PRRSV has haunted the swine industry for over 30 years since its emergence. Besides the limited efficacy of PRRSV modified live vaccines (MLVs) against heterogeneous PRRSV isolates, rapid induction of nonneutralizing antibodies (non-NAbs) against PRRSV NSPs after MLV immunization or wild-strain infection is one of the reasons why development of an effective vaccine has been hampered. By using in vitro-generated BMDCs as models to understand the antigen presentation process of PRRSV, we obtained data indicating that PRRSV infection of BMDCs promotes functional SLA-DR upregulation to present PRRSV NSP-derived immunopeptides for evoking a non-NAb response in vivo. Our work not only uncovered a novel mechanism for interference in host antigen presentation by PRRSV but also revealed a novel insight for understanding the rapid production of nonneutralizing antibodies against PRRSV NSPs, which may have benefit for developing an effective vaccine against PRRSV in the future.
KEYWORDS: MHC-II molecules, mass spectrometry, PRRSV, SLA-DR, antigen presentation, nonneutralizing antibodies
ABSTRACT
The humoral immune response against porcine reproductive and respiratory syndrome virus (PRRSV) infection is characterized by a rapid induction of nonneutralizing antibodies (non-NAbs) against nonstructural proteins (NSPs). Here, we systematically investigated the potential mechanism for the induction of PRRSV NSP-specific non-NAbs. Our data suggested that PRRSV NSP-specific antibodies appeared within 10 days after PRRSV infection in vivo. In the in vitro model, functional upregulation of swine leukocyte antigen (SLA)-DR was observed in bone marrow-derived dendritic cells (BMDCs) and porcine alveolar macrophages (PAMs), whereas remarkable inhibition at the mRNA level was observed after infection by both PRRSV-1 and PRRSV-2 isolates. Notably, the inconsistency in SLA-DR expression between the mRNA and protein levels resulted from deubiquitination of SLA-DR via the ovarian tumor (OTU) domain of PRRSV NSP2, which inhibited ubiquitin-mediated degradation. Moreover, mass spectrometry-based immunopeptidome analysis identified immunopeptides originating from multiple PRRSV NSPs within SLA-DR of PRRSV-infected BMDCs. Meanwhile, these PRRSV NSP-derived immunopeptides could be specifically recognized by serum from PRRSV-infected piglets. Notably, certain NSP-derived immunopeptides characterized in vitro could be identified from PAMs or hilar lymph nodes from PRRSV-infected piglets. More importantly, an in vitro neutralizing assay indicated that serum antibodies against NSP immunopeptides were unable to neutralize PRRSV in vitro. Conversely, certain structural protein (SP)-derived immunopeptides were identified and could be recognize by pig hyperimmune serum against PRRSV, which further indicates that the NSP-derived antibody response is nonprotective in vivo. In conclusion, our data suggested that PRRSV infection interferes with major histocompatibility complex class II (MHC-II) molecule-mediated antigen presentation in antigen-presenting cells (APCs) via promoting SLA-DR expression to present immunopeptides from PRRSV NSPs, which contributes to the induction of non-NAbs in vivo.
IMPORTANCE PRRSV has haunted the swine industry for over 30 years since its emergence. Besides the limited efficacy of PRRSV modified live vaccines (MLVs) against heterogeneous PRRSV isolates, rapid induction of nonneutralizing antibodies (non-NAbs) against PRRSV NSPs after MLV immunization or wild-strain infection is one of the reasons why development of an effective vaccine has been hampered. By using in vitro-generated BMDCs as models to understand the antigen presentation process of PRRSV, we obtained data indicating that PRRSV infection of BMDCs promotes functional SLA-DR upregulation to present PRRSV NSP-derived immunopeptides for evoking a non-NAb response in vivo. Our work not only uncovered a novel mechanism for interference in host antigen presentation by PRRSV but also revealed a novel insight for understanding the rapid production of nonneutralizing antibodies against PRRSV NSPs, which may have benefit for developing an effective vaccine against PRRSV in the future.
INTRODUCTION
Porcine reproductive and respiratory syndrome virus (PRRSV) is a positive-sense, single-stranded, enveloped RNA virus which belongs to the genus Porartevirus (1, 2). Based on the new taxonomic scheme, there are two species within the genus Porartevirus, PRRSV-1 and PRRSV-2 (1, 2). Although PRRSV-1 and PRRSV-2 strains share only approximately 60% nucleotide sequence identity and exhibit serotype differences (3, 4), the overall disease phenotype, gross clinical signs, and genomic organization are similar between the two species (5). Unlike other members of Arteriviridae that have relatively broad cell tropisms (6), PRRSV infection is highly restricted to cells originating from the monocyte/macrophage lineage, such as porcine alveolar macrophages (7, 8), macrophages from the spleen, tonsils, and lymph nodes, and peritoneal macrophages from the blood and progenitor cells in the bone marrow (9–12). Moreover, bone marrow-derived dendritic cells (BMDCs) and bone marrow-derived macrophages are susceptible to PRRSV infection in vitro as well (13, 14). Generally, porcine alveolar macrophages (PAMs) are considered to be the primary target of PRRSV in vivo (7, 8).
Current control of PRRSV is inadequate despite the substantial efforts that have been dedicated to managing it. Since the first report in 1987, PRRSV remains one of the major challenges for the swine industry globally, and it continuously evolves to cause new outbreaks (15, 16). The typical immune features in hosts after PRRSV infection include persistent viremia, a strong inhibition of innate cytokines (interferon alpha/beta [IFN-α/β], tumor necrosis factor alpha [TNF-α], and interleukin-1β [IL-1β]), dysregulation of NK cell function, rapid induction of nonneutralizing antibodies (non-NAbs), delayed appearance of neutralizing antibodies (NAbs), a late and low CD8+ T cell response, and induction of regulatory T cells (8, 17, 18). Tremendous effort has been made to elucidate the virus-host interaction between PRRSV and the host immune system to understand the viral pathogenesis. For the innate immune response, it has been confirmed that the PRRSV genome encodes several IFN antagonists to block either IFN induction or IFN-activated JAK/STAT signaling (19–21), such as nonstructural protein 1α (NSP1α), NSP1β (22), NSP2 (23, 24), NSP4 (25), NSP11 (26), and the nucleocapsid (N) protein (27). On the other hand, the understanding of how PRRSV interferes with the adaptive immune response remains incomplete.
It has been previously reported that PRRSV infection leads to the downregulation of swine leukocyte antigen class I (SLA-I) molecules (28, 29). These in vitro observations are consistent with the delayed, low-level cytotoxic T lymphocyte (CTL) response observed in pigs infected by PRRSV in vivo, which is thought to contribute to persistent infections of PRRSV in swine herds (28). However, the humoral response to PRRSV infection is more complicated and remains not fully understood. Generally, PRRSV-specific NAbs typically appear 28 days postinoculation (30, 31), but nonprotective antibodies against PRRSV can be rapidly induced within the first week postinfection. These early nonneutralizing antibodies are specific and recognize certain structural proteins (SPs), such as the N protein, as well as NSPs (30, 32). PRRSV N, NSP1, NSP2, and NSP7 have been demonstrated to be highly immunogenic (33, 34).
Induction of PRRSV-specific antibodies requires antigen-specific B cell activation with assistance from CD4+ T helper cells (35, 36), while the activation of CD4+ T helper cells relies on the engagement of T cell receptors (TCRs) with major histocompatibility complex class II (MHC-II)–immunopeptide complexes presented by antigen-presenting cells (APCs). However, available reports conflict with each other regarding the understanding of the expression of swine MHC-II molecules (swine leukocyte antigen class II [SLA-II]) on macrophages or dendritic cells (DCs) after PRRSV infection (37–40). Thus, the correlation between the function of SLA-II molecules on APCs and humoral immunity after PRRSV infection in the host remains elusive, especially regarding the induction of PRRSV NSP-specific antibodies.
In our previous report, we developed a modified assay based on luciferase immunoprecipitation (IP) systems (LIPS) to quantify PRRSV-specific antibodies from serum and referred to it as the luciferase-linked antibody capture assay (LACA) (41). Based on the analysis of early antibody profiles in PRRSV-infected hosts, it was suggested that antibodies against PRRSV NSP1α could reach an even higher level than antibodies to the PRRSV N protein, as determined by the sample-to-negative ratio (S/N ratio) (41). Meanwhile, by employing a homemade monoclonal antibody (MAb) against the swine leukocyte antigen II DR α chain (SLA-DRα), we noticed that PRRSV infection of BMDCs induces upregulation of SLA-DR at the protein level and promotes surface expression of SLA-DR in PRRSV-infected BMDCs (42). These data imply that SLA-II-mediated antigen presentation is not impaired by PRRSV infection but may contribute to the rapid production of nonneutralizing antibodies at the early stage of infection.
In the present study, we systematically investigated the antibody response to PRRSV at the early stage of infection and elucidated the potential mechanism for the induction of non-NAbs recognizing PRRSV NSPs. Notably, PRRSV infection promoted the expression of total SLA-DR protein and functional SLA-DR expressed on the cellular surfaces of BMDCs, although mRNA levels of SLA-DR were inhibited by PRRSV infection regardless of the genotype or strain of PRRSV. Meanwhile, upregulation of the SLA-DR protein in PRRSV-infected BMDCs appears to be caused by the deubiquitination of the SLA-DR protein by the PRRSV NSP2 ovarian tumor (OTU) domain (24), which inhibits ubiquitin-mediated degradation of SLA-DR. More importantly, mass spectrometry-based analysis for peptides eluted from SLA-DR–immunopeptide complexes obtained both in vivo and in vitro indicated that immunopeptides derived from PRRSV NSPs were presented by APCs, and these immunopeptides were recognized by corresponding antibodies from the serum of PRRSV-infected pigs but lacked the ability to neutralize PRRSV in an in vitro virus neutralization assay. For the first time, our data suggest that PRRSV infection could interfere with MHC-II-mediated (SLA-DR in swine) antigen presentation in BMDCs for presenting PRRSV NSPs to further produce corresponding non-NAbs in the early stage of PRRSV infection.
RESULTS
Serum of PRRSV-infected piglets contained antibodies recognizing multiple PRRSV NSPs and appeared within 10 days after PRRSV infection in vivo.
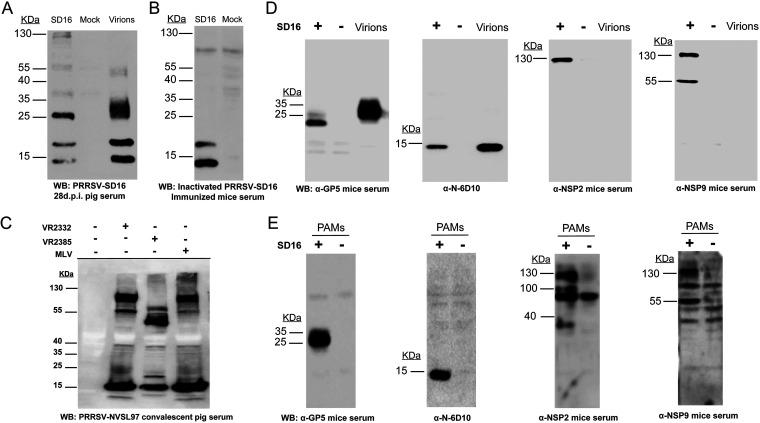
It has long been observed that the early antibody response in PRRSV-infected hosts is mainly specific to recognize the N protein and certain NSPs (30, 32), while antibodies recognizing PRRSV NSPs can persist for months, as PRRSV NSP1, NSP2, and NSP7 are highly immunogenic (33, 34). To confirm the existence of PRRSV NSP antibodies in the serum of PRRSV-infected pigs at 28 days postinoculation (28-dpi serum), purified PRRSV virions (strain SD16) and PRRSV SD16-infected MARC-145 cells were used to probe the serum. As shown by the results in Fig. 1A, the molecular weight (MW) of the largest protein in the purified virions (containing structural proteins [SPs] only) recognized by PRRSV SD16 28-dpi serum was below 55 kDa. However, there were multiple bands from PRRSV-infected MARC-145 cells (containing both SPs and NSPs) that were higher than 55 kDa and could be recognized by 28-dpi serum, suggesting that these virus-specific proteins with MWs higher than 55 kDa might be PRRSV NSPs and could be recognized by PRRSV-infected-pig serum. Since PRRSV-infected hosts contain polyclonal antibodies recognizing both NSPs and SPs, to further confirm the above-described results, we immunized mice (completely resistant to PRRSV) with inactivated PRRSV three times and confirmed seroconversion of the mice using the Idexx PRRS X3 enzyme-linked immunosorbent assay (ELISA). When serum from immunized mice that did not contain antibodies recognizing PRRSV NSPs was used to probe the PRRSV SD16-infected MARC-145 cells, only two bands below 25 kDa were visualized, suggesting that the viral proteins with higher MWs in infected MARC-145 cells were PRRSV NSPs and that PRRSV replication in the host is required for the immune system to develop antibodies that recognize PRRSV NSPs.
FIG 1.
Serum of PRRSV-infected pigs contains antibodies recognizing multiple nonstructural proteins (NSPs) of PRRSV. (A) MARC-145 cells infected by PRRSV strain SD16 at an MOI of 1 for 24 h were subjected to Western blotting using 28-day-postinoculation (28-dpi) serum from PRRSV SD16-infected pigs. Noninfected MARC-145 cells and 5 μg purified PRRSV SD16 virions were included as the negative control and positive control, respectively. (B) MARC-145 cells infected by PRRSV SD16 at an MOI of 1 for 24 h and noninfected MARC-145 cells were subjected to Western blotting using mouse serum from mice immunized with inactivated PRRSV SD16. (C) MARC-145 cells infected by PRRSV strain VR2332, a modified live virus vaccine strain (Ingelvac MLV), and PRRSV strain VR2385 at an MOI of 1 for 24 h were subjected to Western blotting using convalescent-phase serum from pigs infected with PRRSV strain NVSL97. Noninfected MARC-145 cells without PRRSV infection were included as a negative control. (D) MARC-145 cells infected by PRRSV SD16 at an MOI of 1 for 24 h were subjected to Western blotting using anti-GP5 mouse antiserum, MAb 6D10 (PRRSV N protein-specific MAb), and anti-NSP2 and anti-NSP9 mouse antisera. MARC-145 cells without PRRSV infection and 5 μg purified PRRSV SD16 virions were included as controls. (E) Porcine alveolar macrophages (PAMs) infected by PRRSV SD16 at an MOI of 1 for 24 h were subjected to Western blotting using anti-GP5 mouse antiserum, MAb 6D10, and anti-NSP2 and anti-NSP9 mouse antisera. Noninfected PAMs were included as controls.
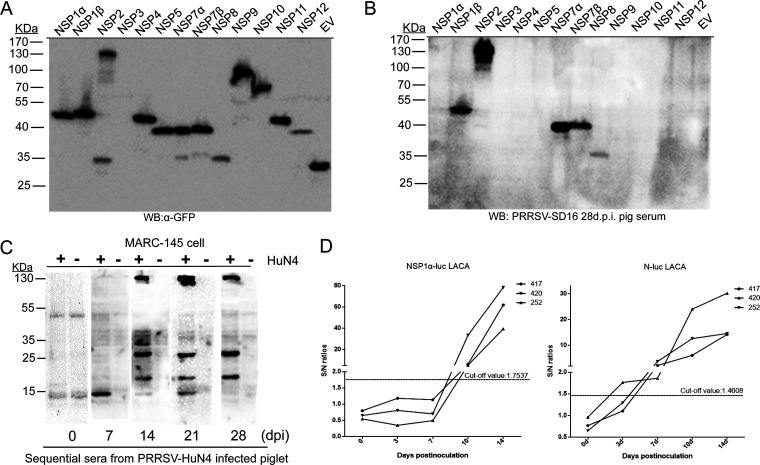
Meanwhile, since deletion has frequently been reported in PRRSV NSP2, we used PRRSV convalescent-phase serum from pigs infected with PRRSV strain NVSL97 to probe MARC-145 cells infected with three different PRRSV strains: VR2332 (PRRSV-2 prototype strain), Ingelvac modified live vaccine (MLV; attenuated vaccine strain based on VR2332), and VR2385 (containing a spontaneous 435-nucleotide deletion in NSP2). Based on the Western blotting results, MARC-145 cells infected with VR2332 and the MLV demonstrated similar band patterns (Fig. 1C), while bands with reduced MWs were observed for VR2385-infected cells, suggesting that the highest band represented PRRSV NSP2 or its precursor pp1a/pp1ab. Moreover, the PRRSV SD16-infected MARC-145 cells or PAMs and purified PRRSV virions were also probed separately using anti-glycoprotein 5 (GP5) mouse antiserum, anti-N protein MAb 6D10, and anti-NSP2 and anti-NSP9 mouse antisera (Fig. 1D and E). The data suggested that PRRSV NSPs only existed in PRRSV-infected MARC-145 cells and PAMs, with molecular weights ranging from 55 to 130 kDa. To further determine the exact NSPs recognized by PRRSV SD16 28-dpi pig serum, all PRRSV NSPs were ligated to the VenusC1 vector to be expressed as green fluorescent protein (GFP)-fused proteins and were then used to probe the PRRSV SD16 28-dpi serum. All PRRSV NSPs except NSP3 were successfully expressed in HEK-293T cells during transient transfection (Fig. 2A), and several NSPs (NSP1β, -2, -7α, -7β, and -8) could be recognized by 28-dpi serum in the Western blot analysis (Fig. 2B). Therefore, these data suggested that PRRSV-infected pigs developed antibodies recognizing multiple PRRSV NSPs and that the 130-kDa band (possibility NSP2) could be used as an indicator for the existence of antibodies recognizing PRRSV NSPs from PRRSV-infected pig serum.
