Abstract
Emerging evidence has suggested that circular RNAs (circRNAs) are involved in multiple physiological processes and participate in a variety of human diseases. However, the underlying biological function of circRNAs in pulmonary hypertension (PH) is still ambiguous. Herein, we investigated the implication and regulatory effect of a typical circRNA, CDR1as, in the pathological process of vascular calcification in PH. Human pulmonary artery smooth muscle cell (HPASMC) calcification was analyzed by western blotting, immunofluorescence, alizarin red S staining, alkaline phosphatase activity analysis, and calcium deposition quantification. CDR1as targets were identified by bioinformatics analysis and validated by dual-luciferase reporter and RNA antisense purification assays. We identified that CDR1as was upregulated in hypoxic conditions and promoted a phenotypic switch of HPASMCs from a contractile to an osteogenic phenotype. Moreover, microRNA (miR)-7-5p was shown to be a target of CDR1as, and calcium/calmodulin-dependent kinase II-delta (CAMK2D) and calponin 3 (CNN3) were suggested to be the putative target genes and regulated by CDR1as/miR-7-5p. The results showed that the CDR1as/miR-7-5p/CNN3 and CAMK2D regulatory axis mediates HPASMC osteoblastic differentiation and calcification induced by hypoxia. This evidence reveals an approach to the treatment of PH.
Keywords: CDR1as, miR-7-5p, pulmonary hypertension, human pulmonary artery smooth muscle cells, calcification
Graphical Abstract
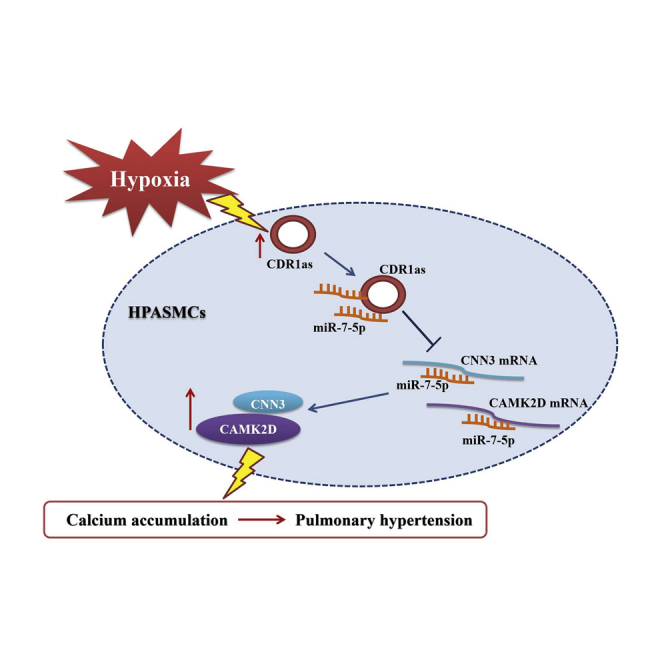
Zhu and colleagues investigate the implication of circular RNA CDR1as in the pathological process of pulmonary hypertension. miR-7-5p is shown to be a target of CDR1as. Moreover, CAMK2D and CNN3 are regulated by CDR1as and miR-7-5p and play an important role in mediating vascular calcification in pulmonary hypertension.
Introduction
Pulmonary hypertension (PH) is a hemodynamic disorder characterized by increased pulmonary blood flow, pulmonary vascular resistance, pulmonary artery pressure, pulmonary vasoconstriction, pulmonary vascular remodeling, and finally, right ventricular afterload.1,2 Although promising therapies consist of approaches to relieve vasoconstriction and inflammation, decrease pressure, and avoid heart failure, PH patients still face a poor prognosis.3 Histologically, in patients with PH, pulmonary vascular injury is markedly increased, as evidenced by endothelial cell impairment and vascular smooth muscle cell (VSMC) proliferation and hypertrophy.4,5 In the pulmonary vasculature, pulmonary artery smooth muscle cells (PASMCs) are important in the physiological regulation and pathological modulation of pulmonary vascular reactivity.6 The switch of PASMCs from the contractile phenotype to the synthetic phenotype plays a vital role in hypertrophy of the pulmonary vascular media and neointimal hyperplasia.7
One central factor linked to the pathologies of vascular lesions is vascular calcification caused by increased secretion of matrix components and osteogenic differentiation of VSMCs.8 Recently, studies have shown that vascular calcification is a common complication of atherosclerosis, chronic renal failure, and diabetes and is associated with cardiovascular morbidity and mortality.9, 10, 11, 12 Hypoxia plays a contributory role in triggering arterial calcification by increasing the expression of osteogenic markers and decreasing the expression of VSMC lineage markers.13,14 It remains unclear how pathologic arterial calcification is activated by hypoxia in PH, specifically in human PASMCs (HPASMCs), where it has localized paracrine effects on the pulmonary vascular beds.
Circular RNAs (circRNAs) are a group of transcripts characterized by covalently closed loop structures without polarities and polyadenylated tails.15 Most of the circRNAs are endogenous noncoding RNAs and show a higher degree of stability than linear mRNAs.16 Studies have highlighted important roles of circRNAs in both physiological and pathological settings, such as cellular metabolism and almost all types of cancers.17,18 CDR1as (also named as ciRS-7 or CDR1NAT) is 1,500 nt long and is formed by reverse splicing of the antisense strand of the cerebellar degeneration-associated antigen 1 (CDR1) gene.19,20 The classic regulatory mechanisms of circRNAs involve their role as competitive endogenous RNAs (ceRNAs); for example, CDR1as has more than 70 microRNA (miR)-7 binding sites that inhibit its binding to target genes.21 Until now, it has been unclear whether circRNAs can act as ceRNAs to regulate the functions of HPASMCs as well as the pathological process of vascular calcification in PH.
In the current study, we hypothesized that circRNAs serve as central regulators in the progression of vascular cell calcification in hypoxia. We identified a representative circRNA (hsa_circ_0001946, termed CDR1as) in hypoxic vascular cell calcification and systemically investigated its role in HPASMCs.
