.L., Parker M., et al. A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genet. 2015;16:138. doi: 10.1186/s12863-015-0299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Cho M.H., Zhou X. What do polymorphisms tell us about the mechanisms of COPD? Clin. Sci. 2017;131:2847–2863. doi: 10.1042/CS20160718. [DOI] [PubMed] [Google Scholar]
- 23.Kim D.K., Cho M.H., Hersh C.P., Lomas D.A., Miller B.E., Kong X., Bakke P., Gulsvik A., Agustí A., Wouters E., et al. Genome-wide association analysis of blood biomarkers in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2012;186:1238–1247. doi: 10.1164/rccm.201206-1013OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manichaikul A., Hoffman E.A., Smolonska J., Gao W., Cho M.H., Baumhauer H., Budoff M., Austin J.H., Washko G.R., Carr J.J., et al. Genome-wide study of percent emphysema on computed tomography in the general population. The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am. J. Respir. Crit. Care. Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W., Brehm J.M., Manichaikul A., Cho M.H., Boutaoui N., Yan Q., Burkart K.M., Enright P.L., Rotter J.I., Petersen H., et al. A genome-wide association study of chronic obstructive pulmonary disease in Hispanics. Ann. Am. Thorac. Soc. 2015;12:340–348. doi: 10.1513/AnnalsATS.201408-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burkart K.M., Sofer T., London S.J., Manichaikul A., Hartwig F.P., Yan Q., Soler Artigas M., Avila L., Chen W., Davis Thomas S., et al. A Genome-Wide Association Study in Hispanics/Latinos Identifies Novel Signals for Lung Function. The Hispanic Community Health Study/Study of Latinos. Am. J. Respir. Crit. Care. Med. 2018;198:208–219. doi: 10.1164/rccm.201707-1493OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosgood Iii H.D., Diaz-Pena R., Blansky D., Jaime S., Parra V., Boekstegers F., Bermejo J.L., Garcia-Valero J., Montes J.F., Valdivia G., et al. PRDM15 Is Associated with Risk of Chronic Obstructive Pulmonary Disease in a Rural Population in Chile. Respiration. 2020;99:307–315. doi: 10.1159/000506649. [DOI] [PubMed] [Google Scholar]
- 28.Hindorff L.A., Bonham V.L., Brody L.C., Ginoza M.E.C., Hutter C.M., Manolio T.A., Green E.D. Prioritizing diversity in human genomics research. Nat. Rev. Genet. 2018;19:175–185. doi: 10.1038/nrg.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Díaz A.A., Celli B., Celedón J.C. Chronic Obstructive Pulmonary Disease in Hispanics. A 9-Year Update. Am. J. Respir. Crit. Care. Med. 2018;197:15–21. doi: 10.1164/rccm.201708-1615PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.GOLD . 2019 Global Strategy for the Diagnosis, Management and Prevention of COPD. Global Initiative for Chronic Obstructive Lung Disease (GOLD); Fontana, WI, USA: 2019. [Google Scholar]
- 31.Miller M.R., Hankinson J., Brusasco V., Burgos F., Casaburi R., Coates A., Crapo R., Enright P., van der Grinten C.P.M., Gustafsson P., et al. Standardisation of spirometry. Eur. Respir. J. 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 32.Olloquequi J., Jaime S., Parra V., Cornejo-Cordova E., Valdivia G., Agusti A., Silva O.R. Comparative analysis of COPD associated with tobacco smoking, biomass smoke exposure or both. Respir. Res. 2018;19:13. doi: 10.1186/s12931-018-0718-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardin M., Cho M.H., McDonald M.L., Wan E., Lomas D.A., Coxson H.O., MacNee W., Vestbo J., Yates J.C., Agusti A., et al. A genome-wide analysis of the response to inhaled beta2-agonists in chronic obstructive pulmonary disease. Pharmacogenomics J. 2016;16:326–335. doi: 10.1038/tpj.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hobbs B.D., de Jong K., Lamontagne M., Bosse Y., Shrine N., Artigas M.S., Wain L.V., Hall I.P., Jackson V.E., Wyss A.B., et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat. Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obeidat M., Zhou G., Li X., Hansel N.N., Rafaels N., Mathias R., Ruczinski I., Beaty T.H., Barnes K.C., Pare P.D., et al. The genetics of smoking in individuals with chronic obstructive pulmonary disease. Respir. Res. 2018;19:59. doi: 10.1186/s12931-018-0762-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorenzo Bermejo J., Boekstegers F., Gonzalez Silos R., Marcelain K., Baez Benavides P., Barahona Ponce C., Muller B., Ferreccio C., Koshiol J., Fischer C., et al. Subtypes of Native American ancestry and leading causes of death: Mapuche ancestry-specific associations with gallbladder cancer risk in Chile. PLoS Genet. 2017;13:e1006756. doi: 10.1371/journal.pgen.1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reich D., Patterson N., Campbell D., Tandon A., Mazieres S., Ray N., Parra M.V., Rojas W., Duque C., Mesa N., et al. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindo J., Haas R., Hofman C., Apata M., Moraga M., Verdugo R.A., Watson J.T., Viviano Llave C., Witonsky D., Beall C., et al. The genetic prehistory of the Andean highlands 7000 years BP though European contact. Sci. Adv. 2018;4:eaau4921. doi: 10.1126/sciadv.aau4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Gen. