Feline leukemia virus (FeLV) can infect a variety of felid species. Only the primary domestic cat host and related small cat species harbor a related endogenous virus in their genomes. Previous studies noted a negative association between the endogenous virus copy number and exogenous virus infection in domestic cats. This report shows that puma cells, which lack endogenous FeLV, produce more virus more rapidly than domestic cat fibroblasts following cell culture challenge. We document a strong association between domestic cat cell susceptibility and FeLV long terminal repeat (LTR) copy number, similar to observations in natural FeLV infections. Viral replication does not, however, correlate with FeLV env copy number, suggesting that this effect is specific to FeLV-LTR elements. This discovery indicates a protective capacity of the endogenous virus against the exogenous form, either via direct interference or indirectly via gene regulation, and may suggest evolutionary outcomes of retroviral endogenization.
KEYWORDS: endogenous retroviruses, FeLV, Felis catus, Puma concolor, retroviral interactions
ABSTRACT
While feline leukemia virus (FeLV) has been shown to infect felid species other than the endemic domestic cat host, differences in FeLV susceptibility among species has not been evaluated. Previous reports have noted a negative correlation between endogenous FeLV (enFeLV) copy number and exogenous FeLV (exFeLV) infection outcomes in domestic cats. Since felids outside the genus Felis do not harbor enFeLV genomes, we hypothesized absence of enFeLV results in more severe disease consequences in felid species lacking these genomic elements. We infected primary fibroblasts isolated from domestic cats (Felis catus) and pumas (Puma concolor) with FeLV and quantitated proviral and viral antigen loads. Domestic cat enFeLV env and long terminal repeat (LTR) copy numbers were determined for each individual and compared to FeLV viral outcomes. FeLV proviral and antigen levels were also measured in 6 naturally infected domestic cats and 11 naturally infected Florida panthers (P. concolor coryi). We demonstrated that puma fibroblasts are more permissive to FeLV than domestic cat cells, and domestic cat FeLV restriction was highly related to enFeLV-LTR copy number. Terminal tissues from FeLV-infected Florida panthers and domestic cats had similar exFeLV proviral copy numbers, but Florida panther tissues have higher FeLV antigen loads. Our work indicates that enFeLV-LTR elements negatively correlate with exogenous FeLV replication. Further, Puma concolor samples lacking enFeLV are more permissive to FeLV infection than domestic cat samples, suggesting that endogenization can play a beneficial role in mitigating exogenous retroviral infections. Conversely, presence of endogenous retroelements may relate to new host susceptibility during viral spillover events.
IMPORTANCE Feline leukemia virus (FeLV) can infect a variety of felid species. Only the primary domestic cat host and related small cat species harbor a related endogenous virus in their genomes. Previous studies noted a negative association between the endogenous virus copy number and exogenous virus infection in domestic cats. This report shows that puma cells, which lack endogenous FeLV, produce more virus more rapidly than domestic cat fibroblasts following cell culture challenge. We document a strong association between domestic cat cell susceptibility and FeLV long terminal repeat (LTR) copy number, similar to observations in natural FeLV infections. Viral replication does not, however, correlate with FeLV env copy number, suggesting that this effect is specific to FeLV-LTR elements. This discovery indicates a protective capacity of the endogenous virus against the exogenous form, either via direct interference or indirectly via gene regulation, and may suggest evolutionary outcomes of retroviral endogenization.
INTRODUCTION
The vast majority of vertebrate genomes, including up to 8% of the human genome, harbor remnants of ancient viral infections made up predominantly of retroviral genetic material (1–3). During infection, the retroviral RNA genome is reverse transcribed to form double-stranded DNA, which is in turn integrated into the host’s genome (4). While most of these infections target somatic cells, these viruses are capable of infecting and integrating into germ cells (5). The consequences of viral integration into the germ line is profound, and ultimately the virus is vertically transmitted as permanent genetic elements inherited in a Mendelian fashion (6). Fixation of the retroviral content in host genomes is a process termed endogenization and leads to new host genetic elements called endogenous retroviruses (ERVs) (7). ERVs in their early stages are believed to undergo massive changes during host cell transcription, and the foreign, potentially deleterious genetic material accumulates mutations and deletions that often render the newly endogenized virus defunct (8). While typically unable to produce infectious virions, many ERVs are still capable of undergoing transcription and may even produce functional viral proteins. Certain ERVs are known to function in important physiologic, cellular, or biological processes, including placentation, oncogenesis, immune modulation, and infectious disease progression (9, 10).
Endogenous feline leukemia virus (enFeLV) is an example of an ERV which has a horizontally transmitted retroviral counterpart (feline leukemia virus [FeLV]). Only members of the Felis genus harbor enFeLV, as endogenization is believed to have originated after the Felis genus split off from other members of the Felidae family (11, 12). FeLV can infect felid species that harbor enFeLV (i.e., domestic cats) as well as species that lack enFeLV (i.e., puma). FeLV epizootics have been documented in multiple non-Felis species populations including the North American puma (Puma concolor) (13–19).
FeLV represents an endogenous-exogenous retroviral system that has perhaps been best studied with regard to disease biology and outcome during naturally occurring infections in an outbred, highly dispersed mammalian host. Thus, evaluation of this system provides opportunities to better understand ERV-exogenous viral interactions that are highly relevant to virus and host evolution and ecology. FeLV epizootics in wild felids are characterized by serious disease of epidemic proportions (20, 21), whereas FeLV infection in adult cats frequently results in regressive and abortive infections (22). It has been hypothesized that enFeLV may be associated with differences in infection outcome. We previously demonstrated that enFeLV long terminal repeat (LTR) copy number was associated with better infection outcomes during a natural FeLV outbreak in a multicat household (23). Here, we evaluate FeLV infection of puma and domestic cat cells in vitro and in situ to examine the susceptibility of endemic and novel hosts to FeLV infection with respect to enFeLV to provide further evaluation of this relationship.
