Abstract
Lactic acid is an organic compound produced via fermentation by different microorganisms that are able to use different carbohydrate sources. Lactic acid bacteria are the main bacteria used to produce lactic acid and among these, Lactobacillus spp. have been showing interesting fermentation capacities. The use of Bacillus spp. revealed good possibilities to reduce the fermentative costs. Interestingly, lactic acid high productivity was achieved by Corynebacterium glutamicum and E. coli, mainly after engineering genetic modification. Fungi, like Rhizopus spp. can metabolize different renewable carbon resources, with advantageously amylolytic properties to produce lactic acid. Additionally, yeasts can tolerate environmental restrictions (for example acidic conditions), being the wild-type low lactic acid producers that have been improved by genetic manipulation. Microalgae and cyanobacteria, as photosynthetic microorganisms can be an alternative lactic acid producer without carbohydrate feed costs. For lactic acid production, it is necessary to have substrates in the fermentation medium. Different carbohydrate sources can be used, from plant waste as molasses, starchy, lignocellulosic materials as agricultural and forestry residues. Dairy waste also can be used by the addition of supplementary components with a nitrogen source.
Keywords: Biotechnology, Microbiology, Lactic acid, Fermentation, Microorganisms, Agricultural waste, Industrial waste
Biotechnology; Microbiology; Lactic acid; fermentation; Microorganisms; agricultural; Industrial waste
1. Introduction
Lactic acid as an organic acid is authorized by the U.S. Food and Drug Administration as GRAS (generally regarded as safe). It provides leading roles in the food and non-food industry. i) It is utilized in the food industry including beverage industry (as food preservative, fermentation agent, acidulant, flavour enhancer, and decontaminant), antioxidant, prebiotic activity, cryoprotectant, viscosifier, ii) chemical industry mainly mosquito repellent, descaling agents, pH regulator, neutralizers, green solvent, cleaning agents, metal complexing agents, substitution of synthetic plastics derived from petro-chemically compounds and environmentally friendly alternative due to production of poly-lactic acid as biodegradable polymers for commercial uses such as fibers and films, production of propylene glycol, lactate esters, acrylic acid, propylene oxide, propanoic acidacetaldehyde, 2,3-pentanedione, and dilactide; iii) cosmetic industry as moisturizers, skin-lightening agents, skin rejuvenating agents, anti-acne agents, humectants, anti tartar agents; iv) medicine and pharmaceuticals industry as a building-block molecule, dialysis solution, mineral preparations, tablettings, prostheses, surgical sutures, controlled drug delivery system, immune-stimulant and manufacture of hygiene and esthetic products [1, 2]. Lactic acid is commonly sold as an 88% solution. The price varies with the application (e.g., food, pharmaceuticals, and PLA) and also depends on the price of commodity starch and sugar feedstocks used for fermentation. A range of around $3.0-$4.0/kg was reported in 2019 (https://www.pharmacompass.com). Upon annual growth of 16.2%, the global lactic acid market increased from 1,220.0 kilotons in 2016 to 1,960.1 kilotons in 2025. This should display USD 9.8 billion in the global market. Market studies mention that the major growth will be for medicines and cosmetics in the Latin America and the Asia Pacific region [2].
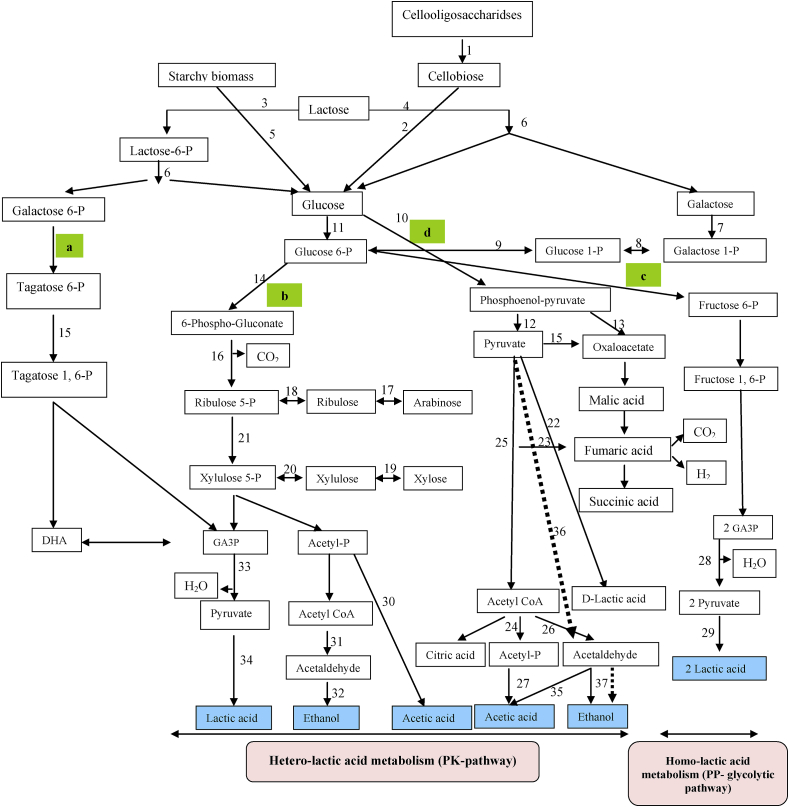
The direct conversion of complex compounds to lactic acid can be categorized mainly into Four groups. a) The lactic acid producing fungi such as Rhizopus oryzae. b) amylolytic lactobacilli namely Lb. amylovorous, Lb. manihotivorans, Lb. amylophilus etc. c) The simultaneously degradation of substrate further treat with enzymes. d) glycolysis pathway in E. coli, K. lactis and S. cerevisiae [3, 4] (Figure 1).
