Abstract
Background
The aim of this study was to explore the utility of circulating free DNA (cfDNA) in the evaluation of clinical tumor burden and survival in Chinese patients with metastatic colorectal cancer (mCRC) and to preliminarily summarize some metastatic characteristics associated with mutational status.
Methods
A panel covering a total of 197 hotspot mutations of KRAS, NRAS, BRAF and PIK3CA was used to evaluate the mutational status in plasma by next-generation sequencing (NGS) technology in 126 patients with mCRC. An amplification-refractory mutation system (ARMS) was used to analyze genomic DNA from matched tissue samples. Clinical markers including carcinoembryonic antigen (CEA), carbohydrate antigen 199 (CA199), carbohydrate antigen 125 (CA125), neuron-specific enolase (NSE) and lactate dehydrogenase (LDH) in serum and the sum of all tumor diameters on CT or PET/CT were collected to indicate clinical tumor burden. The correlations between cfDNA and clinical tumor burden were analyzed using Pearson correlation and linear regression models. The median progression-free survival (PFS) and 1-year overall survival (OS) rates were calculated by Kaplan-Meier (K-M) survival analysis.
Results
Of the 126 enrolled patients, patients who were tested positive for mutations in plasma accounted for 45.2% (57/126). Mutations in KRAS, NRAS, BRAF and PIK3CA were detected in 37.3% (47/126), 1.6% (2/126), 3.2% (4/126) and 13.5% (17/126) of patients, respectively. The overall concordance rate of mutational status between plasma and matched tissues was 78.6% (99/126). Sixteen patients had mutations in plasma that were not detected in tissue, including some rare hotspot mutations. The cfDNA concentration was significantly correlated with the levels of clinical markers, especially CEA (P < 0.0001, Pearson r = 0.81), LDH (P < 0.0001, Pearson r = 0.84) and the sum of tumor diameters (P < 0.0001, Pearson r = 0.80). Patients with a high cfDNA concentration (> 17.91 ng/ml) had shorter median progression-free survival (6.6 versus 11.7 months, P < 0.0001) and lower 1-year overall survival rate (56% versus 94%, P < 0.0001) than those with a low cfDNA concentration (≤17.91 ng/ml). The most common metastatic site was the liver (77.8%), followed by the lymph nodes (62.7%), lung (40.5%), peritoneum (14.3%) and bone (10.3%), in all patients. There was no significant difference in metastasis between different mutational statuses.
Conclusion
Analyzing mutations in plasma could provide a more comprehensive overview of the mutational landscape than analyzing mutations in tissue. The cfDNA concentration could be a quantitative biomarker of tumor burden and could predict survival in Chinese patients with mCRC.
Keywords: Metastatic colorectal Cancer (mCRC), Circulating free DNA, Tumor burden, Survival, Metastasis
Background
Colorectal cancer (CRC) is the fourth leading cause of cancer-related death worldwide, and the incidence has increased in the past decade with an aging and growing population [1]. With the development of liquid biopsy strategies, circulating free DNA (cfDNA) derived from plasma, consisting of healthy and tumor DNA (circulating tumor DNA, ctDNA), has been increasingly investigated as a promising biomarker for cancer management [2]. Sequencing of plasma samples may improve diagnostic efficiency and preemptively predict recurrence and treatment response in CRC [3]. Furthermore, monitoring genomic alterations in RAS, BRAF and other cancer-related genes in plasma can guide targeted therapeutic strategies and especially optimize anti-epidermal growth factor receptor (EGFR) therapy for metastatic CRC (mCRC) [4, 5].
The change in tumor burden is an essential feature in the clinical evaluation of cancer therapy. However, there is no clear definition of or uniform assessment method for determining overall tumor burden. The imaging-based Response Evaluation Criteria in Solid Tumors (RECIST) are widely used to measure partial disease load and to evaluate sequential treatment responses based on the assessment of anatomical tumor burden [6]. In addition, circulating biomarkers and clinical symptoms are also significant for the effective surveillance of tumor burden in clinical practice. Such serum biomarkers include carcinoembryonic antigen (CEA), carbohydrate antigen 199 (CA199), carbohydrate antigen 125 (CA125), neuron-specific enolase (NSE), lactate dehydrogenase (LDH), etc. The potential of exploiting quantified levels and mutational load of cfDNA to monitor tumor burden in CRC patients has also been reported in a few studies [7, 8].
The aims of this study were to further explore the mutational signature in plasma and the utility of cfDNA in the evaluation of tumor burden and survival in Chinese patients with mCRC. We also summarized and preliminarily analyzed some metastatic characteristics according to mutations in plasma.
Methods
Patients
Patients with mCRC treated in the Oncology Department, ZhongShan Hospital, Fudan University, Shanghai, China, from April to November 2018 were enrolled in this study. The inclusion criteria were adenocarcinoma in the colon or rectum, recurrent or primary metastatic disease, planned treatment with first-line chemotherapy, life expectancy more than 3 months, age > 18 years, and Eastern Cooperative Oncology Group performance status 0–1. Plasma samples were collected at baseline. This study was approved by the ethics committee of Zhongshan Hospital. Written informed consent was obtained from all patients.
