Abstract
Human chorionic gonadotropin (hCG) is a well-known hormone produced by the trophoblast during pregnancy as well as by both trophoblastic and non-trophoblastic tumors. hCG is built from two subunits: α (hCGα) and β (hCGβ). The hormone-specific β subunit is encoded by six allelic genes: CGB3, CGB5, CGB6, CGB7, CGB8, and CGB9, mapped to the 19q13.32 locus. This gene cluster also encompasses the CGB1 and CGB2 genes, which were originally considered to be pseudogenes, but as documented by several studies are transcriptionally active. Even though the protein products of these genes have not yet been identified, based on The Cancer Genome Atlas (TCGA) database analysis we showed that the mutual presence of CGB1 and CGB2 transcripts is a characteristic feature of cancers of different origin, including bladder urothelial carcinoma, cervical squamous cell carcinoma, esophageal carcinoma, head and neck squamous cell carcinoma, ovarian serous cystadenocarcinoma, lung squamous cell carcinoma, pancreatic adenocarcinoma, rectum adenocacinoma, testis germ cell tumors, thymoma, uterine corpus endometrial carcinoma and uterine carcinosarcoma.
Keywords: human chorionic gonadotropin, TCGA, cancer, CGB1, CGB2
1. Introduction
Human chorionic gonadotropin (hCG) is a heterodimeric hormone, which comprises two non-covalently linked subunits: alpha (α) and beta (β). The α subunit is common to all gonadotropic hormones, such as follicle stimulating hormone (FSH), thyrotropin (TSH) and luteinizing hormone (LH), while the β subunit is hormone specific and determines its biological properties [1,2,3]. Most gonadotropins are synthesized by the anterior pituitary gland, however hCG synthesis takes place in the placenta as a result of syncytiotrophoblast cells activity [4].
Human chorionic gonadotropin participates in and regulates many physiological processes related to the normal course of pregnancy, which includes: maintenance of progesterone production by the corpus luteum, chorionic villi development, embryo implantation, or angiogenesis [5]. In addition, hCG determines the mother’s immunotolerance to the antigens presented by the developing fetus [6,7].
Human chorionic gonadotropin, especially its β subunit (hCGβ) is also secreted in gestational trophoblastic disease, and by a large group of non-trophoblastic tumors. hCGβ expression has been observed in tumors of different origin such as: breast, cervix, prostate, lung, colon, kidney, bladder, pancreas, anus, vulva, ovary, brain, endometrium and mouth [8,9].
The mechanisms regulating hCG and hCGβ expression, as well as their role in cancer development and growth, are not fully understood.
The formation of functional human chorionic gonadotropin is associated with the activation of genes encoding both subunits of the hormone (α and β). The α subunit is encoded by a single CGA gene, located on the long arm of the sixth chromosome at the 6q14-q21 locus. The β subunit is encoded by eight allelic genes: CGB1, CGB2, CGB3, CGB5, CGB6, CGB7, CGB8, and CGB9, located in the luteinizing hormone beta subunit/chorionic gonadotropin beta subunit (LHB/CGB) cluster on the long arm of the nineteenth chromosome at the 19q13.32 locus [10]. The cluster also holds the CGB4 gene encoding the β subunit of luteotropic hormone [11,12].
Interestingly, sequencing of the nineteenth chromosome revealed that the CGB7 and CGB3 genes are allelic forms of the CGB6 and CGB9 genes, respectively [13,14].
The protein products of individual CGB genes show amino acid differences at position 117. This led to the division of the genes into two groups. The first group, which includes the CGB7 and CGB9 genes, was categorized as type I genes and gives rise to protein products with alanine in position 117. Type II genes, which include CGB3, CGB5, CGB6, and CGB8, encode a protein with aspartic acid in position 117 [10,12,15].
It is hypothesized that in the course of evolution, the CGB3, CGB5, CGB6, CGB7, CGB8, and CGB9 genes arose as a result of multiple duplications of the gene encoding the β subunit of luteinizing hormone (CGB4) [16]. In contrast, the CGB1 and CGB2 genes evolved as a consequence of the insertion of a DNA fragment into the duplicated genes. In the case of the CGB1 gene, this fragment included 736 base pairs, while for CGB2, it was 724 base pairs. The insertion led to the deletion of a 52-base long segment of the proximal promoter, as well as the entire 5′ untranslated region (5′UTR) region of the CGB gene. The consequence of this mutation was the creation of a new promoter sequence for CGB1 and CGB2, a new 5′UTR region with an alternative start codon, and a new first exon. This led to a shift in the open reading frame for exons 2 and 3 [14,17]. The development of systemic and molecular biology methods made it possible to show that the nucleotide sequences of these genes in humans and chimpanzee differ by only 0.5% of their total composition. This high degree of conservatism of CGB1 and CGB2 confirms their common evolutionary path of origin [18].
Until recently, CGB1 and CGB2 were considered to be pseudogenes. Their biological activity was, however, demonstrated at the level of transcription. The genes’ transcripts have been detected in healthy tissues, such as placenta, testes and the pituitary gland [14,19,20,21,22]. CGB1 and CGB2 expression has also been confirmed in breast and bladder cancer cell lines, as well as in ovarian cancer tissue [9,23,24,25].