Results
Hypoxia Induces Osteochondrogenic Differentiation and CDR1as Expression in HPASMCs
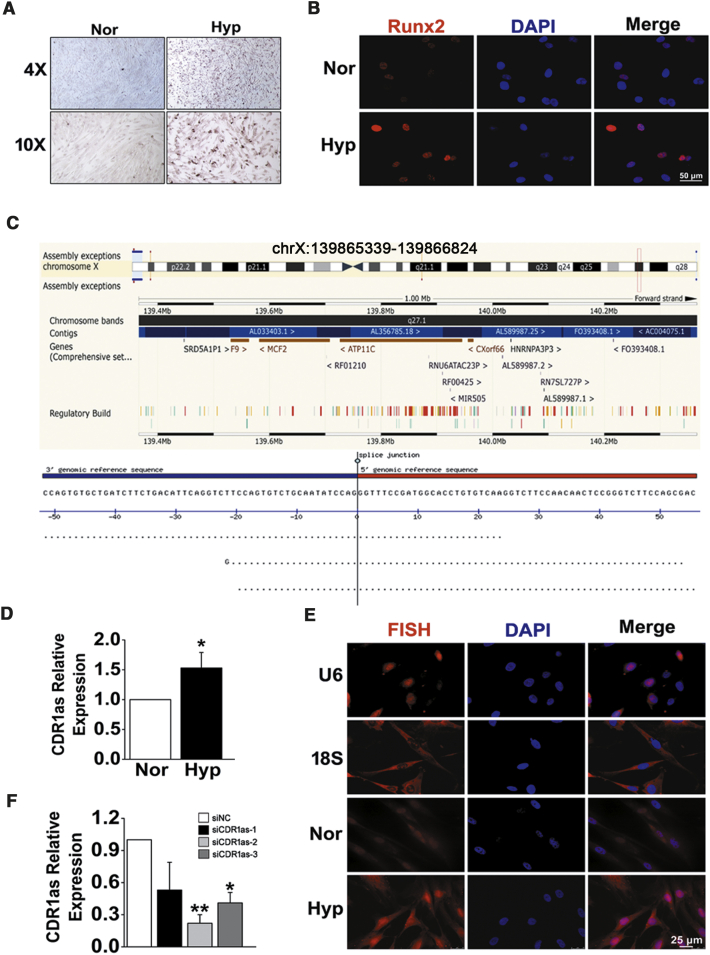
To confirm the role of hypoxia in osteochondrogenic reprogramming of HPASMCs, calcium deposition was first assessed by Alizarin Red S (ARS) staining. After treating HPASMCs with pro-calcifying media containing 2.5 mM inorganic phosphate, we found that compared with normoxia, hypoxia augmented calcium deposition in cultured HPASMCs (Figure 1A). Parallel with these changes, hypoxia also caused elevated expression of runt-related transcription factor-2 (Runx2), a key regulator of osteoblast differentiation and bone development (Figure 1B). CDR1as is generated from the CDR1 gene and located on human chromosome (chr) X: 139865340–139866824 (Figure 1C). To investigate the significance of CDR1as in hypoxic HPASMC osteogenesis, the expression of CDR1as was determined by qPCR. The results showed that the CDR1as level was elevated in HPASMCs exposed to hypoxia (approximately 1.5-fold increases) compared with those in normoxic conditions (Figure 1D). The results were further confirmed by fluorescence in situ hybridization (FISH) analysis. The expression of CDR1as in HPASMCs was found to be distributed in both the cytoplasm and the nucleus using U6 and 18S staining for nuclear and cytoplasmic areas, respectively (Figure 1E). Then, CDR1as-specific small interfering RNA (siRNA [siCDR1as]) was constructed and transfected into HPASMCs. Quantitative RT-PCR (qRT-PCR) analysis confirmed that the CDR1as expression level was significantly downregulated by siCDR1as-2 instead of siCDR1as-1 and siCDR1as-3 (Figure 1F). Therefore, we chose siCDR1as-2 for the following experiments.
Figure 1.
Osteogenic Differentiation and Expression Patterns of CDR1as in HPASMCs during Hypoxia (Hyp)
(A) HPASMCs were treated with pro-calcifying media containing 2.5 mM inorganic phosphate, and cell calcification was assessed by ARS staining. (B) Immunofluorescence analysis of Runx2 in HPASMCs from normoxia (Nor) and Hyp. Scale bar, 50 μm. (C) Genomic source of CDR1as. (D) Relative expression of CDR1as in Hyp was increased as determined by qPCR analysis. (E) FISH assay was performed to detect CDR1as distribution and expression in cultured cells. Scale bar, 25 μm. (F) The expression level of CDR1as in small interfering RNA (siRNA)- and negative control (siNC)-transfected cells. ∗p < 0.05, ∗∗p < 0.01. All tests were performed at least three times, and the values are presented as the mean ± SEM.
Downregulation of CDR1as Inhibits HPASMC Calcification Induced by Hypoxia
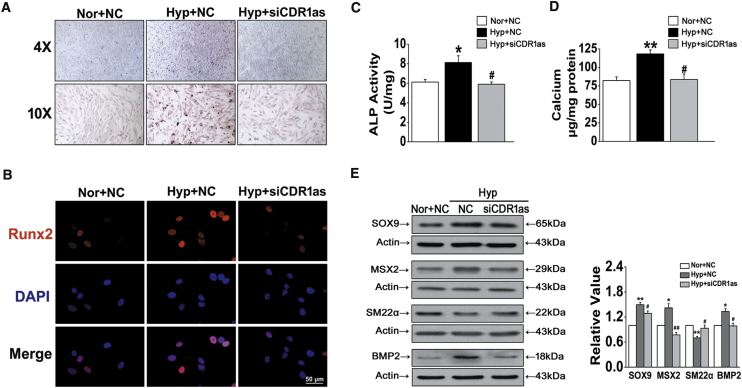
We next evaluated the functional role of CDR1as during the development of HPASMC calcification. We found that hypoxia-triggered calcium deposition in HPASMCs was inhibited by siRNA-CDR1as, as indicated by ARS staining (Figure 2A). In addition, siCDR1as attenuated hypoxia-mediated upregulation of the osteochondrogenic transcription factor Runx2 and diminished hypoxia-driven increases in alkaline phosphatase (ALP) activity (Figures 2B and 2C). Furthermore, specific knockdown of CDR1as markedly decreased hypoxia-triggered calcium accumulation in HPASMCs (Figure 2D). The expression of the bone markers SOX9, MSX2, and BMP2 was dramatically increased in hypoxia-treated cultures compared to normoxic cultures; in contrast, the expression of the VSMC marker SM22α was decreased gradually during the osteogenic differentiation of VSMCs in hypoxic conditions, and siCDR1as reversed the above effects (Figure 2E). These results suggest that CDR1as is an essential and sufficient player in regulating hypoxia-induced HPASMC calcification.
Figure 2.
siRNA Targeting CDR1as (siCDR1as) Prevents HPASMC Calcification in Hyp
(A) Representative images of ARS staining in Nor and NC, Hyp and NC, and Hyp and siCDR1as groups (B) Immunofluorescent staining of Runx2 in fixed HPASMCs. Scale bar, 50 μm. (C) ALP activity was measured in HPASMCs after siCDR1as. (D) Quantification of calcium content in HPASMCs. The results shown were normalized to the total protein amount. (E) HPASMCs were exposed to Hyp, and the expression of SOX9, MSX2, SM22α, and BMP2 was evaluated by western blot with β-actin as a standard. ∗p < 0.05, ∗∗p < 0.01 versus Nor + NC; #p < 0.05, ##p < 0.01 versus Hyp + NC. All tests were performed at least three times, and the values are presented as the mean ± SEM.
CDR1as Acts as an Efficient MicroRNA (miRNA) Sponge for miR-7-5p
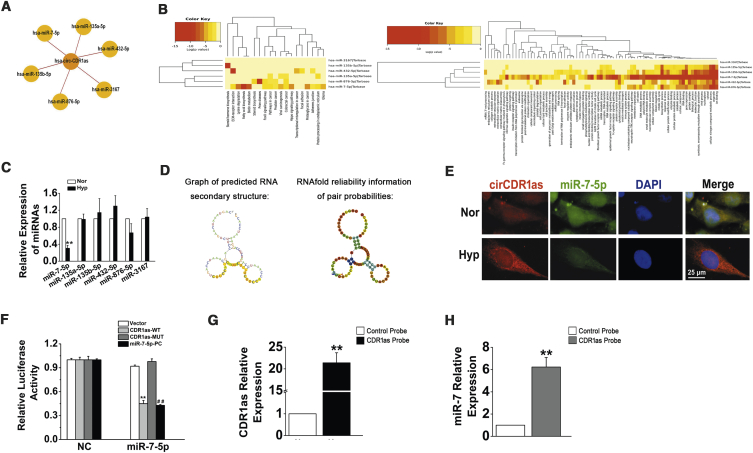
We further investigated the downstream targets of CDR1as in HPASMCs. Several miRNAs (miR-7-5p, miR-135a-5p, miR-135b-5p, miR-432-5p, miR-876-5p, and miR-3167) that potentially interact with CDR1as were chosen based on the miRanda, PITA, and RNAhybrid databases (Figure 3A). Then, we evaluated miR-7-5p, miR-135a-5p, miR-135b-5p, miR-432-5p, miR-876-5p, and miR-3167 by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis with DIANA TOOLS (Figure 3B).22 qRT-PCR assays revealed that only miR-7-5p was negatively modulated by hypoxia in HPASMCs and was negatively correlated with CDR1as (Figure 3C). Subsequently, the potential binding sites between CDR1as and miR-7-5p were predicted by RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/detection.html), and the representative putative target sites of miR-7-5p are shown in Figure 3D.23 Previous studies have shown that CDR1as adsorbs miR-7-5p in hepatocellular carcinoma and colorectal and lung cancer.24, 25, 26 To validate whether miR-7-5p bound to CDR1as in HPASMCs, we used a FISH assay, and colocalization between CDR1as and miR-7-5p was observed in the cytoplasm of HPASMCs (Figure 3E). Dual-luciferase reporter assays were performed to verify whether there was an interaction between CDR1as and miR-7-5p. The firefly luciferase reporter activity was significantly decreased in the hsa-miR-7-5p mimics and the CDR1as wild-type (WT) transfection group, whereas the CDR1as mutant (MUT) group showed no notable changes in luciferase reporter activity (Figure 3F). Moreover, an RNA antisense purification (RAP) assay revealed that miR-7-5p was significantly enriched in RNAs retrieved from the CDR1as complex (Figures 3G and 3H). These results indicate that CDR1as serves as a sponge for miR-7-5p in HPASMCs.
