Intestinal organoids are a newly developed culture system for investigating pathogen-host interactions. Intestinal organoid models have been widely used since their development, because the results obtained from this type of culture model better represent physiological conditions than those from well-established cell lines. The three-dimensional (3D) porcine intestinal organoid model was reported in 2018 and 2019 for the investigation of intestinal pathogens. However, those organoid culture models were basal-out intestinal organoids, which are not suitable for porcine enteric virus research because they invade the intestines via the apical side of epithelial cells on villi. In this study, we developed a porcine apical-out intestinal organoid culture system and verified its infectivity, type I and type III interferon (IFN) antiviral responses, and inflammatory responses following infection by a swine enteric virus. Our results imply that this apical-out porcine intestinal organoid culture system is an ideal model for the investigation of interactions between swine enteric viruses and the intestines.
KEYWORDS: apical-out, TGEV, crypt cell, intestinal organoids, pigs
ABSTRACT
The intestinal organoid culture system is a pathbreaking working model for investigating pathogen-host interactions in the intestines. However, due to the limitations of the first generation of intestinal organoids, basal-out structure and growth in Matrigel, most pathogens can rarely attach to the apical membrane directly and hardly initiate infection. In this study, we first developed a next-generation porcine intestinal organoid culture system, characterized by an apical membrane on the surface, called apical-out. To investigate the infectivity and antiviral immune responses of this apical-out porcine intestinal organoid, a swine enteric virus, transmissible gastroenteritis virus (TGEV), was employed to inoculate the culture system. Both reverse transcription-quantitative PCR (RT-qPCR) and immunofluorescence assay (IFA) analysis demonstrated that TGEV replicated in the apical-out porcine intestinal organoid culture system. Additionally, our results illustrated that TGEV infection significantly upregulated the expression levels of alpha interferon (IFN-α), IFN-λ1, interferon-stimulated gene 15 (ISG15), ISG58, tumor necrosis factor alpha (TNF-α), and interleukin 6 (IL-6) in this culture system. Hence, we successfully developed a porcine intestinal apical-out organoid culture system, which will facilitate the investigation of pathogen-host interactions in pig intestines.
IMPORTANCE Intestinal organoids are a newly developed culture system for investigating pathogen-host interactions. Intestinal organoid models have been widely used since their development, because the results obtained from this type of culture model better represent physiological conditions than those from well-established cell lines. The three-dimensional (3D) porcine intestinal organoid model was reported in 2018 and 2019 for the investigation of intestinal pathogens. However, those organoid culture models were basal-out intestinal organoids, which are not suitable for porcine enteric virus research because they invade the intestines via the apical side of epithelial cells on villi. In this study, we developed a porcine apical-out intestinal organoid culture system and verified its infectivity, type I and type III interferon (IFN) antiviral responses, and inflammatory responses following infection by a swine enteric virus. Our results imply that this apical-out porcine intestinal organoid culture system is an ideal model for the investigation of interactions between swine enteric viruses and the intestines.
INTRODUCTION
Swine enteric coronaviruses, such as porcine epidemic diarrheal virus (PEDV), transmissible gastroenteritis virus (TGEV), porcine deltacoronavirus (PDCoV), and swine acute diarrhea syndrome coronavirus (SADS-CoV), have caused severe economic losses in the pig industry in the past decade (1). However, these viruses can rarely infect and propagate in vitro culture systems. Some investigators have used nonintestinal cells, including nonporcine cells, for swine enteric coronavirus research (2). However, these cells may not reveal the actual virus-host interaction. Therefore, some other researchers developed swine enteric epithelial cell lines, such as IPEC-J2 and IP-21 (3). Unfortunately, the viral infectivity in IPEC-J2 and IP-21 cells is quite low, and huge variations exist in the findings reported from different research groups using these cell lines (4). Thus, the development of a more physiological and robust culture system is urgently needed for investigations of porcine coronaviruses.
Intestinal organoids were first established in 2009 by differentiation from Lgr5+ stem cells in crypts (5). Stem cells possess pluripotency and can differentiate into many cell types. Thus, intestinal organoids differentiated from crypt Lgr5+ stem cells include many intestinal cell types, e.g., enterocytes, goblet cells, and Paneth cells (6). This organoid culture system was gradually employed to investigate the interactions between the epithelial cells and bacterium or viruses (7). The fact that the intestinal basal membrane was exhibited on the surface of this three-dimensional (3D) intestinal organoid led to difficulties with infection because most intestinal pathogens recognize receptors expressed on the apical membrane. Although some strategies, such as repeated pipetting or microinjection, were used to facilitate the invasion of pathogens into organoids (7), these methods are still difficult to operate and cause considerable variations in subsequent results when the 3D culture system is used to investigate pathogen-host cross talk. Co et al. developed an apical-out intestinal organoid model in 2019 to study the invasion of intestinal pathogens (8). These researchers reversed the polarity of conventional human intestinal organoids in a suspension culture system and analyzed the invasion process of Salmonella enterica serovar Typhimurium and Listeria monocytogenes using this system (8). These investigations illuminated the interaction studies between swine enteric viruses and the host.
