Forest and agroecosystems, as well as animal and human health, are threatened by emerging pathogens. Following decimation of chestnuts in the United States, the fungal pathogen Cryphonectria parasitica colonized Europe. After establishment, the pathogen population gave rise to a highly successful lineage that spread rapidly across the continent. Core to our understanding of what makes a successful pathogen is the genetic repertoire enabling the colonization and exploitation of host species. Here, we have assembled >100 genomes across two related genera to identify key genomic determinants leading to the emergence of chestnut blight. We found subtle yet highly specific changes in the transition from saprotrophy to latent pathogenicity mostly determined by enzymes involved in carbohydrate metabolism. Large-scale genomic analyses of genes underlying key nutrition modes can facilitate the detection of species with the potential to emerge as pathogens.
KEYWORDS: Cryphonectria, comparative genomics, lifestyle evolution, tree pathogen
ABSTRACT
Emerging fungal pathogens are a threat to forest and agroecosystems, as well as animal and human health. How pathogens evolve from nonpathogenic ancestors is still poorly understood, making the prediction of future outbreaks challenging. Most pathogens have evolved lifestyle adaptations, which were enabled by specific changes in the gene content of the species. Hence, understanding transitions in the functions encoded by genomes gives valuable insight into the evolution of pathogenicity. Here, we studied lifestyle evolution in the genus Cryphonectria, including the prominent invasive pathogen Cryphonectria parasitica, the causal agent of chestnut blight on Castanea species. We assembled and compared the genomes of pathogenic and putatively nonpathogenic Cryphonectria species, as well as sister group pathogens in the family Cryphonectriaceae (Diaporthales, Ascomycetes), to investigate the evolution of genome size and gene content. We found a striking loss of genes associated with carbohydrate metabolism (CAZymes) in C. parasitica compared to other Cryphonectriaceae. Despite substantial CAZyme gene loss, experimental data suggest that C. parasitica has retained wood colonization abilities shared with other Cryphonectria species. Putative effectors substantially varied in number, cysteine content, and protein length among species. In contrast, secondary metabolite gene clusters show a high degree of conservation within the genus. Overall, our results underpin the recent lifestyle transition of C. parasitica toward a more pathogenic lifestyle. Our findings suggest that a CAZyme loss may have promoted pathogenicity of C. parasitica on Castanea species. Analyzing gene complements underlying key nutrition modes can facilitate the detection of species with the potential to emerge as pathogens.
IMPORTANCE Forest and agroecosystems, as well as animal and human health, are threatened by emerging pathogens. Following decimation of chestnuts in the United States, the fungal pathogen Cryphonectria parasitica colonized Europe. After establishment, the pathogen population gave rise to a highly successful lineage that spread rapidly across the continent. Core to our understanding of what makes a successful pathogen is the genetic repertoire enabling the colonization and exploitation of host species. Here, we have assembled >100 genomes across two related genera to identify key genomic determinants leading to the emergence of chestnut blight. We found subtle yet highly specific changes in the transition from saprotrophy to latent pathogenicity mostly determined by enzymes involved in carbohydrate metabolism. Large-scale genomic analyses of genes underlying key nutrition modes can facilitate the detection of species with the potential to emerge as pathogens.
INTRODUCTION
Across the fungal kingdom, species have evolved the ability to persist as either symbionts, commensals, or pathogens on a wide range of living insect, animal, and plant hosts. This variety of fungal lifestyles requires complex adaptations encoded in the genome. Lifestyle-associated adaptations have been of particular interest as pathogen emergence is frequently associated with a significant gain in virulence of a formerly weak pathogen (1). This has been shown for Pyrenophora tritici-repentis, a former saprophyte or weak pathogen on grass species including wheat, which became highly pathogenic on wheat through acquisition of the virulence gene ToxA from the wheat pathogen Stagonospora nodorum (2). Moreover, pathogen emergence can be promoted through host jumps or geographic range expansions (3) or complete host shifts (1). Such host shifts can occur across kingdoms, as shown for insect pathogens from the genus Metarhizium, which likely evolved from plant endophytes or pathogens (4). Interestingly, phylogenomic analyses have shown that pathogens can emerge repeatedly within fungal clades such as Dothideomycetes or even at the genus level (e.g., Aspergillus) (5, 6). Hence, many pathogenic fungi have nonpathogenic ancestors. This suggests that the emergence and evolution of pathogenic lifestyles are coupled with the acquisition of specific traits distinct from nonpathogenic relatives.
To be successful, pathogens must overcome physical and chemical barriers deployed by the host (7). Plant-pathogenic fungi have evolved specific lifestyles (i.e., biotrophy, hemibiotrophy, and necrotrophy) to exploit the host, and each lifestyle requires distinct sets of genes (8–11). The gene repertoire of pathogens evolved through gene gains or losses and proliferation of transposable elements, as well as expansions or contractions of entire gene families, sometimes resulting in increased genome sizes, compared to related nonpathogenic species (12, 13). Gene families notably associated with fungal plant pathogenicity include enzymes for cell wall degradation, small secreted proteins (i.e., effectors), and secondary metabolite gene clusters (14–19). Cell walls are an important physical barrier against pathogens but can be broken down and used as carbon sources by a variety of fungi. Carbohydrate-active enzymes (CAZymes) specific for cellulose, hemicellulose, or pectin degradation are typically classified into the superfamilies of glycoside hydrolases (GHs), glycosyl transferases (GTs), polysaccharide lyases (PLs), and carbohydrate esterases (CEs), as well as enzymes with auxiliary activities (AAs) and carbohydrate-binding modules (CBMs) (20). The types and number of CAZyme-encoding genes vary among species and likely reflect adaptation to different nutritional niches (21). Most notably, necrotrophic pathogens tend to deploy cell wall-degrading enzymes to promote host damage and colonization (22). In contrast, biotrophic pathogens tend to have fewer enzymes involved in cell wall degradation (20, 22). Saprotrophic fungi feeding on decaying plant matter often show an overall reduced CAZyme complement compared to necrotrophic fungi (23) but specific expansions in CAZymes related to cellulose degradation (24).
The emergence of pathogenic lifestyles has often required the ability to secrete effector proteins and secondary metabolites during contact with the host. Effectors are characterized as quickly evolving small, cysteine-rich secreted proteins, which are produced to manipulate plant host immune responses (25, 26). Biotrophic and hemibiotrophic pathogens secrete effector proteins to suppress host immunity and manipulate host cell physiology (27). Necrotrophs deploy effectors also as host-specific toxins (27, 28). However, small secreted proteins resembling effectors are also expressed by saprophytic fungi and may be involved in degradative processes (29). Virulence factors in pathogenic fungi can also include secondary metabolites, which are often low-molecular-weight compounds not essential for fungal growth. Polyketides, nonribosomal peptides, terpenes, and indole alkaloids are the main bioactive compounds acting as cytotoxins, antimicrobials, or enzyme inhibitors (30). Genes underlying secondary metabolite biosynthesis pathways are often clustered in the genome (31). Secondary metabolites are produced by fungi of various lifestyles but may be more relevant virulence factors for necrotrophs, while biotrophs tend to lose the underlying genes (8). Beyond pathogenicity-related functions, saprophytic or endophytic fungi produce secondary metabolites with important antimicrobial activity (32, 33).
The family Cryphonectriaceae (Diaporthales, Ascomycetes) includes mainly bark-inhabiting species ranging from weak to severe pathogens (34, 35). The most aggressive pathogens include Chrysoporthe species affecting hosts in the order Myrtales (e.g., Eucalyptus spp.), as well as Cryphonectria parasitica (Murr.) Barr., the causal agent of chestnut blight on Castanea (Fagaceae) species (36, 37). C. parasitica is native to East Asia (i.e., China, Korea, and Japan), where it occurs as a weak pathogen on Chinese (Castanea mollissima Blume) and Japanese (Castanea crenata Siebold & Zucc.) chestnuts. However, C. parasitica was first described after its discovery in 1904 on American chestnut [Castanea dentata (Marsh.) Borkh.] in the United States (37). The rapid spread of the pathogen following its introduction resulted in the ecological extinction of Ca. dentata throughout its native distribution range in North America (38). In Europe, chestnut blight was first observed in the 1930s and is nowadays present in all major chestnut-growing areas (37). Following the colonization of Europe, C. parasitica has rapidly spread through most of southeastern Europe, driven by the emergence of a highly successful lineage (39). The invasion success likely stems from the establishment of a highly diverse European bridgehead population and a switch to asexual reproduction (39). Besides host species in the genus Castanea, C. parasitica has been occasionally reported on oaks (Quercus spp.), maples (Acer spp.), and European hornbeam (Carpinus betulus L.) (37).
Both in the native and in the invasive range, C. parasitica has closely related sister species, which are considered weak pathogens or saprophytes (40). Among these, Cryphonectria japonica Tak. Kobay. & Kaz. Itô (previously named Cryphonectria nitschkei) was isolated from Ca. crenata in Japan (41, 42) and from oaks in China, on which it causes bark cankers (43). The European species Cryphonectria naterciae M.H. Bragança (syn. Cryphonectria decipiens [44]) was isolated from Castanea sativa and Quercus spp. in Portugal, Sardinia, and Algeria (45–47). Inoculation experiments showed that both C. japonica and C. naterciae are significantly less virulent on Ca. sativa, Quercus robur L., and Fagus sylvatica L. than C. parasitica (40, 43). Two other Cryphonectria species occurring in Europe are C. radicalis and C. carpinicola. The former is also present in North America and considered to be a saprophyte on dead wood of Castanea and Quercus species (48). Interestingly, the low prevalence may be the result of a displacement that occurred when the pathogenic sister species C. parasitica was first introduced to both continents (48). C. carpinicola is a recently described species isolated from declining European hornbeams in Austria, Georgia, Italy, and Switzerland (C. Cornejo, personal communication). The diversity of lifestyles within the Cryphonectriaceae, including the emergence of new pathogens, raises important questions of whether genetic factors facilitate pathogenic lifestyles.
