Accumulating evidence suggests that some endogenous viral elements (EVEs), including endogenous retroviruses and endogenous nonretroviral virus elements, have acquired functions in the host as a result of long-term coevolution. Recently, an endogenous bornavirus-like element (itEBLN) found in the ground squirrel genome has been shown to have antiviral activity against exogenous bornavirus infection. In this study, we first quantified bornavirus spread in cultured cells and then calculated the antiviral activity of itEBLN on bornavirus infection. The calculated antiviral activity of itEBLN suggests its suppression of multiple processes in the viral life cycle. To our knowledge, this is the first study quantifying the antiviral activity of EVEs and speculating on a model of how some EVEs have acquired antiviral activity during host-virus arms races.
KEYWORDS: Borna disease virus, endogenous bornavirus-like nucleoproteins, mathematical modeling, data analysis, endogenous bornavirus-like element
ABSTRACT
Endogenous retroviruses have demonstrated exaptation during long-term evolution with hosts, e.g., resulting in acquisition of antiviral effect on related extant viral infections. While empirical studies have found that an endogenous bornavirus-like element derived from viral nucleoprotein (itEBLN) in the ground squirrel genome shows antiviral effect on virus replication and de novo infection, the antiviral mechanism, dynamics, and quantitative effect of itEBLN remain unknown. In this study, we experimentally and theoretically investigated the dynamics of how an extant bornavirus, Borna disease virus 1 (BoDV-1), spreads and replicates in uninfected, BoDV-1-infected, and itEBLN-expressing cultured cells. Quantifying antiviral effect based on time course data sets, we found that the antiviral effects of itEBLN are estimated to be 75% and 34% on intercellular virus spread and intracellular virus replication, respectively. This discrepancy between intercellular virus spread and intracellular viral replication suggests that viral processes other than the replication of viral ribonucleoprotein complex (RNP) contributed to the suppression of virus spread in itEBLN-expressing cells. Because itEBLN binds to the BoDV-1 RNP, the suppression of viral RNP trafficking can be an attractive candidate explaining this discrepancy.
IMPORTANCE Accumulating evidence suggests that some endogenous viral elements (EVEs), including endogenous retroviruses and endogenous nonretroviral virus elements, have acquired functions in the host as a result of long-term coevolution. Recently, an endogenous bornavirus-like element (itEBLN) found in the ground squirrel genome has been shown to have antiviral activity against exogenous bornavirus infection. In this study, we first quantified bornavirus spread in cultured cells and then calculated the antiviral activity of itEBLN on bornavirus infection. The calculated antiviral activity of itEBLN suggests its suppression of multiple processes in the viral life cycle. To our knowledge, this is the first study quantifying the antiviral activity of EVEs and speculating on a model of how some EVEs have acquired antiviral activity during host-virus arms races.
INTRODUCTION
Endogenous retroviruses (ERVs) have accumulated in the genomes of many organisms during evolution and account for around 8% of the human genome (1). Although the significance of ERV sequences remains unclear, some have been proposed to exert antiviral, reproduction-related, or immune-modulating functions, that is, inhibiting infection with related viruses (2–4), controlling cell-to-cell fusion during placentation (5), or regulating the immune system (6, 7). In addition to ERVs, nonretroviruses have also left their sequences in eukaryotic genomes (8). Among nonretroviral endogenous viral elements (EVEs), endogenous bornavirus-like elements derived from nucleoprotein (EBLNs) have been intensively studied (9–11). As species with EBLNs appear relatively protected against symptoms caused by infection with Borna disease virus 1 (BoDV-1), a current mammalian orthobornavirus, it has been speculated that EBLNs play roles in antiviral host responses (9, 10). Consistent with this, we have recently demonstrated that an EBLN in the thirteen-lined ground squirrel genome (itEBLN) inhibits BoDV-1 replication and infection (12). Although the expression of itEBLN was confirmed in thirteen-lined ground squirrel tissues, including the brain, a main target tissue of bornaviruses, the antiviral effect of itEBLN in the target cells has not been evaluated because of the lack of available cultured cells of thirteen-lined ground squirrels (12).
BoDV-1 is a nonsegmented, negative-stranded RNA virus that causes fatal encephalitis in various species, including humans (13–19). BoDV-1 replicates in the nucleus without causing overt cytopathic effects and readily establishes a persistent infection (20). BoDV-1 ribonucleoprotein complex (RNP) is the replication unit of BoDV-1, consisting of BoDV-1 genomic RNA, nucleoprotein (N), phosphoprotein (P), and large protein (L) (20). Because BoDV-1 spreads in cultured cells in the absence of detectable extracellular virus or syncytium formation (21), it has been proposed that cell-to-cell spread is the main route of BoDV-1 transmission. BoDV-1 glycoprotein (G), a viral surface protein responsible for the viral entry, is required for viral spread, including a primary infection and subsequent cell-to-cell spread (22–25). During spread of the virus to adjacent cells, BoDV-1 RNP needs to be exported to the cytoplasm from the nuclei of infected cells, where BoDV-1 RNA synthesis occurs, then transported to uninfected cells via an as-yet-unknown mechanism, and, finally, enter the nuclei of uninfected cells to replicate (20). Although itEBLN was shown to inhibit BoDV-1 RNA synthesis (both genomic RNA replication and mRNA transcription) and cell-to-cell spread, the experiments were conducted using different cell lines and evaluated inhibition only at the endpoint (12). Thus, the relationship between inhibition of BoDV-1 RNA synthesis and spread and the difference in dynamics of BoDV-1 spread in the presence and absence of itEBLN have not been comprehensively or systematically investigated.