FIG 2.
PRRSV infection causes rapid induction of antibodies against PRRSV NSPs. (A) The cDNAs of individual PRRSV NSPs from PRRSV SD16 were cloned into the VenusC1 vector and transfected into HEK-293T cells for transient expression. Cell lysates from 293T cells with transfection of Venus-fused PRRSV NSPs were harvested and subjected to Western blotting using anti-GFP MAb to confirm the expression. (B) Cell lysates from 293T cells with transfection of PRRSV NSPs were harvested and subjected to Western blotting using 28-dpi serum from PRRSV SD16-infected pigs to probe the existence of antibodies recognizing NSPs. (C) MARC-145 cells infected by PRRSV strain HuN4 at an MOI of 1 were subjected to Western blotting using sequential serum samples collected at indicated days postinoculation (dpi) from PRRSV HuN4-infected pigs. MARC-145 cells without PRRSV infection were included as negative controls. (D) Sequential serum samples collected at the indicated dpi from three PRRSV HuN4-infected pigs (animals 417, 420, and 252) were subjected to luciferase-linked antibody capture assay (LACA) using Renilla luciferase-fused NSP1α and N proteins to quantify antibody levels of PRRSV NSP1α and N. Quantification of antibodies was determined by sample-to-negative (S/N) ratio. Cutoff values for S/N ratios of PRRSV NSP1α LACA and PRRSV N LACA were determined using confirmed anti-PRRSV antibody-negative and -positive serum samples.
To understand the dynamic changes of PRRSV-specific antibodies in hosts, sequential serum samples obtained from piglets experimentally infected by PRRSV strain HuN4 were used to probe PRRSV HuN4-infected MARC-145 cells. It appeared that N-specific antibodies were developed within 7 dpi (Fig. 2C), which is consistent with the time point of serum conversion in infected pigs, as determined by Idexx PRRSV ELISA in our previous report (41). Meanwhile, antibodies recognizing NSPs could be detected within 14 dpi as determined by the visualization of 130-kDa bands by Western blotting; it appeared that anti-NSP antibodies in pig serum persisted at a much higher level until 28 dpi, as the relative signal strength observed in the Western blot for the 15-kDa band (potential N protein) became much weaker than the signal strength for the 130-kDa band (Fig. 2C).
To further quantify the kinetics of antibodies against PRRSV NSPs and SPs in detail, a previously described luciferase-linked antibody capture assay (LACA) was carried out to evaluate the levels of antibodies against PRRSV NSP1α (the first autoclaved protein from pp1a polyprotein during replication) and PRRSV N (the first structural protein translated from PRRSV subgenomic RNA during replication). Based on the quantification data (sample-to-negative [S/N] ratio), although the S/N ratio of antibodies against NSP1α was below the defined cutoff value before day 7, it increased drastically on day 10 and reached 80 on day 14, whereas the maximum S/N ratio of antibodies against the PRRSV N protein only reached 30 during the sampling period (Fig. 3D). Taken together, the above-described data suggested that PRRSV-infected piglets developed antibodies recognizing multiple PRRSV NSPs and that NSP-specific antibodies appeared concurrently with antibodies against PRRSV N but increased to more drastic levels in hosts during the early stage (before 28 days) of PRRSV infection. This observation implies that the host humoral immune response against PRRSV infection preferentially developed antibodies against PRRSV NSPs, which are potentially nonprotective against PRRSV infection.
FIG 3.
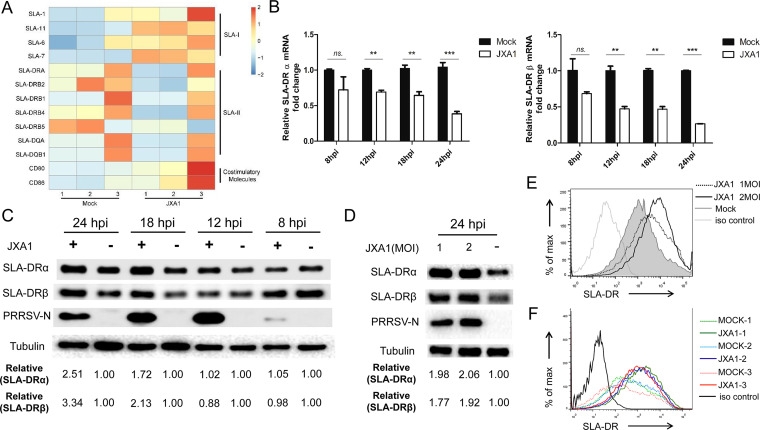
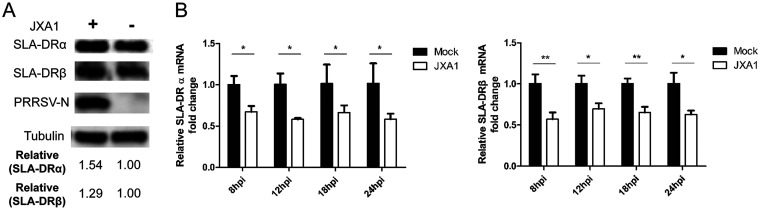
PRRSV infection inhibits mRNA expression of SLA-DR but promotes its protein expression in bone marrow-derived dendritic cells (BMDCs). (A) BMDCs developed by in vitro culture and granulocyte-macrophage colony-stimulating factor (GM-CSF) stimulation of bone marrow cells from 3 different piglets were collected and infected with PRRSV strain JXA1 at an MOI of 1 for 12 h. The cDNAs from PRRSV JXA1-infected and noninfected BMDCs were subjected to transcriptome analysis to reveal the effect of PRRSV infection on the mRNA expression levels of costimulatory molecules (CD80 and CD86) and SLA-I and SLA-II molecules. The mRNA expression of each molecule is presented in a heat map. (B) BMDCs generated in vitro were infected with PRRSV JXA1 and collected at indicated time points for qPCR to evaluate mRNA levels of SLA-DRα and SLA-DRβ. All experiments were repeated at least three times. Error bars show standard deviations. Significant differences between indicated groups are marked as follows: **, P < 0.01; ***, P < 0.001; ns, nonsignificant. (C) BMDCs generated in vitro were infected with PRRSV JXA1 and collected at indicated time points (hours postinoculation [hpi]) for Western blot analysis using a homemade anti-SLA-DRα MAb and anti-SLA-DRβ mouse antiserum. BMDCs without PRRSV infection but harvested at the same time points were included as controls. Replication of PRRSV virus in infected BMDCs was determined using anti-PRRSV N protein MAb 6D10. (D) BMDCs generated in vitro were infected with PRRSV JXA1 at MOIs of 1 and 2 for 24 h, and then the cells were harvested for Western blot analysis to evaluate SLA-DRα and SLA-DRβ expression. BMDCs without PRRSV infection were included as a control. Replication of PRRSV virus in BMDCs was confirmed using anti-PRRSV N MAb 6D10. (E) BMDCs were infected with PRRSV JXA1 at MOIs of 1 and 2 for 24 h and stained with anti-SLA-DR antibodies, followed by visualization of allophycocyanin-labeled goat anti-mouse IgG. Then, the cells were subjected to flow cytometry analysis to evaluate cell surface expression of SLA-DR. BMDCs without PRRSV infection and cells stained with noninfected-mouse IgG were included as a negative control and antibody isotype control, respectively. (F) BMDCs generated from 3 different pigs (animals 1, 2, and 3) were infected with PRRSV JXA1 at an MOI of 1 for 24 h and stained with anti-SLA-DR antibodies, followed by visualization of allophycocyanin-labeled goat anti-mouse IgG. Then, the cells were subjected to flow cytometry analysis to evaluate cell surface expression of SLA-DR. BMDCs without PRRSV infection and cells stained with noninfected-mouse IgG were included as a negative control and antibody isotype control, respectively.
PRRSV infection of BMDCs promoted the expression of SLA-DR at the protein level.
Induction of PRRSV-specific antibodies requires antigen-specific B cell activation with assistance from activated CD4+ T helper cells (35, 36), which is engaged by T cell receptor (TCR) recognition of immunopeptides presented by major histocompatibility complex class II (MHC-II) molecules on the surface of APCs. It has been confirmed that PRRSV can infect various types of APCs, including macrophages, monocytes, and dendritic cells isolated in vivo and generated in vitro (9–14). Here, bone marrow-derived dendritic cells (BMDCs) were developed using a previously described protocol (42). To analyze the antigen presentation function of BMDCs after PRRSV infection, a transcriptome analysis of the mRNA levels of antigen presentation-related molecules in PRRSV-infected BMDCs (using BM isolated from different piglets) was conducted. As demonstrated by the results shown in Fig. 3A, the transcription levels of costimulation molecules (CD80/CD86) and certain members of the swine leukocyte antigen class I (SLA-I) gene class (SLA-1, -7, and -11) were upregulated in PRRSV-infected BMDCs, while the levels of SLA-6 and -8 were similar between noninfected BMDCs and PRRSV-infected BMDCs. However, mRNAs of the SLA-II gene class (DQ and DR) were significantly reduced in PRRSV-infected cells (Fig. 3A). Since human leukocyte antigen-DR (HLA-DR), the counterpart of SLA-DR in humans, is a major player responsible for foreign-antigen presentation and cross talk with CD4+ T helper cells, which enables activation of the adaptive immune response (43, 44), SLA-DR was selected for quantitative real-time PCR (qPCR) verification. Our data suggested that mRNA of both the α chain and β chain of SLA-DR is strongly inhibited by PRRSV replication in BMDCs during the time points analyzed (Fig. 3B). Meanwhile, the SLA-DR protein levels were examined in detail using a previously reported MAb against SLA-DRα (42), along with our in-house-made mouse antiserum against SLA-DRβ. It appeared that no obvious changes in SLA-DRα and -β could be detected within 12 h after PRRSV infection (Fig. 3C). However, nearly 2-fold upregulation of both the SLA-DRα and -β protein level in PRRSV-infected BMDCs was observed at 18 h postinfection (hpi), and the levels increased continually to more than 3-fold at 24 hpi (Fig. 3C). Moreover, if higher doses of PRRSV were used to inoculate BMDCs, higher SLA-DRα and SLA-DRβ protein levels were detected in infected cells (Fig. 3D).
Since only surface-expressed SLA-II is able to activate CD4+ T helper cells, the cell surface expression of SLA-DR in PRRSV-infected BMDCs was analyzed as well. Much higher levels of surface SLA-DR expression were observed, correlated with increased multiplicities of infection (MOIs) used to inoculate BMDCs (Fig. 3E). Conversely, BMDCs developed from three different piglets were further analyzed to ensure that the PRRSV infection-induced surface upregulation of SLA-DR was universal (Fig. 3F). Collectively, the above-described data demonstrated that functional SLA-DR expression in PRRSV-susceptible BMDCs was not impaired by PRRSV infection and that SLA-DR protein expression on the surface of BMDCs could be promoted by PRRSV JXA1 infection.
Evaluation showed surface SLA-DR expression in PRRSV-infected BMDCs was not strain or genotype specific and could be observed in PAMs.
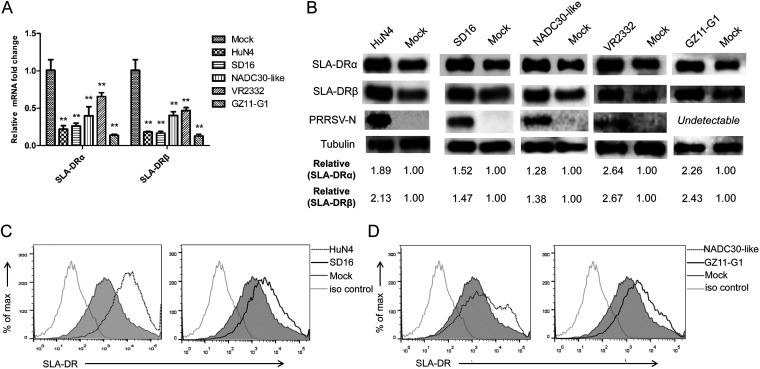
Based on the literature, the available data investigating the expression and function of SLA-II in PRRSV-infected APCs were controversial (37–40). Considering the heterogeneous nature of different PRRSV isolates, we further investigated whether the SLA-DR expression promoted by PRRSV in BMDCs is universal or strain and genotype specific. Therefore, heterogeneous PRRSV isolates, including the classical PRRSV isolate (VR2332), highly pathogenic (HP) PRRSV isolates other than JXA1 (SD16 and HuN4), an NADC30-like Chinese isolate, and PRRSV-1 Chinese isolate GZ11 were used to inoculate BMDCs. Similar to the results for BMDCs infected by PRRSV JXA1, significant reductions of SLA-DRα and -β mRNA levels were observed for all strains tested (Fig. 4A). Meanwhile, the protein levels of SLA-DRα and -β were investigated, and all heterogeneous PRRSV isolates tested promoted the expression of both SLA-DRα and -β in infected BMDCs compared to their expression in noninfected cells (Fig. 4B). Replication of these PRRSV isolates was determined by examining the N protein level in infected cells by using the anti-PRRSV N MAb 6D10, except for PRRSV-1 strain GZ11, due to the antigenic variation of its N protein. Moreover, upregulation of SLA-DR on the cell surface of BMDCs infected by heterogeneous PRRSV isolates was confirmed as well (Fig. 4C and D). Conversely, since BMDCs are in vitro-developed APCs, the natural in vivo targets of PRRSV, PAMs, were infected with PRRSV to evaluated the mRNA and protein levels of SLA-DR. Similar to the observations in BMDCs, upregulation of the SLA-DR protein level (Fig. 5A) and downregulation of SLA-DR mRNA (Fig. 5B) caused by PRRSV infection in PAMs were observed as well. Therefore, these data indicated that the expression of SLA-DR on cell surfaces of PRRSV-infected APCs (such as BMDCs or PAMs) was not strain or genotype specific; instead, it was universal for both PRRSV-1 and PRRSV-2, which might infer a biological function of antigen presentation of PRRSV proteins during natural infection.
FIG 4.
PRRSV-induced surface expression of SLA-DR in bone marrow-derived dendritic cells (BMDCs) is not strain or genotype specific. (A) BMDCs generated in vitro were infected with heterogeneous isolates (PRRSV strains HuN4, SD16, NADC30-like, and PRRSV-1 GZ11) at an MOI of 1 for 24 h. Then, the cells were harvested for qPCR to evaluate the mRNA levels of SLA-DRα and SLA-DRβ. BMDCs without PRRSV infection were included as a control. Error bars show standard deviations. All experiments were repeated at least three times. Significant differences between indicated groups are marked as follows: **, P < 0.01. (B) BMDCs generated in vitro were infected with heterogeneous isolates (PRRSV-HuN4, SD16, NADC30-like, and PRRSV-1 GZ11) at an MOI of 1 for 24 h. Then, the cells were harvested for Western blot analysis using a homemade anti-SLA-DRα MAb and anti-SLA-DRβ mouse antiserum to evaluate the expression of SLA-DRα and -β. Replication of PRRSV was confirmed by probing the same PVDF membrane with anti-PRRSV N MAb 6D10. (C) BMDCs were infected with PRRSV HuN4 and SD16 at an MOI of 1 for 24 h and stained with anti-SLA-DR antibody, followed by visualization of allophycocyanin-labeled goat anti-mouse IgG. Then, the cells were subjected to flow cytometry analysis for evaluating cell surface expression of SLA-DR. BMDCs without PRRSV infection and cells stained with noninfected-mouse IgG were included as a negative control and antibody isotype control. (D) BMDCs were infected with PRRSV isolate NADC30-like and PRRSV-1 isolate GZ11 at an MOI of 1 for 24 h, followed by staining for flow cytometry analysis to evaluate cell surface expression of SLA-DR. Noninfected BMDCs and cells stained with noninfected-mouse IgG were included as a negative control and antibody isotype control, respectively.
FIG 5.
