Abstract
Simple Summary
To overcome the potential threat of drug resistance or limit of potency, the combination treatment of drugs is a promising strategy. Around 22% of patients with GCB-DLBCL carry EZH2 gain-of-function mutations and several PRC2 inhibitors are under clinical trials. Herein, we demonstrate that combination of GSK126 with pomalidomide synergistically inhibit tumor growth through inducing B-cell maturation and apoptosis in EZH2 gain-of-function mutant DLBCL. Our study provides the molecular basis of the combination strategy of PRC2 inhibitors and IMiDs in DLBCLs harboring EZH2 hyperactive mutation.
Abstract
Enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), the catalytic subunit of polycomb repressive complex 2 (PRC2), regulates genes involved in cell lineage and differentiation through methylating lysine 27 on histone H3 (H3K27me3). Recurrent gain-of-function mutations of EZH2 have been identified in various cancer types, in particular, diffuse large B-cell lymphoma (DLBCL), through large-scale genome-wide association studies and EZH2 depletion or pharmacological inhibition has been shown to exert an antiproliferative effect on cancer cells, both in vitro and in vivo. In the current study, a combination of pomalidomide and GSK126 synergistically inhibited the growth of EZH2 gain-of-function mutant Diffuse large B-cell lymphoma (DLBCL) cells. Furthermore, this synergistic effect appeared to be dependent on cereblon (CRBN), a cellular receptor of pomalidomide, but not degradation of IKAROS family zinc finger 1 (IKZF1) or IKAROS family zinc finger 3 (IKZF3). RNA sequencing analyses revealed that co-treatment with GSK126 and pomalidomide induced specific gene sets involved in B-cell differentiation and apoptosis. Synergistic growth inhibition and B-cell differentiation were further validated in xenograft mouse models. Our collective results provide a molecular basis for the mechanisms underlying the combined therapeutic effects of PRC2 inhibitors and pomalidomide on EZH2-mutated DLBCL.
Keywords: PRC2, EZH2, GSK126, pomalidomide, IMiD, DLBCL, synergistic effect, combination therapy
1. Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of lymphoid neoplasm accounting for 30–40% of adult non-Hodgkin lymphoma (NHL) worldwide [1]. Gene expression profiling has led to the identification of germinal center B-cell-like (GCB) and activated B-cell-like (ABC) subgroups of DLBCL according to cell origin [2]. Further genetic analyses have revealed recurrent alterations in EZH2, a catalytic component of polycomb repressive complex 2 (PRC2). In particular, gain-of-function mutations in the Su(var)3-9, Enhancer-of-zeste and Trithorax (SET) domain, including tyrosine 641 (Y641), account for 22% of GCB DLBCL, supporting the utility of EZH2 intervention for the treatment of this subtype [3]. The PRC2 complex constituting EZH2 together with suppressor of zeste 12 (SUZ12), Retinoblastoma (RB)-binding protein 4 (RBBP4), and embryonic ectoderm development (EED) displays intrinsic histone methyltransferase activity catalyzing lysine 27 methylation of histone H3 (H3K27) [4]. PRC2 plays essential roles in silencing of genes involved in cellular differentiation and maintenance of stem cells [5]. The hyperactive EZH2Y646 mutant, commonly identified in DLBCL, induces an aberrant increase in the repressive histone marker, H3K27me3, in turn, affecting the regulation of genes associated with cell differentiation and tumor suppression [6,7,8]. Several EZH2 and allosteric EED inhibitors have been shown to impede tumor growth, both in vivo and in vitro. The potent EZH2 inhibitor, EPZ005687, suppresses H3K27me3 in EZH2Y641/A677-mutant lymphoma in a dose-dependent manner [9]. Another EZH2 inhibitor, GSK126, is reported to significantly impair the growth of lymphoma with EZH2 gain-of-function mutations in vivo [10]. Allosteric EED inhibitors directly interact with the H3K27me3-binding pocket of EED, thereby preventing the activation of PRC2 [11]. Moreover, EZH2 overexpression has been documented in several human cancer types, including liver [12], lung [13], bladder [14], breast [15], melanoma [15], and colorectal cancers [16]. Dysregulated EZH2 is reported to inhibit the expression of tumor suppressors, thus promoting uncontrollable growth [8].
Immunomodulatory drugs (IMiDs), including thalidomide, lenalidomide, and pomalidomide, possess antimyeloma, anti-inflammatory, and antiproliferative properties [17,18]. Once IMiDs engage the cellular receptor cereblon (CRBN), a ubiquitin E3 ligase, the Cul4-RBX1-DDB1-CRBN (CRL4CRBN) complex promotes the degradation of neosubstrates, such as IKAROS family zinc finger 1 (IKZF1) and IKAROS family zinc finger 3 (IKZF3) [19,20]. Recent global proteomic analyses have demonstrated that individual IMiDs additionally induce the degradation of different zinc finger proteins other than IKZF1 and IKZF3 [21]. In addition to its E3 ligase function, CRBN epigenetically regulates the Kv1.3 potassium channel required for CD4+ T-cell activation. CRBN binds directly adjacent to the Kv1.3 regulatory region, promoting H3K27me3 through interactions with EZH1/2. Moreover, thalidomide treatment competes out CRBN from the Kv1.3 locus, leading to a decrease in the H3K27me3 level [22]. From this point of view, it is possible that IMiDs influence EZH2 or the methylation status of H3K27 [22].