Sample collection, DNA extraction and sequencing of plasma samples
A total of 20 ml of venous blood was collected from each patient and then centrifuged at 1900×g for 10 min at 4 °C to separate the plasma from peripheral blood cells within 2 h of receipt. Plasma was then further centrifuged at 16,000×g for 10 min at 4 °C to pellet any remaining cells. The cfDNA was extracted from 8.0 mL of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, 55,114) according to the manufacturer’s instructions and quantified using the Qubit Fluorometer 3.0 (Life Technologies, Grand Island, NY) and 2100 bioanalyzer (Technologies, Palo Alto, CA). The quality control criteria for cfDNA were as follows: the concentration of the cfDNA was less than 0.9 ng/μl as measured by the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Q32854); the total amount of cfDNA was greater than 20 ng; the fragment distribution feature detected by the 2100 bioanalyzer had a typical peak at 160–180 bp and a small peak at 320–360 bp. The extracted cfDNA was immediately subjected to the next step or cryopreservation. The cfDNA was stored at − 20 °C for no more than 1 week; for long-term storage, the DNA was placed at − 80 °C for no more than 6 months. Attempts were made to keep the number of repeated freeze-thaw cycles of cfDNA samples ≤3.
Plasma cfDNA (20 ng) was subjected to Firefly™ amplicon-based next-generation sequencing (NGS) technology using Accu-Kit™ CRC-01 (AccuraGen, Shanghai, China), as described in a previous study [9]. The panel covered a total of 197 hotspot mutations in exons 2, 3 and 4 of KRAS; exons 2, 3 and 4 of NRAS; exons 9 and 20 of PIK3CA; and exon 15 of BRAF (Supplemental Table 1). NGS libraries were generated using the KAPA Sequencing Library Construction Kit (Kapa Biosystems, Boston, MA, USA). The quality control criteria for the cfDNA libraries were as follows: the concentration of the library was less than 0.5 ng/μl as measured by the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Q32854); the fragment distribution feature detected by the 2100 bioanalyzer had a typical peak at 600–1300 bp, and the average length was 900–1000 bp. Libraries were then sequenced using 2 × 250 pair-end reads on an Illumina MiSeq Dx sequencer (Illumina, San Diego, CA, USA). Unique sequencing reads were determined by using an AccuraGen proprietary algorithm including quality control, read collapse, read alignment, and variant calling. The average coverage depth for all probes in plasma was nearly 30,000×, and the sensitivity of variant detection in this study was 0.2%. For details of this laboratory-developed test, refer to the reported articles [10]. Generally, sequence reads were aligned to the hg19/GRCh37 human reference sequence, and background noise introduced by random NGS error was removed by AccuraGen proprietary algorithms. The firefly algorithm used a 2-proportion Z-test [11] to reduce false positive mutations from background noise, and mutations passing the Z-test were reported. The cfDNA concentration was determined by the Qubit Fluorometer 3.0.
Identification of mutations in tissue samples
The isolation and purification of genomic DNA from formalin-fixed, paraffin-embedded (FFPE) tissue sections was performed using an AmoyDx® FFPE DNA Kit (Xiamen, China). FFPE specimen tissue sections were first deparaffinized with the xylene/ethanol method and then incubated in buffer DTL and proteinase K solution to release DNA from the sections. A short incubation in DES buffer at a high temperature partially reversed formalin crosslinking of the released nucleic acids, improving DNA yield and quality as well as DNA performance in downstream assays. The lysate was mixed with DTB buffer and ethanol to provide appropriate binding conditions for DNA. Then, the mixture was applied to a DNA spin column, where the DNA bound to the membrane and impurities were removed with wash buffer. The DNA was eluted in DTE buffer. A human KRAS/NRAS/BRAF/PIK3CA gene mutation fluorescence polymerase chain reaction (PCR) diagnostic kit (Amoy Diagnostics, Xiamen, China) based on amplification refractory mutation system (ARMS) technology approved by the China Food and Drug Administration was used to analyze genomic DNA from tissue samples. The list of mutations detectable with this panel is shown in Supplemental Table 2. The assay was carried out according to the manufacturer’s protocol for the ABI7500 real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). We defined a cut-off of 1% mutation content as a sample quality check according to the minimum requirement of ARMS technology (approximately 1% analytical sensitivity).
Assessment of serum markers
Carcinoembryonic antigen (CEA) was measured using an enzyme-linked immunosorbent assay (CanAg CEA EIA). Carbohydrate antigen 199 (CA199), carbohydrate antigen 125 (CA125), and neuron-specific enolase (NSE) were measured by electrochemical fluoroimmunoassay (Roche Elecsys series). Lactate dehydrogenase (LDH) was detected by an LDH Assay Kit (Abcam).
Statistical methods
The correlations between clinical factors and cfDNA levels were analyzed using Pearson correlation and linear regression models. The optimal cutoff level for cfDNA was determined by receiver operating curve (ROC) analysis. Median progression-free survival (PFS) and 1-year overall survival (OS) were calculated by Kaplan-Meier (K-M) survival analysis, and the significance was evaluated by the log-rank test. Tumor metastatic sites with different gene statuses were compared using the chi square test. Statistical tests provided two-sided P values, and a significance level of P < 0.05 was used. Statistical analyses were carried out using SPSS 24.
