In work with pathogenic viruses, it is useful to have rapid quantitative tests for viral infectivity that can be performed without strict biocontainment restrictions. A common way of accomplishing this is to generate viral pseudoparticles that contain the surface glycoprotein from the pathogenic virus incorporated into a replication-defective viral particle that contains a sensitive reporter system. These pseudoparticles enter cells using the glycoprotein from the pathogenic virus, leading to a readout for infection. Conditions that block entry of the pathogenic virus, such as neutralizing antibodies, will also block entry of the viral pseudoparticles. However, viral glycoproteins often are not readily suited for generating pseudoparticles. Here, we describe a series of modifications that result in the production of relatively high-titer SARS-COV-2 pseudoparticles that are suitable for the detection of neutralizing antibodies from COVID-19 patients.
KEYWORDS: SARS-COV-2, COVID-19, pseudotypes, glycoprotein, lentiviral vector, neutralization assay
ABSTRACT
The severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) Spike glycoprotein is solely responsible for binding to the host cell receptor and facilitating fusion between the viral and host membranes. The ability to generate viral particles pseudotyped with SARS-COV-2 Spike is useful for many types of studies, such as characterization of neutralizing antibodies or development of fusion-inhibiting small molecules. Here, we characterized the use of a codon-optimized SARS-COV-2 Spike glycoprotein for the generation of pseudotyped HIV-1, murine leukemia virus (MLV), and vesicular stomatitis virus (VSV) particles. The full-length Spike protein functioned inefficiently with all three systems but was enhanced over 10-fold by deleting the last 19 amino acids of the cytoplasmic tail. Infection of 293FT target cells was possible only if the cells were engineered to stably express the human angiotensin-converting enzyme 2 (ACE2) receptor, but stably introducing an additional copy of this receptor did not further enhance susceptibility. Stable introduction of the Spike-activating protease TMPRSS2 further enhanced susceptibility to infection by 5- to 10-fold. Replacement of the signal peptide of the Spike protein with an optimal signal peptide did not enhance or reduce infectious particle production. However, modifications D614G and R682Q further enhanced infectious particle production. With all enhancing elements combined, the titer of pseudotyped HIV-1 particles reached almost 106 infectious particles/ml. Finally, HIV-1 particles pseudotyped with SARS-COV-2 Spike were successfully used to detect neutralizing antibodies in plasma from coronavirus disease 2019 (COVID-19) patients, but not in plasma from uninfected individuals.
IMPORTANCE In work with pathogenic viruses, it is useful to have rapid quantitative tests for viral infectivity that can be performed without strict biocontainment restrictions. A common way of accomplishing this is to generate viral pseudoparticles that contain the surface glycoprotein from the pathogenic virus incorporated into a replication-defective viral particle that contains a sensitive reporter system. These pseudoparticles enter cells using the glycoprotein from the pathogenic virus, leading to a readout for infection. Conditions that block entry of the pathogenic virus, such as neutralizing antibodies, will also block entry of the viral pseudoparticles. However, viral glycoproteins often are not readily suited for generating pseudoparticles. Here, we describe a series of modifications that result in the production of relatively high-titer SARS-COV-2 pseudoparticles that are suitable for the detection of neutralizing antibodies from COVID-19 patients.
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic is a severe threat to human health and the global economy. COVID-19 is caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-COV-2), which is a highly pathogenic betacoronavirus (1, 2). A critical tool for the study of pathogenic viruses such as SARS-COV-2 is a rapid and sensitive assay for agents that block viral entry, such as neutralizing antibodies or small-molecule inhibitors. A safe and powerful technique for generating such an assay is to generate viral pseudotyped particles where the surface fusion protein of the pathogen of interest is assembled onto the surface of a replication-defective virus that contains a sensitive reporter protein.
The Spike glycoprotein from coronaviruses facilitates binding to the host cell receptor and fusion between viral and cellular membranes. The SARS-COV-2 Spike protein is a large, 1,274-amino-acid protein that contains an N-terminal S1 receptor binding domain and a C-terminal S2 fusion domain. Fusion by SARS-COV-2 Spike requires binding to the host receptor angiotensin-converting enzyme 2 (ACE2) (3–6) as well as proteolytic cleavage at the S1/S2 and S2′ positions by host cysteine proteases cathepsin B and L (CatB/L) or serine protease TMPRSS2 (3, 6). Depending on the cell line, inhibition of one or both of these proteases is sufficient to block viral entry. An interesting difference between the SARS and SARS-COV-2 Spike proteins is the presence of a furin cleavage site near the S1/S2 cleavage site (6, 7). This cleavage site was found to be essential for infection of human lung cells (3). Surprisingly, passaging of SARS-COV-2 in Vero E6 cells selects for modifications that alter the furin cleavage site, resulting in viruses that produce larger plaque sizes (8). Similar modifications in the furin cleavage site have also been reported upon passaging of vesicular stomatitis virus (VSV) chimeras containing the SARS-COV-2 Spike (9, 10), as well as in human patients (11). Another SARS-COV-2 Spike modification that has become very prominent in circulating strains and has been reported to enhance viral titers is the modification D614G (12–14). Whether these modify viral transmission or the pathogenicity of SARS-COV-2 is currently still debated.
