Baculoviruses are nonpathogenic viruses that infect mammals, which, among other applications, are used as vehicles for gene delivery. Here, we demonstrated that the cytosolic DNA sensor cGAS recognizes baculoviral DNA and that the cGAS-STING axis is primarily responsible for the attenuation of transduction in human and mouse cell lines through type I and type III IFNs. Furthermore, we identified DNA-dependent protein kinase (DNA-PK) as a cGAS-independent and alternative DNA cytosolic sensor that contributes less to the antiviral state in baculovirus infection in human epithelial cells than cGAS. Knowledge of the pathways involved in the response of mammalian cells to baculovirus infection will improve the use of this vector as a tool for gene therapy.
KEYWORDS: STING, baculovirus, cGAS, gene delivery, interferons
ABSTRACT
The baculovirus Autographa californica multiple nucleopolyhedrovirus is an insect virus with a circular double-stranded DNA genome, which, among other multiple biotechnological applications, is used as an expression vector for gene delivery in mammalian cells. Nevertheless, the nonspecific immune response triggered by viral vectors often suppresses transgene expression. To understand the mechanisms involved in that response, in the present study, we studied the cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) pathway by using two approaches: the genetic edition through CRISPR/Cas9 technology of genes encoding STING or cGAS in NIH/3T3 murine fibroblasts and the infection of HEK293 and HEK293 T human epithelial cells, deficient in cGAS and in cGAS and STING expression, respectively. Overall, our results suggest the existence of two different pathways involved in the establishment of the antiviral response, both dependent on STING expression. Particularly, the cGAS-STING pathway resulted in the more relevant production of beta interferon (IFN-β) and IFN-λ1 in response to baculovirus infection. In human epithelial cells, IFN-λ1 production was also induced in a cGAS-independent and DNA-protein kinase (DNA-PK)-dependent manner. Finally, we demonstrated that these cellular responses toward baculovirus infection affect the efficiency of transduction of baculovirus vectors.
IMPORTANCE Baculoviruses are nonpathogenic viruses that infect mammals, which, among other applications, are used as vehicles for gene delivery. Here, we demonstrated that the cytosolic DNA sensor cGAS recognizes baculoviral DNA and that the cGAS-STING axis is primarily responsible for the attenuation of transduction in human and mouse cell lines through type I and type III IFNs. Furthermore, we identified DNA-dependent protein kinase (DNA-PK) as a cGAS-independent and alternative DNA cytosolic sensor that contributes less to the antiviral state in baculovirus infection in human epithelial cells than cGAS. Knowledge of the pathways involved in the response of mammalian cells to baculovirus infection will improve the use of this vector as a tool for gene therapy.
INTRODUCTION
The baculovirus (BV) Autographa californica multiple nucleopolyhedrovirus (AcMNPV), which infects arthropods, mainly from the order Lepidoptera, has a circular double-stranded DNA (dsDNA) genome of 150 kb which encodes more than 150 viral open reading frames (ORFs) (1). One of the main characteristics of this virus is that it comprises two phenotypes throughout its viral cycle: the occlusion-derived virus and the budded virus, responsible for the primary infection and the viral dissemination, respectively (2).
Since the 1980s, researchers have described the interactions between BVs and mammalian cells (3). BVs are able to enter mammalian cells but cannot produce viral progeny (4–8). BVs can transduce different cell types, such as epithelial, hepatic, immune, and fibroblast cells, under mammalian promoters (3, 9, 10). This allows the use of BVs as expression vectors in different research fields, including gene therapy, cellular reprogramming, and genomic edition (3). Importantly, BV transduction efficiency is regulated by the ability of virions to escape from the endosome and by intracellular trafficking to the nucleus (11).
Several studies have reported that the nonspecific immune response triggered by viral vectors, particularly BVs, might suppress transgene expression (12–14). BVs induce high levels of type I interferon (IFN-I) in vivo and in vitro (15–19). The antiviral response generated by BVs in vivo is strong enough to nonspecifically protect animals from lethal doses of different viruses, such as foot-and-mouth disease virus, encephalomyocarditis virus, influenza virus, and infectious bronchitis virus (15, 18, 20, 21). In fact, another potential application field of BVs is their use as immunomodulators and adjuvants for vaccines (22–24).
During a viral infection, the first line of defense is the innate immune response produced by several cell types, such as dendritic cells, macrophages, and nonimmune cells (i.e., fibroblast, endothelial, and epithelial cells) (25). The response of these cells depends on pattern recognition receptors (PRRs), which detect and interact with specific pathogen sequences called pathogen-associated molecular patterns (26). PRRs include several proteins, such as Toll-like receptors (TLRs), which are expressed on the cell surface and in endosomal compartments, and cytosolic PRRs, which sense pathogen nucleic acids in the cytoplasm (26). The DNA sensor family includes DNA-dependent activator of IRFs (DAI), IFI16, DDX41, DNA-dependent protein kinase (DNA-PK), RNA polymerase III (RNA Pol III), and cyclic GMP-AMP synthase (cGAS), among others (26). Among them, the most relevant is cGAS, which promotes the synthesis of 2′3′-cGAMP, a strong activator of the stimulator of interferon genes (STING), involved in the detection of different DNA pathogens producing IFN-I through the STING-TBK1-IRF3 axis (27–31). STING is a key adaptor protein for most DNA- and RNA-sensing pathways (32, 33). On the other hand, the cytosolic DNA sensor RNA Pol III transcribes poly(dA-dT) DNAs in 5′-ppp RNA to induce IFNs through the retinoic acid-inducible gene I (RIG-I) pathway (34, 35). This pathway is involved in the detection of several organisms, including Legionella pneumophila and Epstein-Barr virus (34, 35). The DNA-PK complex, which comprises Ku70, Ku80, and the catalytic subunit of DNA-PK (DNA-PKcs), mediates the antiviral response triggered by cytosolic DNA dependent and independent of STING. Particularly, Ku70 may participate in the induction of type III IFN production in response to exogenous DNA (36).
The mechanisms involved in the innate immune response triggered by BVs are unclear. In 2005, Abe et al. described a mechanism mediated by TLR9 in response to the CpG motifs of the BV genome, which results in the production of proinflammatory cytokines, IFN-I, and IFN-inducible chemokines (17). A TLR9-independent mechanism, however, also exists, suggesting the importance of cytoplasmic DNA-sensing receptors (37). Ono et al., in 2014, showed that STING signaling is involved in the sensing of BVs in mouse embryonic fibroblasts (MEFs), as STING-deficient cells show more efficient transduction and lower production of IFN-β and proinflammatory cytokines than wild-type STING-expressing cells (12). To date, no study has reported the involvement of a cytosolic DNA detector in the cellular response to BV infection.
The aim of the present study was to evaluate the role of the cGAS-STING pathway in BV-mediated IFN production in nonimmune mammalian cells by using two approaches: the genetic edition through clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 technology of genes encoding STING or cGAS in NIH/3T3 murine fibroblasts and the infection of HEK293 and HEK293 T human epithelial cells, deficient in cGAS or in cGAS and STING expression, respectively. We also evaluated the effect of the antiviral state and of the sensors involved in the efficiency of BV transduction.
RESULTS
Baculovirus DNA is available in the cytosol of infected mammalian cells.
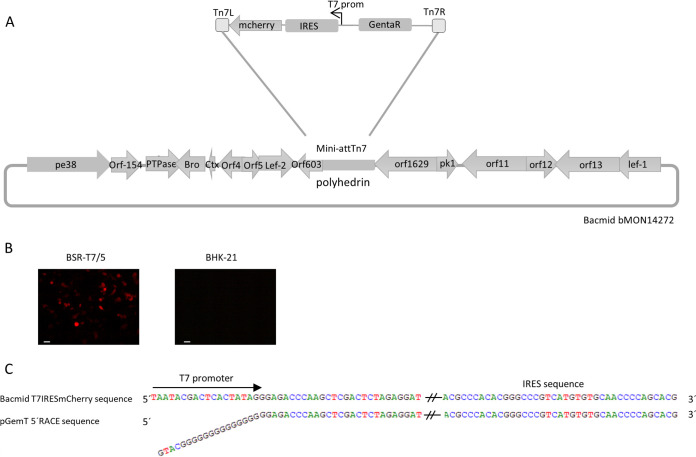
To determine whether BV DNA localizes in the cytosol during infection, available to interact with nucleic acid cytosolic sensors, we produced an mCherry-reporter recombinant BV (Fig. 1A). The BV AcT7IRESmCherry carries mCherry cDNA downstream of the T7 promoter sequence and an internal ribosomal entry site (IRES) element. This BV was used to infect BSR-T7/5 cells, a BHK-21-derived cell line that stably expresses T7 RNA polymerase in the cytoplasm (38), and BHK-21 cells were used as a control. The expression of mCherry was detectable 24 h postinfection (hpi) in BSR-T7/5 cells but not in BHK-21 cells (Fig. 1B), showing that expression of the cytoplasmic T7 RNA polymerase was necessary for expression. To determine whether mCherry transcripts were produced from the T7 promoter and not from any BV promoter upstream of it, a 5′ rapid amplification of cDNA ends (5′ RACE) of this transcript was performed in BSR-T7/5-infected cells. The results (Fig. 1C) show that transcripts started immediately after the T7 promoter sequence. This indicates that the viral DNA was available for cytosolic transcription and can thus be recognized by DNA cytosolic sensors.