Li et al. developed a traditional porcine intestinal organoid (basal-out) culture system and used this 3D organoid for swine enteric virus infection. Instead, these researchers transformed this 3D organoid into a two-dimensional (2D) monolayer culture system to investigate infection by PEDV (9), a typical porcine enteric virus that causes severe diarrhea in piglets (10). This working model did not mimic the infection within the real gut environment, since it was basal-out and the inoculation was carried out in a 2D monolayer phase. To develop a more physiological model for swine enteric viral infection studies, we developed polarity-reversed porcine intestinal organoids (apical-out) maintained in suspension culture and analyzed their infectivity and host antiviral immune responses. Our results demonstrated that this apical-out porcine intestinal organoid is an ideal model for virus-host interaction studies of swine enteric viruses.
RESULTS
Development of porcine intestinal organoids.
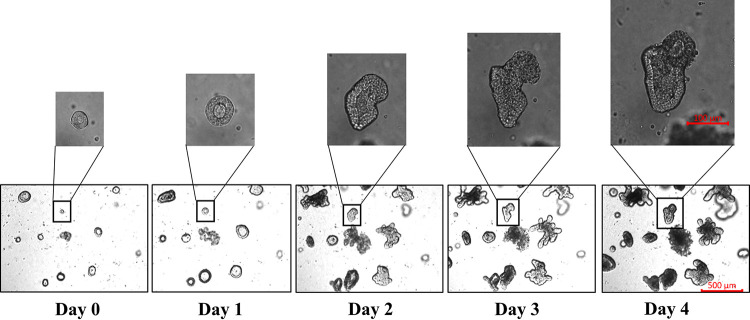
To establish a more physiological infection model for swine enteric virus investigation, we first prepared basal-out porcine intestinal organoids from crypts of the jejunum. The crypts were isolated from swine jejunum and cultured in Matrigel supplemented with organoid growth medium (OGM). Over time, the sizes of each crypt in the culture became larger. At 3 to 4 days postculture, most of the crypts differentiated and budded into organoids with villi, and some of the organoids showed a symmetrical structure (Fig. 1). We next developed a freeze-thaw protocol for the cryopreservation of these organoids. These results demonstrated that a porcine intestinal organoid culture system was successfully established.
FIG 1.
Development of porcine intestinal organoids. Crypts were isolated from the jejuna of 1- to 3-month-old pigs, cultured in Matrigel matrix with organoid growth medium, and monitored daily under a microscope to verify the formation diameter of the organoids. The developed organoids were subjected to freeze-thaw cycling, and then their daily growth was observed under a microscope. Images in the top panels are enlarged representative organoids. Images were obtained with a Zeiss Vert A1 microscope.
Generation of apical-out porcine intestinal organoids.
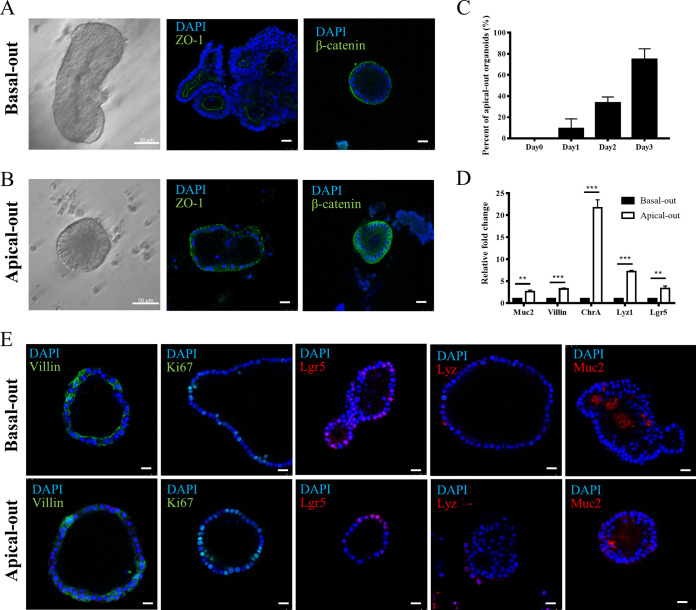
Generally, most intestinal pathogens invade tissue from the apical membrane of intestinal epithelial cells. However, traditional intestinal organoids cultured in Matrigel are all basal-out. To expose the apical membrane of epithelial cells to the pathogens for better attachment, traditional porcine intestinal organoids were first cultured in Matrigel and then were induced in a suspension culture system for polarity reversal, apical-out. From the morphology analysis under a differential interference contrast (DIC) microscope, the epithelial cells were more columnar on the apical-out organoids (Fig. 2A and B). To further confirm the formation of apical-out organoids, they were fixed and stained with the apical protein marker ZO-1 and basal