In this study, we assembled and analyzed 104 genomes of the Cryphonectriaceae family including the major representatives C. parasitica, C. radicalis, C. naterciae, and C. japonica and a recently detected European Cryphonectria species named C. carpinicola (Cornejo, personal communication). We analyzed orthology among the gene sets of the species and constructed a robust phylogenomic tree. We find that Cryphonectriaceae share similar trophic lifestyle traits. However, the chestnut pathogen C. parasitica has a substantially reduced complement in CAZymes. In contrast, the capacity to produce secondary metabolites is reduced among Cryphonectria species but is broadly conserved within the genus. Effector candidate proteins show genus and species specificity consistent with faster evolvability of the underlying genes.
RESULTS
Genome assemblies for the Cryphonectria genus.
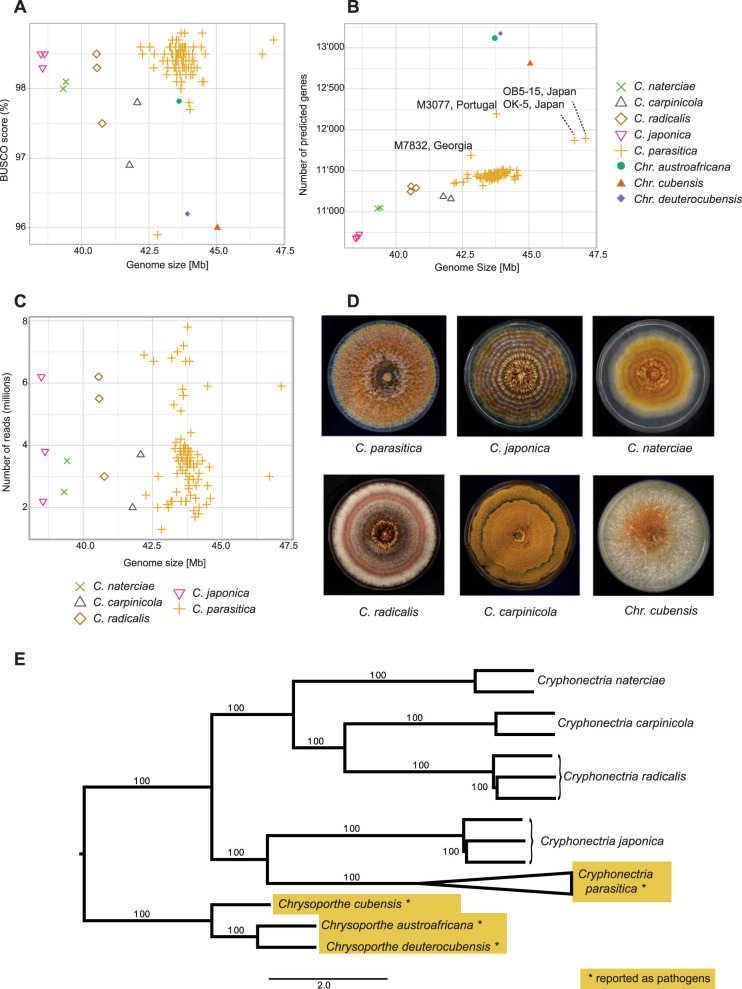
We assembled draft genomes of 100 Cryphonectria species isolates of Asian, European, and North American origin, in addition to the previously assembled genome of C. parasitica reference genome EP155. As a near outgroup to the genus Cryphonectria, we analyzed previously assembled draft genomes of 3 Chrysoporthe species from South Africa, Colombia, and Indonesia. To assemble Cryphonectria genomes de novo, we used Illumina sequencing data at 9 to 53× coverage (Table 1). All Cryphonectria and Chrysoporthe genome assemblies showed >95% completeness for BUSCO genes (ascomycota_odb9 database) with the C. parasitica isolate M7832 having the lowest score at 95.9% (Table 1 and Fig. 1A). Based on the assembly size, we estimated that nonpathogenic species had smaller genomes ranging from 38.6 Mb (C. japonica) to 41.9 Mb (C. carpinicola). Pathogenic species had slightly larger genomes ranging from 43.7 Mb (C. parasitica and Chrysoporthe austroafricana) to 45 Mb (Chrysoporthe cubensis) (Table 1; Fig. 1A). We found no apparent correlation between the estimated genome size and the completeness in BUSCO genes (Fig. 1A and B). Similarly, we detected no correlation between the sequencing depth and the assembled genome size (Fig. 1C). This shows that the short-read-based assemblies are expected to reliably represent the gene content across species.
TABLE 1.
Genome assembly statistics for Cryphonectria spp. and Chrysoporthe spp.a
| Species | Mean size (Mb) | Mean N50 | Mean complete BUSCO (%) | Coverage (min–max) | No. of predicted genes (min–max) | No. of isolates |
|---|---|---|---|---|---|---|
| C. parasitica* | 43.7 | 125,044 | 98.40 | 9–53× | 11,321–12,195 | 91 |
| C. japonica | 38.6 | 364,390 | 98.43 | 17–48× | 10,680–10,729 | 3 |
| C. radicalis | 40.6 | 197,843 | 98.1 | 22–46× | 11,247–11,312 | 3 |
| C. naterciae | 39.4 | 120,703 | 98.05 | 19–27× | 11,041–11,050 | 2 |
| C. carpinicola | 41.9 | 58,390 | 97.35 | 14–26× | 11,159–11,187 | 2 |
| Chr. austroafricana* | 43.7 | 48,708 | 97.8 | NA | 13,125 | 1 |
| Chr. cubensis* | 45.0 | 345,702 | 96 | NA | 12,807 | 1 |
| Chr. deuterocubensis* | 43.9 | 83,661 | 96.2 | NA | 13,174 | 1 |
The C. parasitica reference genome is not included in the summary of C. parasitica genomes. Asterisks indicate pathogenic species. NA, not available.
FIG 1.
Assembly statistics and phylogenetic reconstruction. (A to C) Estimated genome size in megabases correlated with assembly completeness assessed by BUSCO scores (A), number of predicted genes (B), and sequencing depth of assembled Cryphonectriaceae genomes (C). (D) Culture morphology of the studied Cryphonectriaceae (cultures of Chr. deuterocubensis and Chr. austroafricana are not shown, as isolates were unavailable for documentation). (E) Maximum-likelihood consensus tree based on 6,770 single-copy ortholog genes showing the phylogenetic relationship of Cryphonectriaceae species.
Gene annotation and phylogenetic reconstruction.
We predicted between ∼10,700 and 12,200 genes in genomes of Cryphonectria species compared to ∼12,800 to 13,170 genes in Chrysoporthe spp. (Table 1 and Fig. 1B). Overall, gene content among species was correlated with genome size except for C. carpinicola and Chr. cubensis, which have fewer predicted genes as expected from their genome size (Fig. 1B). Among C. parasitica isolates, M3077 had a higher gene content than isolates of similar genome size (Fig. 1B). Moreover, assembled genomes of C. parasitica isolates OB5-15 and OK-7 showed increased genome sizes while having only slightly higher gene content than other C. parasitica isolates (Fig. 1B).
The gene ortholog analyses revealed 6,770 single-copy orthologs among all species. We found 85 species-specific orthologs, of which 22 were specific for C. parasitica. Additionally, we found between 1 and 10 isolate-specific orthologs among the C. parasitica isolates TA51, M7832, DU5, OB5-15, OK-17, and M4030. Moreover, one ortholog was specific for C. carpinicola, while no species-specific orthologs were detected in all other Cryphonectria species. Within Chrysoporthe, we found 19 orthologs specific to Chysoporthe deuterocubensis, as well as 12 and 5 orthologs specific to Chr. cubensis and Chr. austroafricana, respectively. To reconstruct the evolutionary history of Cryphonectria and Chrysoporthe species, we generated a consensus maximum-likelihood tree based on 6,770 single-copy ortholog genes. We found 100% bootstrap branch support between species and a clear divergence at the genus level (Fig. 1E). Furthermore, Cryphonectria species were grouping by geographic origin, with C. naterciae, C. radicalis, and C. carpinicola being of European origin and C. japonica and C. parasitica being of Asian descent. Overall, our consensus tree is in accordance with phylogenetic studies on the genera Cryphonectria and Chrysoporthe (49; C. Cornejo, personal communication).
Lifestyle prediction and capacity for carbohydrate metabolism across species.
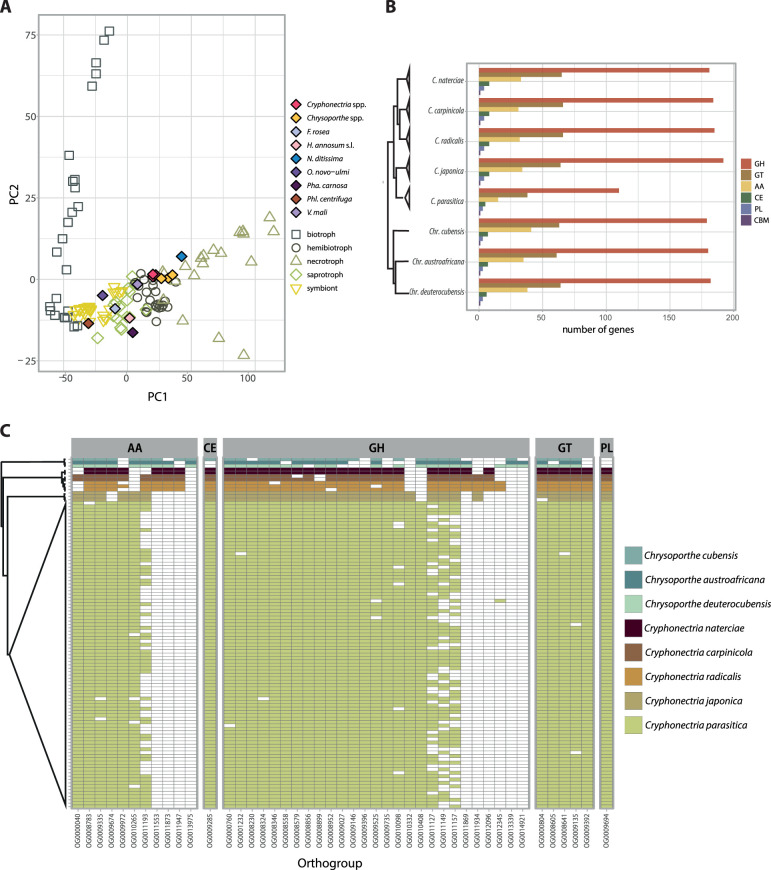
In order to degrade plant cell walls for nutrition or infection, fungi produce a variety of enzymes involved in carbohydrate metabolism (CAZymes) (50). We analyzed the predicted proteome of Cryphonectriaceae species and other tree-associated fungi to assess trophic lifestyles according to CAZyme content. All Cryphonectria species were identified as hemibiotrophs by CATAStrophy, while Chrysoporthe species were classified as necrotrophs. However, the principal-component analysis (PCA) shows close proximity of analyzed Cryphonectria and Chrysoporthe species, clustering at the verge with other hemibiotrophic and necrotrophic species (Fig. 2A). Lifestyles of most fungi outside the Cryphonectriaceae family matched with predicted lifestyles according to CAZyme content, except for Valsa mali (Fig. 2A).