FIG 1.
Cytoplasmic transduction in BSR-T7/5 cells by AcT7IRESmCherry. (A) Schematic representation of the recombinant bacmid BacT7IRESmCherry, showing the insertion site of the reporter gene, its orientation, and the surrender genes. mCherry was under the control of the T7 promoter and downstream of the IRES of foot-and-mouth disease virus. (B) BSR-T7/5 and BHK-21 cells were infected with AcT7IRESmCherry at an MOI of 100 for 4 h and then incubated for 20 h. mCherry expression was analyzed with a Zeiss microscope. The scale bar represents 20 μm. (C) Transcriptional characterization of the mCherry gene in BSR-T7/5 cells infected with AcT7IRESmCherry. The sequenced DNA from a 5´ RACE assay (pGemT 5´ RACE sequence) was compared with the theoretical sequence of the constructed bacmid.
Baculoviruses induce an antiviral response mediated by STING and the production of IFN-I through the cGAS-STING pathway in murine fibroblasts.
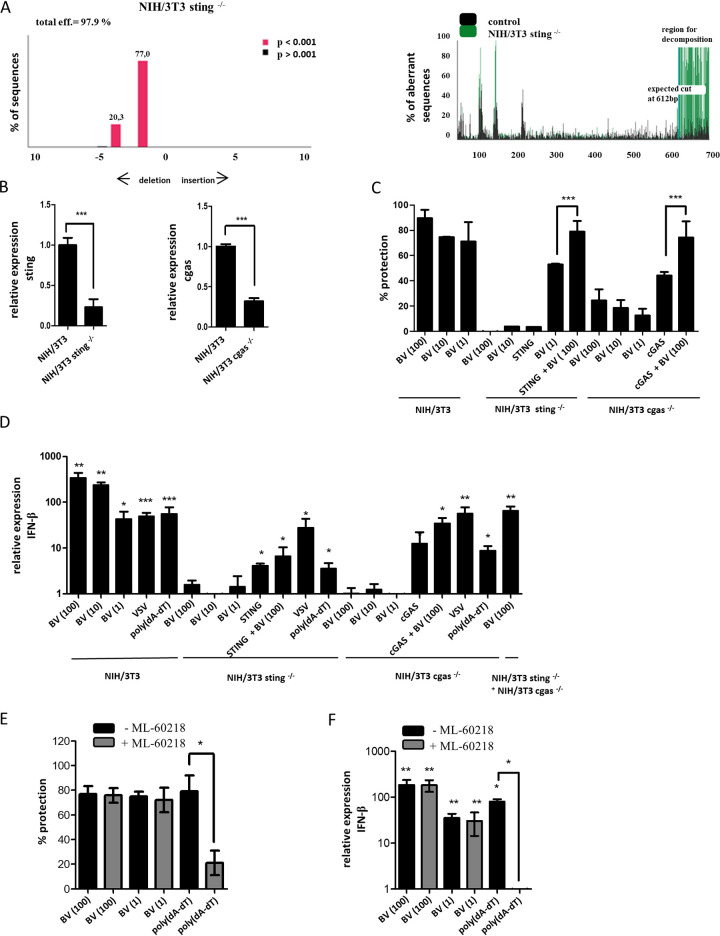
After demonstrating that BV DNA was in the cytosol, we assessed the role of cytosolic DNA sensors in the antiviral response. First, we investigated the cGAS-STING pathway in murine fibroblasts after BV infection. For this purpose, our approach was the edition of genes encoding STING or cGAS through CRISPR/Cas9 technology in NIH/3T3 murine fibroblasts. By means of tracking of indels by decomposition (TIDE) analysis, we corroborated a knockout efficiency of 97.9% for the genetic edition of STING in NIH/3T3 sting−/− cells (Fig. 2A). The targeted gene had deletions of two or four nucleotides, thus resulting in frameshift mutations. Unfortunately, a high GC content and repeats in the sequence flanking the targeted cGAS impeded confirmation of the knockout efficiency by TIDE analysis for this gene. Still, as demonstrated by other authors (39), reverse transcriptase quantitative PCR (RT-qPCR) data (Fig. 2B) revealed that the expression of cGAS or STING in the cell lines generated was efficiently decreased.
FIG 2.
Antiviral activity and IFN-I mRNA induction by BV infection in murine fibroblasts. (A) TIDE analysis to quantify indel frequencies and composition in NIH/3T3 sting−/− cells. (B) STING and cGAS mRNA depletion assayed by RT-qPCR, with gapdh used as a housekeeping gene. ***, P < 0.001 by permutation test for comparison of edited to control cells. (C and D) NIH/3T3, NIH/3T3 sting−/−, or NIH/3T3 cgas−/− cells were transfected with pcDNA3.1-Hygro(+)-mscGAS (cGAS) or pcDNAmSTING (STING). After 48 h, cells were transfected with poly(dA-dT) (1 μg/ml) or infected with BV (MOI of 100, 10, or 1) or with VSV at an MOI of 10. After 4 hpi, cells were infected with VSV at an MOI of 1 and the percentage of protection from VSV infection was calculated as described in Materials and Methods (C) or RNA was extracted and RT-qPCR assays were performed (D). (E and F) NIH/3T3 cells were treated with ML-60218 (20 μM) for 2 h and then transfected with poly(dA-dT) (1 μg/ml) or infected with BV at an MOI of 100 or 1. At 4 hpi, cells were infected with VSV at an MOI of 1 and the percentage of protection was calculated as described in Materials and Methods (E) or RNA was extracted and RT-qPCR assays were performed (F). All the data from three independent experiments in biological triplicates were averaged and are shown as means ± SD. gapdh was used as the housekeeping gene. The permutation test was used for comparisons of each treatment to untreated cells (indicated above each column: *, P < 0.05; **, P < 0.01; ***, P < 0.001). Statistical analyses among treatments were performed by a t test or one-way analysis of variance (ANOVA) followed by Bonferroni’s test (*, P < 0.05; ***, P < 0.001).
We next evaluated the antiviral activity triggered by BV infection at different multiplicities of infection (MOIs) in NIH/3T3 cells (Fig. 2C). The BV-mediated protection against a vesicular stomatitis virus (VSV) infection varied between 89% and 71% when BV MOIs of 100, 10, and 1 were used. In NIH/3T3 cgas−/− cells, the antiviral response detected after BV infection was reduced (24%, 19%, and 13% of cell protection when using BV MOIs of 100, 10, and 1, respectively) and completely abrogated by STING knockout in comparison to that of control cells (Fig. 2C). Trans-complementation of the edited cells with expression plasmids coding for murine cGAS or STING restored the ability to trigger an antiviral activity after BV infection, which was significantly higher than that observed with transfection of the plasmid alone. These data suggest the involvement of both molecules and the essentiality of STING, but not of cGAS, for the establishment of an antiviral state by BV infection.
We next assessed the role of the cGAS-STING pathway in the production of IFN-β after BV infection by infecting NIH/3T3 murine fibroblasts with BV for 4 h and performing RT-qPCR analyses. BV-infected NIH/3T3 cells showed a 344.4 ± 93.5-fold increase (MOI, 100), a 236.0 ± 39.1-fold increase (MOI, 10), and a 42.7 ± 19.9-fold increase (MOI, 1) in IFN-β mRNA levels in comparison to noninfected cells (Fig. 2D). In contrast, neither NIH/3T3 sting−/− nor NIH/3T3 cgas−/− cells produced IFN-β under these conditions (Fig. 2D). The transcription of this cytokine was detected in cGAS or STING trans-complemented edited cells and also in these transfected cells after infection.
Furthermore, when NIH/3T3 sting−/− and NIH/3T3 cgas−/− cells were coincubated and then infected with BV, IFN-β mRNA was significantly increased (Fig. 2D). This was probably due to the 2′3′-cGAMP production of infected NIH/3T3 sting−/− cells, which activated STING in NIH/3T3 cgas−/− cells. These data indicate that in murine fibroblasts, the stimulation of IFN-β production by BV infection depends on the complete cGAS-STING pathway. As expected, edited fibroblasts produced IFN-β mRNA in response to VSV infection, showing that RNA-sensing pathways were not altered in these cells. Poly(dA-dT) transfection of these cells induced the production of IFN-β mRNA, which suggests that the RNA Pol III sensor is functional and does not participate in BV-mediated IFN-β induction (Fig. 2D).
To study the RNA Pol III pathway in a deeper way and because of the high A-T genomic content of BV, we used the chemical inhibitor of RNA Pol III, ML-60218, in NIH/3T3 cells. This analysis revealed that pretreatment of the cells with the inhibitor did not modify the antiviral activity or the levels of IFN-β mRNA triggered by BV infection at an MOI of 100 or 1 (Fig. 2E and F). As a control for the ML-60218 treatment, we used the inhibition of the antiviral effect and the IFN-β mRNA production induced by transfection with poly(dA-dT). The results indicated that RNA Pol III is not involved in BV DNA detection.
To evaluate whether the antiviral response detected in the absence of cGAS expression was related to the induction of another type of IFN, we evaluated type III IFN mRNA production. Although BV triggered a significant increase in IFN-λ2/3 mRNA in the macrophage cell line RAW 264.7, murine fibroblasts showed no alterations in the transcription levels of this cytokine after BV infection (data not shown).