In this study, we employed an experimental and theoretical investigation based on viral infection experiments. We first quantitatively evaluated how BoDV-1 spreads and replicates in cultured cell lines. With these quantitative BoDV-1 infection dynamics, we found that the antiviral effects of itEBLN are estimated to be 75% and 34% on intercellular virus spread and intracellular virus replication, respectively. This is the first report to quantitatively evaluate BoDV-1 spread. Based on our calculation, we speculated on the mode of antiviral effect conferred by itEBLN.
RESULTS
Modeling intercellular BoDV-1 spread in cultured cells.
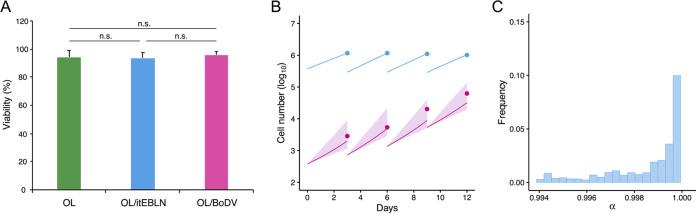
To quantify the early phase of BoDV-1 spread in OL cells (a human oligodendrocyte cell line), we employed a combined experimental-theoretical approach (26, 27). As described in Table 1, we cocultured OL cells with OL/BoDV-GFP cells, cells infected with BoDV-1 carrying a green fluorescent protein (GFP) reporter (ratio of target cells to OL/BoDV-GFP cells, 1,000:1). We then passaged and measured the numbers of uninfected and infected cells every 3 days. BoDV-1 carrying a GFP reporter was expected to spread from OL/BoDV-GFP cells to neighboring uninfected cells through cell-to-cell contacts without producing detectable virions. Thus, the increase of GFP-positive cells was determined by the rates of the cell-to-cell spread of BoDV-1 and the division of infected cells in the culture. We also measured BoDV-1 spread in OL/itEBLN cells, OL cells stably expressing itEBLN, and OL/BoDV cells, OL cells persistently infected with BoDV-1, to evaluate the antiviral effect of itEBLN and the potential for superinfection, respectively. First, we confirmed that the viabilities of OL, OL/itEBLN, and OL/BoDV cells were comparable (Fig. 1A). Then, since the number of cells did not reach carrying capacity of cell culture system, we assumed that the total number of target cells increased exponentially every 3 days (i.e., there were four periods of 0 to 3, 3 to 6, 6 to 9, and 9 to 12 days), described by , where is the number of target cells and is the growth rate for each period (). Therefore, we estimated and fixed them (Table 1).
TABLE 1.
Time course of experimental data from BoDV-1 spread assay
| Value on measurement day: |
|||||||
|---|---|---|---|---|---|---|---|
| Cell line name | Cell type | Type of data | 0 | 3 | 6 | 9 | 12 |
| OL | Uninfected | Data used for fitting | 1,161,800 | 1,166,627 | 1,105,046 | 1,016,170 | |
| Fixed values for fitting | 377,242 | 294,127 | 293,491 | 289,532 | |||
| Infected | Data used for fitting | 2,852 | 5,396 | 20,294 | 62,484 | ||
| Fixed values for fitting | 378 | 1,357 | 1,357 | 5,317 | |||
| OL/itEBLN | Uninfected | Data used for fitting | 653,486 | 695,830 | 654,621 | 697,053 | |
| Fixed values for fitting | 374,134 | 167,203 | 166,999 | 289,531 | |||
| Infected | Data used for fitting | 958 | 1,872 | 2,614 | 3,440 | ||
| Fixed values for fitting | 375 | 246 | 449 | 666 | |||
| OL/BoDV | Uninfected | Data used for fitting | 1,352,390 | 1,189,718 | 1,095,837 | 962,687 | |
| Fixed values for fitting | 382,571 | 301,089 | 301,195 | 300,916 | |||
| Infected | Data used for fitting | 1,615 | 1,003 | 1,937 | 1,948 | ||
| Fixed values for fitting | 383 | 360 | 254 | 532 | |||
FIG 1.
Cell viability and density dependency for intercellular BoDV-1 dynamics. Viabilities of OL, OL/itEBLN, and OL/BoDV cells were evaluated and are expressed as the means + five independent experiments in panel A. BoDV dynamics in OL cells are shown in panel B. Dots show the time course of experimental data for the numbers of uninfected cells (blue) and infected cells (red). The shaded region corresponds to 95% posterior predictive intervals, and the solid lines yield the mean for the full mathematical model with best-fit parameter estimates. Using all accepted MCMC parameter estimates from the time course experimental data sets, the distribution of is shown in panel C. n.s., no significance (t test).