There have been several reports on generating SARS (15–18) and SARS-COV-2 (3–5, 10, 13, 19, 20) viral pseudotypes with glycoprotein-defective murine leukemia virus (MLV), HIV, and VSV particles. Although pseudoparticles could be generated with full-length SARS Spike, pseudotyping efficiency was shown to be enhanced by about 100-fold by deleting the last 19 amino acids of the cytoplasmic tail (15), which removed a presumptive endoplasmic reticulum (ER) retention sequence. Modification of the presumptive ER retention site in the cytoplasmic tail of SARS-COV-2 (K1269A H1271A) was not found to enhance pseudotyping efficiency (20). Here, we develop optimized genetic conditions for generating SARS-COV-2 Spike pseudotyped particles.
RESULTS
Truncation of SARS-COV-2 Spike enhances viral pseudotyping.
A codon-optimized, full-length Spike gene was synthesized and introduced in place of the green fluorescent protein (GFP) gene in plasmid pEGFP-N1. Because truncation of the cytoplasmic tail of SARS Spike was shown to enhance production of viral pseudotypes (15), we also generated a SARS-COV-2 Spike subclone with the last 19 amino acids deleted (Fig. 1). Pseudotyped particles were generated with the two Spike proteins as well as VSV-G with glycoprotein-defective MLV, HIV-1, and VSV particles (Fig. 1). To avoid potential false-positive signals, a reporter system was utilized where the Cre recombinase gene is expressed by HIV-1 and MLV particles. When these particles transduce the Cre reporter cell line, the protein causes a recombination in an engineered reporter element that results in GFP expression. Because the virus-producing cells do not express GFP, they are eliminated as a source of false-positive signals. This 293FT-based reporter cell line was further engineered to express human ACE2. Infectious particle production with all three types of particles was very low with the full-length Spike protein, but the Δ19 Spike produced viral titers approaching 104 infectious particles/ml with both HIV-1 and VSV particles. The Δ19 truncation also enhanced infectivity with MLV particles, although the viral titer remained near background. Infectious particle production remained over 1,000-fold lower than that with the control glycoprotein, VSV-G.
FIG 1.
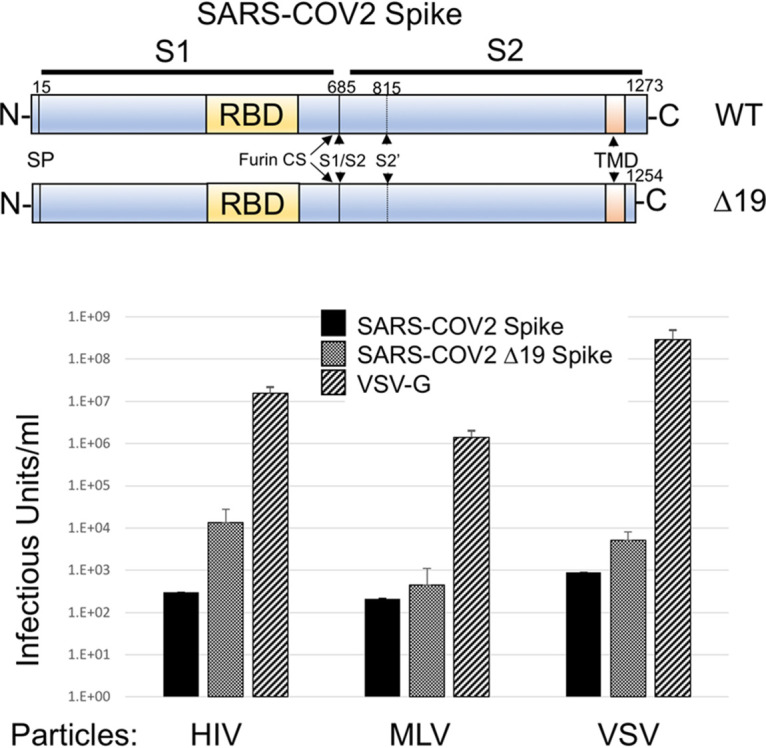
SARS-COV-2 pseudotyped particles. (Top) Schematic of SARS-COV-2 Spike protein. RBD, receptor binding domain; SP, signal peptide; TMD, transmembrane domain; furin CS, furin cleavage site; WT, wild type. (Bottom) Infectious particle production by glycoprotein-defective HIV-1, MLV, and VSV pseudoparticles with SARS-COV-2 Spike, SARS-COV-2 Δ19 Spike, or VSV-G. HIV-1 and MLV particles contained a Cre reporter and were scored on 293FT/ACE2 cells containing a Cre-inducible GFP reporter. VSV-G particles directly contained GFP (VSVΔG-GFP). Data are averages and standard errors for three independent experiments.