Results
Patient characteristics
A total of 128 patients were enrolled in this study. Because the cfDNA or the library did not meet the quality control standards in 2 patients, blood samples from 126 patients were collected for analysis of four genes (KRAS, NRAS, BRAF and PIK3CA) before treatment. All patients had gene results from matched tissues. The flow chart is shown in Fig. 1. The clinical characteristics are summarized in Table 1. The median age was 58 years (26–84 years). There were 90 males and 36 females. The most common locations of primary lesions were the sigmoid colon and rectum (38.9 and 35.7%, respectively). Ninety-one (72.2%) patients had received surgery before.
Fig. 1.
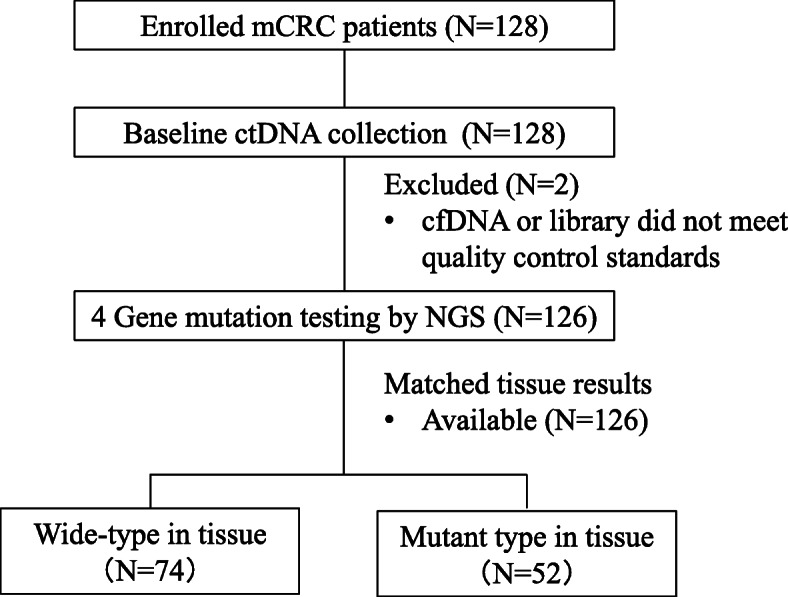
Overview of patient enrollment
Table 1.
Baseline Characteristics
| Characteristics | No. (%) (N = 126) |
|---|---|
| Median Age (years,range) | 58 (26–84) |
| Gender | |
| Male | 90 (71.4) |
| Female | 36 (28.6) |
| Primary site | |
| Ascending colon | 18 (14.3) |
| Transverse colon | 7 (5.6) |
| Descending colon | 7 (5.6) |
| Sigmoid colon | 49 (38.9) |
| Rectum | 45 (35.7) |
| Surgery | |
| No surgery | 35 (27.8) |
| Primary tumor resection | 48 (38.1) |
| Primary tumor and liver metastase resection | 43 (34.1) |
| Metastatic site | |
| Liver | 98 (77.8) |
| Lymph node | 79 (62.7) |
| Lung | 51 (40.5) |
| Peritoneum | 18 (14.3) |
| Bone | 13 (10.3) |
| Others | 13 (10.3) |
| Metastatic organ number | |
| One | 46 (36.5) |
| Multi | 80 (63.5) |
Mutations in plasma and tissues
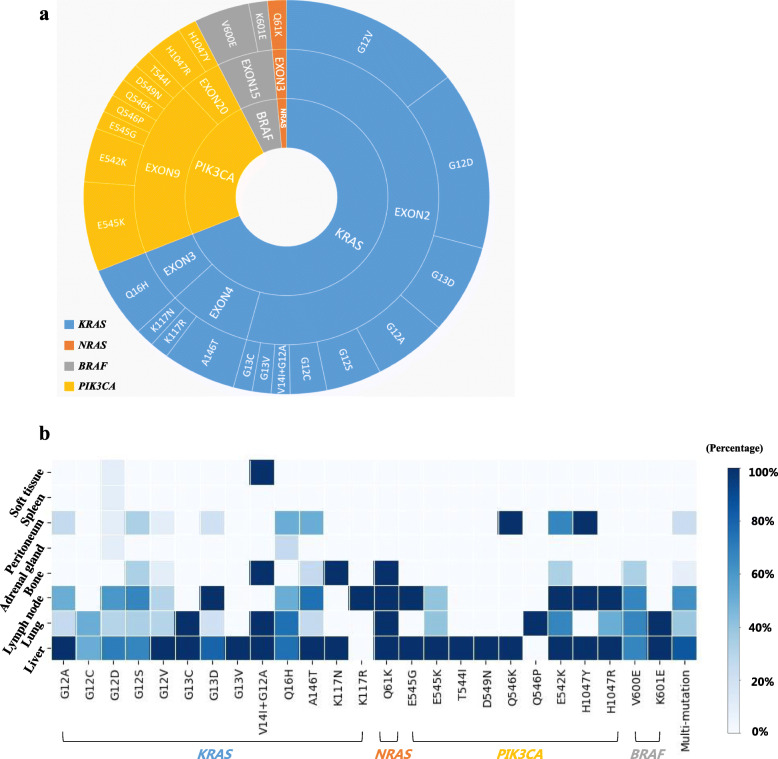
The results showed that 69 patients (54.8%) had wild-type DNA and 57 patients (45.2%) had gene mutations in plasma (Table 2). Mutations of KRAS, NRAS, BRAF and PIK3CA were detected in 37.3% (47/126), 1.6% (2/126), 3.2% (4/126) and 13.5% (17/126) of patients, respectively. Polyclonal mutations were detected in 13 patients (10.3%), which were present mostly in the form of double mutations in KRAS/PIK3CA or