In this study, we examined the potential cooperativity between PRC2 inhibitors and pomalidomide, with a view to determine the efficacy of their combined administration as anticancer therapy. PRC2 inhibitors and pomalidomide synergistically and selectively inhibited the proliferation of EZH2-mutant DLBCL cells. Notably, this synergistic effect was dependent on CRBN, but not on the degradation of IKZF1 or IKZF3. RNA sequencing analyses revealed that co-treatment with GSK126 and pomalidomide induced B-cell differentiation in EZH2-mutant DLBCL cells. Our collective results support the utility of combination therapy involving EZH2 inhibition and pomalidomide for EZH2-mutated DLBCL and provide insights into the mechanisms underlying tumor suppression activity.
2. Results
2.1. Pomalidomide Enhances the Cytotoxic Effect of GSK126 in EZH2-Mutant DLBCL Cells
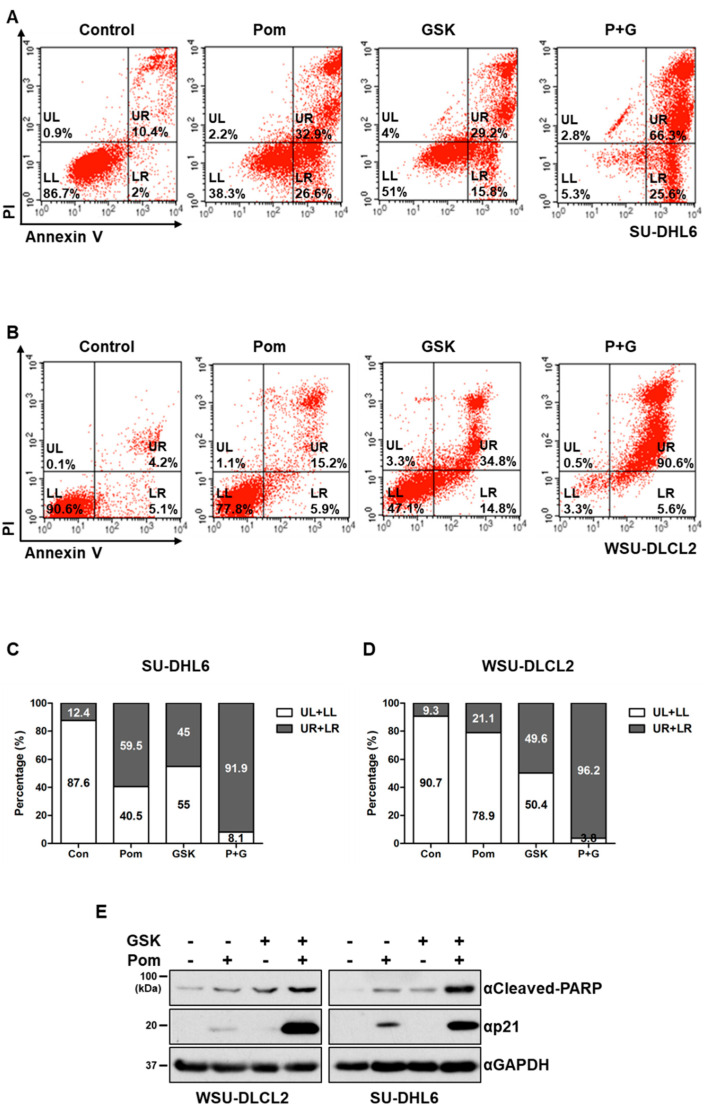
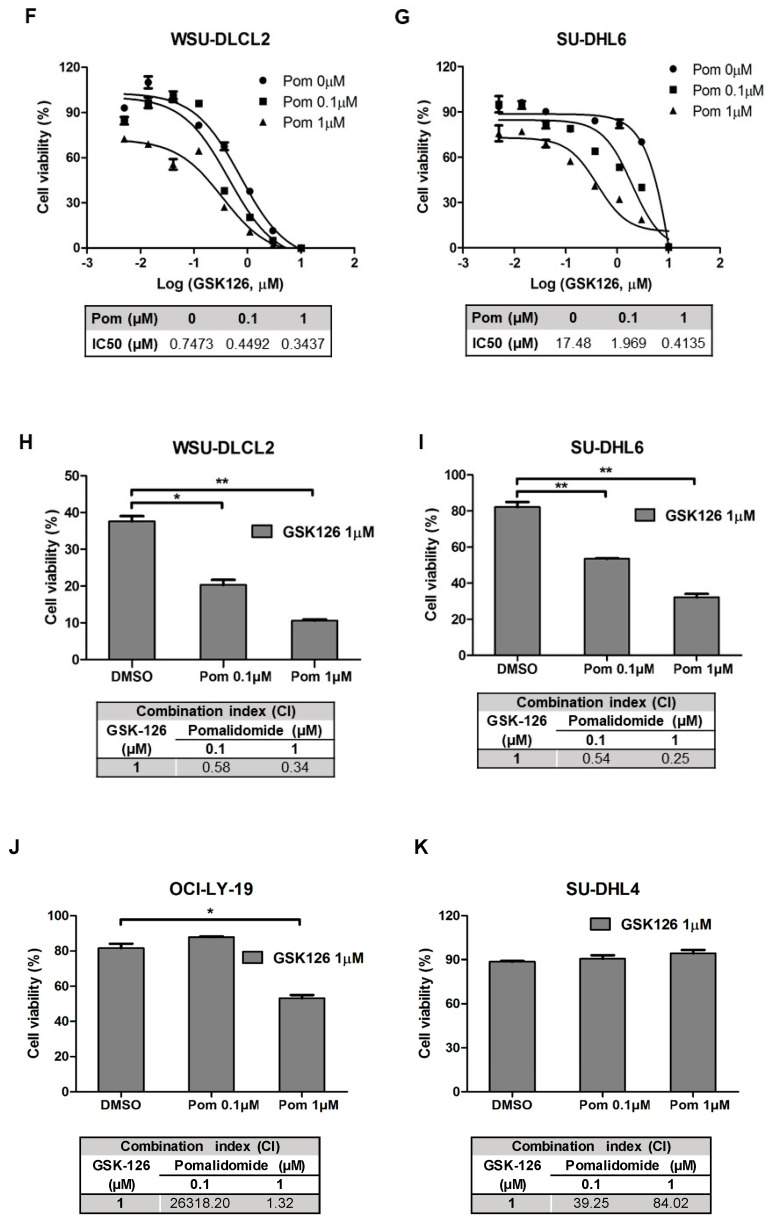
To determine the specific impacts of pomalidomide and GSK126 on the viability of DLBCL cells harboring EZH2 mutations, propidium iodide (PI)/annexin V staining was performed to monitor cell death. Co-treatment with pomalidomide and GSK126 induced significant SU-DHL6 (EZH2Y641N) and WSU-DLCL2 (EZH2Y641F) cell death, compared with either pomalidomide or GSK126 alone (Figure 1A–D). Consistently, cleavage of Poly (ADP-ribose) polymerase (PARP), a representative apoptotic marker, and protein levels of p21, a Cyclin-dependent kinase (CDK) inhibitor, increased in the presence of both GSK126 and pomalidomide (Figure 1E and Figure S6A). In the terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay, co-treatment induced apoptosis in WSU-DLBCL2 cells (Figure S1A,B). To establish whether these two compounds acted synergistically, we performed a cell viability assay at a range of doses. Three EZH2Y641-mutant DLBCL cell lines (WSU-DLCL2, SU-DHL6, and SU-DHL10) were treated with increasing concentrations of GSK126, along with a fixed concentration of pomalidomide (0.1 or 1 μM), and cell viability was measured using the CellTiter-Glo assay. Treatment with GSK126 in conjunction with pomalidomide induced a marked decrease in viability of all three cell lines (Figure 1F,G and Figure S1C). The Chou–Talalay drug combination index (CI) was calculated to establish whether the effects of pomalidomide and GSK126 were synergistic [23]. Based on the dose-response results, CI values of the two compounds were determined as 0.58 and 0.34 in WSU-DLCL2, 0.54 and 0.25 in SU-DHL6, and 5.432 and 0.249 in SU-DHL10 in the presence of 0.1 μM and 1 μM pomalidomide, respectively (Figure 1H,I and Figure S1D). As CI values <1 signify a synergistic effect [23], we concluded that pomalidomide acted synergistically with GSK126 to exert cytotoxicity in EZH2-mutant DLBCL. In contrast, no synergistic effect was observed in wild-type EZH2 DLBCL cell lines (OCI-LY-19, SU-DHL4, and Toledo) (Figure 1J,K and Figure S1E–H) and other lymphoma types, such as mantle cell lymphoma (JeKo-1, Rec-1, Mino, and Z138) and Burkitt’s lymphoma (NC-37) (Figure S1I–M). Moreover, the combination treatment did not exert additional or synergistic effects on the viability of multiple myeloma cell lines (IM-9, RPMI8226, and U266; Figure S1N–P), indicating that the synergistic antiproliferative effects of GSK126 and pomalidomide are specific for DLBCLs harboring gain-of-function mutations.
Figure 1.
Pomalidomide enhances the cytotoxic effect of GSK126 on enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2)-mutant diffuse large B-cell lymphoma cells. (A,B) Annexin V-FITC (annexin V)/propidium iodide (PI) staining of SU-DHL6 and WSU-DLCL2 cells treated with dimethyl sulfoxide (DMSO, control), pomalidomide alone (Pom), GSK126 alone (GSK), or pomalidomide and GSK126 combination (P+G). LL, viable cells (annexin V-negative/PI-negative); LR, early apoptotic cells (annexin V-positive/PI-negative); UL, late necrotic cells (annexin V-negative/PI-positive); and UR, late apoptotic/necrotic cells (annexin V-positive/PI-positive). (C,D) Quantification of results from Figure 1A,B. (E) WSU-DLCL2 and SU-DHL6 cells were treated with DMSO, pomalidomide, GSK126, or GSK126 and pomalidomide for 48 h, and levels of cleaved PARP, p21, and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were analyzed via western blot. (F,G) Effects of pomalidomide and GSK126 on cell viability. WSU-DLCL2 and SU-DHL6 cells were treated with the indicated concentrations of GSK126 and pomalidomide (Pom) for 6 days. (H–K) Combination index (CI) values obtained for indicated cells treated with different concentrations of pomalidomide in combination with GSK126 (1 μM). (* p-value < 0.05, ** p-value < 0.005, Student’s t-test).