PRRSV infection upregulates SLA-DR expression in its in vivo target porcine alveolar macrophages (PAMs). (A) PAMs were infected with PRRSV strain JXA1 at an MOI of 1 for 24 h. Then, the cells were harvested for Western blot analysis using a homemade anti-SLA-DRα MAb and anti-SLA-DRβ mouse antiserum to evaluate the expression of SLA-DRα and -β. Replication of PRRSV was confirmed by probing the same PVDF membrane with anti-PRRSV N protein MAb 6D10. (B) PAMs were infected with PRRSV JXA1 at an MOI of 1 for 24 h. Then, the cells were harvested for qPCR to evaluate the mRNA levels of SLA-DRα and SLA-DRβ. PAMs without PRRSV infection were included as a control. All experiments were repeated at least three times. Error bars show standard deviations. Significant differences between indicated groups are marked as follows: *, P < 0.05; **, P < 0.01.
PRRSV infection extended the protein half-life of SLA-DR by inhibiting ubiquitination of SLA-DR.
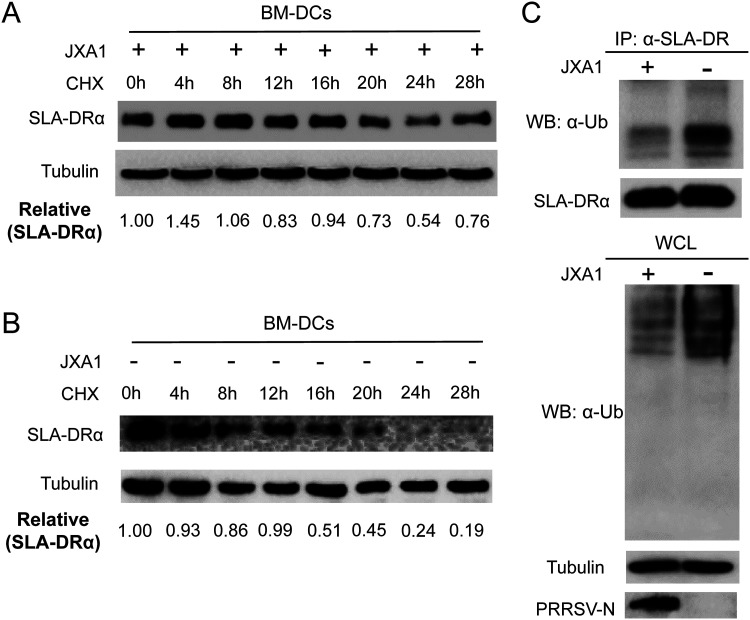
As the above-mentioned data suggested an inconsistency between SLA-DR protein and mRNA levels in PRRSV-infected BMDCs, we sought to understand how the replication of PRRSV promotes higher SLA-DR protein levels while inhibiting the mRNA expression of SLA-DR. To figure out the mechanism, cycloheximide (CHX), a universal protein translation inhibitor, was used to treat PRRSV-infected BMDCs 8 h after PRRSV inoculation; CHX and PRRSV cannot be added to BMDCs simultaneously because the protein translation inhibition caused by CHX inhibits PRRSV replication as well (data not shown). Based on the remaining SLA-DRα protein levels in PRRSV-infected BMDCs following CHX treatment for different times, it appeared that 24 h of CHX treatment was required for a 50% reduction of SLA-DRα protein (Fig. 6A). Conversely, when the same experiment was carried out in BMDCs without PRRSV infection, a 50% reduction in SLA-DRα protein could be observed as early as 8 to 12 h after CHX treatment (Fig. 6B). Therefore, our data suggested that the level of expressed SLA-DRα protein in BMDCs after PRRSV infection was a consequence of an extended protein half-life of SLA-DRα, which may be caused by PRRSV replication.
FIG 6.
PRRSV infection promotes the protein half-life of SLA-DR via inhibition of ubiquitination-mediated protein degradation of SLA-DR. (A) BMDCs generated in vitro were infected with PRRSV JXA1 at an MOI of 1 for 8 h, followed by treatment with cycloheximide (CHX) to inhibit universal protein translation. Cells were harvested at indicated time points after CHX treatment and subjected to Western blotting with anti-SLA-DRα MAb to evaluate the protein half-life of SLA-DRα in PRRSV-infected BMDCs. (B) Noninfected BMDCs treated with CHX were harvested at indicated time points and subjected to Western blotting with anti-SLA-DRα Mab to evaluate the protein half-life of SLA-DRα in noninfected BMDCs. (C) BMDCs were infected with PRRSV JXA1 at an MOI of 1 and lysed with NP-40 cell lysis buffer for immunoprecipitation (IP) of SLA-DR by anti-SLA-DR MAb to enrich the SLA-DR molecules. The immune complexes containing IP-enriched SLA-DR were subjected to Western blotting using antiubiquitin antibody to analyze the ubiquitination status of SLA-DR in PRRSV-infected BMDCs. The immune complexes obtained from noninfected BMDCs were included as controls. The universal ubiquitination level of whole-cell lysate (WCL) was analyzed as well.
The ubiquitin-proteasome system is involved in the turnover of most cellular proteins, and several reports demonstrated that PRRSV interfered with the ubiquitin-proteasome system in infected cells to accelerate or inhibit the degradation of host proteins like KPNA6 (45), STAT3 (21), and SLA-I molecules (28). Thus, we expected that the extended half-life of SLA-DR in PRRSV-infected BMDCs was caused by the alteration of SLA-DR ubiquitination as well. As demonstrated by the results shown in Fig. 5C, the ubiquitination status of SLA-DRα enriched in the immune complex precipitated from PRRSV JXA1-infected BMDCs was significantly reduced compared to that of noninfected cells; meanwhile, the total SLA-DRα levels within the immune complexes pulled down by immunoprecipitation (IP) were similar between the two groups of BMDCs. Also, the total protein ubiquitination of the whole-cell lysate was examined, and it appeared that PRRSV infection inhibited host protein ubiquitination universally in BMDCs (Fig. 6C), which is consistent with our previous observation that PRRSV infection inhibits universal protein ubiquitination in MARC-145 cells (45). Taken together, these data suggested that PRRSV infection in BMDCs blocked the ubiquitin-proteasome-mediated degradation of SLA-DR to extend the protein half-life of SLA-DR, which resulted in accumulation of SLA-DR proteins along with increased surface expression of SLA-DR in PRRSV-infected BMDCs.
OTU domain of PRRSV NSP2 blocked SLA-DR ubiquitination and led to accumulation of SLA-DR.
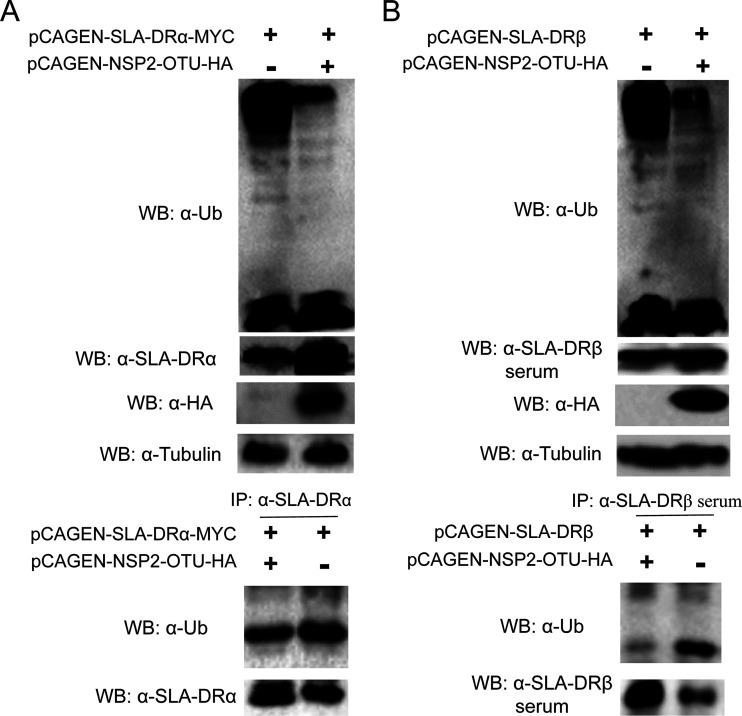
It was previously reported that the cysteine protease domain of PPRSV NSP2 belongs to the ovarian tumor (OTU) protease superfamily and acts as a deubiquitinase to antagonize interferon induction (24). Therefore, we sought to understand whether the OTU domain of NSP2 acts as a deubiquitinase for SLA-DR, as well as blocking SLA-DR ubiquitination. To confirm this, HEK-293T cells were cotransfected with plasmids encoding the PRRSV NSP2 OTU domain and SLA-DRα. Compared with cells transfected with SLA-DRα alone, cotransfection of the PRRSV NSP2 OTU domain significantly increased protein levels of SLA-DRα (Fig. 7A), which is similar to the observations in PRRSV-infected BMDCs. It was also notable that the presence of the OTU domain strongly inhibited universal ubiquitination in HEK-293T cells as well (Fig. 7A), which is consisted with the results for PRRSV-infected BMDCs. Meanwhile, HEK-293T cells were transfected with SLA-DRα alone or cotransfected with plasmids encoding both the OTU domain and SLA-DRα; ubiquitination analysis of IP-enriched SLA-DRα from these cells suggested that the presence of the NSP2 OTU domain blocked SLA-DRα ubiquitination (Fig. 7A). Similar experiments were conducted to examine whether the NSP2 OTU domain inhibits ubiquitination of SLA-DRβ. As demonstrated by the results shown in Fig. 7B, although to a lesser extent, the protein level of SLA-DRβ was upregulated with the presence of the NSP2 OTU domain, along with a reduced ubiquitination status of SLA-DRβ, which was similar to that of SLA-DRα. Collectively, we concluded that the OTU domain of PRRSV NSP2 acts as a deubiquitinase to block the ubiquitination and proteasome-mediated degradation of both the SLA-DRα and -β chain, thus leading to the accumulation of SLA-DR protein in PRRSV-infected cells.
FIG 7.
The ovarian tumor (OTU) domain of PRRSV NSP2 inhibits both SLA-DRα and -β chain ubiquitination. (A) HEK-293T cells were transfected with plasmid encoding SLA-DRα and the PRRSV NSP2 OTU domain for 48 h. Then, the cells were harvested for Western blotting using anti-MYC, antiubiquitin, and anti-SLA-DRα antibodies to confirm the transient expression of SLA-DRα and the NSP2 OTU domain, as well as the cellular ubiquitination level. Next, HEK-293T cells transfected with the indicated plasmids were lysed by NP-40 buffer, SLA-DRα was enriched by IP using anti-SLA-DRα MAb, and the immune complexes were analyzed by Western blotting using antiubiquitin antibodies to evaluate the ubiquitination status of SLA-DRα with or without presentation of the PRRSV NSP2 OTU domain. (B) HEK-293T cells were transfected with plasmid encoding SLA-DRβ and the PRRSV NSP2 OTU domain for 48 h. Then, the cells were harvested for Western blotting using anti-HA and antiubiquitin antibodies and anti-SLA-DRβ mouse antiserum to confirm the transient expression of SLA-DRβ chain and the NSP2 OTU domain, along with cellular ubiquitination status. Next, HEK-293T cells transfected with the indicated plasmids were lysed by NP-40 buffer, SLA-DRβ was enriched by IP using anti-SLA-DRβ mouse antiserum, and the immune complexes were analyzed by Western blotting using antiubiquitin antibodies to evaluate the ubiquitination status of SLA-DRβ with or without presentation of the PRRSV NSP2 OTU domain.
SLA-DR in PRRSV-infected BMDCs presents immunopeptides derived from PRRSV NSPs.
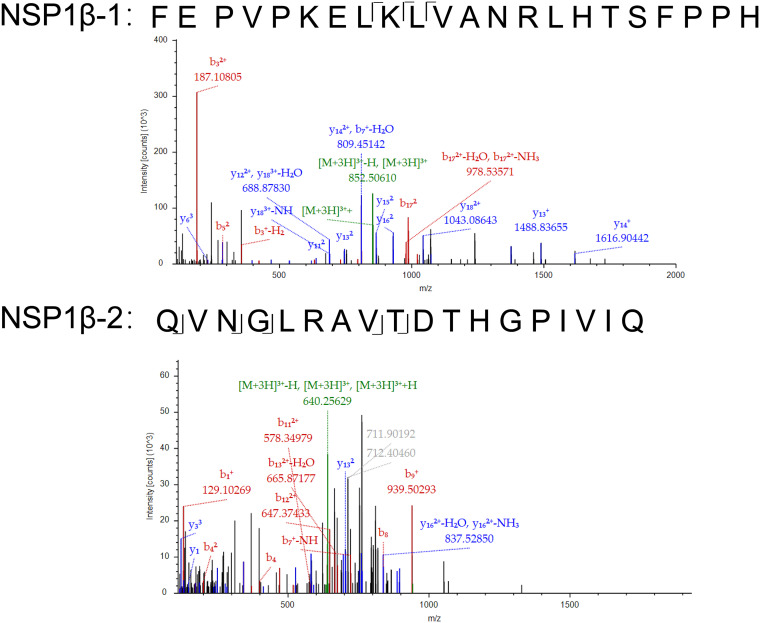
All the above-described data suggested that PRRSV infection promoted the functional expression of SLA-DR on the cell surfaces of BMDCs; however, it remained uncertain whether the SLA-DRs presented immunopeptides derived from either PRRSV NSPs or SPs to stimulate T cells (as a T epitope), which could potentially evoke the humoral immune response in vivo. To address this question, a mass spectrometry-based immunopeptidome analysis was conducted. The SLA-DRs from BMDCs infected by either PRRSV-2 JXA1 or PRRSV-1 GZ11 were extracted using membrane protein isolation kits to avoid contamination with cytoplasm proteins, and therefore, only membrane-bound SLA-DRs were further used for immunopeptide elution. Next, the immunopeptide–SLA-DR complex was enriched by anti-SLA-DR MAbs covalently conjugated to agarose beads, and immunopeptides presented by SLA-DR were further eluted for mass spectrometry (MS) analysis. To meet the minimum peptide requirement for MS analysis to ensure the credibility of the MS spectrum, trifluoroacetic acid (TFA)-eluted immunopeptide samples obtained from 3 parallel experiments using BMDCs originating from 3 piglets were pooled for MS analysis. The representative mass spectrum for the identified immunopeptides from NSP1β obtained from PRRSV JXA1-infected BMDCs is shown in Fig. 8. As listed in Table 1, more peptides derived from PRRSV NSPs were identified by mass spectrometry; these NSPs, such as NSP1, -2, and -7α, were reported to be especially highly immunogenic in PRRSV-2. For PRRSV-1 strain GZ11, four peptides located in the NSP2 region were identified, while peptides from NSP3, -8, -9, and -12 were identified as well (Table 2). In addition, peptides derived from certain structural proteins, such as GP5, matrix protein (M), N, and GP2 (for PRRSV-1 GZ11), were identified (Tables 1 and 2). Interestingly, differences were observed between PRRSV-1 and PRRSV-2 NSP- and SP-derived immunopeptides, which may partially explain the lack of cross-protection of the immune response between PRRSV-1 and PRRSV-2, as the PRRSV proteins presented by SLA-DR were different between the two genotypes of PRRSV. Taken together, these data suggest that PRRSV NSPs are presented by SLA-DR molecules, which may potentially stimulate CD4+ T cells to evoke PRRSV NSP-specific antibodies.
FIG 8.
Representative mass spectrometry spectra for immunopeptides originating from PRRSV NSP1β. Two typical mass spectrometry spectra for immunopeptide analysis are presented, based on the mass-to-charge ratios. The proposed amino acid sequences of the two spectra originated from PRRSV NSP1β.
TABLE 1.
Immunopeptide sequences identified from PRRSV-1 strain JXA1-infected BMDCs
| Peptidea | Amino acid sequence | PRRSV location |
|---|---|---|
| NSP1β-1 | FEPVPKELKLVANRLHTSFPPH | NSP1β |
| NSP1β-2 | QVNGLRAVTDTHGPIVIQ | NSP1β |
| NSP2-1 | RSGATTMVAHHASSA | NSP2 |
| NSP2-2 | PAKDPRMSPRESDESMIA | NSP2 |
| NSP3 | LTILWLVFFLISVN | NSP3 |
| NSP7α | IELAQLVQVDKVRGT | NSP7α |
| NSP10 | MHVEQGLTPLDPGRYQTRRGL | NSP10 |
| GP5 | VLDGSAATPLTRV | GP5 |
| M-1 | APQKVLLAFSITYTPV | M |
| M-2 | KQGVVNLVKYAK | M |
| N-1 | FSLPTQHTVRLIRATASPSA | N |
| N-2 | TVSFSLPTQHTVRLIRATASPSA | N |
Boldface indicates peptides used for further LACAs and ELISAs in this study.
TABLE 2.