Next, to describe intercellular BoDV-1 spread in cell culture, we divided the total cell count by the number of uninfected cells, , and infected cells, . itEBLN has been proposed to function as a dominant negative mutant of BoDV-1 N, suppressing viral replication and de novo infection, although the detailed mechanism remains unknown (12). Superinfection with homologous viruses in virus-infected cells is, in general, rarely successful. Because of this, BoDV-1 spread was expected to be inhibited in OL/itEBLN and OL/BoDV cells in our experimental setting. Assuming that the rate constant for infection is and the antiviral effects on intercellular virus spread in OL/itEBLN and OL/BoDV cells are ( or ), we have the following mathematical model:
Here, is a scaling parameter describing how the infection rate depends on total cell density; for example, and if is and 0 (i.e., and ), respectively. Interestingly, in our data fitting, our estimation indicated (Fig. 1B and C). Without loss of generality, here, we assumed the following simple model:
| (1) |
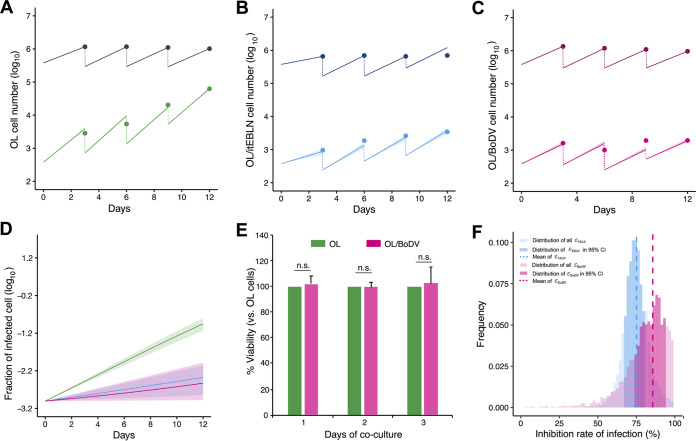
To assess the variability of kinetic parameters and model predictions, we performed Bayesian estimation for the whole data set and simultaneously fitted equation 1 above to the numbers of uninfected and infected target cells from OL, OL/itEBLN, and OL/BoDV cell cultures as described previously (26, 27). These estimated parameter and initial values are listed in Table 2. The typical behaviors of the model using these best-fit parameter estimates are shown together with the data in Fig. 2A to C for OL, OL/itEBLN, and OL/BoDV cells, respectively, which reveal that equation 1 describes these in vitro data very well. The shaded regions correspond to 95% posterior predictive intervals, the blue and pink solid lines give the best-fit solution (mean) for the uninfected and infected cells, respectively, and the dots show the experimental data sets. In addition, using estimated parameters in Table 2, we performed in silico analysis for the predicted fraction of infected cells among total cells under the condition that there is no cell growth in Fig. 2D, to decompose two processes, de novo infection and cell division, which maintain the number of infected cells. Because the viabilities of OL and OL/BoDV cells were comparable during 3 days of coculture with OL/BoDV-GFP cells (Fig. 2E) and the infected cell fraction in OL/BoDV cells is not changed but almost in a steady state, we speculate that the infected cells are mainly maintained by cell division but not de novo infection. Conversely, the fraction slightly increases, and therefore, de novo infection still contributes in OL/itEBLN cells. In Fig. 2F, we show the distributions of antiviral effects on cell-to-cell spread, and , in OL/itEBLN and OL/BoDV cells. Comparing these parameters for OL/itEBLN and OL/BoDV cells, we found that there was a significant difference between the antiviral effects of (mean, 0.75; 95% credible interval [CI], 0.70 to 0.79) and (mean, 0.94; 95% CI, 0.82 to 0.99) (P = 0.0017 by repeated bootstrap t test). Taken together, these estimates reveal the antiviral effect of itEBLN and demonstrate that BoDV-1 superinfection to the BoDV-1-infected cells is almost completely inhibited.
TABLE 2.
Parameter values for BoDV-1 spread assay
| Parameter | Symbol (unit) | Value (95% CI) for: |
||
|---|---|---|---|---|
| OL cells | OL/itEBLN cells | OL/BoDV cells | ||
| Parameters obtained simultaneous fit to full in vitro data set | ||||
| Rate constant for infections | (no./well)−1 (day)−1 | 0.40 (0.39–0.42) | 0.40 (0.39–0.42) | 0.40 (0.39–0.42) |
| Antiviral effect % of OL/itEBLN cell line | — | 0.75 (0.70–0.79) | — | |
| Antiviral effect % of OL/BoDV cell line | — | — | 0.83 (0.76–0.91) | |
| Parameters obtained fit to each cell type in vitro data set | ||||
| Growth rate from day 0–day 3 | (day−1) | 0.375 | 0.186 | 0.421 |
| Growth rate from day 3–day 6 | (day−1) | 0.460 | 0.476 | 0.458 |
| Growth rate from day 6–day 9 | (day−1) | 0.446 | 0.456 | 0.431 |
| Growth rate from day 9–day 12 | (day−1) | 0.432 | 0.477 | 0.388 |
FIG 2.
Quantification of intercellular BoDV-1 dynamics in OL, OL/itEBLN, and OL/BoDV cells and the antiviral effect of itEBLN. OL, OL/itEBLN, and OL/BoDV cells were cocultured with OL/BoDV-GFP cells (target-to-seed ratio, 1,000:1). OL/itEBLN cells are OL cells stably expressing itEBLN. OL/BoDV cells are OL cells persistently infected with BoDV-1 strain huP2br. These cells were passaged once every 3 days and cultured in new wells at 4 × 105 cells/well. BoDV-1 dynamics in OL, OL/itEBLN, and OL/BoDV cells are shown in panels A, B, and C, respectively. Dots show the time course of experimental data (the averaged data of at least four independent experiments) for the numbers of uninfected cells (darker) and infected cells (lighter). The shaded region corresponds to 95% posterior predictive intervals, and the solid lines indicate the mean for equation 1 with best-fit parameter estimates. In addition, the solid lines and shaded regions show the mean and 95% posterior predictive intervals of the predicted fraction of infected cells among total cells in panel D. Viabilities of OL and OL/BoDV cells during 3 days of coculture with OL/BoDV-GFP cells were evaluated and are expressed as the means + five independent experiments in panel E. Using all accepted MCMC parameter estimates from the time course experimental data sets, the distributions of the antiviral effect on BoDV-1 infection in OL/itEBLN and OL/BoDV cells (i.e., and ) are shown in panel F. The dotted lines and dark and light bars show the mean, 95% credible interval, and whole estimations of the antiviral effects, respectively.