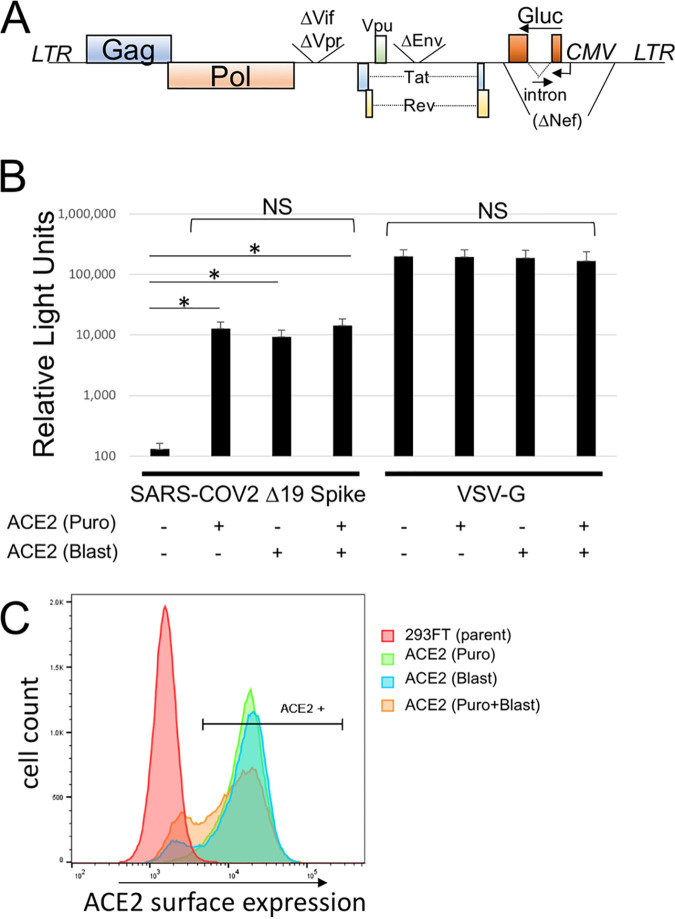
To allow rapid quantitation with minimum background, we proceeded with an Env-defective HIV-1 provirus containing a Gaussia luciferase (Gluc) gene in the reverse orientation containing a forward intron (Fig. 2A) (21). This viral construct produces very low background because it is not capable of producing a luciferase signal unless the gene is reverse transcribed in the target cell. Infectivity with SARS-COV-2 Spike requires expression of the host receptor ACE2. Our starting cell line was generated using a retroviral transfer vector containing ACE2 and a puromycin selection cassette. To determine if introduction of an additional stable copy of ACE2 would enhance susceptibility to infection, we generated a second retroviral transfer vector with a blasticidin resistance cassette. Retroviral particles were generated with this vector and used to stably transduce 293FT or 293FT/ACE2 (Puro) cells. Each of the cell lines was transduced with HIV-1-Gluc particles pseudotyped with SARS-COV-2 Δ19 Spike (Fig. 2B). As expected, 293FT cells were not susceptible to infection with the HIV-1/SARS-COV-2 Δ19 Spike. However, both cell lines containing a single introduction of ACE2, and the cell line containing two introductions, had approximately 1,000-fold increases in the luciferase signal, but the surface ACE2 expression and viral susceptibility were roughly equivalent among the three cell lines (Fig. 2B and C).
FIG 2.
Introduction of ACE2 is required for infection of 293FT cells with SARS-COV-2 Spike. (A) Schematic of the HIV-1 Gaussia luciferase vector. (B) Transduction of 293FT cells transduced with different numbers of ACE2 genes with HIV-1-Gluc pseudoparticles pseudotyped with SARS-COV-2 Δ19 Spike or VSV-G. Transductions with VSV-G used 100 times less viral supernatant. Puro, vector with puromycin selection cassette; Blast, vector with blasticidin resistance cassette. Data are averages and standard errors for three independent experiments. Asterisks indicate significant differences (P < 0.05) by a paired Student t test; NS, no significant difference. (C) ACE2 surface expression of ACE2-expressing cell lines. Shown is an example representative of the results of three experiments.
TMPRSS2 expression enhances susceptibility of cells to SARS-COV-2 pseudotypes.
The SARS-COV-2 Spike requires proteolytic priming during infection by either cysteine proteases CatB/L or serine protease TMPRSS2, produced in the target cell (3, 6). To determine whether introduction of TMPRSS2 would enhance susceptibility to infection, we synthesized a codon-optimized TMPRSS2 gene, introduced this gene into a retroviral transfer vector, and stably transduced 293FT or 293FT/ACE2 cells. The stable introduction of TMPRSS2 into 293FT cells did not impart sensitivity to transduction with HIV-1 particles pseudotyped with SARS-COV-2 Δ19 Spike (Fig. 3). However, stable introduction of TMPRSS2 into 293FT/ACE2 cells increased the Gluc signal from target cells by 5- to 10-fold. Neither ACE2 expression nor TMPRSS2 expression affected the Gluc signal from particles pseudotyped with VSV-G.