RESULTS
Primary fibroblasts were successfully propagated from ear punches from three free-ranging puma (two kittens of unknown sex and one adult male) and 1 abdominal skin incision from a mature adult female captive puma. Primary fibroblasts were cultured from 7 domestic cat abdominal full-skin biopsies from necropsied cats at Colorado State University (6 male, 1 female).
Fibroblast infections.
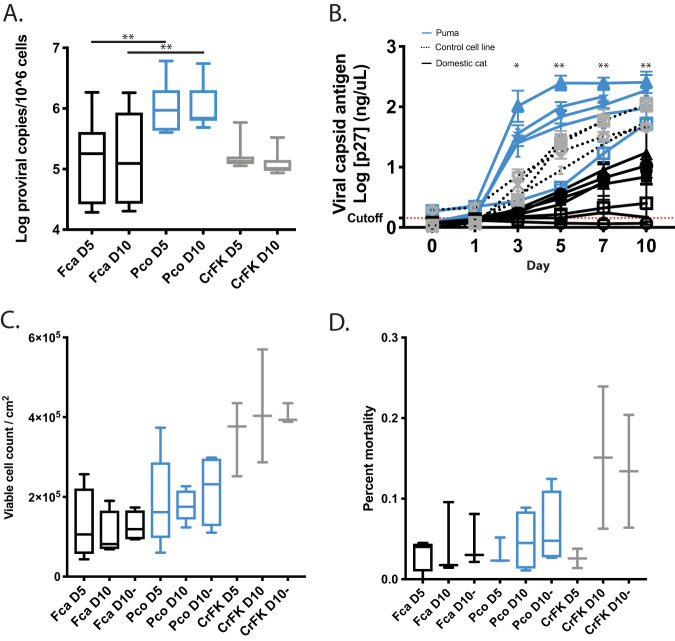
Citing a negative correlation between enFeLV-LTR and infection levels in previous literature, we hypothesized that enFeLV would negatively correlate to viral replication measured by antigen load following experimental cell culture infections. At days 5 and 10, puma cells were infected with approximately 1 FeLV provirus per cell, whereas domestic cat cell infections were approximately 1 log lower. Experimental control FeLV infections of Crandell-Rees feline kidney (CrFK) cells displayed reproducible levels of infections with tight interquartile ranges, while domestic cat and puma fibroblast infections displayed greater individual variance (Fig. 1A). Exogenous FeLV proviral load was significantly higher in puma versus domestic cat fibroblasts (Fig. 1A, Kruskal-Wallis test—Fca D5 versus Pco D5, H = −29.8, P < 0.01; Fca D10 versus Pco D10, H = −32.55, P < 0.01). FeLV viral antigen production was similarly greater in puma fibroblasts (Fig. 1B). Puma cell-generated FeLV antigen detected in the supernatant was greater than in domestic cat cultures beginning on day 3 and continued to be greater for the rest of the experiment (repeated measures analysis of variance [ANOVA]; F = 8.78; P = 0.001).
FIG 1.
(A) Puma fibroblasts exposed to FeLV have greater FeLV proviral load compared to that of domestic cat fibroblasts. On both day 5 and day 10, puma cells (Pco) demonstrated significantly higher proviral copy numbers than domestic cat primary fibroblasts (Kruskal-Wallis; **, P < 0.01). Variance between domestic cat (Fca) cells was greater than that of puma cells, while CrFK cell infections displayed the least amount of variation. (B) FeLV viral capsid antigen (p27) production is higher in puma than in domestic cat cells. Beginning day 3 postinfection, puma cells replicated more virus than domestic cat cells (repeated measure ANOVA individual test statistics, *, P < 0.05; **, P < 0.01). One puma, one domestic cat, and one control cat experiment were ended at day 7. Cutoff values were established by 3× standard error above average value for negative control wells (red line). Viable cell numbers and cell mortality increase over time and are similar in puma and domestic cat cultures. (C) Puma (blue) and domestic cat (black) fibroblast cell counts increased over time and reached 100% confluence by day 5. Initial seeding density was 5 × 104 cells per 2 cm2. Differences in domestic cat and puma fibroblast cell counts were not significant between day 5 and day 10. Minus signs denote uninfected cell cultures. (D) Puma (blue) and domestic cat (black) fibroblast mortality increased over time following 100% confluence at day 5. Differences in domestic cat and puma fibroblast cell counts were not significant between day 5 and day 10. Minus signs denote uninfected cell cultures.
Domestic cat and puma viable cell counts were equivalent on day 5 or 10, averaging 1.28 × 105 cells per 2 cm2 for domestic cat cells and 1.92 × 105 cells per 2 cm2 for puma cells (Fig. 1C). CrFK cells are smaller than domestic cat and puma primary fibroblasts, and therefore, cell density was higher in these cultures (3.54 × 105 cells per 2 cm2 at day 5 and 4.13 × 105 cells per 2 cm2 at day 10). Percent dead cells (as measured by trypan blue exclusion) on days 5 and 10 ranged between 3.94 ± 0.70% for domestic cat cells and 4.73 ± 1.45% for puma cells regardless of infection status (Fig. 1D). There was a consistent trend for lower percent mortality at day 5 when cells first reached confluence. CrFK cells experienced greater cell mortality over the course of the infection regardless of infection status (2.59% at day 5 and 14.26% at day 10).
enFeLV-exFeLV correlation.