Figure 3.
CDR1as Functions as a ceRNA to Sequester miR-7-5p
(A) A schematic drawing showing the putative miRNAs associated with CDR1as. (B) GO and KEGG analysis of miR-7-5p, miR-135a-5p, miR-135b-5p, miR-432-5p, miR-876-5p, and miR-3167. (C) HPASMCs were subjected to Hyp, and qPCR was used to detect the expression of miR-7-5p, miR-135a-5p, miR-135b-5p, miR-432-5p, miR-876-5p, and miR-3167. ∗∗p < 0.01 compared with Nor. (D) The secondary structures of CDR1as and miR-7-5p. (E) The colocalization of CDR1as and miR-7-5p in the cytoplasm of HPASMCs was observed by FISH assay. Scale bar, 25 μm. (F) Luciferase reporter assay for the luciferase activity of HPASMCs cotransfected with scrambled miRNA (NC) and vector (VE)/CDR1as-wild-type (WT)/CDR1as-mutant (MUT)/complementary sequences of miR-7-5p (miR-7-5p-positive control [PC]) or miR-7-5p mimics with VE/CDR1as-WT/CDR1as-MUT complementary sequences of miR-7-5p (miR-7-5p-PC). ∗∗p < 0.01 compared with NC + CDR1as-WT; ##p < 0.01 compared with NC + miR-7-5p-PC. (G) The enrichment efficiency of the CDR1as-specific probe was detected by qRT-PCR. ∗∗p < 0.01. (H) The fold enrichment of miR-7-5p on the biotinylated CDR1as-specific probe. ∗∗p < 0.01. All tests were performed at least three times, and the values are presented as the mean ± SEM.
miR-7 Overexpression Aggravates the Osteogenic Transition of HPASMCs
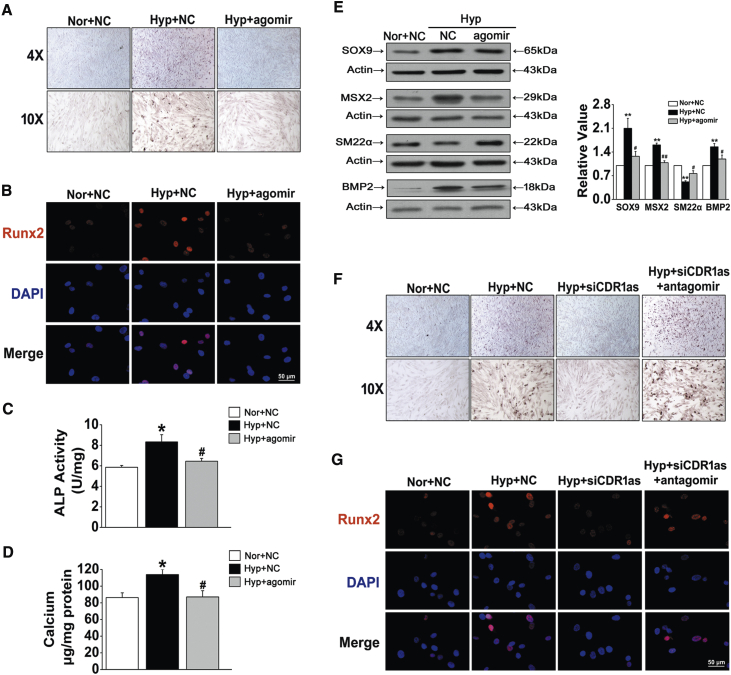
Overexpression or silencing of miR-7-5p by using the double-stranded agomir (miRNA mimics) and the antagomir (antisense oligonucleotides) complementary to the mature miRNAs was performed to further understand its role in the progression of HPASMC osteogenic transition. ARS staining showed that, compared with the control, transfection with miR-7-5p agomir significantly inhibited the mineralization of the cell matrix under hypoxic conditions (Figure 4A). Similar trends in Runx2 immunofluorescent staining and ALP activity were found in HPASMCs after miR-7-5p agomir treatment (Figures 4B and 4C). Moreover, the hypoxia-induced increase in total calcium content in HPASMCs was blocked by miR-7-5p recovery (Figure 4D). Western blot analysis confirmed that the expression of osteogenic markers, including SOX9, MSX2, and BMP2, was inhibited by overexpression of miR-7-5p, whereas miR-7-5p overexpression increased the protein level of SM22α (Figure 4E). To further confirm the effects of the CDR1as/miR-7 axis in mediating HPASMC calcification promoted by hypoxia, we cotransfected siCDR1as and miR-7-5p inhibitor (antagomir) into HPASMCs. Compared with that in cells transfection with siCDR1as alone, calcification was increased in cells cotransfected with siCDR1as and miR-7-5p antagomir (Figure 4F). Likewise, silencing CDR1as decreased the hypoxia-induced expression of Runx2, as revealed by immunofluorescence analysis, and this effect was antagonized by miR-7 inhibition (Figure 4G). Taken together, these results demonstrate that CDR1as suppressed HPASMC osteogenic progression by sponging miR-7-5p.
Figure 4.
miR-7-5p Regulates HPASMC Osteogenic Transition
(A) Representative images from sections stained with ARS. (B) Runx2 expression was detected by immunofluorescence staining. (C) ALP activity in the cells transfected with NC or miR-7-5p mimics was measured. (D) The calcium content in calcified HPASMCs was assayed. (E) Western blot analysis of osteogenic markers in HPASMCs exposed to Hyp with or without agomir. (F) Calcium deposition was determined after cotransfecting cells with siCDR1as and antagomir of miR-7-5p. (G) The expression level of Runx2 in HPASMCs of the indicated groups was determined by immunofluorescence assay. ∗p < 0.05, ∗∗p < 0.01 versus Nor + NC; #p < 0.05, ##p < 0.01 versus Hyp + NC. All tests were performed at least three times, and the values are presented as the mean ± SEM.