Immunopeptide sequences identified from PRRSV-1 strain GZ11-G1-infected BMDCs
| Peptide | Amino acid sequence | PRRSV location |
|---|---|---|
| NSP2-1 | LPAAIVRNRACPNAK | NSP2 |
| NSP2-2 | FWTLDKMLTSPSPE | NSP2 |
| NSP2-3 | CPSSKQAMALLAKIKA | NSP2 |
| NSP2-4 | ANLDEKKISAQTV | NSP2 |
| NSP3 | DIHQYTSGPRGA | NSP3 |
| NSP8 | DLTASEVEKLKRIISQL | NSP8 |
| NSP9 | TPSMFTKHGTSKAAAEDL | NSP9 |
| NSP12 | DWRADLAVTPYDYGAQN | NSP12 |
| GP2 | LAVGNVSLQYNTTLDQV | GP2 |
| M-1 | KLVLAFSITYTPIMIYALKVSRGRLLGLLHIL | M |
| M-2 | HHVESAAGLHSIPA | M |
| N-1 | LIRVTSTSASQGAN | N |
| N-2 | IRVTSTSASQGAN | N |
PRRSV NSP immunopeptides eluted from SLA-DR could be recognized by serum samples obtained from PRRSV-infected piglets.
Immunopeptidome analysis clearly demonstrated that PRRSV NSP-derived immunopeptides were presented by SLA-DR molecules in PRRSV-infected BMDCs; however, it remained elusive whether these immunopeptides could activate CD4+ T cells to evoke an antibody response in vivo, which might serve as the major cause of nonneutralizing antibodies in the early stage of PRRSV infection. To verify this, PRRSV-positive antiserum samples (confirmed by Idexx ELISA and immunofluorescence assay [IFA]; data not shown) obtained from PRRSV JXA1-infected piglets at 21 dpi (the same strain used for the in vitro immunopeptidome analysis, with virus-neutralizing titers below 1:8) and PRRSV-negative antiserum samples were used for LACA analysis by using NanoLuc luciferase (NanoLuc)-fused immunopeptides. All cDNA sequences for NSP immunopeptides were synthesized and cloned to the 3′ end of the cDNA of NanoLuc and expressed in HEK-293T cells (Fig. 9A) according to the LACA protocol described previously (41). LACA analysis revealed that for immunopeptides originating from NSP1β, NSP2, and NSP10, the S/N ratios of serum samples obtained from PRRSV-infected piglets increased from 5- to 10-fold and demonstrated statistical significance compared to the results for negative controls (Fig. 9B), indicating that these immunopeptides truly evoke an antibody response in vivo. Meanwhile, although statistical significance among PRRSV-infected samples and noninfected samples in LACA was observed for NSP3 immunopeptides (Fig. 9B), the S/N ratio was too low (below 2) to confirm. Conversely, no significant difference or clearly evaluable S/N ratio was observed for the NSP7α immunopeptide by LACA. As a result, to further confirm the reactivity of these immunopeptides to PRRSV-positive antiserum samples, artificially synthesized NSP3 and NSP7α immunopeptides were used to coat ELISA plates for peptide ELISA. Based on the ELISA results, increased optical density (OD) values with statistical significance were observed in PRRSV-infected-pig serum samples for the NSP3 peptide, but not for NSP7α (Fig. 9C).
FIG 9.
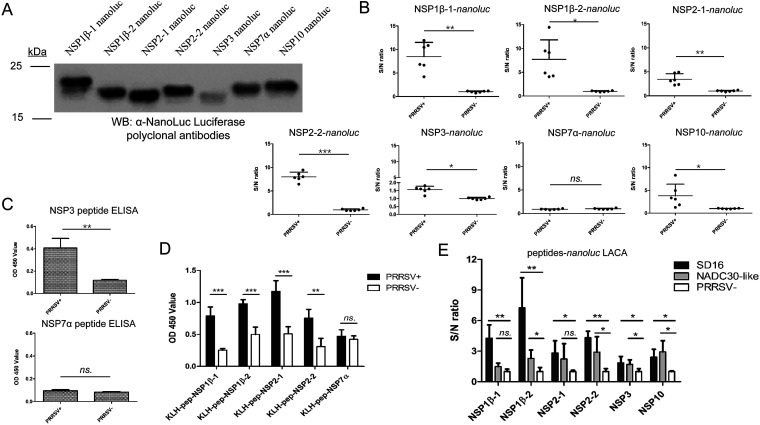
SLA-DR-presented NSP immunopeptides evoke antibody response in vivo. (A) cDNAs of PRRSV NSP immunopeptides from PRRSV strain JXA1 were cloned into the pCAGEN vector and fused with NanoLuc luciferase, followed by transfection of HEK-293T cells for transient expression. Cell lysates from 293T cells with transfection of indicated plasmids were harvested and subjected to Western blotting using rabbit anti-NanoLuc polyclonal antibodies to confirm the expression of NanoLuc-fused NSP immunopeptides. (B) Serum samples collected at 21 days postinoculation (dpi) from PRRSV JXA1-infected pigs (n = 6) and noninfected piglets (n = 6) were subjected to luciferase-linked antibody capture assay (LACA) using NanoLuc luciferase fused with indicated immunopeptides from different NSPs to identify corresponding antibody levels as determined by sample-to-negative (S/N) ratio. (C) Amounts of 1 μg of indicated artificial immunopeptides were used for coating ELISA plates. After blocking with 5% skim milk, indirect ELISA was conducted using 21-dpi serum from PRRSV JXA1-infected pigs (n = 6) or noninfected piglets (n = 6) (1-to-200 dilution in PBS), along with visualization by HRP-conjugated goat anti-swine IgG and TMB substrate. (D) Amounts of 400 ng of indicated KLH-conjugated artificial immunopeptides were used for coating ELISA plates. After blocking with 5% skim milk, indirect ELISA was conducted using 21-dpi serum from PRRSV JXA1-infected pigs (n = 6) or noninfected piglets (n = 6) (1 to 200 dilution in PBS), along with visualization by HRP-conjugated goat anti-swine IgG and TMB substrate. (E) Serum samples collected at 21 dpi from PRRSV SD16- and PRRSV NADC30-like strain-infected pigs (n = 5 for each) and noninfected piglets (n = 5) were subjected to LACA using NanoLuc luciferase fused with indicated immunopeptides from different NSPs (JXA1 strain) and evaluated by S/N ratio. OD 450, optical density at 450 nm. All data are expressed as mean values ± standard deviations and were subjected to Student’s t test. Error bars show standard deviations. Significant differences between indicated groups are marked as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant.
We noticed that it was difficult to coat ELISA plates with the free peptides, and amino acid sequence analysis suggested that these artificially synthesized immunopeptides are highly hydrophilic, except for NSP3. Therefore, to avoid a false-negative result from the NSP7α immunopeptide, artificial immunopeptides of NSP1β, NSP2, and NSP7α were conjugated to keyhole limpet hemocyanin (KLH) for ELISA. As shown by the results in Fig. 9D, all KLH-conjugated artificial immunopeptides from NSP1β and NSP2 could be recognized by 21-dpi serum from PRRSV JXA1-infected piglets with statistically significant increased OD values compared to the OD values for the negative samples. However, if the KLH-NSP7α immunopeptide was used to coat the plates, no significant change in OD values was observed between serum samples obtained from JXA1-infected-piglets and noninfected piglets. Taken together, these data demonstrated that all identified SLA-DR immunopeptides derived from the above-named NSPs except NSP7α (which may be a falsely discovered peptide by MS) were capable of evoking an antibody response in vivo, indicating that SLA-DR-mediated antigen presentation of PRRSV NSPs plays a key role in the host immune system to induce antibodies specific for PRRSV NSPs in the early stage of PRRSV infection.
We further sought to determine whether antibodies evoked by these NSP immunopeptides presented by SLA-DR are common or not during the natural PRRSV infection course in hosts. Therefore, serum samples were obtained from piglets inoculated with the PRRSV SD16 strain (an HP PRRSV strain similar to JXA1) and the PRRSV NADC30-like strain (the most divergent heterogenous PRRSV-2 strain) were subjected to LACA analysis for JXA1 NSP immunopeptides. Both types of serum samples contained low level of NAbs (virus-neutralizing titer below 1:8) for the corresponding strain. The deduced amino acid sequences for the corresponding peptides originating from the JXA1, SD16, and NADC30-like strains are listed in Table 3. As shown by the results in Fig. 9E, it appeared that the reactivity of PRRSV SD16 serum samples to JXA1 NSP immunopeptides (as determined by the S/N ratio) was similar to that of JXA1 in LACA, as the amino acid sequences for the corresponding NSP immunopeptides were nearly identical (Fig. 9E). In contrast, serum samples from NADC-30-like-infected piglets demonstrated reduced and variable reactivities to the immunopeptides of NSP1β-2, NSP2-1, and NSP2-2 of JXA1 in LACA due to amino acid sequence variation, although the reactivities to the NSP3 and NSP10 immunopeptides, with amino acid sequences identical to those of JXA1, were similar. Collectively, these data suggested that SLA-DR-mediated NSP presentation and the development of corresponding antibodies is a common phenomenon during natural PRRSV infection and is not strain specific.
TABLE 3.
Immunopeptide sequences from PRRSV strains SD16 and NADC30-like
| Immunopeptide | Strain | Amino acid sequencea |
|---|---|---|
| NSP1β-1 | JXA1 | FEPVPKELKLVANRLHTSFPPH |
| SD16 | FEPVPKELKLVANRLHTSFPPH | |
| NADC30-like | FETVPEELRLVAERLYTSFPPH | |
| NSP1β-2 | JXA1 | QVNGLRAVTDTHGPIVIQ |
| SD16 | QVNGLRAVTDTHGPIVIQ | |
| NADC30-like | QINGLRAVVDPTGPIVVQ | |
| NSP2-1 | JXA1 | RSGATTMVAHHASSA |
| SD16 | RSGATTMVAHHASSA | |
| NADC30-like | RASAITAVAGRAPSA | |
| NSP2-2 | JXA1 | PAKDPRMSPRESDESMIA |
| SD16 | PAKDPRMSPRESDESIIA | |
| NADC30-like | PAGEPLTQAPPASAGGVG | |
| NSP3 | JXA1 | LTILWLVFFLISVN |
| SD16 | LTILWLVFFLISVN | |
| NADC30-like | LTILWLVFFLISVN | |
| NSP10 | JXA1 | MHVEQGLTPLDPGRYQTRRGL |
| SD16 | MHVEQGLTPLDPGRYQTRRGL | |
| NADC30-like | MHVEQGLTPLDPGRYQTRRGL |
Underlining indicates amino acid variations from JXA1.
PRRSV NSP-derived immunopeptides evoked a nonneutralizing humoral response in vivo.
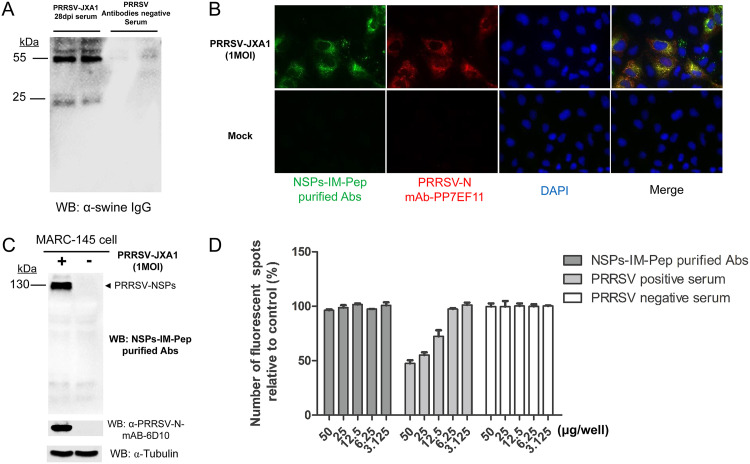
It has long been noticed that PRRSV-infected hosts rapidly developed a nonprotective antibody response, and most of these nonneutralizing antibodies are specific to recognize PRRSV NSPs (30, 32). The above-mentioned data suggested that SLA-DR-mediated antigen presentation of NSPs is responsible for evoking corresponding antibody responses in vivo; therefore, we sought to understand whether these NSP immunopeptide-specific antibodies are neutralizing or not against PRRSV. To confirm this, all artificial NSP immunopeptides except NSP7α were conjugated to CNBr-activated Sepharose 4B resin to purify corresponding antibodies from 28-dpi serum of PRRSV JXA1-infected pigs (containing both virus-neutralizing antibodies with virus-neutralizing titers of about 1:16 and NSP-specific antibodies) or serum from noninfected piglets. Our results demonstrated that swine IgG could only be detected from the NSP immunopeptide-conjugated Sepharose 4B resin elution of 28-dpi serum from PRRSV JXA1-infected pigs and not from PRRSV antibody-negative serum samples (Fig. 10A). Meanwhile, these purified NSP immunopeptide Abs pulled from 28-dpi serum were tested for their activity with PRRSV. In the IFA, when PRRSV-infected MARC-145 cells were coincubated with anti-N MAb PP7EF11 and purified NSP immunopeptide Abs, there was a costaining pattern for two types of antibodies, suggesting that these purified NSP immunopeptide Abs recognized PRRSV-infected cells (Fig. 10B). Moreover, Western blotting demonstrated that these purified NSP immunopeptide Abs recognized the 130-kDa protein (PRRSV NSP2 or its precursor) from PRRSV-infected cells as well (Fig. 10C). Furthermore, an in vitro virus-neutralizing assay was conducted to evaluate the protection by these purified NSP immunopeptide Abs against PRRSV. Protein G-purified total swine IgG from the same serum samples (28 dpi, with a neutralizing titer of 1:16) and noninfected piglets’ serum samples were included as the positive control and negative control, respectively, for comparison with purified NSP immunopeptide Abs in virus-neutralizing assays. Based on our data, little viral neutralization activity was observed in the negative-control groups or purified NSP immunopeptide Ab groups (Fig. 10D), while total IgG purified from the same 28-dpi serum samples demonstrated neutralizing activity for PRRSV JXA1 infection when 25 μg total IgG was used for incubation with virus. These data indicated that NSP-specific antibodies within the serum samples from PRRSV-infected piglets were not neutralizing against PRRSV, while the serum samples truly contained NAbs for PRRSV.
FIG 10.
SLA-DR-mediated antigen presentation of PRRSV NSPs evokes a nonneutralizing antibody response in vivo. (A) Swine serum (28 dpi) from PRRSV JXA1-infected swine or noninfected swine was used to incubate Sepharose 4B resin conjugated with mixed NSP immunopeptides listed in Table 1, except NSP7α, for 2 h at RT, followed by washing resin with PBS. Next, immunopeptide-specific antibodies were eluted and subjected to SDS-PAGE, and the existence of immunopeptide-specific antibodies was visualized using HRP-labeled goat anti-swine IgG. (B) MARC-145 cells were infected by PRRSV JXA1 at an MOI of 1 for 24 h. After cells were fixed and permeabilized, PRRSV-infected cells were costained by anti-PRRSV N protein MAb PP7EF11 and purified PRRSV NSP immunopeptide (IM-Pep) antibodies, followed by visualization via Alexa Fluor 555-labeled goat anti-mouse IgG(H+L) (red channel) and FITC-labeled goat anti-swine IgG (green channel). (C) MARC-145 cells were infected by PRRSV JXA1 at an MOI of 1 for 24 h and harvested for SDS-PAGE. PVDF membrane was blotted using purified PRRSV NSP immunopeptide swine antibodies or anti-PRRSV N MAb 6D10, followed by visualization via HRP-labeled goat anti-mouse IgG(H+L) or goat anti-swine IgG. (D) PRRSV JXA1 (MOI of 0.1) was preincubated with indicated doses of purified PRRSV NSP immunopeptide swine antibodies at 37°C for 1 h and then used to inoculate MARC-145 cells for 4 h. After removing antibodies and unbound virions by washing cells with PBS, virus-infected cells were incubated at 37°C for another 14 h. Replication of PRRSV was visualized by IFA using anti-PRRSV N MAb PP7EF11 for counting of fluorescent spots. All data are expressed as mean values ± standard deviations from at least three independent experiments.
