Abstract
We investigated whether diurnal differences in muscle force output are associated with the post-translational state of muscle proteins. Ten physically active men (mean ± SD; age 26.7 ± 3.7 y) performed experimental sessions in the morning (08:00 h) and evening (17:00 h), which were counterbalanced in order of administration and separated by at least 72 h. Knee extensor maximal voluntary isometric contraction (MVIC) force and peak rate of force development (RFD) were measured, and samples of vastus lateralis were collected immediately after exercise. MVIC force was greater in the evening (mean difference of 67 N, 10.2%; p < 0.05). Two-dimensional (2D) gel analysis encompassed 122 proteoforms and discovered 6 significant (p < 0.05; false discovery rate [FDR] = 10%) diurnal differences. Phosphopeptide analysis identified 1693 phosphopeptides and detected 140 phosphopeptides from 104 proteins that were more (p < 0.05, FDR = 22%) phosphorylated in the morning. Myomesin 2, muscle creatine kinase, and the C-terminus of titin exhibited the most robust (FDR < 10%) diurnal differences. Exercise in the morning, compared to the evening, coincided with a greater phosphorylation of M-band-associated proteins in human muscle. These protein modifications may alter the M-band structure and disrupt force transmission, thus potentially explaining the lower force output in the morning.
Keywords: maximum voluntary isometric contraction, muscle contraction, phosphopeptide, phosphoproteomic, protein processing, post-translational, proteomics, rate-of-force development, sarcomere, time of day
1. Introduction
The force output of muscle is a key component of athletic performance, and adequate levels of muscle mass and function are critical to health and well-being in the general population [1]. For example, the power output of muscle becomes a key determinant of independence and quality of life in older individuals [2]. The sarcomere is the basic contractile unit of muscle and consists of an ordered arrangement of thick and thin myofilaments in a three-dimensional lattice. Sarcomeres are arranged in series, bounded by Z-disks that connect with the muscle cytoskeleton, and have a central M-band that connects the anti-parallel thick filaments at the center [3]. Myosin heavy-chain and filamentous actin are the main proteins of the thick and thin filaments, respectively. In addition, the myofibrillar proteome consists of numerous other major proteins, e.g., myosin light chains, troponins, and tropomyosins, as well as lesser-known ancillary proteins whose functions remain enigmatic or are only recently coming to light. Despite its highly ordered structure, striated muscle is renowned for its plasticity and can adapt both quantitatively, through hypertrophy or atrophy, and qualitatively by changing the relative proportion of myofibrillar protein isoforms. Remarkably, the maximum force that can be produced by muscle varies throughout the day and is significantly greater in the evening than the morning [4,5]. The difference in maximum isometric force between the morning and evening is typically approximately 10% [6,7]. For context, this magnitude of difference is equivalent to the gain in muscle force achieved after several weeks of resistance exercise training [8].
Time-of-day differences in maximal isometric force appear to be intrinsic to muscle rather than secondary to fluctuations in neuromuscular drive [9] or body temperature [10]. In the last decades, significant progress has been made in understanding the molecular workings of the central (suprachiasmatic nucleus) and peripheral tissue clocks and their roles in biology [11]. At the molecular level, the circadian clock is a well-defined gene regulatory network composed of transcriptional–translational feedback loops involving proteins such as circadian promotor output cycles kaput (CLOCK) and brain and muscle ARNT like-1 (BMAL1; [12]). Early work in human muscle [13] reported a diurnal difference in the expression of clock genes and the muscle linage gene, myogenic factor 6. More detailed analysis of circadian gene expression in animals [14] identified further muscle-specific genes, including myogenic differentiation protein 1 (MyoD), which exhibits a circadian rhythm. The genetic deletion of key components of the molecular clock continues to be a powerful tool for investigating cellular processes that are dependent on circadian regulation. Muscle-specific disruption of the molecular clock results in impairments to glucose metabolism and insulin homeostasis [15] in mice, and exercise at different times of day is associated with different metabolic responses in muscle [16]. The deletion of BMAL1, whether from germline [17] or induced in adult muscle [18], is associated with losses in muscle contractile force, which may be relevant to the diurnal differences in maximum isometric force observed in humans. However, it is not yet clear how circadian oscillations in gene expression affect hour-by-hour differences in the force output of muscle. The majority of myofibrillar proteins have a half-life of approximately 30 days [19], so it is unlikely that changes in gene expression and protein abundance account for diurnal differences in muscle force production. Instead, the post-translational modification of proteins may be responsible. Modifications such as phosphorylation can occur on second or minute timescales and are known to change the functional properties of myofibrillar proteins [20].
Currently, fewer than 12 articles report analyses of human muscle biopsy samples in the context of chronobiology. Moreover, a proteomic analysis of human muscle in the context of diurnal or circadian rhythm has not yet been reported. Maximal contractions evoke acute changes in muscle protein phosphorylation [21], but it is not yet clear whether an interaction exists between exercise and time of day that could contribute to differences in the force output of muscle or the Zeitgeber effect of exercise on muscle function. Therefore, we used proteomic techniques to investigate the post-translational state of muscle proteins immediately after maximum force contractions in either the morning or evening. Two-dimensional gel-electrophoresis (2DGE) is a useful ‘top–down’ proteomic method for the non-targeted analysis of proteoforms. Proteoforms arise through specific combinatorial patterns of post-translational modification and the splice-variation of protein isoforms. The post-translational modification of specific myosin light chain isoforms alters the contractile properties of striated muscle [22] and could contribute to time-of-day differences in maximal isometric force. Myosin light chain isoforms are readily resolved by 2DGE into proteoforms, which include the post-translational modification of specific amino acid residues that can be identified using mass spectrometry [23]. In addition, the selective enrichment of modified proteins or peptides prior to mass spectrometry is a well-established ‘bottom–up’ proteomic approach to studying post-translational modifications. Often, bottom–up methods offer more comprehensive and higher throughput analysis than 2DGE; however, the isolated analysis of site-specific modifications does not provide proteoform-specific information [24]. Nevertheless, bottom–up analyses can be useful in identifying modifications to key protein residues that may be associated with the difference in muscle force output between morning and evening. Phosphorylation is perhaps the most widely studied post-translational modification. Robust methods for the enrichment of phosphopeptides are established [25]; however, currently, fewer than 6 articles report the phosphoproteome analysis of human skeletal muscle. The phosphoproteome of human muscle has been mapped [26], and the acute response to endurance exercise has been reported [27]. Herein, we investigate whether differences also exist in the muscle phosphoproteome after exercise in the morning versus evening.
2. Materials and Methods
Pilot experiments recruited n = 20 males with age (mean ± SD): 26.0 ± 4.4 years; body mass: 75.1 ± 8.2 kg; height: 177.3 ± 6.8 cm; and normative retiring and rising times: 23:36 ± 0:36 h:min and 06:24 ± 0:18 h:min, respectively. Verbal explanation of the experimental procedure was provided to each individual, including the aims of the study, the possible risks associated with participation, and the experimental procedures. Individuals provided written informed consent before participating in the study. The experimental procedures were approved (14/WM/0065) by the National Research Ethics Committee, UK and conformed to the standards set by the Declaration of Helsinki, except for registration in a database.
Participants were selected if they met all the inclusion requirements, including habitual sleeping duration of approximately 8 h per day, no reported sleep problems, did not routinely nap, had at least 2 years experience in strength or weight training, and habitually trained at any time of the day during a typical week. Participants were also required to avoid alcohol 48 h prior to each test. None of the participants had a history of bone fractures or musculoskeletal abnormalities, and none were prescribed pharmacological treatment during the study. Participants were excluded from participating if they had (i) recently (within the last 14 days) undertaken shift work or rapid travel across time zones; (ii) declared health issues in the Physical Activity Readiness Questionnaire (PARQ); or (iii) had a mean chronotype score <22 or >43 based on the Composite Morningness questionnaire (CMQ) [28]. The mean chronotype score of the selected participants was intermediate (35.2 ± 4.5). Sleep Flexibility/Rigidity [F/R] score 43.4 ± 5.6 and Languid/Vigour [L/V] score 43.4 ± 9.3. All experimental tests were conducted in the United Kingdom between the months of March and May.
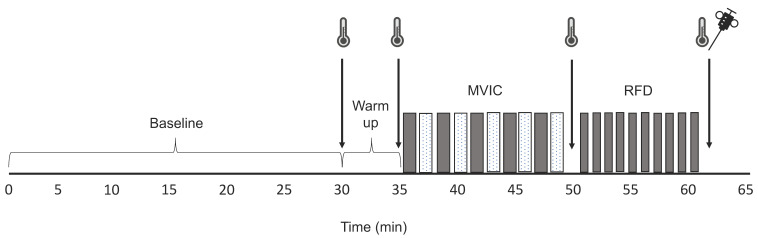
Each participant completed 5 familiarization sessions (which occurred between 12:00 and 13:00 h), separated by 3-d intervals, to ensure full acquaintance with the experimental protocol. During each familiarization session, participants performed maximal voluntary isometric contractions (MVIC) of m. quadriceps femoris with electrical stimulation to assess the percentage activation (i.e., twitch interpolation technique) and trials designed to measure the peak rate of force development (RFD) of the same muscle. Thereafter, each participant completed a morning (07:30 h) and an evening (17:30 h) experimental session (Figure 1) separated by 3 d and assigned in a randomized counter-balanced order. Ten participants elected to undergo percutaneous needle biopsies, which were taken immediately after RFD measurements. Participants retired at 23:00 h and woke at 06:00 h to be in the laboratories for 07:00 h. All sessions took place under standard laboratory conditions (lighting, room temperature, and humidity = 200–250 lux, 19–20 °C, and 38–52%, respectively). Participants reported to the laboratory in a fasted (≥7 h) state for the morning session and were instructed not to consume food for ≥4 h prior to the evening session. Participants verbally stated they had complied with these restrictions on entry to the laboratory and recorded their dietary habits throughout the day including amount, type, and timing of food and fluid intake.
Figure 1.
Experimental protocol. Schematic diagram of the experimental protocol undertaken by participants in the morning (07:30 h) and evening (17:30 h) interspersed by a 72-h wash-out period. Temperature data were collected from the rectum, muscle, and skin after a 30 min acclimatization period (baseline), after a cycle ergometry warm up (5 min at 150 W) and after periods of data collection, including the measurement of quadriceps maximal voluntary isometric contraction (MVIC) force and peak rate of force development (RFD). Participants performed 5 blocks of MVIC (dark bars) consisting of 1 unstimulated and 3 stimulated contractions, which were interspersed by rest periods (light bars). The peak RFD of the quadriceps muscle was assessed by 10 attempts (dark bars) interspersed by 30 s recovery periods. Samples of vastus lateralis muscle were taken by percutaneous needle biopsy immediately after RFD measurements.
At the beginning of each experimental session, participants inserted a rectal probe approximately 10 cm past their external anal sphincter (Grant, Squirrel, Cambridge, UK). Skin thermistors (Grant, Cambridge, UK) were attached to four points of the body: sternum, upper arm (bicep mid-point), thigh (quadriceps mid-point), and calf (mid-point). Recordings of skin temperature (Tsk) and rectal temperature (Trec) were taken every minute for 30 min (data used for analysis were averaged over the last 5 min), whilst participants remained awake in a semi-supine position. Muscle temperature (Tm) was assessed in the vastus lateralis of the right leg using a needle thermistor (13050; ELAB, Rodovre, Denmark). Trec, Tsk, and Tm were recorded after the active warm-up and after each MVIC and RFD attempt. Participants self-rated their perceived effort (0–10 Likert scale; “0” representing no effort and “10” maximal) after each MVIC and RFD exercise.
