Zika virus belongs to the Flavivirus genus, which comprises several important human pathogens. There is currently neither an approved vaccine nor antiviral drugs available to prevent infection by ZIKV. The nonstructural protein 5 (NS5) polymerase, which is responsible for replicating the viral RNA genome, represents one of the most promising targets for antiviral drug development. Starting from compounds recently developed against dengue virus NS5, we designed and synthesized inhibitors targeting Zika virus NS5. We show that these novel compounds inhibit viral replication by targeting the polymerase activity. High-resolution X-ray crystallographic structures of protein-inhibitor complexes demonstrated specific binding to an allosteric site within the polymerase, called the N pocket. This work paves the way for the future structure-based design of potent compounds specifically targeting ZIKV RNA polymerase activity.
KEYWORDS: nonstructural protein 5, polymerase inhibitors, RNA polymerases, Zika virus
ABSTRACT
Zika virus (ZIKV) remains a potentially significant public health concern because it can cause teratogenic effects, such as microcephaly in newborns and neurological disease, like Guillain-Barré syndrome. Together with efforts to develop a vaccine, the discovery of antiviral molecules is important to control ZIKV infections and to prevent its most severe symptoms. Here, we report the development of small nonnucleoside inhibitors (NNIs) of ZIKV RNA-dependent RNA polymerase (RdRp) activity. These NNIs target an allosteric pocket (N pocket) located next to a putative hinge region between the thumb and the palm subdomains that was originally described for dengue virus (DENV) RdRp. We first tested the activity of DENV RdRp N-pocket inhibitors against ZIKV RdRp, introduced chemical modifications into these molecules, and assessed their potency using both enzymatic and cell-based assays. The most potent compound had a 50% inhibitory concentration value of 7.3 μM and inhibited ZIKV replication in a cell-based assay with a 50% effective concentration value of 24.3 μM. Importantly, we report four high-resolution crystal structures detailing how these NNIs insert into the N pocket of ZIKV RdRp. Our observations point to subtle differences in the size, shape, chemical environment, and hydration of the N pocket from ZIKV RdRp from those of the N pocket from DENV RdRp that are crucial for the design of improved antiviral inhibitors with activity against ZIKV.
IMPORTANCE Zika virus belongs to the Flavivirus genus, which comprises several important human pathogens. There is currently neither an approved vaccine nor antiviral drugs available to prevent infection by ZIKV. The nonstructural protein 5 (NS5) polymerase, which is responsible for replicating the viral RNA genome, represents one of the most promising targets for antiviral drug development. Starting from compounds recently developed against dengue virus NS5, we designed and synthesized inhibitors targeting Zika virus NS5. We show that these novel compounds inhibit viral replication by targeting the polymerase activity. High-resolution X-ray crystallographic structures of protein-inhibitor complexes demonstrated specific binding to an allosteric site within the polymerase, called the N pocket. This work paves the way for the future structure-based design of potent compounds specifically targeting ZIKV RNA polymerase activity.
INTRODUCTION
Zika virus (ZIKV), a member of the Flavivirus genus in the family Flaviviridae, was first identified in Uganda in 1947 (1). Flaviviruses include several other major human pathogens, such as dengue virus (DENV), yellow fever virus, West Nile virus, and Japanese encephalitis virus (2). ZIKV outbreaks emerged in 2007 in the South Pacific (3) and in 2016 in Latin America, causing severe pathologies, such as Guillain-Barré syndrome and microcephaly in newborns from infected mothers (3, 4). ZIKV is mainly transmitted by Aedes aegypti mosquitoes, but blood, sexual, or maternal-fetal modes of transmission have also been observed (5). The potential severity of diseases caused by ZIKV triggered a rapid response from the international community, and drug repurposing efforts led to the identification of FDA-approved drugs showing antiviral activity against ZIKV infection (6, 7). Nonstructural protein 5 (NS5), the largest and most conserved flavivirus enzyme, constitutes a prime target in the development of pan-flavivirus antiviral inhibitors (8–12). NS5 is the result of gene fusion between the gene for a methyltransferase domain at the N terminus of the protein (30 kDa) and the gene for an RNA-dependent RNA polymerase (RdRp) domain of 75 kDa at the C-terminal end, with the two domains connected by a linker region (9, 12, 13). The RdRp replicates viral RNA, and its inhibition could prevent severe symptoms associated with infection (14, 15). Both nucleoside inhibitors (NI) and nonnucleoside inhibitors (NNIs) of the preclinical stage of DENV RdRp have been developed (16, 17); however, they have failed so far to reach the market (8, 18). During replication, the RdRp transitions from a narrow RNA tunnel conformation, adopted during initiation, to a wide RNA tunnel conformation, adopted in the elongation step. In the narrow tunnel conformation, the RdRp is able to accommodate only the single-stranded template RNA, with the priming loop that plays a key role for de novo pppAG dinucleotide synthesis adopting an extended conformation. The enzyme is thus believed to undergo dynamic conformational transitions, whereby the fingers and thumb domains move apart and the priming loop retracts to accommodate the double-stranded RNA produced during the elongation phase. In this work, we rapidly obtained high-resolution structures of ZIKV RdRp that showed high overall structural similarity to DENV RdRp. We tested the inhibition of ZIKV RdRp with compounds designed to bind the allosteric pocket (N pocket) of DENV RdRp (19) and found that compound NITD29 also inhibits ZIKV NS5 polymerase activity, although with a potency much lower than that for DENV NS5 polymerase activity. We designed more potent derivatives of NITD29 that showed inhibition of the enzymatic activity of ZIKV RdRp at low-micromolar concentrations and promising activity in cell culture. To validate the mode of action of these inhibitors, we also determined the high-resolution crystal structures of four inhibitors bound to ZIKV NS5 RdRp. Taken together, these data should inform further development of antiviral molecules targeting ZIKV RdRp.
RESULTS
Crystal structure of free ZIKV RdRp and assay development.
The novel robust crystallization condition identified in this study is suitable for structure-based drug discovery; therefore, we first determined the structure of the free ZIKV RdRp at a resolution of 1.4 Å (Fig.1A; Table 1). The structure of the RdRp domain is similar to the published structures of either the ZIKV NS5 full-length protein or its RdRp domain (20–23), with root mean square deviation (RMSD) values being in the range of 0.35 to 0.65 Å, while comparisons with the DENV serotype 3 (DENV3) RdRp (PDB accession number 2J7U) and DENV2 RdRp (PDB accession number 5K5M) structures (9, 19) returned slightly higher RMSD values of 0.80 to 1.20 Å. The present ZIKV RdRp structure adopts a conformation slightly more compact than that of the DENV3 RdRp structure: the distance between equivalent residues of the fingers and the thumb subdomains is 50.1 Å for ZIKV RdRp (the distance between residue 434 from the fingers domain and residue 874 in the thumb domain), while it is 52.1 Å for DENV3 RdRp (the distance between residues 432 and 872). The priming loop, which serves as a proteinaceous platform to initiate RNA synthesis, is formed by residues 785 to 810 of ZIKV NS5 (Fig. 1A). This loop connects two α helices from the thumb region, and in the preinitiation form that was crystallized, it largely obstructs the RNA binding tunnel (20, 24). ZIKV RdRp is capable of primer-independent (de novo) initiation (10, 25). Overall, the closed conformation observed in the present crystal structure conforms with the report by Duan et al. of a closed preinitiation-like conformation for the ZIKV RdRp, with a narrow and positively charged RNA binding tunnel being partially obstructed by the priming loop (20).
FIG 1.
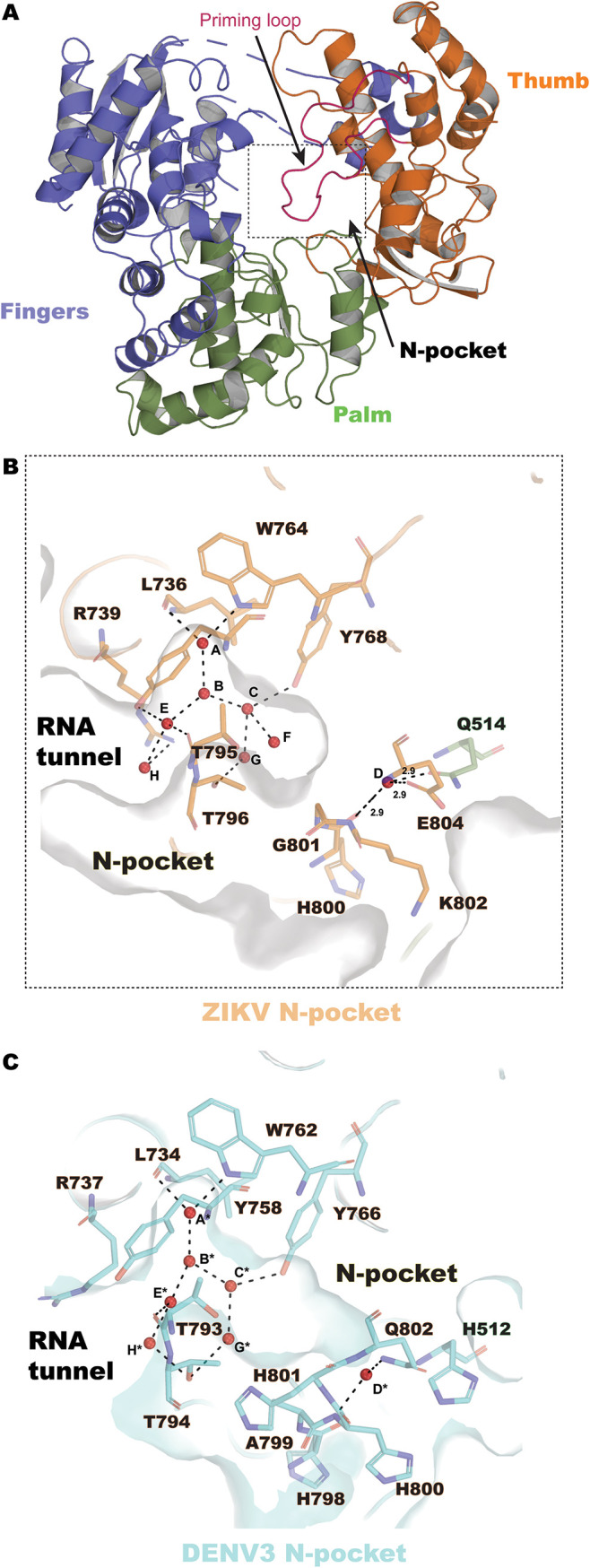
ZIKV RdRp structure at 1.4 Å and view of the N pocket. (A) Overall structure of ZIKV RdRp, represented as ribbons and colored according to subdomain (fingers in slate, residues 273 to 498 and 544 to 607; palm in green, residues 499 to 543 and 607 to 709; and thumb in orange, residues 710 to 887), and the priming loop region (residues 785 to 810) is shown in purple. (B) The hydrated N pocket of ZIKV RdRp is represented as a molecular surface and colored in gray. Eight water molecules (named A to H, red spheres) are inserted in the N pocket and are coordinated by residues from the thumb subdomain (represented as sticks, in orange). (C) The hydrated N pocket of DENV3 RdRp (PDB accession number 4HHJ) is represented as a molecular surface and is in cyan. Seven water molecules (named A* to H*, red spheres) are present in the N pocket and are coordinated by protein residues (represented as sticks).