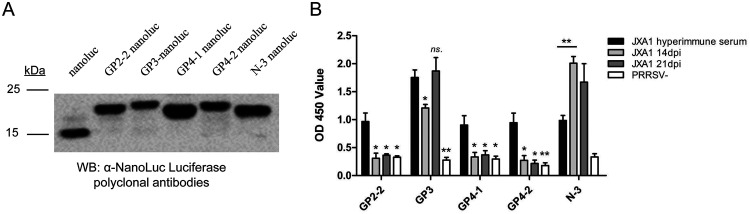
It is notable that no peptides were identified from other GPs of PRRSV, such as GP2, -3, and -4, in PRRSV JXA1-infected BMDCs, which was possibly caused by the internal limitations of Proteome Discoverer software (not suitable for peptide searching from glycolyzed protein such as PRRSV GPs). Therefore, the MS data were further analyzed using the more powerful PEAKS studio search engine to locate the potential immunopeptide sequences originating from GP2, -3, and -4. Based on our results, a total of 6 peptides originating from GP2, -3, and -4 were identified, along with another N protein-derived peptide (Table 4). Brief screening of these peptides suggested that only four of them (GP2-2, GP3, GP4-1, and GP4-2) demonstrated reactivity to PRRSV JXA1 hyperimmune serum (containing the full spectrum of both PRRSV NAbs and non-NAbs), while the remaining peptides could be falsely discovered by MS (data not shown) and did not show clear reactivity to PRRSV JXA1 hyperimmune serum. Since these peptide sequences originate from GPs that could be potential neutralizing targets, the NanoLuc-fused peptides were expressed and purified for ELISA (Fig. 11A). Both early serum samples (14 dpi and 21 dpi, with virus neutralization titers below 1:8) from PRRSV JXA1-infected piglets and hyperimmune serum standard samples (with a minimum virus neutralization titer of 1:32 for JXA1) developed against the same strain were used to evaluate their reactivities with these immunopeptides. As shown by the results in Fig. 11B, except for N- and GP3-derived peptides, immunopeptides identified from GP2 and GP4 were unable to be recognized by 14-dpi and 21-dpi serum samples but could be recognized by hyperimmune serum, implying that these immunopeptides potentially contain neutralizing B epitopes, although this requires further investigation. Taken together, these data suggested that the early humoral response developed against SLA-DR-presented NSP immunopeptides is unable to neutralize PRRSV and could be nonprotective against PRRSV in vivo.
TABLE 4.
Immunopeptide sequences originating from structural proteins identified in PRRSV-2 JXA1-infected BMDCsa
| Peptide | Amino acid sequence | PRRSV location |
|---|---|---|
| GP2-1 | ASLTKLANFLWML | GP2 |
| GP2-2 | KVSTLIDEMVSRRMYR | GP2 |
| GP3 | PLCPTRQAAAEILEPGK | GP3 |
| GP4-1 | MAASFLFLLVGF | GP4 |
| GP4-2 | TIRKISQCRTAIGTPV | GP4 |
| GP4-3 | RTAIGTPVYITITAN | GP4 |
| N-3 | AQQNQSRGKGPGKKNRKKNP | N |
Analyzed using PEAKS studio.
FIG 11.
PRRSV structural protein-derived immunopeptides could be recognized by PRRSV hyperimmune serum but not early serum samples from PRRSV-infected pigs. (A) cDNAs of PRRSV structural protein (SP) immunopeptides from PRRSV strain JXA1 were synthesized and ligated into the pET-28a-His-NanoLuc expression vector to fuse with NanoLuc luciferase, followed by expression in E. coli cells. After dialysis and purification, 10 μg of each recombinant protein was subjected to SDS-PAGE and Western blot analysis using rabbit anti-NanoLuc polyclonal antibodies to confirm the expression. (B) Amounts of 200 ng of indicated NanoLuc-fused immunopeptides were used for coating ELISA plates. After blocking with 5% skim milk, indirect ELISA was conducted using 14-dpi or 21-dpi serum from PRRSV JXA1-infected pigs (n = 4) or PRRSV hyperimmune serum samples (n = 4) (20-fold dilution in PBS), along with visualization by HRP-conjugated goat anti-swine IgG and TMB substrate. OD 450, optical density at 450 nm. All data are expressed as mean values ± standard deviations and were subjected to Student’s t test. Significant differences between indicated groups are marked as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant.
Antigen presentation of PRRSV NSP-derived immunopeptides occurred in vivo in PAMs and hilar lymph nodes during the early stage of PRRSV infection.
Since the above-described immunopeptide analysis for NSPs in PRRSV-infected APCs was conducted in an in vitro BMDC model, to further strengthen these in vitro data, a preliminary in vivo study was conducted by infecting two piglets with the PRRSV JXA1 strain to harvest APCs. Unfortunately, one piglet died 6 days after virus challenge due to the virulence phenotype of JXA1. The PAMs, peripheral blood mononuclear cells (PBMCs), and hilar lymph nodes were harvested 10 days after challenge from the remaining piglet as in vivo APCs for immunopeptide analysis. However, expression of SLA-DR was undetectable in freshly isolated PBMCs from the PRRSV-infected piglet (data not shown), and thus, the PBMCs were not applicable for enriching SLA-DR complexes. As a result, the PAM (in vivo target) and hilar lymph node (primary sites for antigen presentation after PRRSV infection) samples were subjected to plasma membrane isolation, enrichment of SLA-DR–peptide complexes, and MS-based immunopeptide analysis. To increase the credibility of MS analysis, PEAKS studio software was applied for searching peptides. Based on our result, the majority of peptides identified from SLA-DR complexes from in vivo-isolated PAMs and hilar lymph nodes originated from NSPs (Tables 5 and 6). Only a limited number of peptides could be identified even using the PEAKS search engine (Tables 5 and 6), which could be a consequence of low percentages of PRRSV-positive cells in hilar lymph nodes, as evidenced by IFA of cryostat sections of hilar lymph node samples using MAb-recognized PRRSV N (data not shown). Even so, it is notable that certain NSP immunopeptides identified in vivo share similar locations with these verified NSP immunopeptides identified in vitro, such as NSP1β-1, NSP2-1, NSP2-2, and NSP10. Therefore, although preliminary, these in vivo observations not only confirmed that antigen presentation of PRRSV NSPs by SLA-DR occurs in vivo, as evidenced by elution of corresponding immunopeptides from APCs or lymph organs isolated from PRRSV-infected piglets, but also provided proof-of-concept data demonstrating the applicability of BMDCs combined with LACA or ELISA as an in vitro model for screening PRRSV-derived immunopeptides evoking antibody responses in vivo.
TABLE 5.
Immunopeptide sequences identified in vivo from PAMs of PRRSV-2 JXA1-infected pigleta
| Viral protein | Amino acid sequence | Similar peptide identified in vitro |
|---|---|---|
| NSP1α | WLSAIFPIARMTSGNLNF | None |
| NSP1β | VKFEPVPKELKLVANRLHTSFPPH | NSP1β-1 |
| NSP2 | PSEPMTPMSEPVLVPASR | None |
| NSP2 | DPATQEWLSRMWDRVDMLTWRNTS | None |
| NSP2 | KDPRMSPRESDESMIAPP | NSP2-2 |
| NSP10 | LPAKGTPVNLAVP | None |
| GP2 | EATLSRISGLDVV | None |
| GP3 | PTRQAAAEILEPGK | GP3 |
| N | KIIAQQNQSRGKGPGKKNRKKNP | N |
Analyzed using PEAKS studio.
TABLE 6.
Immunopeptide sequences identified in vivo from hilar lymph nodes of PRRSV-2 JXA1-infected pigletsa
| Viral protein | Amino acid sequence | Similar peptide identified in vitro |
|---|---|---|
| NSP2 | SGATTMVAHHASSAHETRQATKH | NSP2-1 |
| NSP2 | KDPRMSPRESDESMIAPPAD | NSP2-2 |
| NSP2 | LEECLAKLERVSP | None |
| NSP3 | GPGSLCTSRLCISQHG | None |
| NSP7α | DPTPAPPPVPVPIPLP | None |
| NSP10 | DPGRYQTRRGLVSVR | NSP10 |
| GP2 | EATLSRISGLDVV | None |
| GP3 | CPTRQAAAEILEPGKS | GP3 |
Analyzed using PEAKS studio.
In conclusion, our data demonstrated that PRRSV infection interferes with the MHC-II molecule (SLA-DR)-mediated antigen presentation process in professional APCs via a functional upregulation of SLA-DR, which is due to inhibition of ubiquitin-proteasome degradation of SLA-DR molecules by the PRRSV NSP2 OTU domain. Notably, SLA-DR presented immunopeptides from PRRSV NSPs, evoking an antibody response that potentially contributed to the rapid induction of nonneutralizing antibodies at an early stage during PRRSV infection in vivo.
DISCUSSION
In recent years, researchers have believed that PRRSV-specific neutralizing antibodies (NAbs) are an essential component of adaptive immunity against PRRSV (31, 46), which was evidenced by the fact that the inactivated PRRSV vaccines failed to induce NAbs and were not protective (31, 46). Meanwhile, antibody kinetics suggested that the onset of NAbs after experimental infection correlated with viral clearance from circulation and tissues (17, 47). Moreover, passive transfer of PRRSV NAbs protected pregnant sows against reproductive failure and conferred sterilizing immunity in herd and offspring in a dose-dependent manner (48). Therefore, all these observations underline the importance of the protective antibody response in PRRSV prevention.
During the natural course of PRRSV infection in pigs, PRRSV-specific NAbs typically appear 28 days postinoculation (30, 31), comparatively later than the appearance of specific NAbs in porcine epidemic diarrhea virus (PEDV), which is a member of Nidovirales, like PRRSV, but which induces neutralizing antibodies within 2 weeks after infection (49). Besides the later onset of NAbs, it is notable that nonneutralizing antibodies (non-NAbs) against PRRSV N proteins and other NSPs can be rapidly induced in the early stage of PRRSV infection regardless of PRRSV infection or MLV immunization (30, 32). However, the mechanism behind this remains undiscovered due to technical difficulties, such as lacking enough surface markers to isolate in vivo immune cells (for instance, peripheral DCs) for in vitro investigation or evaluation of T or B cell activation in swine species.
Induction of PRRSV-specific antibodies (either NAbs or non-NAbs) requires activation of antigen-specific B cells with assistance from CD4+ T helper cells (35, 36), which relies on TCR-based recognition of SLA-II-mediated antigen presentation by APCs. Considering the natural role SLA-II molecules played during the development of humoral immunity and rapid onset of non-NAbs against PRRSV in vivo, it is unlikely that PRRSV infection should impair the function of SLA-II in APCs, which appears to be different from that of SLA-I (28, 29). In fact, upregulation of SLA-DR was observed in in vitro-generated BMDCs after PRRSV infection (42). In this study, further investigation revealed that infection of heterogeneous isolates from either PRRSV-1 or PRRSV-2 in BMDCs promoted the surface expression of SLA-DR while it inhibited SLA-DR at the mRNA level, as evidenced by Western blotting and fluorescence-activated cell sorting (FACS) analysis for SLA-DR protein expression and transcriptome and qPCR analysis of the mRNA. Notably, SLA-DR promoted by PRRSV infection occurred at the posttranslational level, which is mainly due to the extended protein half-life of SLA-DR in PRRSV-infected BMDCs. Furthermore, our data also indicated that the PRRSV NSP2 ovarian tumor (OTU) domain (a well-defined deubiquitinating enzyme) is responsible for inhibiting ubiquitination-mediated protein degradation of SLA-DR, which leads to an extended protein half-life and the accumulation of SLA-DR in PRRSV-infected BMDCs.
The downregulation of SLA-I molecules by PRRSV infection in vitro correlated with delayed, low-level in vivo cytotoxic T lymphocyte (CTL) responses in PRRSV-infected pigs (28, 29). The biological significance of upregulated SLA-DR levels in PRRSV-infected BMDCs or monocyte-derived dendritic cells is less characterized. Theoretically, impaired SLA-II function should result in a delayed antibody response in vivo regardless of whether the antibodies stimulated by PRRSV are NAbs or non-NAbs. Therefore, we proposed that SLA-DR upregulation on the cell surface of BMDCs may correlate with rapid induction of PRRSV-specific non-NAbs, especially for non-NAbs specific for PRRSV NSPs. To confirm this, a mass spectrometry-based immunopeptidome analysis was conducted, and immunopeptides derived from PRRSV NSPs were identified in SLA-DR–peptide complexes from either PRRSV-1- or PRRSV-2-infected BMDCs. Notably, the majority of these immunopeptides originated from PRRSV NSPs such as NSP1, -2, -3, -7, and -10, which are confirmed to be highly immunogenic by previous studies (33, 34). Notably, by employing immunopeptide-based LACA and ELISA to probe serum samples obtained from PRRSV-infected piglets, a direct correlation between SLA-DR-presented NSP immunopeptides and the existence of corresponding antibodies was observed. Most importantly, certain immunopeptides recognized by PRRSV-infected-pig serum were identified from PAMs and hilar lymph node samples isolated from PRRSV-infected piglets as well. Therefore, our study provides streamlined evidence demonstrating that SLA-DR-mediated antigen presentation of PRRSV NSPs in PRRSV-infected APCs is responsible for the induction of antibodies against PRRSV NSPs during the early stage of PRRSV infection.
As the peptide sequences identified from mass spectrometry in this study are relatively few in number due to the fact that Proteome Discoverer software does not support searching of PRRSV immunopeptides bearing point mutations generated from quasispecies virus or peptides bearing amino acid modifications resulting from posttranslational modification such as glycosylation, we are currently working on de novo analysis of available MS data using the PEAKS engine to identify more PRRSV-derived immunopeptide sequences. It is worth noting that preliminary searching based on PEAKS identified immunopeptides originating from GP2, -3, and -4, which are minor envelope proteins of PRRSV but form heterotrimers in the virion and are required for interaction with CD163 (50, 51), the essential cellular receptor for PRRSV infection in vitro and in vivo (52, 53). The ELISA results suggested that antibodies recognizing immunopeptides of GP2, -3, and -4 mainly existed in PRRSV hyperimmune serum with higher virus-neutralizing activity and not in early serum samples of PRRSV-infected piglets (low virus-neutralizing activity, before 21 dpi). These results were consistent with a previous observation in PRRSV-1 which demonstrated that GP2, -3, and -4 contain neutralizing epitopes (54). Moreover, a recent study screening for conserved neutralizing epitopes from GP2, -3, and -4 ectodomains suggested that conserved regions located in GP2, -3, and -4 ectodomains were unable to evoke NAbs in vivo (55). However, none of the immunopeptides originating from GP2, -3, and -4 characterized here fall in previously analyzed conserved ectodomains of GP2, -3, and -4. In contrast, the traditional conception of the GP5-M heterodimer as a major neutralizing target of PRRSV has frequently been challenged in recent years, although exceptions have been reported (54, 56, 57). Therefore, an MS-based immunopeptide analysis technique combined with feasible software like PEAKS, along with in vivo verification, may provide novel tools for understanding PRRSV neutralization and mapping corresponding epitopes.
To date, the antigen presentation process in PRRSV-infected APCs regarding PRRSV proteins remains elusive. Based on the pathways of MHC class I and II molecule-mediated antigen presentation, immunopeptide-loaded MHC class II molecules (peptide–MHC-II complexes) are constitutively expressed on the surface of APCs (including dendritic cells, B cells, macrophages, and thymic epithelial cells) and are presented to antigen-specific CD4+ T cells (58). Briefly, exogenous antigens are internalized by several pathways, including phagocytosis, macropinocytosis, and endocytosis, and eventually traffic to a mature or late endosomal compartment where they are processed and loaded onto MHC-II molecules (58, 59). As endogenously synthesized cytosolic proteins, PRRSV NSPs should be processed primarily by the action of the proteasome to generate shorter peptides, which are then transported into the endoplasmic reticulum (ER) by TAP (transporter associated with antigen processing) for subsequent assembly with MHC-I molecules (58, 59) and are then recognized by cytotoxic T cells to evoke cell-mediated immunity. However, our data suggested that PRRSV NSP-derived immunopeptides were assembled to MHC class II molecules (SLA-DR) to stimulate the B cell response during PRRSV infection by unknown pathways. Conversely, available data suggested that autophagy, a highly regulated process originally considered to recycle unnecessary or dysfunctional components for cellular homeostasis, is involved in engulfing cytosolic macromolecules and organelles to form autophagosomes; this is followed by membrane fusion with lysosomal compartments to form autophagolysosomes and, finally, the generation of peptide–MHC-II complexes (58, 59). Several reports had confirmed that PRRSV infection promotes autophagy in infected cells both in vitro and in vivo (60–62), which is consistent with our preliminary findings that PRRSV infection in BMDCs induced autophagy, as evidenced by the enhanced conversion of LC3-I to LC3-II (data not shown). It is possible that in vivo PRRSV infection in APCs such as DCs will lead to enhanced autophagy in order to promote the loading of cytosolic PRRSV NSP-originated immunopeptides to SLA-II-mediated antigen presentation pathways and finally evoke an antibody response against PRRSV NSPs in the early stage of PRRSV infection, but this speculation requires further investigation.
In conclusion, our study demonstrated that PRRSV infection of in vitro-generated BMDCs promotes functional SLA-DR upregulation on the cell surface to present PRRSV NSP-derived immunopeptides. SLA-DR-mediated presentation of PRRSV NSP-derived immunopeptides could be observed from in vivo-isolated APCs (PAMs or APCs in lymph node tissue) as well. These discoveries are consistent with the induction of the corresponding non-NAbs against PRRSV in vivo. Our results not only reveal a novel mechanism of interference in the host antigen presentation process by PRRSV but also provide novel insight for understanding the rapid induction of nonneutralizing antibodies against PRRSV NSPs in vivo. Moreover, based on the literature, our study provides proof-of-concept evidence indicating that MS-based SLA-DR immunopeptidome analysis is feasible in swine species to analyze MHC-II-mediated antigen presentation for swine pathogens. Employing such a technique combined with applicable antibody screening methods could be used to monitor the humoral immune response in vivo, which may serve as a novel path for future development of vaccines against swine pathogens.