Modeling of intracellular BoDV-1 replication in cultured cells.
Although our quantitative data analysis in cell cultures revealed the antiviral effect on cell-to-cell spread of BoDV-1, it is still not fully understood how intracellular BoDV-1 replication is inhibited in OL/itEBLN and OL/BoDV cell cultures. To address this point, we used a minigenome assay and measured viral polymerase activity in OL, OL/itEBLN, and OL/BoDV cells at several time points (Table 3). In the BoDV-1 minigenome assay, BoDV-1 minigenome RNP was reconstituted by coexpression of the N, P, and L proteins with a BoDV-1 minigenome RNA containing the leader and trailer sequences and the Gaussia luciferase (GLuc) gene. As BoDV-1 RNP is the replication unit of BoDV-1, the luciferase activity is considered correlated with BoDV-1 polymerase activity (28). We calculated the expected number of OL, OL/itEBLN, and OL/BoDV cells with our estimated growth rate ( in Table 2) and normalized luciferase activity per cell. Therefore, we assumed that the variable is the luciferase activity per cell proportional to the amount of viral RNA synthesis from the minigenome (28), which becomes measurable after , and the parameters r, d, and μ represent the production rate of luciferase activity by the viral RNA synthesis, the delay of the viral RNA synthesis, and the decay of the viral RNA synthesis product, respectively. Considering that the inhibition rates of viral polymerase activity in OL/itEBLN and OL/BoDV cells are ( or ), the luciferase activities are described by the following equation:
| (2) |
TABLE 3.
Time course of experimental normalized data from minigenome assay
| Cell line name | Value on measurement day: |
|||
|---|---|---|---|---|
| 0.75 | 1 | 2 | 3 | |
| OL | 0.005 | 0.128 | 9.524 | 18.426 |
| OL/itEBLN | 0.012 | 0.117 | 6.638 | 10.320 |
| OL/BoDV | 0.004 | 0.029 | 0.454 | 0.471 |
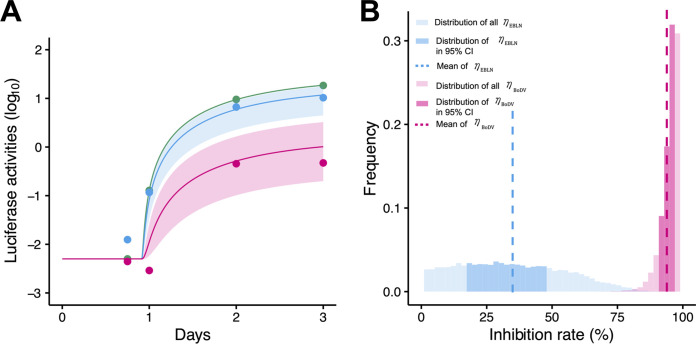
By fitting equation 2 above to the luciferase activities in OL cells, we first estimated kinetic parameters of , , , and and initial values. Then, fixing these values, we further independently estimated the inhibition rates and in OL/itEBLN and OL/BoDV cells. All estimated parameter and initial values are listed in Table 4. The typical behaviors of equation 2 using these best-fit parameter estimates are shown together with the data in Fig. 3A. The shaded regions correspond to 95% posterior predictive intervals, the solid lines give the best-fit solution (mean), and the dots show the experimental data sets. In Fig. 3B, we show the distributions of inhibition rates (mean, 0.34; 95% CI, 0.18 to 0.49) and (mean, 0.83; 95% CI, 0.76 to 0.91); we found that there was a significant difference between them (P = 0.00029 by repeated bootstrap t test). In OL/BoDV cells, the antiviral effect on cell-to-cell BoDV-1 spread, , and that on intracellular BoDV-1 replication, , were in similar ranges, consistent with the idea that the infected cells were maintained almost entirely by cell division. Interestingly and converse to this, comparing Fig. 2F and 3B, we found the antiviral effect on intracellular BoDV-1 replication, , to be much smaller than that on cell-to-cell BoDV-1 spread, , in OL/itEBLN cells, although the minigenome assay, which does not consider transmission chain of BoDV-1, might not be able to capture the mild antiviral effect because of its flat distribution. This discrepancy between intercellular virus spread and intracellular viral replication is discussed below in detail.
TABLE 4.
Parameter values for minigenome assay
| Parameter | Symbol (unit) | Value (95% CI) for: |
||
|---|---|---|---|---|
| OL cells | OL/itEBLN cells | OL/BoDV cells | ||
| Production rate of luciferase activity in OL cell line | (day−1) | 33.31 | 33.31 | 33.31 |
| Antiviral effect for viral RNA synthesis in OL/itEBLN cell line | — | 0.34 (0.18–0.49) | — | |
| Antiviral effect for viral RNA synthesis in OL/BoDV cell line | — | — | 0.95 (0.94–0.97) | |
| Delay of viral RNA synthesis | (day−1) | 1.03 | 1.03 | 1.03 |
| Clearance rate of viral RNA synthesis | (day−1) | 1.21 | 1.21 | 1.21 |
| Time for detection of viral RNA synthesis | (day−1) | 0.91 | 0.91 | 0.91 |
| Initial condition of viral RNA synthesis | 0.005 | 0.005 | 0.005 | |
FIG 3.