Baculoviruses induce the establishment of a STING-dependent antiviral response, IFN-I and IFN-III production through the cGAS-STING pathway, and induction of IFN-III through a DNA-PK sensor in human epithelial cells.
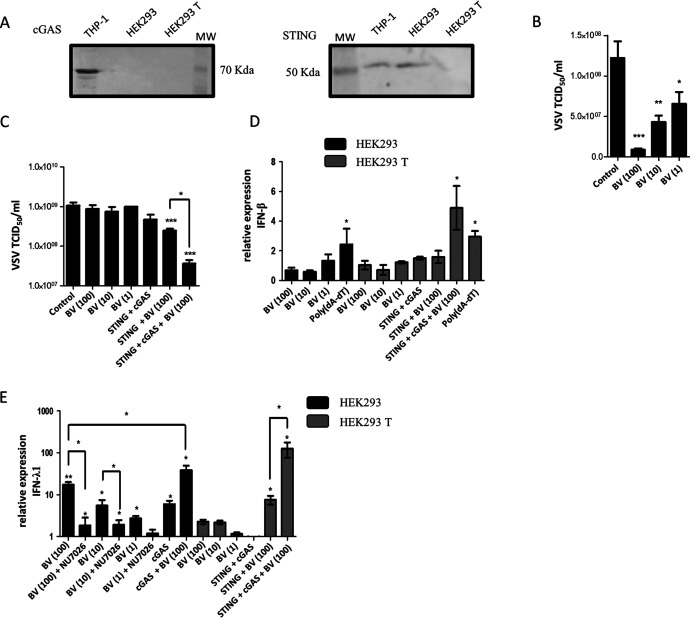
We next studied BV infection of HEK293 and HEK293 T human epithelial cells, which are deficient in cGAS or in cGAS and STING expression, respectively (Fig. 3A). First, we evaluated the antiviral activity induced by BV by measuring the VSV titer 16 hpi. In the absence of cGAS, only human epithelial cells that express STING (HEK293) displayed antiviral activity in response to BV infection at MOIs of 100, 10, and 1 (Fig. 3B and C). In HEK293 cells, trans-complementation showed no informative results due to the strong antiviral activity triggered by transfection with a cGAS expression plasmid (data not shown). In concordance with the results obtained in infected HEK293 cells, trans-complementation of HEK293 T cells with a STING expression plasmid before BV infection induced a decrease in VSV production (Fig. 3C). Cotransfection with both cGAS and STING expression plasmids before BV infection resulted in higher antiviral activity (Fig. 3C).
FIG 3.
Antiviral activity and IFN-I and -III mRNA induction by BV infection in human epithelial cells. (A) Characterization of cGAS and STING expression in HEK293 and HEK293 T cells by Western blotting. (B and C) HEK293 (B) or HEK293 T (C) cells were transfected with pTRIP-CMV-Puro-2A-cGAS (cGAS) or pcDNAhSTING (STING) and, after 48 h, infected with BV at an MOI of 100, 10, or 1. At 4 hpi, cells were infected with VSV at an MOI of 0.1. Sixteen hours later, VSV production was determined in Vero cells. (D and E) As indicated in the figure, some HEK293 cells were transfected only with poly(dA-dT) (1 μg/ml) or were pretreated with NU7026 (30 μM) for 16 h, and some HEK293 T cells were transfected with pTRIP-CMV-Puro-2A-cGAS (cGAS) or pcDNAhSTING (STING). After 48 h, cells were infected with BV at an MOI of 100, 10, or 1. After 4 h, RNA was extracted and RT-qPCR assays were performed. All the data from three independent experiments in biological triplicate were averaged and are shown as means ± SD. β actin was used as the housekeeping gene. The permutation test was used for comparisons of each treatment to untreated cells (indicated above each column: *, P < 0.05; **, P < 0.01; ***, P < 0.001). Statistical analyses among treatments were performed by a t test or one-way ANOVA followed by Bonferroni’s test (*, P < 0.05).
In concordance with the results obtained in murine fibroblasts, neither HEK293 nor HEK293 T cells produced IFN-β mRNA after BV infection at any of the MOIs evaluated (Fig. 3D). These findings highlight the essential role of cGAS in this response and confirm that the RNA Pol III pathway, which is functional in these cells, is not involved in BV detection. As observed for the antiviral activity, trans-complementation of HEK293 cells with the cGAS expression plasmid triggered high levels of IFN-β mRNA, thus preventing the measurement of the BV infection effect (data not shown). In HEK293 T cells, BV infection stimulated IFN-β mRNA production only after cotransfection with cGAS and STING expression plasmids (Fig. 3D).
Next, we studied IFN-III production in infected HEK293 and HEK293 T cells. As shown in Fig. 3E, IFN-λ1 mRNA was detectable only in the presence of STING, either in HEK293 cells or in transfected HEK293 T cells with the STING expression plasmid. The induction of IFN-λ1 mRNA in HEK293 cells ranged from 17.3 ± 2.8 to 2.7 ± 0.4 for BV MOIs of 100, 10, and 1. Pretreatment with NU7026, a DNA-PKcs kinase inhibitor, significantly reduced IFN-λ1 mRNA induction in HEK293 cells. Trans-complementation with cGAS in HEK293 cells or with cGAS and STING in HEK293 T cells potentiated the BV-mediated IFN-λ1 mRNA production, which reached 38.9 ± 10.5- and 127.0 ± 50.6-fold increased levels, respectively, using a BV MOI of 100 (Fig. 3E).
These results indicate that the cGAS-STING pathway is relevant in the production of IFN-λ1 in response to BV infection. In the same way as observed for antiviral activity, where STING is essential, cGAS reinforces the type III IFN response after BV infection. In human epithelial cells, although not in murine fibroblasts, the antiviral effect observed in the presence of STING and the absence of cGAS may be explained by type III IFN production after DNA-PK sensing.
The cGAS-STING pathway is involved in transduction efficiency in mammalian cell lines.
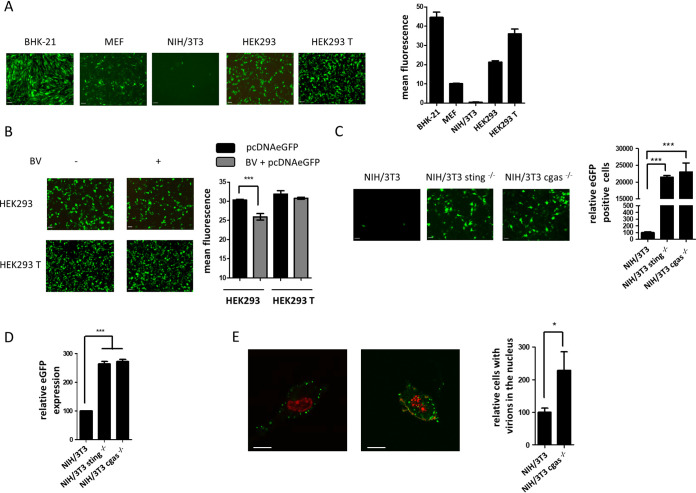
BVs can transduce several types of mammalian cells with variable efficiency (3, 10). To analyze the performance of some nonimmune cells to express a transgene driven by a BV vector, we compared different cell types: HEK293 T, HEK293, BHK-21, MEFs, and NIH/3T3 cells. The cells were infected with the recombinant BV AcCAGeGFP, which carries an egfp reporter gene under the control of the CAG promoter (a strong synthetic mammalian promoter), to assess the expression of enhanced green fluorescent protein (eGFP) 24 hpi. The different cells evaluated showed high variability of transduction efficiency (Fig. 4A). BHK-21 and HEK293 T cells displayed the most efficient transduction, whereas NIH/3T3 cells showed relatively low efficiency. This difference might arise from intrinsic cell physiologies influencing viral trafficking to the nucleus but also from diverse cellular responsiveness to viral infection. In accordance with the antiviral state induced by BV in HEK293 cells, this cell type showed lower transduction than HEK293 T cells. Thus, the expression of eGFP driven by a plasmid was studied in BV-infected and noninfected HEK293 and HEK293 T cells. Only infected HEK293 cells displayed significantly lower mean fluorescence intensity at 16 hours posttransfection than noninfected cells (Fig. 4B). These results indicate that even in the absence of cGAS, a condition that does not allow IFN-β production, BV infection was able to induce an antiviral state through STING, which modulated the efficiency of a transgene expression independently of how it was vectorized.
FIG 4.