2.2. EED Inhibition or EZH2 Knockdown Combined with Pomalidomide Synergistically Suppresses Proliferation of EZH2-Mutant DLBCL Cells
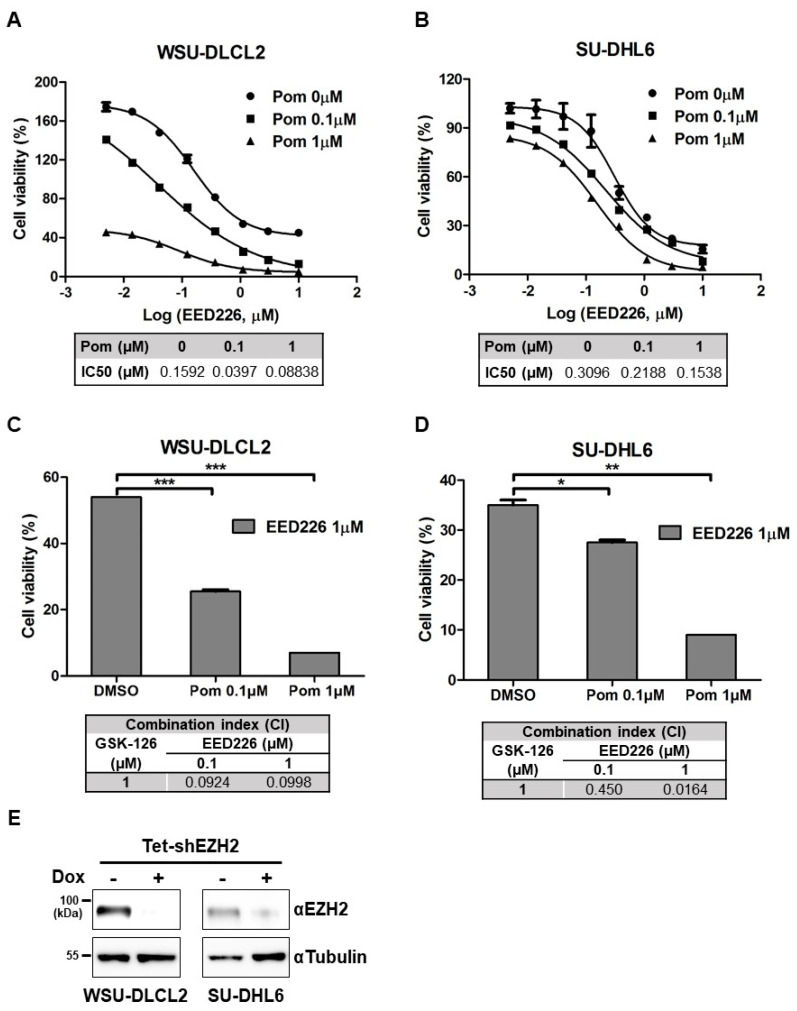
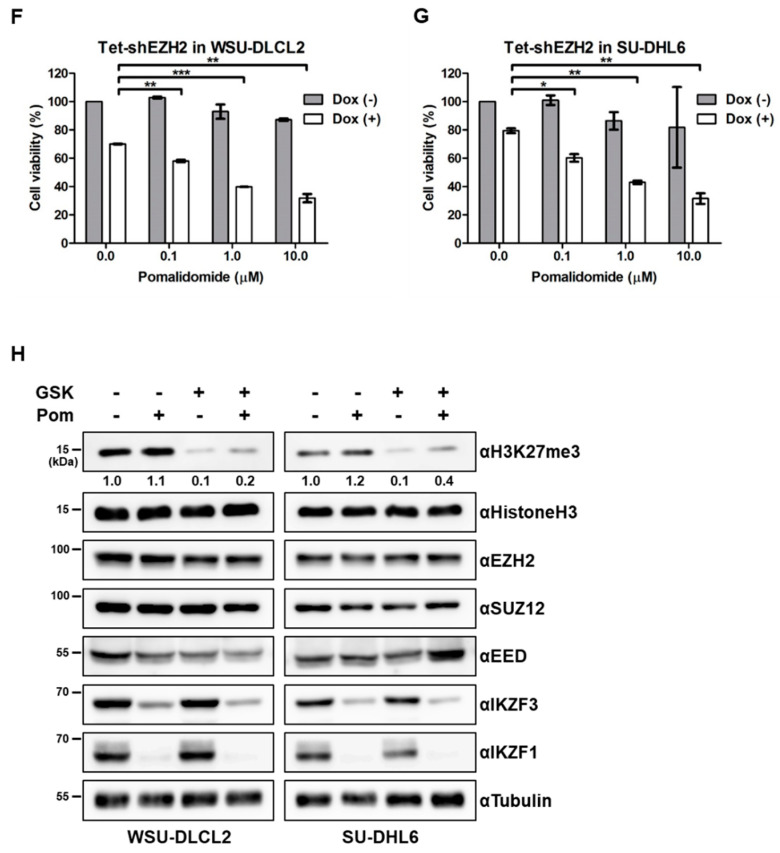
Given that EZH2 is a component of PRC2 and allosteric activation of EED is a prerequisite for its activity [24], we examined cellular toxicity of the EED inhibitor, EED226 [11], in combination with pomalidomide. Consistent with the data obtained using GSK126, co-treatment with EED226 and pomalidomide induced enhanced cytotoxicity against WSU-DLCL2, SU-DHL6, and SU-DHL10 cells (Figure 2A,B and Figure S2A). CI values were determined as 0.0998, 0.0164, and 0.00319 with 1 μM EED226 and pomalidomide in WSU-DLCL2, SU-DHL6, and SU-DHL10, respectively (Figure 2C,D and Figure S2B), indicating synergistic effects. We further investigated whether knockdown of EZH2 would also exert a synergistic effect with pomalidomide. For this purpose, WSU-DLCL2 and SU-DHL6 cell lines with conditional depletion of EZH2 were established using a Tet-inducible shRNA. In both cell lines, doxycycline treatment suppressed EZH2 expression (Figure 2E, Figures S2C and S6B). Upon knockdown of EZH2 with doxycycline, the viability of both cell lines was decreased in a pomalidomide dose-dependent manner (Figure 2F,G and Figure S2D,E). Our collective results implied that, similar to EZH2 inhibition, EZH2 knockdown or treatment with EED inhibitors, in combination with pomalidomide, can effectively induce synergistic antiproliferative activity.
Figure 2.
Inhibition or EZH2 knockdown combined with pomalidomide synergistically inhibits proliferation of EZH2-mutant DLBCL cells. (A,B) Effects of pomalidomide and EED226 on cell viability. WSU-DLCL2 and SU-DHL6 cells were treated with the indicated concentrations of EED226 and pomalidomide (Pom) for 6 days. (C,D) Combination index (CI) values obtained for WSU-DLCL2 and SU-DHL6 cells treated with different concentrations of pomalidomide in combination with EED226 (1 μM). (E) EZH2 levels in Tetracycline (Tet)-inducible shEZH2 WSU-DLCL2 or SU-DHL6 cells. Cells were treated with 1 μg/mL doxycycline (Dox) for 6 days and extracts were analyzed via western blot. (F,G) Effects of EZH2 knockdown and pomalidomide on the viability of WSU-DLCL2 and SU-DHL6 cells. (H) WSU-DLCL2 and SU-DHL6 cells were treated with GSK126 (GSK, 1 μM), pomalidomide (Pom, 1 μM), or both for 48 h, and cell lysates were analyzed via western blot using the indicated antibodies. (* p-value < 0.05, ** p-value < 0.005, *** p-value < 0.0001, Student’s t-test).
In view of the above findings, we hypothesized that co-treatment with GSK126 and pomalidomide affected H3K27 methylation levels, thereby inducing cell death in DLBCL with EZH2 hyperactive mutations. To examine this theory, H3K27 methylation status was examined to determine EZH2 activity in WSU-DLCL2 and SU-DHL6 cells treated with GSK126, pomalidomide, or both compounds (Figure 2H and Figure S6C). As expected, GSK126 treatment induced a decrease in the H3K27Me3 level. However, co-treatment did not affect H3K27Me3 levels, compared with GSK126 alone. In addition, co-treatment did not influence the expression of PRC2 subunits (SUZ12, EED, and EZH2), suggesting that synergistic activity is not attributable to changes in histone H3 methylation or PRC2 complex integrity. Moreover, GSK126 had no effect on IKZF1/3 degradation induced by pomalidomide.
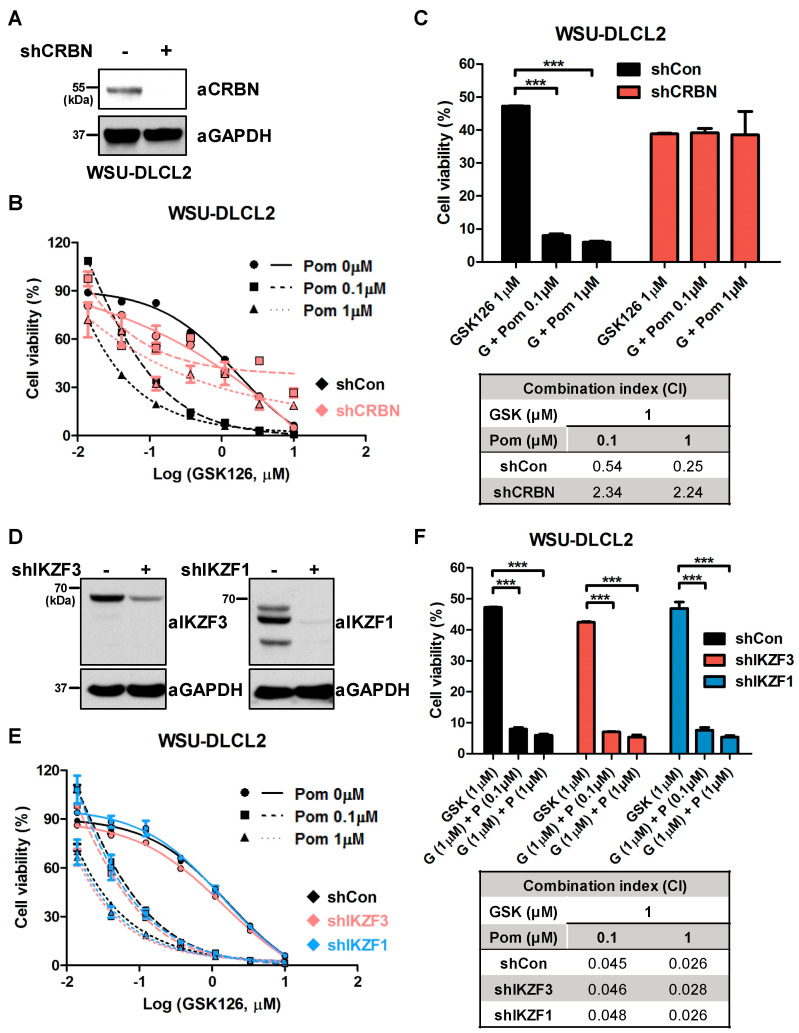
2.3. Synergistic Effects of GSK126 and Pomalidomide Are Dependent on CRBN, But Not on the Degradation of IKZF1 or IKZF3
As pomalidomide belongs to the IMiD family together with thalidomide and lenalidomide, we examined whether other IMiDs, such as lenalidomide, affect proliferation activity in combination with GSK126. Similar to pomalidomide, co-treatment with lenalidomide and GSK126 synergistically inhibited WSU-DLCL2 proliferation (Figure S3A,B). IMiDs interact directly with the cellular receptor, CRBN, inducing the degradation of IKZF1 and IKZF3, thereby exerting antimyeloma activity [19]. To ascertain whether the synergistic effects of GSK126 and pomalidomide are dependent on CRBN, we generated CRBN-depleted WSU-DLCL2 cell lines with the aid of specific shRNAs (Figure 3A and Figure S6D). Compared with shControl WSU-DLCL2, pomalidomide did not enhance GSK126-induced cytotoxicity in shCRBN WSU-DLCL2 cells (Figure 3B). The CI value of shControl in the co-treatment group was <1, whereas that of shCRBN was >1, indicating that CRBN is required for the synergistic effects of GSK126 and pomalidomide on EZH2-mutant DLBCL (Figure 3C). We further addressed whether pomalidomide-induced degradation of IZKF1 or IKZF3 contributed to this effect. To this end, IKZF1 or IKZF3-depleted WSU-DLCL2 cells were generated using shRNA, and IKZF1 and IKZF3 levels were examined (Figure 3D and Figure S6E). As shown in Figure 3E,F, knockdown of IKZF1 or IKZF3 did not induce differences relative to shControl cells, suggesting that the synergistic effects of pomalidomide and GSK126 are independent of IKZF1 or IKZF3 degradation.