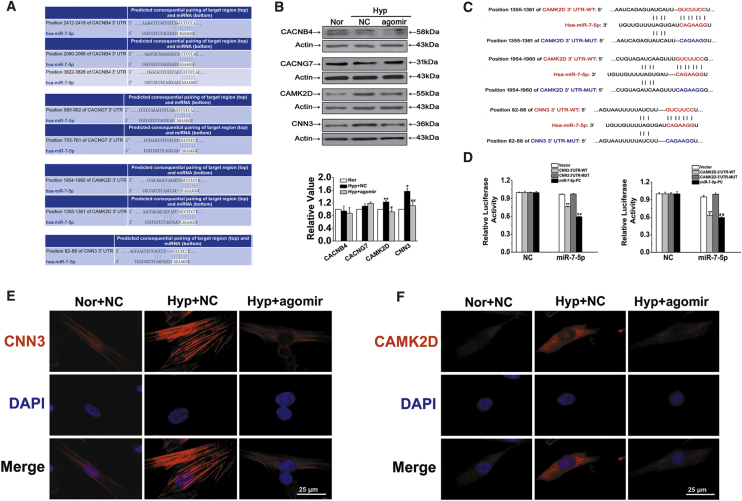
miR-7-5p Targets CAMK2D and CNN3
To assess the effects of CDR1as/miR-7-5p on HPASMC calcification, we selected 4 targets related to calcification from hundreds of miR-7-5p targets predicted by TargetScan, including calcium channel β (4) subunit gene (CACNB4), calcium voltage-gated channel auxiliary subunit gamma 7 (CACNG7), calcium/calmodulin-dependent kinase II-delta (CAMK2D), and calponin 3 (CNN3) (Figure 5A). First, the protein levels of the target genes were detected using western blotting, and the results showed that CAMK2D and CNN3 expression was most significantly affected by hypoxia and miR-7-5p (Figure 5B). To verify whether CAMK2D and CNN3 mRNA is targeted by miR-7-5p, a luciferase reporter gene assay was performed using 3′ UTR sequence fragments, containing the CAMK2D or CNN3 predicted target region (WT) of miR-7-5p and a mutated target region (MUT) inserted downstream of a luciferase reporter (Figure 5C). Cell cotransfection with hsa-miR-7-5p mimics and CAMK2D or CNN3 WT showed a significant reduction in firefly luciferase reporter activity compared with that in the negative control (NC) group, and the CAMK2D and CNN3 MUT groups showed no notable changes (Figure 5D). Moreover, immunofluorescence assays proved that the expression of both CNN3 and CAMK2D was increased in hypoxic conditions and downregulated by miR-7-5p (Figures 5E and 5F). This finding confirms the binding interaction between miR-7-5p and CAMK2D and CNN3.
Figure 5.
Verification of CAMK2D (CA) and CNN3 (CN) but Not CACNB4 and CACNG7 as Cognate Targets of miR-7-5p
(A) TargetScan-predicted binding sites of targets with miR-7-5p. (B) Effects of miR-7-5p on the expression levels of CACNB4, CACNG7, CAMK2D, and CNN3. ∗p < 0.05, ∗∗p < 0.01 versus Nor + NC; #p < 0.05, ##p < 0.01 versus Hyp + NC. (C) Diagrammatic sketch of the binding sites for the WT and MUT binding sites of CAMK2D and CNN3 associated with miR-7-5p. (D) Luciferase reporter assay for the luciferase activity of HPASMCs cotransfected with scrambled miRNA (NC) and VE/CAMK2D and CNN3 3′ UTR-WT/CAMK2D and CNN3 3′ UTR-MUT/complementary sequences of miR-7-5p (miR-7-5p-PC) or miR-7-5p mimics with VE/CAMK2D and CNN3 3′ UTR-WT/CAMK2D and CNN3 3′ UTR-MUT/complementary sequences of miR-7-5p (miR-7-5p-PC). ∗∗p < 0.01 compared with NC + WT; ##p < 0.01 compared with NC + miR-7-5p-PC. (E and F) HPASMCs were labeled with antibodies against CNN3 (E) and CAMK2D (F), and nuclei were stained with DAPI and merged to represent an overlay figure. Scale bars, 25 μm. All tests were performed at least three times, and the values are presented as the mean ± SEM.
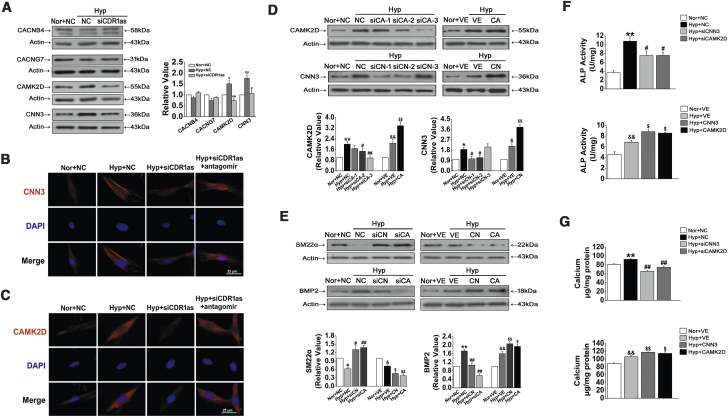
CAMK2D and CNN3 Are Overexpressed in Hypoxia and Regulated by CDR1as/miR-7
To investigate the effects of CDR1as on miR-7-5p-mediated suppression of calcification-associated genes, cells were infected with siCDR1as. Compared with the hypoxia and scramble NC groups, the expression of CAMK2D and CNN3 was reduced by siCDR1as; however, we did not detect significant changes in the expression levels of CACNB4 and CACNG7 (Figure 6A). Further analysis showed that transfection with siCDR1as inhibited the distribution of CNN3 and CAMK2D in the cytoplasm compared with that under hypoxic conditions, and this effect was reversed after cotransfecting cells with siCDR1as and miR-7-5p inhibitor (Figures 6B and 6C). Consequently, these data indicate that there are interactions among CDR1as, miR-7-5p, and CAMK2D/CNN3.
Figure 6.
CNN3 and CAMK2D Are Targets of CDR1as, which Are Required for Osteogenic Transition
(A) The CNN3 and CAMK2D protein levels in the Hyp with the siCDR1as group were evidently downregulated compared with those in the Hyp with NC group. (B and C) siCDR1as reduced the level of CNN3 (B) and CAMK2D (C) in HPASMC cells induced by Hyp, and this effect was reversed by cotransfecting cells with antagomir. Scale bars, 25 μm. (D) Verification of the effect of knockdown and overexpression on CNN3 and CAMK2D. We used siCAMK2D-3 and siCNN3-1 to knock down CAMK2D and CNN3 in the subsequent experiments. (E) Effects of CNN3 and CAMK2D siRNA and overexpression plasmid transfection on the expression levels of SM22α and BMP2. (F) ALP activity was assayed in siNC-, siCNN3-, siCAMK2D-, VE-, CNN3-, and CAMK2D-overexpressing plasmid-transfected HPASMCs. (G) Quantification of the calcium content in Hyp-induced HPASMCs with or without silencing or overexpression of CNN3 and CAMK2D. ∗p < 0.05, ∗∗p < 0.01 compared with Nor + NC; #p < 0.05, ##p < 0.01 compared with Hyp + NC; &p < 0.05, &&p < 0.01 compared with Nor + VE; $p < 0.05, $$p < 0.01 compared with Hyp + VE. All tests were performed at least three times, and the values are presented as the mean ± SEM.
CNN3 and CAMK2D Promote Calcification under Hypoxic Conditions
To verify whether CNN3 and CAMK2D participate in regulating the osteogenic differentiation of HPASMCs, we transfected cells with CNN3 and CAMK2D siRNAs and overexpression plasmids. The transfection specificity and efficiency are shown in Figure 6D, which shows that CNN3 and CAMK2D expression was significantly reduced by siRNAs and enhanced by overexpression plasmids. As expected, siCNN3 and siCAMK2D attenuated the hypoxia-induced upregulation of BMP2 expression and downregulation of SM22α expression; in contrast, the opposite effect was observed after CNN3 and CAMK2D overexpression (Figure 6E). Accordingly, transfection with the siRNAs targeting CNN3 and CAMK2D decreased the levels of ALP activity, whereas CNN3 and CAMK2D overexpression increased ALP activity compared with that under hypoxic conditions (Figure 6F). Furthermore, CNN3 and CAMK2D knockdown and overexpression reduced and increased the hypoxia-induced calcium accumulation, respectively (Figure 6G). These results suggest that CNN3 and CAMK2D might be critical drivers in HPASMC osteogenic differentiation induced by hypoxia.