NRAS/PIK3CA. The most common mutation sites were G12 (52.6%) and G13 (12.3%) in exon 2 of the KRAS gene. Some rare mutations, including the V14I mutation in exon 2 of KRAS and the K117N and K117R mutations in exon 4 of KRAS, were detected in this study (Table 3 and Fig. 3a). In matched tissue samples, 74 patients (58.7%) had no mutations and 52 patients (41.3%) had mutations, including KRAS mutations in 42 patients (33.3%), NRAS mutations in 3 patients (2.4%), BRAF mutations in 3 patients (2.4%) and PIK3CA mutations in 5 patients (4.0%). The concordance rate of mutational status between plasma and tissue was 78.6%. Of the 27 patients with inconsistency, 11 patients had mutations in tissue that were not detected in plasma, and 16 patients had mutations in plasma that were not detected in tissue.
Table 2.
Mutational status in plasma and tissue
| Mutational status | No. (%) | |
|---|---|---|
| Plasma (N = 126) | Tissue (N = 126) | |
| Wild-type | 69 (54.8) | 74 (58.7) |
| KRAS mutation | 35 (27.8) | 41 (32.5) |
| NRAS mutation | 1 (0.8) | 3 (2.4) |
| BRAF mutation | 4 (3.2) | 3 (2.4) |
| PIK3CA mutation | 4 (3.2) | 4 (3.2) |
| KRAS + PIK3CA mutations | 12 (9.5) | 1 (0.9) |
| NRAS + PIK3CA mutations | 1 (0.8) | 0 (0) |
Table 3.
Mutations detected in plasma
| Gene | Exon | Mutational hotspots | No. (%) (N = 57) |
|---|---|---|---|
| KRAS | 2 | G12D | 10 (17.5) |
| G12V | 10 (17.5) | ||
| G13D | 5 (8.8) | ||
| G12A | 4 (7.0) | ||
| G12S | 3 (5.3) | ||
| G12C | 2 (3.5) | ||
| G13C | 1 (1.8) | ||
| G13V | 1 (1.8) | ||
| V14I + G12A | 1 (1.8) | ||
| 3 | Q61H | 4 (7.0) | |
| 4 | A146T | 4 (7.0) | |
| K117N | 1 (1.8) | ||
| K117R | 1 (1.8) | ||
| NRAS | 3 | Q61K | 1 (1.8) |
| PIK3CA | 9 | E545K | 5 (8.8) |
| E542K | 3 (5.3) | ||
| T544I | 1 (1.8) | ||
| D549N | 1 (1.8) | ||
| Q546K | 1 (1.8) | ||
| Q546P | 1 (1.8) | ||
| E545G | 1 (1.8) | ||
| 20 | H1047R | 2 (3.5) | |
| H1047Y | 1 (1.8) | ||
| BRAF | 15 | V600E | 3 (5.3) |
| K601E | 1 (1.8) |
Fig. 3.