Figure 3.
Synergistic effect of GSK126 and pomalidomide is dependent on cereblon (CRBN), but not degradation of IKAROS family zinc finger 1 (IKZF1) or 3 (IKZF3). (A) CRBN levels were examined via western blot in shControl and shCRBN WSU-DLCL2 cells. (B) Effects of pomalidomide and GSK126 on the viability of shControl and shCRBN WSU-DLCL2. Cells were treated with the indicated concentrations of GSK126 and pomalidomide (Pom) for 6 days. (C) Combination index (CI) values obtained for shControl and shCRBN WSU-DLCL2. Cells were treated with different concentrations of pomalidomide in combination with GSK126 (1 μM). (D) Western blot analysis of IKZF3 and IKZF1 levels in shControl, shIKZF1, and shIKZF3 WSU-DLCL2. (E) Effects of pomalidomide and GSK126 on the viability of shControl, shIKZF1, and shIKZF3 WSU-DLCL2. Cells were treated with the indicated concentrations of GSK126 and pomalidomide (Pom) for 6 days. (F) Combination index (CI) values obtained for shControl, shIKZF1, and shIKZF3 WSU-DLCL2. Cells were treated with different concentrations of pomalidomide in combination with GSK126 (1 μM). (*** p-value < 0.0001, Student’s t-test).
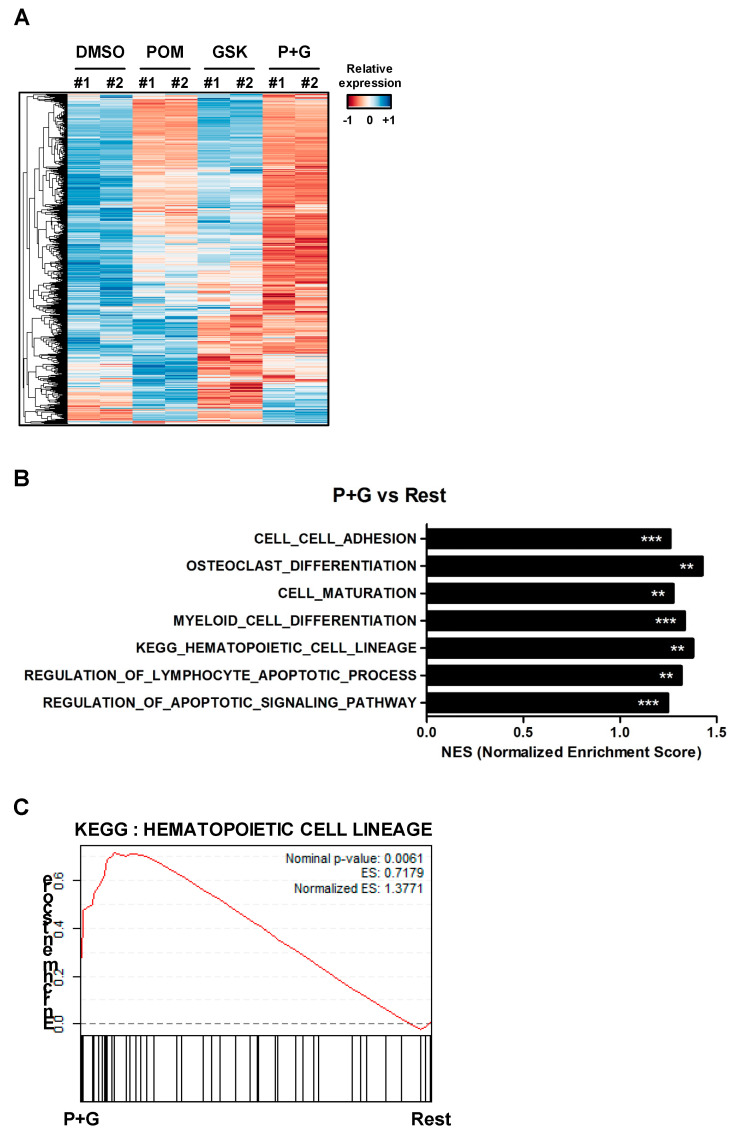
2.4. The GSK126–Pomalidomide Combination Alters Cell Plasticity and Regulates Hematopoietic Cell Lineage
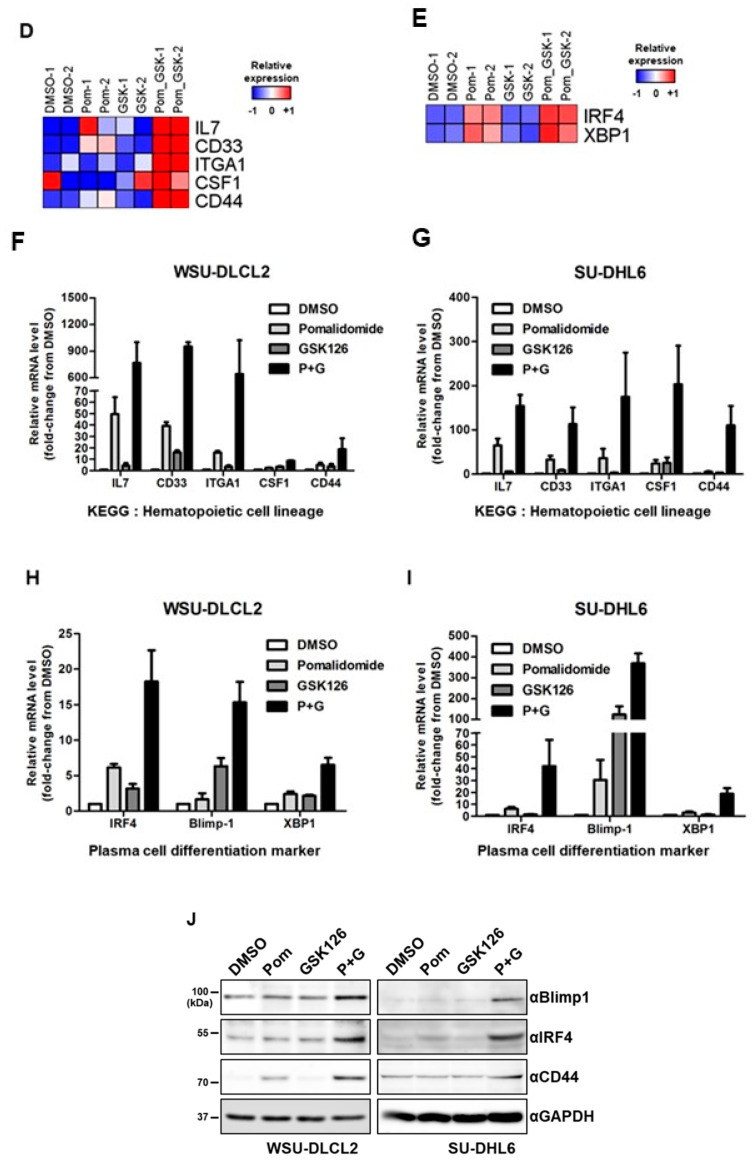
To analyze transcriptional changes induced by the combination of GSK126 and pomalidomide, WSU-DLCL2 cells were treated with dimethyl sulfoxide (DMSO), pomalidomide, GSK126, or a combination of GSK126 and pomalidomide for 24 h, followed by RNA sequencing analysis. Hierarchical clustering and differential expression analysis showed that the pomalidomide–GSK126 combination differentially regulated subsets of genes, compared with DMSO, pomalidomide, or GSK126 alone (Figure 4A and Figure S4A–C). Gene set enrichment analysis (GSEA) revealed enhanced expression of genes involved in cell plasticity, such as hematopoietic cell lineage (Normalized enrichment score (NES) = 1.377) and myeloid cell differentiation (NES = 1.334) as well as lymphocyte apoptotic processes (NES = 1.317) in co-treated cells (Figure 4B–E and Figure S4D,E). Consistent with our cell viability data, genes involved in apoptosis were upregulated in the co-treatment group (Figure S4F). Increased expression of genes related to hematopoietic cell lineage (interleukin 7 (IL7), CD33, integrin subunit alpha 1 (ITGA1), colony stimulating factor 1 (CSF1), and CD44) was validated in WSU-DLCL2 and SU-DHL6 cells using quantitative reverse transcription PCR (RT-qPCR) (Figure 4F,G). IL7, an essential regulator of B-cell development, upregulates cell adhesion molecules and monocyte chemoattractant protein [25,26,27]. The other three genes (ITGA1, CSF1, and CD33) are abundantly expressed in plasma cells [28,29,30]. The results suggested that co-treatment altered cell polarity, thereby inducing plasma cell-like differentiation of -mutant DLBCL. Accordingly, we focused on three representative genes necessary for plasma cell differentiation, specifically, interferon regulatory factor 4 (IRF4), PR/SET domain 1 (PRDM1/Blimp-1), and X-box binding protein 1 (XBP1) [31,32]. RNA sequencing analysis showed that co-treatment enhanced the expression of IRF4 and XBP1 (Figure 4G), consistent with RT-qPCR findings of IRF4, XBP1, and Blimp-1 mRNA upregulation in WSU-DLCL2 and SU-DHL6 (Figure 4H,I). Additionally, we examined protein levels of Blimp-1, IRF4, and CD44 via western blot analysis. CD44, an adhesion molecule, is highly expressed in plasma cells, but barely in GCB cells [33,34,35,36]. Blimp-1 and IRF4 are essential transcriptional regulators in plasma cell differentiation. IRF4 deficiency leads to impaired expression of B-cell activation-induced deaminase and suppression of class-switch recombination [37]. Blimp-1 plays a critical role in the terminal differentiation of B-cells into Ig-secreting plasma cells [38]. The levels of all three proteins were increased in WSU-DLCL2 and SU-DHL6 cells (Figure 4J and Figure S6F), collectively indicating that combined treatment with pomalidomide and GSK126 modulates cell plasticity and promotes plasma cell differentiation in EZH2-mutant DLBCL.