FIG 3.
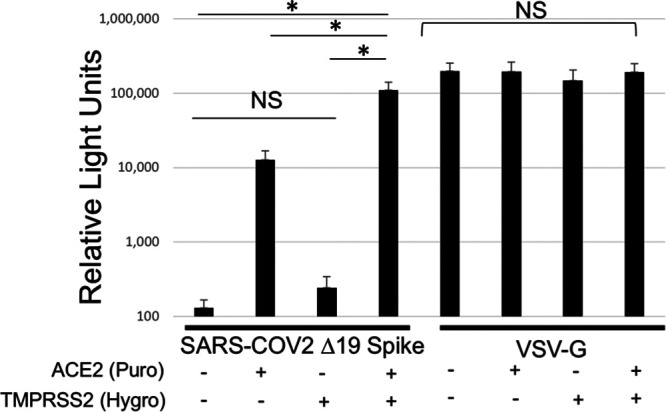
TMPRSS2 expression enhances susceptibility to infection with SARS-COV-2 Spike. 293FT cells stably expressing ACE2, TMPRSS2, or both were transduced with HIV-1-Gluc particles pseudotyped with SARS-COV-2 Δ19 Spike or VSV-G. Transductions with VSV-G used 100 times less viral supernatant. Data are averages and standard errors for three independent experiments. Asterisks indicate significant differences (P < 0.05) by a paired Student t test; NS, no significant difference.
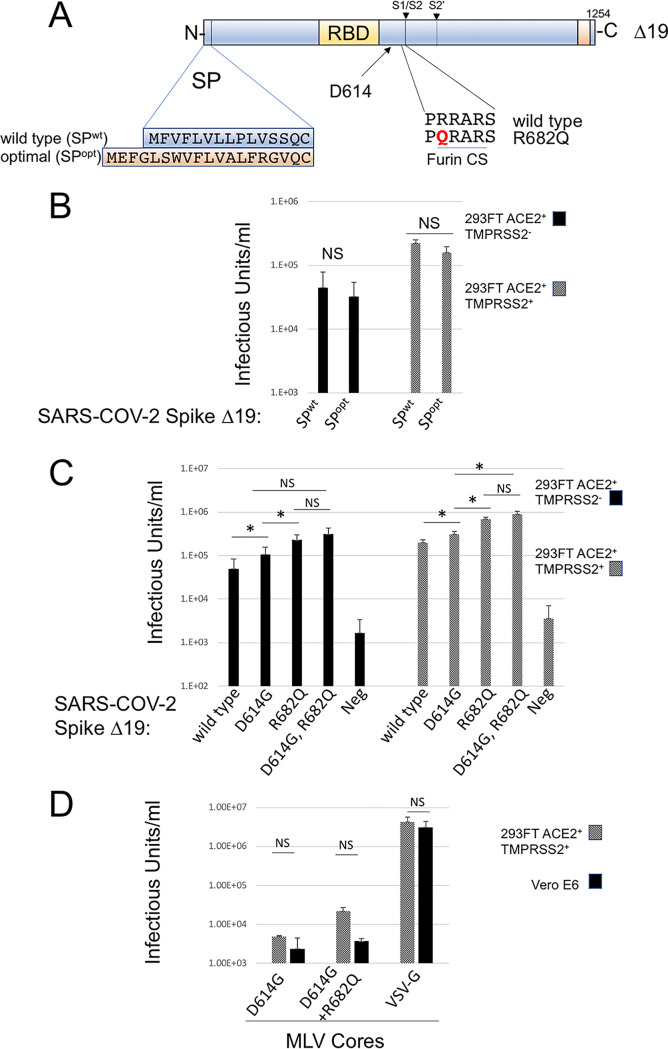
The D614G and R682Q modifications in SARS-COV-2 Δ19 Spike enhance pseudotyping efficiency.
To explore additional genetic modifications in SARS-COV-2 that might enhance infectious particle production, we replaced the endogenous signal peptide with a well-characterized strong signal peptide (22), and separately, we introduced the modifications D614G and R682Q, alone or in combination, into SARS-COV-2 Δ19 Spike. Both of these modifications have been reported previously to increase SARS-COV-2 Spike efficiency (Fig. 4A) (8, 12–14). Infectious particle production was assessed using the Cre reporter system so that a viral titer could be determined, and transductions were performed in cell lines expressing ACE2 with or without TMPRSS2. Replacement of the signal peptide did not increase or decrease infectious particle production (Fig. 4B). However, the D614G and R682Q modifications both significantly enhanced infectious particle production under all conditions. The D614G modification enhanced infectious particle production by 2- or 1.5-fold in cells lacking or containing TMPRSS2, respectively. The R682Q modification enhanced infectious particle production by 5- or 3.5-fold in cells lacking or containing TMPRSS2, respectively. The D614G R682G double modification consistently produced the highest titer of infectious particles, although the difference between D614G R682G and R682G alone was not statistically significant. The highest average titer of 9 × 105 was achieved with the D614G R682G Δ19 Spike in cells expressing both ACE2 and TMPRSS2. To compare this engineered cell to a cell line that is naturally susceptible to SARS-COV-2 infection, we did parallel infections of 293FT (ACE2+ TMPRSS2+) cells and Vero E6 cells. Because Vero E6 cells are resistant to HIV-1 infection, pseudotypes with MLV particles containing a GFP reporter were used. Infectivity in 293FT cells (ACE2+ TMPRSS2+) was consistently higher than that in VeroE6 cells, although the difference was not statistically significant.