TABLE 1.
Data collection and refinement statistics for ZIKV RdRp crystals
| Parametera | Value(s) for the following combinationb
: |
||||
|---|---|---|---|---|---|
| ZRDRP-Apo | ZRDRP-Cpd7 | ZRDRP-Cpd13 | ZRDRP-Cpd14 | ZRDRP-Cpd15 | |
| PDB accession no. | 6LD1 | 6LD2 | 6LD3 | 6LD4 | 6LD5 |
| Data collection statistics | |||||
| Wavelength | 0.9999 | 0.9538 | 0.9538 | 0.9538 | 1.542 |
| Resolution range (Å) | 19.69–1.4 (1.45–1.4) | 55.65–1.4 (1.45–1.4) | 51.18–2.3 (2.38–2.3) | 21.91–1.5 (1.55–1.5) | 34.18–1.94 (2.01–1.94) |
| Space group | P21 | P21 | P21 | P21 | P21 |
| Unit cell dimensions | |||||
| a, b, c (Å) | 64.03, 82.89, 69.43 | 63.94, 85.06, 69.74 | 63.65, 84.99, 69.78 | 64.1, 85.17, 69.81 | 63.47, 84.59, 69.15 |
| α, β, γ (°) | 90, 114.52, 90 | 90, 113.44, 90 | 90, 113.28, 90 | 90, 113.34, 90 | 90, 113.9, 90 |
| No. of reflections | |||||
| Measured | 868,128 (82,587) | 409,259 (30,450) | 105,406 (10,139) | 350,892 (29,638) | 132,120 (7,631) |
| Unique | 126,423 (12,347) | 132,763 (13,223) | 29,894 (2,928) | 109,191 (10,777) | 46,894 (4,079) |
| Multiplicity | 6.9 (6.7) | 3.1 (2.3) | 3.5 (3.5) | 3.2 (2.8) | 2.8 (1.9) |
| Completeness (%) | 97.54 (95.46) | 98.72 (98.27) | 98.15 (97.18) | 99.23 (98.44) | 95.88 (84.16) |
| Mean I/sigma〈I〉 | 15.95 (0.93) | 13.06 (0.69) | 10.22 (1.75) | 11.84 (0.59) | 5.89 (1.41) |
| Refinement statistics | |||||
| Wilson B factor (Å2) | 20.79 | 24.83 | 45.33 | 28.32 | 15.94 |
| Rmerge | 0.05486 (1.707) | 0.03512 (0.8589) | 0.08542 (0.6131) | 0.04154 (1.258) | 0.1362 (0.5432) |
| Rmeas | 0.05936 (1.845) | 0.04193 (1.128) | 0.1007 (0.7268) | 0.04953 (1.561) | 0.168 (0.7425) |
| Rpim | 0.02245 (0.6945) | 0.02267 (0.723) | 0.05254 (0.385) | 0.02672 (0.9115) | 0.09655 (0.503) |
| CC1/2 | 0.999 (0.433) | 0.999 (0.419) | 0.997 (0.656) | 0.999 (0.381) | 0.981 (0.534) |
| CC* | 1 (0.777) | 1 (0.769) | 0.999 (0.89) | 1 (0.742) | 0.995 (0.834) |
| No. of reflections used in refinement | 126,423 (12,344) | 132,761 (13,221) | 29,888 (2,928) | 109,190 (10,772) | 46,890 (4,079) |
| No. of reflections used for Rfree | 6,321 (617) | 6,637 (661) | 1,492 (147) | 5,458 (539) | 2,343 (204) |
| Rwork | 0.1962 (0.3212) | 0.2020 (0.3789) | 0.1916 (0.2657) | 0.1964 (0.3560) | 0.1840 (0.2749) |
| Rfree | 0.2182 (0.3260) | 0.2271 (0.3786) | 0.2489 (0.3294) | 0.2238 (0.3833) | 0.2260 (0.3265) |
| CC | |||||
| Work | 0.956 (0.630) | 0.957 (0.597) | 0.931 (0.758) | 0.958 (0.592) | 0.945 (0.717) |
| Free | 0.953 (0.623) | 0.944 (0.659) | 0.968 (0.582) | 0.953 (0.625) | 0.911 (0.704) |
| No. of: | |||||
| Nonhydrogen atoms | 5,314 | 5,347 | 4,989 | 5,434 | 5,463 |
| Macromolecules | 4,834 | 4,709 | 4,745 | 4,693 | 4,742 |
| Ligands | 14 | 20 | 29 | 60 | 64 |
| Solvent atoms | 466 | 618 | 215 | 681 | 657 |
| Protein residues | 574 | 565 | 578 | 565 | 575 |
| RMSD | |||||
| Bond length (Å) | 0.012 | 0.012 | 0.012 | 0.012 | 0.012 |
| Bond angle (°) | 1.47 | 1.50 | 1.52 | 1.45 | 1.47 |
| Ramachandran plot (%) | |||||
| Favored | 98.23 | 97.65 | 97.18 | 97.28 | 97.17 |
| Allowed | 1.77 | 2.17 | 2.82 | 2.54 | 2.83 |
| Outliers | 0.00 | 0.18 | 0.00 | 0.18 | 0.00 |
| Rotamer outliers (%) | 1.54 | 1.39 | 3.54 | 1.39 | 1.38 |
| Clash score | 2.08 | 3.21 | 2.76 | 3.31 | 2.96 |
| B factor | |||||
| Avg | 29.44 | 34.00 | 51.35 | 37.19 | 20.45 |
| Macromolecules | 28.18 | 32.83 | 51.08 | 35.77 | 18.86 |
| Ligands | 38.54 | 30.31 | 86.65 | 38.63 | 31.48 |
| Solvent | 42.18 | 43.00 | 52.69 | 46.86 | 30.90 |
I, intensity of a reflection; sigma〈I〉, average intensity; CC1/2, percentage of correlation between intensities from random half data sets; CC*, estimate of correlation of the observed data set with the underlying true signal (33).
Statistics for the highest-resolution shell are shown in parentheses. ZRDRP, ZIKV RdRp.
An allosteric pocket (N pocket) located next to a hinge region between the thumb and the palm subdomains that was originally described for DENV RdRp constitutes a prime target for the design of flavivirus inhibitors. With a view to target the N pocket of ZIKV RdRp for inhibition, we compared the volume of the N pocket in the RdRp from DENV with that of the N pocket in the RdRp from DENV ZIKV: the CASTp (26) server returned significantly lower values for the N pocket of ZIKV RdRp (21.9 surface accessible [SA] units) than for those of the N pocket of DENV3 RdRp (50.9 SA units) or DENV2 RdRp (73.3 SA units) (Fig. 1B and C). Moreover, inspection of our high-resolution free ZIKV RdRp structure revealed that the ZIKV RdRp N pocket is significantly hydrated with eight water molecules (Fig. 1B). Inhibitor binding therefore likely displaces several of these bound water molecules. When the present ZIKV RdRp structure was compared with the previously reported DENV3 NS5 structure (Fig. 1C) (PDB accession number 4HHJ) (27) a total of only seven water molecules were found within the N pocket of the free DENV3 RdRp structure. While their positions are equivalent to those of the water molecules present in the ZIKV RdRp N pocket, water F is absent in DENV3 RdRp (Fig. 1B and C). This could be due to the substitution of Gly801 in ZIKV RdRp for His801 in DENV3 RdRp, with the cavity created by the removal of the histidine side chain being filled by an extra water molecule.
Next, we optimized both a de novo initiation (dnI) assay and an elongation enzyme assay (see Materials and Methods) that were previously developed for DENV RdRp (28, 29) and used them to screen for inhibitors targeting the N pocket.
Chemical synthesis of N-pocket binders and their inhibitory effects on ZIKV RdRp.