Although the contribution of CGB1 and CGB2 in the total expression of all CGB genes does not exceed 1/1000, both CGB1 and CGB2 genes were shown to have a considerable expression peak during the first trimester in normal pregnancy, but not in placentas of the second and third trimester, or in recurrent miscarriages. This particular expression pattern of the genes suggests their putative role in the implantation stage of the fetus development. Thus, it may be speculated that their expression drives placental cytotrophoblast cell invasion and growth during implantation. In the same manner, CGB1 and CGB2 may promote the pregnancy-like invasion of cancer cells into specific microenvironments, and later tumor growth and metastasis [26].
In order to verify the transcriptional activity of CGB1 and CGB2 genes in cancer, we analyzed data collected in TCGA.
2. Materials and Methods
Information related to thirty-three different types of tumors deposited in The Cancer Genome Atlas (TCGA) database was evaluated in terms of CGB1 and CGB2 gene expression. Twelve tumors, characterized by the presence of both CGB1 and CGB2 gene transcripts, were selected for further research and the expression level of the studied genes in cancer and corresponding non-malignant tissues was analyzed using UALCAN web-portal [27].
TCGA level 3 RNASeq V2 data equivalent to the expression of CGB1 and CGB2 in non-malignant and primary tumor samples for each gene was presented as a box and whisker plot. All values of gene expression were presented as transcripts per million (TPM)—a normalization method for RNA-seq which represents the relative abundance of a gene or transcript in a sample.
Student’s t-test was used to calculate the level of statistical significance (p-value). In the case of a lack of information regarding the expression of the genes or when the number of available data was extremely small, no statistical test was performed.
The data were presented using figures showing the interquartile range (IQR), including the median, minimum, maximum values, 25th percentile (Lower Q) and 75th (Upper Q) percentile (if available).
3. Results
To evaluate the expression level of CGB1 and CGB2 genes in cancers of different origin, the molecular characterization of thirty-three different tumors deposited in the TCGA database was analyzed. First, the screening of TCGA transcriptome data related to CGB1 and CGB2 gene expression in different cancers was performed.
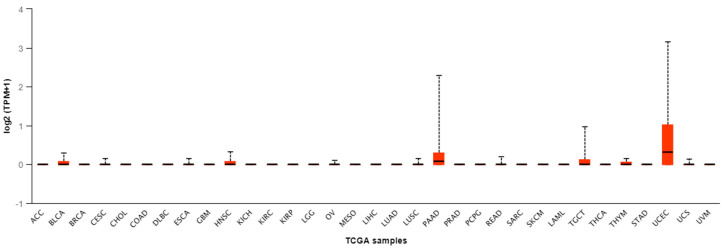
The CGB1 gene transcript was found in 12 types of cancer only, namely: BLCA (Bladder urothelial carcinoma), CESC (Cervical squamous cell carcinoma), ESCA (Esophageal carcinoma), HNSC (Head and Neck squamous cell carcinoma), OV (Ovarian serous cystadenocarcinoma), LUSC (Lung squamous cell carcinoma), PAAD (Pancreatic adenocarcinoma), READ (Rectum adenocacinoma), TGCT (Testis germ cell tumors), THYM (Thymoma), UCEC (Uterine corpus endometrial carcinoma) and USC (Uterine carcinosarcoma); (Figure 1). The highest median values of CGB1 expression were noted for two cancers: PAAD and UCEC (median: 0.078 and 0.317, respectively) (Table 1).
Figure 1.
Expression level of CGB1 in cancer, according to The Cancer Genome Atlas (TCGA) data; TPM—transcripts per million; ACC—Adrenocortical carcinoma; BLCA—Bladder urothelial carcinoma; BRCA—Breast invasive carcinoma; CESC—Cervical squamous cell carcinoma; CHOL—Cholangiocarcinoma; COAD—Colon adenocarcinoma; DLBC—Lymphiod neoplasm diffuse large B-cell lymphoma; ESCA—Esophageal carcinoma; GBM—Glioblastoma multiforme; HNSC—Head and Neck squamous cell carcinoma; KICH—Kidney chromophobe; KIRC—Kidney renal clear cell carcinoma; KIRP—Kidney renal papillary cell carcinoma; LGG—Brain lower grade glioma; OV—Ovarian serous cystadenocarcinoma; MESO—Mesothelioma; LIHC—Liver hepatocellular carcinoma; LUAD—Lung adenocarcinoma; LUSC—Lung squamous cell carcinoma; PAAD—Pancreatic adenocarcinoma; PRAD—Prostate adenocarcinoma; PCPG—Pheochromocytoma and Paraganglioma; READ—Rectum adenocacinoma; SARC—Sarcoma; SKCM—Skin cutaneous melanoma; LAML—Acute Myeloid Leukemia; TGCT—Testis germ cell tumors; THCA—Thyroid carcinoma; THYM—Thymoma; STAD—Stomach adenocarcinomna; UCEC—Uterine corpus endometrial carcinoma; USC—Uterine carcinosarcoma; UVM—Uveal Melanoma.