MATERIALS AND METHODS
Cells, viruses, chemicals, and plasmids.
MARC-145 and HEK-293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Biological Industries, Israel) supplemented with 10% fetal bovine serum (FBS; Biological Industries). Porcine bone marrow-derived dendritic cells (BMDCs) were obtained as previously described by stimulating bone marrow cells with porcine granulocyte-macrophage colony-stimulating factor (GM-CSF; GenScript, Nanjing, Jiangsu, China) to a final concentration of 40 ng/ml for 6 consecutive days, and only the suspended cells (with typical dendritic cell morphology) were collected at day 7 for further experiments (42). Porcine alveolar macrophages (PAMs) were collected from 4-week-old PRRSV-negative pigs as previously described (63). PAMs were maintained in RPMI 1640 medium (Biological Industries) supplemented with 10% FBS (Biological Industries).
PRRSV strain VR2385 (GenBank accession number JX044140.1) was recovered from the infectious clone pIR-VR2385-CA (64) by transfecting the plasmid into MARC-145 cells using FuGENE HD transfection reagent (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Other PRRSV virus isolates used in this study were VR2332 (GenBank accession number EF536003.1) and highly pathogenic PRRSV (HP PRRSV) isolates, including SD16 (GenBank accession number JX087437.1), HuN4 (GenBank accession number EF635006.1), JXA1 (GenBank accession number EF112445.1), NADC30-like HNhx (GenBank accession number KX766379), and PRRSV-1 Chinese isolate GZ11-G1 (GenBank accession number KF001144.1).
All PRRSV isolates were used to inoculate BMDCs or MARC-145 cells at multiplicities of infection (MOI) as indicated in the figures and figure legends. The median tissue culture infectious dose (TCID50) of each PRRSV isolate (except the NADC30-like isolate HNhx) was titrated in MARC-145 cells as previously described (65). Inoculation and titration of the NADC30-like isolate HNhx was conducted in PAMs.
All PRRSV NSPs and the NSP2 ovarian tumor (OTU) domain were cloned from PRRSV SD16 infectious clones, and PRRSV NSPs were ligated to the VenusC1 vector as previously described (66), while the NSP2 OTU domain was ligated to the pCAGEN vector fused with a hemagglutinin (HA) tag. Construction of pCAGEN-SLA-DRα was previously described (42). The cDNA sequence of SLA-DRβ were cloned from total PAM cDNA and ligated to the pCAGEN vector without any tag. The primers and corresponding sequences used for plasmid construction are listed in Tables 7 and 8, respectively. The transfection of indicated plasmids into HEK-293T cells was conducted using FuGENE HD transfection reagent (Promega) according to the manufacturer’s instructions.
TABLE 7.
List of primers used to clone PRRSV SD16 NSPs into VenusC1 expression vector
| Primers | Sequence (5′–3′)a | Description |
|---|---|---|
| NSP1α-F | CGCTCGAGATTCTGGGATACTTGATC | Cloning of NSP1α |
| NSP1α-R | GAATTCTTACATAGCACACTCAAAAGG | Cloning of NSP1α |
| NSP1β-F | CGCTCGAGATGCTGACGTCTATGACA | Cloning of NSP1β |
| NSP1β-R | GAATTCTTAACCGTACCACTTATGAC | Cloning of NSP1β |
| NSP2-F | CGAAGCTTATGCCGGAAAGAGAGCAA | Cloning of NSP2 |
| NSP2-R | GGATCCTTACCCGCCCAGTAACCTGC | Cloning of NSP2 |
| NSP3-F | CGCTCGAGATGCACGCTACATCTGGC | Cloning of NSP3 |
| NSP3-R | GAATTCTTACTCAAGGAGGGACCCG | Cloning of NSP3 |
| NSP4-F | CGCTCGAGATGGTGCTTTCAGAACTC | Cloning of NSP4 |
| NSP4-R | AAGCTTTTATTCCAGTTCGGGTTTGG | Cloning of NSP4 |
| NSP5-F | CGCTCGAGATGGAGGCCTTTCTACAG | Cloning of NSP5 |
| NSP5-R | GAATTCTTACTCAGCAAAGTATCGC | Cloning of NSP5 |
| NSP7α-F | CGCTCGAGATTCGCTGACTGGTGCCC | Cloning of NSP7α |
| NSP7α-R | GAATTCTTACTCCAGAACTTTCGGT | Cloning of NSP7α |
| NSP7β-F | CGCTCGAGATAACGGTCCCAATGCCT | Cloning of NSP7β |
| NSP7β-R | GGATCCTTATTCCCACTGAGCTCTTC | Cloning of NSP7β |
| NSP8-F | CGCTCGAGATGCCGCCAAGCTTTCCG | Cloning of NSP8 |
| NSP8-R | GAATTCTTACTAGCAGTTTAAACACT | Cloning of NSP8 |
| NSP9-F | CGCTCGAGATCTAGCCGCCAGCGGCT | Cloning of NSP9 |
| NSP9-R | AAGCTTTTACTCATGATTGGACCTGA | Cloning of NSP9 |
| NSP10-F | CGCTCGAGATGGGAAGAAGTCCAGAA | Cloning of NSP10 |
| NSP10-R | GGATCCTTATTCCAGATCTGCGCAAAT | Cloning of NSP10 |
| NSP11-F | CGCTCGAGATGGGTCGAGCTCCCCGC | Cloning of NSP11 |
| NSP11-R | GAATTCTTATTCAAGTTGAAAATAGG | Cloning of NSP11 |
| NSP12-F | CGCTCGAGATGGCCGCCATTTTACCT | Cloning of NSP12 |
| NSP12-R | GAATTCTTATCAATTCAGGCCTAAAG | Cloning of NSP12 |
Boldface indicates sequences that overlap with the corresponding NSPs, and italics indicate restriction enzyme cleavage sites.
TABLE 8.
List of primers used in this study
| Primer | Sequence (5′–3′)a | Description |
|---|---|---|
| NSP2-OTU-F | CGCGTCTCGAATTCAAGAGAGCAAGGAAAACA | Cloning of NSP2 OTU |
| NSP2-OTU-R | CGCGTCTCCTCGAGAACGGTTGGAGTCGTCGG | Cloning of NSP2 OTU |
| NSP1β-1-nanoluc-F | GGCGGTGGAGGCAGTGAATTCTTCGAGCCCGTGCCCAAG | Cloning of NSP1β-1–NanoLuc |
| NSP1β-1-nanoluc-R | CTGCACCTGAGGAGTGCGGCCGCTTAGTGGGGGGGGAAG | Cloning of NSP1β-1–NanoLuc |
| NSP1β-2-nanoluc-F | GGCGGTGGAGGCAGTGAATTCCAGGTGAACGGGCTGAGA | Cloning of NSP1β-2–NanoLuc |
| NSP1β-2-nanoluc-R | CTGCACCTGAGGAGTGCGGCCGCTTACTGGATCACGATG | Cloning of NSP1β-2–NanoLuc |
| NSP2-1-nanoluc-F | GGCGGTGGAGGCAGTGAATTCAGAAGCGGCGCCACCACC | Cloning of NSP2-1–NanoLuc |
| NSP2-1-nanoluc-R | CTGCACCTGAGGAGTGCGGCCGCTTAGGCGCTACTAGCG | Cloning of NSP2-1–NanoLuc |
| NSP3-nanoluc-F | GGCGGTGGAGGCAGTGAATTCCTGACCATCCTGTGGCTG | Cloning of NSP3–NanoLuc |
| NSP3-nanoluc-R | CTGCACCTGAGGAGTGCGGCCGCTTAGTTCACGGAGATC | Cloning of NSP3–NanoLuc |
| NSP7α-nanoluc-F | GGCGGTGGAGGCAGTGAATTCATCGAGCTGGCCCAGCTG | Cloning of NSP7α–NanoLuc |
| NSP7α-nanoluc-R | CTGCACCTGAGGAGTGCGGCCGCTTAGGTTCCTCTCACC | Cloning of NSP7α–NanoLuc |
| NSP10-nanoluc-F | GGCGGTGGAGGCAGTGAATTCCACGTGGAGCAGGGGCTG | Cloning of NSP10–NanoLuc |
| NSP10-nanoluc-R | CTGCACCTGAGGAGTGCGGCCGCTTACAGCCCCCTTCTG | Cloning of NSP10–NanoLuc |
| Tubulin-F | GGCTGTCTGCGATATCCCTC | For qPCR detection |
| Tubulin-R | TGCTCTGAGATGCGCTTGAA | For qPCR detection |
| SLA-DRα-F | GCCCTGAAGCCACTCTAA | For qPCR detection |
| SLA-DRα-R | GGAAAGCCAGCACAAGAA | For qPCR detection |
| SLA-DRβ-F | GAGGGCACGGTCTGAATC | For qPCR detection |
| SLA-DRβ-R | AGGGCGTCCTTTCTGATT | For qPCR detection |
Boldface indicates sequences that overlap with the corresponding NSPs, and italics indicate restriction enzyme cleavage sites.
Ethics statement and animal studies.
Six-week-old BALB/c mice were obtained from Dashuo Biotech (Chengdu, Sichuan, China), and the animal protocol was reviewed and approved by the Animal Welfare Committee of Northwest A&F University. All mice were monitored on a daily basis for any clinical signs. All effort was made to minimize the suffering of mice, and euthanasia was performed if a humane endpoint was reached according our protocol.
Purification of PRRSV SD16 virions from cell culture media obtained from PRRSV-infected MARC-145 cells was performed as previously described (67). Preparation of mouse serum against inactivated PRRSV virions was previously described (68). Serum samples of immunized mice were evaluated by using the Idexx PRRS X3 enzyme-linked immunosorbent assay (ELISA) kit by replacing the original horseradish peroxidase (HRP)-labeled anti-swine antibodies with HRP-labeled anti-mouse IgG (Thermo Fisher Scientific) in order to determine the serum conversion as previously described (68).
Preparation of mouse sera against various PRRSV proteins and SLA-DRβ was conducted with protocols as previously described (42, 67). Briefly, the cDNAs of PRRSV NSP2, NSP9, and GP5 were amplified from the infectious clone of the PRRSV SD16 strain and ligated to the pET-28a vector (69). The cDNA of SLA-DRβ was ligated to the pET-28a vector fused with a 6×His tag as well. The pET-28a vectors encoding indicated proteins mentioned above were transformed into Escherichia coli strain BL21(DE3) and cultured in LB medium at 37°C until they were induced with 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG) at 25°C. After IPTG induction, bacterial cells were collected and resuspended in cell lysis buffer for sonication as previously described (42). All recombinant proteins were expressed as inclusion bodies, washed with phosphate-buffered saline (PBS) three times, and then reconstituted in 8 M urea (Sigma-Aldrich). All proteins were further purified using Ni+ affinity chromatography (Transgene, Beijing, China) and eluted by elution buffer for dialysis as previously described (42). Dialysis of recombinant proteins was conducted by a gradient decrease of the urea concentration until the buffer was completely replaced by PBS or reached the minimum urea concentration allowed without visible protein precipitation during dialysis. All purified recombinant proteins were quantified by using a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific) and stored at −80°C until immunization or ELISA plate coating.
Mice were immunized with 100 μg recombinant 6×His-tagged recombinant proteins (2 mg/ml) with an equal volume of adjuvant for a total of three times with 2-week intervals between immunizations. Freund’s complete adjuvant (Sigma-Aldrich) was used for the primary immunization, while Freund’s incomplete adjuvant (Sigma-Aldrich) was used for the rest. Mouse sera were collected on a weekly basis to monitor the antibody levels by ELISA. Serum collected before immunization was included as a negative control.
Four-week-old piglets were obtained from a PRRSV-free pig farm near Yangling, Shaanxi, and further subjected to screening for classical swine fever virus (CSFV), PRRSV, porcine circovirus 2 (PCV2), and African swine fever virus (ASFV), along with corresponding antibodies, by a government-authorized agency (Shaanxi Innolever Biotechnology Co., Ltd., Yangling, Shaanxi, China). Only piglets (n = 2) negative for all examined pathogens and antibodies against PRRSV and ASFV were selected. The animal protocol for the PRRSV challenge experiment was reviewed and approved by the Animal Welfare Committee of Northwest A&F University. Piglets were inoculated intranasally and intramuscularly with 1 ml 1 × 105 TCID50/ml PRRSV JXA1 viral stock. The piglets were monitored on a daily basis for clinical signs. All effort was made to minimize suffering, and euthanasia was performed if a humane endpoint was reached according our protocol. One piglet died at day 6 after PRRSV JXA1 challenge due to the virulence of this HP PRRSV. The surviving piglet was euthanized at 10 days after challenge for peripheral blood mononuclear cell (PBMC), PAM, and hilar lymph node collection.
LACA.
Luciferase-linked antibody capture assays (LACA) were conducted as previously described (41). Briefly, HEK-293T cells were transfected with plasmids coding for Renilla luciferase-fused PRRSV N or NSP1α to obtain the cell lysates for LACA as previously described (41). Black 96-well polystyrene microplates (Corning, Inc., Corning, NY, USA) were coated with 1 μg of recombinant protein G (Smart-Lifesciences, Changzhou, Jiangsu, China) at 4°C overnight, followed by blocking with PBS-T buffer containing 2.5% gelatin (Sigma-Aldrich). Then, 20-fold-diluted serum samples and PBS-diluted HEK-293T cell lysates containing 1 testing unit (TU) Renilla luciferase-fused PRRSV N or PRRSV NSP1α at a final volume of 100 μl were incubated in protein G-coated polystyrene microplates for 2 h at room temperature (RT). Luciferase activities within cell lysates were evaluated using Renilla luciferase substrate (Transgene Biotech) according to the manufacturer’s instructions and quantified using a Victor X multilabel reader (Perkin-Elmer, Wellesley, MA, USA).
In order to increase the sensitivity of LACA for immunopeptides, Renilla luciferase was replaced by NanoLuc luciferase; the cDNA sequence (GenBank accession number KY563063.1) of NanoLuc was synthesized by Tsingke Biological Technology (Beijing, China) and ligated in the pCAGEN vector. Additionally, the cDNA sequences encoding immunopeptides derived from PRRSV NSPs were cloned from cDNA by reverse transcription of RNA obtained from PRRSV JXA1-infected BMDCs and fused to the 3′ end of NanoLuc cDNA. The primers and corresponding DNAs for NanoLuc-fused NSP immunopeptides are listed in Table 8. For NanoLuc LACA, cell lysates from 1 × 106 HEK-293T cells transfected with the indicated plasmids were applied and analyzed using the same Renilla luciferase substrate (Transgene Biotech) and by following the same procedures as for Renilla luciferase-LACA.
Western blot analysis.
Whole-cell lysates of BMDCs, PAMs, HEK-293T cells, and MARC-145 cells were harvested using 1× Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA, USA) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (70). The separated proteins were then transferred to polyvinylidene difluoride (PVDF) membranes as previously described (71). Membranes were probed with swine serum (1:200 dilution in TBS), mouse sera developed against PRRSV GP5, NSP2, NSP4, and NSP9 (1:200 dilution in TBS), homemade MAb 6D10 against PRRSV N protein, homemade MAb against SLA-DRα, homemade polyclonal antibody against SLA-DRβ, anti-GFP MAb (Rockland Immunochemicals, Limerick, PA, USA), anti-Myc MAb (Transgene), anti-β-tubulin MAb (Transgene), and antiubiquitin MAb (Santa Cruz Biotech, Santa Cruz, CA, USA). Specific binding between antibodies and their targets was detected using HRP-conjugated goat anti-swine IgG (Jackson ImmunoResearch) or HRP-conjugated goat anti-mouse IgG (Thermo Fisher Scientific) and visualized using enhanced chemiluminescence (ECL) substrate (Bio-Rad Laboratories). Chemiluminescence signal acquisition was conducted using a ChemiDoc MP imaging system (Bio-Rad Laboratories) and analyzed using Image Lab software (version 5.1; Bio-Rad Laboratories).
For Western blot detection of NanoLuc-fused immunopeptides, the cDNA sequence synthesis, bacterial expression of NanoLuc, and rabbit immunization and antibody purification were conducted by GenScript (Nanjing, China). The purified rabbit anti-NanoLuc polyclonal antibodies were used at a concentration of 1 μg/ml for blotting.
To determine the SLA-DR protein half-life, cycloheximide (Sigma-Aldrich) was used to treat BMDCs at a final concentration of 200 ng/ml to inhibit universal protein translation with or without PRRSV infection. Cell lysate samples were harvested at indicated time points after cycloheximide treatment for Western blotting.