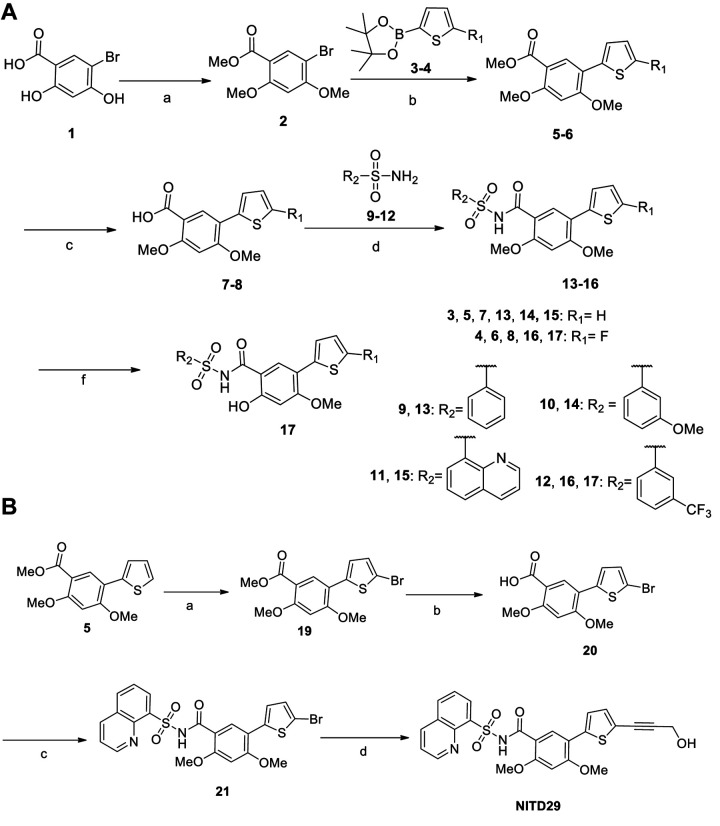
Previous work using fragment-based screening targeting DENV RdRp led to the identification of the N pocket, which is located in the thumb subdomain close to the polymerase active site (19). This pocket can be targeted by NNIs, leading to allosteric inhibition of viral replication in cell-based assays with 50% effective concentration (EC50) values in the low-nanomolar range across the RdRp from the four DENV serotypes (19). We chemically synthesized derivatives of these compounds, as described in Fig. 2 and Materials and Methods.
FIG 2.
(A) General synthesis of thiophene sulfone derivatives, compounds 13 to 17. Reagents and conditions were as follows: (a) MeI, K2CO3, acetone, reflux; (b) compounds 3 and 4, PdCl2(dppf) · CH2Cl2, potassium acetate, dioxane-H2O, 100°C; (c) LiOH, THF-MeOH-H2O, 40°C; (d) sulfonamides (compounds 9 to 12), EDC · HCl, DMAP, CH2Cl2, RT; (f) NH4Cl, DMF, 120°C. (B) Synthesis of NITD29. Reagents and conditions were as follows: (a) NBS, DMF, RT; (b) LiOH, THF-MeOH-H2O, H2O, 40°C; (c) quinoline-8-sulfonamide (Cpd11), EDC · HCl, DMAP, CH2Cl2, RT; (d) propargyl alcohol, PdCl2(dppf) · CH2Cl2, CuI, Et3N, DMF, 80°C. MeO or OMe, methoxy group.
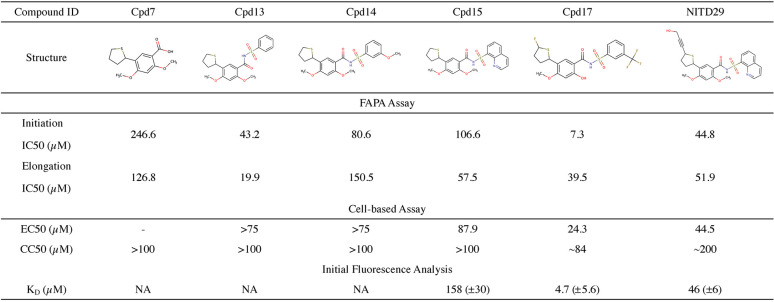
We first tested the activity of NITD29, the NNI compound that was the most potent against DENV RdRp (19), and found that NITD29 inhibits the ZIKV polymerase activity with 50% inhibitory concentration (IC50) values of 44.8 μM and 51.9 μM in initiation and elongation fluorescence-based alkaline phosphatase-coupled polymerase assays (FAPA), respectively (Fig. 3 and 4). Given that the volume of the N pocket is about 50% smaller in the free ZIKV RdRp structure than in the free DENV RdRp structure (Fig. 1B and C), only the core scaffold compound (compound 7 [Cpd7]) was used as a starting template for NITD29 derivative synthesis. We retained only the thiophene ring and the acid moiety projecting from the phenyl ring (Table 2), which were shown to significantly improve compound potency in the DENV study (19). While the core scaffold showed only moderate inhibition, compounds 13, 14, and 15, in which the carboxylic acid in Cpd7 was replaced with arylsulfonamides, showed slightly better inhibitory activities (Table 2). Cocrystal structures in the presence of the compound (discussed below) revealed that the thiophene ring points toward the interior of the ZIKV RdRp N pocket, while the phenyl ring points outward, toward the RNA binding channel, similar to what was observed for DENV RdRp (20). Next, efforts were made to synthesize analogues with various substitutions on the thiophene ring. Cpd17, where the propargyl alcohol moiety of NITD29 was replaced by a fluorine on the thiophene ring, while the quinoline was swapped for a 3-(trifluoromethyl)benzene and the 2-methoxy was changed to 2-hydroxy, had the best enzymatic inhibition activity, with an IC50 value of 7.3 μM in the dnI assay (Fig. 3A and 4A; Table 2).
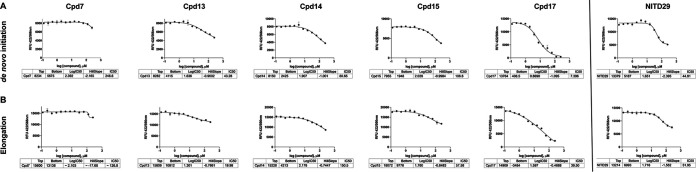
FIG 3.
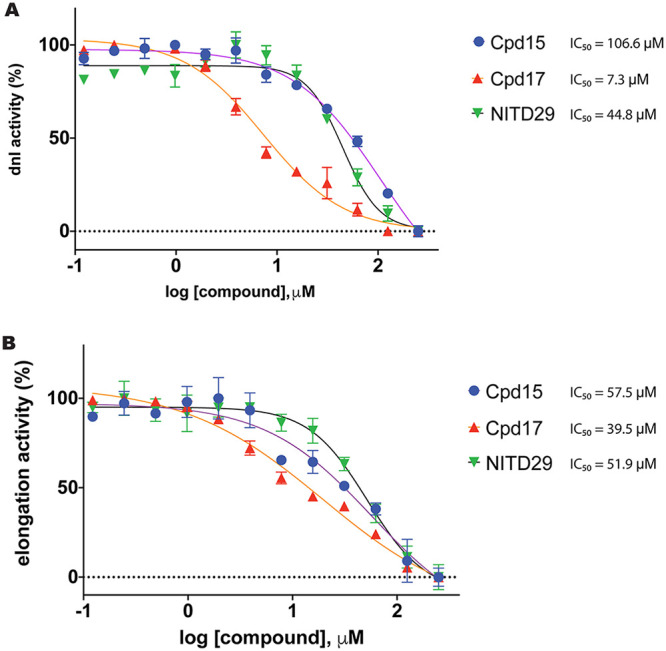
Inhibition of ZIKV RdRp by targeting its N pocket. Inhibition of ZIKV RdRp activity was determined using FAPA. The ZIKV RdRp dnI FAPA (A) and elongation FAPA (B) were performed as described in Materials and Methods.
TABLE 2.
Inhibitory properties of ZIKV RdRp compoundsa
MeO or OMe, methoxy group; NA, not applicable. We observed that the intrinsic fluorescence of some compounds impaired the analysis.
FIG 4.
Representative IC50 curves for ZIKV inhibitors tested in de novo initiation and elongation FAPAs. Compounds (12 concentration points, with the compounds being twofold serially diluted from 250 μM) were incubated at 37°C for 15 min with the enzyme alone in 384-well opaque plates. The reactions were then started with addition of the RNA mix. The reaction was terminated by the addition of stop buffer containing 25 nM CIP after 3 h. The reaction mixtures were incubated at 37°C for 1 h, and the fluorescence was read on a Tecan Spark 10M microplate reader (maximum excitation and emission wavelengths, 422 nm and 566 nm, respectively). Data were fitted to the four-parameter logistic equation, and IC50 curves were plotted using GraphPad Prism software. The tables at the bottom of each panel contain the average IC50 values and the Hill slopes obtained. All data points were measured in triplicate. RFU, relative fluorescence units.
Antiviral activity in a cell-based assay and compound binding.