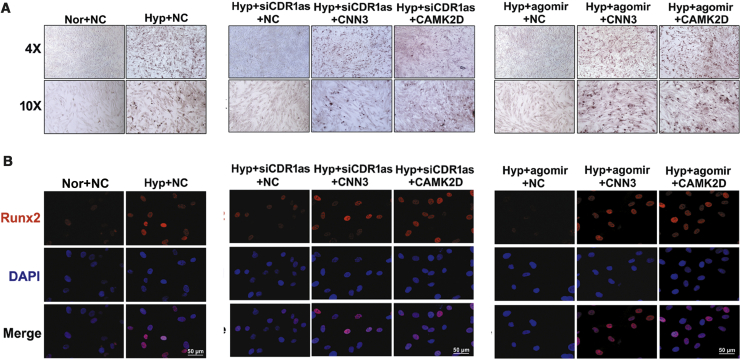
The CDR1as/miR-7/CNN3/CAMK2D Axis Plays a Role in Hypoxia-Induced HPASMC Calcification
Finally, to clarify the link between CDR1as, miR-7-5p, CNN3, and CAMK2D and calcification, we performed rescue experiments in HPASMCs after exposure to hypoxia. As shown in Figure 7A, hypoxia-triggered HPASMC mineralization was decreased in the siCDR1as and miR-7-5p agomir groups, whereas CNN3 and CAMK2D overexpression had the opposite effect. Similarly, we also found that the expression of Runx2 was increased in cells transfected with CNN3- and CAMK2D-overexpressing plasmids compared with those transfected with siCDR1as or miR-7-5p agomir alone (Figure 7B). Together, these data suggest that CDR1as mediates HPASMC mineralization via the miR-7/CNN3 and CAMK2D axis in hypoxia.
Figure 7.
CDR1as/miR-7-5p Aggravates HPASMC Calcification via CNN3 and CAMK2D
(A) HPASMCs were incubated with CNN3- or CAMK2D-overexpressing plasmids with or without siCDR1as or miR-7-5p agomir, and calcium mineral deposits were assessed. (B) Transfection of siCDR1as or miR-7-5p agomir decreased the Runx2 level in Hyp HPASMCs, whereas the opposite phenomenon was observed after CNN3 or CAMK2D overexpression. Scale bars, 50 μm. All tests were performed at least three times.
Discussion
Emerging evidence suggests a link between chronic respiratory diseases, including asthma, chronic obstructive pulmonary disease, and obstructive sleep apnea, and accelerated calcification.13,27,28 Moreover, the combination of hypoxia and vascular calcification in the established PH rat models was identified in our previous study.14 In this research, we confirmed that in HPASMCs, hypoxia treatment promoted the expression of bone-related proteins (Runx2, MSX2, BMP2, and SOX9) during calcification. More importantly, in the current study, we sought to characterize circRNAs involved in the regulation of HPASMC calcification and homeostasis. We identified CDR1as as an activator of HPASMC calcification, whose expression was elicited by hypoxic conditions. We also confirmed that CDR1as can function as a molecular sponge of miR-7-5p, which weakens the inhibitory effect of the miRNA on its downstream target genes CAMK2D and CNN3 to increase HPASMC mineralization in hypoxia.
Increasing evidence suggests that circRNAs are abundantly expressed and evolutionarily conserved across eukaryotic life and exert functions in the occurrence and progression of various multiple diseases.29,30 For example, circ-Foxo3 was shown to bind to the mouse double minute 2 homolog and increase p53 stability and as a result of this interaction, induce cell apoptosis.31 Shen et al.32 reported that circ-SERPINE2 overexpression can alleviate human chondrocyte apoptosis and promote anabolism of the extracellular matrix to regulate the pathological process of osteoarthritis. In inflammatory phenotypic switching of VSMCs, circ-Sirt1 is involved in inhibiting nuclear factor κB (NF-κB) activation by directly interacting with SIRT1 and promoting its expression.33 In this study, we demonstrated that CDR1as is an important circRNA that is upregulated in hypoxic HPASMCs compared to normoxic HPASMCs and showed that high expression of CDR1as was positively correlated with cell calcification in PH. We believe this is an important study to examine the role of circRNA in hypoxia-induced HPASMC calcification in PH.
Although a large number of circRNAs have been identified as ceRNAs that regulate miRNAs, recent studies have shown that circRNAs can function as protein scaffolds, regulators of gene transcription, RNA-binding proteins (RBPs), and templates for protein translation in many different organisms, tissues, and cellular functions.34 For example, Legnini et al.35 revealed that circ-ZNF609 contains an open reading frame and is translated into a protein in a splicing-dependent/cap-independent manner. In addition, intronic circRNA generated from the AGO2 gene (circAGO2) physically interacts with RBP human antigen R (HuR) by facilitating its competitive enrichment in the 3′ UTR of target genes, resulting in a reduction in AGO2 binding and promotion of tumorigenesis.36 In addition, epithelial splicing regulatory protein 1 (ESRP1), a splicing factor for epithelial mesenchymal transformation progression, was identified to target the flanking introns of circUHRF1 and accelerate its circularization in oral squamous cell carcinoma.37 However, compared to that of these circRNAs, the study of CDR1as is still in its preliminary stage. According to the CPC2.0 prediction, CDR1as has a complete open reading frame, and the probability to encode the peptide is 58.3%; the length of the probable encoded peptide is 152 amino acids. Further work will be required to understand whether CDR1as can translate and interact with other functional RNAs or RBPs in response to specific stimuli or signals.
The classic regulatory pathway of CDR1as, which acts as an miRNA sponge, has been confirmed in many diseases. In obese patients with diabetes, the CDR1as/miR-7 pathway was found to regulate insulin content and secretion.38 In esophageal squamous cell carcinoma, CDR1as accelerates cancer progression by acting as a miR-876-5p sponge to enhance melanoma antigen family A expression.39 In ovarian and bladder cancer progression, CDR1as, acting as the sponge of miR-135b-5p or miR-1270, functions as a tumor regulator.40,41 Here, we observed that the expression of miR-7-5p was downregulated in hypoxic conditions compared with the control conditions, but miR-135a-5p, miR-135b-5p, miR-432-5p, miR-876-5p, or miR-3167 were unaffected. Furthermore, hypoxia-induced HPASMC mineralization was eliminated by miR-7-5p agomir treatment and promoted by siCDR1as cotransfection with the miR-7-5p antagomir, suggesting that the regulatory role of CDR1as in HPASMC calcification is associated with the inhibition of miR-7-5p function.
Although many hsa-miR-7-5p target genes were predicted by bioinformatics analysis, only some of them have been experimentally demonstrated to be related to calcium signals; these include CACNB4, CACNG7, CAMK2D, and CNN3. Our data showed that CAMK2D and CNN3 are increased in response to hypoxia and are modulated by miR-7-5p. Then, a luciferase reporter assay validated CAMK2D and CNN3 as direct targets of miR-7-5p. Recent publications have indicated the role of CAMK2D in heart failure and diabetes.42,43 Upregulation of CAMK2D kinases also led to activation of the NF-κB signaling pathway, which protects neurons from ischemic damage.44 As a muscle regulator, CNN3 has been shown to interact with calmodulin and participate in neuronal remodeling and trophoblast cell fusion during embryonic development and morphogenesis.45, 46, 47 In this study, ARS staining and Runx2 immunofluorescence assays indicated that knockdown of CDR1as and miR-7-5p agomir treatment inhibits hypoxia-induced HPASMC mineralization and that these effects are reversed by CAMK2D and CNN3 overexpression, respectively. As one of the highlights of this study, our findings suggest that CAMK2D and CNN3 play critical roles in CDR1as/miR-7-5p-induced HPASMC calcification in hypoxic conditions.