Mutational status and tumor metastasis. a Mutations detected in plasma. b The liver, lung and lymph nodes were the most common metastatic sites in mCRC regardless of the mutational status
Correlation between cfDNA and tumor burden
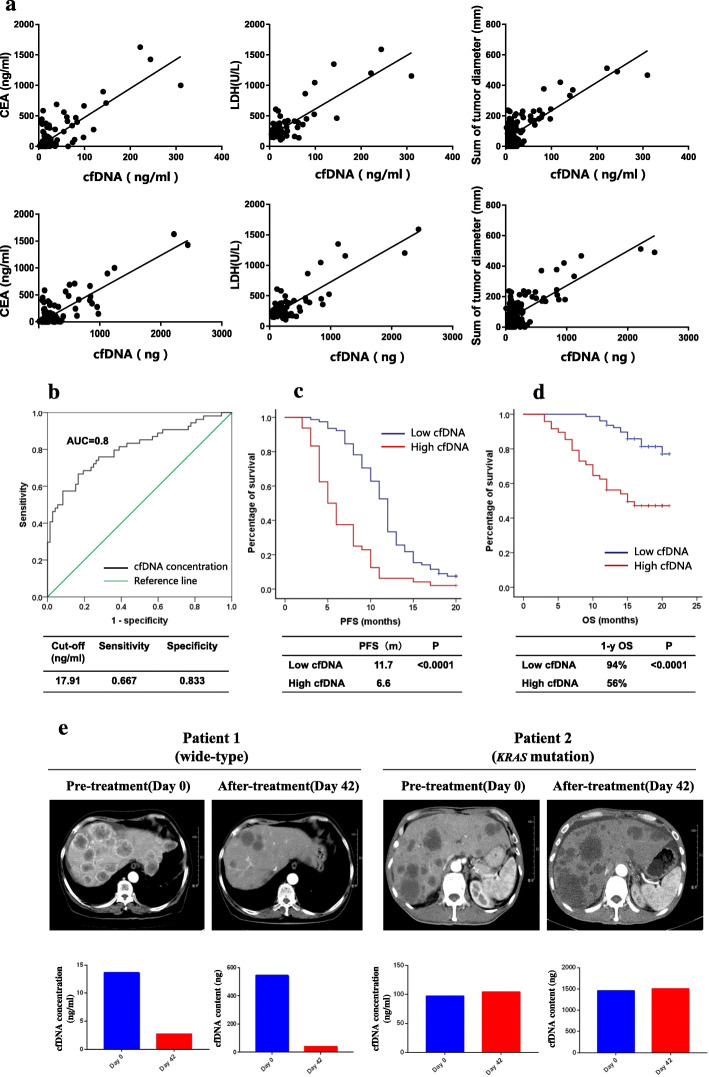
The median concentration of cfDNA was 10.4 ng/ml (2.2–33.5 ng/ml), and the median content of cfDNA was 105 ng (8.8–438.4 ng). The results showed that the concentration of cfDNA was significantly correlated with the levels of CEA, CA199, CA125, NSE, LDH and the sum of tumor diameters, especially CEA (P < 0.0001, Pearson r = 0.81), LDH (P < 0.0001, Pearson r = 0.84) and the sum of tumor diameters (P < 0.0001, Pearson r = 0.80). The cfDNA content was strongly correlated with the levels of CEA (P < 0.0001, Pearson r = 0.84), LDH (P < 0.0001, Pearson r = 0.86) and the sum of tumor diameters (P < 0.0001, Pearson r = 0.76) (Table 4 and Fig. 2a). The mutational frequency in plasma was moderately correlated with the levels of CA199 (P < 0.0001, Pearson r = 0.66), LDH (P < 0.0001, Pearson r = 0.61) and the sum of tumor diameters (P < 0.0001, Pearson r = 0.64) (Table 4). The optimal cutoff level for cfDNA concentration was determined to be 17.91 ng/ml based on the 9-month PFS through ROC curve analysis (Fig. 2b). The patients with a low cfDNA concentration (≤17.91 ng/ml) had a longer PFS (11.7 months versus 6.6 months, P < 0.0001) and a higher 1-year overall survival rate (94% versus 56%, P < 0.0001) than those with a high cfDNA concentration (>17.91 ng/ml) (Fig. 2c and d). Two typical mCRC patients in this study were included, and the changes in cfDNA concentration were correlated with the effect of therapy (Fig. 2e).
Table 4.
Correlation between cfDNA and clinical tumor burden
| Tumor burden | cfDNA concentration (ng/ml) |
cfDNA content (ng) |
Mutational frequency (%) | |||
|---|---|---|---|---|---|---|
| P value | Pearson r(95%CI) | P value | Pearson r(95%CI) | P value | Pearson r(95%CI) | |
| CEA (ng/ml) | < 0.0001 | 0.81 (0.74–0.87) | < 0.0001 | 0.84 (0.78–0.89) | < 0.0001 | 0.56 (0.35–0.72) |
| CA199 (U/ml) | < 0.0001 | 0.66 (0.54–0.75) | < 0.0001 | 0.63 (0.50–0.73) | < 0.0001 | 0.66 (0.48–0.79) |
| CA125 (U/ml) | < 0.0001 | 0.59 (0.44–0.71) | < 0.0001 | 0.65 (0.51–0.75) | 0.0036 | 0.42 (0.15–0.63) |
| NSE (ng/ml) | < 0.0001 | 0.65 (0.49–0.77) | < 0.0001 | 0.67 (0.51–0.78) | 0.0022 | 0.50 (0.20–0.71) |
| LDH (U/L) | < 0.0001 | 0.84 (0.78–0.89) | < 0.0001 | 0.86 (0.80–0.90) | < 0.0001 | 0.61 (0.39–0.76) |
| Sum of diameter (mm) | < 0.0001 | 0.80 (0.72–0.85) | < 0.0001 | 0.76 (0.68–0.83) | < 0.0001 | 0.64 (0.46–0.77) |
Fig. 2.