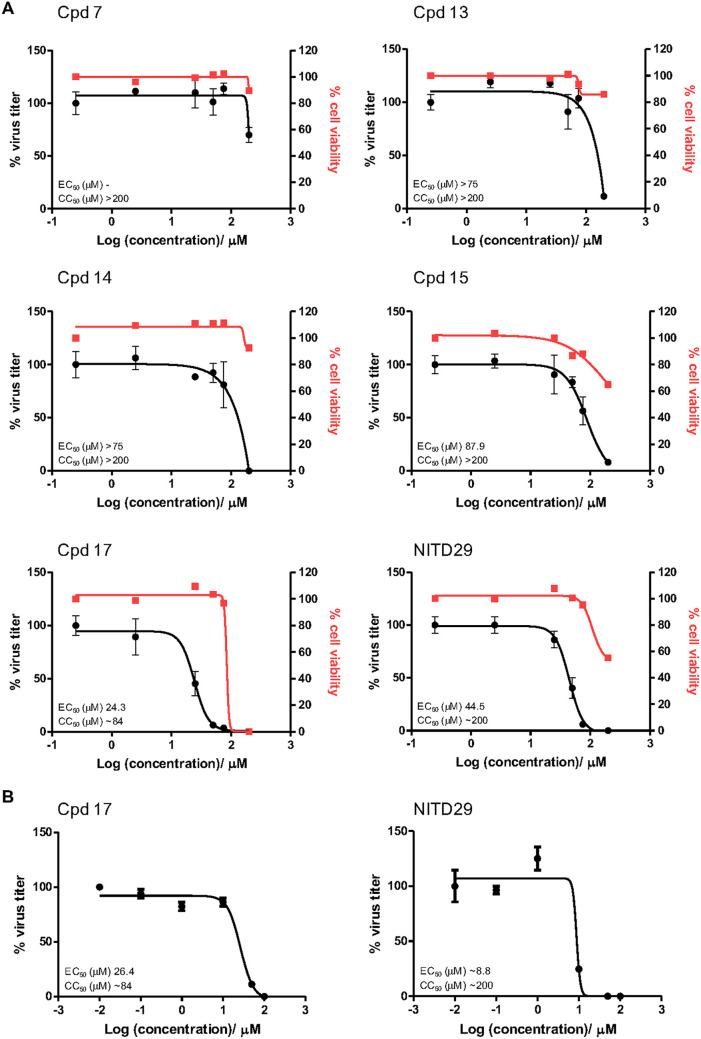
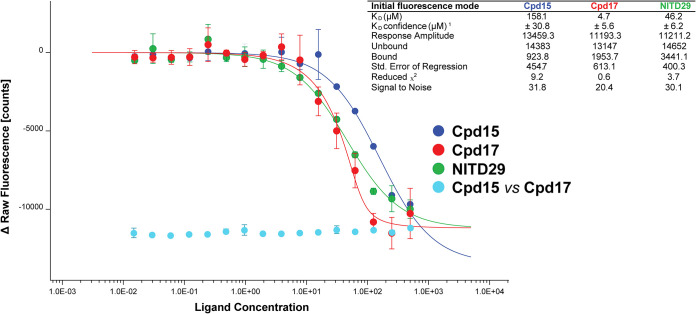
Next, we evaluated the antiviral activity of these compounds in a cell-based assay and compared the activity with that of NITD29, which was also active against ZIKV replication with an EC50 of 44.5 μM (Fig. 5A), a value significantly reduced compared to its potency against DENV (20). Cpd7 was not active in the cell-based assay, while Cpd13, Cpd14, and Cpd15 demonstrated similar EC50 values, ranging from 75 to 87.9 μM (Fig. 6). Cpd17 displayed the highest activity in cell-based assays, with an EC50 of 24.3 μM and moderate cell toxicity (Table 2 and Fig. 5A). Moreover, treatment with 50 μM Cpd17 was able to yield a 1.5-log reduction in viral yield, and its activity was able to halt replication in infected cells (Fig. 5B), findings similar to those achieved by treatment with NITD29. To confirm the direct binding of the compounds to the RdRp, we also determined their affinity (equilibrium dissociation constant [KD] values) for ZIKV RdRp. We used the MicroScale Thermophoresis (MST) experimental setup from NanoTemper Technologies. However, we observed a strong fluorescence change with increased compound concentration that hindered the MST signal usage. The fluorescence intensity change was due to the presence of tryptophan residues within the N pocket. As shown below in cocrystal structures, compound binding was accompanied by water displacement, changing the environment of these fluorescent residues. This intrinsic fluorescence intensity quenching signal was measured and used to assess binding affinities. This method can be used for compounds that do not show any intrinsic fluorescence. As expected from its highest potency in the enzyme and cell-based assays, Cpd17 also showed the highest binding affinity for the polymerase, with a KD value of 4.6 μM (Table 2 and Fig. 7). Taken together, these results demonstrate that Cpd17 may be considered the lead molecule in this series.
FIG 5.
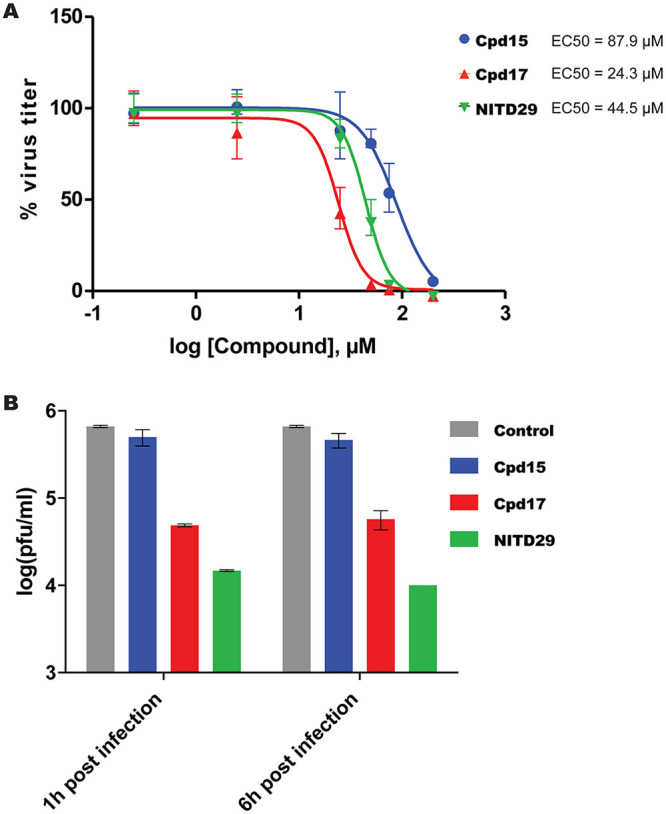
Efficacy of lead compounds against ZIKV infection in HuH-7 cells. (A) EC50 of Cpd15, Cpd17, and NITD29 tested against ZIKV H/PF/2013 infection in HuH-7 cells. (B) Viral plaque reduction assay results. The plaque reduction achieved with a single concentration (50 μM) of Cpd15, Cpd17, NITD29, or the control treatment at 1 or 6 h postinfection is shown.
FIG 6.
Representative EC50 and CC50 curves for ZIKV inhibitors tested in a HuH-7 cell-based assay. (A) Efficacy testing of the compounds against ZIKV. (B) Efficacy testing of the compounds against DENV2.
FIG 7.
Compound KD for ZIKV RdRp determined by initial fluorescence analysis. The protein was applied at a final concentration of 100 nM. A twofold dilution series (16 concentration points) starting from 500 μM was prepared for each compound. The competition between Cpd15 and Cpd17 was performed in a similar way with a fixed concentration of 100 μM Cpd17 with a twofold dilution series of Cpd15 starting from 500 μM. Samples were filled into hydrophilic capillaries (NanoTemper Technologies) and incubated shortly before measurement at 37°C using a Monolith NT.LabelFree instrument (NanoTemper Technologies). The curves were generated using MO.Affinity software by NanoTemper Technologies. The value of the initial fluorescence was plotted against the ligand concentration to derive the dose-response curve. The KD values were obtained with the initial fluorescence mode, measuring the fluorescence intensity variation upon compound binding. The inset table shows the data processing statistics exported from MO.Affinity software.
Crystal structures of ZIKV RdRp bound to inhibitors.
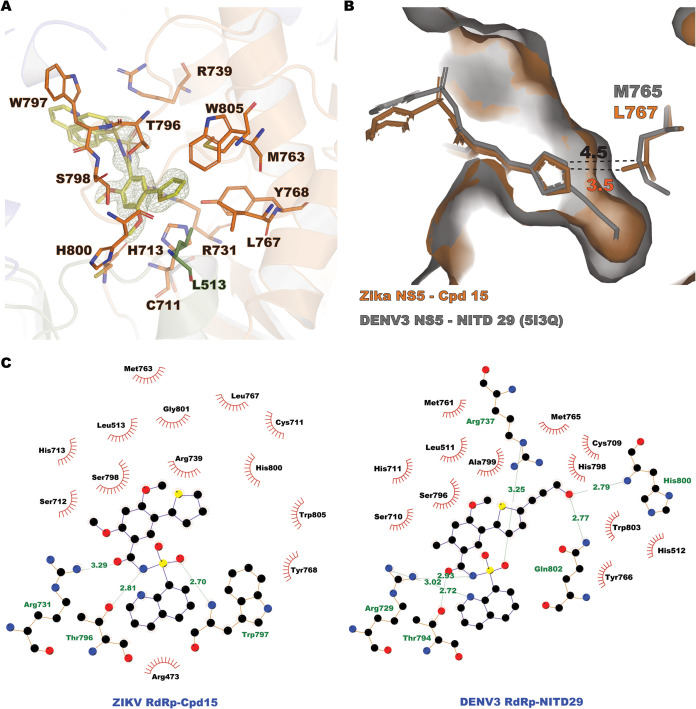
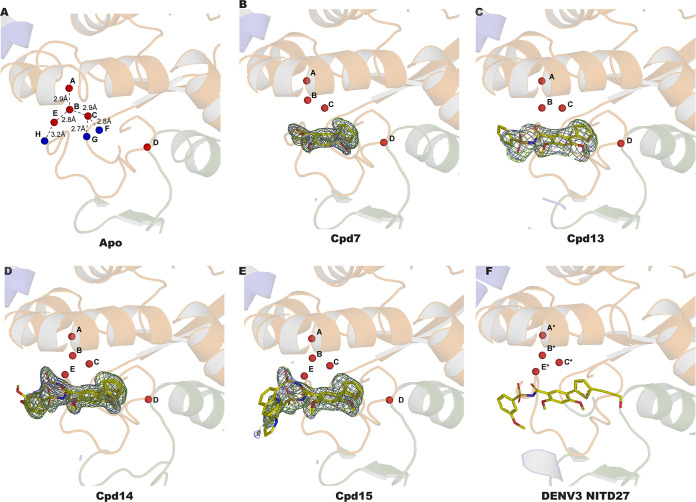
We determined the crystal structures of ZIKV RdRp bound to compounds 7, 13, 14, and 15 (Table 1) but not compound 17. High-resolution structures were obtained by soaking crystals of the free ZIKV RdRp in crystallization solution supplemented with 1 mM each compound for 6 h. Following crystallographic refinement of the RdRp polypeptide chain, continuous residual electron density in the N pocket was visible at a level of 3σ above the mean density, revealing the presence of the soaked compounds (Fig. 8A and 9B to E). Clear electron density was consistently found for the thiophene, benzene, methoxy, and sulfonamide groups of the inhibitors, while the benzyne and methoxybenzene substitutions appeared to be mobile, as little to no electron density was visible in the residual difference maps. This contrasts with the findings for the compounds bound to the DENV2 and DENV3 RdRp N pockets, where the methoxybenzene and 8-quinolyl moieties of NITD27 and NITD29 were clearly defined in the electron density, stabilized by stacking with the W797 indole ring (corresponding to residue W795, using DENV3 numbering) (17). Here, inhibitor stabilization in the N pocket is achieved mainly through hydrophobic interactions (Fig. 8A and C) via residues projecting from the thumb subdomain, except for residue L513, which belongs to the palm subdomain. Three hydrogen bonds were observed between the sulfonamide moiety of the inhibitors with residues R739, W797, R731, and T796, and equivalent interactions were present in the NITD29-DENV3 RdRp complex (Fig. 8B). In contrast, interactions involving the thiophene, the benzene, and the methoxy groups were hydrophobic. Following compound binding, out of eight water molecules present in the N pocket, labile water molecules F, G, and H were displaced by the thiophene, benzene, and sulfonamide moieties, respectively (Fig. 9), while four nonlabile waters (molecules A to D) were present in all the structures obtained. The temperature factors of the labile water molecules (42.4 Å2, on average) were 63% higher in the free RdRp structure than the B factors of the strictly nonlabile water molecules (25.9 Å2, on average), indicating that inhibitor binding displaces the less tightly bound water molecules from the N pocket of the free RdRp structure (Fig. 9A and Table 3). This is also coherent with the observation that displaced water molecules F, G, and H were not stabilized by direct interactions with RdRp residues and were thus intrinsically more mobile. A fourth water molecule, water molecule E, became displaced only when compound 7 or 13 was bound to the RdRp. In the case of the complexes with compounds 14 and 15, a water molecule occupied position E, although its temperature factors (46.4 and 22.1 Å2 in the presence of compounds 14 and 15, respectively) were higher than those of nonlabile water molecules present in these structures (on average, 28.3 Å2 for Cpd14 and 15.1 Å2 for Cpd15) (Fig. 9 and Table 3). This mode of binding of the compounds to the N pocket was also observed with compounds NITD29 and NITD27, where water molecules at positions similar to positions G*, H*, and D* of the DENV3 RdRp N pocket were not observed in the DENV3 NS5-NITD27 complex structure (PDB accession number 5I3P) (Fig. 1C and 9F). The similar mode of binding validates the use of the thiophene and phenyl scaffold as a basis to design compounds targeting the ZIKV NS5 N pocket.