Measurements of quadriceps femoris maximum voluntary isometric contraction (MVIC) force and peak RFD of the right leg were conducted using an isometric chair (Lido Active, Loredan, Davis, CA, USA). The position of the subject in the chair was standardized in accordance with the guidelines set by the manufacturer, and the hips and knee were secured at an angle of 90° with inextensible straps. MVIC was conducted with and without the interpolated twitch technique (ITT) to accustom participants to producing and maintaining MVIC force over the required (4 s) duration. For ITT contractions, the quadriceps femoris was electrically stimulated using two surface electrodes (7 × 12.7 cm Dura-stick II, Chattanooga Group, Hixson, TN, USA,), which were positioned on the anterolateral side of the thigh, on the belly of the rectus femoris and vastus lateralis muscles. An initial familiarization session was used to obtain maximal current (mA) tolerance and establish the supra-maximal current amplitude for superimposition during MVIC. Under resting conditions, the amperage of a 240 V square-wave pulse (100 μs, 1 Hz; Digimeter, DS7, Hertfordshire, UK) was progressively increased until further increases in intensity caused no further gain in twitch force [29]. Familiarization sessions were conducted until MVIC and RFD values reached a plateau, and the average voluntary muscle activation during the ITT was greater than 80%. N.B. ITT was used solely to determine the voluntary activation capacity and not to estimate the force of a maximally activated muscle. Force data were acquired (200 Hz) using a force transducer (KAP, Bienfait B.V. Haarlem, The Netherlands) and analyzed with commercially available software (AcqKnowledge III; Biopac Systems Inc, Aero Camino Goleta, CA, USA), which is consistent with previous work [10].
To measure quadriceps femoris peak RFD, participants performed maximum velocity contractions as forcefully as possible without countermovement. Visual data presentation was used to provide real-time ‘biofeedback’ and ensure no countermovement preceded each kick. Ten kicks were performed interspersed by 30 s rest periods (Figure 1). The peak RFD was defined as the highest positive value from the first derivative of the force signal (i.e., the greatest slope of the force–time curve). The 3 efforts of greatest force and no discernible counter-movement were selected for analysis [30].
A subset of 10 participants underwent a percutaneous needle biopsy of their right quadriceps (vastus lateralis) that was completed within 5 min of cessation of the muscle contraction protocol (Figure 1). Biopsies were performed under local anaesthesia (0.5% Marcaine) using a Bard Monopty 12-gauge × 10 cm length disposable core biopsy instrument (Bard Biopsy System, Tempe, AZ, USA). A 10 mm longitudinal stab incision was made, and the muscle fascia was pierced to enable the biopsy needle to be passed through the incision. Muscle samples (approximately 40 mg) were immediately frozen in liquid nitrogen and stored at −80 °C until further analysis.
Muscle samples were ground under liquid nitrogen using a pestle and mortar. Muscle powders were accurately weighed and homogenized on ice in 10 volumes of 1% (v/v) Triton X-100, 40 mM Tris pH 7.4, including PhosSTOPTM phosphatase inhibitor and cOmpleteTM protease inhibitor cocktail (Roche, Indianapolis, USA) using a glass–Teflon homogenizer. Homogenates were centrifuged at 1000× g, 4 °C for 5 min and supernatants containing soluble proteins were decanted and stored on ice. The pellet, which contains the majority of myofibrillar proteins, was resuspended in homogenization buffer and centrifuged at 1000× g, 4 °C for 5 min. The washed myofibrillar pellet was solubilized in Lysis buffer (7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 30 mM Tris pH 8.5) and cleared by centrifugation at 12,000× g, 4 °C for 45 min. The protein concentrations of each myofibrillar and soluble protein fraction were measured using the Bradford assay (Sigma-Aldrich, Poole, Dorset, UK), and samples were normalized to 5 μg/μL in Lysis buffer.
Top–down analysis of muscle protein modifications was performed by 2DGE as described previously [31]. Myofibrillar samples were resolved on immobilized pH gradient (IPG) strips (GE Healthcare, Little Chalfont, UK) by isoelectric focusing. The protein load was 250 μg and 50 μg per gel for broad-range (pH 3–11) and narrow-range (pH 4–7) gels, respectively. Second-dimension denaturing electrophoresis was performed through linear 11% poly-acrylamide gels. Proteins were stained with colloidal Coomassie blue (Bio-Rad Laboratories, Hercules, CA, USA). Digitized gel images (600 dpi, 16-bit grayscale) of samples taken from each participant in morning and evening conditions (n = 10 participants; total of 20 gels) were normalized by an inter-sample abundance ratio calculated from ‘non-changing’ features and between group differences were analyzed using SameSpots (TotalLab, Newcastle upon Tyne, UK), which is consistent with previous work [32].
Proteins were identified by reverse-phase liquid chromatography (LC)–electrospray ionization (ESI)–tandem mass spectrometry (MS/MS) of tryptic digests, as reported in [33]. Briefly, gel spots were excised from analytical gels (n = 20 biological replicates) or preparative gels of pooled samples and tryptic in-gel digestion was performed using an Xcise robot (Proteome Systems, North Ryde, Australia). LC-ESI-MS/MS was performed using a quadrupole-high capacity ion-trap mass spectrometer (HCT Ultra ETD II; Bruker Daltonics, Billerica, MA, USA) coupled via an ESI source to a nano-flow LC system (Ultimate 3000; Thermo Scientific, Waltham, MA, USA). Data-dependent MS/MS analysis in alternating collision-induced dissociation (CID) and electron-transfer dissociation (ETD) modes was performed, selecting the two most abundant precursor ions with active exclusion (30 s) enabled. Raw data were processed (Data Analysis 4.0, Bruker Daltonics, Billerica, MA, USA) and error tolerant searches of Mascot generic format files were performed against the UniProt database, which was restricted to Human using a locally implemented Mascot (www.matrixscience.com) server (version 2.2.03). The enzyme specificity was trypsin allowing 1 missed cleavage, carbamidomethyl modification of cysteine (fixed), oxidation of methionine (variable), and an m/z error of ± 0.5 Da.
The tryptic digestion of soluble muscle proteins was performed using the filter-aided sample preparation (FASP) method. Aliquots containing 400 µg protein were washed with 200 µL of UA buffer (8 M urea, 100 mM Tris, pH 8.5). Proteins were incubated at 37 °C for 15 min in UA buffer containing 100 mM dithiothreitol followed by incubation (20 min at 4 °C) protected from light in UA buffer containing 50 mM iodoacetamide. UA buffer was exchanged for 50 mM ammonium bicarbonate, and sequencing-grade trypsin (Promega, Madison, WI, USA) was added at an enzyme to protein ratio of 1:50. Digestion was allowed to proceed at 37 °C overnight; then, peptides were collected in 100 μL 50 mM ammonium bicarbonate containing 0.2% trifluoroacetic acid. Peptide solutions were desalted using disposable Toptip C18 spin columns (Glygen Corp., Columbia, MD, USA) and vacuum centrifuged to dryness. Phosphopeptides were selectively enriched by binding to titanium dioxide (TiO2)-spin tips (Pierce), which was consistent with previous work [34]. Enriched phosphopeptide solutions were immediately adjusted to pH 4 with formic acid, vacuum centrifuged to dryness, and then resuspended in 0.1% formic acid (FA)prior to LC-ESI-MS/MS analysis.
The LC-ESI-MS/MS analysis of phosphopeptide-enriched samples was performed using an Ultimate 3000 RSLCTM nano system (Thermo Scientific, Waltham, MA, USA) coupled to a QExactiveTM mass spectrometer (Thermo Scientific, Waltham, MA, USA). Samples (5 μL each) were loaded by partial loop injection onto the trapping column (Thermo Scientific, PepMap100, C18, 75 μm × 20 mm) at a flow rate of 4 μL/min with 0.1% (v/v) trifluoroacetic acid (TFA) for 7 min. Samples were resolved on the analytical column (Easy-Spray C18 75 μm × 500 mm 2 μm column) using a gradient of 97% A (0.1% FA) 3% B (99.9% ACN 0.1% FA) to 60% A 40% B over 90 min at a flow rate of 300 nL/min. The data-dependent program used for data acquisition consisted of a 70,000 resolution full-scan MS scan (AGC set to 106 ions with a maximum fill time of 250 ms); the 10 most abundant peaks were selected for MS/MS using a 17,000 resolution scan (AGC set to 5 × 104 ions with a maximum fill time of 250 ms) with an ion selection window of 3 m/z and a normalized collision energy of 30. To avoid the repeated selection of peptides, the MS/MS program used a 30 s dynamic exclusion window.
MS spectra were aligned using the Progenesis QI for Proteomics (Nonlinear Dynamics, Newcastle, UK). Prominent ion features were used as vectors to match each dataset to a common reference chromatogram. An analysis window of 9–99 min and 200–2000 m/z was selected encompassing charge states of ≤8. Peak lists were searched against the UniProt database and restricted to Human using a locally implemented Mascot (www.matrixscience.com) server (version 2.2.03). The enzyme specificity was trypsin, allowing one missed cleavage, fixed carbamidomethyl of cysteine, and variable oxidation of methionine and phosphorylation of serine (S), threonine (T), and tyrosine (Y). The instrument type was ESI-FTICR, and mass tolerances were 10 ppm for peptides and 0.01 Da for fragment ions. The Mascot output (xml format), which was restricted to non-homologous protein identifications (false discovery rate < 1%), was recombined with MS profile data in Progenesis QI for Proteomics, and peptide abundance data were normalized by the inter-sample abundance ratio calculated from ‘non-changing’ features. Peptide features with molecular weight search score (MOWSE) scores below the identity threshold (Mud-PIT scoring) were excluded.
Data were analyzed in R version 3.5.2 and are presented as mean and standard deviation (SD) unless otherwise stated. Morning and evening MVIC force and peak RFD were compared with paired t-tests. Statistical analysis of proteomic data was conducted by within-subject one-way analysis of variance. Log transformed spot volumes or MS peptide data were normalized by inter-sample abundance ratio and used to investigate within-subject differences between morning and evening conditions. To determine the false discovery rate (FDR), p-value distributions were used to calculate q-values [35]. Functional annotation of proteomic data was conducted using the Search Tool for the Retrieval of Interacting Genes/Proteins [STRING; [36].