In this study, we confirmed that CDR1as and its target genes CAMK2D and CNN3 were overexpressed in hypoxic HPASMCs. CDR1as promoted HPASMC mineralization by upregulating CAMK2D and CNN3 expression by sponging miR-7-5p. Given the importance of HPASMC calcification in the progression of hypoxic PH, our results indicate the importance of the CDR1as/miR-7-5p/CAMK2D/CNN3 axis as a potential therapeutic target in human PH.
Materials and Methods
Materials
The antibodies and reagents used were as follows: Runx2 (ab23981; Abcam, MA, USA); MSX2 (M-70; sc-15396; Santa Cruz Biotechnology, TX, USA); BMP2 (ab14933; Abcam, MA, USA), SOX9 (ab26414; Abcam, MA, USA); SM22α (ab14106; Abcam, MA, USA), CACNB4 (17770-1-AP; Proteintech, IL, USA); CACNG7 (17862-1-AP; Proteintech, IL, USA); CAMK2D (20667-1-AP; Proteintech, IL, USA, and ab52476; Abcam, MA, USA); CNN3 (11509-1-AP; Proteintech, IL, USA); ARS (GMS80046; GENMED, Shanghai, PR China); and the ALP Assay Kit (P0321; Beyotime, Shanghai, PR China).
Cell Culture
HPASMCs and smooth muscle cell medium were obtained from Sciencell (CA, USA; catalog #3110 and #1101). The HPASMCs were plated in smooth muscle cell medium, which contained 15% fetal bovine serum and 1% penicillin and streptomycin and were cultured at 37°C with 5% CO2 in humidified conditions. Cells in hypoxic culture were incubated in a Tri-Gas Incubator (Heal Force, Shanghai, PR China) with a gas mixture containing 92% N2-5% CO2-3% O2.
RNA-FISH
RNA-FISH was carried out according to the manufacturer’s instructions using fluorescence-conjugated probes and a FISH Kit supplied by Genepharma (Shanghai, PR China). Briefly, cells were incubated with hybridization solution overnight at 37°C in the dark containing circ-CDR1as and miR-7-5p probes, and 18S RNA and U6 probes were used as internal references. The nucleotide sequences of the probes were as follows: circ-CDR1as, 5′-ACAGGTGCCATCGGAAACCCTGGATATTGCAGACACTG-3′; miR-7-5p, 5′-AACAACAAAATCACTAGTCTTCCA-3′; 18S RNA, 5′-CTTCCTTGGATGTGGTAGCCGTTTC-3′; U6, 5′-TTTGCGTGTCATCCTTGCG-3′.
Finally the RNA-FISH sections were incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. Images were recorded with a living cell workstation (AF6000; Leica, Germany).
Immunocytochemistry
HPASMCs were cultured on coverslips, and after treatment, cells were washed with PBS and fixed with 4% paraformaldehyde at room temperature. Then, the cells were permeabilized with 0.5% Triton X-100 and blocked with 5% normal bovine serum at 37°C for 30 min. HPASMCs were incubated with Runx2 antibody (1:50) overnight. After washing with PBS, the cells were incubated with Cy3-conjugated secondary antibody (1:100) followed by DAPI away from light. Then, the coverslips were mounted and examined with a living cell workstation (AF6000; Leica, Germany).
siRNA and Plasmid Construction
siRNAs were designed and synthesized by Genepharma (Shanghai, PR China). Negative control siRNA (siNC) was used as a NC. The sequences were as follows, si-hsa-circ-CDR1as: sense, 5′-GCAAUAUCCAGGGUUUCCGTT-3′, antisense, 5′-CGGAAACCCUGGAUAUUGCTT-3′; si-NC: sense, 5′-UUCUUCGAACGUGUCACGUTT-3′, antisense, 5′-ACGUGACACGUUCGGAGAATT-3′; si-CAMK2D (gene ID: 817): sense, 5′-GCACCCAUGGAUCUGUCAATT-3′, antisense, 5′-UUGACAGAUCCAUGGGUGCTT-3′; si-CNN3 (gene ID: 1266): sense, 5′-GCAGAUGGGCACUAAUAAATT-3′, antisense, 5′-UUUAUUAGUGCCCAUCUGCTT-3′; hsa-miR-7-5p agomir (miRNA mimics): sense, 5′-UGGAAGACUAGUGAUUUUGUUGUU-3′, antisense, 5′-CAACAAAAUCACUAGUCUUCCAUU-3′, hsa-miR-7-5p antagomir (antisense oligonucleotides): 5′-AACAACAAAAUCACUAGUCUUCCA-3′. The CAMK2D and CNN3 plasmids were constructed with the GV230 vector. The vector alone was used as a NC. Transfection was implemented according to the manufacturer’s instructions for the Lipofectamine 2000 Reagent (Life Technologies, Carlsbad, CA). Cells were cultured to 50%–70% confluence, and 2–3 μg siRNA or plasmids and 10 μL transfection reagent were diluted in serum-free Opti-MEM-1 medium. Next, we incubated the mixture (siRNA/transfection reagent) for 20 min and then added it directly to the cells. The transfection reagents were removed after 4–6 h, and the cells were incubated for 24 h and then used as required.
Western Blot Analysis
Proteins were solubilized and extracted with cold lysis buffer containing protease inhibitor after sonication and centrifugation at 16,099 × g for 15 min. The protein samples (20–40 μg) were fractionated by SDS-PAGE (8%–12% polyacrylamide gels) and transferred onto nitrocellulose membranes after incubation in a blocking buffer (20 mM Tris, pH 7.6, 150 mM NaCl, and 0.1% Tween 20) containing 5% nonfat dry milk powder. Anti-MSX2 (1:500), anti-BMP2 (1:1,000), anti-SOX9 (1:1,000), anti-SM22α (1:1,000), anti-CACNB4 (1:500), anti-CACNG7 (1:500), anti-CAMK2D (1:500), anti-CNN3 (1:500), and anti-β-actin (1:8,000) were used as primary antibodies and were incubated overnight at 4°C, followed by incubation with appropriate horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence reagents.
qRT-PCR
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from 2 μg of RNA using the SuperScript First-Strand cDNA Synthesis Kit (Invitrogen). cDNA samples were amplified in a DNA thermal cycler (Bio-Rad, CA, USA). Conventional real-time PCR with SYBR Green (Takara, Tokyo, Japan) was carried out with total RNA samples following the manufacturer’s instructions. A Roche LightCycler 480 Instrument II was used for real-time quantitative fluorescence analysis with a two-step method. 18S and U6 were used as the internal controls. Primer sequences were as follows: hsa-miR-7-5p: forward (F), 5′-ACGTTGGAAGACTAGTGATTT-3′, and reverse (R), 5′-TATGGTTGTTCTGCTCTCTGTCTC-3′; hsa-miR-135a-5p: F, 5′-TGCTGCTCTATGGCTTTTTATTC-3′, and R, 5′-TATGGTTGTTCACGACTCCTTCAC-3′; hsa-miR-135b-5p: F, 5′-GCTGCTCTATGGCTTTTCATTC-3′, and R, 5′-TATGGTTGTTCACGACTCCTTCAC-3′; hsa-miR-432-5p: F, 5′-CTGCCGTTCTTGGAGTAGGTCA-3′, and R, 5′-CAGAGCAGGGTCCGAGGTA-3′; hsa-miR-876-5p: F, 5′-GATGCTCTTGGATTTCTTTGTGA-3′, and R, 5′-TATGGTTGTTCACGACTCCTTCAC-3′; hsa-miR-3167: F, 5′-CCGTACCCAGGATTTCAGAAAT-3′, and R, 5′-TATGGTTGTTCACGACTCCTTCAC-3′.