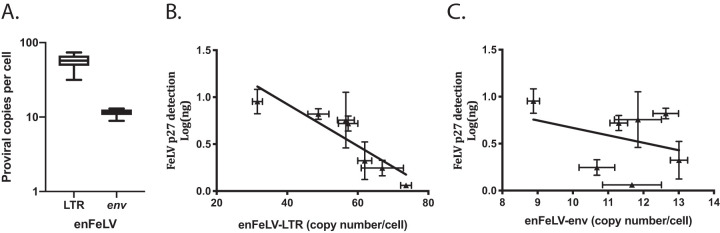
As anticipated, domestic cat cells harbored more enFeLV-LTRs than enFeLV env genes (Fig. 2A). Normalized LTR sequence copy numbers in domestic cat cells ranged from 32 to 74 copies per cell with an average of 57 copies per cell. Copy numbers for enFeLV env were significantly lower, ranging from 9 to 13 copies per cell with an average of 11 copies.
FIG 2.
(A) enFeLV proviral loads vary between individual cats. Domestic cat cells (n = 7) display variation between enFeLV-LTRs and env. In individual cats, LTR copy number was greater than env copy number. enFeLV-LTR ranged between 9 and 13 copies per cell. enFeLV-env was more variable ranging between 32 and 74 copies per cell. (B) Domestic cat enFeLV-LTR copy number was negatively correlated with FeLV antigen production (Pearson’s coefficient = −0.8943; P = 0.0066). (C) Domestic cat enFeLV-env copy number does not correlate with FeLV antigen production (Pearson’s coefficient = −0.1071; P = 0.8397), suggesting a direct role of enLTR in suppression of FeLV replication.
Variation in domestic cat fibroblast enFeLV-LTR copy number correlated to FeLV antigen loads (day 7, Pearson’s correlation coefficient = −0.894; P < 0.05; Fig. 2B), whereas variation in enFeLV env did not (day 7, Pearson’s correlation coefficient = −0.107; P = 0.840; Fig. 2C). enFeLV-exogenous FeLV (exFeLV) correlations were calculated at day 7 since this is the time point that cells reached complete confluence and the rate at which antigen was produced waned after this time point. Linear regression analysis of FeLV proviral load against antigen load showed that only 44% of the variation in antigen production could be explained by proviral load (R2 = 0.438; Fig. 3).
FIG 3.
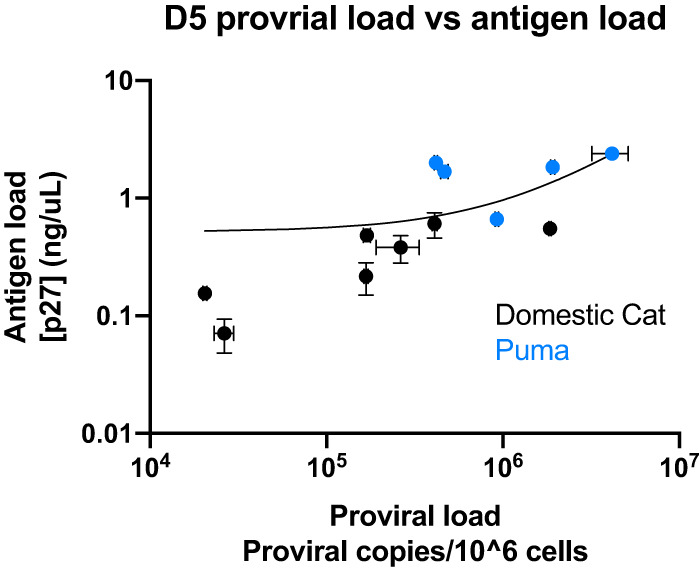
Proviral load does not highly correlate with viral antigen load. Linear regression analysis shows only 44% of variation in antigen production can be explained by proviral load following experimental FeLV infection of domestic cat (black) and puma (blue) cells. This indicates that the amount of viral antigen (a proxy for viral replication) is not governed completely by the degree of infection as measured by proviral load. Horizontal and vertical error bars indicate standard error for proviral load and viral antigen load, respectively.
FeLV proviral and antigen loads during natural infection.
FeLV loads in bone marrow, spleen, thymus, and peripheral lymph nodes were assessed in 6 experimentally infected cats and 11 naturally infected pumas (not all tissues were available for each animal; see Table 1). Both tissue proviral load and tissue antigen load failed the Kolmogorov-Smirnov test for log-normality, indicating a non-normal distribution. FeLV proviral load was greater in domestic cats compared to Florida panther (Puma concolor coryi, a recognized subspecies of Puma concolor) by a median difference of 1.07 by Mann-Whitney test of log-transformed copy numbers per cell (U = 122; P = 0.0001; Fig. 4A). Despite lower mean proviral load in Florida panther tissues, p27 capsid antigen in tissues tended to be higher (median value of 3.41 versus 2.87; Mann-Whitney test, U = 250; P = 0.127; Fig. 4B). There was no difference in antigen or proviral load among tissues, with the exception of bone marrow antigen.
TABLE 1.
Tissues from FeLV-positive domestic cats and pumas used for qPCR and ELISA testing
| Tissue | No. domestic cat samples (n = 6) | No. puma samples (n = 11) |
|---|---|---|
| Bone marrow | 6 | 6 |
| Lymph node | 5 | 9 |
| Spleen | 6 | 10 |
| Thymus | 6 | 4 |
FIG 4.