Table 1.
Expression values of CGB1 in cancer samples deposited in TCGA. Q—quartile.
| Tumor Type | Number of Samples | Minimal Value | Lower Q | Median | Upper Q | Maximal Value |
|---|---|---|---|---|---|---|
| ACC | 79 | 0 | 0 | 0 | 0 | 0 |
| BLCA | 408 | 0 | 0 | 0 | 0.071 | 0.297 |
| BRCA | 1097 | 0 | 0 | 0 | 0 | 0 |
| CESC | 305 | 0 | 0 | 0 | 0 | 0.159 |
| CHOL | 36 | 0 | 0 | 0 | 0 | 0 |
| COAD | 286 | 0 | 0 | 0 | 0 | 0 |
| DLBC | 48 | 0 | 0 | 0 | 0 | 0 |
| ESCA | 184 | 0 | 0 | 0 | 0 | 0.152 |
| GBM | 156 | 0 | 0 | 0 | 0 | 0 |
| HNSC | 520 | 0 | 0 | 0 | 0.075 | 0.321 |
| KICH | 67 | 0 | 0 | 0 | 0 | 0 |
| KIRC | 533 | 0 | 0 | 0 | 0 | 0 |
| KIRP | 290 | 0 | 0 | 0 | 0 | 0 |
| LGG | 513 | 0 | 0 | 0 | 0 | 0 |
| OV | 305 | 0 | 0 | 0 | 0 | 0.103 |
| MESO | 87 | 0 | 0 | 0 | 0 | 0 |
| LIHC | 371 | 0 | 0 | 0 | 0 | 0 |
| LUAD | 515 | 0 | 0 | 0 | 0 | 0 |
| LUSC | 503 | 0 | 0 | 0 | 0 | 0.152 |
| PAAD | 178 | 0 | 0 | 0.078 | 0.302 | 2.291 |
| PRAD | 497 | 0 | 0 | 0 | 0 | 0 |
| PCPG | 179 | 0 | 0 | 0 | 0 | 0 |
| READ | 166 | 0 | 0 | 0 | 0 | 0.198 |
| SARC | 260 | 0 | 0 | 0 | 0 | 0 |
| SKCM | 472 | 0 | 0 | 0 | 0 | 0 |
| LAML | 173 | 0 | 0 | 0 | 0 | 0 |
| TGCT | 150 | 0 | 0 | 0 | 0.124 | 0.997 |
| THCA | 505 | 0 | 0 | 0 | 0 | 0 |
| THYM | 120 | 0 | 0 | 0 | 0.061 | 0.149 |
| STAD | 415 | 0 | 0 | 0 | 0 | 0 |
| UCEC | 546 | 0 | 0 | 0.317 | 1.016 | 3.158 |
| USC | 57 | 0 | 0 | 0 | 0 | 0.134 |
| UVM | 80 | 0 | 0 | 0 | 0 | 0 |
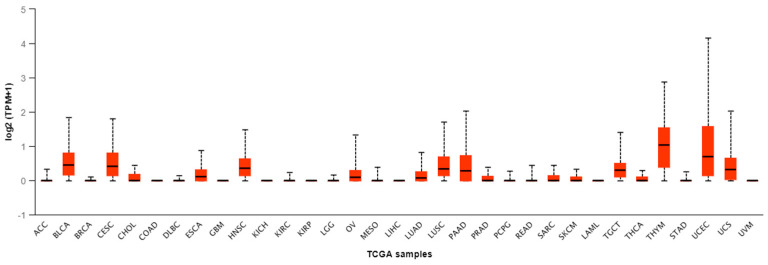
The CGB2 gene transcripts were found in 26 types of cancer: ACC (Adrenocortical carcinoma), BLCA (Bladder urothelial carcinoma), BRCA (Breast invasive carcinoma), CESC (Cervical squamous cell carcinoma), CHOL (Cholangiocarcinoma), DLBC (Lymphiod neoplasm diffuse large B-cell lymphoma), ESCA (Esophageal carcinoma), HNSC (Head and Neck squamous cell carcinoma), KIRC (Kidney renal clear cell carcinoma), LGG (Brain lower grade glioma); OV (Ovarian serous cystadenocarcinoma), MESO (Mesothelioma), LUAD (Lung adenocarcinoma), LUSC (Lung squamous cell carcinoma), PAAD (Pancreatic adenocarcinoma); PRAD (Prostate adenocarcinoma), PCPG (Pheochromocytoma and Paraganglioma), READ (Rectum adenocacinoma), SARC (Sarcoma), SKCM (Skin cutaneous melanoma); TGCT (Testis germ cell tumors), THCA (Thyroid carcinoma), THYM (Thymoma), STAD (Stomach adenocarcinomna), UCEC (Uterine corpus endometrial carcinoma) and USC (Uterine carcinosarcoma); (Figure 2).
Figure 2.