3. Results
3.1. Reliability and Diurnal Variation Data: MVIC, % Activation, RFD, and Subjects Rated Maximal Effort in n = 20 Subjects
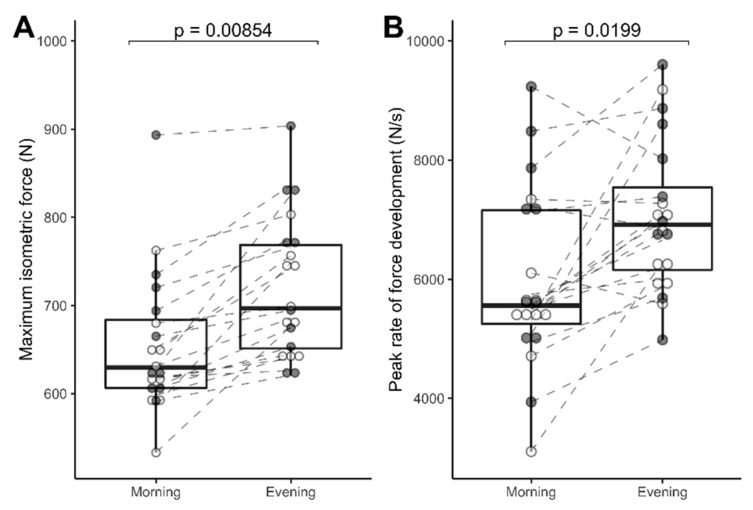
The intra-class correlation coefficient between MVIC force during the 4th and 5th familiarization sessions was 0.96 (95% CI, 0.91–0.99). MVIC force in the evening was 10.2% greater than in the morning (721 ± 80 versus 654 ± 79 N, p = 0.00854; Figure 2A). The intra-class correlation coefficient between peak RFD data collected during the 4th and 5th familiarization sessions was 0.45 (95% CI, 0.03–0.74). Peak RFD in the evening was 18% greater (7052 ± 1261 versus 5972 ± 1501 N/s, p = 0.0199; Figure 2B) than in the morning. The level of voluntary activation in the morning 82.8 ± 7.8% did not differ (p = 0.061) from the evening 86.5 ± 6.4%, and there were no significant difference in the rating of perceived effort. In all cases, participants rated 10/10 for effort regardless of whether the session was conducted in the morning or evening.
Figure 2.
Diurnal differences in muscle force. Maximum voluntary isometric contraction (MVIC) force (A) and peak rate of force development; (B) of the quadriceps femoris muscle in the morning (08:00 h) and evening (18:00 h). Shaded (gray) data points represent data from n = 10 participants that gave muscle biopsy samples, whereas open (white) data points represent data from n = 10 participants that did not undergo biopsy procedures. Within-subject data are connected by dashed lines and group data (median, quartile ranges) are summarized by ‘box and whisker’ plots. Statistical differences were assessed by Student’s paired t-test, n = 20.
3.2. Data Handling: 10 Subjects from the Cohort of 20 Volunteered for Muscle Biopsy Procedures; Hence, Only Their Data (Temperature, MVIC, and RFD) Are Reported below to Align with the Proteomic Analyses
3.2.1. Resting Rectal, Skin, and Muscle Temperatures (n = 10)
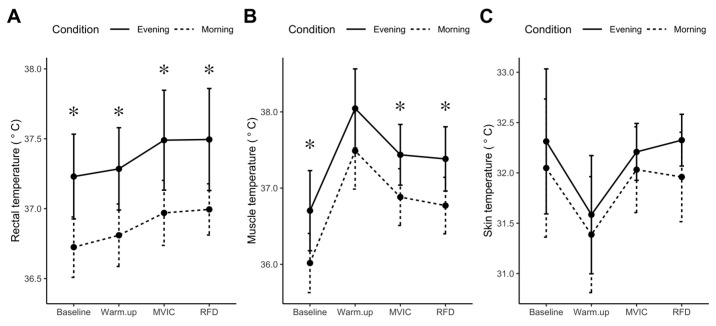
At rest, Trec was greater in the evening (mean difference = 0.50 °C, p = 0.001, 95% CI: 0.27–0.74 °C) than morning (Figure 3A). Tm exhibited a similar diurnal variation (mean difference = 0.69 °C, p = 0.019, 95% CI: 0.14–1.23 °C; Figure 3B), whereas, Tsk exhibited no significant diurnal variation (mean difference = 0.26 °C, p = 0.377; Figure 3C). No significant interactions were found between temperature responses and time of day. The profiles of the temperature measurements (Trec, Tm, and Tsk) rose and fell in parallel over the three time points in both morning and evening conditions.
Figure 3.
Diurnal differences in core, muscle, and skin temperature. Core (rectal) temperature (A), muscle temperature (B), and skin temperature (C), were recorded during the last 5 min of baseline and immediately after cycle ergometry warm-up, maximal voluntary isometric contraction (MVIC), or peak rate of force development (RFD) protocols. Data are presented as mean, standard deviation (n = 10), experimental condition is identified by morning (dashed line) and evening (solid line). * p < 0.05 statistical difference between morning and evening condition.
3.2.2. MVIC, % Activation, Peak RFD, and Subject-Rated Maximal Effort (n = 10)
MVIC force in the evening was 9.2% greater (738 ± 98 versus 676 ± 92 N, p = 0.007) than in the morning (Figure 2A, closed/shaded data points). Peak RFD (7364 ± 1440 versus 6514 ± 1724 N/s, p = 0.064) was 13% greater in the evening (Figure 2B, closed/shaded data points). The level of voluntary activation for MVIC in the morning 79.6% did not differ (p = 0.056) from the evening 85.2%, and there were no significant differences in the rating of perceived effort.
3.2.3. Proteomic Data
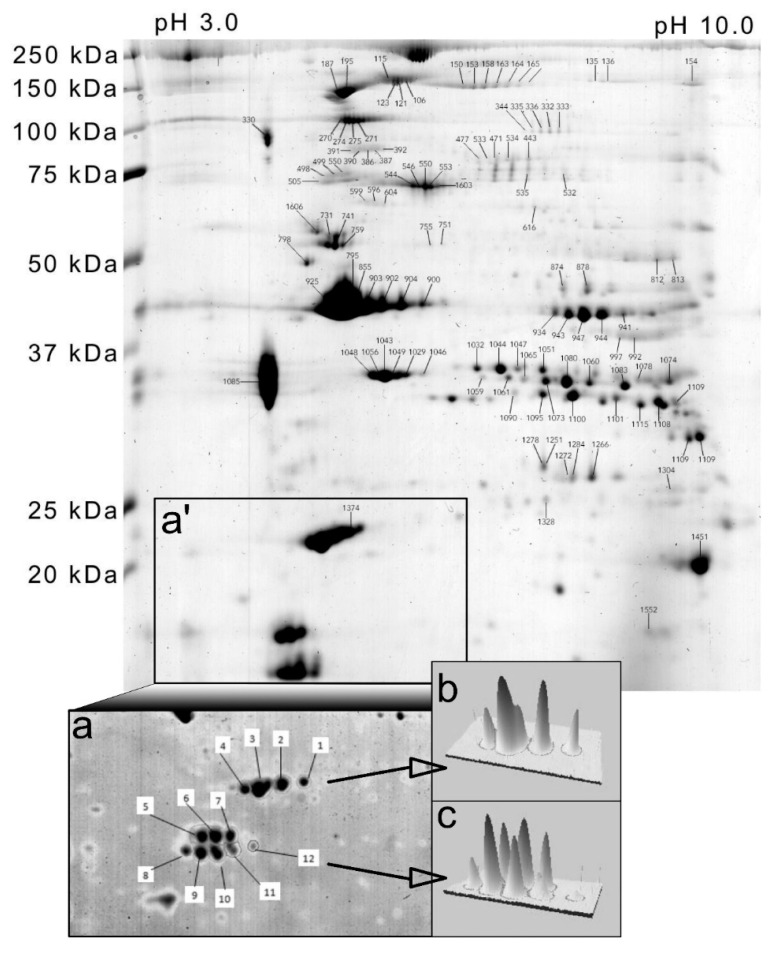
The analysis of human myofibrillar proteins by narrow range (pH 4–7) 2DGE (Figure 4 insets) was used to investigate differences in the relative abundance of proteoforms of essential and regulatory myosin light chains. Essential myosin light chains (MYLC1/3) were resolved as 4 proteoforms, slow-twitch/ventricular regulatory myosin light chains (MLRV) were resolved as 3 proteoforms, and fast-twitch/skeletal muscle myosin regulatory light chains (MLRS) were resolved as 4 proteoforms. There were no statistically significant differences in myosin light chain proteoform abundance between morning and evening conditions (Table 1).
Figure 4.
Analysis of myofibrillar proteoforms by two-dimensional gel electrophoresis. Gel map of human myofibrillar proteins annotated with spot numbers that correspond to protein identities in Table 1 and Table 2. Subpanels illustrate the region of interest (a’) and higher-resolution separation of region (a’) by narrow-range (pH 4–7) isoelectric focusing (a) to resolve myosin light chain isoforms. Panels (b,c) illustrate three-dimensional representations of essential myosin light chains (MyLC1/3) and regulatory myosin light chains MLRV and MLRS spots, respectively.
Table 1.
Myosin light chain proteoform abundances.
| UniProt ID | Description | Spot # | Morning | Evening | p |
|---|---|---|---|---|---|
| MYL1/3 | Essential myosin light chain 1/3 | 1 | 924 (498) | 785 (612) | 0.727 |
| 2 | 4426 (1504) | 3968 (1984) | 0.884 | ||
| 3 | 10,942 (3173) | 9788 (4894) | 0.485 | ||
| 4 | 2378 (808) | 2192 (1293) | 0.431 | ||
| MLRV | Regulatory myosin light chain, slow/ventricular | 5 | 2675 (1524) | 2985 (2179) | 0.498 |
| 6 | 4176 (1837) | 3699 (2182) | 0.459 | ||
| 7 | 1940 (950) | 1600 (10,089) | 0.389 | ||
| MLRS | Regulatory myosin light chain, fast/skeletal | 8 | 1557 (1027) | 2206 (1875) | 0.216 |
| 9 | 4675 (1589) | 5214 (3128) | 0.151 | ||
| 10 | 4420 (1149) | 3762 (1768) | 0.467 | ||
| 11 | 1286 (540) | 982 (569) | 0.426 | ||
| HSPB6 | Heat shock protein 20 | 12 | 282 (214) | 0.00 (0.00) | - |
Spot # correspond with the 2D gel map (Figure 5 inset a). UniProt identifier and protein description based on peptide mass spectrometry of in-gel protein digests. Normalized spot abundances (AU) in the morning and evening are presented as mean (SD) and statistical difference (p) was determined by within-subject ANOVA (n = 10).
A broader range (pH 3–11) of 2DGE resolved 122 protein spots (Figure 4 and Table 2). In total, 40 non-redundant proteins were identified, i.e., the majority of proteins were detected in multiple gel spots that represent different proteoforms. The abundance of 6 proteoforms was statistically (p < 0.05) different between morning and evening conditions (Table 2). In the evening, greater abundances were observed in myosin binding protein C1 (80%), glycogen phosphorylase (43%), and beta enolase (24%), whereas the abundances of myosin binding protein H (−18%), nebulin (−48%), and troponin T slow (−22%) were less in the evening compared to the morning. The greatest difference was in the abundance of the slow isoform of myosin binding protein C (MYBPC1), which was resolved as a series of 6 proteoforms. Therefore, the difference in abundance of spot # 164 reported here may represent a change in post-translational modification. LC-ESI-MS/MS analysis of all MYBPC1 proteoforms (spot numbers: 150, 153, 158, 163, 164, and 165) did not unambiguously identify site-specific modifications that were unique to a particular MYBPC1 proteoform. The phosphorylation of MYBPC1 S178 was detected in two MYBPC1 proteoforms (spots 153 and 158), and therefore, it was not specifically related to time-of-day differences in the pattern of MYBPC1 proteoforms.
Table 2.