Transduction efficiency in nonimmune mammalian cells by infection with BV. (A) Different cell lines were infected with BV AcCAGeGFP at an MOI of 100 for 4 h and then incubated for 20 h. eGFP expression was analyzed with a Zeiss microscope, and mean fluorescence was quantified using ZEN 2.3 software. (B) HEK293 or HEK293 T cells were infected (BV+) or not (BV−) with wild-type BV for 4 h at an MOI of 100 and then transfected with pcDNAeGFP. Mean fluorescence was quantified using ZEN 2.3 software. (C and D) NIH/3T3, NIH/3T3 sting−/−, or NIH/3T3 cgas−/− cells were infected with AcCAGeGFP at an MOI of 100 for 4 h and then incubated for 20 h. eGFP expression was analyzed with a Zeiss microscope and the number of eGFP-positive cells was quantified, and 100% corresponded to NIH/3T3 control cells (C). The relative expression of eGFP mRNA was evaluated by RT-qPCR, and 100% corresponded to NIH/3T3 control cells. gapdh was used as the housekeeping gene (D). (E) NIH/3T3 or NIH/3T3 cgas−/− cells were infected with BV at an MOI of 50, and at 6 hpi, viral particles were evidenced inside cells by immunofluorescence confocal microscopy using anti-VP39 antibody. Nuclei were stained using TOPRO-3. The numbers of cells with viral particles inside the nucleus were compared, and 100% corresponded to NIH/3T3 control cells. Statistical analyses were performed by a t test or one-way ANOVA followed by Bonferroni’s test (*, P < 0.05; ***, P < 0.001). Scale bars, 20 μm (A, B, and C) or 10 μm (E).
Next, the participation of cGAS and STING in the efficiency of a transgene expression driven by BV vectors was evaluated, or confirmed in the case of STING, by measuring eGFP fluorescence in edited NIH/3T3 cells. The expression of eGFP from AcCAGeGFP was dramatically increased in the edited cells (Fig. 4C), thus indicating that both STING and cGAS affect BV transduction.
Next, the transcription of the reporter gene was measured by RT-qPCR in control and edited NIH/3T3 cells at 24 hpi. The relative expression of eGFP mRNA was significantly higher in the knockout cells than in the control cells (264% ± 8% for NIH/3T3 sting−/− cells and 273% ± 6% for NIH/3T3 cgas−/− cells). In contrast, no differences were detected between the two types of edited cells (Fig. 4D).
In addition, the transport of nucleocapsids to the nucleus in NIH/3T3 cgas−/− cells was studied by immunofluorescence confocal microscopy with an antibody directed to the major capsid protein of BV, VP39. In coincidence with the results of RT-qPCR, the immunofluorescence assay showed that at 6 hpi, the number of cells with viral particles detected in the nucleus was significantly higher in NIH/3T3 cgas−/− cells (Fig. 4E). This finding suggests that the cGAS-STING pathway modulates viral trafficking and, consequently, the efficiency of viral transcription. However, the magnitude of these differences does not seem to fully explain the efficiency of BV transduction in edited cells. Thus, we postulate an additional translational effect.
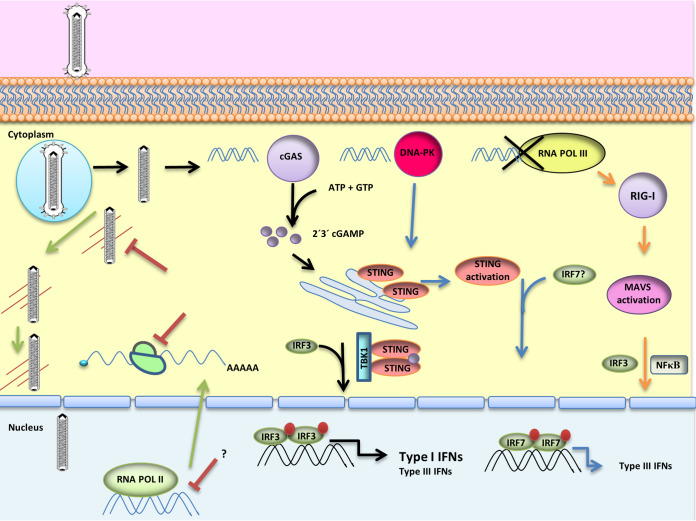
In summary (Fig. 5), our results showed that RNA Pol III is not relevant in the detection of the BV genome and that STING is required for the establishment of an antiviral state in nonimmune mammalian cells. At least two different signaling pathways have an impact on STING and contribute to the antiviral state induced by BV. The detection of the viral genome by the cGAS sensor induces the strongest cellular response, and furthermore, this DNA sensor is necessary for IFN-β production. In addition, the cGAS-independent STING activation produces an antiviral state in human and murine cells. In human epithelial cell lines, the antiviral response correlates with the DNA-PK-mediated production of IFN-λ1.
FIG 5.
Innate immune signaling pathways modulating BV transgene expression. BV infection activates at least two different STING-dependent signaling pathways: the cGAS-dependent signaling pathway (black arrows), which activates the STING-TBK1-IRF3 axis, inducing type I IFN production and exacerbating the production of type III IFN, and the DNA-PK-dependent pathway (blue arrows), which produces IFN-λ1 in human epithelial cells. All of these responses suppress BV transgene expression (red) by affecting the transport of nucleocapsids to the nucleus and the transcription and translation of the transgene. RNA Pol III is not a relevant cytosolic nucleic acid sensor in BV infection (orange arrows).
DISCUSSION
The baculovirus AcMNPV is able to transduce genes under the control of an adequate promoter in mammalian cells, although it cannot replicate its genome in these hosts. Besides this potential for gene delivery, BVs can also be used for vaccine development and immunomodulation (22–24). This work aimed to evaluate the participation of cytosolic DNA sensors in the antiviral response induced by BV infection and its relevance in BV transduction in nonimmune mammalian cells.
The infection of mammalian immune cells by BVs induces a strong innate immune response and can generate a nonspecific antiviral state, triggering the production of IFN-α, IFN-β, and proinflammatory cytokines (17, 37). Furthermore, BV infection produces maturation of dendritic cells and stimulates natural killer cells, generating IFN-γ (19, 40, 41). Because of the high frequency of CpG motifs in the BV genome, TLR9/MyD88 has been the most studied pathway (17, 37). Nevertheless, in vivo studies have shown partial reduction of IFN-γ and interleukin 12 (IL-12) in MyD88−/− mice (17). In addition, IFN-α and IFN-β production are completely or partially independent of TLR9 and MyD88, respectively (17, 37). Thus, researchers began to study other PRRs and, therefore, identified the adaptor molecule STING as a central mediator of systemic IFN-I levels (12). Yet, to date, the mechanisms involved in DNA sensor and STING activation remain uncharacterized.
In this work, we showed the accessibility of BV genomes in the cytosol of mammalian cells. Some evidence shows that most BV nucleocapsids are retained in mammalian cell endosomes (11). However, nuclear importation or cytoplasmic traffic suffers a blockage in these cells (11). Regarding the accessibility to gene transcription, we have previously demonstrated the presence of viral genomes in the cytoplasm of BHK-21 cells; nevertheless, we were not able to rule out a disassembling of inner cellular membranes or disruption of actin microfilaments by the vaccinia virus used as a tool and subsequent detection of endosomal DNA (42). Fluorescent in situ hybridization (FISH) assays in PK1 cells (6) and DNA detection in the cytoplasm of rat chondrocytes (16) by qPCR might allow the detection of endosomal content. In the present work, the BSR-T7/5 system allowed us to evaluate, for the first time, the presence of BV DNA without disrupting cellular structures. In this way, we showed that BV particles, despite being viral vectors with a proven ability to express transgenes in mammalian cell nuclei, are frequently retained in the cytoplasm and that they disassemble and remain available to interact with cellular sensors.
Some nonimmune cells, like fibroblasts, are central in IFN-I production in response to viral infections, in particular by producing high levels of IFN-β. BV infection in MEFs stimulates IFN-I synthesis (37). Ono et al., in 2014, reported that STING-, TBK1-, or IRF3-deficient MEFs showed affected IFN-β induction by BV (12). In addition, they reported the translocation of STING toward the perinuclear zone after BV infection, while Takama et al., in 2017, showed that silencing of RAB2 and GARIL5, two proteins that modulate IRF3 phosphorylation through the STING-TBK1 axis, decreases IFN-β production in BV-infected MEFs (12, 43). In infections with DNA viruses, cGAS is the most relevant detector producing the activation of the STING-TBK1-IRF3 axis; probably for this reason, it has been considered necessary in STING activation by BV infection (43, 44).
In our work, we studied the cGAS-STING pathway by using edited murine fibroblasts and human epithelial cells with deficiencies in cGAS and differential expression of STING. The selection of edition of NIH/3T3 cells over the small interfering RNA (siRNA) strategy was because some researchers detected IFN-stimulated genes 12 h poststimulus despite their being silenced (43). In this analysis, both genes showed an overexpression in BV-infected NIH/3T3 cells (data not shown). In addition, siRNA transfection promotes a nonspecific induction of inflammatory cytokines and type I and III IFNs (45).
Herein, we demonstrated that STING is essential for the establishment of an antiviral state by BVs in murine fibroblasts and human epithelial cells. We also proved the involvement of cGAS in BV detection and the existence of an alternative sensing mechanism. In addition, we showed that the cGAS-independent antiviral activity was unrelated to IFN-β synthesis, since the production of this cytokine kept a strict correlation with the presence of the aforementioned DNA sensor. This research thus shows the importance of the cGAS sensor regarding the strength of the response. In 2018, Costa Franco et al. showed the importance of cGAS in IFN-I production in response to infection by Brucella abortus and demonstrated that STING, but not cGAS, is critical for controlling the infection in macrophages and in vivo (46). As with that previously described for Listeria monocytogenes (47), c-di-GMP produced by Brucella activates STING (46). For DNA viruses, however, no similar cGAS-independent mechanism has been described.