Expression of CGB2 in cancer, according to TCGA data.
In BLCA, CESC, ESCA, HNSC, OV, LUAD, LUSC, PAAD, TGCT, THYM, UCEC, and USC, the median values of CGB2 transcriptional activity were greater than 0, (median: 0.447, 0.412, 0.112, 0.356, 0.087, 0.084, 0.336, 0.279, 0.305, 1.033, 0.703, 0.318, respectively); (Table 2).
Table 2.
Expression values of CGB2 in different tumor types according to TCGA.
| Tumor Type | Number of Samples | Minimal Value | Lower Q | Median | Upper Q | Maximal Value |
|---|---|---|---|---|---|---|
| ACC | 79 | 0 | 0 | 0 | 0 | 0.329 |
| BLCA | 408 | 0 | 0.157 | 0.447 | 0.8 | 1.848 |
| BRCA | 1097 | 0 | 0 | 0 | 0 | 0.105 |
| CESC | 305 | 0 | 0.133 | 0.412 | 0.801 | 1.81 |
| CHOL | 36 | 0 | 0 | 0 | 0.17 | 0.44 |
| COAD | 286 | 0 | 0 | 0 | 0 | 0 |
| DLBC | 48 | 0 | 0 | 0 | 0 | 0.144 |
| ESCA | 184 | 0 | 0 | 0.112 | 0.311 | 0.869 |
| GBM | 156 | 0 | 0 | 0 | 0 | 0 |
| HNSC | 520 | 0 | 0.15 | 0.356 | 0.631 | 1.475 |
| KICH | 67 | 0 | 0 | 0 | 0 | 0 |
| KIRC | 533 | 0 | 0 | 0 | 0 | 0.236 |
| KIRP | 290 | 0 | 0 | 0 | 0 | 0 |
| LGG | 513 | 0 | 0 | 0 | 0 | 0.158 |
| OV | 305 | 0 | 0 | 0.087 | 0.299 | 1.329 |
| MESO | 87 | 0 | 0 | 0 | 0 | 0.396 |
| LIHC | 371 | 0 | 0 | 0 | 0 | 0 |
| LUAD | 515 | 0 | 0 | 0.084 | 0.257 | 0.823 |
| LUSC | 503 | 0 | 0.14 | 0.336 | 0.69 | 1.71 |
| PAAD | 178 | 0 | 0 | 0.279 | 0.725 | 2.024 |
| PRAD | 497 | 0 | 0 | 0 | 0.132 | 0.38 |
| PCPG | 179 | 0 | 0 | 0 | 0 | 0.265 |
| READ | 166 | 0 | 0 | 0 | 0 | 0.434 |
| SARC | 260 | 0 | 0 | 0 | 0.145 | 0.442 |
| SKCM | 472 | 0 | 0 | 0 | 0.098 | 0.329 |
| LAML | 173 | 0 | 0 | 0 | 0 | 0 |
| TGCT | 150 | 0 | 0.106 | 0.305 | 0.502 | 1.414 |
| THCA | 505 | 0 | 0 | 0 | 0.105 | 0.288 |
| THYM | 120 | 0 | 0.384 | 1.033 | 1.537 | 2.871 |
| STAD | 415 | 0 | 0 | 0 | 0 | 0.257 |
| UCEC | 546 | 0 | 0.135 | 0.703 | 1.572 | 4.154 |
| USC | 57 | 0 | 0.033 | 0.318 | 0.644 | 2.019 |
| UVM | 80 | 0 | 0 | 0 | 0 | 0 |
Among the 33 different studied cancers, only 12 were characterized by concomitant expression of CGB1 and CGB2. This group consisted of BLCA, CESC, ESCA, HNSC, OV, LUSC, PAAD, READ, TGCT, THYM, UCEC and USC (Table 3). The highest TPM values noted for both studied CGB genes (CGB1: 3.158 and CGB2: 4.154) characterized UCEC (Table 3). Detailed analysis of the studied gene expression in these tumors revealed differences between cancerous and corresponding non-malignant tissues (Table 4).
Table 3.
The comparison of CGB1 and CGB2 expression in non-malignant and cancerous tissues according to The Cancer Genome Atlas (TCGA) database data.