protein β-catenin. Then, these organoids were observed under a confocal microscope. The results demonstrated that ZO-1 was expressed on the inner lumen of the traditional basal-out organoids (Fig. 2A). However, in the apical-out organoids, ZO-1 was present on the outer membrane of the organoids (Fig. 2B). More β-catenin was expressed on the lumen side of apical-out organoids than that of basal-out organoids (Fig. 2A and B). These results suggested that the polarity of the porcine intestinal organoids was successfully reversed to apical-out by the removal of Matrigel and that they could be cultured in a suspension medium. To determine the organoid’s polarity reversal efficiency, the percentages of apical-out organoids in suspension-cultured organoids were determined from day 0 to day 3. In 3-day cultures, the percentages of apical-out organoids ranged from 0% to 80% from day 0 to day 3 (Fig. 2C). The mRNA levels of goblet cells, enterocytes, enteroendocrine, Paneth cells, intestinal stem cell-related gene mucin 2 (Muc2), villin, chromogranin A (ChrA), lysozyme 1 (Lyz1), and Lgr5 were determined for both basal-out and apical-out organoids. The results suggested that all of these genes were upregulated between 2- and 20-fold (Fig. 2D) and that a lack of Matrigel may promote the differentiation of these cell types on organoids. To verify the cell types in both basal-out and apical-out organoids, the cellular markers villin, Ki67, Lgr5, lysozyme (Lyz), and mucin 2 (Muc2) were stained by indirect immunofluorescence assay (IFA) to identify the enterocytes, proliferating cells, intestinal stem cells, Paneth cells, and goblet cells, respectively. The results illustrated that these types of cells were present on both basal-out and apical-out organoids (Fig. 2E).
FIG 2.
Induction of porcine apical-out intestinal organoids. Porcine basal-out intestinal organoids were first prepared and isolated from the 3D Matrigel culture system. The isolated basal-out organoids were then cultured in ultralow-attachment 24-well tissue culture plates supplemented with organoid growth medium. The morphology of organoids was observed under a microscope daily to check their reversal polarity. (A and B) Representative DIC images of porcine basal-out and apical-out intestinal organoids are shown. The organoids were stained with ZO-1, β-catenin, and DAPI to localize the apical side, basal side, and nucleus, respectively, and then they were analyzed under a Leica TCS SP8 confocal microscope. Scale bars, 25 μm. (C) The percentages of apical-out organoids were analyzed daily by microscopy. (D) The mRNA levels of goblet cells, enterocytes, enteroendocrine cells, Paneth cells, and intestinal stem cell-related genes, such as mucin 2 (Muc2), villin, chromogranin A (ChrA), lysozyme 1 (Lyz1), and Lgr5 on both basal-out and apical-out organoids were compared via RT-qPCR. The RT-qPCR data were calculated using the comparative threshold cycle (2−ΔΔCT) method. **, P < 0.01; ***, P < 0.001. (E) The basal-out and apical-out organoids were subjected to IFA staining for enterocytes (villin), proliferating cells (Ki67), Paneth cells (Lyz), intestinal stem cells (Lgr5), and goblet cells (Muc2). Images were obtained with a Leica TCS SP8 confocal microscope. Scale bars, 25 μm.