ARS Staining
ARS staining was performed as described previously.14 In brief, HPASMCs were cultured with different treatments in pro-calcifying media containing 2.5 mM inorganic phosphate for the induction of calcification. After washing with PBS, the cells were fixed with 95% ethanol for 10 min and stained with ARS dye (pH 4.2) at 37°C for 30 min. Excessive dye was removed by several washes in deionized water.
ALP Activity
ALP activity in HPASMCs was assayed following the manufacturer’s instructions (P0321; Beyotime, Shanghai, PR China). Briefly, ALP yellow liquid substrate was combined with protein samples and incubated at 37°C for 30 min. Finally, after adding 100 μL stop solution, the cleavage of 10 mM p-nitrophenyl phosphate (pNPP) at 405 nm was measured. Values were normalized to the total protein level, and the maximum slope of the kinetic curves was used for calculation.
Quantification of Calcium Deposition
Cells were washed three times with PBS and decalcified with 0.6 M HCl. The calcium content of the HCl supernatants was determined using the QuantiChrom calcium assay kit (QuantiChrom BioAssay Systems, Hayward, CA, USA), according to the manufacturer’s recommendations. The total calcium content was measured with a spectrophotometer set at 612 nm. The protein content of the samples was measured with a bicinchoninic acid (BCA) protein assay kit, and the calcium content of the cells was normalized to the protein content and expressed as micrograms per milligrams protein.
Dual-Luciferase Reporter Assays
circRNA-CDR1as, CAMK2D, and CNN3 mRNAs containing miRNA binding sites were cloned into plasmids expressing luciferase (Genepharma, Shanghai, PR China). Cells were cotransfected with 120 pmol miRNA mimics and 120 ng pmirGLO vector carrying the desired fragment with Lipofectamine 2000 for 48 h. Luciferase activities were measured by the dual-luciferase reporter assay system (Promega), according to its instructions.
RAP
The specific biotinylated probes that hybridized to CDR1as and the control were synthesized by Genepharma (Shanghai, PR China). The sequences were as follows: CDR1as probe, biotin-5′-CAGGUGCCAUCGGAAACCCUGGAUAUUGCAGACACU-3′, and control probe, biotin-5′-UUGUACUACACAAAAGUACUG-3′. The experiment was performed according to the protocol of the RAP kit (Bes5103-1; BersinBio, Guangzhou, PR China). In brief, 4 × 107 cells were washed and homogenized with a 0.4-mm syringe. After removing DNA, the CDR1as-specific biotinylated probes were added to the circRNA-RAP system and incubated for 2 h at room temperature before adding 200 μL streptavidin-coated magnetic beads. After removing nonspecifically bound RNAs, qRT-PCR was used to analyze miRNAs specifically interacting with CDR1as.
Statistical Analysis
All values are presented as the mean ± SEM. Statistical analysis was performed using Student’s t test or one-way ANOVA, followed by Tukey’s test where appropriate. A value of p <0.05 was considered statistically significant.
Author Contributions
D.Z. designed the research. C.M. and R.G. analyzed the data. X.W., S.H., J.B., W.X., Y.J., and F.L. performed the research. C.M., L.Z., and X.Z. wrote the paper. J.Z., Q.L., and L.Q. contributed new reagents or analytic tools. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the following grants: National Natural Science Foundation of China (contract grant numbers 31820103007, 31771276, and 31471095 to D.Z. and 81873412 to C.M.); Excellent Young Talents Fund Program of Higher Education Institutions of Heilongjiang Province (contract grant number UNPYSCT-2017047 to C.M.); Returned Overseas Students Funding of Heilongjiang Province (contract grant number 2017QD0040 to C.M.); Postdoctoral Research Funding of Heilongjiang Province (contract grant number LBH-Q17098 to C.M.); and National Natural Science Foundation of Heilongjiang Province (contract grant numbers YQ2019H006 to C.M. and LH2020H028 to L.Z.).
References
- 1.Bellaye P.S., Yanagihara T., Granton E., Sato S., Shimbori C., Upagupta C., Imani J., Hambly N., Ask K., Gauldie J. Macitentan reduces progression of TGF-β1-induced pulmonary fibrosis and pulmonary hypertension. Eur. Respir. J. 2018;52:1701857. doi: 10.1183/13993003.01857-2017. [DOI] [PubMed] [Google Scholar]
- 2.Budas G.R., Boehm M., Kojonazarov B., Viswanathan G., Tian X., Veeroju S., Novoyatleva T., Grimminger F., Hinojosa-Kirschenbaum F., Ghofrani H.A. ASK1 Inhibition Halts Disease Progression in Preclinical Models of Pulmonary Arterial Hypertension. Am. J. Respir. Crit. Care Med. 2018;197:373–385. doi: 10.1164/rccm.201703-0502OC. [DOI] [PubMed] [Google Scholar]
- 3.Hansmann G. Pulmonary Hypertension in Infants, Children, and Young Adults. J. Am. Coll. Cardiol. 2017;69:2551–2569. doi: 10.1016/j.jacc.2017.03.575. [DOI] [PubMed] [Google Scholar]
- 4.Fayyaz A.U., Edwards W.D., Maleszewski J.J., Konik E.A., DuBrock H.M., Borlaug B.A., Frantz R.P., Jenkins S.M., Redfield M.M. Global Pulmonary Vascular Remodeling in Pulmonary Hypertension Associated With Heart Failure and Preserved or Reduced Ejection Fraction. Circulation. 2018;137:1796–1810. doi: 10.1161/CIRCULATIONAHA.117.031608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coghlan J.G., Wolf M., Distler O., Denton C.P., Doelberg M., Harutyunova S., Marra A.M., Benjamin N., Fischer C., Grünig E. Incidence of pulmonary hypertension and determining factors in patients with systemic sclerosis. Eur. Respir. J. 2018;51:1701197. doi: 10.1183/13993003.01197-2017. [DOI] [PubMed] [Google Scholar]
- 6.Weiss A., Neubauer M.C., Yerabolu D., Kojonazarov B., Schlueter B.C., Neubert L., Jonigk D., Baal N., Ruppert C., Dorfmuller P. Targeting cyclin-dependent kinases for the treatment of pulmonary arterial hypertension. Nat. Commun. 2019;10:2204. doi: 10.1038/s41467-019-10135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C.J., Hsu H.C., Ho W.J., Chang G.J., Pang J.S., Chen W.J., Huang C.C., Lai Y.J. Cathepsin S promotes the development of pulmonary arterial hypertension. Am. J. Physiol. Lung Cell. Mol. Physiol. 2019;317:L1–L13. doi: 10.1152/ajplung.00530.2018. [DOI] [PubMed] [Google Scholar]
- 8.Byon C.H., Javed A., Dai Q., Kappes J.C., Clemens T.L., Darley-Usmar V.M., McDonald J.M., Chen Y. Oxidative stress induces vascular calcification through modulation of the osteogenic transcription factor Runx2 by AKT signaling. J. Biol. Chem. 2008;283:15319–15327. doi: 10.1074/jbc.M800021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demer L.L., Tintut Y. Vascular calcification: pathobiology of a multifaceted disease. Circulation. 2008;117:2938–2948. doi: 10.1161/CIRCULATIONAHA.107.743161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanzer P., Boehm M., Sorribas V., Thiriet M., Janzen J., Zeller T., St Hilaire C., Shanahan C. Medial vascular calcification revisited: review and perspectives. Eur. Heart J. 2014;35:1515–1525. doi: 10.1093/eurheartj/ehu163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z., Huang Y., Du J., Liu A.D., Tang C., Qi Y., Jin H. Endogenous Sulfur Dioxide Inhibits Vascular Calcification in Association with the TGF-β/Smad Signaling Pathway. Int. J. Mol. Sci. 2016;17:266. doi: 10.3390/ijms17030266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C., Xu W., An J., Liang M., Li Y., Zhang F., Tong Q., Huang K. Poly(ADP-ribose) polymerase 1 accelerates vascular calcification by upregulating Runx2. Nat. Commun. 2019;10:1203. doi: 10.1038/s41467-019-09174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balogh E., Tóth A., Méhes G., Trencsényi G., Paragh G., Jeney V. Hypoxia Triggers Osteochondrogenic Differentiation of Vascular Smooth Muscle Cells in an HIF-1 (Hypoxia-Inducible Factor 1)-Dependent and Reactive Oxygen Species-Dependent Manner. Arterioscler. Thromb. Vasc. Biol. 2019;39:1088–1099. doi: 10.1161/ATVBAHA.119.312509. [DOI] [PubMed] [Google Scholar]