Diurnal differences in myofibrillar proteoform abundance.
| Spot # | UniProt ID | Protein Name | MOWSE | % Seq | Anova (p) | FC | Morning Mean | Morning SD | Evening Mean | Evening SD |
|---|---|---|---|---|---|---|---|---|---|---|
| 335 | PYGM | Glycogen phosphorylase, muscle form | 1938 | 53 | 0.0155 | 1.43 | 245 | 44 | 350 | 104 |
| 163 | MYPC1 | Myosin-binding protein C, slow-type | 2135 | 42 | 0.0226 | 1.80 | 343 | 86 | 617 | 357 |
| 703 | MYBPH | Myosin-binding protein H | 425 | 23 | 0.0313 | 1.17 | 133 | 28 | 114 | 26 |
| 874 | ENOB | Beta-enolase | 1040 | 55 | 0.0335 | 1.24 | 814 | 199 | 1007 | 174 |
| 1046 | TNNT1 | Troponin T, slow skeletal muscle | 738 | 32 | 0.0433 | 1.22 | 239 | 71 | 196 | 45 |
| 154 | NEBU | Nebulin | 964 | 6 | 0.0435 | 1.48 | 959 | 352 | 648 | 252 |
| 391 | MYH2 | Myosin-2 | 1510 | 18 | 0.0546 | 1.37 | 239 | 56 | 329 | 158 |
| 881 | KCRM | Creatine kinase M-type | 922 | 54 | 0.0570 | 1.17 | 535 | 74 | 625 | 118 |
| 795 | ACTS | Actin, alpha skeletal muscle | 1011 | 54 | 0.0620 | 1.47 | 271 | 114 | 185 | 56 |
| 742 | ACTN2 | Alpha-actinin-2 | 80 | 4 | 0.0727 | 1.25 | 510 | 102 | 408 | 134 |
| 1059 | TNNT3 | Troponin T, fast skeletal muscle | 394 | 24 | 0.0735 | 1.32 | 244 | 84 | 186 | 54 |
| 992 | ALDOA | Fructose-bisphosphate aldolase A | 733 | 58 | 0.0929 | 1.19 | 472 | 126 | 559 | 138 |
| 903 | ACTS | Actin, alpha skeletal muscle | 1009 | 62 | 0.0931 | 1.67 | 16,872 | 10,694 | 10,109 | 2314 |
| 135 | MYPC2 | Myosin-binding protein C, fast-type | 2015 | 45 | 0.0978 | 1.23 | 168 | 54 | 137 | 41 |
| 1357 | KCRM | Creatine kinase M-type | 466 | 32 | 0.0978 | 1.43 | 2542 | 1135 | 1782 | 577 |
| 686 | NEBU | Nebulin | 2181 | 13 | 0.1017 | 1.17 | 336 | 66 | 394 | 69 |
| 386 | MYH2 | Myosin-2 | 1266 | 13 | 0.1048 | 1.27 | 233 | 78 | 295 | 104 |
| 739 | DESM | Desmin | 1884 | 63 | 0.1072 | 1.20 | 1176 | 181 | 979 | 292 |
| 1043 | TNNT1 | Troponin T, slow skeletal muscle | 599 | 33 | 0.1143 | 1.28 | 884 | 293 | 689 | 278 |
| 596 | MYH7 | Myosin-7 | 1229 | 17 | 0.1321 | 1.15 | 259 | 66 | 298 | 49 |
| 392 | MYH2 | Myosin-2 | 709 | 12 | 0.1328 | 1.17 | 109 | 44 | 128 | 36 |
| 390 | MYH2 | Myosin-2 | 1367 | 14 | 0.1481 | 1.28 | 177 | 40 | 227 | 89 |
| 855 | ACTS | Actin, alpha skeletal muscle | 1319 | 65 | 0.1531 | 1.25 | 18,032 | 6216 | 14,461 | 4631 |
| 158 | MYPC1 | Myosin-binding protein C, slow-type | 2460 | 49 | 0.1606 | 1.32 | 547 | 224 | 721 | 297 |
| 387 | MYH2 | Myosin-2 | 915 | 12 | 0.1636 | 1.24 | 173 | 85 | 214 | 71 |
| 685 | KPYM | Pyruvate kinase isozymes M1/M2 | 1329 | 51 | 0.1662 | 1.17 | 398 | 68 | 466 | 112 |
| 731 | DESM | Desmin | 2161 | 72 | 0.1818 | 1.16 | 2919 | 408 | 2527 | 728 |
| 153 | MYPC1 | Myosin-binding protein C, slow-type | 2648 | 51 | 0.1857 | 1.34 | 518 | 218 | 694 | 314 |
| 344 | PYGM | Glycogen phosphorylase, muscle form | 1560 | 43 | 0.1876 | 1.53 | 198 | 48 | 304 | 242 |
| 741 | DESM | Desmin | 1927 | 67 | 0.1927 | 1.17 | 3290 | 778 | 2811 | 997 |
| 544 | ALBU | Serum albumin | 1539 | 56 | 0.2047 | 1.25 | 682 | 203 | 852 | 348 |
| 389 | TNNT1 | Troponin T, slow skeletal muscle | 153 | 23 | 0.2070 | 1.16 | 558 | 103 | 645 | 155 |
| 1065 | TNNT3 | Troponin T, fast skeletal muscle | 421 | 26 | 0.2103 | 1.18 | 542 | 124 | 460 | 117 |
| 164 | MYPC1 | Myosin-binding protein C, slow-type | 2581 | 51 | 0.2105 | 1.25 | 337 | 57 | 423 | 190 |
| 751 | TBA4A | Tubulin alpha-4A | 467 | 29 | 0.2232 | 1.14 | 312 | 60 | 274 | 43 |
| 997 | ALDOA | Fructose-bisphosphate aldolase A | 812 | 60 | 0.2234 | 1.21 | 652 | 188 | 789 | 289 |
| 546 | ALBU | Serum albumin | 1622 | 59 | 0.2244 | 1.34 | 1381 | 468 | 1850 | 766 |
| 812 | ATPA | ATP synthase subunit alpha, mitochondrial | 1275 | 45 | 0.2274 | 1.15 | 688 | 164 | 790 | 219 |
| 150 | MYPC1 | Myosin-binding protein C, slow-type | 2849 | 50 | 0.2300 | 1.30 | 442 | 236 | 575 | 280 |
| 813 | ATPA | ATP synthase subunit alpha, mitochondrial | 963 | 42 | 0.2328 | 1.10 | 755 | 167 | 829 | 176 |
| 1606 | VIME | Vimentin | 1861 | 69 | 0.2342 | 1.35 | 707 | 339 | 955 | 389 |
| 1105 | TPM2 | Tropomyosin beta chain | 567 | 32 | 0.2501 | 1.19 | 454 | 120 | 381 | 80 |
| 1251 | CAH3 | Carbonic anhydrase 3 | 419 | 44 | 0.2622 | 1.14 | 829 | 240 | 944 | 230 |
| 878 | ENOB | Beta-enolase | 1196 | 53 | 0.2624 | 1.19 | 1109 | 283 | 1325 | 358 |
| 616 | CATA | Catalase | 710 | 38 | 0.2974 | 1.77 | 832 | 103 | 1476 | 1963 |
| 941 | KCRM | Creatine kinase M-type | 890 | 54 | 0.3013 | 1.14 | 1295 | 299 | 1480 | 439 |
| 947 | KCRM | Creatine kinase M-type | 1295 | 57 | 0.3075 | 1.14 | 6801 | 1461 | 7786 | 1689 |
| 1057 | ACTC | Actin, alpha cardiac muscle 1 | 52 | 11 | 0.3596 | 1.16 | 329 | 118 | 383 | 185 |
| 505 | HSP7C | Heat shock cognate 71 kDa protein | 1246 | 39 | 0.3731 | 1.19 | 1386 | 475 | 1643 | 850 |
| 500 | HSP71 | Heat shock 70 kDa protein 1A/1B | 1470 | 46 | 0.3816 | 1.02 | 583 | 99 | 570 | 136 |
| 1085 | TPM3 | Tropomyosin alpha-3 chain | 1478 | 72 | 0.3846 | 1.10 | 37,171 | 11,538 | 33,691 | 9083 |
| 1603 | ALBU | Serum albumin | 1625 | 58 | 0.3869 | 1.22 | 714 | 404 | 867 | 335 |
| 1097 | TNNT1 | Troponin T, slow skeletal muscle | 576 | 32 | 0.3914 | 1.20 | 1448 | 656 | 1207 | 609 |
| 553 | ALBU | Serum albumin | 1838 | 60 | 0.3938 | 1.29 | 4881 | 2765 | 6293 | 3338 |
| 902 | ACTS | Actin, alpha skeletal muscle | 962 | 63 | 0.4326 | 1.25 | 8992 | 5539 | 7196 | 2076 |
| 1374 | MYL3 | Myosin light chain 3 | 856 | 77 | 0.4537 | 1.10 | 8098 | 3473 | 7382 | 2866 |
| 1078 | MYOZ1 | Myozenin-1 | 848 | 85 | 0.4556 | 1.00 | 401 | 76 | 402 | 239 |
| 1336 | TPIS | Triosephosphate isomerase | 929 | 76 | 0.4585 | 1.04 | 316 | 48 | 304 | 100 |
| 1103 | KCRM | Creatine kinase M-type | 418 | 29 | 0.4598 | 1.40 | 623 | 607 | 446 | 123 |
| 944 | KCRM | Creatine kinase M-type | 1100 | 53 | 0.4644 | 1.12 | 3853 | 690 | 4310 | 1195 |
| 900 | ACTS | Actin, alpha skeletal muscle | 415 | 39 | 0.4649 | 1.21 | 3869 | 2169 | 3198 | 713 |
| 1272 | MYOZ1 | Myozenin-1 | 154 | 29 | 0.4785 | 1.05 | 355 | 88 | 373 | 47 |
| 532 | PGM1 | Phosphoglucomutase-1 | 876 | 37 | 0.4889 | 1.09 | 446 | 125 | 409 | 80 |
| 106 | MYH2 | Myosin-2 | 3726 | 33 | 0.4924 | 1.59 | 550 | 340 | 874 | 888 |
| 1090 | TNNT3 | Troponin T, fast skeletal muscle | 520 | 32 | 0.5034 | 1.12 | 591 | 178 | 528 | 89 |
| 333 | PYGM | Glycogen phosphorylase, muscle form | 1611 | 47 | 0.5039 | 1.21 | 164 | 33 | 198 | 95 |
| 1503 | KCRM | Creatine kinase M-type | 151 | 13 | 0.5085 | 1.06 | 1063 | 345 | 1007 | 590 |
| 1032 | TNNT3 | Troponin T, fast skeletal muscle | 491 | 27 | 0.5192 | 1.23 | 1032 | 534 | 837 | 258 |
| 943 | KCRM | Creatine kinase M-type | 1126 | 59 | 0.5267 | 1.06 | 2690 | 574 | 2846 | 551 |
| 1056 | TNNT1 | Troponin T, slow skeletal muscle | 687 | 37 | 0.5340 | 1.13 | 8515 | 4319 | 7506 | 4221 |
| 1049 | TNNT1 | Troponin T, slow skeletal muscle | 652 | 36 | 0.5550 | 1.10 | 1792 | 601 | 1630 | 818 |
| 165 | MYPC1 | Myosin-binding protein C, slow-type | 2366 | 51 | 0.5618 | 1.14 | 323 | 69 | 368 | 161 |