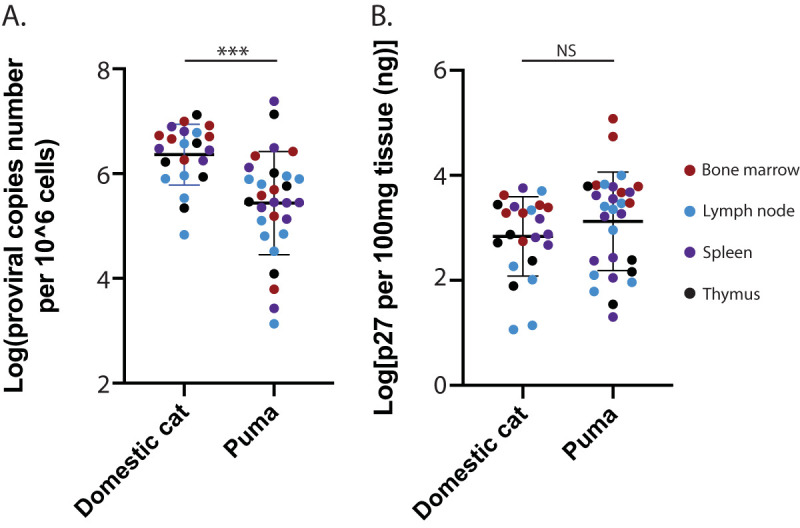
Lymphoid tissues from naturally infected FeLV-positive domestic cats and pumas have varying viral loads at end-stage disease. (A) Domestic cat lymphoid tissues displayed greater FeLV proviral load compared to puma tissues (Mann-Whitney test; ***, P < 0.0001). Puma tissues had a much wider range of viral loads, which is potentially attributable to sample quality. (B) Puma tissues trended to greater amounts of viral antigen (Mann-Whitney test; P = 0.1495).
DISCUSSION
Alterations in infectivity and virulence have been noted following cross-species viral transmission (24). Pathogen spillover into novel species can result in dead-end hosts with no follow-on transmission (infectious hematopoietic necrosis virus [25]; feline immunodeficiency virus strain lru [26]). In other cases, spillover results in active infections that maintain persistent transmission in the new host (mycoplasmosis [27]; feline foamy virus [FFV] [28]). Most problematic are cases of spillover that result in adaptation of the virus in novel hosts leading to increased morbidity and mortality (HIV [29]; SARS-CoV-2 [30]). We have limited understanding of host and ecological factors that underlie these different outcomes. FeLV spillover events have been documented in free-ranging pumas with resultant significant morbidity, suggesting enhanced virulence in the new host (13, 31). Here, we evaluate the hypothesis that the presence of an endogenous viral element may restrict competent viral replication in the primary host as one explanation of this specific spillover outcome.
FeLV replicates more rapidly and productively in puma versus domestic cat fibroblasts. Experimental infections in this study established differences in FeLV replication in puma and domestic cat cells in the absence of immunological and physiological parameters. Thus, at a cellular level, puma cells appear to be more competent at supporting FeLV infection and replication than fibroblasts of the primary host, domestic cats.
The enFeLV-LTR copy number is associated with resistance to FeLV infection and antigen production. Endogenous elements constitute a sizable component of an animal’s genome (2), and solo LTRs vastly outnumber full endogenous genomes/pseudogenomes (7). Solo LTRs are left behind from full genomes following a process that occurs most frequently in early endogenization. The two flanking LTRs in the full genome allow for the intervening genes to be removed by homologous recombination (32). As such, env copy number serves as a proxy for full enFeLV genomes, whereas LTR copy number represents full enFeLV genomes plus a solo LTR remaining following recombination and retrotransposition (32). In this sampling of domestic cat fibroblast enFeLVs, solo LTRs range from 32 to 74 copies per cell, while env ranges from 9 to 13 copies per cell (Fig. 2A). This is consistent with previously reported measures of a full-length enFeLV range of 6 to 26 copies per cell (23, 33, 34) versus 19 to 58 copies of enFeLV-LTR per cell (23). LTR copy number variation, but not full enFeLV genomes, correlated with FeLV replication as evidenced by directly proportional antigen load, suggesting a direct effect of the LTR element in modulation of exFeLV replication.
While FeLV replication may intuitively seem to be correlated to exFeLV proviral load, only 44% of FeLV antigen variation was explained by proviral copy number (Fig. 3). Linear regression analysis suggests that factors other than the number of exFeLV proviral units contribute to host susceptibility. This finding corroborates observations made in vivo in a domestic cat colony naturally infected with FeLV (23), strongly suggesting that enFeLV-LTR, versus other enFeLV elements, interact with exFeLV to mitigate infection.
CrFK cell proviral copy number and viral replication had a much smaller range than primary domestic cat and puma cell infections, with domestic cat variation exceeding individual puma range (Fig. 1). This observation related to domestic cat cells can be explained by the constant enFeLV-LTR copy number of this cell line (44 copies per cell).
One mechanism by which endogenous retroviruses may interact with exogenous retroviral infection relates to LTR enhancer and promoter regions that stimulate proximal host gene transcription. LTRs may activate antiviral or innate immune genes in close proximity to LTR insertion sites, resulting in protection against exogenous retroviruses. This indirect ERV modulation has been documented in other systems and can be mediated through cis activation (promoter) or trans activation (enhancer) of host genes (35). In the case of murine leukemia virus (MuLV), endogenous MuLV-LTRs have been co-opted to transcribe host antiviral genes, including but not limited to APOBEC3, a potent host retroviral restriction factor (36).
Alternatively, it is possible that the enFeLV genetic elements may directly interfere with exFeLV infection by encoding for small interfering RNAs or P-element-induced wimpy (PIWI)-interacting RNAs that activate host endoribonuclease DICER complexes to specifically target FeLV transcripts (37, 38). Our results are more suggestive of direct interference mechanisms due to the linearity of the FeLV restriction afforded by enFeLV-LTRs. It is unlikely that all LTR integration sites are influencing the transcription of host genetic factors and would have an effect in a dose-dependent manner.
While we have made an interesting correlation between enFeLV-LTR and viral replication capacity, other host-related factors undoubtedly influence viral production in puma cells or restriction of viral replication in domestic cats. In support of this, we observed greater variation in viral infection outcome in puma cells than in cat cells, suggesting that other host restriction factors modulate individual viral replication capacity in addition to enFeLV-LTR. Nevertheless, the in vitro system that we have developed here permits a unique interrogation of exFeLV in the presence and absence of an endogenous analogue that can be compared to the biological behavior of FeLV in free-ranging puma and naturally infected domestic cats.