Correlation between cfDNA and tumor burden. a The concentration and content of cfDNA were significantly correlated with the levels of CEA and LDH and the sum of tumor diameters. b Through ROC curve analysis, the optimal cutoff level for cfDNA concentration was determined to be 17.91 ng/ml. c The median progression-free survival (11.7 versus 6.6 months, P < 0.0001) was significantly different between patients with a low cfDNA concentration (≤17.91 ng/ml) and those with a high cfDNA concentration (> 17.91 ng/ml). d Patients with a low cfDNA concentration had a higher 1-year overall survival rates (94% versus 56%, P < 0.0001) than those with a high cfDNA concentration. e An early decrease in cfDNA was related to a good therapeutic effect in two typical mCRC patients
Mutational status and tumor metastasis
The most common metastatic site was the liver (77.8%), followed by the lymph nodes (62.7%), lung (40.5%), peritoneum (14.3%) and bone (10.3%), in all patients. Other rare metastatic sites included the adrenal gland, the spleen and some soft tissues. Whether the gene status was wild-type or mutated, the sites of metastasis were similar (Supplemental Table 3). In the patients with gene mutations, the top three metastatic sites were also the liver, lymph nodes and lung, regardless of the type of mutation. Bone metastasis was often found in patients with G12 mutations in exon 2 of KRAS, A146 and K117 mutations in exon 4 of KRAS, Q61 mutations in exon 3 of NRAS, E542 mutations in exon 9 of PIK3CA and V600 mutations in exon 15 of BRAF. Peritoneal metastasis was mainly found in patients with KRAS or PIK3CA mutations (Fig. 3b).
Discussion
In mCRC, patients harboring RAS and BRAF mutations do not benefit from anti-EGFR therapies, such as cetuximab [12–14]. Due to the heterogeneity of tissue samples, the mutational status may be different intralesionally and interlesionally. In recent years, analyzing mutations in plasma-derived ctDNA has been used to provide a more comprehensive overview of the mutational landscape compared to analyses of primary and/or metastatic tumors. Consequently, ctDNA analysis, as an alternative approach with a minimally invasive nature, has been gradually used to guide management and monitor treatment response in some cancers, including CRC [3, 15]. In this study, mutations in plasma were measured in 126 Chinese patients with mCRC by a panel of 4 genes (197 hotpots), including KRAS, NRAS, BRAF, and PIK3CA, using a NGS approach.
Our results showed that the concordance rate of RAS/BRAF mutations between plasma and tissue was 78.6%, which was in line with that found in previous studies [15–17]. In a subgroup of 92 patients from the CAPRI-GOIM clinical trial, NGS analysis of both tumor tissue and plasma identified a concordance rate of 78.3% [18], similar to our results. In our study, 11 patients had mutations in tissue that were not detected in plasma. Nine of these patients had a low tumor burden and their cfDNA concentrations ranged from 4 to 10 ng/ml, which was less than the median concentration. A low amount of tumor DNA could hypothetically lead to false-negative results in plasma. In the other two patients with high tumor load, their metastatic sites were predominantly the lymph nodes, bone and peritoneum. In such patients, these metastatic tumors might release less DNA into the circulation than metastatic tumors at other sites. For the subgroup of 16 patients with mutations in plasma but no mutations in tissue, we propose three possible reasons. First, the discrepancy may have resulted from the different sampling times and heterogeneity of tissue specimens. Eleven of these patients had their primary tumor removed before blood sample collection. Therefore, the testing results in plasma might have indicated the mutational status of metastases originating from a different clone. Indeed, the mutational signature of KRAS has been reported to be different between primary tumors and metastatic lesions, with an inconsistency rate ranging from 5 to 32% [19]. Second, sequencing analysis was performed by different approaches in our routine clinical work-up. Mutations in plasma were measured by a panel covering 197 hotpot mutations using NGS approach, while mutations in tissue were assayed by a panel containing only 17 hotpots using ARMS technology. Some rare mutations detected in plasma, such as KRAS p.V14I, BRAF p.K601E, and PIK3CA p.E545G, were not covered by the sequencing panel of tissue. Third, the sensitivity of variant detection used in plasma was 0.2%, while the ARMS technology used in tissue had an analytical sensitivity of approximately 1%, which could also have led to false-negative results in tissue.
How can anti-EGFR treatment be guided using the testing results from both plasma and tissue? It is generally believed that patients with any RAS/BRAF mutations in either tumor tissue or plasma have less treatment benefit than patients who have no mutations. A prospective multicenter clinical study [20] reported that 59, 11.8 and 14.4% of KRAS, NRAS and BRAF mutations were found in plasma, while 44, 8.8 and 7.2% were found in tissue respectively. Even patients who showed RAS wild-type in tissue failed to respond to anti-EGFR therapy when mutations were present in their plasma, with the incidence of disease progression as high as 56% [20]. Therefore, in our study, the majority of these patients with RAS/BRAF mutations in tissue and/or in plasma did not receive anti-EGFR treatment, except one (patient No. 25, in Supplemental Table 4). After surgical removal of this patient’s RAS wild-type primary tumor, only a small number of KRAS-mutated subclones were found in his plasma with a mutational frequency of 0.26%. Considering that there were only a few mutated clones in his plasma, we treated him with cetuximab plus chemotherapy, and the PFS was up to 12 months, indicating that he could still have clinical benefit from anti-EGFR therapy. Therefore, whether low frequency mutation in plasma (0.2–1%) is a contraindication for anti-EGFR treatment needs further study. In addition, polyclonal mutations were more frequently detected in plasma than in tissue (10.3% versus 0.9% in our study), which emerged mostly in the form of double mutations in KRAS/PIK3CA or NRAS/PIK3CA. Such polyclonal mutations may influence mechanisms of resistance, as reported in some other studies [21]. Consequently, the baseline assessment of the mutational status in plasma can identify additional mutated patients and further improve patient selection for anti-EGFR treatment.