| 1266 | TPIS | Triosephosphate isomerase | 932 | 65 | 0.5624 | 1.07 | 1389 | 280 | 1489 | 354 |
| 1132 | MYH2 | Myosin-2 | 616 | 8 | 0.5679 | 1.05 | 1154 | 324 | 1209 | 255 |
| 604 | MYH2 | Myosin-2 | 1104 | 13 | 0.5805 | 1.05 | 199 | 43 | 189 | 41 |
| 121 | MYH2 | Myosin-2 | 4044 | 37 | 0.5910 | 1.51 | 691 | 599 | 1041 | 1196 |
| 455 | KPYM | Pyruvate kinase isozymes M1/M2 | 815 | 36 | 0.6060 | 1.06 | 856 | 158 | 810 | 117 |
| 550 | ALBU | Serum albumin | 1852 | 65 | 0.6109 | 1.23 | 2514 | 1360 | 3093 | 1897 |
| 1074 | G3P | Glyceraldehyde-3-phosphate dehydrogenase | 1068 | 54 | 0.6170 | 1.03 | 3635 | 771 | 3532 | 1105 |
| 925 | ACTS | Actin, alpha skeletal muscle | 1277 | 63 | 0.6537 | 1.05 | 73,798 | 17,469 | 77,544 | 17,257 |
| 123 | MYH2 | Myosin-2 | 3325 | 34 | 0.6580 | 1.31 | 819 | 851 | 1072 | 1290 |
| 798 | ATPB | ATP synthase subunit beta, mitochondrial | 1882 | 68 | 0.6678 | 1.13 | 947 | 356 | 1069 | 547 |
| 1083 | TNNT3 | Troponin T, fast skeletal muscle | 583 | 34 | 0.6867 | 1.12 | 2406 | 775 | 2689 | 1066 |
| 136 | MYPC2 | Myosin-binding protein C, fast-type | 1838 | 41 | 0.6868 | 1.07 | 223 | 72 | 207 | 45 |
| 1073 | TNNT3 | Troponin T, fast skeletal muscle | 482 | 26 | 0.6887 | 1.09 | 1620 | 579 | 1492 | 603 |
| 1048 | TNNT1 | Troponin T, slow skeletal muscle | 738 | 32 | 0.6912 | 1.25 | 3355 | 2869 | 2681 | 1052 |
| 471 | MYH2 | Myosin-2 | 1204 | 15 | 0.6940 | 1.09 | 202 | 91 | 186 | 98 |
| 934 | KCRM | Creatine kinase M-type | 1134 | 54 | 0.6981 | 1.03 | 1253 | 233 | 1222 | 210 |
| 1115 | MYOZ1 | Myozenin-1 | 378 | 30 | 0.7011 | 1.05 | 1462 | 416 | 1393 | 415 |
| 904 | ACTS | Actin, alpha skeletal muscle | 877 | 55 | 0.7143 | 1.01 | 9431 | 1361 | 9383 | 3034 |
| 443 | MYH2 | Myosin-2 | 978 | 13 | 0.7267 | 1.05 | 331 | 103 | 348 | 96 |
| 1381 | KCRM | Creatine kinase M-type | 469 | 28 | 0.7267 | 1.08 | 24,048 | 9478 | 22,334 | 6612 |
| 115 | MYH7 | Myosin-7 | 4288 | 35 | 0.7551 | 1.14 | 2646 | 2960 | 3028 | 3141 |
| 1061 | TNNT3 | Troponin T, fast skeletal muscle | 457 | 36 | 0.7595 | 1.28 | 540 | 382 | 424 | 155 |
| 1044 | ACTC | Actin, alpha cardiac muscle 1 | 342 | 28 | 0.7668 | 1.14 | 2912 | 1480 | 2552 | 917 |
| 1080 | ACTC | Actin, alpha cardiac muscle 1 | 183 | 23 | 0.7707 | 1.06 | 4741 | 1932 | 4479 | 2310 |
| 270 | ACTN2 | Alpha-actinin-2 | 3558 | 69 | 0.7779 | 1.08 | 2145 | 422 | 2326 | 812 |
| 271 | ACTN2 | Alpha-actinin-2 | 3354 | 71 | 0.7866 | 1.04 | 679 | 243 | 706 | 256 |
| 274 | ACTN2 | Alpha-actinin-2 | 3445 | 69 | 0.7899 | 1.07 | 1058 | 239 | 1128 | 390 |
| 187 | MYH7 | Myosin-7 | 3534 | 31 | 0.7939 | 1.55 | 6318 | 8061 | 4082 | 3064 |
| 1051 | TNNT3 | Troponin T, fast skeletal muscle | 507 | 36 | 0.8007 | 1.11 | 1454 | 657 | 1312 | 446 |
| 1108 | MYOZ1 | Myozenin-1 | 708 | 50 | 0.8010 | 1.02 | 3435 | 1569 | 3513 | 1097 |
| 332 | PYGM | Glycogen phosphorylase, muscle form | 1651 | 44 | 0.8203 | 1.01 | 295 | 50 | 299 | 93 |
| 330 | TPM1 | Tropomyosin alpha-1 chain | 947 | 47 | 0.8216 | 1.07 | 1964 | 874 | 1834 | 716 |
| 1100 | TNNT3 | Troponin T, fast skeletal muscle | 474 | 37 | 0.8243 | 1.12 | 4641 | 2707 | 4144 | 1391 |
| 1095 | TNNT3 | Troponin T, fast skeletal muscle | 452 | 28 | 0.8373 | 1.09 | 1477 | 719 | 1355 | 374 |
| 499 | HSP71 | Heat shock 70 kDa protein 1A/1B | 1070 | 39 | 0.8419 | 1.02 | 516 | 114 | 508 | 118 |
| 980 | TNNT1 | Troponin T, slow skeletal muscle | 696 | 35 | 0.8427 | 1.01 | 1131 | 271 | 1146 | 265 |
| 1198 | LDB3 | LIM domain-binding protein 3 | 609 | 17 | 0.8445 | 1.01 | 2437 | 827 | 2415 | 1038 |
| 899 | DESM | Desmin | 198 | 11 | 0.8495 | 1.08 | 1329 | 574 | 1230 | 235 |
| 1129 | VDAC1 | Voltage-dependent anion-selective channel protein 1 | 612 | 59 | 0.8774 | 1.10 | 383 | 206 | 347 | 82 |
| 1552 | HERC1 | Probable E3 ubiquitin-protein ligase HERC1 | 52 | 2 | 0.8852 | 1.06 | 1557 | 547 | 1652 | 1207 |
| 1101 | TNNT3 | Troponin T, fast skeletal muscle | 540 | 33 | 0.9155 | 1.01 | 1068 | 440 | 1077 | 383 |
| 477 | MYH2 | Myosin-2 | 2036 | 26 | 0.9173 | 1.08 | 564 | 398 | 609 | 497 |
| 533 | MYH2 | Myosin-2 | 1084 | 13 | 0.9250 | 1.27 | 365 | 409 | 289 | 165 |
| 1397 | ENOB | Beta-enolase | 266 | 26 | 0.9287 | 1.02 | 12,335 | 3415 | 12,540 | 3761 |
| 535 | MYH2 | Myosin-2 | 1195 | 17 | 0.9394 | 1.23 | 456 | 482 | 370 | 163 |
| 1451 | TNNI2 | Troponin I, fast skeletal muscle | 378 | 50 | 0.9545 | 1.02 | 229 | 127 | 225 | 130 |
| 479 | MYH2 | Myosin-2 | 1359 | 18 | 0.9612 | 1.04 | 543 | 385 | 563 | 481 |
| 275 | ACTN2 | Alpha-actinin-2 | 3512 | 71 | 0.9648 | 1.04 | 925 | 221 | 965 | 355 |
| 534 | MYH2 | Myosin-2 | 1360 | 16 | 0.9683 | 1.14 | 588 | 587 | 515 | 327 |
| 336 | PYGM | Glycogen phosphorylase, muscle form | 1778 | 50 | 0.9720 | 1.02 | 310 | 96 | 305 | 69 |
| 1251 | HBB | Hemoglobin subunit beta | 441 | 60 | 0.9780 | 1.00 | 834 | 137 | 837 | 171 |
| 195 | MYH7 | Myosin-7 | 4307 | 32 | 0.9894 | 1.34 | 2446 | 3633 | 1821 | 1188 |
| 1060 | TNNT3 | Troponin T, fast skeletal muscle | 481 | 26 | 0.9915 | 1.01 | 2012 | 749 | 2001 | 801 |
| 1034 | TPM2 | Tropomyosin beta chain | 1450 | 69 | 0.9963 | 1.01 | 22,483 | 6641 | 22,339 | 5236 |
Spot # correspond with the 2D gel map (Figure 4). UniProt identifier and protein name based on peptide mass spectrometry of in-gel protein digests searched against human entries in the SwissProt database using Mascot. The probability-based molecular weight search score (MOWSE) and percentage of protein sequence identified are presented. Spot abundances (AU) in the morning and evening are presented as mean (SD) and statistical difference (P) was determined by within-subject ANOVA (n = 10). Fold change (FC) is spot abundance in the evening compared to the morning condition.
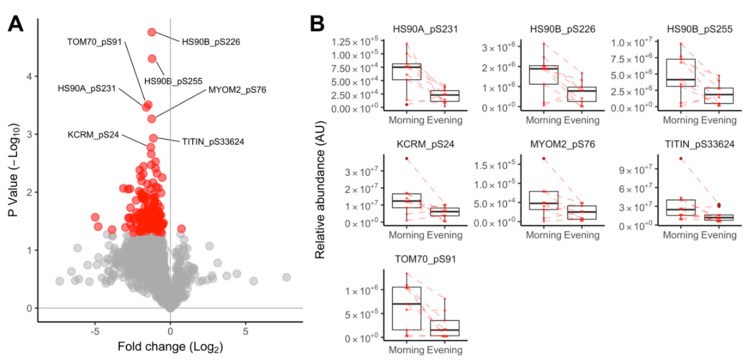
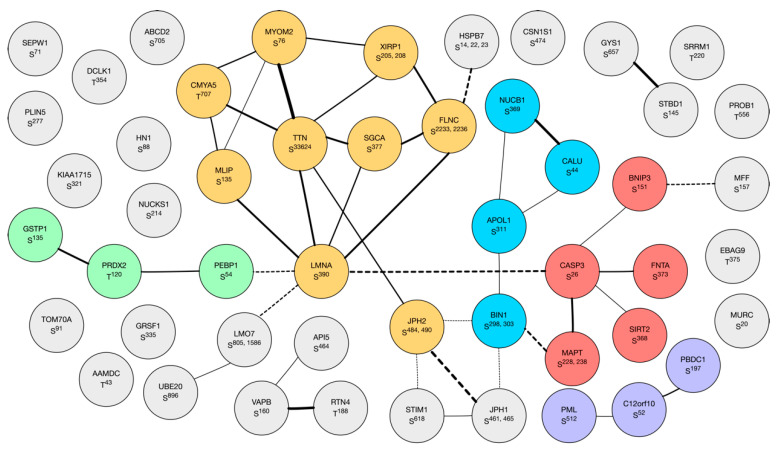
The label-free quantitation of TiO2-enriched human muscle peptides profiled 1693 phosphopeptides (721 proteins) in n = 9 paired samples (1 sample was lost during processing) taken in the morning and evening condition. Generally, the abundance of muscle phosphopeptides was greater in the morning than evening (Figure 5A). One-hundred and forty phosphopeptides from 104 proteins were statistically different (p < 0.05) at a false discovery rate (FDR) of 22% (Figure 5A), and 7 peptides had a false discovery rate of <10% (Figure 5B). Of 141 peptides at p < 0.05, 131 are reported in PhosphositePlus® (https://www.phosphosite.org), 122 have high throughput (HTP) associated papers with PubMed identifiers (PMID), 46 had low throughput (LHP) PMID associated papers, and 10 were new or have not been reported previously. Phosphopeptide abundance data including literature links are available online at 10.6084/m9.figshare.10316624 (Supplementary Material). Network analysis of 104 significant proteins using STRING (Figure 6) highlighted clusters including (i) myofibrillar proteins centering around titin and myomesin; (ii) apoptosis-related proteins centering around caspase; and (iii) redox proteins including glutathione transferase and peroxiredoxin 2.