FIG 4.
D614G and R682Q modifications enhance SARS-COV-2 Δ19 pseudotyping. (A) Schematic illustrating introduction of the optimal signal peptide (SPopt), D614G, and R682Q into SARS-COV-2 Δ19 Spike. SPwt, wild-type signal peptide. (B and C) HIV-CMV-Cre was pseudotyped with SARS-COV-2 Δ19 Spike with SPwt or SPopt (B) or with the indicated modifications (C). Transductions were performed on 293FT/ACE2 and 293FT/ACE2/TMPRSS2 cells, both containing a Cre-inducible GFP reporter. (D) MLV-GFP particles were pseudotyped with VSV-G or the indicated SARS-COV-2 Δ19 modifications and were used to transduce 293FT/ACE2/TMPRSS2 or Vero E6 cells. Data are averages and standard errors for three independent experiments. Asterisks indicate significant differences (P < 0.05) by a paired Student t test; NS, no significant difference.
SARS-COV-2 Spike pseudotypes function in neutralization assays.
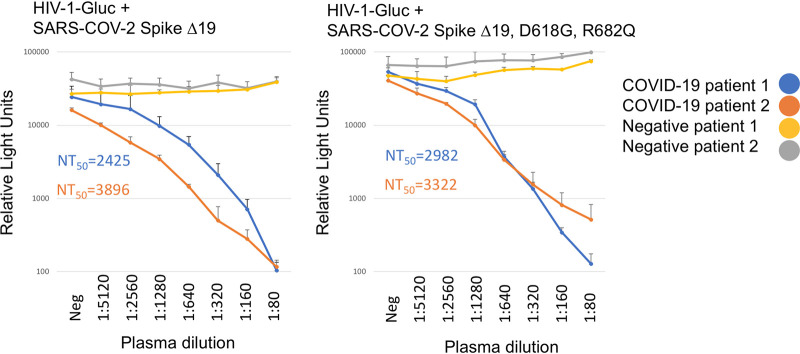
To test if the pseudoparticles produced were suitable for use in a neutralization assay, we incubated HIV-1 particles pseudotyped with SARS-COV-2 Δ19 Spike, with or without the D614G R682Q modifications, with serial dilutions of control plasma and plasma from COVID-19 patients before adding 293FT/ACE2/TMPRSS2 cells. Robust and quantifiable neutralization was detected in plasma from COVID-19 patients, but not in control plasma (Fig. 5). The D614G R682Q modification did change the overall sensitivity to neutralizing antibodies.
FIG 5.
Particles pseudotyped with SARS-COV-2 Spike function in neutralization assays. HIV-1-Gluc particles were pseudotyped with SARS-COV-2 Δ19 Spike (left) or SARS-COV-2 D614G R682Q Δ19 Spike (right). Plasma was obtained from two patients with confirmed COVID-19 infection or from infection-negative patients. Twofold serially diluted plasma was incubated with pseudotypes for 1 h at 37°C in triplicate. The mixtures were subsequently incubated with 293/ACE2/TMPRSS2 cells, and Gluc measurements were taken 3 days postransduction. Shown are the average and standard deviation for each measure. Calculated 50% neutralizing titers (NT50) are indicated.
DISCUSSION
Here, we outline glycoprotein and target cell modifications that enhance pseudotyping efficiency with the SARS-COV-2 Spike glycoprotein. The first and most important changes involve the necessity to express the human ACE2 receptor, which has been defined previously by numerous investigators (3–6), and the enhancement gained from truncation of the Spike cytoplasmic tail. Truncation of the last 19 amino acids of SARS-COV Spike was also shown to significantly enhance pseudotyping with that glycoprotein (15). In that study, the original reason for making the truncation was to eliminate potential ER retention sequences in the cytoplasmic tail. Despite enhancing pseudotyping capacity, the truncation did not enhance surface expression. It is likely that the enhancement in pseudotyping efficiency upon deletion of the cytoplasmic sequence is at least in part due to eliminating steric interference of the cytoplasmic tail with the viral capsid. Such interference has been noted in the pseudotyping of other viral glycoproteins with large cytoplasmic tails, such as HIV-1 Env (23, 24).
The next modification that enhanced efficiency was the introduction of the human protease TMPRSS2. The importance of this protease for SARS-COV-2 Spike glycoprotein entry has been noted previously, so this enhancement was not surprising (3, 6).