FeLV reaches high viral load in lymphoid tissues during natural infections. While exFeLV proviral loads in naturally infected pumas were generally lower than in domestic cats, antigen load was equivalent between the two populations. Field collections were performed opportunistically on Florida panthers when animals were either found deceased or hit by vehicle, often hours to days after death occurred. In contrast, FeLV-positive shelter cats were euthanized prior to death, and tissues were collected rapidly following death. Challenges associated with field collection, including time from death to sample collection and lack of ideal laboratory conditions for sample processing, may have contributed to poorer DNA quality from extracted puma tissues. While normalization to feline C-C chemokine receptor type 5 (CCR5) helped to address these issues for proviral copy number calculations, it is possible that viral antigen loads measured in puma tissues underestimate actual values.
Biological aspects of FeLV transmission that differ between domestic cats and pumas may impact subsequent infection kinetics. The initial spillover events of FeLV to pumas has been associated with predation of domestic cats (13, 39), whereas FeLV in domestic cats is believed to be transmitted in households through social interactions, such as grooming or via antagonistic interactions (11). Domestic cats interact socially, so behaviors like grooming may sustain infection in animals in close contact, and infections may result from repeated exposures. Unlike domestic cats, pumas are much more solitary, and interactions between pumas outside of mother-offspring groups are primarily believed to be antagonistic (40).
No differences exist between cell culture parameters in relation to FeLV infection. Gammaretroviruses require the dissolution of the nucleus during mitosis in order to integrate into cells (41), and therefore, dividing cells are more susceptible to FeLV infection and replication. We measured cell count as a proxy for rate of cell division and percent cell mortality as a measure of viability. Neither measure differed between domestic cat and puma fibroblasts, though CrFK cells displayed greater cell count and cell mortality at confluence (Fig. 1C and D). Immortalized cell lines have accumulated multiple changes that fundamentally alter their morphology and physiologic behaviors, including decreases in contact inhibition (42). FeLV proviral integration and antigen production in CrFK cell infections had far less within- and between-run variation than primary fibroblasts, likely attributable to the clonal nature of CrFK cells versus wild-type-derived primary tissue cultures. Observation of cell culture parameters did not suggest differences in growth characteristics that explain the variant FeLV susceptibly of puma and domestic cat fibroblasts.
In this report, we present information that demonstrates a strong correlation between enFeLV-LTR copy number and protection against exFeLV infection in vitro and examine correlates for naturally occurring infections. We hypothesize that enFeLV-LTR restriction may manifest as direct interference through RNA silencing mechanisms or by indirect enFeLV-LTR-mediated promotion of host antiviral genes. This interesting system provides an opportunity to directly interrogate these mechanisms in an outbred population.
MATERIALS AND METHODS
Sample acquisition.
Full-skin thickness biopsies from puma were collected by ear punch by Colorado Parks and Wildlife under approved IACUC 16-2008. Abdominal skin samples were opportunistically collected from domestic cats during necropsies performed at the Colorado State University College of Veterinary Medicine. Primary fibroblasts were isolated as previously described (43) and cultured in 20% fetal bovine serum (FBS)-supplemented Dulbecco modified Eagle medium (DMEM) high-glucose medium and 1× antibiotic-antimycotic solution (Gibco; penicillin/streptomycin/fungizone). One puma cell culture was infected with feline foamy virus and was treated prophylactically with the antiretroviral drug zidovudine (AZT) (100 μg/ml; Sigma) per manufacturer’s direction in cultures where FFV cytopathic effect (CPE) was detected. Cells were passaged two times for approximately 10 days in medium without AZT washout prior to infection. Primary cultures were expanded for, at most, four passages before being frozen (in 20% dimethyl sulfoxide [DMSO], 10% FBS, 70% serum free DMEM) using a freezing container (Nalgene) and stored at −80°C.
Bone marrow, thymus, spleen, and lymph node from naturally FeLV-infected domestic cats (n = 6) and Florida panthers (P. concolor coryi, n = 11) were collected and shipped to the Colorado State University Feline Retrovirus Research Laboratory for additional testing. Panther tissues were collected by the Florida Fish and Wildlife Conservation Commission from animals that had succumbed to FeLV following the introduction of the virus in two epizootics (13, 21). Domestic cat tissues were obtained from cats with terminal FeLV disease graciously provided by Animal Protective League (Springfield, IL). Puma and domestic cat lymphoid tissues were prepared for enzyme-linked immunosorbent assay (ELISA) and quantitative PCR (qPCR) assays as described below. DNA was extracted by bead-beater disruption and phenol-chloroform extraction using previously reported methods (13) to measure FeLV proviral load. Tissues were homogenized by bead-beater disruption (6.0 m/s for 60 s) in phosphate-buffered saline (PBS) with protease inhibitor (Pierce, Waltham, MA). Homogenates were diluted to 1% and 0.1% tissue for p27 capsid antigen quantification.
Mycoplasma-free Crandell-Rees feline kidney (CrFK) cells were generously obtained by Martin Lochelt. Infections occurred when cells were between passage numbers 4 and 15.
Virus titration.
Crandell-Rees feline kidney cells were plated at a density of 1,250 cells per 0.32 cm2 in a flat-bottomed 96-well plate. FeLV-61E was obtained (gift of Edward Hoover and Candace Mathiason), and CrFK cells were infected in quintuplicate following a 10-fold dilution series. Cells were washed with sterile PBS, were given fresh medium, and were incubated with 5% CO2 at 37°C for 10 days. Titration was repeated three times. FeLV antigen ELISA detection, described below, was used to detect viral capsid antigen p27 in the supernatant. The quantity of virus necessary to infect 50% of tissue cultures (TCID50) was calculated by previously published methods (44).
enFeLV and exFeLV quantification by real-time qPCR.