This study further analyzed the correlation between cfDNA and tumor burden. We found that cfDNA concentration was positively correlated with the serological levels of CEA, CA199, NSE, LDH, and the sum of tumor diameters (including all tumors assessable). However, the correlation might not be tumor-specific because cfDNA comprises both healthy and tumor DNA. Therefore, we further found that the mutational frequency in plasma was also correlated with the levels of CA199 and LDH and the sum of tumor diameters. It was indicated that cfDNA concentration and mutational frequency could serve as quantitative tools for assessing tumor burden, which has been supported by some other studies [4]. In this study, we also found that patients with a higher cfDNA concentration had shorter PFS and OS than those with a lower cfDNA concentration, with an optimal threshold of 17.91 ng/ml. Another potential application of cfDNA testing is to dynamically evaluate therapeutic effects and predict recurrence [7]. As seen in Fig. 2e, we found that an early decrease in cfDNA concentration was related to a good therapeutic effect. Thomsen et al. also reported that a low level of cfDNA after chemotherapy, prior to radiological imaging evaluation, was associated with a low risk of progression [22].
In this study, the liver, lung and lymph nodes were the most common metastatic sites in mCRC regardless of specific mutations. Additionally, we also observed some interesting metastatic patterns in patients harboring rare mutations in this study. For example, one patient (patient No. 40 in Supplemental Table 4) with a high-frequency (28%) Q61K mutation in exon 3 of NRAS had mandible metastasis, which is a rare metastatic site in clinical practice. Patient No. 31, carrying both the KRAS p.V14I and KRAS p.G12A mutations, had metastasis mainly in soft tissues, such as the chest wall, anterior sacrum, and psoas major muscle. Patient No. 39, with the K117R mutation in exon 4 of KRAS, had only systemic lymph node metastases, and his cfDNA concentration was more than 50 ng/ml, but his PFS was more than 20 months. We believe that these patients with rare mutations might have unique mechanisms of metastasis that further influence survival, which might be an interesting problem worthy of further research.
There are some limitations of this study. First, correlations between rare mutations and metastatic sites could not be made because of the small sample size. Second, the mutational status in tissue and plasma were sequenced with panels used in our daily routine practice, covering only four genes, and these panels need to be optimized to obtain more information with low cost for every patient.
Conclusion
In conclusion, our results suggest that analyzing mutations in plasma could provide a more comprehensive overview of the mutational landscape than analyzing mutations in tissue and that cfDNA concentration could be a quantitative biomarker to evaluate tumor burden and predict the survival of mCRC patients. The liver, lung and lymph nodes were the most common metastatic sites in mCRC regardless of the types of mutation.
Supplementary information
Additional file 1: Supplemental Table 1. The testing panel covering a total of 197 hotspots in plasma. Supplemental Table 2. The testing panel covering a total of 17 hotspots in tissue. Supplemental Table 3. Mutational status in plasma and metastatic sites. Supplemental Table 4. Mutational status and metastatic sites of all the 126 patients.
Acknowledgements
Not Applicable.
Abbreviations
- ctDNA
Circulating tumor DNA
- cfDNA
Circulating free DNA
- mCRC
Metastatic colorectal cancer
- NGS
Next-generation sequencing
- ARMS
Amplification-refractory mutation system
- CEA
Carcinoembryonic antigen
- CA199
Carbohydrate antigen 199
- CA125
Carbohydrate antigen 125
- NSE
Neuron-specific enolase
- LDH
Lactate dehydrogenase
- PET/CT
Positron emission tomography/computed tomography
- PFS
Progression-free survival
- OS
Overall survival
- EGFR
Epidermal growth factor receptor
- RECIST
Response evaluation criteria in solid tumors
- FFPE
Formalin-fixed paraffin-embedded
- ROC
Receiver operating curve
Authors’ contributions
XX, WG and TL conceived and designed the study. XX, YY enrolled the patients and MS performed the experiments. SW provided the results of serum markers. ML, LL, FH and CZ collected the clinical information. XX, YY and MS analyzed and wrote the paper. WG and TL reviewed and edited the manuscript. All authors read and approved the manuscript.
Funding
The work was supported by the National Nature Science Foundation of China (81502003, 81772511 and 81602038).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Some important information about the mutational status and metastatic sites of all the 126 patients has been presented in the Supplemental Table 4.
Ethics approval and consent to participate
This study was approved by the ethics committee of Zhongshan Hospital affiliated to Fudan University (Approval No: B2017-165R) (Shanghai, China). Written informed consent was obtained from all patients.
Consent for publication
Not Applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaojing Xu, Yiyi Yu and Minna Shen contributed equally to this work.
Contributor Information
Xiaojing Xu, Email: xu.xiaojing@zs-hospital.sh.cn.
Yiyi Yu, Email: yu.yiyi@zs-hospital.sh.cn.
Minna Shen, Email: shen.minna@zs-hospital.sh.cn.