Figure 4.
Combination treatment with GSK126 and pomalidomide changes cell plasticity and regulates hematopoietic cell lineage. (A) Heatmap depicting differentially expressed genes in WSU-DLCL2 treated with DMSO, pomalidomide (POM), GSK126 (GSK), or pomalidomide and GSK126 (P+G). Genes were hierarchically clustered (rows). In the heatmap, orange denotes upregulation, while blue denotes downregulation. (B) Gene set enrichment analysis (GSEA) of RNA-seq data. Significantly enriched gene sets in P+G relative to the remaining groups are shown as a bar chart. Asterisks indicate significance (** FDR < 0.01, and *** FDR < 0.001). (C) GSEA enrichment plot of the hematopoietic cell lineage pathway. (D) Genes involved in hematopoietic cell lineage were upregulated to a more significant extent in P+G than in other groups. (E) Genes essential for plasma cell differentiation were upregulated to a more significant extent in P+G than in other groups. (F,G) RT-qPCR analysis of mRNA levels of genes involved in hematopoietic cell lineage in WSU-DLCL2 and SU-DHL6 cells. (H,I) RT-qPCR analysis of mRNA levels of genes involved in plasma cell differentiation in WSU-DLCL2 and SU-DHL6. (J) WSU-DLCL2 and SU-DHL6 cells were treated with pomalidomide (Pom, 1 μM), GSK126 (1 μM), and P+G for 48 h. Cell lysates were analyzed via western blot using the indicated antibodies.
2.5. GSK126 and Pomalidomide Synergistically Inhibit Tumor Growth In Vivo
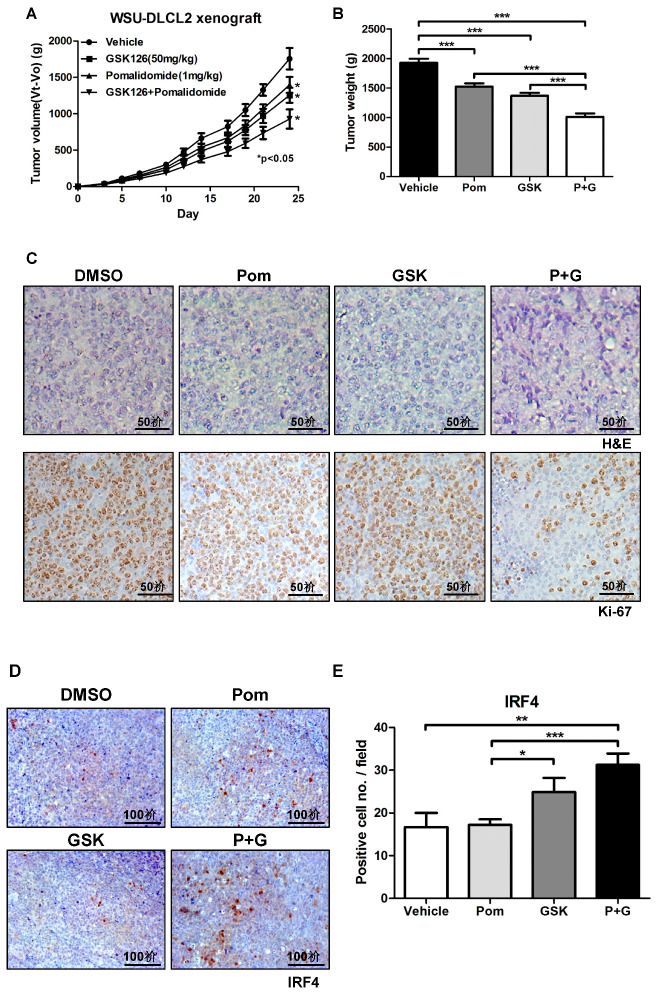
Given the findings from our in vitro analyses that co-treatment with pomalidomide and GSK126 promotes B-cell differentiation, thereby inhibiting the proliferation of EZH2 mutant DLBCL, we further examined the potential synergistic effects of pomalidomide and GSK126 in vivo. To this end, DLBCL xenograft mice implanted with WSU-DLCL2 were intraperitoneally administered vehicle, GSK126, pomalidomide, or both compounds for 25 days. In this model, the combination treatment significantly inhibited tumor volume and weight, compared with individual treatments (Figure 5A,B). No significant weight loss was observed with either the combination or individual treatments (Figure S5A). Immunohistochemical staining additionally revealed reduced expression of Ki-67, a representative proliferative marker, along with sharp and spiky cell morphology in tumor samples treated with both pomalidomide and GSK126 (Figure 5C). Moreover, in line with transcriptome analyses, the combination treatment enhanced the expression of IRF4, a key transcription factor for Blimp-1-dependent plasma cell differentiation [39] (Figure 5D,E), and apoptotic markers, such as cleaved PARP, cleaved caspase 3, and p21 (Figures S5B and S6G). Our collective results indicated that GSK126 and pomalidomide act synergistically to promote plasma differentiation, thereby inhibiting the proliferation of EZH2 gain-of-function mutant DLBCL.
Figure 5.
Combination of GSK126 with pomalidomide synergistically inhibit tumor growth in vivo. Specific pathogen-free CB17/SCID mice (n = 6) were implanted subcutaneously with WSU-DLCL2 cells (1 × 106) and treated with vehicle, GSK126 (50 mg/kg), pomalidomide (1 mg/kg), or GSK126 + pomalidomide. (A) Tumor volumes were measured three times a week for 24 days. (B) Tumor weights were measured at the end of the experimental period. (C) Representative hematoxylin/eosin (H&E) staining (top) and Ki-67 immunostaining (bottom) of xenograft tumors (scale bar, 50 μm). (D) Representative photographs of IRF4 immunostaining (scale bar, 100 μm). (E) Mean numbers of IRF4-positive cells per field in xenograft tumor tissues are shown. (* p-value < 0.05, ** p-value < 0.005, *** p-value < 0.0001, Student’s t-test).
3. Discussion
Several genome studies have revealed that GCB-DLBCL harbors oncogenic events such as t(14;18) translocation, and mutations in EZH2, KMT2D, S1PR2, or GNA13 [40]. Among these, genetically altered EZH2 has been detected in many cases. Given that the 5-year survival rate of GCB-DLBCL is ~59% with currently available immune chemotherapeutic agents, including R-CHOP, R-DHAP, R-ICE, and DA-EPOCH-R [41,42], targeting EZH2 to DLBCL with EZH2 mutations may be more beneficial. Indeed, several clinical trials with EZH2 inhibitors are underway. A phase 1/2 study of tazemetostat, a potent inhibitor of EZH2, either as a single agent or combined with the steroid, prednisolone, is ongoing in DLBCL patients (NCT01897571). While a single EZH2 inhibitor may prove effective, determination of the efficacy of combination treatments with EZH2 inhibitors and other FDA-approved drugs is essential to overcome the potential threat of drug resistance or limitation of potency.