| Tumor Type | Number of Samples | Gene | Minimal Value | Lower Q | Median | Upper Q | Maximal Value |
|---|---|---|---|---|---|---|---|
| BLCA | 408 | CGB1 | 0 | 0 | 0 | 0.071 | 0.297 |
| CGB2 | 0 | 0.157 | 0.447 | 0.8 | 1.848 | ||
| CESC | 305 | CGB1 | 0 | 0 | 0 | 0 | 0.159 |
| CGB2 | 0 | 0.133 | 0.412 | 0.801 | 1.81 | ||
| ESCA | 184 | CGB1 | 0 | 0 | 0 | 0 | 0.152 |
| CGB2 | 0 | 0 | 0.112 | 0.311 | 0.869 | ||
| HNSC | 520 | CGB1 | 0 | 0 | 0 | 0.075 | 0.321 |
| CGB2 | 0 | 0.15 | 0.356 | 0.631 | 1.475 | ||
| OV | 305 | CGB1 | 0 | 0 | 0 | 0 | 0.103 |
| CGB2 | 0 | 0 | 0.087 | 0.299 | 1.329 | ||
| LUSC | 503 | CGB1 | 0 | 0 | 0 | 0 | 0.152 |
| CGB2 | 0 | 0.14 | 0.336 | 0.69 | 1.71 | ||
| PAAD | 178 | CGB1 | 0 | 0 | 0.078 | 0.302 | 2.291 |
| CGB2 | 0 | 0 | 0.279 | 0.725 | 2.024 | ||
| READ | 166 | CGB1 | 0 | 0 | 0 | 0 | 0.198 |
| CGB2 | 0 | 0 | 0 | 0 | 0.434 | ||
| TGCT | 150 | CGB1 | 0 | 0 | 0 | 0.124 | 0.997 |
| CGB2 | 0 | 0.106 | 0.305 | 0.502 | 1.414 | ||
| THYM | 120 | CGB1 | 0 | 0 | 0 | 0.061 | 0.149 |
| CGB2 | 0 | 0.384 | 1.033 | 1.537 | 2.871 | ||
| UCEC | 546 | CGB1 | 0 | 0 | 0.317 | 1.016 | 3.158 |
| CGB2 | 0 | 0.135 | 0.703 | 1.572 | 4.154 | ||
| USC | 57 | CGB1 | 0 | 0 | 0 | 0 | 0.134 |
| CGB2 | 0 | 0.033 | 0.318 | 0.644 | 2.019 |
Table 4.
The difference in CGB1 and CGB2 expression in non-malignant and primary tumor tissues based on data deposited in TCGA database. p ≤ 0.05 was considered as statistically significant. n.a.—not applicable.
| Tumor Type | Number of Tissue Samples | Gene | p | |
|---|---|---|---|---|
| Non-Malignant | Primary Tumor | |||
| BLCA | 19 | 408 | CGB1 | 0.07 |
| CGB2 | 1 × 10−6 | |||
| CESC | 3 | 305 | CGB1 | n.a. |
| CGB2 | n.a. | |||
| ESCA | 11 | 184 | CGB1 | n.a. |
| CGB2 | 0.006 | |||
| HNSC | 44 | 520 | CGB1 | n.a. |
| CGB2 | <1 × 10−12 | |||
| OV | no data | 305 | CGB1 | n.a. |
| CGB2 | n.a. | |||
| LUSC | 52 | 503 | CGB1 | n.a. |
| CGB2 | <1 × 10−12 | |||
| PAAD | 4 | 178 | CGB1 | n.a. |
| CGB2 | n.a. | |||
| READ | 10 | 166 | CGB1 | n.a. |
| CGB2 | n.a. | |||
| TGCT | no data | 150 | CGB1 | n.a. |
| CGB2 | n.a. | |||
| THYM | 2 | 120 | CGB1 | n.a. |
| CGB2 | n.a. | |||
| UCEC | 35 | 546 | CGB1 | 0.0044 |
| CGB2 | 0.0017 | |||
| USC | no data | 57 | CGB1 | n.a. |
| CGB2 | n.a. | |||
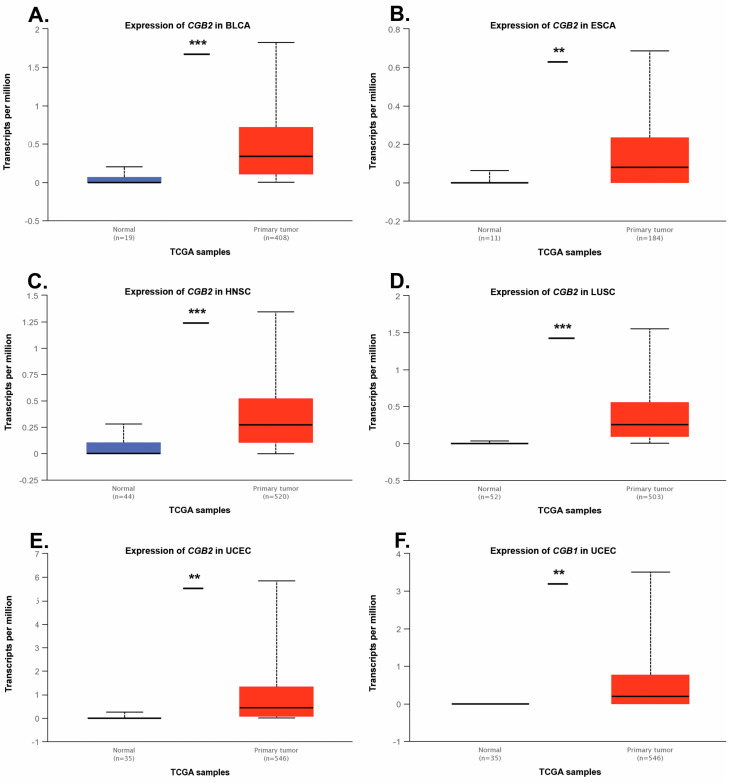
Significant differences in CGB2 expression in non-malignant tissues and primary tumor samples were noted for BLCA, ESCA, HNSC, LUSC and UCEC; p-value: 1 × 10−6, 0.006, <1 × 10−12, <1 × 10−12, 0.0017 respectively. The expression level of CGB1 in primary tumor and healthy tissue differed for UCEC only (p = 0.0044) (Figure 3).