Apical-out porcine intestinal organoids are susceptible to TGEV.
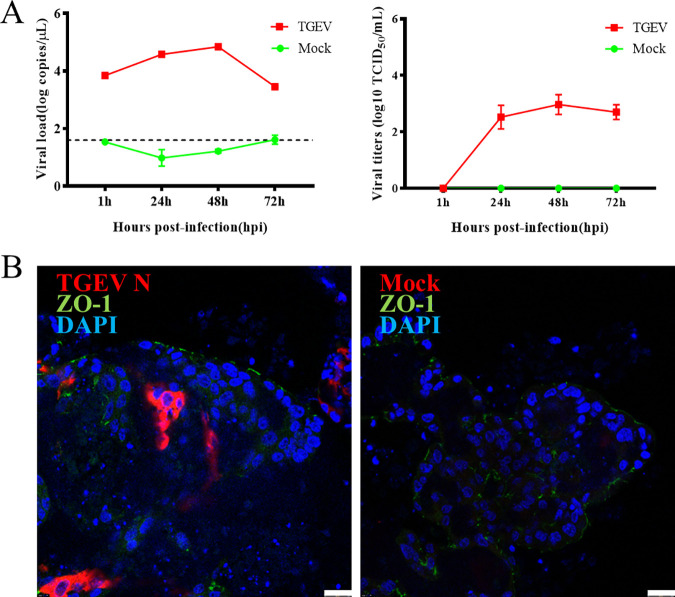
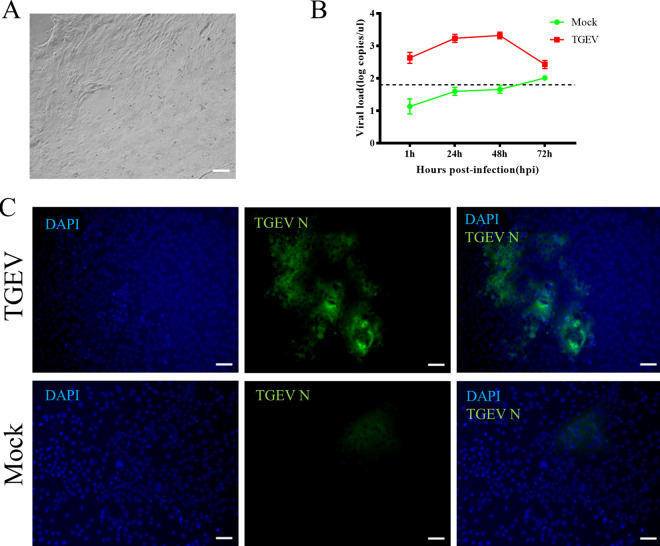
To investigate whether apical-out porcine organoids can be infected by swine enteric virus, a classical and well-characterized virus, TGEV, was selected to inoculate the organoids. Organoids were collected at different time points postinfection for viral load detection by reverse transcription-quantitative (RT-qPCR) and 50% tissue culture infective dose (TCID50) assay on LLC-PK1 cells. The results showed that both the TGEV genome and infectious viral titers remained increased with the culture duration and peaked at 48 h postinfection (hpi) (Fig. 3A). The organoids were collected at 48 hpi and subjected to IFA analysis to confirm TGEV replication. The results illustrated that the epithelial cell junction protein existed on the surfaces of the organoids. In contrast, the TGEV N protein was detected in some cells of the organoids (Fig. 3B), indicating that TGEV successfully invaded the organoids via the apical surface. These results suggested that the apical-out porcine intestinal organoids developed in this study can be used for swine enteric virus infection. In a parallel TGEV infection experiment, a porcine 2D organoid culture system was established (Fig. 4A). The 2D organoids were subjected to TGEV infection at a multiplicity of infection (MOI) of 0.1. Finally, a similar dynamic of viral copies in organoids can be observed (Fig. 4B), and the TGEV N protein was also detected in 2D organoids at 48 hpi (Fig. 4C).
FIG 3.
TGEV infection in porcine apical-out intestinal organoids. Apical-out organoids were inoculated with TGEV and cultured in a suspension system. (A, left) Samples were collected at the indicated time points for viral load detection by RT-qPCR. The dashed line represents the limits of detection. (Right) The infectious TGEV in the supernatant was titrated by TCID50 assay on the LLC-PK1 cell line. (B) TGEV-infected or mock-infected organoids were collected at 48 hpi, fixed, permeabilized, and stained with ZO-1 (green) and TGEV N protein (red). DAPI was used for nuclear staining. Images were obtained with a Leica TCS SP8 confocal microscope. Scale bars, 25 μm.
FIG 4.
TGEV infection in porcine 2D intestinal organoids. (A) Organoids were dissociated and seeded on a Matrigel-precoated 24-well plate, and 2D monolayer organoids formed after 3 days in culture. (B) Then, 2D organoids were inoculated with TGEV, and samples were collected at the indicated time points for viral load detection by RT-qPCR. The dashed line represents the limit of detection. (C) TGEV-infected or mock-infected organoids were fixed at 48 hpi for staining with TGEV N protein (green). DAPI was used for nuclear staining. Images were obtained with a ZEISS Vert A1 microscope. Scale bars, 50 μm.
Antiviral responses are activated in apical-out organoids.