FIG 8.
Compound 15 binds to the ZIKV RdRp N pocket. (A) Compound 15 (yellow sticks) bound to the N pocket formed by residues projecting from the thumb (orange sticks) and the palm (green sticks) subdomains. Cpd15 (omitted from the Fourier transform calculation) is represented in the residual electron density map Fo − Fc contoured at 3σ and colored in green. (B) The N pocket of ZIKV NS5 (represented as a surface and colored in orange) in complex with Cpd15 (orange sticks) superimposed with DENV3 NS5 (represented as a surface and colored in gray) in complex with NITD29 (gray sticks), with residue L767 in ZIKV and residue M765 in DENV3 represented as sticks. The distances between the thiophene moieties of the compounds and the closest atom of the residues are shown. (C) Flat representations of the Cpd15 inhibitor-RdRp contacts (determined using the LigPlot program). (Left) Interactions observed in the ZIKV NS5-Cpd15 complex; (right) interactions observed in the DENV3 NS5-NITD29 complex. The interatomic distances with the spatially equivalent residue L767 in ZIKV and residue M765 in DENV3 are displayed.
FIG 9.
Crystal structures of ZIKV RdRp in its apo form (A) and in complex with Cpd7 (B), Cpd13 (C), Cpd14 (D), and Cpd15 (E) as well as DENV3 in complex with NITD27 (F). Each compound present in the N pocket is represented as a yellow stick. The water molecules (A to H) present in the N pocket are shown as spheres and colored in red (A to D and E, present in all structures) and blue (F to H, displaced by compound binding). The electron density maps are represented with Fourier coefficients and compounds omitted from the calculation: 2Fo − Fc difference maps are contoured at 1s and colored in light blue; Fo − Fc maps are contoured at 3s and colored in green. The water molecules present in the DENV3 N pocket in complex with the NITD27 compound (PDB accession number 5I3P) and equivalent to the water molecules present in the ZIKV N pocket are noted A* to C* and E*.
TABLE 3.
N-pocket water molecule B factors
| Water molecule | B factor (Å2) for the following compound: |
||||
|---|---|---|---|---|---|
| Apo | 7 | 13 | 14 | 15 | |
| A | 20.2 | 22.8 | 32.9 | 25.7 | 11.8 |
| B | 27.4 | 29.1 | 45.1 | 28.2 | 13.14 |
| C | 32.1 | 28.8 | 38.2 | 29.4 | 13.3 |
| D | 24.1 | 27.0 | 34.9 | 29.8 | 15.2 |
| E | 44.8 | 46.4 | 22.1 | ||
| F | 36.4 | ||||
| G | 42.1 | ||||
| H | 48.8 | ||||
Despite several attempts to use various soaking and cocrystallization conditions, no structure of the complex with the most potent inhibitor, Cpd17, could be obtained using the present crystal form of ZIKV RdRp. Nonetheless, we were able to demonstrate the direct binding of Cpd17 to the RdRp, using initial fluorescence analysis to deduce binding affinity information (Fig. 7; see also Materials and Methods). Using the same methodology, a KD value of 158 μM was obtained for Cpd15. To confirm the binding site of compound 17 to the N pocket, competition experiments between Cpd17 and Cpd15 using initial fluorescence analysis were performed by detecting the binding of Cpd15 in the presence of Cpd17. No change in fluorescence was observed in this case (Fig. 7), suggesting that no Cpd15 binding to ZIKV RdRp occurs in the presence of Cpd17. This experiment demonstrated that Cpd17 occupies the same N pocket as Cpd15 and that Cpd15 is not able to displace Cpd17 at the concentrations tested (note that for Cpd15, we have crystallographic evidence that Cpd15 binds to the N pocket [Fig. 9]).
DISCUSSION
Several strategies have been pursued in order to identify NNIs of ZIKV RdRp. For instance, ZINC39563464 was identified via in silico rational drug design and structure-based docking, although its efficacy remains to be validated (30). Another study using inhibitor screening coupled to molecular docking identified a compound that inhibits ZIKV RdRp by binding to its active site (31).
Recently, a novel allosteric pocket, named the N pocket, was identified at the interface of the thumb and palm subdomains of DENV RdRp (19). Compounds that bind to this pocket inhibit DENV polymerase activity. Here, we used compound NITD29, one of the most active compounds generated against DENV, as a starting point to develop novel NNIs targeting ZIKV RdRp. Over a hundred analogues were synthesized, leading to the identification of four compounds, compounds 13, 14, 15, and 17, which displayed promising inhibition profiles. Interestingly, Cpd17 exhibited a 3 times higher affinity and a 1.8 times higher cell activity than NITD29, even though the latter compound lacked the propargyl moiety. The propargyl moiety is critical for displacing a deeply buried water molecule, leading to the most potent compounds, NITD29 and NITD27, against DENV RdRp. Here, replacing the thiophene moiety with highly hydrophobic groups seems to be more appropriate for inhibition of ZIKV RdRp activity. This appears to be due to a smaller volume of the ZIKV RdRp N pocket compared to that of the DENV RdRp N pocket (the ZIKV RdRp N pocket is 2 to 3 times smaller the DENV RdRp N pocket, as measured by the solvent-accessible volume).
The crystallization conditions used in this study are robust and reproducible and are therefore suitable for high-throughput compound screening and structure-based drug design. However, these crystallization conditions select a compact conformation of the protein (and, probably, also of the N pocket) from the range of RdRp conformations that probably coexist in solution. As all flavivirus RdRp structures are obtained in their preinitiation narrow RNA binding tunnel state, we can assume that this conformation corresponds in the absence of RNA to a stable, lower energy level favoring crystal formation. Indeed, protein dynamics in solution where the protein is not in a fixed conformation allows the thumb and fingers subdomains to move and adopt different conformations. While smaller derivatives could be successfully soaked in this crystal form, allowing determination of the structure of the bound inhibitors, soaking the larger compounds, such as Cpd17 and NITD29, did not produce any cocrystal structure. How can we explain the observed differences between ZIKV and DENV RdRp binding, given the chemical similarity between their N pockets? With the exception of Leu767 in the ZIKV RdRp, which replaces Met765 in the DENV3 RdRp, residues from the ZIKV RdRp N pocket interacting with the compounds are shared with residues of DENV3 NS5. This Met-to-Leu substitution nonetheless appears to impact binding: although this substitution between two aliphatic residues is usually classified as conservative, the presence of a branched methyl group in the ZIKV RdRp pocket leads to a reduction in the overall volume of the ZIKV RdRp N pocket compared with that of the DENV3 RdRp N pocket (Fig. 8B). Superimposing the N-pocket residues alone, we observed an RMSD of 0.25 Å, indicating that the backbone atoms of the N pockets align almost perfectly. Only the Met-to-Leu substitution results in a narrowing of the channel, provoking steric hindrance with several substitutions introduced in the thiophene moiety. An in silico L767M variant of ZIKV RdRp increases the measured SA volume of the N pocket by 12% (as measured by the CASTp server). Thus, this subtle structural difference (Met → Leu) provides a potential explanation for the absence of complex formation within the ZIKV NS5 RdRp crystals that we observed with compounds harboring any substitution on the thiophene moiety.