LTR and env enFeLV copy number was quantified in domestic cat cells. env was used as a proxy for full-length endogenous FeLV, and LTR copy number detected both full-length enFeLV as well as solo LTRs. Exogenous FeLV proviral DNA was measured by a third qPCR protocol targeting exFeLV-specific LTRs, which vary from enFeLV (22). enFeLV-env, enFELV-LTR, and exFeLV-LTR primers and probes were previously designed, and reactions were performed as described (45) on a Bio-Rad CFX96 thermocycler. In order to determine enFeLV and exFeLV proviral load, quantified FeLV was normalized against feline CCR5 (C-C chemokine receptor type 5 [46]), recognizing both domestic cat and puma CCR5 sequences. We used the ΔΔCT method accounting for two CCR5 genes per cell (47). Custom DNA oligonucleotides were synthetically constructed with target regions of enFeLV-LTR and env, exFeLV, and CCR5 on one DNA construct for quantification (gBlocks, IDT) (Fig. 5).
FIG 5.
Oligonucleotides synthesized for qPCR quantification. Two gBlocks were synthesized commercially (Integrated DNA Technologies, Coralville, IA). Both concatenated gBlocks contained sequences for FeLV-61E (blue) and CCR5 (yellow). One contained enFeLV-LTR sequence, and the second contained enFeLV-env sequence. Bold sequences represent forward and reverse primer binding sites.
Primer and probe sequences and qPCR thermocycling conditions are reported in Table 2 (22). exFeLV qPCRs contain 400 nM both primers, 80 nM probe, iTaq universal probe supermix (Bio-Rad), water, and DNA template. CCR5 exists as two copies per cell. Puma-specific CCR5 primers and probes adapted from Howard et al. (46) were used to normalize FeLV copy numbers per 106 cells (Table 2). The probe was labeled with FAM (6-carboxyfluorescein) reporter dye at the 5′ end, ZEN (Integrated DNA Technologies [IDT], Coralville, Iowa) internal quencher, and IABkFQ (Iowa Black fluorescein quencher; IDT, Coralville, Iowa) at the 3′ end. CCR5 qPCR mixtures contained 200 nM forward primer, 500 nM reverse primer, and 200 nM probe, iTaq universal probe supermix (Bio-Rad), water, and DNA template. FeLV and CCR5 reactions were run simultaneously on the same plate on a Bio-Rad CFX96 at 95°C for 3 min, followed by 40 cycles of 95°C for 5 s and 60°C for 15 s. The limit of detection for this assay is ≥10 copies per reaction. Standards for this assay were created as custom synthetic oligonucleotides (gBlocks, IDT) containing a relevant fragment of the exogenous FeLV and CCR5 genes (Fig. 5). Standard dilution and controls were run in duplicate, and samples were run in triplicate.
TABLE 2.
Primer sequences and cycling conditions used for qPCRsa
The cycling conditions shown apply to all regions.
Viral infection.
Primary puma and domestic cat fibroblast cultures passaged fewer than five times were cultured in 20% FBS-supplemented DMEM high-glucose medium. Cells were plated at a density of 50,000 cells per 2 cm2 in a 24-well plate and infected with a multiplicity of infection (MOI) of 0.01 FeLV-61E in triplicate and cultured with 1.2 ml medium. A total of 120 μl of supernatant was collected and stored at 80°C on days 0, 1, 3, 5, 7, and 10 for detection of p27 ELISA. On days 5 and 10, cells were harvested to determine cellular viability based on cell number and percent mortality by counting cells stained with trypan blue (Gibco) on a hemocytometer. One domestic cat cell culture, one puma cell culture, and one CrFK cell culture infection were terminated at day 7 due to equipment failure. One puma primary cell culture triplicate infection was repeated twice.
ELISA.
FeLV capsid antigen p27 was measured by sandwich ELISA. Costar Immulon 2HB plates were coated with 600 μg CM1 capture antibody (Custom Monoclonal, Inc., USA) in 100 μl 0.1 M carbonate buffer (7.5 g/liter sodium bicarbonate, 2.0 g/liter sodium carbonate, pH ∼9.5) overnight at 4°C. Plates were blocked with 200 μl 2% bovine serum albumin (BSA) in TRIS-Cl/EDTA/NaCl (TEN) buffer for 2 h. One hundred microliters of samples buffered with 50 μl ELISA diluent were incubated for 2 h on a plate shaker. Six hundred micrograms of biotinylated secondary antibody (CM2-B; Custom Monoclonal, Inc., USA) was incubated in each well, followed by a 1:4,000 dilution of horseradish peroxidase (HRP)-conjugated streptavidin (Thermo Fisher Scientific, MA). Each step following sample incubation was followed with 5× wash with TEN buffer (0.05 M Tris base, 0.001 M EDTA, 0.15 M NaCl, pH 7.2 to 7.4) with 0.1% Tween. All incubations were performed at room temperature. p27 antigen was detected indirectly following the addition of 3, 3′, 5, 5′ tetramethyl benzidine (TMB) substrate and peroxidase (BioLegend, San Diego, CA) at room temperature for 7.5 min before adding 2.5 N H2SO4 and was quantified by Bioanalyzer at 450 nm. Semipurified FeLV p27 diluted in appropriate medium (DMEM or RPMI) was used as a standard curve. Cutoff values for negative samples were three times the standard error over the average optical density (OD) measured for control medium samples.
ACKNOWLEDGMENTS
We thank Mark Cunningham, Dave Onorato, Lisa Wolfe, Mat Aldredge, Mark Fisher, and Ivy Levan for collection and provision of puma tissue samples; Gary Mason, Jennifer Malmberg, Alex Byas, Laura Hoon-Hanks, Lauren Harris, and Devin von Stade for the collection of domestic cat full-skin biopsies; and Kathryn Stutzman-Rodriguez for FeLV-positive domestic cat tissues.