The essentiality of both cGAS and STING for BV-mediated triggering of IFN-β production in murine fibroblasts was confirmed by infection of a mix of edited cells. The second messenger 2′3′-cGAMP, synthesized by the activation of cGAS, is spread from one cell to another by gap junctions (48) and secreted to the extracellular medium, from which it can be taken by nearby cells (49). The reestablishment of IFN-I production after stimulation with BV shows that edited cells were not impeded for transcription of this cytokine and states the critical role of 2′3′-cGAMP in BV-mediated induction of the IFN-I response. In HEK293 T cells, cGAS participation in BV-induced IFN-β production could be demonstrated by trans-complementation with cGAS and STING. Study of the BV effect by complementation assays in these cells could be performed since STING levels achieved by transfection may not be as high as the endogenous levels in HEK293 cells.
The source of STING activation determines the signaling pathway to be triggered (50). STING is subject to multiple posttranslational modifications (50). Although the precise molecular function of these modifications remains unclear, they play together to obtain the appropriate response in each case (50). Our results showed concordance between antiviral activity and IFN-III production in BV-infected human epithelial cells. Although the mechanisms involved in the differential induction of IFN-I or -III are unknown, it is accepted that it depends on the use of different signaling molecules or transcription factors. IFN-λ1 and IFN-λ3 promoters contain binding sites for IRF1, IRF3, IRF7, and NF-κB, but unlike IFN-I synthesis, which requires the joint participation of several transcription factors, IFN-λ transcription may depend mainly on IRF1, IRF7, and NF-κB to a lesser extent (51, 52). Our results using BV as a dsDNA stimulus are in agreement with those described by Zhang et al. in 2011 (36), who found that HEK293 cells, but not HEK293 T cells, produce IFN-λ in response to dsDNA (36). Sui et al., in 2017, further demonstrated the participation of the Ku70 sensor in DNA detection, the interaction of Ku70 with STING, and the STING-dependent increase of IRF1 and IRF7 but not of IRF3 (53). These mechanisms may also be operating in BV genome sensing. Although DNA-PKcs was not necessarily required for the interaction between Ku70 and STING (54), our findings show that the kinase activity of DNA-PKcs is essential for BV-mediated IFN-λ1 production. Trans-complementation with cGAS in HEK293 cells and with cGAS-STING in HEK293 T cells increased the production of IFN-λ1 by BV, probably by the restoration of the IRF3 pathway. Ding et al. demonstrated that 2′3′-cGAMP induces the transcription not only of IFN-I but also of IFN-III (55). On the other hand, the antiviral activity stimulated in NIH/3T3 cgas−/− cells was not the result of inefficient edition of the gene, as evidenced by the total lack of IFN-β induction. However, it coincides with the data obtained in HEK293 cells. In NIH/3T3 cgas−/− cells, unlike in human epithelial cells, BV did not promote IFN-III transcription. In contrast, in RAW 264.7 cells, BV promoted IFN-λ2/3 synthesis, showing that murine cells can produce this cytokine after infection with BV. In addition, the antiviral activity produced by BV in NIH/3T3 cgas−/− cells was not modified when a DNA-PK inhibitor was used (data not shown). These findings suggest an alternative route of a different sensor that induces an antiviral response mediated by other cytokines.
In BV-infected NIH/3T3 cells, the chemical inhibition of RNA Pol III did not affect the antiviral response or induction of IFN-β mRNA transcription. Accordingly, BV did not stimulate IFN-β mRNA production in HEK293 or HEK293 T cells, where the RNA Pol III pathway is active, as shown by the results obtained by poly(dA-dT) transfection. DNA virus activation of mitochondrial antiviral-signaling protein (MAVS), a molecule involved in RNA detection through RIG-I stimulation, has been described through RNA Pol III participation (34, 35). Our results cannot explain why BV-infected MEFs produce lower levels of IFN-β if they are prepared from MAVS-deficient mice (12). We cannot rule out the sensing of dsRNA structures from some viral transcript, which activates RIG-I/MAVS-mediated antiviral signaling in fibroblasts. In HEK293 and HEK293 T cells, this pathway is functional and, as shown in the present study, is not activated by BV infection. The differences in responsiveness to BV infection between cell types can be explained by differential BV transcriptomes in mammalian cells postinfection, as previously reported (56–58).
In accordance with previous results (9), in the present study, nonimmune mammalian cells displayed a very variable efficiency to express transgenes contained in the BV genome. As mentioned before, the endosomal escape has been pointed out as the limiting step for BV transduction (11). However, some evidence suggests that IFN-I production interferes with viral transit into the cell (59), increases intracellular pH (60), downregulates actin mRNA expression (61), and prevents its reorganization (62). Moreover, IFN-I production causes changes in gene transcriptional restriction due to interference with the binding of transcriptional factors and with RNA polymerase II elongation (63, 64). All these cell responses induced by IFNs directly affect transgene expression driven by a viral vector. A high-throughput short hairpin RNA (shRNA) library screening identified 43 host factor genes that restrict BV transduction in A549, a human lung carcinoma-derived cell line (13). In addition, Lee et al. described the relation between BV-induced IFN-β and the transgene expression driven by BV in supertransduction experiments (16). In our work, the efficiency of transgene expression in nonimmune cell lines correlated with the antiviral state induced by the BV carrying the reporter gene. BHK-21, a cell line with proven inability to produce IFNs, was transduced with high efficiency, whereas NIH/3T3 cells displayed a strong antiviral activity and were poorly transduced. BV infection resulted in the reduction of VSV production in HEK293 cells but not in HEK293 T cells (which had a more efficient transduction). Thus, as previously shown in MEFs (12), STING expression in human epithelial cells affected BV transduction. Moreover, the antiviral state induced by BV in this type of cells worsened the reporter expression driven by a different vector. The antiviral activity that modulates transduction efficiency in these epithelial cells is independent of IFN-β and can be explained by the production of IFN-λ1. In murine fibroblasts, we not only confirmed the role of STING in the modulation of transduction efficiency but also demonstrated that cGAS is involved in this process. Indeed, we showed that the cGAS-STING pathway decreases the number of viral particles that reach the cell nucleus and the number of reporter gene transcripts, but mainly reporter fluorescence, thus suggesting a translational effect. These results coincide with a previous report in which a preceding infection with BV affected translation, but not transcription, of a transgene driven by another BV in chondrocytes (16).
In conclusion, the detection of BV genomes by the cGAS-STING nucleic acid sensing pathway plays a decisive role in establishing an antiviral state given by IFN-I and -III, which affects the efficiency of BV as a gene transporter.
MATERIALS AND METHODS
Cell cultures.
The Sf-9 insect cell line, a clonal isolate from the parental Spodoptera frugiperda cell line IPLB-Sf21-AE, was purchased from Invitrogen and maintained at 27°C in TNMFH insect medium (Sigma) supplemented with 10% fetal bovine serum (FBS) (Internegocios). NIH/3T3 (ATCC), Vero (ATCC), HEK293 (ATCC), HEK293 T (ATCC), RAW 264.7 (ATCC), THP-1 (ATCC), and BSR-T7/5 (38) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS in a 5% CO2 atmosphere at 37°C. BSR-T7/5 cells were cultured with Geneticin (Gibco) (1 mg/ml) at every second passage. MEFs were obtained as described by Jozefczuk et al. (65).
Plasmids, bacmid, and viruses.
Wild-type AcMNPV was obtained from Pharmingen (BD Biosciences). The AcCAGeGFP virus was constructed previously (42). AcT7IRESmCherry was generated using the Bac-to-Bac system.
For the preparation of BV stocks, Sf-9 cells grown in 175-cm2 flasks were infected at an MOI of 0.05, and at 4 dpi, the supernatants were collected and clarified. Virus titers were determined by an endpoint dilution assay by using a GFP infection-induced cell line as previously described (66).
VSV Indiana strain stocks were prepared by infecting Vero cells grown in 175-cm2 flasks at an MOI of 0.1 and collecting the supernatants at 2 dpi. Virus titers were determined by an endpoint dilution assay in Vero cells (67).
BacT7IRESmCherry was constructed by first amplifying the T7IRES sequence using primers T7IRESfor, 5′-GCGCATGCTTAATACGACTCACTATAG-3′, and T7IRESrev, 5′-GCCCATGGAGGGTCATTAATTGTAA-3′, and the plasmid pIC-LUC (provided by Encarnación Martínez-Salas, Centro de Biología Molecular Severo Ochoa [CBMSO], Spain), as a template. The linear PCR fragment was gel purified and cloned into pGEM-T Easy vector systems (Promega) to obtain pGemT-T7IRES. This plasmid was digested with the enzymes PvuII and SpHI and cloned into the pFBDmCherry plasmid (42) digested with the same enzymes, to obtain pFBD-T7IRESmCherry.