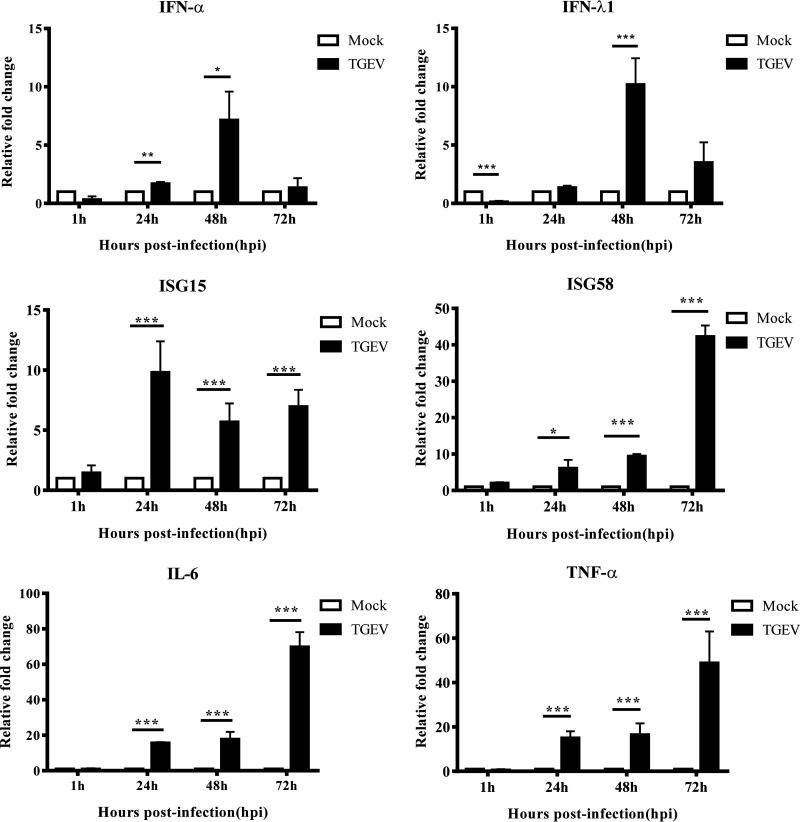
To verify whether immune responses can be activated in the apical-out porcine organoids, certain antiviral and inflammatory factors were selected to be evaluated by RT-qPCR. We found that the mRNA levels of alpha interferon (IFN-α) and IFN-λ1 decreased at 1 hpi, recovered quickly at 24 hpi, and peaked at 48 hpi (Fig. 5). Interferon-stimulated gene 15 (ISG15) was significantly upregulated and remained at a high level throughout the time course, while ISG58 continued to increase and reached a peak at 72 hpi (Fig. 5). These results suggest that type I and type III IFN signaling pathways are activated by TGEV infection. Additionally, the expression levels of interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α) were significantly increased and peaked at 72 hpi (Fig. 5), implying that the inflammatory pathway was also activated in the apical-out porcine organoids by TGEV inoculation. These results indicated that the immune responses of apical-out porcine intestinal organoids can be successfully activated by a viral infection.
FIG 5.
Antiviral and inflammatory responses are activated in apical-out intestinal organoids by TGEV infection. Apical-out organoids were inoculated with TGEV and cultured in a suspension system. Samples were collected at the indicated time points to detect IFN-α, IFN-λ1, IFN responses (ISG15 and ISG58), and inflammatory responses (IL-6 and TNF-α) using relative RT-qPCR. All the data were calculated using the 2−ΔΔCT method. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Since the development of murine intestinal organoids in 2009 (5), human, porcine, and bovine intestinal organoid culture models have emerged from different research groups (6, 7, 9). This newly developed culture model, intestinal organoids, has been widely used to investigate the interactions between pathogens and the host, such as Salmonella Typhimurium, Listeria monocytogenes, human rotavirus, and PEDV (7, 9, 11, 12). It is well known that the epithelial cell is polarized, with a basal side and an apical side. The polarity of intestinal epithelial cells maintains the directional transport function, while the specialization of the plasma membrane leads to structural and functional differences between apical and basal membranes (13, 14). The distributional gap of epithelial adhesion molecules between apical and basal membrane determines the tendency of pathogens. The intestinal organoid systems mentioned above belong to basal-out organoids. Some pathogens, for example, Listeria monocytogenes, exclusively invade the host via receptors on the basal side of intestinal epithelial cells even when they are on the apical membrane (15). However, most enteric pathogens infect the intestines through the apical surface of epithelial cells, limiting their investigation if the basal-out intestinal organoid system is used. Some research groups have attempted to avoid this limitation by establishing high-throughput microinjection platforms to inoculate the organoids (16). However, this protocol cannot be widely applied in conventional laboratories. To solve this problem, Co et al. developed apical-out human intestinal organoids to replace the microinjection method for pathogen-host interaction research (8). In their study, they removed the basement membrane extract from the 3D organoid culture system, since the Matrigel contained extracellular matrix protein that determine the organoid polarity, and then induced polarity reversal in suspension culture medium (8).
Li et al. developed the first basal-out 3D porcine intestinal organoids to investigate PEDV infection and the host innate response (9). However, porcine enteric viruses, e.g., PEDV, TGEV, and reovirus, invade the intestines via the apical side of the epithelial cells on villi. Therefore, PEDV infection and subsequent experiments were carried out on a 2D interface originating from the intestinal organoids that were different from the infection procedures under physiological conditions (17). Our preliminary investigations demonstrated that basal-out organoids were not susceptible to TGEV infection (data not shown). To solve this problem, we developed a porcine apical-out intestinal organoid culture system that facilitates the study of enteric viruses that exclusively infect the apical surface. Subsequently, a traditional swine enteric coronavirus, TGEV, was employed to verify the infectivity of these apical-out porcine intestinal organoids. It produced high infection and replication efficacy. Additionally, TGEV infection successfully activated type I IFN and type III antiviral pathways and inflammatory pathways. These results imply that the apical-out porcine intestinal organoid culture system is an ideal model to investigate interactions between swine enteric viruses and the intestines.
In conclusion, we developed an apical-out porcine intestinal organoid culture system and provided a more physiological research model for porcine virus-epithelial interaction investigations. This new model also provides a reference to establish apical-out intestinal organoid models in other animals.
MATERIALS AND METHODS
Viruses and animals.