In this study, we demonstrated that combined treatment with GSK126 and pomalidomide or lenalidomide synergistically inhibited cell growth and induced apoptosis in EZH2 gain-of-function mutant DLBCL. This synergistic activity was dependent on CRBN, but not on the degradation of the well-known substrates, IKZF1 and IKZF3, highlighting the possibility that proteins other than IKZF1 and IKZF3, which are degraded by pomalidomide and lenalidomide, contribute to the collective effect. Sievers and co-workers defined the human C2H2 zinc finger degrome targeted by IMiDs using whole zinc finger domain degradation reporters [43]. Another group identified multiple proteins degraded by IMiD-dependent CRL4CRBN with the aid of quantitative mass spectrometry [21]. Based on these findings, we speculate that EZH2 inhibition, combined with degradation of one or more proteins targeted by pomalidomide or lenalidomide, may derepress genes related to plasma cell differentiation. On the other hand, CRBN could act as an epigenetic modulator in addition to its role as an E3 ligase receptor. Kang et al. [22] demonstrated that CRBN interacts with EZH1/2, repressing the expression of Kcna3 through direct binding to the Kcna3 locus in murine T-cells. Interestingly, thalidomide treatment blocks recruitment of CRBN to the Kcna3 locus, leading to derepression of Kcna3 transcription. In this regard, the combined activities of EZH2 inhibition with transcriptional regulation by IMiDs may lead to synergistic effects in hyperactive EZH2 DLBCL.
Gain-of-function mutations in EZH2 have been reported in not only GCB-DLBCL, but also in other cancer types, including follicular lymphoma and melanoma [44,45]. Several missense mutations in the catalytic SET domain of EZH2 have been identified in about 7–12% of follicular lymphoma cases [6]. Functional analysis of EZH2 revealed that these mutations promote the levels of H3K27m3, resulting in suppression of specific genes, such as TCF4, FOXP1, TCL1A, BIK, RASSF6P, and CDKN1 [6,7]. In addition, hyperactive EZH2 mutations are reported to control cell growth and metastasis through silencing of distinct tumor suppressors, such as DCK, AMD1, and WDR19, in cutaneous melanoma [46]. Further investigation of these combinational effects on different cancer types should provide a solid foundation for treating patients with hyperactive EZH2 mutations.
Besides acquisition of gain-of-function mutations, EZH2 is overexpressed in various cancer types and its targeting has been shown to inhibit cell proliferation and metastasis [47]. EZH2 with mutations in genes that encode subunits of SWI/SNF chromatin remodeling complexes is synthetically lethal [48,49]. Genetic alterations involving various subunits of SWI/SNF complexes have been detected in ~20% of human tumors [50]. EZH2 knockdown and EZH2 inhibitor treatment are reported to impair proliferation and colony formation in SWI/SNF-mutant cancer cells. Our examination of the combined effects of GSK126 and pomalidomide in the SWI/SNF-mutant A549 (SMARCA4 mutant) and SKOV3 (ARID1A mutant) cell lines revealed no additional effects on cell viability (data not shown).
Our results showed the potential therapeutic opportunity for EZH2 inhibition and IMiDs in DLBCL with EZH2 mutation; however, further studies will be needed to validate this co-treatment. Studies with patient-derived DLBCL cells or patient-derived xenograft (PDX) models will definitely help to prove the efficacy of this therapeutic modality. From a mechanistic point of view, it will also be intriguing to examine whether this combination treatment would promote the differentiation in primary immature B-cells.
4. Materials and Methods
4.1. Western Blot
Cells were lysed in RIPA buffer (150 mM NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH 8.0). Lysates were separated via SDS-PAGE and transferred to a nitrocellulose (NC) membrane (GE Healthcare, Chicago, IL, USA). Blots were probed with the indicated primary antibodies, followed by Horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG antibody, and detection was performed with a chemiluminescent HRP substrate (Thermo, Waltham, MA, USA). Antibodies for western blots are listed in Table S1.
4.2. Cell Cultures and Stable Cell Lines
The lymphoma cell lines, Toledo (ATCC, Cat. No. CRL-2631, RRID: CVCL_3611), SU-DHL6 (ATCC, Cat. No. CRL-2959, RRID: CVCL_2206), and SU-DHL4 (ATCC, Cat. No. CRL-2957, RRID: CVCL_0539), were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). WSU-DLCL2 (DSMZ, Cat. No. ACC-575, RRID: CVCL_1902), SU-DHL10 (DSMZ, Cat. No. ACC-576, RRID: CVCL_1889), and OCI-LY19 (DSMZ, Cat. No. ACC-528, RRID: CVCL_1878) were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). It was confirmed that there was no mycoplasma contamination in the cells upon thawing by using the Universal Mycoplasma Detection Kit (ATCC, Cat. No. 30-1012K). All experiments were performed between passages 6 and 20 upon purchase. Each cell line was cultured in RPMI 1640 (GIBCO, Waltham, MA, USA, Cat# 11875-093) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Cat. No. 16000-044) and 100x Antibiotic-Antimycotic (GIBCO, Cat. No. 15240062) diluted to 1×. Cells were maintained in a humidified incubator at 37 °C and 5% (v/v) CO2. Short hairpin (shRNA) knockdown cell lines were generated using the retroviral infection method. For production of lentiviral particles, HEK293T (ATCC, Cat. No. CRL-3216, RRID: CVCL_0063) cells were transfected with a lentiviral plasmid containing each gene, psPAX2, and pMD2.G at a ratio of 1:0.75:0.25. After 72 h of transfection, supernatant fractions containing lentiviruses were collected and filtered through a 0.45 μm sterile filter to remove cell debris. Cells were treated with the viral supernatant and 8 μg/mL polybrene (Sigma Aldrich, St. Louis, MO, USA, Cat. No. H9268), followed by incubation for 48 h. After viral transduction, cells were subjected to treatments and selected with 1 μg/mL puromycin. Mission shRNA plasmids for CRBN, IKZF1, and IKZF3 were purchased from Sigma-Aldrich. An inducible shRNA Tet-pLKO-puro vector was obtained from Addgene (Watertown, MA, USA) (plasmid #21915), and EZH2 targeting sequences were cloned as reported previously [51]. Each target sequence is listed in Table S2.
4.3. Compounds
Pomalidomide (Cat. No. S1567) and GSK126 (Cat. No. S7061) were purchased from Selleck Chemicals (Houston, TX, USA). EED226 was synthesized as described previously [11]. Chemicals were diluted in dimethyl sulfoxide (DMSO) and stored at −20 °C until use.
4.4. Cell Viability Assay
Cells were cultured in 96-well white opaque plates at a density of 1 × 104/mL in the appropriate medium with FBS. After treatment with the appropriate compounds, assay plates were incubated for 6 days at 37 °C and 5% CO2 under >90% relative humidity. The number of viable cells was determined using the CellTiter-Glo Luminescent Cell Viability Assay (Promega Corp, Madison, WI, USA) based on quantification of cellular ATP as a marker of metabolically active cells using Victor X3 (Perkin Elmer, Waltham, MA, USA). Data were expressed as the mean percentage of duplicate cells and normalized to DMSO. Results from the cell viability assay were analyzed using the CompuSyn 1.0 software (ComboSyn Inc., Paramus, NJ, USA) developed by Chou based on the median-effect principle to determine whether the combined effects of drugs were synergistic, antagonistic, or additive. An antagonistic effect is above the slope line (combination index (CI) > 1), a synergistic effect is under the line (CI < 1), and an additive effect is on the line (CI = 1) [23].
4.5. RNA Sequencing and Data Processing
The RNA sequencing library was prepared using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, USA) and sequencing was performed using the Illumina HiSeq2000 platform to generate 100 bp paired-end reads. The human reference genome was obtained from the NCBI genome and genome indexing was performed using STAR (v.2.5.1) [52]. Sequenced reads were mapped to human genome (hg19) STAR, and gene expression levels were quantified with the count module. The edgeR (v.3.12.1) [53] package was applied to select differentially expressed genes from RNA-seq count data between DMSO, POM, GSK, and POM+GSK samples (fold change > 1.5, p-value < 0.05). The TMM (trimmed mean of M-value normalization)-normalized CPM (counts per million) value of each gene was added to 1 and log2-transformed for further analysis. A heatmap was generated using MultiExperiment Viewer (MeV) [54] and R (v.3.5.0) heatmap package (v.1.0.12). Next generation sequencing (NGS) data were deposited in the NCBI Gene Expression Omnibus under accession number GSE147320. Raw sequence tags were deposited in the NCBI Short Read Archive (SRA) under accession number SRP253527.
For GSEA analysis of RNA-seq data, we used GSEA software [55] from the Broad Institute website (v.4.0.0). Our expression dataset was analyzed against a hallmark Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) gene sets (H, C2, and C5.gmt files from MSigDB v.7.0). Statistical significance (nominal p-value) of the enrichment score (ES) was calculated by running 500 gene set permutations. The normalized enrichment score (NES) accounted for the size of the gene set.