Quantification of intracellular BoDV-1 dynamics in OL, OL/itEBLN, and OL/BoDV cells and antiviral effect of itEBLN. OL, OL/itEBLN, and OL/BoDV cells were transfected with plasmids expressing BoDV-1 N, P, L, and a Pol II-driven minigenome encoding GLuc. The luciferase activities in OL, OL/itEBLN, and OL/BoDV cells are shown together in panel A. Dots show the time course of experimental data for the luciferase activities of OL (green), OL/itEBLN (blue), and OL/BoDV (red) cells (the averaged data of at least three independent experiments), the shaded region corresponds to 95% posterior predictive intervals, and the solid lines indicate the mean for equation 2 with best-fit parameter estimates. The distributions of the inhibition rate on intracellular BoDV-1 replication in OL/itEBLN and OL/BoDV cells (i.e., and ) are shown in panel B. The dotted lines and dark and light bars show the mean, 95% credible interval, and whole estimations of the antiviral effects, respectively.
DISCUSSION
A recent study has demonstrated that itEBLN inhibits BoDV-1 infection (12), but the mode of antiviral effect of itEBLN remains obscure. In this study, we, for the first time, experimentally and theoretically investigated how BoDV-1 spreads and replicates in OL, OL/itEBLN, and OL/BoDV cells. Estimating parameter values from the time course data sets, we found that cell-to-cell virus spread and intracellular virus replication were similarly inhibited in OL/BoDV cells (more than 80%), and infected cells were mainly maintained by cell division but not de novo infection (Fig. 2F and 3B). Although the mechanism of how superinfection of BoDV-1 was suppressed is not clear, we speculate that host factors required for BoDV-1 replication had been occupied by existing viral RNPs, and therefore, de novo infection to the infected cells were suppressed. On the other hand, de novo infection still contributed to maintenance of infected cell numbers in OL/itEBLN cells even though itEBLN inhibited 75% and 34% of cell-to-cell virus spread and intracellular virus replication, respectively (Fig. 2F and 3B).
In a previous study, itEBLN was proposed to exert its antiviral function as a dominant negative mutant of BoDV-1 N (12). A comparison of amino acids in the itEBLN and BoDV-1 N proteins suggested that itEBLN could coassemble with BoDV-1 N into viral RNP and that itEBLN might bind to P, thereby affecting the dynamics of this protein (12). All these assumptions may explain the mechanism of how itEBLN inhibits intracellular replication. In this study, we further noticed that N but not itEBLN contains a nuclear localization signal (NLS) at the N terminus (12). Because of this, NLS-deficient itEBLN may inhibit viral RNP transport from the cytoplasm to the nucleus, similarly to the case of Fv1, a gene derived from an ERV (29). We demonstrated that the antiviral effect of itEBLN on intracellular BoDV-1 replication was much smaller than that on cell-to-cell BoDV-1 spread. At present, actual antiviral mechanisms of itEBLN are not fully understood. However, because itEBLN cannot affect the dynamics of viral RNP in the seed OL/BoDV-GFP cells and amino acid sequence analysis revealed the lack of an NLS in itEBLN, the difference in antiviral effect between intercellular spread and intracellular replication might be explained by the suppression of both viral RNA synthesis and viral RNP transport from the cytoplasm to the nucleus by itEBLN. As a result, itEBLN exerts its antiviral effect on intercellular spread to a greater extent than on intracellular replication, because our minigenome assays considered only the viral RNA synthesis process and not the viral RNP transport process (Fig. 2F and 3B).
Our analyses may also provide insights into the mechanisms of how virus-derived sequences acquire antiviral activity. As described above, itEBLN appears to block at least two different steps in the viral replication cycle. Based on our quantitative calculation, the antiviral effect on intracellular replication was less than 40%, while the antiviral effect in total (i.e., intercellular spread) was 75%. Considering that itEBLN is derived from an ancient bornavirus N protein, the original N sequence integrated into the ancestor species seemingly evolved to become itEBLN by losing its NLS and reducing viral polymerase-supporting activity. If the original sequence has undergone only one of these modifications, the antiviral effect on bornaviruses would be expected to be insufficient. In this case, bornavirus mutants that can escape from restriction mediated by this sequence will readily emerge and such sequences are not maintained in the host genome. Thus, itEBLN may be maintained in the genome because it has uniquely acquired two modifications and the antiviral effect is strong enough to prevent the emergence of bornavirus escape mutants. In other words, multifaceted antiviral effects may be a key mechanism to prevent the emergence of escape viruses, since the probability of viruses to simultaneously acquire multiple mutations to conquer the restriction is very low. Because of this, the sequences with multifaceted antiviral effects are likely maintained in the host genome long term. At present, although bornaviruses seem to have infected ∼3% of squirrels, the infection has not been detected in the subfamily to which the thirteen-lined ground squirrel belongs (30). itEBLN is phylogenetically located within the cluster of extant bornaviruses, suggesting the recent formation of the element. A previous study estimated the integration time of itEBLN as <0.3 million years ago (MYA) via a mechanism of recombination with ERVs and a short interspersed element (31). Based on these lines of information and the antiviral effects of itEBLN we observed, bornavirus infection may still function as a selective pressure for itEBLN-mediated restriction in ground squirrels.