The TGEV H165 strain was recovered from a commercialized attenuated live vaccine. The infections with TGEV were titrated on the LLC-PK1 cell line by TCID50 assay. Pigs were purchased from a pig farm and housed in isolated animal rooms. All experimental procedures and animal care protocols were approved by and in accordance with the guidelines for Care and Use of Laboratory Animals of Lanzhou Veterinary Research Institute (LVRI), Chinese Academy of Agricultural Sciences, China.
Isolation of crypts and culture of porcine intestinal organoids.
Crypts of small intestines were isolated from the jejuna of 3-month-old pigs. Briefly, the partial jejunum was collected and opened longitudinally. Next, the intestinal content, mucus, and part of the villus were removed by gently scraping the intestine wall with a glass slide. After washing with cold 10 mM phosphate buffer (pH 7.4), the jejunum segments were incubated with 5 mM EDTA dissociation buffer for 30 min on ice. After the dissociation buffer was removed and three rounds of washes, the crypts were isolated using a surgical blade to scrape the intestine wall. The crypts were then washed several times in 10 mM cold phosphate buffer (pH 7.4) and cultured in Matrigel matrix (Corning, USA) with organoid growth medium (StemCell, Canada) as approximately 500 organoids in each well of a 24-well tissue culture plate (Corning, USA). The morphology of organoids was observed under a microscope daily.
Freeze and thaw of porcine intestinal organoids.
Organoid freezing medium (OFM) was prepared using 90% fetal bovine serum (FBS; AusGeneX, Australia) and 10% dimethyl sulfoxide (DMSO; Solarbio, China). The porcine intestinal organoids were collected and washed with Dulbecco’s modified Eagle medium (DMEM)/F12 (Sigma, USA). After removing the supernatant, approximately 1,500 organoids were resuspended in 1.2 ml OFM. Next, the organoid mix was subjected to gradient freezing and stored in liquid nitrogen. To thaw the organoids, frozen organoids were heated in a water bath at 37°C until thawed completely. Next, the organoid mix was washed with DMEM/F12 and cultured in Matrigel with OGM as approximately 500 organoids in each well of a 24-well tissue culture plate.
Development of apical-out porcine intestinal organoids.
Porcine basal-out intestinal organoids were cultured in the 3D system with Matrigel for 1 to 2 weeks. They were harvested through incubation with 5 mM cold EDTA dissociation buffer on a rotating platform for 1 h at 4°C. To generate apical-out intestinal organoids, the organoids were then collected by centrifugation at 200 × g for 5 min, washed with DMEM/F12 medium, and cultured in ultralow-attachment 24-well tissue culture plates (Corning, USA) in OGM supplemented with 10 μM ATP-competitive inhibitor of Rho-associated kinases (Y-27632; CST, USA) at 37°C with 5% CO2. The morphology of the organoids was observed under a microscope daily to check their reversal polarity.
Development of 2D porcine intestinal organoids.
Twenty-four-well tissue culture plates were precoated with DMEM/F12 with 1% Matrigel for 1 h at 37°C. Well-developed organoids were collected from Matrigel and dissociated with TrypLE Express (Gibco, USA) at 37°C for 10 min. After removing TrypLE by centrifugation at 300 × g for 5 min, the organoids were resuspended in OGM (with 20% FBS) and seeded in precoated 24-well tissue culture plates at approximately 1,500 organoids per well. After a 3-day culture at 37°C with 5% CO2, 2D organoids were formed.
Virus infection on apical-out and 2D organoids.
Apical-out organoids were collected from culture suspension by centrifugation at 200 × g for 5 min. They were inoculated with the TGEV H165 strain (MOI = 0.1) or mock (DMEM/F12 medium) for 1 h at 37°C. The virus residue was removed by centrifugation and washed 3 times with DMEM/F12. The organoids were incubated in OGM in a 37°C incubator supplied with 5% CO2. They were harvested at 1 h, 24 h, 48 h, and 72 h postinfection for viral load analysis and evaluation of antiviral immune responses.
2D organoids were washed with 10 mM phosphate buffer and incubated with the TGEV H165 strain (MOI = 0.1) or mock (DMEM/F12 medium) for 1 h at 37°C. The virus residue was removed and washed 3 times with DMEM/F12. The organoids were incubated in OGM (with 20% FBS) in a 37°C incubator supplied with 5% CO2 and were harvested at 1 h, 24 h, 48 h, and 72 h postinfection for viral load analysis and immunostaining.
RNA extraction and quantitative real-time PCR.