FIG 2.
Carbohydrate-active enzyme (CAZyme) content among Cryphonectriaceae. (A) Principal-component analysis (PCA) of fungal lifestyle predictions, as inferred by CATAStrophy. The plot incorporates 85 reference species of fungi with different lifestyles (i.e., biotroph, hemibiotroph, nectrotroph, saprotroph, and symbiont) used as a training set by CATAStrophy and shows the CAZyme-inferred phenotypic trophism of Cryphonectriaceae and other pathogenic and nonpathogenic tree-associated fungi. (B) Number of detected CAZyme genes per species grouped according to CAZyme superfamily: glycoside hydrolase (GH), glycosyl transferase (GT), auxiliary activity (AA), carbohydrate esterase (CE), polysaccharide lyase activity (PL), and carbohydrate-binding modules (CBM). (C) Ortholog presence/absence of CAZyme superfamilies for which at least one species is missing an ortholog (the CBM superfamily is not shown, as orthologs were found in all species).
We further assessed CAZyme gene content among Cryphonectriaceae and found a striking gene loss in the chestnut blight pathogen C. parasitica (Fig. 2B). The gene loss particularly affected the group of glycoside hydrolases (GHs), glycosyl transferases, and enzymes with auxiliary activity (AA). Overall, all nonpathogenic Cryphonectria species, as well as the pathogenic Chrysoporthe species, encoded between 38.5 and 42.7% more GH, 37.7 and 42.4% more GT, and 51.6 and 63.4% more AA than C. parasitica (Fig. 2B). We identified gene losses in C. parasitica across most CAZyme categories. GH5 associated with hemicellulose degradation showed a particularly remarkable reduction (see Fig. S2 in the supplemental material). We found between 12 and 13 GH5 genes in saprophytic Cryphonectria and 11 GH5 genes in Chrysoporthe spp., while C. parasitica had only four GH5 genes. Moreover, slightly fewer GH28 genes involved in pectin degradation were detected in C. parasitica (n = 11) than in Chrysoporthe spp. (n = 12 to 14) and saprophytic Cryphonectria (n = 15 to 16). Analyzing CAZymes for which at least one species is missing an ortholog, Cryphonectria species share a relatively conserved set of ortholog CAZyme genes as expected from their short phylogenetic distance (Fig. 2C). We found one PL orthogroup encoding pectate lyase (OG0009694), shared only among Cryphonectria species. Moreover, we detected one GH orthogroup belonging to the sialidase superfamily (OG0010332), which is present only in Asian Cryphonectria species, as well as a single GH orthogroup (OG0012096, GH3) present only in European Cryphonectria species (Fig. 2C). C. parasitica displayed a particularly high degree of intraspecific presence/absence variation for four auxiliary activity (AA) and GH enzymes, which are otherwise well conserved (OG0011193, GMC [glucose-methanol-choline] oxidoreductase; OG0011127, GH76; OG0011149, GH43; OG0011157, GH76) (Fig. 2C). The four orthogroups likely underwent recent gene losses in C. parasitica.
To assess the wood-colonizing capabilities of different Cryphonectria species and a member of the genus Chrysoporthe (Chr. cubensis), we conducted an inoculation experiment on dormant and healthy chestnut stems. We performed the experiment with and without prior removal of the bark. None of the species were able to colonize dormant chestnut logs without artificial wound induction. After 2 weeks of incubation, C. japonica showed signs of mycelial growth on the bark at a maximum of 1 cm beyond the inoculation point. No bark penetration was detected. For inoculations with bark removal, C. parasitica expectedly showed the fastest and most extensive lesion growth. Other Cryphonectria species, with the exception of one C. radicalis isolate (M4733), developed only minimal lesions (Fig. S1). We found intraspecific variance in lesion growth, possibly attributed to varying isolate vigor (e.g., C. radicalis isolate M283 was isolated in 1953) or variable substrate conditions (e.g., state of dormancy and stem thickness) (Fig. S1). The eucalyptus pathogen Chr. cubensis showed growth on nonhost chestnut (Ca. sativa) logs; however, lesions developed at a comparatively slow pace (Fig. S1). After 4 weeks of incubation, mycelial fans were found only in lesions caused by C. parasitica.
Lesion length on dormant chestnut stems (Castanea sativa) with bark removal. Measurements were taken once per week during four weeks. Download FIG S1, PDF file, 1.2 MB (1.2MB, pdf) .
Copyright © 2020 Stauber et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene count of all identified CAZyme families among Cryphonectriaceae. Download FIG S2, PDF file, 0.2 MB (219.4KB, pdf) .
Copyright © 2020 Stauber et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Variation in secondary metabolite production potential among species.
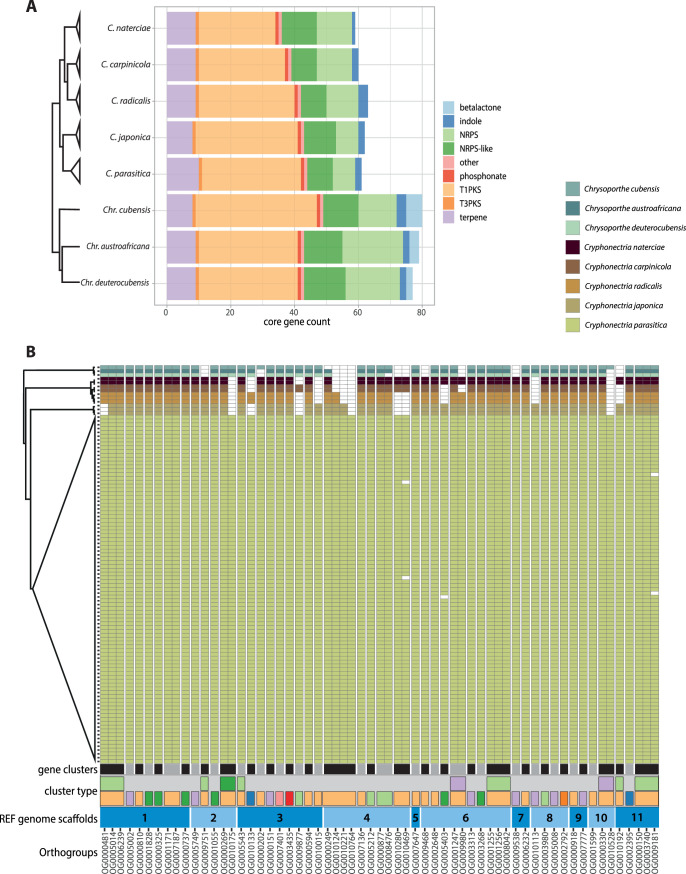
Secondary metabolites (SMs) can play important roles in pathogenicity and the interaction with microbes (51, 52). We investigated variation in biosynthetic core genes as an indicator for metabolite production potential among species. Loss of a biosynthetic core gene from a cluster invariably leads to loss of cluster function. Overall, biosynthetic core gene counts were variable only between genera. Cryphonectria species had comparatively fewer biosynthetic core genes than Chrysoporthe species (Fig. 3A). Among the detected biosynthetic core genes, the two genera shared similar proportions of different gene cluster classes with type 1 polyketide synthase (T1PKS) being the most abundant gene cluster class (Fig. 3A). The class of beta-lactone production clusters, which can produce potent antibacterial and antifungal compounds (53), was exclusively found in Chrysoporthe species (Fig. 3A). The presence/absence analyses of biosynthetic core genes per gene cluster (n = 47) revealed 28 clusters conserved among all analyzed Cryphonectriaceae (Fig. 3B). Additionally, core genes in five clusters were conserved in Cryphonectria. The same clusters showed a partial or complete absence in Chrysoporthe. The largest cluster was found on scaffold 4, containing four T1PKS biosynthetic core genes. All four core orthologs of the cluster were retained in C. parasitica. Other species lost between one (C. japonica) and all four (Chr. cubensis) core genes (Fig. 3B). Overall, core genes were highly conserved among C. parasitica isolates, except for three T1PKS, nonribosomal peptide synthase (NRPS)-like, and NRPS-T1PKS clusters on scaffolds 4, 6, and 11 (OG0010469, OG0005403, and OG0009181) (Fig. 3B). The clusters showed gene losses in C. parasitica isolates from China (TA51), Georgia (M7776 and M7832), Japan (WB-3), and the United States (MD-1). Generally, we identified only weak homology with secondary metabolite clusters in other species. A notable exception includes two gene clusters potentially underlying emodin production (Fig. S3).
FIG 3.
Secondary metabolite core gene content among Cryphonectriaceae. (A) Count of detected biosynthetic core gene categories across species as identified by antiSMASH. (B) Presence/absence of biosynthetic core gene orthologs among species. The plot shows the number of biosynthetic core genes within a gene cluster, the cluster type (color codes are as in panel A), and the location of clusters according to C. parasitica reference genome scaffolds.
Two PKS gene clusters (biosynthetic core gene orthologs OG0009468 and OG0002648) potentially underlying emodin production. Synteny plots of emodin and identified homologous gene clusters among species. Heatmaps show similarity (%) of genes in the C. parasitica reference genome cluster to emodin and orthologs in other Cryphonectriaceae (gray = gene is absent). Download FIG S3, PDF file, 0.4 MB (394.8KB, pdf) .
Copyright © 2020 Stauber et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted effector genes among Cryphonectriaceae, effector orthologs, and cysteine content.