In conclusion, our modeling and quantitative analyses show the impact of antiviral effect by itEBLN on BoDV-1 at the intracellular and intercellular levels. Together with amino acid sequence analysis, these findings lead us to propose that itEBLN effectively blocks BoDV-1 spread by inhibiting viral RNP transport to the nucleus and viral RNA synthesis in the nucleus. Our study emphasizes the importance and usefulness of quantitative analysis to deeply understand the biological processes, including antiviral effects, of EVEs.
MATERIALS AND METHODS
Cells.
OL (a human oligodendrocyte cell line) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Nacalai, Kyoto, Japan) supplemented with 5% fetal bovine serum (FBS). OL cells persistently infected with the huP2Br strain (32) of BoDV-1 (OL/BoDV) and recombinant BoDV-1 carrying the GFP gene cassette (OL/BoDV-GFP [33]) were cultured under the same conditions as the parental cells. OL cells stably expressing itEBLN (OL/itEBLN) were established as described previously (12) and maintained in culture medium with G418 (Invitrogen, Carlsbad, CA).
BoDV-1 spread assay.
OL/BoDV-GFP cells (4 × 102/well) were seeded onto OL, OL/BoDV, or OL/itEBLN cells (4 × 105/well) in 12-well plates. The coculture was passaged every 3 days. At passage, 4 × 105 cells were seeded in new 12-well plates. The spread of BoDV-GFP in the culture was determined by measuring the number of GFP-positive cells using a Tali image-based cytometer (Invitrogen) at the desired time points. Cell viability was determined using a CytoTox-Glo cytotoxicity assay (Promega, Fitchburg, MA).
Minigenome assay.
Minigenome assays were conducted as described previously (28, 34), with some modifications. Briefly, OL cells were transfected with plasmids expressing BoDV-1 N, P, L, and a polymerase II (Pol II)-driven minigenome construct carrying the Gaussia luciferase (GLuc) gene as a reporter gene using Lipofectamine 2000 (Invitrogen). At 18, 24, 48, and 72 h posttransfection, luciferase activities in culture media were measured using a Gaussia luciferase assay kit (New England BioLabs) according to the manufacturer’s instructions. The GLuc activity was normalized to the corresponding cell number.
Data fitting and parameter estimation.
To assess the variability of kinetic parameters and model predictions as described previously (27, 35), we performed Bayesian estimation for the whole data set using Markov chain Monte Carlo (MCMC) sampling (36). To adopt the Bayesian inference model, we assumed a measurement error following normal distribution with mean zero and error variance. A distribution of the error variance following gamma distribution as its prior distribution can be assumed. The MCMC calculation was applied to obtain the distribution of the posterior prediction parameters, which indicate the parameter variability. Distributions of model parameters in equations 1 and 2 were inferred directly by MCMC computations. These estimated parameter values are listed in Tables 2 and 4. We employed a bootstrap t test (37) to quantitatively characterize differences in parameters. In total, 5,000 parameter sets were sampled with replacement from the posterior predictive distributions to calculate the bootstrap t statistics. To avoid potential sampling bias, the bootstrap t test was performed 100 times repeatedly. The averages of the computed P values were used as indicators of differences.
ACKNOWLEDGMENTS
This study was supported in part by Grants-in-Aid for Scientific Research (KAKENHI) Scientific Research B JP18KT0018 (to S.I.), JP18H01139 (to S.I.), JP18H02664 (to T.H.), JP16H04845 (to S.I.), JP17H04083 (to K.T.), Challenging Research (Exploratory) JP18K19449 (to T.H.), JP19K22530 (to K.T.), Scientific Research on Innovative Areas JP19H04839 (to S.I.), JP18H05103 (to S.I.), JP16H06429 (to K.T.), JP16K21723 (to K.T.), and JP16H06430 (to K.T.), AMED CREST JP19gm1310002 (to S.I.), AMED J-PRIDE JP19fm0208006s0103 (to S.I.), JP19fm0208014h (to K.T. and S.I.), and JP19fm0208019h0103 (to S.I.), AMED Research Program on HIV/AIDS JP19fk0410023s0101 (to S.I.), Research Program on Emerging and Re-emerging Infectious Diseases JP19fk0108050h0003 (to S.I.), Program for Basic and Clinical Research on Hepatitis JP19fk0210036h0502 (to S.I.), Program on the Innovative Development and the Application of New Drugs for Hepatitis B JP19fk0310114h0103 (to S.I.), JSPS Core-to-Core Program, JST MIRAI (to S.I.), JST CREST (to S.I.), Mitsui Life Social Welfare Foundation (to S.I.), Shin-Nihon of Advanced Medical Research (to S.I.), Suzuken Memorial Foundation (to S.I.), Life Science Foundation of Japan (to S.I.), SECOM Science and Technology Foundation (to S.I.), The Japan Prize Foundation (to S.I.), Toyota Physical and Chemical Research Institute (to S.I.), the Basic Science Research Program through the National Research Foundation of Korea funded by Ministry of Education grant 2019R1A6A3A12031316 (to K.S.K.), the Shimizu Foundation for Immunology and Neuroscience Grant for 2015 (T.H.), and the Cooperative Research Program (Joint Usage/Research Center program) of the Institute for Frontier Life and Medical Sciences, Kyoto University (to T.H.).