4.6. Quantitative Real-Time PCR
Total RNA was extracted using a Qiagen RNeasy kit (Qiagen, Hilden, Germany, Cat. No. 74104) and cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Cat. No. K1622). Quantitative PCR (qPCR) was conducted using the Solg™ Real-Time PCR Kit, with EvaGreen™ intercalating dye detection (Solgent, Daejeon, Korea, Cat. No. SRH91-R25h). HPRT1 was employed as the internal control. The primers used are listed in Table S3.
4.7. Flow Cytometry and Annexin V Staining
Apoptosis was assessed using the FITC Annexin V Apoptosis Detection Kit (BD Biosciences, Cat. No.556547) according to the manufacturer’s instructions. SU-DHL6 cells were seeded in a 150 mm cell culture dish at a density of 4 × 105/mL cells per plate and treated for 6 days. Harvested cells were resuspended in annexin-binding buffer according to the manufacturer’s protocol. Next, cells were labeled using annexin V-FITC and PI. After incubation for 15 minutes at room temperature in the dark, samples were analyzed on a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA, FACSCalibur) for the detection of annexin V- and PI-positive subpopulations. DMSO-treated cells were used as the control group.
4.8. Immunohistochemistry
Xenograft tumor tissues were fixed in 10% neutral buffered formalin and processed to construct paraffin blocks following standard protocols [56]. Tissues were sliced into 4 μm thick sections and stained with hematoxylin/eosin (H&E). For immunohistochemical analysis, sections were blocked using 5% goat serum and incubated with antibodies against Ki-67 and IRF4. The mean number of IRF4-positive cells per field was calculated. Antibodies are listed in Table S1.
4.9. Xenograft Mouse Experiments
Female CB17/SCID mice (CB17/Icr-Prkdcscid/CrlCrlj, 5 weeks old) were purchased from Charles River Japan (Yokohama, Japan) and housed under specific pathogen-free conditions. Rooms were maintained under a 12 h light–dark cycle at 21 ± 2 °C. Animals were allowed to acclimatize to the local environment for one week before use in experiments. All animal experiments were approved by the Institutional Animal Care and Use Committee of Korea Research Institute of Bioscience and Biotechnology. WSU-DLCL2 cell suspensions (3 × 106 cells) were injected subcutaneously into CB17/SCID mice. When tumor volumes reached ~50 mm3, mice were randomly divided into four groups. GSK126 and pomalidomide were dissolved in 20% Captisol® and saline, respectively. Vehicle, GSK126 (50 mg/kg), and pomalidomide (1 mg/kg) were administered intraperitoneally (5 days a week for 3 weeks). Tumor volumes were measured three times a week using Vernier calipers and calculated using the formula: length (mm) × width (mm) × height (mm)/2. On day 24, tumors were surgically removed, weighed, photographed, divided into two sections, and stored in formalin and liquid nitrogen for further analysis. Body weights of mice were measured three times a week throughout the experimental period. The ethical code number for animal experiments was KRIBB-AEC-19024.
4.10. Statistical Analysis
One-way ANOVA, followed by Dunnett’s multiple comparison test, and two-way ANOVA, followed by Bonferroni multiple comparison test, were applied for statistical analysis using the GraphPad Prism5 software (GraphPad Software Inc., La Jolla, CA, USA). Cell viability and tumor volume data were analyzed using an independent Student’s t-test and significance was indicated (* p-value < 0.05, ** p-value < 0.005, *** p-value < 0.0001).
5. Conclusions
In conclusion, data from our study collectively demonstrated the synergistic effects of PRC2 inhibition and IMiDs for the first time and shed light on the molecular mechanisms by which the combination therapy induced plasma cell differentiation and eventually apoptosis in EZH2 gain-of-function mutant DLBCL.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/9/2541/s1, Figure S1: Cytotoxic effect on various cancer cell lines, Figure S2: Inhibition or EZH2 knockdown combined with pomalidomide synergistically inhibits proliferation of EZH2-mutant DLBCL cells, Figure S3: Cytotoxic effect of GSK126 with lenalidomide in WSU-DLCL2 cells, Figure S4: Whole transcriptomic comparative analysis between POM+GSK and rest, Figure S5: Specific pathogen-free CB17/SCID mice (n = 6) were implanted subcutaneously with WSU-DLCL2 cells, Figure S6: Original images of westernblot, Table S1: Antibodies used for detecting proteins, Table S2: shRNA targeting sequences for knockdown cell lines, and Table S3: Primer sequences for RT-qPCR.
Author Contributions
S.-H.J. and S.P. designed and performed experiments, analyzed data, and co-wrote the paper; J.-H.K. (Jong-Hwan Kim) and S.-Y.K. performed bioinformatic analyses; J.D.H. and J.Y.H. performed chemical analyses; M.Y.L., J.S.K., and T.-S.H. performed animal analyses; S.G.P. and B.C.P. supervised the research; S.K. and J.-H.K. (Jeong-Hoon Kim) designed the experiments and co-wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant (CAP-15-11-KRICT) from the National Research Council of Science and Technology, Ministry of Science, ICT, and Future Planning; a grant (NRF-2019M3E5D4069882) from the National Research Foundation, Ministry of Science, ICT, and Future Planning; and a grant from the KRIBB Initiative Program.
Conflicts of Interest
The authors declare no competing interests.
References
- 1.Jaffe E.S., Harris N.L., Diebold J., Muller-Hermelink H.K. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: Report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J. Clin. Oncol. 1999;17:3835–3849. doi: 10.1200/JCO.1999.17.12.3835. [DOI] [PubMed] [Google Scholar]
- 2.Jaffe E.S., Hans C.P., Weisenburger D.D., Greiner T.C., Gascoyne R.D., Delabie J., Ott G., Müller-Hermelink H.K., Campo E., Braziel R., et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 3.Morin R.D., Johnson N.A., Severson T.M., Mungall A.J., An J., Goya R., Paul J.E., Boyle M., Woolcock B.W., Kuchenbauer F., et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Eynde A.V., Bernard D., Vanderwinden J.-M., et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 5.Aldiri I., Vetter M.L. PRC2 during vertebrate organogenesis: A complex in transition. Dev. Biol. 2012;367:91–99. doi: 10.1016/j.ydbio.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yap D.B., Chu J., Berg T., Schapira M., Cheng S.-W., Grace C., Moradian A., Morin R.D., Mungall A.J., Meissner B., et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo S., Chan J.K., Iqbal J., McKeithan T., Fu K., Meng B., Pan Y., Cheuk W., Luo D., Wang R.S., et al. EZH2 mutations in follicular lymphoma from different ethnic groups and associated gene expression alterations. Clin. Cancer Res. 2014;20:3078–3086. doi: 10.1158/1078-0432.CCR-13-1597. [DOI] [PubMed] [Google Scholar]
- 8.Beguelin W., Popovic R., Teater M., Jiang Y., Bunting K.L., Rosen M., Shen H., Yang S.N., Wang L., Ezponda T., et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knutson S.K., Wigle T.J., Warholic N.M., Sneeringer C.J., Allain C.J., Klaus C.R., Sacks J.D., Raimondi A., Majer C.R., Song J., et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 10.McCabe M.T., Ott H.M., Ganji G., Korenchuk S., Thompson C., Van Aller G.S., Liu Y., Graves A.P., Della Pietra A., 3rd, Diaz E., et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 11.Qi W., Zhao K., Gu J., Huang Y., Wang Y., Zhang H., Zhang M., Zhang J., Yu Z., Li L., et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol. 2017;13:381–388. doi: 10.1038/nchembio.2304. [DOI] [PubMed] [Google Scholar]
- 12.Sudo T., Utsunomiya T., Mimori K., Nagahara H., Ogawa K., Inoue H., Wakiyama S., Fujita H., Shirouzu K., Mori M. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br. J. Cancer. 2005;92:1754–1758. doi: 10.1038/sj.bjc.6602531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain M., Rao M., Humphries A.E., Hong J.A., Liu F., Yang M., Caragacianu D., Schrump D.S. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69:3570–3578. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weikert S., Christoph F., Köllermann J., Müller M., Schrader M., Miller K., Krause H. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int. J. Mol. Med. 2005;16:349–353. doi: 10.3892/ijmm.16.2.349. [DOI] [PubMed] [Google Scholar]
- 15.Bachmann I.M., Halvorsen O.J., Collett K., Stefansson I.M., Straume O., Haukaas S.A., Salvesen H.B., Otte A.P., Akslen L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z., Yang P., Li W., He F., Wei J., Zhang T., Zhong J., Chen H., Cao J. Expression of EZH2 is associated with poor outcome in colorectal cancer. Oncol. Lett. 2018;15:2953–2961. doi: 10.3892/ol.2017.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chanan-Khan A.A., Swaika A., Paulus A., Kumar S.K., Mikhael J.R., Rajkumar S.V., Dispenzieri A., Lacy M.Q. Pomalidomide: The new immunomodulatory agent for the treatment of multiple myeloma. Blood Cancer J. 2013;3:e143. doi: 10.1038/bcj.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shortt J., Hsu A.K., Johnstone R.W. Thalidomide-analogue biology: Immunological, molecular and epigenetic targets in cancer therapy. Oncogene. 2013;32:4191–4202. doi: 10.1038/onc.2012.599. [DOI] [PubMed] [Google Scholar]