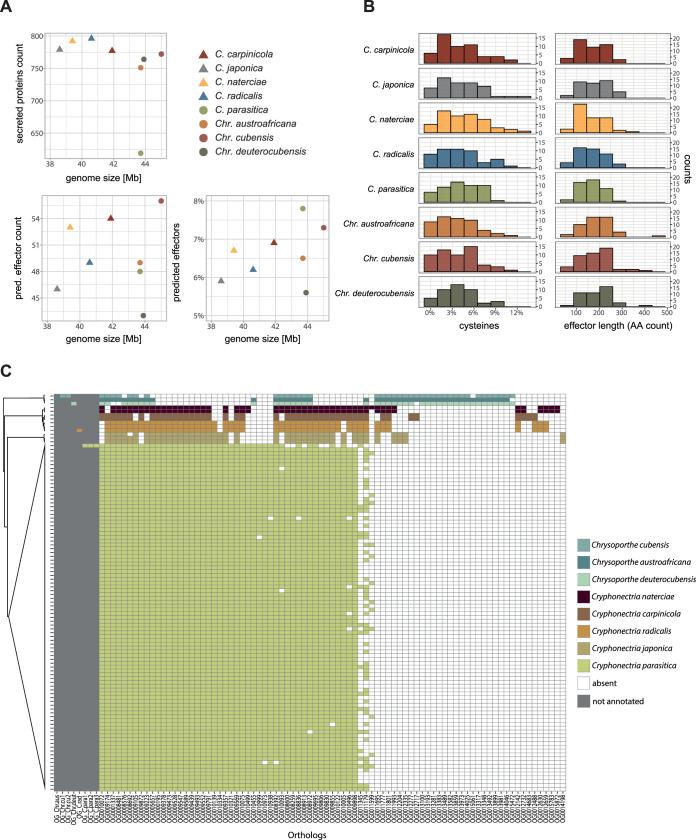
Effectors are mostly secreted, cysteine-rich proteins, which play a major role in fungal virulence to overcome host immune defenses (9). We predicted effector genes with a machine-learning approach and found that neither the number of putative secreted proteins nor the predicted effector content correlated with genome size (Fig. 4A). Saprophytic Cryphonectria species encode slightly more putatively secreted proteins (n = 777 to 796) than pathogenic Chrysoporthe spp. (n = 751 to 772). Surprisingly, C. parasitica encodes markedly fewer secreted proteins (n = 619) than all other species (Fig. 4A). However, despite the small amount of secreted proteins, C. parasitica had the highest ratio of predicted effectors among all species with 7.8% of all secreted proteins predicted to function as effectors (Fig. 4A). Overall, the pathogenic versus saprophytic lifestyle did not correlate with predicted effector content. For example, we found that pathogenic Chr. deuterocubensis encoded the smallest number of predicted effectors of all analyzed species (Fig. 4A). The cysteine content of predicted Cryphonectriaceae effectors ranged from 0 to 12.9% (Fig. 4B). The predicted effectors among Cryphonectria contained 53 to 348 amino acids with one outlier of only 33 amino acids in C. radicalis. Predicted Chrysoporthe effectors contained 67 to 436 amino acids (Fig. 4B). The divergence in candidate effector gene content among Cryphonectriaceae matches the divergence in cysteine content and protein length. Analysis of predicted effector ortholog presence/absence among Cryphonectriaceae revealed 41.5% (n = 59) conserved orthologs in all Cryphonectriaceae, and 91 orthologs showed presence/absence variation among species (Fig. 4C). We found several orthologs unique to a single species (Fig. 4C). Interestingly, the species-specific C. parasitica orthologs OG0010999, OG0010973, and OG0010938 showed presence-absence variation with orthologs missing in isolates from China and South Korea (LB86, M8510, and S35) (Fig. 4C). For eight candidate effectors, we could not find a corresponding ortholog annotation with OrthoFinder (gray area in Fig. 4C).
FIG 4.
Predicted secretome and putative effectors among Cryphonectriaceae. Conserved orthologs (i.e., effector genes shared among all species) were omitted. (A) Genome size correlations with secreted proteins and predicted effectors (identified by EffectorP). Saprophytic species are shown with triangles, and pathogens are shown with circles. (B) Histograms showing the cysteine content (%) and the size of predicted effectors per species. (C) Presence/absence of predicted effector orthologs among species. Areas in gray show orthologs for which we found no corresponding ortholog.
DISCUSSION
We assembled and analyzed genomes of eight bark-inhabiting Cryphonectriaceae species to retrace the evolution of genome size and gene content. Based on CAZyme content, all analyzed species are predicted to share a similar trophic lifestyle. In the genus Cryphonectria, we detected striking CAZyme gene loss in the invasive pathogen C. parasitica. In spite of the substantial CAZyme gene loss, C. parasitica shares wood colonization strategies with the other Cryphonectria species and has retained the ability for early saprotrophic wood decay. In contrast, secondary metabolite gene clusters diverged at the genus level but were largely conserved among Cryphonectria species. Putative effector content varied substantially among species with differences in cysteine content and protein length.
Distinct CAZyme gene loss in a pathogenic species.
The CAZyme profiles of the Cryphonectriaceae species analyzed in this study match those of other hemibiotrophic or necrotrophic fungi. Thus, despite substantial difference in pathogenicity (40), Cryphonectriaceae species seem to share trophic lifestyle traits, which challenges previous classifications of C. japonica, C. naterciae, and C. radicalis as predominantly saprotrophic species (37). Nonetheless, the distinct CAZyme loss in C. parasitica coincides with an increased pathogenicity toward nonnative (i.e., non-Asian) Castanea species, which seems to be absent in other Cryphonectria species. Many CAZymes play a role in plant cell wall degradation and can be important virulence factors in necrotrophic fungi. Reductions in CAZyme genes have been observed in biotrophic pathogens and are thought to be an adaptation to reduce the exposure of molecular patterns, which can trigger host defenses (54, 55). Moreover, CAZyme loss can occur during host shifts, such as from plant to animal or insect hosts (56). In C. parasitica, the CAZyme loss may be an adaptation facilitating an increased pathogenic lifestyle. At the intraspecific level, fewer CAZymes are expressed during pathogenic growth compared to saprotrophic wood decay in the conifer pathogen Heterobasidion annosum sensu lato, which has plastic lifestyles (57). Similarly, C. parasitica may have undergone a transitory phase in the evolution of the predominant pathogenic lifestyle favoring reduced CAZyme expression and ultimately gene losses. Moreover, H. annosum sensu lato produces more secondary metabolites including phytotoxins during the pathogenic lifestyle (57). These findings suggest that necrotrophic pathogens of trees have evolved different wood degradation strategies from those of saprotrophic relatives. The most significant gene loss in C. parasitica was found in the CAZyme subfamily GH5, which underlies hemicellulose degradation. Consistent with this, GH5 expression is lower during pathogenic growth in H. annosum sensu lato (57). In contrast, genomes of saprotrophic wood degraders such as Phanerochaete carnosa have expanded GH5 repertoires (58). Despite extensive CAZyme loss in C. parasitica, our experimental data show that all Cryphonectria species, including C. parasitica, have retained similar wood colonization capabilities through bark wounds. Moreover, C. parasitica appears to have retained CAZymes suitable for early wood decay. This confirms field observations indicating that the fungus is able to survive a few years on the bark of fresh dead chestnut wood (59). In parallel to GH5, pectin-degrading enzymes of GH28 are also slightly reduced in C. parasitica. However, polygalacturonases belonging to the GH28 family are suggested to contribute to virulence in C. parasitica (60). Similarly, in other necrotrophic pathogens GH28 is also associated with pathogenicity showing expansions in the GH28 family (61). Similar to GH5, C. parasitica may have lost GH28 enzymes triggering host defenses through molecular pattern recognition by the host (62).
Potential virulence-associated traits in C. parasitica.
In contrast to the evolution of CAZymes, secondary metabolite production capabilities are largely conserved within the genus Cryphonectria. C. parasitica produces virulence-associated compounds including oxalic acid, tannases, laccases, and phytotoxins such as cryparin and diaporthin, but the genetic basis is only partially resolved (60). The diaporthin production pathway is encoded by a PKS gene cluster in Aspergillus oryzae (63). However, we identified no clearly orthologous cluster in C. parasitica. The conservation of gene clusters across Cryphonectria species suggests that secondary metabolites played no particular role in the evolution of pathogenicity by C. parasitica. However, many fungi can modulate metabolite production depending on environmental conditions (64). Hence, even if all Cryphonectria species share a core set of gene clusters, lifestyle transitions may induce differential expression depending on biotic or abiotic conditions. In addition to secondary metabolites, small secreted proteins (i.e., effectors) can play key roles in the emergence of new pathogens. We identified a broad pool of putative effector orthologs among Cryphonectriaceae. The size of the effector gene pool did not correlate with genome size or lifestyle as seen in other clades of plant pathogens (27, 65). Effector homologs in the orthogroups OG0010999, OG0010973, and OG0010938 are particularly interesting candidates because the genes both are unique to C. parasitica and show presence/absence variation within the species. The recent gene gains in the pathogen lineage and the presence/absence variation within the species could explain variation in pathogenicity between and within the species, respectively. Combining analyses of positive selection, gene expression, and targeted gene deletion assays of effector candidates in C. parasitica will be needed to elucidate the role of effectors in causing chestnut blight.
Lifestyle and the role of hosts.
Cryphonectriaceae species represent a useful model to retrace how lifestyle transitions toward pathogenicity impact the evolution of gene content. On its native Asian hosts (Ca. crenata and Ca. mollissima), the chestnut blight fungus C. parasitica causes only mild symptoms, which has been attributed to host-pathogen coevolution (37). In contrast, on the naive American and European chestnut species (Ca. dentata and Ca. sativa), the pathogen causes lethal bark cankers (66). In the invasive range, C. parasitica might also be a weak pathogen on Quercus spp., Acer spp., or Carpinus betulus (37). This suggests that C. parasitica has the genetic repertoire of a broad-host-range pathogen and that chestnut species may be the least able to resist pathogen invasion. In diverse forest ecosystems, disease incidence is often negatively correlated with host species richness (the “dilution effect” [67 to 69]). Hence, growth on largely resistant hosts may be a bet-hedging strategy of C. parasitica to survive and spread in the absence of the primary host (70). The weak pathogenicity of other Cryphonectria species may be facilitated by environmental conditions, such as abiotic stress on the host or disturbance of the host microbiome (71). Subsequently, these Cryphonectria species may be considered latent pathogens similar to some endophytes (72–74). Latent pathogenicity has been observed in other Cryphonectriaceae. For example, Granados et al. (75) found that the Eucalyptus pathogen Chr. cubensis is an endophyte on Colombian Melastomataceae trees. Moreover, the pathogen Chr. austroafricana occurs as an endophyte in its native range but is pathogenic on nonnative Eucalyptus trees (76). Hence, host jumps likely facilitated the switch from endophytic to pathogenic lifestyle in both species (75, 76). C. parasitica has recently emerged as a major pathogen on non-Asian chestnut species. To what degree the extensive CAZyme loss increased the pathogenic potential prior to the emergence as an invasive pathogen remains to be investigated. Comparative genomics combined with gene function analyses provide a powerful approach to study lifestyle evolution and changes in the underlying genome architecture.