The plasmid pcDNAhSTING was constructed by amplifying the hSTING sequence with primers hSTINGfor, 5′-GCGAATTCATGCCTCATTCCAGCC-3′, and hSTINGrev, 5′-CGTCTAGATTAAGAGAAATCCGTGC-3′, and using DNA extracted from HeLa cells as a template. The PCR fragment was gel purified and cloned into pCR 2.1-TOPO TA (Invitrogen) to obtain pTOPO-hSTING. This plasmid was digested with EcoRI and XbaI (underlined) and cloned into the pcDNAeGFP plasmid digested with the same enzymes to obtain pcDNA-hSTING. pcDNAmSTING was similarly constructed using the DNA of NIH/3T3 cells as a template and the following primers: mSTINGfor, 5′-GCGAATTCATGCCATACTCCAACC-3′, and mSTINGrev, 5′-CGTCTAGATTTAGATGAGGTCAGTG-3′. The insertion was confirmed by sequencing (Unidad de Genómica [IABIMO, Hurlingham, Argentina]). The expression plasmids of murine cGAS [pcDNA3.1-Hygro (+)-mscGAS] and human cGAS (pTRIP-CMV-Puro-2A-cGAS) were gifts from the Nicolas Manel Lab (Addgene plasmid number 102607 [http://n2t.net/addgene:102607] and Addgene plasmid number 102612 [http://n2t.net/addgene:102612]) (68).
Transduction assays.
Cells were seeded into 96-multiwell culture plates at a density of 1 × 105 cells/well (NIH/3T3, NIH/3T3 sting−/−, and NIH/3T3 cgas−/− cells) or 1.6 × 104 cells/well (HEK293, HEK293 T, BHK-21, MEF, and BSR-T7/5 cells) and cultured overnight. They were then incubated with AcCAGeGFP or AcT7IRESmCherry at an MOI of 100 (centrifuged at 13,000 × g and resuspended in TNMFH-phosphate-buffered saline (PBS) [1:4]) at 27°C for 4 h. Subsequently, the supernatants were removed and the cells were washed three times with PBS before the addition of fresh culture medium. After 20 h at 37°C, eGFP or mCherry expression was analyzed using a fluorescence microscope (Zeiss).
5´ RACE.
BSR-T7/5 cells were incubated with AcT7IRESmCherry at an MOI of 100 at 27°C for 4 h and washed three times with PBS before the addition of fresh culture medium. At 16 hpi, total RNA was isolated by using TRIzol reagent (Invitrogen), and cDNA 5′ ends were rapidly amplified using the 5′ RACE system version 2.0 (Invitrogen), in accordance with the manufacturer’s instructions. For cDNA synthesis, the primer was mCherryRT 5′-AGGCCTTCCTTCTCCTTCAC-3′. For the PCR, the primers were Abridged Anchor primer, Abridged universal amplification primer (Invitrogen), and reverse primer IRESrace 5′-CGTGCTGGGGTTGCACAC-3′. Sequences were obtained after cloning in the pGEM-T Easy vector.
Transfections.
The purified bacmid was transfected using 1 μg of DNA per 1 × 106 Sf-9 cells by using Cellfectin II reagent (Invitrogen), in accordance with the manufacturer’s suggestions.
Mammalian cells were transfected using Lipofectamine 3000 transfection reagent (ThermoFisher Scientific), in accordance with the manufacturer’s suggestions.
Generation of STING- and cGAS-deficient cells with CRISPR-Cas9 technology.
A single guide RNA targeting STING exon 1 (5′-GAAGATGAGGGCTACATATT-3′) or cGAS exon 1 (5′-GGGCTGGGGCTCCCGTACGG-3′) (69) was cloned into the pSPgRNA plasmid to generate pSPgSTING or pSPgcGAS. pSPgRNA was a gift from the Charles Gersbach lab (Addgene plasmid number 47108; http://n2t.net/addgene:47108) (70). Then, 5 × 107 NIH/3T3 cells were centrifuged and resuspended in 200 μl of HBS (HEPES, 20 mM; NaCl, 150 mM; Ficoll 400, 0.5%; pH 7.2) and then electroporated (180 V, 2,000 μF, ∞R; 4-mm cuvettes; Bio-Rad) with 10 μg of pSPgSTING, pSPgcGAS, or pSPgRNA (control cells) and 7 μg of pST1374-NLS-flag-linker-Cas9. pST1374-NLS-flag-linker-Cas9 was a gift from the Xingxu Huang lab (Addgene plasmid number 44758; http://n2t.net/addgene:44758) (71). After 24 h, cells were selected with Blasticidin (2.5 μg/ml; Gibco) for 24 h. Single-cell clones were obtained from cells plated at 0.5 cells/well into 96-well plates. For NIH/3T3 sting−/− cells, the STING sequence was amplified using primers STINGKOfor (5′-CAACAGTAGTCCAAGTTCGTG-3′) and STINGKOrev (5′-CTCAGACCTGGTTTATGGAGGC-3′) and genomic DNA from edited and control cells as a template. The purified amplicon was sequenced. Indel frequencies were quantified using TIDE software (72).
Antiviral activity assays.
(i) NIH/3T3 cells. A total of 1 × 105 NIH/3T3, NIH/3T3 sting−/−, or NIH/3T3 cgas−/− cells were seeded onto 96-multiwell culture plates and either transfected or not with 250 ng of pcDNA3.1-Hygro(+)-mscGAS or pcDNAmSTING. After 48 h, the cells were infected with BV at an MOI of 1, 10, or 100 or stimulated with poly(dA-dT) (1 μg/ml; InvivoGen). For the RNA Pol III inhibition assay, cells were pretreated with ML-60218 (Cayman Chemical) at 20 μM for 2 h. After 4 hpi, BV was removed and the cells were washed twice with PBS. The cells were infected with VSV at an MOI of 1 and, after 16 hpi, stained with crystal violet. The absorbance at 595 nm was measured using a Multiskan Spectrum spectrophotometer (Thermo Scientific). The antiviral activity was calculated as the percentage of protection, where 100% corresponded to cells without infection and 0% corresponded to cells infected with VSV.
(ii) HEK293 and HEK293 T cells. A total of 3.2 × 104 cells were seeded into 48-multiwell culture plates and either transfected or not with 250 ng of pTRIP-CMV-Puro-2A-cGAS or pcDNAhSTING. After 48 h, the cells were infected with BV at an MOI of 1, 10, or 100. After 4 hpi, BV was removed and the cells were washed twice with PBS. The cells were then infected with VSV at an MOI of 0.01, and after 16 h, the supernatants were removed and virus titers were determined by an endpoint dilution assay in Vero cells.
RT-qPCRs.
A total of 2 × 105 (NIH/3T3, NIH/3T3 sting−/−, or NIH/3T3 cgas−/− cells) or 3.2 × 104 (HEK293 or HEK293 T cells) cells were seeded into 48-multiwell culture plates and either transfected or not with 500 ng of pcDNA3.1-Hygro(+)-mscGAS, pTRIP-CMV-Puro-2A-cGAS, pcDNAmSTING, or pcDNA-hSTING. After 48 h, the cells were infected with BV at an MOI of 1, 10, or 100 or with VSV at an MOI of 10 or stimulated with poly(dA-dT) (1 μg/ml). For RNA Pol III and DNA-PK inhibition treatments, cells were pretreated with ML-60218 or NU7026 (Abcam) at a concentration of 20 μM for 2 h or 30 μM for 16 h. After 4 hpi, BV was removed and the cells were washed twice with PBS. Total RNA was isolated by using TRIzol reagent (Invitrogen). Residual DNA was removed by using a Turbo DNase-free kit (Life Technologies). Equal amounts of total RNA (1 μg) were used to synthesize cDNA with Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega) and random primers (Biodynamics). Gene expression was analyzed by RT-qPCR. Briefly, FastStart universal SYBR green master (Rox) (Roche) was used and reactions were made on an Applied Biosystem 7000 SDS using standard cycling conditions. All reactions were performed in duplicate, and qPCR data were analyzed using the 2−ΔΔCT method with efficiency correction as described previously (73). Differences in cytokine expression between groups were assessed with gapdh or β actin as control genes, and data from the control group were used for calibration. The primers were as follows: β h-actin F, 5′-TGACGGGGTCACCCACACTG-3´; β h-actin R, 5′-AAGCTGTAGCCGCGCTCGGT-3′; murine GAPDH F, 5′-GCCTTCCGTGTTCCTACCC-3′; murine GAPDH R, 5′-TGCCTGCTTCACCACCTTCT-3′; hIFN β F, 5′-CGCCGCATTGACCATCTA-3′; hIFN β R, 5′-GACATTAGCCAGGAGGTTCTCA-3′; hIFN λ1 F, 5′-CGCCTTGGAAGAGTCACTCA-3′; hIFN λ1 R, 5′-GAAGCCTCAGGTCCCAATTC-3′; mIFN λ2/3 F, 5′-AGCTGCAGGTCCAAGAGCG-3′; mIFN λ2/3 F, 5′-GGTGGTCAGGGCTGAGTCATT-3′; mIFN β F, 5′-ACACCAGCCTGGCTTCCATC-3′; mIFN β R, 5′-TTGGAGCTGGAGCTGCTTATAGTTG-3′; qGFP F, 5′-ATGGTGAGCAAGGGCGGAG-3′; qGFP R, 5´-CACGCTGAACTTGTGGCC-3′; mSTING F, 5′-CTTCAGAGCTTGACTCCAGC-3′; mSTING R, 5′-TGTACAGTCTTCGGCTCCCT-3′; mcGAS F, 5′-AAGGCAGCTGGCCTATTAGT-3′; mcGAS R, 5′-CGCCAGGTCTCTCCTTGAAA-3′.
Western blot analyses.