The final modification that enhanced SARS-COV-2 Δ19 Spike pseudotyping was the introduction of the D614G and R682Q modifications, the latter of which is predicted to eliminate the furin cleavage site. D614G is a naturally occurring modification in SARS-COV-2 Spike that has become prevalent among circulating strains (12). The R682Q modification was naturally selected for upon culturing SARS-COV-2 in Vero cells, and viruses containing the modification were reported to have larger plaque sizes (8). In addition, modifications that eliminate the furin cleavage site have also been reported upon passaging VSV chimeras containing the SARS-COV-2 Spike (9, 10), as well as in human patients (11). This observation is slightly at odds with a recent publication by Hoffmann et al. that found that the furin cleavage site was essential for infecting the lung cell line Calu-3 but not for infecting Vero cells (3). The Calu-3 cell line is reported to express TMPRSS2 but to lack sufficient CatB/L to promote glycoprotein priming (3, 25); Vero cells are reported to be TMPRSS2 negative but CatB/L positive. Thus, the furin cleavage site appeared to be required only when the virus was dependent on TMPRSS2 for entry. A possible explanation is that furin cleavage enhances TMPRSS2-mediated entry but suppresses CatB/L-mediated entry. Consistent with this, the enhancement we observed from the R682Q modification was less pronounced in cells that express TMPRSS2. It should be noted that the modification described by Hoffman et al. replaced the furin cleavage site from SARS-COV-2 with the equivalent sequence from SARS-COV, which effectively causes a 4-amino-acid deletion. Thus, the loss of infectivity observed by Hoffmann et al. could in part have been the result of secondary effects of the deletion that are independent of furin cleavage.
Finally, we demonstrate that HIV-1-Gluc particles pseudotyped with SARS-COV-2 Δ19 Spike could be used for detecting neutralizing antibodies from COVID-19 patient plasma. Inclusion of the D614G R682Q modifications in Spike did not obviously change the neutralization results in this assay. Thus, this appears to be a suitable system for studying inhibitors of SARS-COV-2 entry.
MATERIALS AND METHODS
Plasmids.
The gammaretroviral transfer plasmids pQCXIP (puromycin resistance) and pQCXIH (hygromycin resistance) were obtained from Clontech. The blasticidin gammaretroviral transfer vector (pQCXIB) was generated by replacing the puromycin resistance cassette from pQCXIP with a blasticidin resistance cassette. The gammaretroviral vector for generating the Cre sensor cell line (MLV lox-mTomato-lox-GFP-Blast) was engineered into pQCXIB and contained the monomeric Tomato gene flanked by Lox sequences (ATAACTTCGTATAGCATACATTATACGAAGTTAT), which were followed immediately by enhanced GFP (eGFP). The ACE2 transfer vectors were generated by engineering the human ACE2 gene (NCBI reference sequence NM_021804.3) into pQCXIP and pQCXIH. The TMPRSS2 transfer vector was generated by synthesizing a codon-optimized version of the human TMPRSS2 gene (NCBI reference sequence NP_001128571.1) and engineering it into the pQCXIH vector. The HIV-1-CMV-Cre vector was an NL4-3-derived provirus that is defective in Vif, Vpr, and Env and has the Nef gene replaced with cytomegalovirus (CMV) Cre. The plasmid was generated by replacing the GFP gene from the previously described plasmid HIV-CMV-GFP+Vpu (26) with the Cre gene from plasmid MLV-Cre (27), provided by Alan Rein (NCI at Frederick). The MLV+ CMV Cre vector was generated by replacing the GFP gene from MLV-GFP (provided by Shan-Lu Liu, The Ohio State University) with the Cre gene from MLV-Cre (27), provided by Alan Rein (NCI at Frederick). The MLV GagPol expression construct was provided by Walther Mothes (Yale University). The HIV-1-Gluc vector was described previously (28). The VSV-G expression construct was obtained from the NIH AIDS Reagent Program (29). The SARS-COV-2 expression vector was generated by synthesizing a codon-optimized version of the SARS-COV-2 Spike (GenBank accession number MN985325.1). The Δ19 Spike and the Δ19 R682Q Spike were generated by PCR mutagenesis. The Spike subclone with the alternative signal peptide contained the previously described H7 peptide (22) and was generated by synthesizing the 5′ end of the Spike gene with the alternative signal peptide and introducing it into the SARS-COV-2 Δ19 Spike clone.
Virus production and infectivity assays.
All transfections were performed in 6-well plates. 293FT cells were transfected with a total of 1 μg of plasmid and 4 μg of polyethyleneimine (PEI) (30). For HIV-1-CMV-Cre particles, cells were transfected with 900 ng of provirus and 100 ng of glycoprotein expression vector (Fig. 1) or with 800 ng of provirus and 200 ng of glycoprotein expression vector (subsequent figures). For HIV-1-Gluc particles, cells were transfected with 800 ng of provirus and 200 ng of glycoprotein expression vector. For MLV particles, cells were transfected with 500 ng of the MLV GagPol expression vector, 400 ng of MLV-CMV-Cre, and 100 ng of glycoprotein expression vector. For VSV particles, cells were transfected with 1 μg of glycoprotein expression vector and were infected 2 days posttransfection with >107 infectious units/well of VSVΔG-GFP (Kerafast) (31). Cells were rinsed with phosphate-buffered saline (PBS) 1 h after infection, and the medium was replaced with complete medium supplemented with 2 μl of mouse hybridoma supernatant containing anti-VSV-G antibody I1 (Kerafast) to neutralize input virus. Neutralizing antibody was excluded from samples pseudotyped with VSV-G. VSV pseudoparticles were collected 24 h later.