Figure 3.
Expression levels of CGB1 and CGB2 in primary tumor samples and corresponding healthy tissues according to data deposited in the UALCAN database. (A) Expression of CGB2 in BLCA, (B) expression of CGB2 in ESCA, (C) expression of CGB2 in HNSC, (D) expression of CGB2 in LUSC, (E) expression of CGB2 in UCEC, (F) expression of CGB1 in UCEC. ** p ≤ 0.01; *** p ≤ 0.001.
4. Discussion
The synthesis of the β subunit of human chorionic gonadotropin occurs as a result of the activity of the tissue-dependent genes mapped to chromosome 19 (19q13.3), called CGB1–CGB9. The expression level of individual CGB genes is not equal. The most transcriptionally active gene, with up to twenty times higher expression compared to other CGB genes in both placenta and many cancerous tissues, is CGB5. The genes with lower transcriptional activity are CGB3, CGB6, CGB7, CGB8, and CGB9, as well as CGB1 and CGB2 [18].
For a long time, the methods used for quantifying gene expression were insufficiently sensitive to detect the presence of CGB1 and CGB2 transcripts in cells and tissues. In fact, for this reason, the CGB1 and CGB2 genes were considered pseudogenes.
Although the expression levels of CGB1 and CGB2 are a thousand times lower compared to the remaining CGBs, it has been shown that these genes’ transcripts play an important role in chorionic villi invasion and embryo implantation [26]. It was suggested that their expression may drive both placental cytotrophoblast cell invasion, as well as the invasion of cancer cells.
Indeed, it was reported that in bladder cancer cell lines, it is CGB2 expression, but not other CGB genes, which correlates with the amount of functional free hCGβ protein secreted by cancer cells and with cancer growth. It was also demonstrated that specific targeting of CGB1 and CGB2 with siRNA was much more effective in reducing cancer cell numbers than silencing other CGB genes [28]. Thus, it was suggested that CGB1 and CGB2 products give rise to tumor-driving molecules.
Since the transcriptional activity of CGB1 and CGB2 in cancer was mainly shown in cell lines, the aim of the present study was to demonstrate the expression of these genes in human cancer tissues of different origin. The data deposited in TCGA database was used in order to verify CGB1 and CGB2 transcriptional activity status in malignant and corresponding non-malignant tissue.
The results of the study showed that out of thirty-three available records related to the studied genes’ expression, CGB1 transcriptional activity characterized twelve types of cancer only, namely: BLCA, CESC, ESCA, HNSC, OV, LUSC, PAAD, READ, TGCT, THYM, UCEC, and USC.
At the same time CGB2 transcripts were detected in twenty six cancers, including: ACC, BLCA, BRCA, CESC, CHOL, DLBC, ESCA, HNSC, KIRC, LGG, OV, MESO, LUAD, LUSC, PAAD, PRAD, PCPG, READ, SARC, SKCM, TGCT, THCA, THYM, STAD, UCEC, and USC.
Interestingly, only twelve cancer types were characterized by the concomitant expression of CGB1 and CGB2. This group included bladder urothelial carcinoma, cervical squamous cell carcinoma, esophageal carcinoma, head and neck squamous cell carcinoma, ovarian serous cystadenocarcinoma, lung squamous cell carcinoma, pancreatic adenocarcinoma, rectum adenocacinoma, testis germ cell tumors, thymoma, uterine corpus endometrial carcinoma and uterine carcinosarcoma.
Despite the fact that the studied genes’ expression levels in cancer tissues were low, with the median often equaling zero, the transcriptional activity of the CGB1 and CGB2 genes in cancerous tissues of different origin was confirmed. Significant differences between CGB2 expression levels in healthy and cancerous tissues were shown. Such differences, which characterized bladder urothelial carcinoma, esophageal carcinoma, head and neck squamous cell carcinoma, lung squamous cell carcinoma, and uterine corpus endometrial carcinoma, may have diagnostic implications. In the case of the CGB1 gene, such a difference was recognized for uterine corpus endometrial carcinoma only. The analysis of the effect of CGB1 and CGB2 gene expression on patient survival did not show any significant correlation.
As mentioned earlier, CGB1 and CGB2 gene products are believed to drive placental cytotrophoblast cell invasion and growth during implantation. In the same way, they may promote the pregnancy-like invasion of cancer cells into specific microenvironments, and later tumor growth and metastasis. In fact, the results of TCGA database analysis show that a crucial factor in carcinogenesis may be CGB2 gene expression. The gene’s products were detected in 26 out of 33 studied samples, and statistically significant differences in CGB2 expression between malignant and non-malignant tissues were noted.