- 14.Mao M., Zhang M., Ge A., Ge X., Gu R., Zhang C., Fu Y., Gao J., Wang X., Liu Y., Zhu D. Granzyme B deficiency promotes osteoblastic differentiation and calcification of vascular smooth muscle cells in hypoxic pulmonary hypertension. Cell Death Dis. 2018;9:221. doi: 10.1038/s41419-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 16.Yang F., Fang E., Mei H., Chen Y., Li H., Li D., Song H., Wang J., Hong M., Xiao W. Cis-Acting circ-CTNNB1 Promotes β-Catenin Signaling and Cancer Progression via DDX3-Mediated Transactivation of YY1. Cancer Res. 2019;79:557–571. doi: 10.1158/0008-5472.CAN-18-1559. [DOI] [PubMed] [Google Scholar]
- 17.Li Q., Wang Y., Wu S., Zhou Z., Ding X., Shi R., Thorne R.F., Zhang X.D., Hu W., Wu M. CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell Metab. 2019;30:157–173.e7. doi: 10.1016/j.cmet.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Huang G., Li S., Yang N., Zou Y., Zheng D., Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18. doi: 10.1016/j.canlet.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 20.Pan H., Li T., Jiang Y., Pan C., Ding Y., Huang Z., Yu H., Kong D. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J. Cell. Biochem. 2018;119:440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 21.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 22.Vlachos I.S., Hatzigeorgiou A.G. Functional Analysis of miRNAs Using the DIANA Tools Online Suite. Methods Mol. Biol. 2017;1517:25–50. doi: 10.1007/978-1-4939-6563-2_2. [DOI] [PubMed] [Google Scholar]
- 23.Chang T.H., Huang H.Y., Hsu J.B., Weng S.L., Horng J.T., Huang H.D. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu L., Gong X., Sun L., Zhou Q., Lu B., Zhu L. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS ONE. 2016;11:e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Zhang X., Yang D., Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. OncoTargets Ther. 2018;11:3979–3987. doi: 10.2147/OTT.S158316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang W., Ji M., He G., Yang L., Niu Z., Jian M., Wei Y., Ren L., Xu J. Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. OncoTargets Ther. 2017;10:2045–2056. doi: 10.2147/OTT.S131597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams M.C., Murchison J.T., Edwards L.D., Agustí A., Bakke P., Calverley P.M., Celli B., Coxson H.O., Crim C., Lomas D.A., Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) investigators Coronary artery calcification is increased in patients with COPD and associated with increased morbidity and mortality. Thorax. 2014;69:718–723. [Google Scholar]
- 28.Green F.H., Butt J.C., James A.L., Carroll N.G. Abnormalities of the bronchial arteries in asthma. Chest. 2006;130:1025–1033. doi: 10.1378/chest.130.4.1025. [DOI] [PubMed] [Google Scholar]
- 29.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 30.Cheng X., Zhang L., Zhang K., Zhang G., Hu Y., Sun X., Zhao C., Li H., Li Y.M., Zhao J. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. 2018;77:770–779. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen S., Wu Y., Chen J., Xie Z., Huang K., Wang G., Yang Y., Ni W., Chen Z., Shi P. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann. Rheum. Dis. 2019;78:826–836. doi: 10.1136/annrheumdis-2018-214786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong P., Yu Y., Wang L., Dou Y.Q., Zhang X.H., Cui Y., Wang H.Y., Yong Y.T., Liu Y.B., Hu H.J. circ-Sirt1 controls NF-κB activation via sequence-specific interaction and enhancement of SIRT1 expression by binding to miR-132/212 in vascular smooth muscle cells. Nucleic Acids Res. 2019;47:3580–3593. doi: 10.1093/nar/gkz141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H.M., Ma X.L., Li H.G. Intriguing circles: Conflicts and controversies in circular RNA research. Wiley Interdiscip. Rev. RNA. 2019;10:e1538. doi: 10.1002/wrna.1538. [DOI] [PubMed] [Google Scholar]
- 35.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao W., Cui Y., Liu L., Qi X., Liu J., Ma S., Hu X., Zhang Z., Wang Y., Li H. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2020;27:919–933. doi: 10.1038/s41418-019-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H., Guo S., Li W., Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sang M., Meng L., Sang Y., Liu S., Ding P., Ju Y., Liu F., Gu L., Lian Y., Li J. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 2018;426:37–46. doi: 10.1016/j.canlet.2018.03.049. [DOI] [PubMed] [Google Scholar]
- 40.Chen H., Mao M., Jiang J., Zhu D., Li P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. OncoTargets Ther. 2019;12:3869–3879. doi: 10.2147/OTT.S207938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan W., Zhou R., Wang J., Han J., Yang X., Yu H., Lu H., Zhang X., Li P., Tao J. Circular RNA Cdr1as sensitizes bladder cancer to cisplatin by upregulating APAF1 expression through miR-1270 inhibition. Mol. Oncol. 2019;13:1559–1576. doi: 10.1002/1878-0261.12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erickson J.R., Pereira L., Wang L., Han G., Ferguson A., Dao K., Copeland R.J., Despa F., Hart G.W., Ripplinger C.M., Bers D.M. Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature. 2013;502:372–376. doi: 10.1038/nature12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniels L.J., Wallace R.S., Nicholson O.M., Wilson G.A., McDonald F.J., Jones P.P., Baldi J.C., Lamberts R.R., Erickson J.R. Inhibition of calcium/calmodulin-dependent kinase II restores contraction and relaxation in isolated cardiac muscle from type 2 diabetic rats. Cardiovasc. Diabetol. 2018;17:89. doi: 10.1186/s12933-018-0732-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye J., Das S., Roy A., Wei W., Huang H., Lorenz-Guertin J.M., Xu Q., Jacob T.C., Wang B., Sun D., Wang Q.J. Ischemic Injury-Induced CaMKIIδ and CaMKIIγ Confer Neuroprotection Through the NF-κB Signaling Pathway. Mol. Neurobiol. 2019;56:2123–2136. doi: 10.1007/s12035-018-1198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang Z., Liang R., Zhao S., Wang R., Huang R., Li K. CNN3 is regulated by microRNA-1 during muscle development in pigs. Int. J. Biol. Sci. 2014;10:377–385. doi: 10.7150/ijbs.8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Junghans D., Herzog S. Cnn3 regulates neural tube morphogenesis and neuronal stem cell properties. FEBS J. 2018;285:325–338. doi: 10.1111/febs.14338. [DOI] [PubMed] [Google Scholar]
- 47.Shibukawa Y., Yamazaki N., Kumasawa K., Daimon E., Tajiri M., Okada Y., Ikawa M., Wada Y. Calponin 3 regulates actin cytoskeleton rearrangement in trophoblastic cell fusion. Mol. Biol. Cell. 2010;21:3973–3984. doi: 10.1091/mbc.E10-03-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]