RNA isolation, transcriptomic analysis, and qPCR.
Total RNA was extracted from BMDCs or PAMs using TRIzol reagent (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. Transcriptomic analysis of BMDCs with or without PRRSV infection was conducted by Genewiz (Suzhou, Jiangsu, China), with three biological duplicates representing three independent experiments. Reverse transcription and quantitative real-time PCR (qPCR) were conducted using a PrimeScript RT reagent kit (TaKaRa, Dalian, China) and 2× RealStar power SYBR mixture (Genstar, Beijing, China), respectively, following the manufacturer’s instructions. Transcripts of tubulin were amplified from the same cDNA to normalize total RNA input. Primers used for qPCR and corresponding DNA sequences are listed in Table 8. Relative quantification of target genes was calculated using the cycle threshold (2−ΔΔCT) method.
IP and protein ubiquitination assay.
The immunoprecipitation (IP)-based protein ubiquitination assay for SLA-DR was conducted as previously described with modifications (66). Briefly, 1 × 107 BMDCs infected with PRRSV JXA1 or mock-infected cells were harvested with 200 μl radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) supplemented with protease inhibitor cocktail (Sigma-Aldrich) and N-ethylmaleimide (NEM; Sigma-Aldrich) to a final concentration of 50 μM. The lysate was clarified by centrifugation at 14,000 × g for 5 min at 4°C. Next, 3 μg of commercial anti-SLA-DR MAb (BD Bioscience, San Jose, CA, USA) was added to the cell lysate and incubated at 4°C for 3 h to enrich SLA-DR; 25 μl of protein G-agarose beads (Genscript) was then added to the cell lysate and incubated for another hour to pull down the immune complex. The IP supernatant was removed by centrifugation at 10,000 × g at 4°C, and then the IP pellet was further washed three times with RIPA buffer supplemented with protease inhibitor cocktail and NEM before harvesting the beads with 1× Laemmli sample buffer for Western blot analysis. The IP samples were subjected to Western blot analysis with antibodies against ubiquitin to analyze the ubiquitination of SLA-DR.
In order to analyze the ubiquitination of the SLA-DRα and -β chains expressed in HEK-293T cells, HEK-293T cells were transfected with pCAGEN-SLA-DRα or -β plasmids along with vector encoding the PRRSV NSP2 OTU domain for 48 h. The cells were then harvested following the same protocol described above, and IP was conducted using homemade SLA-DRα MAbs or anti-SLA-DRβ mouse serum before ubiquitination analysis.
Mass spectrometry-based immunopeptide analysis.
Mass spectrometry-based immunopeptide analysis was conducted as previously described with modifications (72). Briefly, CNBr-activated Sepharose 4B resin (GE Healthcare, Chicago, IL, USA) was washed with a cold solution of 1 mM HCl and conjugated with anti-SLA-DR MAb (BD Bioscience) following the manufacturer’s instructions at a ratio of 1 ml (Sepharose 4B) to 2 mg (SLA-DR MAb). Next, to avoid contamination with cytoplasm proteins (both viral and cellular) and analyze only immunopeptides presented on the cell surface of BMDCs, 1 × 108 BMDCs infected with an MOI of 1 of PRRSV JXA1 or PRRSV-1-GZ11 for 20 h were harvested for plasma membrane protein extraction using a Minute plasma membrane protein isolation kit (Invent Biotechnologies, Inc., Plymouth, MN, USA) according to the manufacturer’s instructions. For PAM and hilar lymph node samples collected from piglets challenged with PRRSV JXA1, plasma membrane protein extraction was conducted using the same kit. Next, plasma membrane proteins were incubated with anti-SLA-DR MAb conjugated to Sepharose 4B resin overnight at 4°C, and the supernatant was removed by centrifugation. The immunopeptides were eluted by 0.2% trifluoroacetic acid (TFA; Sigma-Aldrich), centrifuged, and filtered through 10-kDa-cutoff ultrafiltration tubes (EMD Millipore, Burlington, MA, USA). After desalting and lyophilizing using Pierce C18 tips (Thermo Fisher Scientific), all samples were resuspended and analyzed using the Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). To ensure the eluted peptides met the minimum requirement for MS detection and data analysis, peptides generated from three individual experiments (for in vitro studies) were pooled as a set for MS analysis. The MS data were analyzed using Proteome Discoverer software (Thermo Fisher Scientific) or the PEAKS Studio X package (Bioinformatics Solutions, Inc., Waterloo, ON, Canada) to locate putative peptides with identical sequences to deduce the amino acids of all open reading frames (ORFs) of PRRSV-2 JXA1 and PRRSV-1 GZ11.
ELISA.
Sequential PRRSV HuN4- and PRRSV SD16-infected pig serum samples (21 dpi and 28 dpi) were obtained from our previous animal experiments (42, 73). The 14-dpi and 21-dpi sera from piglets infected with PRRSV JXA1 and 21-dpi serum samples from piglets infected with NADC30-like isolate were obtained as by-products from another animal experiments with piglets inoculated with PRRSV JXA1 and the NADC30-like isolate (whole experiments terminated at 21 dpi, these animals were not the source for 28-dpi serum samples for PRRSV JXA1 used in this study) with the approval of the Animal Welfare Committee of Northwest A&F University. The 28-dpi serum samples from piglets challenged with PRRSV JXA1 were kindly provided by Kegong Tian of National Research Center for Veterinary Medicine (Luoyang, Henan, China). The standard HP PRRSV hyperimmune pig sera (developed against PRRSV JXA1) with a minimum virus neutralization titer of 1:32 were obtained from the China Veterinary Culture Collection Center of the China Institute of Veterinary Drug Control (Beijing, China). To evaluate the PRRSV-specific antibodies of the PRRSV-infected pigs, the Idexx PRRS X3 ELISA kit (Idexx, Westbrook, ME, USA) was used for analysis as instructed by the manufacturer. Standard PRRSV neutralization assays using the above-named serum samples were conducted to measure the virus neutralization titers.
For the evaluation of mouse sera containing antibodies against PRRSV proteins, recombinant proteins (400 ng) in a volume of 100 μl PBS (pH 8.0) were used to coat 96-well polystyrene microplates (Corning, Inc.) overnight at 4°C. Plates were further blocked with 5% skim milk in PBS containing 0.5% Tween 20 (Sigma-Aldrich). Diluted mouse serum was added to the wells and incubated for 1 h at 37°C, followed by washing with PBS containing 0.5% Triton X-100 (Sigma-Aldrich). The binding of an antibody to the corresponding antigen was detected by using HRP-conjugated goat anti-mouse IgG antibodies (GenScript) and visualized by using a TMB (3,3′,5,5′-tetramethylbenzidine) substrate kit (Tiangen Biotech, Beijing, China). The absorbance of each well was measured using a Victor X5 multilabel plate reader (Perkin Elmer) at 450 nm. For peptide ELISA or keyhole limpet hemocyanin (KLH)-conjugated peptide ELISA, the corresponding peptides or KLH-conjugated peptides were artificially synthesized as coating antigens by GenScript, and the ELISA protocols were conducted as stated above.
To evaluate the pig serum using bacterially expressed NanoLuc luciferase-fused immunopeptides as the coating antigens in ELISAs, the cDNA sequence of NanoLuc with a 6×His tag fused at the N terminus was artificially synthesized and ligated to the pET-28a vector. The cDNAs of immunopeptides were artificially synthesized, ligated at the C terminus of the NanoLuc sequence, and expressed as NanoLuc-fused proteins in Escherichia coli strain BL21(DE3). Recombinant protein expression, purification, and ELISA procedures were conducted as mentioned above.
FACS.
To analyze the surface SLA-DR expression in BMDCs after PRRSV infection, 1 × 106 BMDCs were infected with the indicated PRRSV-1 and PRRSV-2 isolates for 24 h and stained with anti-SLA-DR MAbs as the primary antibody, as well as allophycocyanin-conjugated goat anti-mouse cross-adsorbed secondary antibody (Thermo Fisher Scientific). Flow cytometry analyses were performed on a FACSAria III cell sorter (BD Biosciences) or Attune NxT flow cytometer (Thermo Fisher Scientific) using FlowJo software, version 10.0.7 (TreeStar, Ashland, OR, USA) for FACS result analysis.
Swine antibody purification.
In order to purify polyclonal antibodies specific for PRRSV NSP-derived immunopeptides, 0.8 mg of each artificially synthesized NSP immunopeptide listed in Table 1, except NSP7α, were mixed together in the conjugating buffer and conjugated to 0.25 g CNBr-activated Sepharose 4B powder according to the manufacturer’s instructions. Next, 15 ml of swine serum (28 dpi) from PRRSV JXA1-infected swine or noninfected swine were used to incubate Sepharose 4B beads conjugated with immunopeptides for 2 h at RT, and then the beads were washed with PBS. Following this, immunopeptide-specific antibodies were eluted by glycine buffer (pH 2.4) and neutralized by 1 M Tris-HCl (pH 8.0). The eluted antibodies were further dialyzed using DMEM and concentrated using 50-kDa-cutoff ultrafiltration tubes (EMD Millipore). Protein G-agarose-purified polyclonal antibodies from the same amount of serum obtained from PRRSV JXA1-infected swine or noninfected swine were included as control antibodies.
Virus neutralization assay.
The virus neutralization assay to distinguish PRRSV-neutralizing antibodies from purified swine antibodies was performed on MARC-145 cells as previously described, with modifications (74). Briefly, PRRSV JXA1 was used as the target virus in the assay at an MOI of 0.1 for each reaction. Purified antibodies from swine serum samples were diluted with DMEM and incubated with the virus. An immunofluorescence assay with the N protein-specific monoclonal antibody PP7EF11 was conducted 18 h after inoculation of the cells. Compared to the results for the antibody samples from mock-infected pigs, the reciprocal of the antibody dose that reduced PRRSV replication by 50% was counted as the virus neutralization dose.
IFA.
PRRSV JXA1-infected or noninfected MARC-145 cells in cell culture plates were fixed with 4% paraformaldehyde (Sigma-Aldrich), permeabilized with PBS containing 0.5% Triton X-100 (Sigma-Aldrich), and blocked with PBS containing 1% bovine serum albumin (BSA; Sigma-Aldrich). Then, the cells were stained with MAb against PRRSV N protein (clone number PP7EF11) or purified swine PRRSV NSP immunopeptide antibodies. Specific binding between the antibodies and their targets was detected using Alexa Fluor 555-labeled goat anti-mouse IgG (Thermo Fisher Scientific) or fluorescein isothiocyanate (FITC)-labeled goat anti-swine secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA). Cellular nuclei were counterstained with DAPI (4,6-diamidino-2-phenylindole; Thermo Fisher Scientific) at 37°C for 10 min and observed under a Leica DM1000 fluorescence microscope (Leica Microsystems, Wetzlar, Germany). All images were captured and processed using Leica Application Suite X (Leica Microsystems).
Statistical analysis.
Results were analyzed using GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA, USA). Statistical significance was determined using either Student’s t test for the comparison of two groups or one-way analysis of variance (ANOVA) for testing of more than two groups. A two-tailed P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Xiang-Jin Meng from Virginia Polytechnic Institute and State University for generously providing the pIR-VR2385-CA plasmid, Hanchun Yang of China Agriculture University for generously providing the PRRSV-1 isolate GZ11, and Gaiping Zhang of Henan Agriculture University for generously providing the PRRSV-2 NADC30-like isolate. We also thank the staff of the Life Science Research Core Service, Northwest A&F University, for their assistance in mass spectrometry analysis.
This work was supported by a grant from the National Key Research and Development Program of China awarded to Y.N. and C.W. (grant no. 2017YFD0501004).
REFERENCES
- 1.Kuhn JH, Lauck M, Bailey AL, Shchetinin AM, Vishnevskaya TV, Bào Y, Ng TFF, LeBreton M, Schneider BS, Gillis A, Tamoufe U, Diffo JLD, Takuo JM, Kondov NO, Coffey LL, Wolfe ND, Delwart E, Clawson AN, Postnikova E, Bollinger L, Lackemeyer MG, Radoshitzky SR, Palacios G, Wada J, Shevtsova ZV, Jahrling PB, Lapin BA, Deriabin PG, Dunowska M, Alkhovsky SV, Rogers J, Friedrich TC, O’Connor DH, Goldberg TL. 2016. Reorganization and expansion of the nidoviral family Arteriviridae. Arch Virol 161:755–768. doi: 10.1007/s00705-015-2672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams MJ, Lefkowitz EJ, King AM, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR, Nibert M, Sabanadzovic S, Sanfacon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Gorbalenya AE, Davison AJ. 2016. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016). Arch Virol 161:2921–2949. doi: 10.1007/s00705-016-2977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forsberg R. 2005. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol Biol Evol 22:2131–2134. doi: 10.1093/molbev/msi208. [DOI] [PubMed] [Google Scholar]
- 4.van Woensel PA, Liefkens K, Demaret S. 1998. Effect on viraemia of an American and a European serotype PRRSV vaccine after challenge with European wild-type strains of the virus. Vet Rec 142:510–512. doi: 10.1136/vr.142.19.510. [DOI] [PubMed] [Google Scholar]
- 5.Kappes MA, Faaberg KS. 2015. PRRSV structure, replication and recombination: origin of phenotype and genotype diversity. Virology 479–480:475–486. doi: 10.1016/j.virol.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q, Yoo D. 2015. PRRS virus receptors and their role for pathogenesis. Vet Microbiol 177:229–241. doi: 10.1016/j.vetmic.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Morgan SB, Frossard JP, Pallares FJ, Gough J, Stadejek T, Graham SP, Steinbach F, Drew TW, Salguero FJ. 2016. Pathology and virus distribution in the lung and lymphoid tissues of pigs experimentally inoculated with three distinct type 1 PRRS virus isolates of varying pathogenicity. Transbound Emerg Dis 63:285–295. doi: 10.1111/tbed.12272. [DOI] [PubMed] [Google Scholar]
- 8.Albina E, Carrat C, Charley B. 1998. Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J Interferon Cytokine Res 18:485–490. doi: 10.1089/jir.1998.18.485. [DOI] [PubMed] [Google Scholar]
- 9.Duan X, Nauwynck HJ, Pensaert MB. 1997. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV). Vet Microbiol 56:9–19. doi: 10.1016/S0378-1135(96)01347-8. [DOI] [PubMed] [Google Scholar]
- 10.Duan X, Nauwynck HJ, Pensaert MB. 1997. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch Virol 142:2483–2497. doi: 10.1007/s007050050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sur JH, Cooper VL, Galeota JA, Hesse RA, Doster AR, Osorio FA. 1996. In vivo detection of porcine reproductive and respiratory syndrome virus RNA by in situ hybridization at different times postinfection. J Clin Microbiol 34:2280–2286. doi: 10.1128/JCM.34.9.2280-2286.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Li L, Yu Y, Tu Y, Tong J, Zhang C, Liu Y, Li Y, Han Z, Jiang C, Wang S, Zhou EM, He X, Cai X. 2016. Highly pathogenic porcine reproductive and respiratory syndrome virus infection and induction of apoptosis in bone marrow cells of infected piglets. J Gen Virol 97:1356–1361. doi: 10.1099/jgv.0.000454. [DOI] [PubMed] [Google Scholar]
- 13.Chang HC, Peng YT, Chang HL, Chaung HC, Chung WB. 2008. Phenotypic and functional modulation of bone marrow-derived dendritic cells by porcine reproductive and respiratory syndrome virus. Vet Microbiol 129:281–293. doi: 10.1016/j.vetmic.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhuri S, McKenna N, Balce DR, Yates RM. 2016. Infection of porcine bone marrow-derived macrophages by porcine respiratory and reproductive syndrome virus impairs phagosomal maturation. J Gen Virol 97:669–679. doi: 10.1099/jgv.0.000384. [DOI] [PubMed] [Google Scholar]
- 15.Rowland RR, Lunney J, Dekkers J. 2012. Control of porcine reproductive and respiratory syndrome (PRRS) through genetic improvements in disease resistance and tolerance. Front Genet 3:260. doi: 10.3389/fgene.2012.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labarque GG, Nauwynck HJ, Van Reeth K, Pensaert MB. 2000. Effect of cellular changes and onset of humoral immunity on the replication of porcine reproductive and respiratory syndrome virus in the lungs of pigs. J Gen Virol 81:1327–1334. doi: 10.1099/0022-1317-81-5-1327. [DOI] [PubMed] [Google Scholar]