A total of 1 × 106 HEK293, HEK293 T, and THP-1 cells were boiled for 5 min in disruption buffer (120 mM Tris-HCl, pH 6.8, 4% SDS, 0.02% bromophenol blue, 1.4 M β-mercaptoethanol, 20% glycerol), and proteins were resolved in 12% acrylamide-bisacrylamide (30:0.8) SDS-PAGE minigels (Bio-Rad). The proteins were blotted onto nitrocellulose membranes, and the blots were probed with rabbit polyclonal anti-cGAS (1:300) or anti-STING (1:300) antibodies (Sigma catalog numbers PA5-76367 and PA5-70420, respectively), followed by anti-rabbit horseradish peroxidase (HRP)-conjugated antibody (1:1,000; Sigma catalog number A6154). The blots were revealed by using SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific).
Image analysis.
The fluorescence intensity in HEK293 and HEK293 T cells was analyzed using ZEN 2.3 software (Zeiss). Six randomly selected fields of each treatment were analyzed, and the mean intensity was calculated according to predetermined software conditions.
Confocal images were acquired in a Leica TCS-SP5 spectral laser confocal microscope (Laboratorio Integral de Microscopía, CNIA, INTA, Hurlingham, Argentina), and 40 images were analyzed using Fiji software (Thornwood, NY, USA).
Immunofluorescence microscopy.
Indirect immunofluorescence analysis was performed as previously described (74).
Statistical analyses.
Statistical analyses were performed with GraphPad Prism (La Jolla, CA, USA). At least three independent experiments were performed; error bars represent means ± standard deviations. Statistical comparisons were evaluated by using Student's t test (unpaired, two-tailed) or one-way analysis of variance (ANOVA) and by Bonferroni’s test. Differences between groups were considered significant when the P value was <0.05.
ACKNOWLEDGMENTS
M.G.L., R.J.B., O.T., and V.A. are career members of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina. S.A. and G.N.M. hold postdoctoral fellowships from CONICET. This work was supported by FONCyT (grant number PICT1334), CONICET (grant number PIP 11220130100258CO), and INTA (grant number PNBIO 1131034). We thank María José Mónaco and Julia Sabio y García for technical assistance.
Author contributions were as follows: conceptualization, V.A and O.T.; methodology, S.A., V.A., and R.B; investigation, S.A., M.G.L., and G.N.M.; resources, R.B.; writing—original draft, S.A.; writing—reviewing and editing, V.A., M.G.L., R.B., and O.T.; funding acquisition, V.A.; supervision, O.T. and V.A.
REFERENCES
- 1.Chen YR, Zhong S, Fei Z, Hashimoto Y, Xiang JZ, Zhang S, Blissard GW. 2013. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J Virol 87:6391–6405. doi: 10.1128/JVI.00194-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blissard GW, Theilmann DA. 2018. Baculovirus entry and egress from insect cells. Annu Rev Virol 5:113–139. doi: 10.1146/annurev-virology-092917-043356. [DOI] [PubMed] [Google Scholar]
- 3.Mansouri M, Berger P. 2018. Baculovirus for gene delivery to mammalian cells: past, present and future. Plasmid 98:1–7. doi: 10.1016/j.plasmid.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Carbonell LF, Miller LK. 1987. Baculovirus interaction with nontarget organisms: a virus-borne reporter gene is not expressed in two mammalian cell lines. Appl Environ Microbiol 53:1412–1417. doi: 10.1128/AEM.53.7.1412-1417.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbonell LF, Klowden MJ, Miller LK. 1985. Baculovirus-mediated expression of bacterial genes in dipteran and mammalian cells. J Virol 56:153–160. doi: 10.1128/JVI.56.1.153-160.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Loo ND, Fortunati E, Ehlert E, Rabelink M, Grosveld F, Scholte BJ. 2001. Baculovirus infection of nondividing mammalian cells: mechanisms of entry and nuclear transport of capsids. J Virol 75:961–970. doi: 10.1128/JVI.75.2.961-970.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kost TA, Condreay JP. 2002. Recombinant baculoviruses as mammalian cell gene-delivery vectors. Trends Biotechnol 20:173–180. doi: 10.1016/s0167-7799(01)01911-4. [DOI] [PubMed] [Google Scholar]
- 8.Chen CY, Lin CY, Chen GY, Hu YC. 2011. Baculovirus as a gene delivery vector: recent understandings of molecular alterations in transduced cells and latest applications. Biotechnol Adv 29:618–631. doi: 10.1016/j.biotechadv.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyce FM, Bucher NL. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc Natl Acad Sci U S A 93:2348–2352. doi: 10.1073/pnas.93.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Airenne KJ, Hu YC, Kost TA, Smith RH, Kotin RM, Ono C, Matsuura Y, Wang S, Ylä-Herttuala S. 2013. Baculovirus: an insect-derived vector for diverse gene transfer applications. Mol Ther 21:739–749. doi: 10.1038/mt.2012.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu L, Li Y, Ning YJ, Deng F, Vlak JM, Hu Z, Wang H, Wang M. 2019. The major hurdle for effective baculovirus transduction into mammalian cells is passing early endosomes. J Virol 93. doi: 10.1128/JVI.00709-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono C, Ninomiya A, Yamamoto S, Abe T, Wen X, Fukuhara T, Sasai M, Yamamoto M, Saitoh T, Satoh T, Kawai T, Ishii KJ, Akira S, Okamoto T, Matsuura Y. 2014. Innate immune response induced by baculovirus attenuates transgene expression in mammalian cells. J Virol 88:2157–2167. doi: 10.1128/JVI.03055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang CH, Naik NG, Liao LL, Wei SC, Chao YC. 2017. Global screening of antiviral genes that suppress baculovirus transgene expression in mammalian cells. Mol Ther Methods Clin Dev 6:194–206. doi: 10.1016/j.omtm.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knaän-Shanzer S, Van Der Velde I, Havenga MJ, Lemckert AA, De Vries AA, Valerio D. 2001. Highly efficient targeted transduction of undifferentiated human hematopoietic cells by adenoviral vectors displaying fiber knobs of subgroup B. Hum Gene Ther 12:1989–2005. doi: 10.1089/104303401753204562. [DOI] [PubMed] [Google Scholar]
- 15.Gronowski AM, Hilbert DM, Sheehan KC, Garotta G, Schreiber RD. 1999. Baculovirus stimulates antiviral effects in mammalian cells. J Virol 73:9944–9951. doi: 10.1128/JVI.73.12.9944-9951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HP, Matsuura Y, Chen HC, Chen YL, Chuang CK, Abe T, Hwang SM, Shiah HC, Hu YC. 2009. Baculovirus transduction of chondrocytes elicits interferon-alpha/beta and suppresses transgene expression. J Gene Med 11:302–312. doi: 10.1002/jgm.1299. [DOI] [PubMed] [Google Scholar]
- 17.Abe T, Hemmi H, Miyamoto H, Moriishi K, Tamura S, Takaku H, Akira S, Matsuura Y. 2005. Involvement of the Toll-like receptor 9 signaling pathway in the induction of innate immunity by baculovirus. J Virol 79:2847–2858. doi: 10.1128/JVI.79.5.2847-2858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abe T, Takahashi H, Hamazaki H, Miyano-Kurosaki N, Matsuura Y, Takaku H. 2003. Baculovirus induces an innate immune response and confers protection from lethal influenza virus infection in mice. J Immunol 171:1133–1139. doi: 10.4049/jimmunol.171.3.1133. [DOI] [PubMed] [Google Scholar]
- 19.Hervas-Stubbs S, Riezu-Boj JI, Mancheño U, Rueda P, Lopez L, Alignani D, Rodríguez-García E, Thieblemont N, Leclerc C. 2014. Conventional but not plasmacytoid dendritic cells foster the systemic virus-induced type I IFN response needed for efficient CD8 T cell priming. J Immunol 193:1151–1161. doi: 10.4049/jimmunol.1301440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molinari P, García-Nuñez S, Gravisaco MJ, Carrillo E, Berinstein A, Taboga O. 2010. Baculovirus treatment fully protects mice against a lethal challenge of FMDV. Antiviral Res 87:276–279. doi: 10.1016/j.antiviral.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Niu M, Han Y, Li W. 2008. Baculovirus up-regulates antiviral systems and induces protection against infectious bronchitis virus challenge in neonatal chicken. Int Immunopharmacol 8:1609–1615. doi: 10.1016/j.intimp.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Molinari P, Crespo MI, Gravisaco MJ, Taboga O, Morón G. 2011. Baculovirus capsid display potentiates OVA cytotoxic and innate immune responses. PLoS One 6:e24108. doi: 10.1371/journal.pone.0024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavarone E, Molina GN, Amalfi S, Peralta A, Molinari P, Taboga O. 2017. The localization of a heterologous displayed antigen in the baculovirus-budded virion determines the type and strength of induced adaptive immune response. Appl Microbiol Biotechnol 101:4175–4184. doi: 10.1007/s00253-017-8183-y. [DOI] [PubMed] [Google Scholar]
- 24.Huang H, Xiao S, Qin J, Jiang Y, Yang S, Li T, Gao Y, Li Z, Li T, Su X, Ruan Y, Xu F, Wang H, Chen H, Xia X. 2011. Construction and immunogenicity of a recombinant pseudotype baculovirus expressing the glycoprotein of rabies virus in mice. Arch Virol 156:753–758. doi: 10.1007/s00705-010-0909-4. [DOI] [PubMed] [Google Scholar]