Nonetheless, we were able to demonstrate that Cpd17 binds to the protein and competes with Cpd15 and that the latter binds to the N pocket. Cpd17 is able to inhibit RNA elongation when the polymerase adopts a more open conformation, suggesting that it binds to the RdRp when it adopts a conformation more open than the one when it is trapped in the crystal structure, possibly corresponding to a conformation able to bind an RNA duplex.
Finally, despite an obvious overall structural similarity with DENV RdRp, ZIKV RdRp harbors significant idiosyncratic features, such as a higher thermostability, which plays an important role in ligand binding. Although the compounds tested in this study showed moderate inhibition of ZIKV RdRp, we hope that the first (to our knowledge) high-resolution structures of ZIKV RdRp bound to inhibitors presented in this work constitute a good starting point for further improvements and can also serve as useful standards for testing inhibitors targeting other regions of the protein. One crucial aspect, we believe, will be to introduce substitutions on the thiophene ring of Cpd15 (Fig. 7) to attempt to mimic water molecule D, with the hope that it will lead to improvements in the affinity and specificity of antiviral inhibitors.
MATERIALS AND METHODS
Protein expression and purification.
The RdRp domain (with a 6-histidine tag) from Zika virus nonstructural protein 5 (French Polynesia strain, GenBank accession number ARU07075.1) encompassing residues 270 to 89 was synthesized and cloned into a pNIC-Bsa4 plasmid (Biobasics, Singapore). The plasmid was transformed into Escherichia coli BL21 cells, and the cells were grown at 37°C in LB medium (Beckton Dickinson) until an optical density at 600 nm of 1.0 was reached. The cells were then grown at 16°C for 1 h, prior to addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to induce protein expression for 16 h. Cells were harvested by centrifugation for 30 min at 4,000 × g. The pellet was resuspended in a lysis buffer [20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole, 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP)] and sonicated for 5 min. The cell lysate was cleared by centrifugation for 1 h at 20,000 × g. The lysate was injected onto a HisTrap 5-ml column (GE Life Sciences). After a wash step using lysis buffer supplemented with 40 mM imidazole, the protein was eluted using a gradient of 50 to 500 mM imidazole. The fractions containing the NS5 protein were pooled and dialyzed against 20 mM Na HEPES, 300 mM NaCl, and 10% (vol/vol) glycerol in the presence of tobacco etch virus protease overnight at 4°C. The protein was injected onto a HisTrap 5-ml column (GE Life Sciences) that had been preequilibrated with lysis buffer to eliminate uncleaved protein and the histidine tag, and the flowthrough was collected, concentrated, and injected onto a Hi Load S200 16/60 (GE Life Sciences) size-exclusion chromatography (SEC) column that had been preequilibrated with 20 mM Na HEPES, pH 7.5, 0.3 M NaCl, 2 mM TCEP. Fraction content and purity were checked by SDS-PAGE. The elution volume from the SEC indicated that the protein eluted as a monomer. Fractions containing NS5 were then pooled and concentrated to 5 mg · ml−1, prior to flash freezing at −80°C.
Crystallization, data collection, and structure determination.
Zika virus RdRp at 5 mg/ml was screened for crystallization conditions using JCSG+ and Morpheus commercial kits (Molecular Dimensions). Crystals suitable for diffraction appeared in 20% (wt/vol) polyethylene glycol (PEG) 6000 and 0.2 M Na HEPES, pH 8. Crystals were then mounted on a cryo-loop and flash frozen in liquid nitrogen. For complex structures, crystals of Zika virus RdRp were transferred to a drop containing 20% (wt/vol) PEG 6000 and 0.2 M Na HEPES, pH 8, containing 1 mM the compound and soaked for 6 h, prior to flash freezing. Diffraction data were collected at 100 K on the Proxima 2A beamline at the Soleil synchrotron (compounds 7, 13, and 14) or on an in-house diffractometer (Rigaku FR-X equipped with a Dectris Pilatus 300 K detector). Data processing was performed using the XDS program (32). Data collection information and statistics for each structure are shown in Table 1. The structure was solved by molecular replacement using the Zika virus RdRp domain structure (PDB accession number 5WZ3) as a search probe in the Molrep program (CCP4). The solution comprised one molecule per asymmetric unit. Refinement was performed using the Buster TNT program (GlobalPhasing), and manual corrections were introduced into the model using Coot (CCP4) software. Structure analysis and figure production were realized using PyMOL software (Schroedinger). Refinement statistics are presented in Table 1.
Primer and template annealing.
RNA primer-template complexes were generated as previously described (29). 6-Carboxyfluorescein (FAM)-labeled RNA primer (1 μM) and unlabeled RNA template (1 μM) were mixed in 50 mM NaCl in RNase-free water and then incubated at 95°C for 10 min, before being cooled to room temperature and subsequently stored in the dark at −20°C before use. The respective sequences of the primer and the template used in the biochemistry assays are 5′-6FAM-AGAACCUGUUGAACAAAAGC-3′ and 5′-CUUAUUCGAGCUUUUGUUCAACAGGUUCU-3′.
De novo initiation FAPA.
ZIKV RdRp dnI assays were performed as described previously for DENV (29). Briefly, 250 nM ZIKV RdRp protein was dispensed in 384-well black opaque plates (Corning), after which compounds twofold serially diluted from a maximum concentration of 250 μM were added. The plates were spun for 1 min at 1,000 rpm and then incubated for 15 min at 37°C in the dark. A reaction mixture (50 mM Tris-HCl, pH 7.5, 0.3 mM MnCl2, 0.001% [vol/vol] Triton X-100, 10 mM KCl, 1 mM MgCl2, 10 μM cysteine) containing the RNA template (at 100 nM), CTP, GTP, and UTP (2 μM each), and Atto-ATP (0.5 μM) were then added to start the reaction, and the plates were incubated for 3 h at 37°C. Reactions were stopped by the addition of the stop solution (200 mM NaCl, 25 mM MgCl2, 1.5 M diethylaminoethyl [DEA], pH 10) containing 25 nM ciprofloxacin (CIP). The plates were then incubated for an additional hour at 37°C, after which the fluorescence was read on a Tecan Spark 10M microplate reader. All measurements were performed in triplicate wells.
Elongation assay.
ZIKV RdRp elongation assays were performed as described previously (29). Briefly, 250 nM ZIKV RdRp protein was dispensed in 384-well black opaque plates (Corning), after which compounds twofold serially diluted from a maximum concentration of 250 μM were added. The plates were spun for 1 min at 1,000 rpm and then incubated for 15 min at 37°C in the dark. A reaction mixture (50 mM Tris-HCl, pH 7.5, 5 mM MnCl2, 0.001% Triton X-100, 10 mM KCl, 10 μM cysteine) containing the primer-template complex (100 nM), CTP, GTP, and UTP (2 μM each), and Atto-ATP (0.5 μM) was then added to start the reaction, and the plates were incubated for 3 h at 37°C. The reactions were stopped by the addition of the stop solution (200 mM NaCl, 25 mM MgCl2, 1.5 M DEA, pH 10) containing 25 nM CIP. The plates were then incubated for an additional hour at 37°C, after which they were read on a Tecan Spark 10M microplate reader. All measurements were performed in triplicate wells. All IC50s reported in this study were calculated using Prism software (GraphPad).
Compound synthesis characterization.