The TGEV-infected organoids, mock-infected organoids, well-developed basal-out organoids, and apical-out organoids were collected and subjected to total RNA extraction using RNAiso reagent (TaKaRa, Japan). The total RNA was quantified and used for cDNA preparation using Honor II 1st Strand cDNA Synthesis SuperMix (Novogene, China) with hexamer random primers. The TGEV virus copy number was determined using the TaqMan probe-based RT-qPCR previously developed in our laboratory (18). To evaluate the antiviral immune responses of apical-out organoids and compare the differentiation of epithelial cells on basal-out organoids and apical-out organoids, the relative expression levels of IFN-α, IFN-λ1, ISG15, ISG58, TNF-α, IL-6, Muc2, Villin, ChrA, Lyz1, and Lgr5 were determined by RT-qPCR using the Unique Aptamer qPCR SYBR green master mix (Novogene, China). The primers and probe used in this study are listed in Table 1.
TABLE 1.
Primers and probes used in this study
| Target gene | Primer or probe | Sequence (5′→3′) |
|---|---|---|
| TGEV N | Forward | TGCCATGAACAAACCAAC |
| Reverse | GGCACTTTACCATCGAAT | |
| Probe | HEX-TAGCACCACGACTACCAAGC-BHQ1a | |
| TNF-α | Forward | GCCCTTCCACCAACGTTTTC |
| Reverse | CAAGGGCTCTTGATGGCAGA | |
| ISG15 | Forward | GGTGAGGAACGACAAGGGTC |
| Reverse | GGCTTGAGGTCATACTCCCC | |
| ISG58 | Forward | CCGTGGAGCGAGACTCTATG |
| Reverse | ATGTTCTCCAGACGAAGGGC | |
| IL-6 | Forward | TGCAATAACCACCCCTGACC |
| Reverse | ATTTGCCGAAGAGCCCTCAG | |
| GAPDH | Forward | GATGGGCGTGAACCATGAGA |
| Reverse | CATGGACCGTGGTCATGAGT | |
| IFN-α | Forward | CTGCTGCCTGGAATGAGAGCC |
| Reverse | TGACACAGGCTTCCAGGTCCC | |
| IFN-λ1 | Forward | CCACGTCGAACTTCAGGCTT |
| Reverse | ATGTGCAAGTCTCCACTGGT | |
| Muc2 | Forward | GGCTGCTCATTGAGAGGAGT |
| Reverse | ATGTTCCCGAACTCCAAGG | |
| Villin | Forward | TTGTAGCGGAGATGAGCGGGAGA |
| Reverse | CGGGGAGTGATGACCAGGGTTTC | |
| ChrA | Forward | GACCTCGCTCTCCAAGGAGCCA |
| Reverse | TGTGCGCCTGGGCGTTTCTT | |
| Lyz1 | Forward | GGTCTATGATCGGTGCGAGT |
| Reverse | AACTGCTTTGGGTGTCTTGC | |
| Lgr5 | Forward | GAGCCTGGGAAAGCAAACC |
| Reverse | GGACAAATGCCACGGAAGA |
HEX, 6-carboxy-2,4,4,5,7,7-hexachlorofluorescein; BHQ1, black hole quencher 1.
Indirect immunofluorescent assay.
Organoids were collected and fixed with 4% paraformaldehyde for 20 min at 4°C and then were blocked and permeabilized with 10 mM phosphate buffer (with 3% bovine serum albumin [BSA] and 1% Triton X-100) for 12 h at 4°C. The primary antibodies, mouse anti-β-catenin monoclonal antibody (Abcam, U.K.), rabbit anti-ZO-1 polyclonal antibody (Invitrogen, USA), mouse anti-villin monoclonal antibody (Santa Cruz Biotechnology, USA), mouse anti-Ki67 monoclonal antibody (BD Pharmingen, USA), rabbit anti-Lgr5 polyclonal antibody (Invitrogen, USA), rabbit anti-Lyz polyclonal antibody (Invitrogen, USA), rabbit anti-Muc2 polyclonal antibody (Abbexa, U.K.), and mouse anti-TGEV N protein monoclonal antibody (kindly provided by L. Feng), were diluted at 1:100 in 10 mM phosphate buffer with 3% BSA and 1% Triton X-100. They were applied to the organoids for a 24-h incubation at 4°C. After washes, the secondary antibodies, goat anti-rabbit IgG Alexa Fluor 488 (Invitrogen, USA), goat anti-rabbit IgG phycoerythrin (PE) (Southern Biotech, USA), goat anti-mouse IgG Alexa Fluor 488 (Southern Biotech, USA), and goat anti-mouse IgG Alexa Fluor 647 (Abcam, U.K.), were diluted at 1:500 and were incubated with the organoids for another 24 h at 4°C. Next, the organoids were subjected to 4′,6-diamidino-2-phenylindole (DAPI) staining followed by analysis by confocal microscopy (Leica TCS SP8).
Statistical analysis.
The data from triplicate experiments were pooled and presented as the means ± standard deviations (SDs). The significance between groups was analyzed by one-way analysis of variance or Student’s t test with GraphPad Prism 7 software and are shown as P values.
ACKNOWLEDGMENTS
We thank Manuel R. Amieva and Julia Y. Co from Stanford University for the technical advice on polarity reversal of porcine intestinal organoids and Li Feng and Pinghuang Liu from Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, for providing a TGEV monoclonal antibody and anti-ChrA antibody. We also thank Qinghua Yu from Nanjing Agricultural University for providing an anti-mucin 2 antibody.