- 18.Xiao Z, Batista L, Dee S, Halbur P, Murtaugh MP. 2004. The level of virus-specific T-cell and macrophage recruitment in porcine reproductive and respiratory syndrome virus infection in pigs is independent of virus load. J Virol 78:5923–5933. doi: 10.1128/JVI.78.11.5923-5933.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel D, Nan Y, Shen M, Ritthipichai K, Zhu X, Zhang YJ. 2010. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J Virol 84:11045–11055. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang R, Zhang YJ. 2014. Antagonizing interferon-mediated immune response by porcine reproductive and respiratory syndrome virus. Biomed Res Int 2014:315470. doi: 10.1155/2014/315470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Wang R, Ma Z, Xiao Y, Nan Y, Wang Y, Lin S, Zhang YJ. 2017. Porcine reproductive and respiratory syndrome virus antagonizes JAK/STAT3 signaling via nsp5, which induces STAT3 degradation. J Virol 91:e02087-16. doi: 10.1128/JVI.02087-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Lawson S, Sun Z, Zhou X, Guan X, Christopher-Hennings J, Nelson EA, Fang Y. 2010. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology 398:87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Zheng Z, Zhou P, Zhang B, Shi Z, Hu Q, Wang H. 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus non-structural protein 2 antagonizes interferon regulatory factor 3 activation. J Gen Virol 91:2947–2958. doi: 10.1099/vir.0.025205-0. [DOI] [PubMed] [Google Scholar]
- 24.Sun Z, Chen Z, Lawson SR, Fang Y. 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J Virol 84:7832–7846. doi: 10.1128/JVI.00217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, Zhang Q, Guo XK, Yu ZB, Xu AT, Tang J, Feng WH. 2014. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 antagonizes beta interferon expression by targeting the NF-kappaB essential modulator. J Virol 88:10934–10945. doi: 10.1128/JVI.01396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi X, Wang L, Li X, Zhang G, Guo J, Zhao D, Chai S, Deng R. 2011. Endoribonuclease activities of porcine reproductive and respiratory syndrome virus nsp11 was essential for nsp11 to inhibit IFN-beta induction. Mol Immunol 48:1568–1572. doi: 10.1016/j.molimm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sagong M, Lee C. 2011. Porcine reproductive and respiratory syndrome virus nucleocapsid protein modulates interferon-beta production by inhibiting IRF3 activation in immortalized porcine alveolar macrophages. Arch Virol 156:2187–2195. doi: 10.1007/s00705-011-1116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du J, Ge X, Liu Y, Jiang P, Wang Z, Zhang R, Zhou L, Guo X, Han J, Yang H. 2016. Targeting swine leukocyte antigen class I molecules for proteasomal degradation by the nsp1alpha replicase protein of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus strain JXwn06. J Virol 90:682–693. doi: 10.1128/JVI.02307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao QM, Subramaniam S, Ni YY, Cao D, Meng XJ. 2016. The non-structural protein Nsp2TF of porcine reproductive and respiratory syndrome virus down-regulates the expression of swine leukocyte antigen class I. Virology 491:115–124. doi: 10.1016/j.virol.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 30.Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, Renukaradhya GJ. 2016. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci 4:129–154. doi: 10.1146/annurev-animal-022114-111025. [DOI] [PubMed] [Google Scholar]
- 31.Lopez OJ, Osorio FA. 2004. Role of neutralizing antibodies in PRRSV protective immunity. Vet Immunol Immunopathol 102:155–163. doi: 10.1016/j.vetimm.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 32.de Lima M, Pattnaik AK, Flores EF, Osorio FA. 2006. Serologic marker candidates identified among B-cell linear epitopes of Nsp2 and structural proteins of a North American strain of porcine reproductive and respiratory syndrome virus. Virology 353:410–421. doi: 10.1016/j.virol.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 33.Brown E, Lawson S, Welbon C, Gnanandarajah J, Li J, Murtaugh MP, Nelson EA, Molina RM, Zimmerman JJ, Rowland RR, Fang Y. 2009. Antibody response to porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural proteins and implications for diagnostic detection and differentiation of PRRSV types I and II. Clin Vaccine Immunol 16:628–635. doi: 10.1128/CVI.00483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langenhorst RJ, Lawson S, Kittawornrat A, Zimmerman JJ, Sun Z, Li Y, Christopher-Hennings J, Nelson EA, Fang Y. 2012. Development of a fluorescent microsphere immunoassay for detection of antibodies against porcine reproductive and respiratory syndrome virus using oral fluid samples as an alternative to serum-based assays. Clin Vaccine Immunol 19:180–189. doi: 10.1128/CVI.05372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Figueiredo MM, Costa PAC, Diniz SQ, Henriques PM, Kano FS, Tada MS, Pereira DB, Soares IS, Martins-Filho OA, Jankovic D, Gazzinelli RT, Antonelli L. 2017. T follicular helper cells regulate the activation of B lymphocytes and antibody production during Plasmodium vivax infection. PLoS Pathog 13:e1006484. doi: 10.1371/journal.ppat.1006484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thornhill JP, Fidler S, Klenerman P, Frater J, Phetsouphanh C. 2017. The role of CD4+ T follicular helper cells in HIV infection: from the germinal center to the periphery. Front Immunol 8:46. doi: 10.3389/fimmu.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Eaton M, Mayer M, Li H, He D, Nelson E, Christopher-Hennings J. 2007. Porcine reproductive and respiratory syndrome virus productively infects monocyte-derived dendritic cells and compromises their antigen-presenting ability. Arch Virol 152:289–303. doi: 10.1007/s00705-006-0857-1. [DOI] [PubMed] [Google Scholar]
- 38.Flores-Mendoza L, Silva-Campa E, Reséndiz M, Osorio FA, Hernández J. 2008. Porcine reproductive and respiratory syndrome virus infects mature porcine dendritic cells and up-regulates interleukin-10 production. Clin Vaccine Immunol 15:720–725. doi: 10.1128/CVI.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva-Campa E, Cordoba L, Fraile L, Flores-Mendoza L, Montoya M, Hernández J. 2010. European genotype of porcine reproductive and respiratory syndrome (PRRSV) infects monocyte-derived dendritic cells but does not induce Treg cells. Virology 396:264–271. doi: 10.1016/j.virol.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 40.Rodríguez-Gómez IM, Käser T, Gómez-Laguna J, Lamp B, Sinn L, Rümenapf T, Carrasco L, Saalmüller A, Gerner W. 2015. PRRSV-infected monocyte-derived dendritic cells express high levels of SLA-DR and CD80/86 but do not stimulate PRRSV-naive regulatory T cells to proliferate. Vet Res 46:54. doi: 10.1186/s13567-015-0186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Wang G, Yang D, Zhao B, Zhao Y, Liu Y, Cai X, Nan Y, Zhou EM, Wu C. 2018. Development of luciferase-linked antibody capture assay based on luciferase immunoprecipitation systems for antibody detection of porcine reproductive and respiratory syndrome virus. BMC Biotechnol 18:73. doi: 10.1186/s12896-018-0483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhai T, Wu C, Wang N, Shi B, Li J, Chen R, Dong J, Zhang Y, Zhou EM, Nan Y. 2019. Development of a monoclonal antibody against swine leukocyte antigen (SLA)-DR alpha chain and evaluation of SLA-DR expression in bone marrow-derived dendritic cells after PRRSV infection. Vet Immunol Immunopathol 211:19–24. doi: 10.1016/j.vetimm.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Tamulyte S, Kopplin J, Brenner T, Weigand MA, Uhle F. 2019. Monocyte HLA-DR assessment by a novel point-of-care device is feasible for early identification of ICU patients with complicated courses—a proof-of-principle study. Front Immunol 10:432. doi: 10.3389/fimmu.2019.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lunney JK, Ho CS, Wysocki M, Smith DM. 2009. Molecular genetics of the swine major histocompatibility complex, the SLA complex. Dev Comp Immunol 33:362–374. doi: 10.1016/j.dci.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 45.Yang L, Wang R, Yang S, Ma Z, Lin S, Nan Y, Li Q, Tang Q, Zhang YJ. 2018. Karyopherin alpha 6 is required for replication of porcine reproductive and respiratory syndrome virus and Zika virus. J Virol 92:e00072-18. doi: 10.1128/JVI.00072-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charerntantanakul W. 2012. Porcine reproductive and respiratory syndrome virus vaccines: immunogenicity, efficacy and safety aspects. World J Virol 1:23–30. doi: 10.5501/wjv.v1.i1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson SR, Li J, Nelson EA, Murtaugh MP. 2015. Broadly neutralizing antibodies against the rapidly evolving porcine reproductive and respiratory syndrome virus. Virus Res 203:56–65. doi: 10.1016/j.virusres.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 48.Lopez OJ, Oliveira MF, Garcia EA, Kwon BJ, Doster A, Osorio FA. 2007. Protection against porcine reproductive and respiratory syndrome virus (PRRSV) infection through passive transfer of PRRSV-neutralizing antibodies is dose dependent. Clin Vaccine Immunol 14:269–275. doi: 10.1128/CVI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okda F, Liu X, Singrey A, Clement T, Nelson J, Christopher-Hennings J, Nelson EA, Lawson S. 2015. Development of an indirect ELISA, blocking ELISA, fluorescent microsphere immunoassay and fluorescent focus neutralization assay for serologic evaluation of exposure to North American strains of porcine epidemic diarrhea virus. BMC Vet Res 11:180. doi: 10.1186/s12917-015-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das PB, Dinh PX, Ansari IH, de Lima M, Osorio FA, Pattnaik AK. 2010. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J Virol 84:1731–1740. doi: 10.1128/JVI.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian D, Wei Z, Zevenhoven-Dobbe JC, Liu R, Tong G, Snijder EJ, Yuan S. 2012. Arterivirus minor envelope proteins are a major determinant of viral tropism in cell culture. J Virol 86:3701–3712. doi: 10.1128/JVI.06836-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burkard C, Lillico SG, Reid E, Jackson B, Mileham AJ, Ait-Ali T, Whitelaw CB, Archibald AL. 2017. Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog 13:e1006206. doi: 10.1371/journal.ppat.1006206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, Samuel MS, Lightner JE, McLaren DG, Mileham AJ, Wells KD, Prather RS. 2016. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat Biotechnol 34:20–22. doi: 10.1038/nbt.3434. [DOI] [PubMed] [Google Scholar]
- 54.Vanhee M, Van Breedam W, Costers S, Geldhof M, Noppe Y, Nauwynck H. 2011. Characterization of antigenic regions in the porcine reproductive and respiratory syndrome virus by the use of peptide-specific serum antibodies. Vaccine 29:4794–4804. doi: 10.1016/j.vaccine.2011.04.071. [DOI] [PubMed] [Google Scholar]
- 55.Robinson SR, Rahe MC, Gray DK, Martins KV, Murtaugh MP. 2018. Porcine reproductive and respiratory syndrome virus neutralizing antibodies provide in vivo cross-protection to PRRSV1 and PRRSV2 viral challenge. Virus Res 248:13–23. doi: 10.1016/j.virusres.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Murtaugh MP. 2012. Dissociation of porcine reproductive and respiratory syndrome virus neutralization from antibodies specific to major envelope protein surface epitopes. Virology 433:367–376. doi: 10.1016/j.virol.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 57.Trible BR, Popescu LN, Monday N, Calvert JG, Rowland RR. 2015. A single amino acid deletion in the matrix protein of porcine reproductive and respiratory syndrome virus confers resistance to a polyclonal swine antibody with broadly neutralizing activity. J Virol 89:6515–6520. doi: 10.1128/JVI.03287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roche PA, Furuta K. 2015. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol 15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blum JS, Wearsch PA, Cresswell P. 2013. Pathways of antigen processing. Annu Rev Immunol 31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang G, Yu Y, Tu Y, Tong J, Liu Y, Zhang C, Chang Y, Wang S, Jiang C, Zhou EM, Cai X. 2015. Highly pathogenic porcine reproductive and respiratory syndrome virus infection induced apoptosis and autophagy in thymi of infected piglets. PLoS One 10:e0128292. doi: 10.1371/journal.pone.0128292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Q, Fang L, Wang D, Wang S, Li P, Li M, Luo R, Chen H, Xiao S. 2012. Induction of autophagy enhances porcine reproductive and respiratory syndrome virus replication. Virus Res 163:650–655. doi: 10.1016/j.virusres.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun MX, Huang L, Wang R, Yu YL, Li C, Li PP, Hu XC, Hao HP, Ishag HA, Mao X. 2012. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy 8:1434–1447. doi: 10.4161/auto.21159. [DOI] [PubMed] [Google Scholar]
- 63.Xiao S, Zhang A, Zhang C, Ni H, Gao J, Wang C, Zhao Q, Wang X, Wang X, Ma C, Liu H, Li N, Mu Y, Sun Y, Zhang G, Hiscox JA, Hsu WH, Zhou E-M. 2014. Heme oxygenase-1 acts as an antiviral factor for porcine reproductive and respiratory syndrome virus infection and over-expression inhibits virus replication in vitro. Antiviral Res 110:60–69. doi: 10.1016/j.antiviral.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 64.Ni YY, Huang YW, Cao D, Opriessnig T, Meng XJ. 2011. Establishment of a DNA-launched infectious clone for a highly pneumovirulent strain of type 2 porcine reproductive and respiratory syndrome virus: identification and in vitro and in vivo characterization of a large spontaneous deletion in the nsp2 region. Virus Res 160:264–273. doi: 10.1016/j.virusres.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 65.Zhang YJ, Stein DA, Fan SM, Wang KY, Kroeker AD, Meng XJ, Iversen PL, Matson DO. 2006. Suppression of porcine reproductive and respiratory syndrome virus replication by morpholino antisense oligomers. Vet Microbiol 117:117–129. doi: 10.1016/j.vetmic.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nan Y, Ma Z, Wang R, Yu Y, Kannan H, Fredericksen B, Zhang YJ. 2014. Enhancement of interferon induction by ORF3 product of hepatitis E virus. J Virol 88:8696–8705. doi: 10.1128/JVI.01228-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L, Xue B, Sun W, Gu G, Hou G, Zhang L, Wu C, Zhao Q, Zhang Y, Zhang G, Hiscox JA, Nan Y, Zhou EM. 2018. Recombinant MYH9 protein C-terminal domain blocks porcine reproductive and respiratory syndrome virus internalization by direct interaction with viral glycoprotein 5. Antiviral Res 156:10–20. doi: 10.1016/j.antiviral.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 68.Wu C, Gu G, Zhai T, Wang Y, Yang Y, Li Y, Zheng X, Zhao Q, Zhou EM, Nan Y. 2020. Broad neutralization activity against both PRRSV-1 and PRRSV-2 and enhancement of cell mediated immunity against PRRSV by a novel IgM monoclonal antibody. Antiviral Res 175:104716. doi: 10.1016/j.antiviral.2020.104716. [DOI] [PubMed] [Google Scholar]
- 69.Wang C, Huang B, Kong N, Li Q, Ma Y, Li Z, Gao J, Zhang C, Wang X, Liang C, Dang L, Xiao S, Mu Y, Zhao Q, Sun Y, Almazan F, Enjuanes L, Zhou EM. 2013. A novel porcine reproductive and respiratory syndrome virus vector system that stably expresses enhanced green fluorescent protein as a separate transcription unit. Vet Res 44:104. doi: 10.1186/1297-9716-44-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel D, Opriessnig T, Stein DA, Halbur PG, Meng XJ, Iversen PL, Zhang YJ. 2008. Peptide-conjugated morpholino oligomers inhibit porcine reproductive and respiratory syndrome virus replication. Antiviral Res 77:95–107. doi: 10.1016/j.antiviral.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mu Y, Li L, Zhang B, Huang B, Gao J, Wang X, Wang C, Xiao S, Zhao Q, Sun Y, Zhang G, Hiscox JA, Zhou EM. 2015. Glycoprotein 5 of porcine reproductive and respiratory syndrome virus strain SD16 inhibits viral replication and causes G2/M cell cycle arrest, but does not induce cellular apoptosis in Marc-145 cells. Virology 484:136–145. doi: 10.1016/j.virol.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 72.Dengjel J, Rammensee HG, Stevanovic S. 2005. Glycan side chains on naturally presented MHC class II ligands. J Mass Spectrom 40:100–104. doi: 10.1002/jms.780. [DOI] [PubMed] [Google Scholar]
- 73.Gao J, Xiao S, Xiao Y, Wang X, Zhang C, Zhao Q, Nan Y, Huang B, Liu H, Liu N, Lv J, Du T, Sun Y, Mu Y, Wang G, Syed SF, Zhang G, Hiscox JA, Goodfellow I, Zhou EM. 2016. MYH9 is an essential factor for porcine reproductive and respiratory syndrome virus infection. Sci Rep 6:25120. doi: 10.1038/srep25120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma Z, Yu Y, Xiao Y, Opriessnig T, Wang R, Yang L, Nan Y, Samal SK, Halbur PG, Zhang YJ. 2016. Sustaining interferon induction by a high-passage atypical porcine reproductive and respiratory syndrome virus strain. Sci Rep 6:36312. doi: 10.1038/srep36312. [DOI] [PMC free article] [PubMed] [Google Scholar]