Supernatants containing virus were frozen at –80°C for at least 4 h, thawed, and spun at 3,200 × g for 5 min, and the same volume of medium was added to target cells with 20 μg of hexadimethrine bromide per ml (H9268; Sigma). For assays with a fluorescent readout, infected cells were collected at about 2 to 3 days postinfection, fixed with 4% paraformaldehyde (PFA), washed with PBS, and analyzed on an Accuri C6 flow cytometer. For infections with a Gluc readout, transductions were allowed to proceed for 2 to 3 days, and 20 μl of supernatant from each well was transferred to a black 96-well plate for measuring Gaussia luciferase activity with 50 μl of 10 μm coelenterazine in 0.1 M Tris (pH 7.4) and 0.3 M sodium ascorbate (NanoLight Technology). Luminescence, representing infectivity, was measured from the supernatant using a PerkinElmer EnSpire 2300 multilabel reader.
Cell culture.
The 293FT cell line was obtained from Invitrogen. All cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 10 mM nonessential amino acids, and 1% minimal essential medium (MEM) vitamins.
Surface labeling.
Cells were washed with PBS and treated with 10 mM EDTA. Cells were then collected with PBS, centrifuged at 300 × g for 5 min, and supernatant removed. Cells were blocked with 5% chicken serum in PBS for 30 min on ice. Cells were centrifuged at 300 × g for 5 min and supernatant removed. A goat anti-hACE2 ectodomain antibody (R&D Systems) was applied to cells at 1:100 in 1% chicken serum in PBS for 1 h. Cells were then washed 3 times with PBS, suspended in an Alexa Fluor 488-conjugated chicken anti-goat antibody solution (Thermo Fisher), and diluted 1:500 in 400 μl 1% chicken serum in PBS for 1 h. Cells were then washed 3 times with PBS and suspended in 100 μl of 4% PFA. After incubation for 20 min on ice, cells were centrifuged at 300 × g for 5 min, and the supernatant was removed. Cells were resuspended in 200 μl of PBS and analyzed via flow cytometry (Accuri).
Cell line generation.
The Cre sensor cell line was generated by transfecting 293FT cells with 500 ng MLV GagPol expression vector, 400 ng of retroviral transfer vector MLV lox-mTomato-lox-GFP-Blast, and 100 ng of VSV-G expression vector. Viral medium was used to transduce 293FT cells, and cells were selected with blasticidin (5 μg/ml) beginning 2 days postransduction and were maintained until control treated cells were all eliminated. A clonal isolate from these cells was selected that expressed mTomato but no GFP. The ACE2 cell lines were generated by transfecting 293FT cells with 500 ng MLV GagPol expression vector, 400 ng of retroviral transfer vector pQCXIP-ACE2 or pQCXIH-ACE2, and 100 ng of VSV-G expression vector. Viral medium was used to transduce 293FT cells or the 293FT sensor cell line, and cells were selected with puromycin (1 μg/ml) or hygromycin (200 μg/ml) beginning 2 days postransduction and were maintained until control treated cells were all eliminated. The TMPRSS2 cell line was generated by transfecting 293FT cells with 500 ng MLV GagPol expression vector, 400 ng of retroviral transfer vector pQCXIH-TMPRSS2, and 100 ng of VSV-G expression vector. Viral medium was used to transduce 293FT, 293FT/ACE2, or 293/Cre-sensor/ACE2 cells, which were selected with hygromycin (200 μg/ml) beginning 2 days postransduction and were maintained until control treated cells were all eliminated.
Neutralization assay.
Deidentified patient plasma samples were obtained from the MU Health Care clinical laboratory. Plasma was heat inactivated for 30 min at 58°C. Twofold serially diluted plasma was incubated with HIV-1/SARS-COV-2 Spike pseudotypes for 1 h at 37°C. The mixture was subsequently incubated with 293/ACE2/TMPRSS2 cells, approximately 20,000 cells per well in a 96-well plate. Gluc measurements were taken 3 days postransduction. Neutralization assays were performed in triplicate, and titers were calculated using regression analyses to correspond to 50% inhibition.