- 19.Krönke J., Udeshi N.D., Narla A., Grauman P., Hurst S.N., McConkey M., Svinkina T., Heckl D., Comer E., Li X., et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu G., Middleton R.E., Sun H., Naniong M., Ott C.J., Mitsiades C.S., Wong K.-K., Bradner J.E., Kaelin W.G. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donovan K.A., An J., Nowak R.P., Yuan J.C., Fink E.C., Berry B.C., Ebert B.L., Fischer E.S. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. Elife. 2018;7:e38430. doi: 10.7554/eLife.38430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang J.A., Park S.H., Jeong S.P., Han M.H., Lee C.R., Lee K.M., Kim N., Song M.R., Choi M., Ye M., et al. Epigenetic regulation of Kcna3-encoding Kv1.3 potassium channel by cereblon contributes to regulation of CD4+ T-cell activation. Proc. Natl. Acad. Sci. USA. 2016;113:8771–8776. doi: 10.1073/pnas.1502166113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 24.Lee C.H., Yu J.R., Kumar S., Jin Y., LeRoy G., Bhanu N., Kaneko S., Garcia B.A., Hamilton A.D., Reinberg D. Allosteric Activation Dictates PRC2 Activity Independent of Its Recruitment to Chromatin. Mol. Cell. 2018;70:422–434. doi: 10.1016/j.molcel.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corfe S.A., Paige C.J. The many roles of IL-7 in B cell development; mediator of survival, proliferation and differentiation. Semin. Immunol. 2012;24:198–208. doi: 10.1016/j.smim.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Dias S., Silva H., Jr., Cumano A., Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J. Exp. Med. 2005;201:971–979. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R., Paul A., Ko K.W., Sheldon M., Rich B.E., Terashima T., Dieker C., Cormier S., Li L., Nour E.A., et al. Interleukin-7 induces recruitment of monocytes/macrophages to endothelium. Eur. Heart J. 2012;33:3114–3123. doi: 10.1093/eurheartj/ehr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos A., Tsafou K., Stolte C., Pletscher-Frankild S., O’Donoghue S.I., Jensen L.J. Comprehensive comparison of large-scale tissue expression datasets. PeerJ. 2015;3:e1054. doi: 10.7717/peerj.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamane F., Nishikawa Y., Matsui K., Asakura M., Iwasaki E., Watanabe K., Tanimoto H., Sano H., Fujiwara Y., Stanley E.R., et al. CSF-1 receptor-mediated differentiation of a new type of monocytic cell with B cell-stimulating activity: Its selective dependence on IL-34. J. Leukoc. Biol. 2014;95:19–31. doi: 10.1189/jlb.0613311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terstappen L.W., Johnsen S., Segers-Nolten I.M., Loken M.R. Identification and characterization of plasma cells in normal human bone marrow by high-resolution flow cytometry. Blood. 1990;76:1739–1747. doi: 10.1182/blood.V76.9.1739.1739. [DOI] [PubMed] [Google Scholar]
- 31.Tellier J., Shi W., Minnich M., Liao Y., Crawford S., Smyth G.K., Kallies A., Busslinger M., Nutt S.L. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat. Immunol. 2016;17:323–330. doi: 10.1038/ni.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minnich M., Tagoh H., Bonelt P., Axelsson E., Fischer M., Cebolla B., Tarakhovsky A., Nutt S.L., Jaritz M., Busslinger M. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat. Immunol. 2016;17:331–343. doi: 10.1038/ni.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manz R.A., Löhning M., Cassese G., Thiel A., Radbruch A. Survival of long-lived plasma cells is independent of antigen. Int. Immunol. 1998;10:1703–1711. doi: 10.1093/intimm/10.11.1703. [DOI] [PubMed] [Google Scholar]
- 34.Medina F., Segundo C., Campos-Caro A., González-García I., Brieva J.A. The heterogeneity shown by human plasma cells from tonsil, blood, and bone marrow reveals graded stages of increasing maturity, but local profiles of adhesion molecule expression. Blood. 2002;99:2154–2161. doi: 10.1182/blood.V99.6.2154. [DOI] [PubMed] [Google Scholar]
- 35.Jalkanen S.T., Bargatze R.F., Herron L.R., Butcher E.C. A lymphoid cell surface glycoprotein involved in endothelial cell recognition and lymphocyte homing in man. Eur. J. Immunol. 1986;16:1195–1202. doi: 10.1002/eji.1830161003. [DOI] [PubMed] [Google Scholar]
- 36.de los Toyos J., Jalkanen S., Butcher E.C. Flow cytometric analysis of the Hermes homing-associated antigen on human lymphocyte subsets. Blood. 1989;74:751–760. doi: 10.1182/blood.V74.2.751.751. [DOI] [PubMed] [Google Scholar]
- 37.Klein U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro-Shelef M., Lin K.-I., McHeyzer-Williams L.J., Liao J., McHeyzer-Williams M.G., Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/S1074-7613(03)00267-X. [DOI] [PubMed] [Google Scholar]
- 39.Sciammas R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Sehn L.H., Gascoyne R.D. Diffuse large B-cell lymphoma: Optimizing outcome in the context of clinical and biologic heterogeneity. Blood. 2015;125:22–32. doi: 10.1182/blood-2014-05-577189. [DOI] [PubMed] [Google Scholar]
- 41.Thieblemont C., Briere J., Mounier N., Voelker H.U., Cuccuini W., Hirchaud E., Rosenwald A., Jack A., Sundstrom C., Cogliatti S., et al. The germinal center/activated B-cell subclassification has a prognostic impact for response to salvage therapy in relapsed/refractory diffuse large B-cell lymphoma: A bio-CORAL study. J. Clin. Oncol. 2011;29:4079–4087. doi: 10.1200/JCO.2011.35.4423. [DOI] [PubMed] [Google Scholar]
- 42.Delarue R., Tilly H., Mounier N., Petrella T., Salles G., Thieblemont C., Bologna S., Ghesquières H., Hacini M., Fruchart C., et al. Dose-dense rituximab-CHOP compared with standard rituximab-CHOP in elderly patients with diffuse large B-cell lymphoma (the LNH03-6B study): A randomised phase 3 trial. Lancet Oncol. 2013;14:525–533. doi: 10.1016/S1470-2045(13)70122-0. [DOI] [PubMed] [Google Scholar]
- 43.Sievers Q.L., Petzold G., Bunker R.D., Renneville A., Slabicki M., Liddicoat B.J., Abdulrahman W., Mikkelsen T., Ebert B.L., Thoma N.H. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science. 2018;362:6414. doi: 10.1126/science.aat0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bodor C., O’Riain C., Wrench D., Matthews J., Iyengar S., Tayyib H., Calaminici M., Clear A., Iqbal S., Quentmeier H., et al. EZH2 Y641 mutations in follicular lymphoma. Leukemia. 2011;25:726–729. doi: 10.1038/leu.2010.311. [DOI] [PubMed] [Google Scholar]
- 45.Ryan R.J., Nitta M., Borger D., Zukerberg L.R., Ferry J.A., Harris N.L., Iafrate A.J., Bernstein B.E., Sohani A.R., Le L.P. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS ONE. 2011;6:e28585. doi: 10.1371/journal.pone.0028585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zingg D., Debbache J., Schaefer S.M., Tuncer E., Frommel S.C., Cheng P., Arenas-Ramirez N., Haeusel J., Zhang Y., Bonalli M. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 47.Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat. Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim K.H., Kim W., Howard T.P., Vazquez F., Tsherniak A., Wu J.N., Wang W., Haswell J.R., Walensky L.D., Hahn W.C., et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 2015;21:1491–1496. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bitler B.G., Aird K.M., Garipov A., Li H., Amatangelo M., Kossenkov A.V., Schultz D.C., Liu Q., Shih Ie M., Conejo-Garcia J.R., et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat. Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadoch C., Hargreaves D.C., Hodges C., Elias L., Ho L., Ranish J., Crabtree G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiederschain D., Wee S., Chen L., Loo A., Yang G., Huang A., Chen Y., Caponigro G., Yao Y.M., Lengauer C., et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 52.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howe E.A., Sinha R., Schlauch D., Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27:3209–3210. doi: 10.1093/bioinformatics/btr490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuda Y., Fujii T., Suzuki T., Yamahatsu K., Kawahara K., Teduka K., Kawamoto Y., Yamamoto T., Ishiwata T., Naito Z. Comparison of fixation methods for preservation of morphology, RNAs, and proteins from paraffin-embedded human cancer cell-implanted mouse models. J. Histochem. Cytochem. 2011;59:68–75. doi: 10.1369/jhc.2010.957217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.