This work was supported by NSF-EID award 1413925 and by the Office of the Director, National Institutes of Health, under award numbers T32OD012201 and F30OD023386.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet 6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Bock M, Stoye JP. 2000. Endogenous retroviruses and the human germline. Curr Opin Genet Dev 10:651–655. doi: 10.1016/s0959-437x(00)00138-6. [DOI] [PubMed] [Google Scholar]
- 4.Coffin JM. 2004. Evolution of retroviruses: fossils in our DNA. Proc Am Philos Soc 148:264–280. [PubMed] [Google Scholar]
- 5.Xu X, Zhao H, Gong Z, Han GZ. 2018. Endogenous retroviruses of non-avian/mammalian vertebrates illuminate diversity and deep history of retroviruses. PLoS Pathog 14:e1007072. doi: 10.1371/journal.ppat.1007072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenwood AD, Ishida Y, O'Brien SP, Roca AL, Eiden MV. 2017. Transmission, evolution, and endogenization: lessons learned from recent retroviral invasions. Microbiol Mol Biol Rev 82:e00044-17. doi: 10.1128/MMBR.00044-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke JD, Stoye JP. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p 343–346. In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 8.Lober U, Hobbs M, Dayaram A, Tsangaras K, Jones K, Alquezar-Planas DE, Ishida Y, Meers J, Mayer J, Quedenau C, Chen W, Johnson RN, Timms P, Young PR, Roca AL, Greenwood AD. 2018. Degradation and remobilization of endogenous retroviruses by recombination during the earliest stages of a germ-line invasion. Proc Natl Acad Sci U S A 115:8609–8614. doi: 10.1073/pnas.1807598115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knerr I, Huppertz B, Weigel C, Dotsch J, Wich C, Schild RL, Beckmann MW, Rascher W. 2004. Endogenous retroviral syncytin: compilation of experimental research on syncytin and its possible role in normal and disturbed human placentogenesis. Mol Hum Reprod 10:581–588. doi: 10.1093/molehr/gah070. [DOI] [PubMed] [Google Scholar]
- 10.Li F, Karlsson H. 2016. Expression and regulation of human endogenous retrovirus W elements. APMIS 124:52–66. doi: 10.1111/apm.12478. [DOI] [PubMed] [Google Scholar]
- 11.Willett BJ, Hosie MJ. 2013. Feline leukaemia virus: half a century since its discovery. Vet J 195:16–23. doi: 10.1016/j.tvjl.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Polani S, Roca AL, Rosensteel BB, Kolokotronis SO, Bar-Gal GK. 2010. Evolutionary dynamics of endogenous feline leukemia virus proliferation among species of the domestic cat lineage. Virology 405:397–407. doi: 10.1016/j.virol.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Chiu ES, Kraberger S, Cunningham M, Cusack L, Roelke M, VandeWoude S. 2019. Multiple introductions of domestic cat feline leukemia virus in endangered Florida panthers. Emerg Infect Dis 25:92–101. doi: 10.3201/eid2501.181347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meli ML, Cattori V, Martinez F, Lopez G, Vargas A, Simon MA, Zorrilla I, Munoz A, Palomares F, Lopez-Bao JV, Pastor J, Tandon R, Willi B, Hofmann-Lehmann R, Lutz H. 2009. Feline leukemia virus and other pathogens as important threats to the survival of the critically endangered Iberian lynx (Lynx pardinus). PLoS One 4:e4744. doi: 10.1371/journal.pone.0004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luria BJ, Levy JK, Lappin MR, Breitschwerdt EB, Legendre AM, Hernandez JA, Gorman SP, Lee IT. 2004. Prevalence of infectious diseases in feral cats in Northern Florida. J Feline Med Surg 6:287–296. doi: 10.1016/j.jfms.2003.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yilmaz H, Ilgaz A, Harbour DA. 2000. Prevalence of FIV and FeLV infections in cats in Istanbul. J Feline Med Surg 2:69–70. doi: 10.1053/jfms.2000.0066. [DOI] [PubMed] [Google Scholar]
- 17.Muirden A. 2002. Prevalence of feline leukaemia virus and antibodies to feline immunodeficiency virus and feline coronavirus in stray cats sent to an RSPCA hospital. Vet Rec 150:621–625. doi: 10.1136/vr.150.20.621. [DOI] [PubMed] [Google Scholar]
- 18.Bandecchi P, Matteucci D, Baldinotti F, Guidi G, Abramo F, Tozzini F, Bendinelli M. 1992. Prevalence of feline immunodeficiency virus and other retroviral infections in sick cats in Italy. Vet Immunol Immunopathol 31:337–345. doi: 10.1016/0165-2427(92)90020-q. [DOI] [PubMed] [Google Scholar]
- 19.Gleich SE, Krieger S, Hartmann K. 2009. Prevalence of feline immunodeficiency virus and feline leukaemia virus among client-owned cats and risk factors for infection in Germany. J Feline Med Surg 11:985–992. doi: 10.1016/j.jfms.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luaces I, Doménech A, García-Montijano M, Collado VM, Sánchez C, Tejerizo JG, Galka M, Fernández P, Gómez-Lucía E. 2008. Detection of feline leukemia virus in the endangered Iberian lynx (Lynx pardinus). J Vet Diagn Invest 20:381–385. doi: 10.1177/104063870802000325. [DOI] [PubMed] [Google Scholar]