Mengling Liu, Email: liu_mengl@163.com.
Shengchao Wu, Email: wu.shengchao@zs-hospital.sh.cn.
Li Liang, Email: liang.li@zs-hospital.sh.cn.
Fei Huang, Email: huang.fei@zs-hospital.sh.cn.
Chenlu Zhang, Email: zhang.chenlu@zs-hospital.sh.cn.
Wei Guo, Email: guo.wei@zs-hospital.sh.cn.
Tianshu Liu, Email: liutianshu1969@126.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12885-020-07516-7.
References
- 1.Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. Jama Oncol. 2017;3(4):524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan J, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD. Caldas Cet al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 3.Thierry AR, Mouliere F, El MS, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20(4):430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 4.Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28(6):1325–1332. doi: 10.1093/annonc/mdx125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Emburgh BO, Arena S, Siravegna G, Lazzari L, Crisafulli G, Corti G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun. 2016;7:13665. doi: 10.1038/ncomms13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Reinert T, Scholer LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65(4):625–634. doi: 10.1136/gutjnl-2014-308859. [DOI] [PubMed] [Google Scholar]
- 8.Yi X, Ma J, Guan Y, Chen R, Yang L, Xia X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int J Cancer. 2017;140(12):2642–2647. doi: 10.1002/ijc.30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang B, Wu S, Huang F, Shen M, Jiang H, Yu Y, et al. Analytical and clinical validation of a novel amplicon-based NGS assay for the evaluation of circulating tumor DNA in metastatic colorectal cancer patients. Clin Chem Lab Med. 2019;57(10):10. doi: 10.1515/cclm-2019-0142. [DOI] [PubMed] [Google Scholar]
- 10.Kastrisiou M, Zarkavelis G, Pentheroudakis G, Magklara A. Clinical application of next-generation sequencing as a liquid biopsy technique in advanced colorectal Cancer: a trick or a treat? Cancers. 2019;11(10):1573. doi: 10.3390/cancers11101573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Cutsem E, Kohne CH, Lang I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29(15):2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 13.Qin S, Li J, Wang L, Xu J, Cheng Y, Bai Y, et al. Efficacy and tolerability of first-line Cetuximab plus Leucovorin, fluorouracil, and Oxaliplatin (FOLFOX-4) versus FOLFOX-4 in patients with RAS wild-type metastatic colorectal Cancer: the open-label, randomized, phase III TAILOR trial. J Clin Oncol. 2018;36(30):3031–3039. doi: 10.1200/JCO.2018.78.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22(7):1535–1546. doi: 10.1093/annonc/mdq632. [DOI] [PubMed] [Google Scholar]
- 15.Schmiegel W, Scott RJ, Dooley S, Lewis W, Meldrum CJ, Pockney P, et al. Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol. 2017;11(2):208–219. doi: 10.1002/1878-0261.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grasselli J, Elez E, Caratu G, Matito J, Santos C, Macarulla T, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2017;28(6):1294–1301. doi: 10.1093/annonc/mdx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachet JB, Bouche O, Taieb J, Dubreuil O, Garcia ML, Meurisse A, et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann Oncol. 2018;29(5):1211–1219. doi: 10.1093/annonc/mdy061. [DOI] [PubMed] [Google Scholar]
- 18.Normanno N, Esposito AR, Lambiase M, Forgione L, Cardone C, Iannaccone A, et al. RAS testing of liquid biopsy correlates with the outcome of metastatic colorectal cancer patients treated with first-line FOLFIRI plus cetuximab in the CAPRI-GOIM trial. Ann Oncol. 2018;29(1):112–118. doi: 10.1093/annonc/mdx417. [DOI] [PubMed] [Google Scholar]
- 19.Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16(3):790–799. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 20.Thierry AR, El MS, Mollevi C, Raoul JL, Guimbaud R, Pezet D, et al. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment. Ann Oncol. 2017;28(9):2149–2159. doi: 10.1093/annonc/mdx330. [DOI] [PubMed] [Google Scholar]
- 21.van Helden EJ, Angus L, Menke-van der Houven van Oordt CW, Heideman DAM, Boon E, van Es SC, et al. RAS and BRAF mutations in cell-free DNA are predictive for outcome of cetuximab monotherapy in patients with tissue-tested RAS wild-type advanced colorectal cancer. Mol Oncol. 2019;1311(11):2361–2374. doi: 10.1002/1878-0261.12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomsen CB, Hansen TF, Andersen RF, Lindebjerg J, Jensen LH, Jakobsen A. Monitoring the effect of first line treatment in RAS/RAF mutated metastatic colorectal cancer by serial analysis of tumor specific DNA in plasma. J Exp Clin Cancer Res. 2018;37(1):55. doi: 10.1186/s13046-018-0723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental Table 1. The testing panel covering a total of 197 hotspots in plasma. Supplemental Table 2. The testing panel covering a total of 17 hotspots in tissue. Supplemental Table 3. Mutational status in plasma and metastatic sites. Supplemental Table 4. Mutational status and metastatic sites of all the 126 patients.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Some important information about the mutational status and metastatic sites of all the 126 patients has been presented in the Supplemental Table 4.