This work was supported by the National Key R&D Program of China (2016YFD0500103), National Natural Science Foundation of China (31972689 and 31572498), China Central Public-interest Scientific Institution Basal Research Fund (1610312020020), and the ULg-CAAS joint PhD Program.
Y.L., N.Y., N.Z., Guo Liu, and Guangliang Liu conceived the study. Y.L., N.Y., and Guangliang Liu designed the experiments. Y.L., N.Y., and X.H. performed the experiments. Y.L., N.Y., and S.Y. analyzed data. J.C. prepared viral stock. Y.L., S.Y., and Guangliang Liu wrote the draft. Guangliang Liu edited the manuscript.
REFERENCES
- 1.Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofmann M, Wyler R. 1988. Propagation of the virus of porcine epidemic diarrhea in cell culture. J Clin Microbiol 26:2235–2239. doi: 10.1128/JCM.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung K, Miyazaki A, Hu H, Saif LJ. 2018. Susceptibility of porcine IPEC-J2 intestinal epithelial cells to infection with porcine deltacoronavirus (PDCoV) and serum cytokine responses of gnotobiotic pigs to acute infection with IPEC-J2 cell culture-passaged PDCoV. Vet Microbiol 221:49–58. doi: 10.1016/j.vetmic.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Fang L, Liu S, Ke W, Wang D, Peng G, Xiao S. 2019. Susceptibility of porcine IPI-2I intestinal epithelial cells to infection with swine enteric coronaviruses. Vet Microbiol 233:21–27. doi: 10.1016/j.vetmic.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 6.Derricott H, Luu L, Fong WY, Hartley CS, Johnston LJ, Armstrong SD, Randle N, Duckworth CA, Campbell BJ, Wastling JM, Coombes JL. 2019. Developing a 3D intestinal epithelium model for livestock species. Cell Tissue Res 375:409–424. doi: 10.1007/s00441-018-2924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena K, Blutt SE, Ettayebi K, Zeng XL, Broughman JR, Crawford SE, Karandikar UC, Sastri NP, Conner ME, Opekun AR, Graham DY, Qureshi W, Sherman V, Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Donowitz M, Estes MK. 2016. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J Virol 90:43–56. doi: 10.1128/JVI.01930-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Co JY, Margalef-Catala M, Li X, Mah AT, Kuo CJ, Monack DM, Amieva MR. 2019. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep 26:2509.e4–2520.e4. doi: 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Fu F, Guo S, Wang H, He X, Xue M, Yin L, Feng L, Liu P. 2018. Porcine intestinal enteroids: a new model for studying enteric coronavirus porcine epidemic diarrhea virus infection and the host innate response. J Virol 93:e01682-18. doi: 10.1128/JVI.01682-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Liu Y, Ji C-M, Yang Y-L, Liang Q-Z, Zhao P, Xu L-D, Lei X-M, Luo W-T, Qin P, Zhou J, Huang Y-W. 2018. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J Virol 92:e00318-18. doi: 10.1128/JVI.00318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forbester J, Goulding D, Vallier L, Hannan N, Hale C, Pickard D, Mukhopadhyay S, Dougan G. 2015. Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun 83:2926–2934. doi: 10.1128/IAI.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. 2016. Organoid models of human gastrointestinal development and disease. Gastroenterology 150:1098–1112. doi: 10.1053/j.gastro.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin K, Fogg V, Margolis B. 2006. Tight junctions and cell polarity. Annu Rev Cell Dev Biol 22:207–235. doi: 10.1146/annurev.cellbio.22.010305.104219. [DOI] [PubMed] [Google Scholar]
- 14.Schlüter M, Margolis B. 2009. Apical lumen formation in renal epithelia. J Am Soc Nephrol 20:1444–1452. doi: 10.1681/ASN.2008090949. [DOI] [PubMed] [Google Scholar]
- 15.Hamon M, Bierne H, Cossart P. 2006. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol 4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 16.Williamson IA, Arnold JW, Samsa LA, Gaynor L, DiSalvo M, Cocchiaro JL, Carroll I, Azcarate-Peril MA, Rawls JF, Allbritton NL, Magness ST. 2018. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell Mol Gastroenterol Hepatol 6:301–319. doi: 10.1016/j.jcmgh.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Hee B, Loonen LMP, Taverne N, Taverne-Thiele JJ, Smidt H, Wells JM. 2018. Optimized procedures for generating an enhanced, near physiological 2D culture system from porcine intestinal organoids. Stem Cell Res 28:165–171. doi: 10.1016/j.scr.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, Chen J, Yao G, Guo Q, Wang J, Liu G. 2019. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl Microbiol Biotechnol 103:4943–4952. doi: 10.1007/s00253-019-09835-7. [DOI] [PMC free article] [PubMed] [Google Scholar]