S.I. and T.H. designed the study. Y.Y., K.T., and T.H. conducted the experiments. K.S.K., S.N., and S.I. carried out the computational analysis. S.I. and T.H. supervised the project. All authors wrote and approved the manuscript.
We declare no competing interests.
REFERENCES
- 1.Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 2.Taylor GM, Gao Y, Sanders DA. 2001. Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus. J Virol 75:11244–11248. doi: 10.1128/JVI.75.22.11244-11248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mura M, Murcia P, Caporale M, Spencer TE, Nagashima K, Rein A, Palmarini M. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci U S A 101:11117–11122. doi: 10.1073/pnas.0402877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aswad A, Katzourakis A. 2012. Paleovirology and virally derived immunity. Trends Ecol Evol 27:627–636. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Mi S, Lee X, Li XP, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 6.Kassiotis G, Stoye JP. 2016. Immune responses to endogenous retroelements: taking the bad with the good. Nat Rev Immunol 16:207–219. doi: 10.1038/nri.2016.27. [DOI] [PubMed] [Google Scholar]
- 7.Honda T, Takemoto K, Ueda K. 2019. Identification of a retroelement-containing human transcript induced in the nucleus by vaccination. Int J Mol Sci 20:2875. doi: 10.3390/ijms20122875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, Tomonaga K. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda T, Tomonaga K. 2016. Endogenous non-retroviral RNA virus elements evidence a novel type of antiviral immunity. Mob Genet Elements 6:e1165785. doi: 10.1080/2159256X.2016.1165785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horie M. 2017. The biological significance of bornavirus-derived genes in mammals. Curr Opin Virol 25:1–6. doi: 10.1016/j.coviro.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Honda T. 2017. Potential links between hepadnavirus and bornavirus sequences in the host genome and cancer. Front Microbiol 8:2537. doi: 10.3389/fmicb.2017.02537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujino K, Horie M, Honda T, Merriman DK, Tomonaga K. 2014. Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc Natl Acad Sci U S A 111:13175–13180. doi: 10.1073/pnas.1407046111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staeheli P, Sauder C, Hausmann J, Ehrensperger F, Schwemmle M. 2000. Epidemiology of Borna disease virus. J Gen Virol 81:2123–2135. doi: 10.1099/0022-1317-81-9-2123. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann B, Tappe D, Höper D, Herden C, Boldt A, Mawrin C, Niederstraßer O, Müller T, Jenckel M, van der Grinten E, Lutter C, Abendroth B, Teifke JP, Cadar D, Schmidt-Chanasit J, Ulrich RG, Beer M. 2015. A variegated squirrel bornavirus associated with fatal human encephalitis. N Engl J Med 373:154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- 15.Schlottau K, Forth L, Angstwurm K, Höper D, Zecher D, Liesche F, Hoffmann B, Kegel V, Seehofer D, Platen S, Salzberger B, Liebert UG, Niller HH, Schmidt B, Matiasek K, Riemenschneider MJ, Brochhausen C, Banas B, Renders L, Moog P, Wunderlich S, Seifert CL, Barreiros A, Rahmel A, Weiss J, Tappe D, Herden C, Schmidt-Chanasit J, Schwemmle M, Rubbenstroth D, Schlegel J, Pietsch C, Hoffmann D, Jantsch J, Beer M. 2018. Fatal encephalitic Borna disease virus 1 in solid-organ transplant recipients. N Engl J Med 379:1377–1379. doi: 10.1056/NEJMc1803115. [DOI] [PubMed] [Google Scholar]
- 16.Korn K, Coras R, Bobinger T, Herzog SM, Lücking H, Stöhr R, Huttner HB, Hartmann A, Ensser A. 2018. Fatal encephalitis associated with Borna disease virus 1. N Engl J Med 379:1375–1377. doi: 10.1056/NEJMc1800724. [DOI] [PubMed] [Google Scholar]
- 17.Liesche F, Ruf V, Zoubaa S, Kaletka G, Rosati M, Rubbenstroth D, Herden C, Goehring L, Wunderlich S, Wachter MF, Rieder G, Lichtmannegger I, Permanetter W, Heckmann JG, Angstwurm K, Neumann B, Märkl B, Haschka S, Niller HH, Schmidt B, Jantsch J, Brochhausen C, Schlottau K, Ebinger A, Hemmer B, Riemenschneider MJ, Herms J, Beer M, Matiasek K, Schlegel J. 2019. The neuropathology of fatal encephalomyelitis in human Borna virus infection. Acta Neuropathol 138:653–665. doi: 10.1007/s00401-019-02047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honda T. 2020. Relaunching human bornavirus research from encephalitis cases with unclear cause. Lancet Infect Dis 20:389–391. doi: 10.1016/S1473-3099(19)30740-6. [DOI] [PubMed] [Google Scholar]
- 19.Niller HH, Angstwurm K, Rubbenstroth D, Schlottau K, Ebinger A, Giese S, Wunderlich S, Banas B, Forth LF, Hoffmann D, Höper D, Schwemmle M, Tappe D, Schmidt-Chanasit J, Nobach D, Herden C, Brochhausen C, Velez-Char N, Mamilos A, Utpatel K, Evert M, Zoubaa S, Riemenschneider MJ, Ruf V, Herms J, Rieder G, Errath M, Matiasek K, Schlegel J, Liesche-Starnecker F, Neumann B, Fuchs K, Linker RA, Salzberger B, Freilinger T, Gartner L, Wenzel JJ, Reischl U, Jilg W, Gessner A, Jantsch J, Beer M, Schmidt B. 2020. Zoonotic spillover infections with Borna disease virus 1 leading to fatal human encephalitis, 1999–2019: an epidemiological investigation. Lancet Infect Dis 20:467–477. doi: 10.1016/S1473-3099(20)30379-0. [DOI] [PubMed] [Google Scholar]