However, taking into account the limited data available, it cannot be excluded that both CGB1 and CGB2 gene products drive tumor development.
Further studies are needed in order to verify the hypothesis that CGB1 and CGB2 genes are involved in tumor growth, invasion and metastasis.
Abbreviations
| ACC | Adrenocortical carcinoma |
| BLCA | Bladder urothelial carcinoma |
| BRCA | Breast invasive carcinoma |
| CESC | Cervical squamous cell carcinoma |
| CHOL | Cholangiocarcinoma |
| COAD | Colon adenocarcinoma |
| DLBC | Lymphoid neoplasm diffuse large B-cell lymphoma |
| ESCA | Esophageal carcinoma |
| GBM | Glioblastoma multiforme |
| HNSC | Head and Neck squamous cell carcinoma |
| KICH | Kidney chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| LGG | Brain lower grade glioma |
| OV | Ovarian serous cystadenocarcinoma |
| MESO | Mesothelioma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| PAAD | Pancreatic adenocarcinoma |
| PRAD | Prostate adenocarcinoma |
| PCPG | Pheochromocytoma and Paraganglioma |
| READ | Rectum adenocarcinoma |
| SARC | Sarcoma |
| SKCM | Skin cutaneous melanoma |
| LAML | Acute Myeloid Leukemia |
| TGCT | Testis germ cell tumors |
| THCA | Thyroid carcinoma |
| THYM | Thymoma |
| STAD | Stomach adenocarcinoma |
| UCEC | Uterine corpus endometrial carcinoma |
| USC | Uterine carcinosarcoma |
| UVM | Uveal Melanoma |
| Mdn | Median |
Author Contributions
Conceptualization, A.J. and P.B.; methodology, P.B.; software, P.B.; validation, A.Ś. and A.S.; formal analysis, A.J.; investigation, P.B.; resources, A.J.; data curation, A.J.; writing—original draft preparation, P.B.; writing—review and editing, A.J. and A.Ś.; visualization, P.B. and A.S.; supervision, A.J.; project administration, A.J. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Science Centre, Poland, Miniatura grant 2019/03/X/NZ4/01447 (A.Ś.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pierce J.G., Parsons T.F. Glycoprotein Hormones: Structure and Function. Annu. Rev. Biochem. 1981;50:465–495. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- 2.Fiddes J.C., Goodman H.M. The gene encoding the common α subunit of the four human glycoprotein hormones. J. Mol. Appl. Genet. 1981;1:3–18. [PubMed] [Google Scholar]
- 3.Policastro P.F., Daniels-McQueen S., Carle G., Boime I. A map of the hCG β-LH β gene cluster. J. Biol. Chem. 1986;261:5907–5916. [PubMed] [Google Scholar]
- 4.Nwabuobi C., Arlier S., Schatz F., Guzeloglu-Kayisli O., Lockwood C.J., Kayisli U.A. hCG: Biological Functions and Clinical Applications. Int. J. Mol. Sci. 2017;18:2037. doi: 10.3390/ijms18102037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herr F., Baal N., Reisinger K., Lorenz A., McKinnon T., Preissner K.T., Zygmunt M. hCG in the Regulation of Placental Angiogenesis. Results of an In Vitro Study. Placenta. 2007;28:S85–S93. doi: 10.1016/j.placenta.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Bansal A.S., Bora S.A., Saso S., Smith J.R., Johnson M.R., Thum M.-Y. Mechanism of human chorionic gonadotrophin-mediated immunomodulation in pregnancy. Expert Rev. Clin. Immunol. 2012;8:747–753. doi: 10.1586/eci.12.77. [DOI] [PubMed] [Google Scholar]
- 7.Poloski Z., Linzke N., Woidacki K., Anne Schumacher A.C., Heinze K., Witte J. Human Chorionic Gonadotropin as a Central Regulator of Pregnancy Immune Tolerance. J. Immunol. Ref. 2013;190:2650–2658. doi: 10.4049/jimmunol.1202698. [DOI] [PubMed] [Google Scholar]
- 8.Iles R.K., Delves P.J., Butler S.A. Does hCG or hCGβ play a role in cancer cell biology? Mol. Cell. Endocrinol. 2010;329:62–70. doi: 10.1016/j.mce.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Hotakainen K., Lintula S., Jarvinen R., Paju A., Stenman J., Rintala E., Stenman U.H. Overexpression of human chorionic gonadotropin β genes 3, 5 and 8 in tumor tissue and urinary cells of bladder cancer patients. Tumor Biol. 2007;28:52–56. doi: 10.1159/000097703. [DOI] [PubMed] [Google Scholar]
- 10.Talmadge K., Boorstein W.R., Vamvakopoulos N.C., Gething M.J., Fiddes J.C. Only three of the seven human chorionic gonadotropin β subunit genes can be expressed in the placenta. Nucleic Acids Res. 1984;12:8415–8436. doi: 10.1093/nar/12.22.8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julier C., Weil D., Couillin P., Côté J.C., Nguyen V.C., Foubert C., Boué A., Thirion J.P., Kaplan J.C., Junien C. The β chorionic gonadotropin-β luteinizing gene cluster maps to human chromosome 19. Hum. Genet. 1984;67:174–177. doi: 10.1007/BF00272995. [DOI] [PubMed] [Google Scholar]