Figure 5.
Diurnal differences in muscle protein phosphorylation. Volcano plot (A) presenting the log2 fold change in phosphopeptide abundance between evening and morning conditions and the statistical significance determined by within-subject ANOVA (n = 9 participants). Phosphopeptides that were statistically different (p < 0.05) and had a false discovery rate <10% are annotated with their unique UniProt identifier and phosphorylation site; phosphopeptide-specific data (B) illustrating within-subject data connected by dashed lines and group data (median, quartile ranges) summarized by ‘box and whisker’ plots for the statistically significant (p < 0.05, FDR < 10%) phosphopeptides.
Figure 6.
Network of phosphorylated proteins. Network of known and predicted interactions between proteins (n = 141) with phosphopeptides that exhibited statistically significant (p < 0.05) differences in phosphorylation between morning and evening conditions. The network was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), and subsets of proteins were highlighted by k-means clustering (6 groups). Node annotations represent gene names and phosphorylation sites. The width of the vectors indicate levels of evidence for protein interconnections (e.g., interaction, co-expression, or bibliometric data). Group 1 (orange) contains proteins associated with the myofibrillar apparatus and muscle cytoskeleton. Group 2 (red) is characterized by proteins involved in apoptosis (e.g., caspase 3, CASP3). Group 1 and 2 are interconnected by myc box-dependent-interacting protein 1 (BIN1; blue) which may be a key component of the muscle M-Band in addition to its role associated with t-tubules. Smaller groups are characterized by calcium-handling proteins (purple) or redox enzymes (green).
4. Discussion
We report widespread differences in the post-translational state of human muscle proteins after exercise in the morning (08:00 h) and evening (18:00 h). In particular, our data indicate differences in the phosphorylation of proteins within or close to the muscle M-band that could relate to the well-established morning versus evening differences in muscle isometric force. Our novel analysis also adds to emerging evidence regarding the daily timing of exercise and the use of exercise as a Zeitgeber for optimal muscle function [37]. The current investigation was limited to the interaction between exercise and time of day; therefore, further work is required to investigate the main effects of exercise or time of day in isolation. For example, it will be important to resolve which of the differences in protein post-translational modification are intrinsic diurnal differences and occur in the absence of the exercise stimulus.
The unsupervised network analysis of proteins that contained significant time-of-day differences in phosphorylation status (Figure 6) highlighted a cluster of myofibrillar proteins, including prominent links between titin (TTN) and myomesin 2 (MYOM2). TTN and MYOM2 were also amongst the most highly significant (p < 0.05, FDR < 10%) differences (Figure 5B) alongside muscle creatine kinase (KCRM), heat shock 90 proteins (HS90A and HS90B), and the mitochondrial importer receptor subunit (TOM70). With the exception of TOM70, a commonality shared amongst these proteins is their involvement in, or localization to, the sarcomeric M-band (Figure 7).
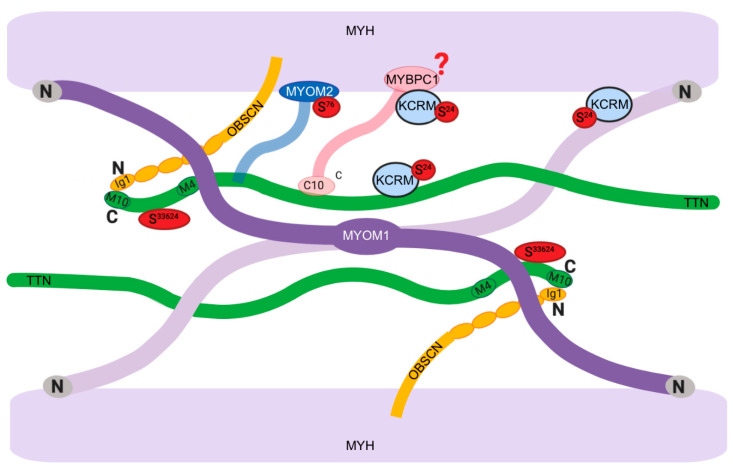
Figure 7.
Diurnal differences in M-band phosphorylation. Diagrammatic representation of M-band protein interactions redrawn from [38]. Myomesin 1 (MYOM1) and obscurin (OBSCN) are principal M-band proteins [39]. The N-terminal regions of MYOM1 interact with myosin heavy chain (MYH), titin (TTN), and muscle creatine kinase (KCRM), whereas the C-terminal regions of MYOM1 dimerize in an anti-parallel arrangement that corresponds with the M-bridges [40]. MYOM1 interacts with the C-terminal regions of TTN within the M-band. The TTN kinase domain (resides 32,178–32,432) includes the regulatory Y32341 residue [41], whereas we discovered that the phosphorylation of S33624 of TTN is greater in the morning than evening. Myomesin 2 (MYOM2, formally known as M-Protein) interacts with the light meromyosin (LMM) region of MYH and the C-terminal region of TTN [42]. The phosphorylation of MYOM2 S76 disrupts the binding of MYOM2, and LMM [43] and is more prevalent after exercise in the morning compared to evening. Myosin binding protein C (MYBPC1) localizes to the M-band through its interaction with obscurin [44]. MYBPC1 is required for the binding of muscle creatine kinase (KCRM) to the M-band region. MYBC1 exhibits differences in post-translational state between morning and evening, which are likely to be phosphorylation [45]. KCRM S24 phosphorylation is greater in the morning than evening. The N-terminal region of KCRM is necessary for the creatine/phosphocreatine- and pH-dependent interaction of KCRM with the M-band [46].
The M-band is a key component of the sarcomeric structure that distributes force across opposing myosin thick filaments [38] and is well-placed to mediate mechanical signals to the nucleus and wider muscle proteome. MYOM2, formerly known as 165 kDa M-protein, is a major component of the M-Band and is specifically expressed in cardiac and fast-twitch skeletal muscle [47]. MYOM2 binds to the light meromyosin (LMM) tail of myosin heavy chains in the M-band region and the phosphorylation of MYOM2 S76 by protein kinase A (PKA) disrupts the binding of MYOM2 and LMM in vitro [43]. We report a significantly greater phosphorylation of MYOM2 S76 in the morning, which could indicate alteration to the M-band structure and the disruption of force transmission in vivo. The circadian regulation of this effect is supported by data from animals carrying clock gene mutations that exhibit a decrement in myofiber-specific force and disruption to the hexagonal structure of thick filament proteins [17]. In the future it will be of interest to investigate whether differences in protein phosphorylation also exist between the muscle of wild-type and clock mutant mice.
We report that the phosphorylation of TTN S33624 near the C-terminus of titin is greater in the morning than evening in human muscle. The kinase domain of human titin includes Y32341 [41]. The TTN S33624 site reported here has been previously detected in phosphoproteome maps of human skeletal muscle [26], but as of yet, no function is assigned to S33624 phosphorylation. Titin is involved in sensing mechanical strain via its C-terminal kinase domain located at the sarcomeric M-band [48]. Mechanical strain causes unfolding of the kinase domain, which occurs prior to unfolding of the structural titin Ig/Fn domains. The kinase domain of titin is involved in gene expression of muscle E3 ligases in response to mechanical strain [49]. If TTN S33624 phosphorylation alters the stretch responsiveness of titin kinase, this could raise questions regarding potential responsiveness to training in morning versus evening.
Contrary to our expectation, in-depth analysis of myosin light chain proteoforms found no differences between morning and evening conditions (Table 1). In skeletal muscle, the phosphorylation of myosin regulatory light chain (MLRS) contributes to twitch potentiation in fast-twitch fibers [20]. However, we report that the post-translational modification of neither essential nor regulatory myosin light chain isoforms is associated with diurnal differences in maximum isometric force in human muscle. Further analysis of myofibrillar proteoforms using broader range (pH 3–10) isoelectric focusing (Figure 4) discovered putative post-translational modifications of myofibrillar proteins and metabolic enzymes in human muscle (Table 2) that raise new hypotheses regarding the differences in muscle force output. MYBPC1 is of interest because it is a major component of the muscle thick filament [50]. Proteomic analysis of hearts from wild-type and cardiac clock mutant mice [45] reported circadian differences in the level of phosphorylation of MYBPC1 using the phosphor-specific, Pro-Q diamond, stain. The phosphorylation of MYBPC1 is associated with enhanced contractility in response to beta-adrenergic receptor stimulation [51], and MYBPC1 can be phosphorylated at numerous sites by protein kinase A (PKA) and protein kinase C [52]. We were unable to unambiguously identify the site-specific modification of MYBPC1 that differed in human muscle between the morning and evening conditions. Nevertheless, the consistency in findings between the heart [45] and human skeletal (current work) muscle indicate that further targeted analysis on MYBPC1 proteoforms is warranted.
Muscle creatine kinase (KCRM, previously known as MM-CK) is a prominent M-band protein and key enzyme in high-energy phosphate metabolism [53]. MYBPC1 is necessary for the recruitment of KCRM to myosin, and this interaction is dependent on both the ratio of creatine/phosphocreatine and intracellular pH [53]. The interaction between KCRM and the M-band weakens as the intracellular pH rises and the ratio of creatine/phosphocreatine increases [53]. KCRM also interacts with immunoglobulin-like domains near the C-terminal of myomesin and M-protein (MYOM2; [46]). The site (S24) of KCRM phosphorylation reported here (Figure 5) is immediately adjacent to K25, which is one of 4 lysine residues near the N-terminus of KCRM that are required for the pH-dependent interaction between KCRM and M-band proteins [46]. The phosphorylation of KCRM S24 has been previously detected in phosphoproteome maps of human muscle [26], but the upstream kinase and functional consequences of KCRM S24 phosphorylation remain to be elucidated. Based on our current findings, it may be hypothesized that KCRM S24 phosphorylation has a disruptive influence and may weaken the interaction between KCRM and the M-band region.
In addition to sarcomeric proteins, isoforms of heat shock protein 90 (HSP90) and the mitochondrial import receptor protein (TOM70) also exhibited a diurnal difference in phosphorylation status. HSP90 does not localize to the M-band, but it is required for the chaperone-mediated assembly of myosin in muscle [54]. HSP90 modulates the myosin replacement rate into myofilaments and influences the expression of myosin genes [55]. Post-translational modifications, including the phosphorylation, of HSP90 are recognized to modulate its chaperone activity [56], but an in-depth understanding of the effects of different site-specific modifications is not yet available. The phosphorylation of S226 and S255 of HS90B and S231 HS90A reported here (Figure 5) is mediated by casein kinase 2 (CK2), which is known to modulate the molecular clock through the phosphorylation of Period 2 S53 [57]. TOM70 S91 is reported to exhibit a circadian rhythm in mouse liver [58], and the deletion of BMAL1 causes muscle mitochondrial dysfunction [17]. The phosphorylation of TOM70 S174 by PKA inhibits its receptor activity [59], but the effect of S91 phosphorylation, reported here, remains to be determined. The future phosphoproteomic analysis of mitochondria may give further insight to time-of-day differences in the response of muscle metabolism to exercise [16].