- 20.Honda T, Tomonaga K. 2013. Nucleocytoplasmic shuttling of viral proteins in Borna disease virus infection. Viruses 5:1978–1990. doi: 10.3390/v5081978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosztonyi G, Ludwig H. 1995. Borna disease—neuropathology and pathogenesis. Curr Top Microbiol Immunol 190:39–73. [PubMed] [Google Scholar]
- 22.Lennartz F, Bayer K, Czerwonka N, Lu Y, Kehr K, Hirz M, Steinmetzer T, Garten W, Herden C. 2016. Surface glycoprotein of Borna disease virus mediates virus spread from cell to cell. Cell Microbiol 18:340–354. doi: 10.1111/cmi.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clemente R, Juan C. 2007. Cell-to-cell spread of Borna disease virus proceeds in the absence of the virus primary receptor and furin-mediated processing of the virus surface glycoprotein. J Virol 81:5968–5977. doi: 10.1128/JVI.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujino K, Yamamoto Y, Daito T, Makino A, Honda T, Tomonaga K. 2017. Generation of a non‐transmissive Borna disease virus vector lacking both matrix and glycoprotein genes. Microbiol Immunol 61:380–386. doi: 10.1111/1348-0421.12505. [DOI] [PubMed] [Google Scholar]
- 25.Honda T, Horie M, Daito T, Ikuta K, Tomonaga K. 2009. Molecular chaperone BiP interacts with Borna disease virus glycoprotein at the cell surface. J Virol 83:12622–12625. doi: 10.1128/JVI.01201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwami S, Sato K, De Boer RJ, Aihara K, Miura T, Koyanagi Y. 2012. Identifying viral parameters from in vitro cell cultures. Front Microbiol 3:319. doi: 10.3389/fmicb.2012.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwami S, Takeuchi JS, Nakaoka S, Mammano F, Clavel F, Inaba H, Kobayashi T, Misawa N, Aihara K, Koyanagi Y, Sato K. 2015. Cell-to-cell infection by HIV contributes over half of virus infection. Elife 4:e08150. doi: 10.7554/eLife.08150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanai H, Hayashi Y, Watanabe Y, Ohtaki N, Kobayashi T, Nozaki Y, Ikuta K, Tomonaga K. 2006. Development of a novel Borna disease virus reverse genetics system using RNA polymerase II promoter and SV40 nuclear import signal. Microbes Infect 8:1522–1529. doi: 10.1016/j.micinf.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Sanz-Ramos M, Stoye JP. 2013. Capsid-binding retrovirus restriction factors: discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol 94:2587–2598. doi: 10.1099/vir.0.058180-0. [DOI] [PubMed] [Google Scholar]
- 30.Schlottau K, Jenckel M, van den Brand J, Fast C, Herden C, Höper D, Homeier-Bachmann T, Thielebein J, Mensing N, Diender B, Hoffmann D, Ulrich RG, Mettenleiter TC, Koopmans M, Tappe D, Schmidt-Chanasit J, Reusken CBEM, Beer M, Hoffmann B. 2017. Variegated squirrel bornavirus 1 in squirrels, Germany and the Netherlands. Emerg Infect Dis 23:477–481. doi: 10.3201/eid2303.161061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki Y, Kobayashi Y, Horie M, Tomonaga K. 2014. Origin of an endogenous bornavirus-like nucleoprotein element in thirteen-lined ground squirrels. Genes Genet Syst 89:143–148. doi: 10.1266/ggs.89.143. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura Y, Takahashi H, Shoya Y, Nakaya T, Watanabe M, Tomonaga K, Iwahashi K, Ameno K, Momiyama N, Taniyama H, Sata T, Kurata T, de la Torre JC, Ikuta K. 2000. Isolation of Borna disease virus from human brain tissue. J Virol 74:4601–4611. doi: 10.1128/.74.10.4601-4611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daito T, Fujino K, Honda T, Matsumoto Y, Watanabe Y, Tomonaga K. 2011. A novel Borna disease virus vector system that stably expresses foreign proteins from an intercistronic noncoding region. J Virol 85:12170–12178. doi: 10.1128/JVI.05554-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kojima S, Honda T, Matsumoto Y, Tomonaga K. 2014. Heat stress is a potent stimulus for enhancing rescue efficiency of recombinant Borna disease virus. Microbiol Immunol 58:636–642. doi: 10.1111/1348-0421.12193. [DOI] [PubMed] [Google Scholar]
- 35.Iwanami S, Kakizoe Y, Morita S, Miura T, Nakaoka S, Iwami S. 2017. A highly pathogenic simian/human immunodeficiency virus effectively produces infectious virions compared with a less pathogenic virus in cell culture. Theor Biol Med Model 14:9. doi: 10.1186/s12976-017-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soetaert K, Petzoldt T. 2010. Inverse modelling, sensitivity and Monte Carlo analysis in R using package FME. J Stat Soft 33:1–28. doi: 10.18637/jss.v033.i03. [DOI] [Google Scholar]
- 37.Efron B, Tibshirani R. 1993. An introduction to the bootstrap. Chapman & Hall, New York, NY. [Google Scholar]