- 12.Policastro P., Ovitt C.E., Hoshina M., Fukuoka H., Boothby M.R., Boime I. The β subunit of human chorionic gonadotropin is encoded by multiple genes. J. Biol. Chem. 1983;258:11492–11499. [PubMed] [Google Scholar]
- 13.Bidart J.M., Baudin E., Troalen F., Bellet D., Schlumberger M. Eutopic and ectopic production of glycoprotein hormones α and β subunits. Ann. D’endocrinol. 1997;58:125–128. [PubMed] [Google Scholar]
- 14.Bo M., Boime I. Identification of the transcriptionally active genes of the chorionic gonadotropin β gene cluster in vivo. J. Biol. Chem. 1992;267:3179–3184. [PubMed] [Google Scholar]
- 15.Fiddes J.C., Goodman H.M. The cDNA for the β-subunit of human chorionic gonadotropin suggests evolution of a gene by readthrough into the 3′-untranslated region. Nature. 1980;286:684–687. doi: 10.1038/286684a0. [DOI] [PubMed] [Google Scholar]
- 16.Maston G.A., Ruvolo M. Chorionic Gonadotropin Has a Recent Origin Within Primates and an Evolutionary History of Selection. Mol. Biol. Evol. 2002;19:320–335. doi: 10.1093/oxfordjournals.molbev.a004085. [DOI] [PubMed] [Google Scholar]
- 17.Hollenberg A.N., Pestell R.G., Albanese C., Boers M.E., Jameson J.L. Multiple promoter elements in the human chorionic gonadotropin β subunit genes distinguish their expression from the luteinizing hormone β gene. Mol. Cell. Endocrinol. 1994;106:111–119. doi: 10.1016/0303-7207(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 18.Hallast P., Rull K., Laan M. The evolution and genomic landscape of CGB1 and CGB2 genes. Mol. Cell. Endocrinol. 2007;260–262:2–11. doi: 10.1016/j.mce.2005.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rull K., Hallast P., Uusküla L., Jackson J., Punab M., Salumets A., Campbell R.K., Laan M. Fine-scale quantification of HCG β gene transcription in human trophoblastic and non-malignant non-trophoblastic tissues. Mol. Hum. Reprod. 2008;14:23–31. doi: 10.1093/molehr/gam082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madersbacher S., Kratzik C., Gerth R., Dirnhofer S., Berger P. Human chorionic gonadotropin (hCG) and its free subunits in hydrocele fluids and neoplastic tissue of testicular cancer patients: Insights into the in vivo hCG-secretion pattern. Cancer Res. 1994;54:5096–5100. [PubMed] [Google Scholar]
- 21.Parrott A.M., Sriram G., Liu Y., Mathews M.B. Expression of type II chorionic gonadotropin genes supports a role in the male reproductive system. Mol. Cell. Biol. 2011;31:287–299. doi: 10.1128/MCB.00603-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dirnhofer S., Hermann M., Hittmair A., Hoermann R., Kapelari K., Berger P. Expression of the human chorionic gonadotropin-β gene cluster in human pituitaries and alternate use of exon 1. J. Clin. Endocrinol. Metab. 1996;81:4212–4217. doi: 10.1210/jcem.81.12.8954017. [DOI] [PubMed] [Google Scholar]
- 23.Berger P., Gruschwitz M., Spoettl G., Dirnhofer S., Madersbacher S., Gerth R., Merz W.E., Plas E., Sampson N. Human chorionic gonadotropin (hCG) in the male reproductive tract. Mol. Cell. Endocrinol. 2007;260–262:190–196. doi: 10.1016/j.mce.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 24.Giovangrandi Y., Parfait B., Asheuer M., Olivi M., Lidereau R., Vidaud M., Bièche I. Analysis of the human CGB/LHB gene cluster in breast tumors by real-time quantitative RT-PCR assays. Cancer Lett. 2001;168:93–100. doi: 10.1016/S0304-3835(01)00496-7. [DOI] [PubMed] [Google Scholar]
- 25.Kubiczak M., Walkowiak G.P., Nowak-Markwitz E., Jankowska A. Human chorionic gonadotropin β subunit genes CGB1 and CGB2 are transcriptionally active in ovarian cancer. Int. J. Mol. Sci. 2013;14:12650–12660. doi: 10.3390/ijms140612650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rull K., Laan M. Expression of β-subunit of HCG genes during normal and failed pregnancy. Hum. Reprod. 2005;20:3360–3368. doi: 10.1093/humrep/dei261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V.S.K., Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burczynska B.B., Kobrouly L., Butler S.A., Naase M., Iles R.K. Novel insights into the expression of CGB1 1-2 genes by epithelial cancer cell lines secreting ectopic free. Anticancer Res. 2014;34:2239–2248. [PubMed] [Google Scholar]