- 25.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 27.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. 2013. Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, Oh BH, Jung JU. 2014. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma F, Li B, Liu SY, Iyer SS, Yu Y, Wu A, Cheng G. 2015. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J Immunol 194:1545–1554. doi: 10.4049/jimmunol.1402066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, Nagarajan UM. 2014. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-β during Chlamydia trachomatis infection. J Immunol 193:2394–2404. doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN. 2013. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell 50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zevini A, Olagnier D, Hiscott J. 2017. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol 38:194–205. doi: 10.1016/j.it.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol 10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, Veenstra TD, Young HA, Lane HC, Imamichi T. 2011. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol 186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abe T, Kaname Y, Wen X, Tani H, Moriishi K, Uematsu S, Takeuchi O, Ishii KJ, Kawai T, Akira S, Matsuura Y. 2009. Baculovirus induces type I interferon production through toll-like receptor-dependent and -independent pathways in a cell-type-specific manner. J Virol 83:7629–7640. doi: 10.1128/JVI.00679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. doi: 10.1128/JVI.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biolatti M, Dell'Oste V, Pautasso S, Gugliesi F, von Einem J, Krapp C, Jakobsen MR, Borgogna C, Gariglio M, De Andrea M, Landolfo S. 2018. Human cytomegalovirus tegument protein pp65 (pUL83) dampens type I interferon production by inactivating the DNA sensor cGAS without affecting STING. J Virol 92:e01774-17. doi: 10.1128/JVI.01774-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hervas-Stubbs S, Rueda P, Lopez L, Leclerc C. 2007. Insect baculoviruses strongly potentiate adaptive immune responses by inducing type I IFN. J Immunol 178:2361–2369. doi: 10.4049/jimmunol.178.4.2361. [DOI] [PubMed] [Google Scholar]
- 41.Moriyama T, Suzuki T, Chang MO, Kitajima M, Takaku H. 2017. Baculovirus directly activates murine NK cells via TLR9. Cancer Gene Ther 24:175–179. doi: 10.1038/cgt.2017.2. [DOI] [PubMed] [Google Scholar]
- 42.Alfonso V, Amalfi S, López MG, Taboga O. 2017. Effects of deletion of the ac109 gene of Autographa californica nucleopolyhedrovirus on interactions with mammalian cells. Arch Virol 162:835–840. doi: 10.1007/s00705-016-3142-y. [DOI] [PubMed] [Google Scholar]
- 43.Takahama M, Fukuda M, Ohbayashi N, Kozaki T, Misawa T, Okamoto T, Matsuura Y, Akira S, Saitoh T. 2017. The RAB2B-GARIL5 complex promotes cytosolic DNA-induced innate immune responses. Cell Rep 20:2944–2954. doi: 10.1016/j.celrep.2017.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ono C, Okamoto T, Abe T, Matsuura Y. 2018. Baculovirus as a tool for gene delivery and gene therapy. Viruses 10:510. doi: 10.3390/v10090510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sui H, Zhou M, Chen Q, Lane HC, Imamichi T. 2014. siRNA enhances DNA-mediated interferon lambda-1 response through crosstalk between RIG-I and IFI16 signalling pathway. Nucleic Acids Res 42:583–598. doi: 10.1093/nar/gkt844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costa Franco MM, Marim F, Guimarães ES, Assis NRG, Cerqueira DM, Alves-Silva J, Harms J, Splitter G, Smith J, Kanneganti TD, de Queiroz N, Gutman D, Barber GN, Oliveira SC. 2018. Brucella abortus triggers a cGAS-independent STING pathway to induce host protection that involves guanylate-binding proteins and inflammasome activation. J Immunol 200:607–622. doi: 10.4049/jimmunol.1700725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodward JJ, Iavarone AT, Portnoy DA. 2010. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. 2013. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carozza JA, Brown JA, Böhnert V, Fernandez D, AlSaif Y, Mardjuki RE, Smith M, Li L. 28 July 2020. Structure-aided development of small-molecule inhibitors of ENPP1, the extracellular phosphodiesterase of the immunotransmitter cGAMP. Cell Chem Biol doi: 10.1016/j.chembiol.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jønsson KL, Jakobsen MR, Nevels MM, Bowie AG, Unterholzner L. 2018. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol Cell 71:745–760.e5. doi: 10.1016/j.molcel.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, Kagan JC. 2014. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol 15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thanos D, Maniatis T. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 53.Sui H, Zhou M, Imamichi H, Jiao X, Sherman BT, Lane HC, Imamichi T. 2017. STING is an essential mediator of the Ku70-mediated production of IFN-λ1 in response to exogenous DNA. Sci Signal 10:eaah5054. doi: 10.1126/scisignal.aah5054. [DOI] [PubMed] [Google Scholar]
- 54.Burleigh K, Maltbaek JH, Cambier S, Green R, Gale M Jr, James RC, Stetson DB. 2020. Human DNA-PK activates a STING-independent DNA sensing pathway. Sci Immunol 5:eaba4219. doi: 10.1126/sciimmunol.aba4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding Q, Gaska JM, Douam F, Wei L, Kim D, Balev M, Heller B, Ploss A. 2018. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc Natl Acad Sci U S A 115:E6310–E6318. doi: 10.1073/pnas.1803406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujita R, Matsuyama T, Yamagishi J, Sahara K, Asano S, Bando H. 2006. Expression of Autographa californica multiple nucleopolyhedrovirus genes in mammalian cells and upregulation of the host beta-actin gene. J Virol 80:2390–2395. doi: 10.1128/JVI.80.5.2390-2395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Efrose R, Swevers L, Iatrou K. 2010. Baculoviruses deficient in ie1 gene function abrogate viral gene expression in transduced mammalian cells. Virology 406:293–301. doi: 10.1016/j.virol.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 58.Liu CY, Wang CH, Wang JC, Chao YC. 2007. Stimulation of baculovirus transcriptome expression in mammalian cells by baculoviral transcriptional activators. J Gen Virol 88:2176–2184. doi: 10.1099/vir.0.82664-0. [DOI] [PubMed] [Google Scholar]
- 59.Ankel H, Chany C, Galliot B, Chevalier MJ, Robert M. 1973. Antiviral effect of interferon covalently bound to Sepharose. Proc Natl Acad Sci U S A 70:2360–2363. doi: 10.1073/pnas.70.8.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maheshwari RK, Sidhu GS, Bhartiya D, Friedman RM. 1991. Primary amines enhance the antiviral activity of interferon against a membrane virus: role of intracellular pH. J Gen Virol 72:2143–2152. doi: 10.1099/0022-1317-72-9-2143. [DOI] [PubMed] [Google Scholar]
- 61.Nedelec B, Shen YJ, Ghahary A, Scott PG, Tredget EE. 1995. The effect of interferon alpha 2b on the expression of cytoskeletal proteins in an in vitro model of wound contraction. J Lab Clin Med 126:474–484. [PubMed] [Google Scholar]
- 62.Duménil G, Olivo JC, Pellegrini S, Fellous M, Sansonetti PJ, Nhieu GT. 1998. Interferon alpha inhibits a Src-mediated pathway necessary for Shigella-induced cytoskeletal rearrangements in epithelial cells. J Cell Biol 143:1003–1012. doi: 10.1083/jcb.143.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smale ST. 2010. Selective transcription in response to an inflammatory stimulus. Cell 140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bell O, Tiwari VK, Thomä NH, Schübeler D. 2011. Determinants and dynamics of genome accessibility. Nat Rev Genet 12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 65.Jozefczuk J, Drews K, Adjaye J. 2012. Preparation of mouse embryonic fibroblast cells suitable for culturing human embryonic and induced pluripotent stem cells. J Vis Exp doi: 10.3791/3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hopkins R, Esposito D. 2009. A rapid method for titrating baculovirus stocks using the Sf-9 Easy Titer cell line. Biotechniques 47:785–788. doi: 10.2144/000113238. [DOI] [PubMed] [Google Scholar]
- 67.Reed LJ, and, Muench H. 1938. A simple method of estimation of 50% end points. Am J Hyg 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 68.Gentili M, Kowal J, Tkach M, Satoh T, Lahaye X, Conrad C, Boyron M, Lombard B, Durand S, Kroemer G, Loew D, Dalod M, Théry C, Manel N. 2015. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science 349:1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 69.Doi A, Iijima K, Kano S, Ishizaka Y. 2015. Viral protein R of HIV type-1 induces retrotransposition and upregulates glutamate synthesis by the signal transducer and activator of transcription 1 signaling pathway. Microbiol Immunol 59:398–409. doi: 10.1111/1348-0421.12266. [DOI] [PubMed] [Google Scholar]
- 70.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. 2013. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. 2013. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brinkman EK, Chen T, Amendola M, van Steensel B. 2014. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alfonso V, Maroniche GA, Reca SR, López MG, del Vas M, Taboga O. 2012. AcMNPV core gene ac109 is required for budded virion transport to the nucleus and for occlusion of viral progeny. PLoS One 7:e46146. doi: 10.1371/journal.pone.0046146. [DOI] [PMC free article] [PubMed] [Google Scholar]