MATERIALS AND METHODS
Genome sequencing.
We sequenced whole genomes of 90 C. parasitica, 3 C. japonica, 3 C. radicalis, 2 C. naterciae, and 2 C. carpinicola isolates covering the global distribution range (see Table S1 in the supplemental material). All isolates were prepared for sequencing as described in reference 39. Sequencing was conducted using the Illumina HiSeq4000 and Illumina NovaSeq 6000 platforms (Illumina, San Diego, CA, USA) at the Functional Genomics Center Zurich (FGCZ). By choosing the Illumina NovaSeq SP flow cell, NovaSeq reads were compatible with HiSeq4000 reads for downstream analysis.
Overview of genomes assembled in the genus Cryphonectria. Country, region, and host identify the location and host species of collection. Download Table S1, XLS file, 0.05 MB (54KB, xls) .
Copyright © 2020 Stauber et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome assembly and gene prediction.
All 100 Cryphonectria sequences were assembled with SPAdes v3.13.0 (77), using the –careful option and choosing the k-mers 21, 33, 45, 57, and 69 for the iterative assembly process. Genome sizes and assembly quality of Cryphonectria de novo assemblies, as well as Chrysoporthe draft genomes, were assessed with QUAST v5.0.2 and BUSCO v3.0.2 (78, 79). Gene models were predicted using BRAKER2 v2.1.4 (80–83). Briefly, we set up gene annotation training using the existing C. parasitica v2 reference genome annotation (available from http://jgi.doe.gov/ [84]) using the BRAKER2 options –alternatives-from-evidence=false, –fungus, –gff3, and –skip_fixing_broken_genes. For splice site hints, intron information was extracted from the reference genome annotation using the construct_introns function from the R package gread v0.99.3 (85). After the training, genes were predicted in all assembled genomes using BRAKER2 adding coding sequence hints of the C. parasitica reference genome obtained using gffread v0.11.0 (86) and EMBOSS v6.6.0 tool transseq (87). We set the BRAKER 2 options –alternatives-from-evidence=false, –gff3, –useexisting, –prg=gth, and –trainFromGth.
Identification of orthologs and secondary metabolite gene clusters.
To identify orthologs among all Cryphonectria and Chrysoporthe isolates, we used OrthoFinder v2.3.7 (88). We selected all single-copy ortholog groups and generated sequence alignments using MAFFT v7.429 (89). Aligned sequences were used for phylogenetic tree building by generating 100 maximum-likelihood (ML) trees using the GTRCAT model with RAxML v8.2.12 (90). The RAxML-generated tree and bootstrap files were subsequently used to build a consensus tree with Astral v5.14.2 (91). The obtained consensus tree was visualized with FigTree v1.4.3 (92). We used the antiSMASH fungal version v5.1.0 (93) to identify secondary metabolite gene clusters using one isolate per species: the reference genome EP155 for C. parasitica, IF-6 for C. japonica, M283 for C. radicalis, M3664 for C. naterciae, CS3 for C. carpinicola, and the three NCBI Chrysoporthe draft genomes. We used a custom Python script to extract the biosynthetic core genes from the antiSMASH regions.js file. The number of core genes per species was then plotted in R with the packages tidyverse (94), reshape2 (95), and ggplot2 (96). Additionally, we identified biosynthetic core genes per cluster of the EP155 genome and searched for orthologs in all species. Secondary metabolite core gene ortholog presence/absence in each cluster was plotted in R, using the packages reshape2, stringr (97), and ggplot2.
Classification of fungal lifestyles according to CAZyme content.
We inferred trophic lifestyles of Cryphonectriaceae according to carbohydrate-active enzyme (CAZyme) gene content using the CAZyme-Assisted Training And Sorting of -trophy (CATAStrophy) prediction tool (23). CATAStrophy annotates CAZymes with HMMER 3.0 (98) and dbCAN (99) and predicts trophic classes based on a multivariate analysis (23). To run CATAStrophy, we selected the same Cryphonectriaceae isolates as described above and added additional tree-associated fungi of different lifestyles. For nonpathogenic saprophytes associated with wood degradation, we selected the proteomes of Fomitopsis rosea (BioProject accession no. PRJNA518053), Phanerochaete carnosa (58), and Phlebia centrifuga (100). Moreover, we included Heterobasidion annosum sensu lato (57), associated with both saprotrophic and pathogenic lifestyles, and the bark pathogens Neonectria ditissima (Nectria canker on apple and pear trees) (101), Ophiostoma novo-ulmi (Dutch elm disease) (102, 103), and Valsa mali (Valsa canker on apple trees) (104). For trophic lifestyle inferences, we used the CATASTrophy pipeline (https://github.com/ccdmb/catastrophy-pipeline), choosing the options -profile conda and –dbcan_version 8. The CATAStrophy literature-derived nomenclature (i.e., classification into biotrophs, hemibiotrophs, nectrotrophs, saprotrophs, and symbionts) was used for defining trophic lifestyles of species included in the CATAStrophy training set. Moreover, we selected principal components PC1 and PC2, which separate most training set species according to lifestyle for visualization (23).
Analysis of carbohydrate-active enzyme genes (CAZymes) and inoculation experiments.
For the identification of CAZyme genes, we ran dbCAN v2.0.0 (99) on the same isolates as in the secondary metabolite analysis (i.e., one isolate per species). Only CAZymes which were identified by all three tools (HMMER, diamond, and hotpep) were then selected for further analysis. CAZyme orthologs were extracted using Python, and plots were generated in R.
To analyze the wood colonization capabilities of the different species, we set up an inoculation experiment. We selected 26 dormant chestnut logs (Ca. sativa; length, 50 cm; diameter, 3.3 to 6.7 cm), which were cut in a healthy state during winter from chestnut stands in Ticino, Switzerland, a week prior to the experiment. The logs were surface sterilized with 70% ethanol and sealed on both ends with paraffin to prevent desiccation. We selected 3 C. parasitica (XA19, CR03, and EP155), 2 C. japonica (M9249 and IF-6), 2 C. naterciae (M3664 and M3656), 2 C. radicalis (M4733 and M283), and 2 C. carpinicola (M9290 and CS3) isolates and 1 Chr. cubensis (CBS115724) isolate for inoculation. For all isolates except CBS115724, full genome sequences were available for this study. Prior to inoculation, all isolates were freshly inoculated from glycerol stocks onto potato dextrose agar (PDA; 39 ml/liter; BD Becton, Dickinson & Company, Franklin Lakes, NJ, USA) and incubated at 25°C in complete darkness for 5 days to induce mycelial growth. For inoculation of the first batch of chestnut logs (n = 13), we removed the bark on 5 equally distanced spots (diameter, 4 mm) on each log, placed a mycelial plug (diameter, 4 mm) into the wound, and sealed it with tape. For the second batch of chestnut logs (n = 13), we directly placed five mycelial plugs onto the bark of each log with equal distance (i.e., no wound induction) and sealed the inoculation spots with tape. For each treatment batch with and without wound, we selected 5 replicates per isolate and 5 negative controls (mycelium-free agar plugs), resulting in a total of 130 completely randomized inoculation spots (n = 65 per treatment). All chestnut logs were randomly placed onto racks in plastic containers, separated by treatment, filled with 2 liters of demineralized water to avoid drying out, and sealed with plastic lids (40). Incubation was at 20°C for both treatments. Logs with wounds were incubated for 4 weeks in complete darkness, and longitudinal lesion size was assessed once a week. Logs without wounds were incubated for 12 weeks, and lesion size was assessed at the end of the experiment.
Prediction of effector genes.
We performed effector gene prediction with the same isolates as in the secondary metabolite and CAZyme analysis, using a machine-learning approach. First, secreted proteins were predicted with SignalP v5.0b (105), choosing the options -org euk and -format short. Only proteins with a likelihood probability >0.5 were selected for further analysis. Next, protein sequences with a predicted secretion signal were extracted with SAMtools v1.9 (106) and used as input for effector prediction with EffectorP v2.0 (107). Presence/absence variation analyses of predicted effector gene orthologs across species and plotting were performed in Python and R as described above. The cysteine content and protein length of predicted effector genes in all species were determined with EMBOSS pepstats v6.6.0.
Data availability.
All sample accession numbers for the NCBI Short Read Archive, as well as NCBI accession numbers for all de novo assemblies of Cryphonectria genomes, are available in Table S1 in the supplemental material. Contigs of <400 bp were trimmed prior to NCBI genome submission. Outgroup genomes to the genus Cryphonectria were obtained for Chrysoporthe cubensis, Chr. deuterocubensis, and Chr. austroafricana from NCBI BioProjects PRJNA279968, PRJNA265023, and PRJNA263707, respectively (108, 109).
ACKNOWLEDGMENTS
We are grateful to Thomas Badet for helpful suggestions on a previous version of the manuscript. We thank Eva Augustiny, Silvia Kobel, Aria Minder, Quirin Kupper, and Hélène Blauenstein for laboratory assistance. We acknowledge the Genetic Diversity Centre (GDC), ETH Zurich, and the Functional Genomics Center Zurich (FGCZ) for technical support and facility access. We also thank Martin Wrann for helping with photographic documentation. Paolo Cortesi, Michael Milgroom, Kiril Sotirovski, Mihajlo Risteski, Marin Ježić, Sang Hyun Lee, and Seçil Akilli kindly provided samples. Michael and Brenda Wingfield gave valuable insight on phylogenetic relationships among Cryphonectriaceae. We kindly thank Sabina Moser Tralamazza for insightful discussions on secondary metabolites and for sharing scripts.