ACKNOWLEDGMENT
Funding for this work was provided by NIAID through grant 143363 to M.C.J.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogando NS, Dalebout TJ, Zevenhoven-Dobbe JC, Limpens RWAL, van der Meer Y, Caly L, Druce J, de Vries JJC, Kikkert M, Bárcena M, Sidorov I, Snijder EJ. 2020. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, Wei Y, Atwal GS, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. 2020. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt F, Weisblum Y, Muecksch F, Hoffmann H-H, Michailidis E, Lorenzi JCC, Mendoza P, Rutkowska M, Bednarski E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Caskey M, Robbiani DF, Nussenzweig MC, Rice CM, Hatziioannou T, Bieniasz PD. 2020. Measuring-SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med 217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JS, Jang JH, Kim JM, Chung YS, Yoo CK, Han MG. 2020. Genome-wide identification and characterization of point mutations in the SARS-CoV-2 genome. Osong Public Health Res Perspect 11:101–111. doi: 10.24171/j.phrp.2020.11.3.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, Keeley AJ, Lindsey BB, Parsons PJ, Raza M, Rowland-Jones S, Smith N, Tucker RM, Wang D, Wyles MD. 2020. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, Zhao C, Zhang Q, Liu H, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Zhang L, Li X, Huang W, Wang Y. 2020. The impact of mutations in SARS-CoV-2 Spike on viral infectivity and antigenicity. Cell doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maitra A, Sarkar MC, Raheja H, Biswas NK, Chakraborti S, Singh AK, Ghosh S, Sarkar S, Patra S, Mondal RK, Ghosh T, Chatterjee A, Banu H, Majumdar A, Chinnaswamy S, Srinivasan N, Dutta S, Das S. 2020. Mutations in SARS-CoV-2 viral RNA identified in Eastern India: possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. J Biosci 45:76. doi: 10.1007/s12038-020-00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giroglou T, Cinatl J Jr, Rabenau H, Drosten C, Schwalbe H, Doerr HW, von Laer D. 2004. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J Virol 78:9007–9015. doi: 10.1128/JVI.78.17.9007-9015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han DP, Kim HG, Kim YB, Poon LL, Cho MW. 2004. Development of a safe neutralization assay for SARS-CoV and characterization of S-glycoprotein. Virology 326:140–149. doi: 10.1016/j.virol.2004.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci U S A 101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang ZY, Huang Y, Ganesh L, Leung K, Kong WP, Schwartz O, Subbarao K, Nabel GJ. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol 78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, Wei P, Ge J, Gou M, Li X, Sun L, Cao T, Wang P, Zhou C, Zhang R, Liang P, Guo H, Wang X, Qin CF, Chen F, Dong C. 2020. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD. 2020. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 Spike protein for neutralization assays. Viruses 12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janaka SK, Gregory DA, Johnson MC. 2013. Retrovirus glycoprotein functionality requires proper alignment of the ectodomain and the membrane proximal cytoplasmic tail. J Virol 87:12805–12813. doi: 10.1128/JVI.01847-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haryadi R, Ho S, Kok YJ, Pu HX, Zheng L, Pereira NA, Li B, Bi X, Goh LT, Yang Y, Song Z. 2015. Optimization of heavy chain and light chain signal peptides for high level expression of therapeutic antibodies in CHO cells. PLoS One 10:e0116878. doi: 10.1371/journal.pone.0116878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mammano F, Salvatori F, Indraccolo S, De Rossi A, Chieco-Bianchi L, Göttlinger HG. 1997. Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J Virol 71:3341–3345. doi: 10.1128/JVI.71.4.3341-3345.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnierle BS, Stitz J, Bosch V, Nocken F, Merget-Millitzer H, Engelstadter M, Kurth R, Groner B, Cichutek K. 1997. Pseudotyping of murine leukemia virus with the envelope glycoproteins of HIV generates a retroviral vector with specificity of infection for CD4-expressing cells. Proc Natl Acad Sci U S A 94:8640–8645. doi: 10.1073/pnas.94.16.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JE, Li K, Barlan A, Fehr AR, Perlman S, McCray PB Jr, Gallagher T. 2016. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A 113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucas TM, Lyddon TD, Cannon PM, Johnson MC. 2010. Pseudotyping incompatibility between HIV-1 and gibbon ape leukemia virus Env is modulated by Vpu. J Virol 84:2666–2674. doi: 10.1128/JVI.01562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahi YS, Zhang S, Thappeta Y, Denman A, Feizpour A, Gummuluru S, Reinhard B, Muriaux D, Fivash MJ, Rein A. 2016. Functional interplay between murine leukemia virus glycogag, Serinc5, and surface glycoprotein governs virus entry, with opposite effects on gammaretroviral and ebolavirus glycoproteins. mBio 7:e01985-16. doi: 10.1128/mBio.01985-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janaka SK, Lucas TM, Johnson MC. 2011. Sequences in gibbon ape leukemia virus envelope that confer sensitivity to HIV-1 accessory protein Vpu. J Virol 85:11945–11954. doi: 10.1128/JVI.05171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang LJ, Urlacher V, Iwakuma T, Cui Y, Zucali J. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther 6:715–728. doi: 10.1038/sj.gt.3300895. [DOI] [PubMed] [Google Scholar]
- 30.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A 92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitt MA. 2010. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods 169:365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]