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez J.R., Armengol L., Sole X., Guino E., Mercader J.M., Estivill X., Moreno V. SNPassoc: An R package to perform whole genome association studies. Bioinformatics. 2007;23:644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 45.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 46.Galesanu R.G., Bernard S., Marquis K., Lacasse Y., Poirier P., Bourbeau J., Maltais F. Obesity in chronic obstructive pulmonary disease: Is fatter really better? Can. Respir. J. 2014;21:297–301. doi: 10.1155/2014/181074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montes de Oca M., Talamo C., Perez-Padilla R., Jardim J.R., Muino A., Lopez M.V., Valdivia G., Pertuze J., Moreno D., Halbert R.J., et al. Chronic obstructive pulmonary disease and body mass index in five Latin America cities: The PLATINO study. Respir. Med. 2008;102:642–650. doi: 10.1016/j.rmed.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 48.Perez-Padilla R., Schilmann A., Riojas-Rodriguez H. Respiratory health effects of indoor air pollution. Int. J. Tuberc. Lung. Dis. 2010;14:1079–1086. [PubMed] [Google Scholar]
- 49.Lisspers K., Larsson K., Janson C., Stallberg B., Tsiligianni I., Gutzwiller F.S., Mezzi K., Bjerregaard B.K., Jorgensen L., Johansson G. Gender differences among Swedish COPD patients: Results from the ARCTIC, a real-world retrospective cohort study. NPJ Prim. Care Respir. Med. 2019;29:45. doi: 10.1038/s41533-019-0157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsiligianni I., Rodriguez M.R., Lisspers K., LeeTan T., Infantino A. Call to action: Improving primary care for women with COPD. NPJ Prim. Care Respir. Med. 2017;27:11. doi: 10.1038/s41533-017-0013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkins C.R., Chapman K.R., Donohue J.F., Roche N., Tsiligianni I., Han M.K. Improving the Management of COPD in Women. Chest. 2017;151:686–696. doi: 10.1016/j.chest.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 52.Hansel N.N., McCormack M.C., Belli A.J., Matsui E.C., Peng R.D., Aloe C., Paulin L., Williams D.L., Diette G.B., Breysse P.N. In-home air pollution is linked to respiratory morbidity in former smokers with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2013;187:1085–1090. doi: 10.1164/rccm.201211-1987OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mzoughi S., Zhang J., Hequet D., Teo S.X., Fang H., Xing Q.R., Bezzi M., Seah M.K.Y., Ong S.L.M., Shin E.M., et al. PRDM15 safeguards naive pluripotency by transcriptionally regulating WNT and MAPK-ERK signaling. Nat. Genet. 2017;49:1354–1363. doi: 10.1038/ng.3922. [DOI] [PubMed] [Google Scholar]
- 54.Ito M., Nakano T., Erdodi F., Hartshorne D.J. Myosin phosphatase: Structure, regulation and function. Mol. Cell Biochem. 2004;259:197–209. doi: 10.1023/B:MCBI.0000021373.14288.00. [DOI] [PubMed] [Google Scholar]
- 55.Freidin M.B., Polonikov A.V. Validation of PPP1R12B as a candidate gene for childhood asthma in Russians. J. Genet. 2013;92:93–96. doi: 10.1007/s12041-013-0210-x. [DOI] [PubMed] [Google Scholar]
- 56.Agustí A., MacNee W., Donaldson K., Cosio M. Hypothesis: Does COPD have an autoimmune component? Thorax. 2003;58:832–834. doi: 10.1136/thorax.58.10.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giallourakis C.C., Benita Y., Molinie B., Cao Z., Despo O., Pratt H.E., Zukerberg L.R., Daly M.J., Rioux J.D., Xavier R.J. Genome-wide analysis of immune system genes by expressed sequence Tag profiling. J. Immunol. 2013;190:5578–5587. doi: 10.4049/jimmunol.1203471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park H.Y., Lee S.-B., Yoo H.-Y., Kim S.-J., Kim W.-S., Kim J.-I., Ko Y.-H. Whole-exome and transcriptome sequencing of refractory diffuse large B-cell lymphoma. Oncotarget. 2016;7:86433–86445. doi: 10.18632/oncotarget.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polverino F., Seys L.J.M., Bracke K.R., Owen C.A. B cells in chronic obstructive pulmonary disease: Moving to center stage. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016;311:L687–L695. doi: 10.1152/ajplung.00304.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Östensson M., Montén C., Bacelis J., Gudjonsdottir A.H., Adamovic S., Ek J., Ascher H., Pollak E., Arnell H., Browaldh L., et al. A possible mechanism behind autoimmune disorders discovered by genome-wide linkage and association analysis in celiac disease. PLoS ONE. 2013;8:e70174. doi: 10.1371/journal.pone.0070174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lynch M., Ackerman M.S., Gout J.F., Long H., Sung W., Thomas W.K., Foster P.L. Genetic drift, selection and the evolution of the mutation rate. Nat. Rev. Genet. 2016;17:704–714. doi: 10.1038/nrg.2016.104. [DOI] [PubMed] [Google Scholar]
- 62.Gurdasani D., Barroso I., Zeggini E., Sandhu M.S. Genomics of disease risk in globally diverse populations. Nat. Rev. Genet. 2019;20:520–535. doi: 10.1038/s41576-019-0144-0. [DOI] [PubMed] [Google Scholar]
- 63.Baran Y., Pasaniuc B., Sankararaman S., Torgerson D.G., Gignoux C., Eng C., Rodriguez-Cintron W., Chapela R., Ford J.G., Avila P.C., et al. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics. 2012;28:1359–1367. doi: 10.1093/bioinformatics/bts144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.