L.S. was supported by the Swiss National Science Foundation (grant 170188 to S.P.).
REFERENCES
- 1.Giraud T, Gladieux P, Gavrilets S. 2010. Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol Evol 25:387–395. doi: 10.1016/j.tree.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, Faris JD, Rasmussen JB, Solomon PS, McDonald BA, Oliver RP. 2006. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet 38:953–956. doi: 10.1038/ng1839. [DOI] [PubMed] [Google Scholar]
- 3.Byrnes EJ, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6:e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moonjely S, Barelli L, Bidochka MJ. 2016. Insect pathogenic fungi as endophytes. Adv Genet 94:107–135. doi: 10.1016/bs.adgen.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Haridas S, Albert R, Binder M, Bloem J, LaButti K, Salamov A, Andreopoulos B, Baker SE, Barry K, Bills G, Bluhm BH, Cannon C, Castanera R, Culley DE, Daum C, Ezra D, González JB, Henrissat B, Kuo A, Liang C, Lipzen A, Lutzoni F, Magnuson J, Mondo SJ, Nolan M, Ohm RA, Pangilinan J, Park H-J, Ramírez L, Alfaro M, Sun H, Tritt A, Yoshinaga Y, Zwiers L-H, Turgeon BG, Goodwin SB, Spatafora JW, Crous PW, Grigoriev IV. 2020. 101 Dothideomycetes genomes: a test case for predicting lifestyles and emergence of pathogens. Stud Mycol 96:141–153. doi: 10.1016/j.simyco.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rokas A, Mead ME, Steenwyk JL, Oberlies NH, Goldman GH. 2020. Evolving moldy murderers: Aspergillus section Fumigati as a model for studying the repeated evolution of fungal pathogenicity. PLoS Pathog 16:e1008315. doi: 10.1371/journal.ppat.1008315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardham AR. 2001. Cell biology of fungal infection of plants, p 91–123. In Howard R, Gow NAR (ed), Biology of the fungal cell. Springer, Berlin, Germany. [Google Scholar]
- 8.Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stüber K, Ver Loren van Themaat E, Brown JKM, Butcher SA, Gurr SJ, Lebrun M-H, Ridout CJ, Schulze-Lefert P, Talbot NJ, Ahmadinejad N, Ametz C, Barton GR, Benjdia M, Bidzinski P, Bindschedler LV, Both M, Brewer MT, Cadle-Davidson L, Cadle-Davidson MM, Collemare J, Cramer R, Frenkel O, Godfrey D, Harriman J, Hoede C, King BC, Klages S, Kleemann J, Knoll D, Koti PS, Kreplak J, López-Ruiz FJ, Lu X, Maekawa T, Mahanil S, Micali C, Milgroom MG, Montana G, Noir S, O’Connell RJ, Oberhaensli S, Parlange F, Pedersen C, Quesneville H, Reinhardt R, Rott M, Sacristán S, Schmidt SM, Schön M, Skamnioti P, Sommer H, Stephens A, Takahara H, Thordal-Christensen H, Vigouroux M, Wessling R, Wicker T, Panstruga R. 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330:1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- 9.Stergiopoulos I, de Wit PJGM. 2009. Fungal effector proteins. Annu Rev Phytopathol 47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 10.O’Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, Damm U, Buiate EA, Epstein L, Alkan N, Altmüller J, Alvarado-Balderrama L, Bauser CA, Becker C, Birren BW, Chen Z, Choi J, Crouch JA, Duvick JP, Farman MA, Gan P, Heiman D, Henrissat B, Howard RJ, Kabbage M, Koch C, Kracher B, Kubo Y, Law AD, Lebrun M-H, Lee Y-H, Miyara I, Moore N, Neumann U, Nordström K, Panaccione DG, Panstruga R, Place M, Proctor RH, Prusky D, Rech G, Reinhardt R, Rollins JA, Rounsley S, Schardl CL, Schwartz DC, Shenoy N, Shirasu K, Sikhakolli UR, Stüber K, Sukno SA, Sweigard JA, Takano Y, Takahara H, Trail F, van der Does HC, Voll LM, Will I, Young S, Zeng Q, Zhang J, Zhou S, Dickman MB, Schulze-Lefert P, Ver Loren van Themaat E, Ma L-J, Vaillancourt LJ. 2012. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet 44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Wit PJGM, van der Burgt A, Ökmen B, Stergiopoulos I, Abd-Elsalam KA, Aerts AL, Bahkali AH, Beenen HG, Chettri P, Cox MP, Datema E, de Vries RP, Dhillon B, Ganley AR, Griffiths SA, Guo Y, Hamelin RC, Henrissat B, Kabir MS, Jashni MK, Kema G, Klaubauf S, Lapidus A, Levasseur A, Lindquist E, Mehrabi R, Ohm RA, Owen TJ, Salamov A, Schwelm A, Schijlen E, Sun H, van den Burg HA, van Ham RCHJ, Zhang S, Goodwin SB, Grigoriev IV, Collemare J, Bradshaw RE. 2012. The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet 8:e1003088. doi: 10.1371/journal.pgen.1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sánchez-Vallet A, Fouché S, Fudal I, Hartmann FE, Soyer JL, Tellier A, Croll D. 2018. The genome biology of effector gene evolution in filamentous plant pathogens. Annu Rev Phytopathol 56:21–40. doi: 10.1146/annurev-phyto-080516-035303. [DOI] [PubMed] [Google Scholar]
- 13.Raffaele S, Kamoun S. 2012. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol 10:417–430. doi: 10.1038/nrmicro2790. [DOI] [PubMed] [Google Scholar]
- 14.Lyu X, Shen C, Fu Y, Xie J, Jiang D, Li G, Cheng J. 2015. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci Rep 5:15565. doi: 10.1038/srep15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lelwala RV, Korhonen PK, Young ND, Scott JB, Ades PK, Gasser RB, Taylor PWJ. 2019. Comparative genome analysis indicates high evolutionary potential of pathogenicity genes in Colletotrichum tanaceti. PLoS One 14:e0212248. doi: 10.1371/journal.pone.0212248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Jonge R, Bolton MD, Thomma BPHJ. 2011. How filamentous pathogens co-opt plants: the ins and outs of fungal effectors. Curr Opin Plant Biol 14:400–406. doi: 10.1016/j.pbi.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Gaulin E, Pel MJC, Camborde L, San-Clemente H, Courbier S, Dupouy M-A, Lengellé J, Veyssiere M, Le Ru A, Grandjean F, Cordaux R, Moumen B, Gilbert C, Cano LM, Aury J-M, Guy J, Wincker P, Bouchez O, Klopp C, Dumas B. 2018. Genomics analysis of Aphanomyces spp. identifies a new class of oomycete effector associated with host adaptation. BMC Biol 16:43. doi: 10.1186/s12915-018-0508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chooi Y-H, Solomon PS. 2014. A chemical ecogenomics approach to understand the roles of secondary metabolites in fungal cereal pathogens. Front Microbiol 5:640. doi: 10.3389/fmicb.2014.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macheleidt J, Mattern DJ, Fischer J, Netzker T, Weber J, Schroeckh V, Valiante V, Brakhage AA. 2016. Regulation and role of fungal secondary metabolites. Annu Rev Genet 50:371–392. doi: 10.1146/annurev-genet-120215-035203. [DOI] [PubMed] [Google Scholar]
- 20.Kubicek CP, Starr TL, Glass NL. 2014. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol 52:427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- 21.Glass NL, Schmoll M, Cate JHD, Coradetti S. 2013. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol 67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Moreno L, Ebert MK, Bolton MD, Thomma BPHJ. 2018. Tools of the crook—infection strategies of fungal plant pathogens. Plant J 93:664–674. doi: 10.1111/tpj.13810. [DOI] [PubMed] [Google Scholar]
- 23.Hane JK, Paxman J, Jones DAB, Oliver RP, de Wit P. 2020. “CATAStrophy,” a genome-informed trophic classification of filamentous plant pathogens–how many different types of filamentous plant pathogens are there? Front Microbiol 10:3088. doi: 10.3389/fmicb.2019.03088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Barry KW, Condon BJ, Copeland AC, Dhillon B, Glaser F, Hesse CN, Kosti I, LaButti K, Lindquist EA, Lucas S, Salamov AA, Bradshaw RE, Ciuffetti L, Hamelin RC, Kema GHJ, Lawrence C, Scott JA, Spatafora JW, Turgeon BG, de Wit PJGM, Zhong S, Goodwin SB, Grigoriev IV. 2012. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog 8:e1003037. doi: 10.1371/journal.ppat.1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stukenbrock EH, Bataillon T, Dutheil JY, Hansen TT, Li R, Zala M, McDonald BA, Wang J, Schierup MH. 2011. The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res 21:2157–2166. doi: 10.1101/gr.118851.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Wang S. 2017. Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu Rev Entomol 62:73–90. doi: 10.1146/annurev-ento-031616-035509. [DOI] [PubMed] [Google Scholar]
- 27.Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R. 2015. Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66:513–545. doi: 10.1146/annurev-arplant-043014-114623. [DOI] [PubMed] [Google Scholar]
- 28.Tan K-C, Oliver RP, Solomon PS, Moffat CS. 2010. Proteinaceous necrotrophic effectors in fungal virulence. Funct Plant Biol 37:907–912. doi: 10.1071/FP10067. [DOI] [Google Scholar]
- 29.Feldman D, Kowbel DJ, Glass NL, Yarden O, Hadar Y. 2017. A role for small secreted proteins (SSPs) in a saprophytic fungal lifestyle: ligninolytic enzyme regulation in Pleurotus ostreatus. Sci Rep 7:14553. doi: 10.1038/s41598-017-15112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller NP, Turner G, Bennett JW. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol 3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen JC, Grijseels S, Prigent S, Ji B, Dainat J, Nielsen KF, Frisvad JC, Workman M, Nielsen J. 2017. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat Microbiol 2:17044. doi: 10.1038/nmicrobiol.2017.44. [DOI] [PubMed] [Google Scholar]