All reactions requiring anhydrous conditions were carried out under a nitrogen atmosphere using oven-dried glassware (100°C), which was cooled under vacuum. Anhydrous dichloromethane (DCM) was dried by distillation from CaH2 immediately prior to use under nitrogen. All other solvents and reagents were used as received. Analytical thin-layer chromatography was carried out on precoated plates (silica gel 60, F254). Column chromatography was performed with silica gel 60 (230 to 400 mesh). 1H nuclear magnetic resonance (NMR) spectra were recorded at 400 or 500 MHz. 13C NMR spectra were recorded at 100 or 125 MHz. Chemical shifts were recorded in parts per million, and coupling constants (J) were recorded in hertz. The purities of the final compounds were determined to be >95% with a Shimadzu Prominence high-performance liquid chromatograph using a Gemini C18 column (100 by 4.6 mm; particle size, 3 μm; 110 Å) or YMC-Triart C18 column (150 by 4.6 mm; particle size, 3 μm; 12 nm), monitored by determination of the UV spectra at 254 nm. Mass spectra were recorded on a Thermo Finnigan LCQ Fleet mass spectrometer (MS). High-resolution mass spectra were recorded on a Waters Q-TOF Premier MS. Compound synthesis is depicted in Fig. 2. To a solution of 5-bromo-2,4-dihydroxybenzoic acid (Cpd1) (3.0 g, 12.8 mmol) in acetone (50 ml) were added potassium carbonate (10.6 g, 77.2 mmol) and methyl iodide (4.8 ml, 77.2 mmol). The reaction mixture was heated to reflux overnight. The reaction mixture was cooled to room temperature and concentrated in vacuo. Water was added to the reaction mixture, and the resulting solid was filtered and dried to afford methyl-5-bromo-2,4-dimethoxybenzoate (Cpd2) (3.3 g, 94%) as a white solid. To a solution of 5-bromo-2,4-dimethoxybenzoate (Cpd2) (800 mg, 2.90 mmol) in tetrahydrofuran (THF; 25 ml) and water (5 ml) were added 4,4,5,5-tetramethyl-2-(thiophen-2-yl)-1,3,2-dioxaborolane (Cpd3) (916 mg, 4.36 mmol) and potassium phosphate tribasic (1.856 g, 8.72 mmol), and the mixture was purged with nitrogen for 5 min. Then, the [1,1′-bis(diphenylphosphino)ferrocene]dichloropalladium(II) complex with dichloromethane [Pd(dppf)Cl2] · CH2Cl2 (475 mg, 0.58 mmol) was added and the reaction mixture was heated by reflux for 2 h. The reaction mixture was cooled to room temperature and filtered through Celite kieselguhr. The Celite kieselguhr was washed with ethyl acetate (EA), and the organic layer was washed with water, brine, dried over sodium sulfate, and concentrated in vacuo. The crude residue was purified by column chromatography (25% EA/hexane) to afford methyl 2,4-dimethoxy-5-(thiophen-2-yl)benzoate (Cpd5) (652 mg, 81%) as a beige solid. Lithium hydroxide (225 mg, 5.39 mmol) was added to a solution of methyl 2,4-dimethoxy-5-(thiophen-2-yl)benzoate (Cpd5) (500 mg, 1.79 mmol) in THF (4 ml), methanol (MeOH) (4 ml), and water (4 ml) at room temperature (RT). The reaction mixture was stirred at 40°C for 4 h. The reaction mixture was cooled to room temperature and concentrated in vacuo. To the reaction mixture, 20 ml of water was added, and the mixture was acidified with 1 N HCl at 0°C. The resulting solid was filtered and dried to afford 2,4-dimethoxy-5-(thiophen-2-yl)benzoic acid (Cpd7) (400 mg, 85%) as a white solid. To a solution of the carboxylic acid (1.0 eq) in dry dichloromethane were added N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) (3.0 eq) and 4-(dimethylamino)pyridine (DMAP) (3.0 eq), and the reaction mixture was stirred for 30 min. Then, the sulfonamide (1.0 eq) was added and the reaction mixture was stirred at RT for overnight. The reaction mixture was diluted with ethyl acetate and washed with 1 N HCl, water, brine. The organic layer was dried over sodium sulfate, filtered, and concentrated in vacuo. The residue was purified by column chromatography (MeOH/DCM) to afford the desired product. Ammonium chloride (208 mg, 3.88 mmol) was added to a solution of 5-(5-fluorothiophen-2-yl)-2,4-dimethoxy-N-((3-(trifluoromethyl)phenyl)sulfonyl)benzamide (Cpd16) (190 mg, 0.38 mmol) in DMF at RT. The reaction mixture was stirred at 120°C for 18 h and then cooled to room temperature. To the reaction mixture water was added and extracted with ethyl acetate. The organic layer was washed with water, brine, dried over sodium sulfate, and concentrated in vacuo. The crude residue was purified by column chromatography (5% MeOH/DCM, 1 drop of acetic acid) to afford 5-(5-fluorothiophen-2-yl)-2-hydroxy-4-methoxy-N-((3-(trifluoromethyl)phenyl)sulfonyl)benzamide (Cpd17) (80 mg, 43%) as an off-white solid. In order to see the amide peak (13.97 ppm) in the 1H NMR spectrum, after purification the off-white solid was dissolved in 100 ml of ethyl acetate and washed with 0.05 M sodium bicarbonate solution (100 ml), saturated NH4Cl solution (100 ml), water (100 ml), brine, dried over sodium sulfate, and concentrated in vacuo, to afford an off-white solid (Cpd17). To a solution of Cpd17 (30 mg, 0.06 mmol) in dry dichloromethane (2 ml) were added acetic anhydride (32 μl, 0.315 mmol) and DMAP (38 mg, 0.315 mmol) at RT. The reaction mixture was stirred at 40°C for 16 h. The reaction mixture was diluted with ethyl acetate and washed with water, brine. The organic layer was dried over sodium sulfate, filtered, and concentrated in vacuo. The residue was purified by column chromatography (1% MeOH/DCM) to afford the desired product (Cpd18) (15 mg, 46%) in white. The X-ray crystallograph of Cpd18 is deposited in CCDC (deposition number 1994169). The synthesis of NITD29 (Cpd22) is depicted in Fig. 2. Monobromination of thiophene 5 using N-bromosuccinimide (NBS) provided the α-brominated thiophene derivative (Cpd19). Hydrolysis of the methyl ester (Cpd19) gave thiophenyl carboxylic acid (Cpd20), which was coupled with quinoline-8-sulfonamide (Cpd11) to afford thiophenyl-sulfonamide (Cpd21). Subsequent Sonogashira coupling of Cpd21 with propargyl alcohol furnished NITD29 (Cpd22). NBS (320 mg, 1.8 mmol) was added to a solution of methyl 2,4-dimethoxy-5-(thiophen-2-yl)benzoate (Cpd5) (500 mg, 1.8 mmol) in DMF (10 ml) at 0°C. The reaction mixture was warmed to room temperature and stirred for 2 h. The reaction mixture was poured into water. The solid was filtered and washed with water, diethyl ether and dried in vacuo to afford methyl 5-(3,5-dibromothiophen-2-yl)-2,4-dimethoxybenzoate (Cpd19) (490 mg, 76%) as an off-white solid. Lithium hydroxide (254 mg, 6.05 mmol) was added to a solution of methyl 5-(5-bromothiophen-2-yl)-2,4-dimethoxybenzoate (Cpd19) (720 mg, 2.01 mmol) in THF (4 ml), MeOH (4 ml), and water (4 ml) at RT. The reaction mixture was stirred at 40°C for 4 h. The reaction mixture was cooled to room temperature and concentrated in vacuo. To the reaction mixture, 10 ml of water was added, and the mixture was acidified with 1 N HCl at 0°C. The resulting solid was filtered, washed with water, and dried to afford 5-(5-bromothiophen-2-yl)-2,4-dimethoxybenzoic acid (Cpd20) (580 mg, 84%) as a white solid. To a solution of 5-(5-bromothiophen-2-yl)-2,4-dimethoxy-N-(quinolin-8-lsulfonyl)benzamide (Cpd21) (50 mg, 0.093 mmol) in DMF (2 ml) were added propargyl alcohol (16 μl, 0.28 mmol), triethylamine (Et3N) (39 μl, 0.28 mmol), and CuI (5 mg, 0.03 mmol). After the mixture was purged with nitrogen for 5 min, PdCl2(dppf) · CH2Cl2 (8 mg, 0.01 mmol) was added and the mixture was stirred at 80°C for 2 h. After cooling to room temperature, the mixture was diluted with ethyl acetate and filtered through a pad of Celite kieselguhr. The organic layer was washed with water, brine, dried over sodium sulfate, and concentrated in vacuo. The crude residue was purified by column chromatography (5% MeOH/DCM) to afford 5-(5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl)-2,4-dimethoxy-N-(quinolin-8-ylsulfonyl)benzamide (NITD29) (20 mg, 42%) as an off-white solid.
Compound testing in ZIKV replicons. (i) Cells and viruses.
Baby hamster kidney fibroblast (BHK-21) cells (ATCC) and human hepatocarcinoma (HuH-7) cells (ATCC) were grown in RPMI 1640 medium (Gibco) and 4.5-g/liter glucose Dulbecco modified Eagle medium (Gibco), respectively, at 37°C in a humidified CO2 incubator. All media were supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 1% (vol/vol) penicillin-streptomycin (P/S). Cells of C6/36, an Aedes albopictus cell line (ATCC), were maintained in RPMI 1640 medium containing 25 mM Na HEPES, 10% (vol/vol) FBS, and 1% (vol/vol) P/S at 28°C in the absence of CO2. The ZIKV strain used in this study, H/PF/2013 (GenBank accession number KJ776791.2), was amplified in C6/36 cells, titers were determined by a standard BHK-21 cell plaque assay, and the virus-infected cells were stored at −80°C.
(ii) Ethics statement.
ZIKV H/PF/2013 was a gift from Cécile Baronti at Aix Marseille Université. Institutional approval to perform experiments with ZIKV at Duke-NUS was granted by the Duke-NUS Medical School.
(iii) Cell viability assay.
HuH-7 cells were seeded at a density of 2 × 104 cells/ml in a 96-well white opaque plate (Grenier). Compounds were diluted to the concentrations indicated above in medium and assayed onto the cells, and the cells were incubated for 24 h at 37°C. Cytotoxicity was determined by use of a CellTiter-Glo luminescent assay kit (Promega) according to the manufacturer’s instructions. The cell viability curve is presented as the luminescence derived from treated samples as a percentage of that derived from the untreated control. The 50% cytotoxic concentration (CC50) was defined as the concentration at which the luminescence decreased by 50% compared to that of the untreated control.
(iv) Plaque reduction assay.
HuH-7 cells were plated at a density of 1 × 105 cells/ml in a 24-well plate. Cells were infected with ZIKV H/PF/2013 at a multiplicity of infection of 0.5 for 1 h at 37°C. After infection, the inocula were removed and the cells were further incubated with medium containing the concentrations of the compound indicated above for 24 h. Supernatants were collected after 24 h and clarified for scoring of plaques by a standard BHK-21 cell plaque assay. The dose-response curve, presented as the number of PFU per milliliter against the concentration and the 50% effective concentration (EC50; defined as the concentration at which the virus level [in numbers of PFU per milliliter] is reduced to 50%), was then determined with GraphPad Prism software.
Initial fluorescence quenching analysis.