Findings from our non-targeted analysis of protein phosphorylation suggest a connection to PKA. Several of the phosphorylation sites on myofibrillar proteins have been reported to be targets of PKA, and we detected greater phosphorylation of the catalytic subunit of PKA on threonine (T)198 in morning samples. PKA is a key second messenger of muscle beta-adrenergic receptor signaling, and adrenal hormones, including catecholamines and glucocorticoids, have a role in synchronizing circadian rhythms across peripheral tissues in vivo [60]. Glucocorticoids are regarded as the main synchronizing hormones [61], but the circadian control of liver metabolism in mice [62] has been linked with adrenergic receptor signaling. The acrophase (approximately mid-day) in plasma catecholamines in humans [63] does not coincide with the morning–evening differences in muscle isometric force. However, plasma catecholamine response to exercise is greater in the morning than evening [63]. There is a well-established interaction between glucocorticoids and adrenergic receptor signaling [64], and the early morning peak in cortisol could potentiate responses to adrenaline (consistent with our data). Glucocorticoid stimulation may enhance the intracellular 3’,5’-cyclic adenosine monophosphate (cAMP) response to beta-adrenergic receptor stimulation, but this might be expected to result in greater muscle force output [65] in the morning. The acrophase of plasma cortisol early in the morning coincides with the bathyphase of muscle force production. The phosphorylation of PKA T198 is mediated by phosphatidylinositol 3-kinase [66], which opens the possibility of upstream regulation by interaction between the glucocorticoid receptor and p85α subunit of phosphatidylinositol 3-kinase [67]. Our data provide the basis for investigating the role of glucocorticoids in the regulation of cAMP in the context of time-of-day differences in muscle force.
The primary aim of this study was to investigate whether modifications occur to the contractile proteins of skeletal muscle that associate with its ability to generate force at the two different times of day. Biopsies were performed to investigate whether modifications co-occur to the contractile proteins of skeletal muscle that affect its ability to produce force at each time of day. A limitation of our work is that baseline samples were not collected prior to exercise, so it remains to be shown whether the time-of-day differences in protein post-translational modification exist in muscle that has not performed exercise. Overall, the phosphorylation of muscle proteins was greater after exercise in the morning than in the evening, which may indicate a diurnal difference in muscle protein phosphatase activity. The modulation of protein phosphatase 1 activity alters circadian rhythms in flies [68], but the selective dephosphorylation of proteins in human muscle remains to be investigated. We principally studied phosphorylation, but other post-translational modifications may also be important. The majority of covalent protein modifications change the isoelectric point of proteins, which makes 2DGE an elegant tool for the non-targeted discovery of protein modifications. However, the relatively small samples extracted from 2DGE spots, particularly in human biopsy samples where the starting material is also limited, make this work challenging. A third limitation concerning our study was the selection of participants for biopsies; in our case, we biopsied only those who were happy to give consent. Even though significant diurnal differences were found in expected parameters, future work should establish diurnal variation first within the population who have consented to have biopsies and select those participants who have the greatest variation.
5. Conclusions
Exercise in the morning, compared to the evening, resulted in the greater phosphorylation of proteins in human muscle that are components of or are associated with the M-band region. The phosphorylation of these proteins may alter the M-band structure and disrupt force transmission, thus potentially explaining the lower force output in the morning compared to evening. Our findings in humans add to data from animal models where deletion of the clock protein, BMAL1, results in disorganization of the 3D myofibrillar lattice and a loss in muscle force. The data generated in human muscle provide new targets to investigate the modulation of muscle force output or the application of exercise as a Zeitgeber of muscle function.
Supplementary Materials
Phosphopeptide data: https://www.mdpi.com/2227-7382/8/3/22/s1.
Author Contributions
Conceptualization, J.G.B. and B.J.E.; methodology, S.A.P., R.M.E., S.O.S., Z.I.; validation, Z.A.M., K.A.B.D., E.C.R.H.; formal analysis, Z.A.M., S.A.P., J.G.B., R.M.E., B.J.E.; investigation, Z.A.M., S.A.P., K.A.B.D., E.C.R.H., S.O.S., Z.I.; data curation, J.B.; writing—original draft preparation, Z.A.M., J.B.; writing—review and editing, J.G.B., B.J.E., S.A.P., R.M.E.; visualization, J.G.B., J.B.; supervision, J.G.B. and B.J.E.; funding acquisition, Z.A.M. and J.G.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Universiti Pendidikan Sultan Idris, Perak, Malaysia; Grant 2018-0164-103-01.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.McLeod M., Breen L., Hamilton D.L., Philp A. Live strong and prosper: The importance of skeletal muscle strength for healthy ageing. Biogerontology. 2016;17:497–510. doi: 10.1007/s10522-015-9631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foldvari M., Clark M., Laviolette L.C., Bernstein M.A., Kaliton D., Castaneda C., Pu C.T., Hausdorff J.M., Fielding R.A., Singh M.A.F. Association of Muscle Power With Functional Status in Community-Dwelling Elderly Women. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2000;55:M192–M199. doi: 10.1093/gerona/55.4.M192. [DOI] [PubMed] [Google Scholar]
- 3.Solaro R.J., De Tombe P.P. Review focus series: Sarcomeric proteins as key elements in integrated control of cardiac function. Cardiovasc. Res. 2008;77:616–618. doi: 10.1093/cvr/cvn004. [DOI] [PubMed] [Google Scholar]
- 4.Drust B., Waterhouse J., Atkinson G., Edwards B., Reilly T. Circadian Rhythms in Sports Performance—An Update. Chronobiol. Int. 2005;22:21–44. doi: 10.1081/CBI-200041039. [DOI] [PubMed] [Google Scholar]
- 5.Reilly T., Waterhouse J. Sports performance: Is there evidence that the body clock plays a role? Eur. J. Appl. Physiol. 2009;106:321–332. doi: 10.1007/s00421-009-1066-x. [DOI] [PubMed] [Google Scholar]
- 6.Martin A., Carpentier A., Guissard N., van Hoecke J., Duchateau J. Effect of time of day on force variation in a human muscle. Muscle Nerve. 1999;22:1380–1387. doi: 10.1002/(SICI)1097-4598(199910)22:10<1380::AID-MUS7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 7.Robinson W.R., Pullinger S.A., Kerry J.W., Giacomoni M., Robertson C.M., Burniston J.G., Waterhouse J.M., Edwards B.J. Does lowering evening rectal temperature to morning levels offset the diurnal variation in muscle force production? Chronobiol. Int. 2013;30:998–1010. doi: 10.3109/07420528.2013.793197. [DOI] [PubMed] [Google Scholar]
- 8.Erskine R.M., Fletcher G., Folland J.P. The contribution of muscle hypertrophy to strength changes following resistance training. Eur. J. Appl. Physiol. 2014;114:1239–1249. doi: 10.1007/s00421-014-2855-4. [DOI] [PubMed] [Google Scholar]
- 9.Sedliak M., Finni T., Cheng S., Haikarainen T., Häkkinen K. Diurnal Variation in Maximal and Submaximal Strength, Power and Neural Activation of Leg Extensors in Men: Multiple Sampling Across Two Consecutive Days. Int. J. Sports Med. 2008;29:217–224. doi: 10.1055/s-2007-965125. [DOI] [PubMed] [Google Scholar]
- 10.Edwards B.J., Pullinger S.A., Kerry J.W., Robinson W.R., Reilly T.P., Robertson C.M., Waterhouse J.M. Does raising morning rectal temperature to evening levels offset the diurnal variation in muscle force production? Chronobiol. Int. 2013;30:486–501. doi: 10.3109/07420528.2012.741174. [DOI] [PubMed] [Google Scholar]
- 11.Albrecht U. Timing to Perfection: The Biology of Central and Peripheral Circadian Clocks. Neuron. 2012;74:246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zambon A.C., McDearmon E.L., Salomonis N., Vranizan K.M., Johansen K.L., Adey D., Takahashi J.S., Schambelan M., Conklin B.R. Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol. 2003;4:R61. doi: 10.1186/gb-2003-4-10-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy J.J., Andrews J.L., McDearmon E.L., Campbell K.S., Barber B.K., Miller B.H., Walker J.R., Hogenesch J.B., Takahashi J.S., Esser K.A. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol. Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyar K.A., Ciciliot S., Wright L.E., Biensø R.S., Tagliazucchi G.M., Patel V.R., Forcato M., Paz M.I.P., Gudiksen A., Solagna F., et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 2014;3:29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezagouri S., Zwighaft Z., Sobel J., Baillieul S., Doutreleau S., Ladeuix B., Golik M., Verges S., Asher G. Physiological and Molecular Dissection of Daily Variance in Exercise Capacity. Cell Metab. 2019;30:78–91.e4. doi: 10.1016/j.cmet.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Andrews J.L., Zhang X., McCarthy J.J., McDearmon E.L., Hornberger T.A., Russell B., Campbell K.S., Arbogast S., Reid M.B., Walker J.R., et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl. Acad. Sci. USA. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroder E.A., Harfmann B.D., Zhang X., Srikuea R., England J.H., Hodge B.A., Wen Y., Riley L.A., Yu Q., Christie A., et al. Intrinsic muscle clock is necessary for musculoskeletal health. J. Physiol. 2015;593:5387–5404. doi: 10.1113/JP271436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camera D.M., Burniston J.G., Pogson M.A., Smiles W.J., Hawley J.A. Dynamic proteome profiling of individual proteins in human skeletal muscle after a high-fat diet and resistance exercise. FASEB J. 2017;31:5478–5494. doi: 10.1096/fj.201700531R. [DOI] [PubMed] [Google Scholar]
- 20.Ryder J.W., Lau K.S., Kamm K.E., Stull J.T. Enhanced skeletal muscle contraction with myosin light chain phosphorylation by a calmodulin-sensing kinase. J. Biol. Chem. 2007;282:20447–20454. doi: 10.1074/jbc.M702927200. [DOI] [PubMed] [Google Scholar]