- 32.Santos MS, Orlandelli RC, Polonio JC, dos Santos Ribeiro MA, Sarragiotto MH, Azevedo JL, Pamphile JA. 2017. Endophytes isolated from passion fruit plants: molecular identification, chemical characterization and antibacterial activity of secondary metabolites. J Appl Pharm Sci 7:38–43. [Google Scholar]
- 33.Schalchli H, Hormazabal E, Becerra J, Birkett M, Alvear M, Vidal J, Quiroz A. 2011. Antifungal activity of volatile metabolites emitted by mycelial cultures of saprophytic fungi. Chem Ecol 27:503–513. doi: 10.1080/02757540.2011.596832. [DOI] [Google Scholar]
- 34.Gryzenhout M, Myburg H, Wingfield BD, Wingfield MJ. 2006. Cryphonectriaceae (Diaporthales), a new family including Cryphonectria, Chrysoporthe, Endothia and allied genera. Mycologia 98:239–249. doi: 10.3852/mycologia.98.2.239. [DOI] [PubMed] [Google Scholar]
- 35.Jiang N, Fan X, Tian C, Crous PW. 2020. Reevaluating Cryphonectriaceae and allied families in Diaporthales. Mycologia 112:267–292. doi: 10.1080/00275514.2019.1698925. [DOI] [PubMed] [Google Scholar]
- 36.Gryzenhout M, Myburg H, der Merwe NA, Wingfield BD, Wingfield MJ. 2004. Chrysoporthe, a new genus to accommodate Cryphonectria cubensis. Stud Mycol 50:119–142. [Google Scholar]
- 37.Rigling D, Prospero S. 2018. Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Mol Plant Pathol 19:7–20. doi: 10.1111/mpp.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elliott KJ, Swank WT. 2008. Long-term changes in forest composition and diversity following early logging (1919–1923) and the decline of American chestnut (Castanea dentata). Plant Ecol 197:155–172. doi: 10.1007/s11258-007-9352-3. [DOI] [Google Scholar]
- 39.Stauber L, Badet T, Prospero S, Croll D. 2020. Emergence and diversification of a highly invasive chestnut pathogen lineage across south-eastern Europe. bioRxiv doi: 10.1101/2020.02.15.950170. [DOI] [PMC free article] [PubMed]
- 40.Dennert F, Rigling D, Meyer JB, Schefer C, Augustiny E, Prospero S. 2020. Testing the pathogenic potential of Cryphonectria parasitica and related species on three common European Fagaceae. Front Glob Chang 3:52. doi: 10.3389/ffgc.2020.00052. [DOI] [Google Scholar]
- 41.Liu Y-C, Linder-Basso D, Hillman BI, Kaneko S, Milgroom MG. 2003. Evidence for interspecies transmission of viruses in natural populations of filamentous fungi in the genus Cryphonectria. Mol Ecol 12:1619–1628. doi: 10.1046/j.1365-294x.2003.01847.x. [DOI] [PubMed] [Google Scholar]
- 42.Myburg H, Gryzenhout M, Wingfield BD, Milgroom MG, Kaneko S, Wingfield MJ. 2004. DNA sequence data and morphology define Cryphonectria species in Europe, China, and Japan. Can J Bot 82:1730–1743. doi: 10.1139/b04-135. [DOI] [Google Scholar]
- 43.Jiang N, Fan XL, Tian CM. 2019. Identification and pathogenicity of Cryphonectriaceae species associated with chestnut canker in China. Plant Pathol 68:1132–1145. doi: 10.1111/ppa.13033. [DOI] [Google Scholar]
- 44.Gryzenhout M, Wingfield BD, Wingfield MJ. 2009. Taxonomy, phylogeny, and ecology of bark-inhabiting and tree-pathogenic fungi in the Cryphonectriaceae. American Phytopathological Society (APS Press), St. Paul, MN. [Google Scholar]
- 45.Bragança H, Rigling D, Diogo E, Capelo J, Phillips A, Tenreiro R. 2011. Cryphonectria naterciae: a new species in the Cryphonectria–Endothia complex and diagnostic molecular markers based on microsatellite-primed PCR. Fungal Biol 115:852–861. doi: 10.1016/j.funbio.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Pinna C, Linaldeddu BT, Deiana V, Maddau L, Montecchio L, Lentini A. 2019. Plant pathogenic fungi associated with Coraebus florentinus (Coleoptera: Buprestidae) attacks in declining oak forests. Forests 10:488. doi: 10.3390/f10060488. [DOI] [Google Scholar]
- 47.Smahi H, Belhoucine-Guezouli L, Bouhraoua RT, Franceschini A, Linaldeddu BT. 2018. First report of branch canker and dieback caused by Cryphonectria naterciae on Quercus suber in Algeria. Plant Dis 102:251. doi: 10.1094/PDIS-07-17-1130-PDN. [DOI] [Google Scholar]
- 48.Hoegger PJ, Rigling D, Holdenrieder O, Heiniger U. 2002. Cryphonectria radicalis: rediscovery of a lost fungus. Mycologia 94:105–115. doi: 10.1080/15572536.2003.11833253. [DOI] [PubMed] [Google Scholar]
- 49.van der Merwe NA, Gryzenhout M, Steenkamp ET, Wingfield BD, Wingfield MJ. 2010. Multigene phylogenetic and population differentiation data confirm the existence of a cryptic species within Chrysoporthe cubensis. Fungal Biol 114:966–979. doi: 10.1016/j.funbio.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Z, Liu H, Wang C, Xu J-R. 2014. Correction: comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 15:6. doi: 10.1186/1471-2164-15-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brakhage AA. 2013. Regulation of fungal secondary metabolism. Nat Rev Microbiol 11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 52.Scharf DH, Heinekamp T, Brakhage AA. 2014. Human and plant fungal pathogens: the role of secondary metabolites. PLoS Pathog 10:e1003859. doi: 10.1371/journal.ppat.1003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson SL, Christenson JK, Wackett LP. 2019. Biosynthesis and chemical diversity of β-lactone natural products. Nat Prod Rep 36:458–475. doi: 10.1039/c8np00052b. [DOI] [PubMed] [Google Scholar]
- 54.Duplessis S, Spanu PD, Schirawski J. 2014. Biotrophic fungi (powdery mildews, rusts and smuts), p 149–168. In Martin F (ed), The ecological genomics of fungi. Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 55.Goulet KM, Saville BJ. 2017. Carbon acquisition and metabolism changes during fungal biotrophic plant pathogenesis: insights from Ustilago maydis. Can J Plant Pathol 39:247–266. doi: 10.1080/07060661.2017.1354330. [DOI] [Google Scholar]
- 56.Zhang W, Zhang X, Li K, Wang C, Cai L, Zhuang W, Xiang M, Liu X. 2018. Introgression and gene family contraction drive the evolution of lifestyle and host shifts of hypocrealean fungi. Mycology 9:176–188. doi: 10.1080/21501203.2018.1478333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olson A, Aerts A, Asiegbu F, Belbahri L, Bouzid O, Broberg A, Canbäck B, Coutinho PM, Cullen D, Dalman K, Deflorio G, van Diepen LTA, Dunand C, Duplessis S, Durling M, Gonthier P, Grimwood J, Fossdal CG, Hansson D, Henrissat B, Hietala A, Himmelstrand K, Hoffmeister D, Högberg N, James TY, Karlsson M, Kohler A, Kües U, Lee Y-H, Lin Y-C, Lind M, Lindquist E, Lombard V, Lucas S, Lundén K, Morin E, Murat C, Park J, Raffaello T, Rouzé P, Salamov A, Schmutz J, Solheim H, Ståhlberg J, Vélëz H, de Vries RP, Wiebenga A, Woodward S, Yakovlev I, Garbelotto M, Martin F, Grigoriev IV, Stenlid J. 2012. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol 194:1001–1013. doi: 10.1111/j.1469-8137.2012.04128.x. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki H, MacDonald J, Syed K, Salamov A, Hori C, Aerts A, Henrissat B, Wiebenga A, VanKuyk PA, Barry K, Lindquist E, LaButti K, Lapidus A, Lucas S, Coutinho P, Gong Y, Samejima M, Mahadevan R, Abou-Zaid M, de Vries RP, Igarashi K, Yadav JS, Grigoriev IV, Master ER. 2012. Comparative genomics of the white-rot fungi, Phanerochaete carnosa and P chrysosporium, to elucidate the genetic basis of the distinct wood types they colonize. BMC Genomics 13:444. doi: 10.1186/1471-2164-13-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prospero S, Conedera M, Heiniger U, Rigling D. 2006. Saprophytic activity and sporulation of Cryphonectria parasitica on dead chestnut wood in forests with naturally established hypovirulence. Phytopathology 96:1337–1344. doi: 10.1094/PHYTO-96-1337. [DOI] [PubMed] [Google Scholar]
- 60.Lovat C-A, Donnelly DJ. 2019. Mechanisms and metabolomics of the host-pathogen interactions between Chestnut (Castanea species) and Chestnut blight (Cryphonectria parasitica). For Pathol 49:e12562. doi: 10.1111/efp.12562. [DOI] [Google Scholar]
- 61.Sprockett DD, Piontkivska H, Blackwood CB. 2011. Evolutionary analysis of glycosyl hydrolase family 28 (GH28) suggests lineage-specific expansions in necrotrophic fungal pathogens. Gene 479:29–36. doi: 10.1016/j.gene.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 62.Mengiste T. 2012. Plant immunity to necrotrophs. Annu Rev Phytopathol 50:267–294. doi: 10.1146/annurev-phyto-081211-172955. [DOI] [PubMed] [Google Scholar]
- 63.Chankhamjon P, Tsunematsu Y, Ishida-Ito M, Sasa Y, Meyer F, Boettger-Schmidt D, Urbansky B, Menzel K-D, Scherlach K, Watanabe K, Hertweck C. 2016. Regioselective dichlorination of a non-activated aliphatic carbon atom and phenolic bismethylation by a multifunctional fungal flavoenzyme. Angew Chem Int Ed Engl 55:11955–11959. doi: 10.1002/anie.201604516. [DOI] [PubMed] [Google Scholar]
- 64.Shwab EK, Keller NP. 2008. Regulation of secondary metabolite production in filamentous ascomycetes. Mycol Res 112:225–230. doi: 10.1016/j.mycres.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 65.Lowe RGT, Howlett BJ. 2012. Indifferent, affectionate, or deceitful: lifestyles and secretomes of fungi. PLoS Pathog 8:e1002515. doi: 10.1371/journal.ppat.1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anagnostakis SL. 1992. Measuring resistance of chestnut trees to chestnut blight. Can J For Res 22:568–571. doi: 10.1139/x92-075. [DOI] [Google Scholar]