- 21.Cunningham MW, Brown MA, Shindle DB, Terrell SP, Hayes KA, Ferree BC, McBride RT, Blankenship EL, Jansen D, Citino SB, Roelke ME, Kiltie RA, Troyer JL, O'Brien SJ. 2008. Epizootiology and management of feline leukemia virus in the Florida puma. J Wildl Dis 44:537–552. doi: 10.7589/0090-3558-44.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres AN, Mathiason CK, Hoover EA. 2005. Re-examination of feline leukemia virus: host relationships using real-time PCR. Virology 332:272–283. doi: 10.1016/j.virol.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 23.Powers JA, Chiu ES, Kraberger SJ, Roelke-Parker M, Lowery I, Erbeck K, Troyer R, Carver S, VandeWoude S. 2018. Feline leukemia virus disease outcomes in a domestic cat breeding colony: relationship to endogenous FeLV and other chronic viral infections. J Virol 92:e00649-18. doi: 10.1128/JVI.00649-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geoghegan JL, Holmes EC. 2018. The phylogenomics of evolving virus virulence. Nat Rev Genet 19:756–769. doi: 10.1038/s41576-018-0055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garver KA, Batts WN, Kurath G. 2006. Virulence comparisons of infectious hematopoietic necrosis virus U and M genogroups in sockeye salmon and rainbow trout. J Aquat Anim Health 18:232–243. doi: 10.1577/H05-038.1. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Malmberg JL, Wood BA, Hladky S, Troyer R, Roelke M, Cunningham M, McBride R, Vickers W, Boyce W, Boydston E, Serieys L, Riley S, Crooks K, VandeWoude S. 2017. Feline immunodeficiency virus cross-species transmission: implications for emergence of new lentiviral infections. J Virol 91:e02134-16. doi: 10.1128/JVI.02134-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellner A, Carver S, Scorza V, McKee CD, Lappin M, Crooks KR, VandeWoude S, Antolin MF. 2018. Transmission pathways and spillover of an erythrocytic bacterial pathogen from domestic cats to wild felids. Ecol Evol 8:9779–9792. doi: 10.1002/ece3.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraberger S, Fountain-Jones NM, Gagne RB, Malmberg J, Dannemiller NG, Logan K, Alldredge M, Varsani A, Crooks KR, Craft M, Carver S, VandeWoude S. 2020. Frequent cross-species transmissions of foamy virus between domestic and wild felids. Virus Evol 6:vez058. doi: 10.1093/ve/vez058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sohrabi C, Alsafi Z, O'Neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C, Agha R. 2020. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19). Int J Surgery 76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown MA, Cunningham MW, Roca AL, Troyer JL, Johnson WE, O'Brien SJ. 2008. Genetic characterization of feline leukemia virus from Florida panthers. Emerg Infect Dis 14:252–259. doi: 10.3201/eid1402.070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitte C, Panaud O. 2003. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol Biol Evol 20:528–540. doi: 10.1093/molbev/msg055. [DOI] [PubMed] [Google Scholar]
- 33.Roca AL, Nash WG, Menninger JC, Murphy WJ, O'Brien SJ. 2005. Insertional polymorphisms of endogenous feline leukemia viruses. J Virol 79:3979–3986. doi: 10.1128/JVI.79.7.3979-3986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koshy R, Gallo RC, Wong-Staal F. 1980. Characterization of the endogenous feline leukemia virus-related DNA sequences in cats and attempts to identify exogenous viral sequences in tissues of virus-negative leukemic animals. Virology 103:434–445. doi: 10.1016/0042-6822(80)90202-0. [DOI] [PubMed] [Google Scholar]
- 35.Thompson PJ, Macfarlan TS, Lorincz MC. 2016. Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol Cell 62:766–776. doi: 10.1016/j.molcel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanville B, Dolan MA, Wollenberg K, Yan Y, Martin C, Yeung ML, Strebel K, Buckler-White A, Kozak CA. 2010. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog 6:e1000974. doi: 10.1371/journal.ppat.1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam JK, Chow MY, Zhang Y, Leung SW. 2015. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids 4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratt AJ, MacRae IJ. 2009. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem 284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith JA, Wang Y, Wilmers CC. 2016. Spatial characteristics of residential development shift large carnivore prey habits. Jour Wild Mgmt 80:1040–1048. doi: 10.1002/jwmg.21098. [DOI] [Google Scholar]
- 40.Elbroch LM, Levy M, Lubell M, Quigley H, Caragiulo A. 2017. Adaptive social strategies in a solitary carnivore. Sci Adv 3:e1701218. doi: 10.1126/sciadv.1701218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita M, Emerman M. 2006. Retroviral infection of non-dividing cells: old and new perspectives. Virology 344:88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Herbert B-S, Pitts AE, Baker SI, Hamilton SE, Wright WE, Shay JW, Corey DR. 1999. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc Natl Acad Sci U S A 96:14276–14281. doi: 10.1073/pnas.96.25.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vangipuram M, Ting D, Kim S, Diaz R, Schüle B. 2013. Skin punch biopsy explant culture for derivation of primary human fibroblasts. JoVE 77:e3779. doi: 10.3791/3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hierholzer JC, Killington RA. 1996. Virus isolation and quantitation, p 25–46. In Mahy BWJ, Kangro HO (ed), Virology methods manual. Academic Press Inc, San Diego, CA. [Google Scholar]
- 45.Tandon R, Cattori V, Willi B, Lutz H, Hofmann-Lehmann R. 2008. Quantification of endogenous and exogenous feline leukemia virus sequences by real-time PCR assays. Vet Immunol Immunopathol 123:129–133. doi: 10.1016/j.vetimm.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 46.Howard KE, Reckling SK, Egan EA, Dean GA. 2010. Acute mucosal pathogenesis of feline immunodeficiency virus is independent of viral dose in vaginally infected cats. Retrovirology 7:2. doi: 10.1186/1742-4690-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfaffl MW. 2006. Relative quantification, p 63–82. In Dorak T. (ed), Real-time PCR. International University Line, San Diego, CA. [Google Scholar]