Fluorescence quenching experiments were performed on a Monolith NT.LabelFree instrument (NanoTemper Technologies) using 20% light-emitting diode and 40% laser power. The laser on and off times were set at 30 s and 5 s, respectively. The protein was applied at a final concentration of 100 nM. A twofold dilution series of the compounds (as a stock at 10 mM in 95% dimethyl sulfoxide [DMSO]) was prepared in the MST optimized buffer (NanoTemper Technologies) containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.05% Tween 20 supplemented with 200 nM ZIKV RdRp and 4.75% (vol/vol) DMSO, thus maintaining the final protein concentration constant at 100 nM and the DMSO concentration constant at 4.75% (vol/vol) from sample to sample. Samples were filled into standard Monolith NT.115 capillaries (NanoTemper Technologies) for measurement at a temperature of 37°C. As ligand-induced fluorescence changes were observed upon measurement, data from three independent measurements were analyzed (MO.Affinity Analysis software NT Control, version 2.0.2.29; NanoTemper Technologies), using the initial fluorescence analysis mode of the MO.Affinity Analysis software. The value of initial fluorescence was plotted against the ligand concentration to derive the dose-response curve. The KD values were obtained with the initial fluorescence mode, measuring the fluorescence intensity variation upon compound binding. For competition assays, the same experimental setup described above was used. The protein was applied at a final concentration of 100 nM and in the presence of 100 μM of Cpd17. A twofold dilution series of Cpd15 (as a stock at 10 mM in 95% DMSO) was prepared in the MST optimized buffer. The protein was applied at a final concentration of 100 nM. A twofold dilution series of the compounds (as a stock at 10 mM in 95% DMSO) was prepared in the MST optimized buffer (NanoTemper Technologies) containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.05% Tween 20 supplemented with 200 nM ZIKV RdRp, 200 μM Cpd17, and 4.75% (vol/vol) DMSO, thus maintaining the final protein concentration constant at 100 nM, the Cpd17 concentration constant at 100 μM, and the DMSO concentration constant at 4.75% (vol/vol) from sample to sample. Samples were filled into standard Monolith NT.115 capillaries (NanoTemper Technologies) for measurement at a temperature of 37°C. Measurements were done in triplicate. No drastic change in the fluorescence intensity was observed.
Data availability.
The coordinates of the free RdRp and RdRp bound to compounds have been deposited in the Protein Data Bank under accession numbers 6LD1, 6LD2, 6LD3, 6LD4, and 6LD5 (Table 1).
ACKNOWLEDGMENTS
We thank Chen MingWei and the Protein Production Platform at the NTU School of Biological Sciences for their help in ZIKV RdRp protein purification. We acknowledge the Soleil synchrotron for the provision of beamtime (proposal 20170003) and thank William Shepard and Martin Savko for expert assistance in using beamline Proxima 2A. We thank the NTU Institute of Structural Biology for access to the in-house Rigaku FR-X diffractometer and NanoTemper MicroScale Thermophoresis and Liew Chong Wai for expert help.
We declare that we have no competing financial interests.
This work was supported by grant NRF2016-CRP001-063 to the laboratories of J.L. and S.G.V. and in part by NMRC grant MOH-000086 (grant MOH-OFIRG18may-0006).
REFERENCES
- 1.Dick GW, Kitchen SF, Haddow AJ. 1952. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Gould EA, Solomon T. 2008. Pathogenic flaviviruses. Lancet 371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- 3.Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. 2014. Zika virus infection complicated by Guillain-Barre syndrome—case report, French Polynesia, December 2013. Euro Surveill 19(9):pii=20720. doi: 10.2807/1560-7917.ES2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- 4.Oliveira Melo AS, Malinger G, Ximenes R, Szejnfeld PO, Alves Sampaio S, Bispo de Filippis AM. 2016. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: tip of the iceberg? Ultrasound Obstet Gynecol 47:6–7. doi: 10.1002/uog.15831. [DOI] [PubMed] [Google Scholar]
- 5.Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D. 2017. An update on Zika virus infection. Lancet 390:2099–2109. doi: 10.1016/S0140-6736(17)31450-2. [DOI] [PubMed] [Google Scholar]
- 6.Delvecchio R, Higa L, Pezzuto P, Valadão A, Garcez P, Monteiro F, Loiola E, Dias A, Silva F, Aliota M, Caine E, Osorio J, Bellio M, O’Connor D, Rehen S, de Aguiar R, Savarino A, Campanati L, Tanuri A. 2016. Chloroquine, an endocytosis blocking agent, inhibits Zika virus infection in different cell models. Viruses 8:322. doi: 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbosa-Lima G, da Silveira Pinto LS, Kaiser CR, Wardell JL, De Freitas CS, Vieira YR, Marttorelli A, Cerbino Neto J, Bozza PT, Wardell SV, de Souza MVN, Souza TML. 2017. N-(2-(Arylmethylimino)ethyl)-7-chloroquinolin-4-amine derivatives, synthesized by thermal and ultrasonic means, are endowed with anti-Zika virus activity. Eur J Med Chem 127:434–441. doi: 10.1016/j.ejmech.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 8.El Sahili A, Lescar J. 2017. Dengue virus non-structural protein 5. Viruses 9:91. doi: 10.3390/v9040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yap TL, Xu T, Chen Y-L, Malet H, Egloff M-P, Canard B, Vasudevan SG, Lescar J. 2007. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol 81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malet H, Masse N, Selisko B, Romette JL, Alvarez K, Guillemot JC, Tolou H, Yap TL, Vasudevan SG, Lescar J, Canard B. 2008. The flavivirus polymerase as a target for drug discovery. Antiviral Res 80:23–35. doi: 10.1016/j.antiviral.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Lim SP, Noble CG, Shi PY. 2015. The dengue virus NS5 protein as a target for drug discovery. Antiviral Res 119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Soh TS, Zheng J, Chan KWK, Phoo WW, Lee CC, Tay MYF, Swaminathan K, Cornvik TC, Lim SP, Shi PY, Lescar J, Vasudevan SG, Luo D. 2015. A crystal structure of the dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog 11:e1004682. doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, Vizjak A, Pižem J, Petrovec M, Avšič Županc T. 2016. Zika virus associated with microcephaly. N Engl J Med 374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 15.Ventura CV, Maia M, Bravo-Filho V, Góis AL, Belfort R. 2016. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet 387:228. doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- 16.Yin Z, Chen Y-L, Schul W, Wang Q-Y, Gu F, Duraiswamy J, Kondreddi RR, Niyomrattanakit P, Lakshminarayana SB, Goh A, Xu HY, Liu W, Liu B, Lim JYH, Ng CY, Qing M, Lim CC, Yip A, Wang G, Chan WL, Tan HP, Lin K, Zhang B, Zou G, Bernard KA, Garrett C, Beltz K, Dong M, Weaver M, He H, Pichota A, Dartois V, Keller TH, Shi P-Y. 2009. An adenosine nucleoside inhibitor of dengue virus. Proc Natl Acad Sci U S A 106:20435–20439. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niyomrattanakit P, Chen Y-L, Dong H, Yin Z, Qing M, Glickman JF, Lin K, Mueller D, Voshol H, Lim JYH, Nilar S, Keller TH, Shi P-Y. 2010. Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J Virol 84:5678–5686. doi: 10.1128/JVI.02451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim SP, Wang QY, Noble CG, Chen YL, Dong H, Zou B, Yokokawa F, Nilar S, Smith P, Beer D, Lescar J, Shi PY. 2013. Ten years of dengue drug discovery: progress and prospects. Antiviral Res 100:500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Lim SP, Noble CG, Seh CC, Soh TS, El Sahili A, Chan GK, Lescar J, Arora R, Benson T, Nilar S, Manjunatha U, Wan KF, Dong H, Xie X, Shi PY, Yokokawa F. 2016. Potent allosteric dengue virus NS5 polymerase inhibitors: mechanism of action and resistance profiling. PLoS Pathog 12:e1005737. doi: 10.1371/journal.ppat.1005737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan W, Song H, Wang H, Chai Y, Su C, Qi J, Shi Y, Gao GF. 2017. The crystal structure of Zika virus NS 5 reveals conserved drug targets. EMBO J 36:919–933. doi: 10.15252/embj.201696241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godoy AS, Lima GM, Oliveira KI, Torres NU, Maluf FV, Guido RV, Oliva G. 2017. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat Commun 8:14764. doi: 10.1038/ncomms14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Upadhyay AK, Cyr M, Longenecker K, Tripathi R, Sun C, Kempf DJ. 2017. Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallogr F Struct Biol Commun 73:116–122. doi: 10.1107/S2053230X17001601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B, Tan XF, Thurmond S, Zhang ZM, Lin A, Hai R, Song J. 2017. The structure of Zika virus NS5 reveals a conserved domain conformation. Nat Commun 8:14763–14766. doi: 10.1038/ncomms14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu G, Gong P. 2017. A structural view of the RNA-dependent RNA polymerases from the Flavivirus genus. Virus Res 234:34–43. doi: 10.1016/j.virusres.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 26.Tian W, Chen C, Lei X, Zhao J, Liang J. 2018. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noble CG, Lim SP, Chen Y-L, Liew CW, Yap L, Lescar J, Shi P-Y. 2013. Conformational flexibility of the dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J Virol 87:5291–5295. doi: 10.1128/JVI.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niyomrattanakit P, Wan KF, Chung KY, Abas SN, Seh CC, Dong H, Lim CC, Chao AT, Lee CB, Nilar S, Lescar J, Shi PY, Beer D, Lim SP. 2015. Stabilization of dengue virus polymerase in de novo initiation assay provides advantages for compound screening. Antiviral Res 119:36–46. doi: 10.1016/j.antiviral.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Niyomrattanakit P, Abas SN, Lim CC, Beer D, Shi PY, Chen YL. 2011. A fluorescence-based alkaline phosphatase-coupled polymerase assay for identification of inhibitors of dengue virus RNA-dependent RNA polymerase. J Biomol Screen 16:201–210. doi: 10.1177/1087057110389323. [DOI] [PubMed] [Google Scholar]
- 30.Ramharack P, Soliman MES. 2018. Zika virus NS5 protein potential inhibitors: an enhanced in silico approach in drug discovery. J Biomol Struct Dyn 36:1118–1133. doi: 10.1080/07391102.2017.1313175. [DOI] [PubMed] [Google Scholar]
- 31.Lin Y, Zhang H, Song W, Si S, Han Y, Jiang J. 2019. Identification and characterization of Zika virus NS5 RNA-dependent RNA polymerase inhibitors. Int J Antimicrob Agents 54:502–506. doi: 10.1016/j.ijantimicag.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karplus PA, Diederichs K. 2012. Linking crystallographic model and data quality. Science 336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The coordinates of the free RdRp and RdRp bound to compounds have been deposited in the Protein Data Bank under accession numbers 6LD1, 6LD2, 6LD3, 6LD4, and 6LD5 (Table 1).