- 67.Haas SE, Hooten MB, Rizzo DM, Meentemeyer RK. 2011. Forest species diversity reduces disease risk in a generalist plant pathogen invasion. Ecol Lett 14:1108–1116. doi: 10.1111/j.1461-0248.2011.01679.x. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen D, Boberg J, Cleary M, Bruelheide H, Hönig L, Koricheva J, Stenlid J. 2017. Foliar fungi of Betula pendula: impact of tree species mixtures and assessment methods. Sci Rep 7:41801–41811. doi: 10.1038/srep41801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prospero S, Cleary M. 2017. Effects of host variability on the spread of invasive forest diseases. Forests 8:80. doi: 10.3390/f8030080. [DOI] [Google Scholar]
- 70.Trapero-Casas A, Kaiser WJ. 2009. Alternative hosts and plant tissues for the survival, sporulation and spread of the Ascochyta blight pathogen of chickpea. Eur J Plant Pathol 125:573–587. doi: 10.1007/s10658-009-9507-2. [DOI] [Google Scholar]
- 71.Brader G, Compant S, Vescio K, Mitter B, Trognitz F, Ma L-J, Sessitsch A. 2017. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu Rev Phytopathol 55:61–83. doi: 10.1146/annurev-phyto-080516-035641. [DOI] [PubMed] [Google Scholar]
- 72.Sakalidis ML, Hardy GES, Burgess TI. 2011. Endophytes as potential pathogens of the baobab species Adansonia gregorii: a focus on the Botryosphaeriaceae. Fungal Ecol 4:1–14. doi: 10.1016/j.funeco.2010.06.001. [DOI] [Google Scholar]
- 73.Zakaria L, Yaakop AS, Salleh B, Zakaria M. 2010. Endophytic fungi from paddy. Trop Life Sci Res 21:101–107. [PMC free article] [PubMed] [Google Scholar]
- 74.Vaz ABM, Fonseca PLC, Badotti F, Skaltsas D, Tomé LMR, Silva AC, Cunha MC, Soares MA, Santos VL, Oliveira G, Chaverri P, Góes-Neto A. 2018. A multiscale study of fungal endophyte communities of the foliar endosphere of native rubber trees in Eastern Amazon. Sci Rep 8:16151. doi: 10.1038/s41598-018-34619-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Granados GM, McTaggart AR, Rodas CA, Roux J, Wingfield MJ. 2020. Species of Cryphonectriaceae occupy an endophytic niche in the Melastomataceae and are putative latent pathogens of Eucalyptus. Eur J Plant Pathol 156:273–283. doi: 10.1007/s10658-019-01887-9. [DOI] [Google Scholar]
- 76.Mausse-Sitoe SND, Rodas CA, Wingfield MJ, Chen S, Roux J. 2016. Endophytic Cryphonectriaceae on native Myrtales: possible origin of Chrysoporthe canker on plantation-grown Eucalyptus. Fungal Biol 120:827–835. doi: 10.1016/j.funbio.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 77.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. 2018. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34:i142–i150. doi: 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol 35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. 2016. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32:767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoff KJ, Lomsadze A, Borodovsky M, Stanke M. 2019. Whole-genome annotation with BRAKER. Methods Mol Biol 1962:65–95. doi: 10.1007/978-1-4939-9173-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stanke M, Schöffmann O, Morgenstern B, Waack S. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 84.Crouch JA, Dawe A, Aerts A, Barry K, Churchill ACL, Grimwood J, Hillman B, Milgroom MG, Pangilinan J, Smith M, Salamov A, Schmutz J, Yadav JS, Grigoriev IV, Nuss DL. 2020. Genome sequence of the chestnut blight fungus Cryphonectria parasitica EP155: a fundamental resource for an archetypical invasive plant pathogen. Phytopathology 110:1180–1188. doi: 10.1094/PHYTO-12-19-0478-A. [DOI] [PubMed] [Google Scholar]
- 85.Srinivasan A. 2019. gread: fast reading and processing of common gene annotation and next generation sequencing format files.
- 86.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rice P, Bleasby A, Ison J. 2011. EMBOSS user’s guide: practical bioinformatics.
- 88.Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yin J, Zhang C, Mirarab S. 2019. ASTRAL-MP: scaling ASTRAL to very large datasets using randomization and parallelization. Bioinformatics 35:3961–3969. doi: 10.1093/bioinformatics/btz211. [DOI] [PubMed] [Google Scholar]
- 92.Rambaut A, Drummond AJ. 2016. FigTree v1. 4.3.
- 93.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wickham H. 2017. tidyverse: easily install and load the “Tidyverse.”
- 95.Wickham H. 2007. Reshaping data with the reshape package. J Stat Softw 21:1–20. [Google Scholar]
- 96.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 97.Wickham H. 2018. stringr: simple, consistent wrappers for common string operations.
- 98.Eddy S. 2010. HMMER3: a new generation of sequence homology search software. http://hmmer.org.
- 99.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mäkelä MR, Peng M, Granchi Z, Chin-A-Woeng T, Hegi R, van Pelt SI, Ahrendt S, Riley R, Hainaut M, Henrissat B, Grigoriev IV, de Vries RP, Hildén KS. 2018. Draft genome sequence of the Basidiomycete white-rot fungus Phlebia centrifuga. Genome Announc 6:e01414-17. doi: 10.1128/genomeA.01414-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gómez-Cortecero A, Harrison RJ, Armitage AD. 2015. Draft genome sequence of a European isolate of the apple canker pathogen Neonectria ditissima. Genome Announc 3:e01243-15. doi: 10.1128/genomeA.01243-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Forgetta V, Leveque G, Dias J, Grove D, Lyons R, Genik S, Wright C, Singh S, Peterson N, Zianni M, Kieleczawa J, Steen R, Perera A, Bintzler D, Adams S, Hintz W, Jacobi V, Bernier L, Levesque R, Dewar K. 2013. Sequencing of the Dutch elm disease fungus genome using the Roche/454 GS-FLX Titanium System in a comparison of multiple genomics core facilities. J Biomol Tech 24:39–49. doi: 10.7171/jbt.12-2401-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Comeau AM, Dufour J, Bouvet GF, Jacobi V, Nigg M, Henrissat B, Laroche J, Levesque RC, Bernier L. 2014. Functional annotation of the Ophiostoma novo-ulmi genome: insights into the phytopathogenicity of the fungal agent of Dutch elm disease. Genome Biol Evol 7:410–430. doi: 10.1093/gbe/evu281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yin Z, Liu H, Li Z, Ke X, Dou D, Gao X, Song N, Dai Q, Wu Y, Xu J-R, Kang Z, Huang L. 2015. Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol 208:1202–1216. doi: 10.1111/nph.13544. [DOI] [PubMed] [Google Scholar]
- 105.Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 106.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM. 2018. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol Plant Pathol 19:2094–2110. doi: 10.1111/mpp.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wingfield BD, Ades PK, Al-Naemi FA, Beirn LA, Bihon W, Crouch JA, de Beer ZW, De Vos L, Duong TA, Fields CJ, Fourie G, Kanzi AM, Malapi-Wight M, Pethybridge SJ, Radwan O, Rendon G, Slippers B, Santana QC, Steenkamp ET, Taylor PWJ, Vaghefi N, van der Merwe NA, Veltri D, Wingfield MJ. 2015. Draft genome sequences of Chrysoporthe austroafricana, Diplodia scrobiculata, Fusarium nygamai, Leptographium lundbergii, Limonomyces culmigenus, Stagonosporopsis tanaceti, and Thielaviopsis punctulata. IMA Fungus 6:233–248. doi: 10.5598/imafungus.2015.06.01.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wingfield BD, Barnes I, Wilhelm de Beer Z, De Vos L, Duong TA, Kanzi AM, Naidoo K, Nguyen HDT, Santana QC, Sayari M, Seifert KA, Steenkamp ET, Trollip C, van der Merwe NA, van der Nest MA, Markus Wilken P, Wingfield MJ. 2015. Draft genome sequences of Ceratocystis eucalypticola, Chrysoporthe cubensis, C. deuterocubensis, Davidsoniella virescens, Fusarium temperatum, Graphilbum fragrans, Penicillium nordicum, and Thielaviopsis musarum. IMA Fungus 6:493–506. doi: 10.5598/imafungus.2015.06.02.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lesion length on dormant chestnut stems (Castanea sativa) with bark removal. Measurements were taken once per week during four weeks. Download FIG S1, PDF file, 1.2 MB (1.2MB, pdf) .
Copyright © 2020 Stauber et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene count of all identified CAZyme families among Cryphonectriaceae. Download FIG S2, PDF file, 0.2 MB (219.4KB, pdf) .
Copyright © 2020 Stauber et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Two PKS gene clusters (biosynthetic core gene orthologs OG0009468 and OG0002648) potentially underlying emodin production. Synteny plots of emodin and identified homologous gene clusters among species. Heatmaps show similarity (%) of genes in the C. parasitica reference genome cluster to emodin and orthologs in other Cryphonectriaceae (gray = gene is absent). Download FIG S3, PDF file, 0.4 MB (394.8KB, pdf) .
Copyright © 2020 Stauber et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overview of genomes assembled in the genus Cryphonectria. Country, region, and host identify the location and host species of collection. Download Table S1, XLS file, 0.05 MB (54KB, xls) .
Copyright © 2020 Stauber et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All sample accession numbers for the NCBI Short Read Archive, as well as NCBI accession numbers for all de novo assemblies of Cryphonectria genomes, are available in Table S1 in the supplemental material. Contigs of <400 bp were trimmed prior to NCBI genome submission. Outgroup genomes to the genus Cryphonectria were obtained for Chrysoporthe cubensis, Chr. deuterocubensis, and Chr. austroafricana from NCBI BioProjects PRJNA279968, PRJNA265023, and PRJNA263707, respectively (108, 109).