- 21.Potts G.K., McNally R.M., Blanco R., You J.-S., Hebert A.S., Westphall M.S., Coon J.J., Hornberger T.A. A map of the phosphoproteomic alterations that occur after a bout of maximal-intensity contractions. J. Physiol. 2017;595:5209–5226. doi: 10.1113/JP273904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kampourakis T., Sun Y., Irving M. Myosin light chain phosphorylation enhances contraction of heart muscle via structural changes in both thick and thin filaments. Proc. Natl. Acad. Sci. USA. 2016;113:E3039–E3047. doi: 10.1073/pnas.1602776113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burniston J.G., Hoffman E.P. Proteomic responses of skeletal and cardiac muscle to exercise. Expert Rev. Proteom. 2011;8:361–377. doi: 10.1586/epr.11.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burniston J.G. How many phosphoproteins does it take to make muscle grow? J. Physiol. 2017;595:5009–5010. doi: 10.1113/JP274672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen M.R., Thingholm T.E., Jensen O.N., Roepstorff P., Jørgensen T.J.D. Highly Selective Enrichment of Phosphorylated Peptides from Peptide Mixtures Using Titanium Dioxide Microcolumns. Mol. Cell. Proteom. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Højlund K., Bowen B.P., Hwang H., Flynn C.R., Madireddy L., Geetha T., Langlais P., Meyer C., Mandarino L.J., Yi Z. In vivo phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC-ESI-MS/MS. J. Proteome Res. 2009;8:4954–4965. doi: 10.1021/pr9007267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman N.J., Parker B.L., Chaudhuri R., Fisher-Wellman K.H., Kleinert M., Humphrey S.J., Yang P., Holliday M., Trefely S., Fazakerley D.J., et al. Global Phosphoproteomic Analysis of Human Skeletal Muscle Reveals a Network of Exercise-Regulated Kinases and AMPK Substrates. Cell Metab. 2015;22:922–935. doi: 10.1016/j.cmet.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith C.S., Reilly C., Midkiff K. Evaluation of three circadian rhythm questionnaires with suggestions for an improved measure of morningness. J. Appl. Psychol. 1989;74:728–738. doi: 10.1037/0021-9010.74.5.728. [DOI] [PubMed] [Google Scholar]
- 29.Morton J.P., Atkinson G., MacLaren D.P.M., Cable N.T., Gilbert G., Broome C., McArdle A., Drust B. Reliability of maximal muscle force and voluntary activation as markers of exercise-induced muscle damage. Eur. J. Appl. Physiol. 2005;94:541–548. doi: 10.1007/s00421-005-1373-9. [DOI] [PubMed] [Google Scholar]
- 30.Murtagh C.F., Nulty C., Vanrenterghem J., O’Boyle A., Morgans R., Drust B., Erskine R.M. The Neuromuscular Determinants of Unilateral Jump Performance in Soccer Players Are Direction-Specific. Int. J. Sports Physiol. Perform. 2018;13:604–611. doi: 10.1123/ijspp.2017-0589. [DOI] [PubMed] [Google Scholar]
- 31.Burniston J.G. Changes in the rat skeletal muscle proteome induced by moderate-intensity endurance exercise. Biochim. Biophys. Acta Proteins Proteom. 2008;1784:1077–1086. doi: 10.1016/j.bbapap.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Holloway K.V., Gorman M.O., Woods P., Morton J.P., Evans L., Cable N.T., Goldspink D.F., Burniston J.G. Proteomic investigation of changes in human vastus lateralis muscle in response to interval-exercise training. Proteomics. 2009;9:5155–5174. doi: 10.1002/pmic.200900068. [DOI] [PubMed] [Google Scholar]
- 33.Burniston J.G., Kenyani J., Gray D., Guadagnin E., Jarman I.H., Cobley J.N., Cuthbertson D.J., Chen Y.-W., Wastling J.M., Lisboa P.J., et al. Conditional independence mapping of DIGE data reveals PDIA3 protein species as key nodes associated with muscle aerobic capacity. J. Proteom. 2014;106:230–245. doi: 10.1016/j.jprot.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo H., Isserlin R., Emili A., Burniston J.G. Exercise-responsive phosphoproteins in the heart. J. Mol. Cell. Cardiol. 2017;111:61–68. doi: 10.1016/j.yjmcc.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., Von Mering C., et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolff C.A., Esser K.A. Exercise timing and circadian rhythms. Curr. Opin. Physiol. 2019;10:64–69. doi: 10.1016/j.cophys.2019.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lange S., Pinotsis N., Agarkova I., Ehler E. The M-band: The underestimated part of the sarcomere. Biochim. Biophys. Acta Mol. Cell Res. 2019 doi: 10.1016/j.bbamcr.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pernigo S., Fukuzawa A., Beedle A.E.M., Holt M., Round A., Pandini A., Garcia-Manyes S., Gautel M., Steiner R.A. Binding of Myomesin to Obscurin-Like-1 at the Muscle M-Band Provides a Strategy for Isoform-Specific Mechanical Protection. Structure. 2017 doi: 10.1016/j.str.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lange S., Himmel M., Auerbach D., Agarkova I., Hayess K., Fürst D.O., Perriard J.C., Ehler E. Dimerisation of myomesin: Implications for the structure of the sarcomeric M-band. J. Mol. Biol. 2005;345:289–298. doi: 10.1016/j.jmb.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 41.Mayans O., van der Ven P.F.M., Wilm M., Mues A., Young P., Fürst D.O., Wilmanns M., Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- 42.Obermann W.M.J., Gautel M., Weber K., Fürst D.O. Molecular structure of the sarcomeric M band: Mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J. 1997 doi: 10.1093/emboj/16.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obermann W.M.J., van der Ven P.F.M., Steiner F., Weber K., Fürst D.O. Mapping of a Myosin-binding Domain and a Regulatory Phosphorylation Site in M-Protein, a Structural Protein of the Sarcomeric M Band. Mol. Biol. Cell. 1998;9:829–840. doi: 10.1091/mbc.9.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ackermann M.A., Hu L.Y.R., Bowman A.L., Bloch R.J., Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol. Biol. Cell. 2009 doi: 10.1091/mbc.e08-12-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Podobed P., Pyle W.G., Ackloo S., Alibhai F.J., Tsimakouridze E.V., Ratcliffe W.F., Mackay A., Simpson J., Wright D.C., Kirby G.M., et al. The day/night proteome in the murine heart. Am. J. Physiol. Integr. Comp. Physiol. 2014;307:R121–R137. doi: 10.1152/ajpregu.00011.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hornemann T., Kempa S., Himmel M., Hayeß K., Fürst D.O., Wallimann T. Muscle-type Creatine Kinase Interacts with Central Domains of the M-band Proteins Myomesin and M-protein. J. Mol. Biol. 2003;332:877–887. doi: 10.1016/S0022-2836(03)00921-5. [DOI] [PubMed] [Google Scholar]
- 47.Carlsson E., Grove B.K., Wallimann T., Eppenberger H.M., Thornell L.E. Myofibrillar M-band proteins in rat skeletal muscles during development. Histochemistry. 1990;95:27–35. doi: 10.1007/BF00737225. [DOI] [PubMed] [Google Scholar]
- 48.Puchner E.M., Alexandrovich A., Kho A.L., Hensen U., Schafer L.V., Brandmeier B., Grater F., Grubmuller H., Gaub H.E., Gautel M. Mechanoenzymatics of titin kinase. Proc. Natl. Acad. Sci. USA. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lange S. The Kinase Domain of Titin Controls Muscle Gene Expression and Protein Turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 50.McGrath M.J., Cottle D.L., Nguyen M.-A., Dyson J.M., Coghill I.D., Robinson P.A., Holdsworth M., Cowling B.S., Hardeman E.C., Mitchell C.A., et al. Four and a Half LIM Protein 1 Binds Myosin-binding Protein C and Regulates Myosin Filament Formation and Sarcomere Assembly. J. Biol. Chem. 2006;281:7666–7683. doi: 10.1074/jbc.M512552200. [DOI] [PubMed] [Google Scholar]
- 51.Gresham K.S., Stelzer J.E. The contributions of cardiac myosin binding protein C and troponin I phosphorylation to β-adrenergic enhancement of in vivo cardiac function. J. Physiol. 2016;594:669–686. doi: 10.1113/JP270959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ackermann M.A., Kontrogianni-Konstantopoulos A. Myosin Binding Protein-C Slow is a Novel Substrate for Protein Kinase A (PKA) and C (PKC) in Skeletal Muscle. J. Proteome Res. 2011;10:4547–4555. doi: 10.1021/pr200355w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Z., Zhao T.-J., Li J., Gao Y.-S., Meng F.-G., Yan Y.-B., Zhou H.-M. Slow skeletal muscle myosin-binding protein-C (MyBPC1) mediates recruitment of muscle-type creatine kinase (CK) to myosin. Biochem. J. 2011;436:437–445. doi: 10.1042/BJ20102007. [DOI] [PubMed] [Google Scholar]
- 54.Srikakulam R., Winkelmann D.A. Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 2004 doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- 55.Ojima K., Ichimura E., Suzuki T., Oe M., Muroya S., Nishimura T. HSP90 modulates the myosin replacement rate in myofibrils. Am. J. Physiol. Physiol. 2018;315:C104–C114. doi: 10.1152/ajpcell.00245.2017. [DOI] [PubMed] [Google Scholar]
- 56.Li J., Soroka J., Buchner J. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta Mol. Cell Res. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 57.Tsuchiya Y., Akashi M., Matsuda M., Goto K., Miyata Y., Node K., Nishida E. Involvement of the Protein Kinase CK2 in the Regulation of Mammalian Circadian Rhythms. Sci. Signal. 2009;2:ra26. doi: 10.1126/scisignal.2000305. [DOI] [PubMed] [Google Scholar]
- 58.Robles M.S., Humphrey S.J., Mann M. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab. 2017;25:118–127. doi: 10.1016/j.cmet.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt O., Harbauer A.B., Rao S., Eyrich B., Zahedi R.P., Stojanovski D., Schönfisch B., Guiard B., Sickmann A., Pfanner N., et al. Regulation of Mitochondrial Protein Import by Cytosolic Kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 60.Tahara Y., Aoyama S., Shibata S. The mammalian circadian clock and its entrainment by stress and exercise. J. Physiol. Sci. 2017;67:1–10. doi: 10.1007/s12576-016-0450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oster H., Challet E., Ott V., Arvat E., de Kloet E.R., Dijk D.-J., Lightman S., Vgontzas A., Van Cauter E. The functional and clinical significance of the 24-h rhythm of circulating glucocorticoids. Endocr. Rev. 2017;38:3–45. doi: 10.1210/er.2015-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terazono H., Mutoh T., Yamaguchi S., Kobayashi M., Akiyama M., Udo R., Ohdo S., Okamura H., Shibata S. Adrenergic regulation of clock gene expression in mouse liver. Proc. Natl. Acad. Sci. USA. 2003;100:6795–6800. doi: 10.1073/pnas.0936797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheer F.A.J.L., Hu K., Evoniuk H., Kelly E.E., Malhotra A., Hilton M.F., Shea S.A. Impact of the human circadian system, exercise, and their interaction on cardiovascular function. Proc. Natl. Acad. Sci. USA. 2010;107:20541–20546. doi: 10.1073/pnas.1006749107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davies A. Regulation of Beta-Adrenergic Receptors by Steroid Hormones. Annu. Rev. Physiol. 1984;46:119–130. doi: 10.1146/annurev.ph.46.030184.001003. [DOI] [PubMed] [Google Scholar]
- 65.Cairns S.P., Borrani F. β-Adrenergic modulation of skeletal muscle contraction: Key role of excitation-contraction coupling. J. Physiol. 2015;593:4713–4727. doi: 10.1113/JP270909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng X., Ma Y., Moore M., Hemmings B.A., Taylor S.S. Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 1998;95:9849–9854. doi: 10.1073/pnas.95.17.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Limbourg F.P., Huang Z., Plumier J.-C., Simoncini T., Fujioka M., Tuckermann J., Schütz G., Moskowitz M.A., Liao J.K. Rapid nontranscriptional activation of endothelial nitric oxide synthase mediates increased cerebral blood flow and stroke protection by corticosteroids. J. Clin. Investig. 2002;110:1729–1738. doi: 10.1172/JCI0215481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang Y., Sathyanarayanan S., Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.