Figure 1.
Pathways of lactic acid production from agro-industrial residues. Number on arrow catalyzed by enzyme and other reaction. 1: Exo β1,4 Glucanase, 2: β -Glucosidase, 3: lactose phosphotransferase system (Lac-PTS), 4: permease, 5: Amylase, 6: β-galactosidase, 7: ATP→ADP, 8: galactose-1-phosphate uridylyltransferase, 9: phosphoglucomutase, 10: NAD→ NADH, 11: ATP→ADP, 12: ATP→ADP, 13: Phosphoenolpyruvate carboxylase, 14: ATP→ADP, 15: ATP→ADP, 16: NAD+→NADH, 17: arabinose isomerase, 18: ribulokinase and ATP→ADP, 19: xylose reductase and xylitol dehydrogenase, 20: ATP→ADP, 21: ribulose 5-phosphate 3-epimerase, 22: D-lactic acid Dehydrogenase, 23: Pyruvate-fomarate lyase, 24: Pta, 25: Pyruvate dehydrogenase complex, 26: Aldehyde dehydrogenase, 2NADH→ 2NAD+, 27: Acetate kinase, 28: 4 ADP→ 4ATP, 2 NAD+→2NADH, 29: 2NADH→2NAD+, 30: ADP→ ATP, 31: NADH→ NAD+, 32: NADH→ NAD+, 33: 2ADP→ATP, NAD+→NADH, 34: Lactate dehydrogenase, NADH→NAD+, 35: Acetaldehyde dehydrogenase, 36: Pyruvate decarboxylase. 37: Alcohol dehydrogenase. GA3P: glyceraldehyde-3-P, DHAP: Dihydroxyacetone-P. A route: D-tagatose 6-phosphate pathway. B route: Pentose phosphoketolase (PK) pathway: for Hetero lactic acid metabolism. C route: Embden-Meyerhof-Parnas (EMP) pathway: for Homo lactic acid metabolism. D route: Glycolysis pathway in E. coli, K. lactis and S. cerevisiae.
The fermentation capacity by several LAB has been studied in order to produce LA. Plenty of lactic acid bacteria have amylase activity were originated from various plant and animal. Main obstructions lactic acid bacteria is that they require complex nutrients and slightly lower fermentation temperatures (˂ 45 °C) than other microorganism, which lead to increased costs and contamination risk and are also poor productivity due to the amylase production in the initial step, causing a long lag phase. Otherwise they require partially hydrolyzed substrates. Certain fungi including Rhizopus sp. can generate high content of lactic acid. They also specify with advantages compared with the bacterial process such as i) the consumption of a chemically defined medium (including inorganic nitrogen sources), which can facilitate product separation and purification, ii) consume both complex carbohydrates and pentose sugars iii) high product concentrations of pure L-lactic acid owing to metabolize high amount of glucose which is preferred for poly-lactide manufacture. For instance, fungal species of R. oryzae 2062 and R. arrhizus 36017 produce lactic acid in a single-stage simultaneous saccharification and fermentation process. In contrast, homofermentative lactic acid bacteria have highly more efficiencies than the fungi to convert sugars to lactic acid because production other byproducts such as ethanol and fumaric acid by R. oryzae-based process. Some researcher tried to enhance lactic acid production using a mutant of R. oryzae with declined alcohol dehydrogenase activity under oxygen limiting conditions. This strain generated almost 10-fold more lactic acid production when compared to the parent strain [3, 4]. Bacillus spp., allows reducing the LA production cost due to fewer nutrition demands and a high temperature of fermentation. Relatively to the use of fungi, the low LA productivity disadvantage of using wild-type yeasts can be overcome by engineering genetic modification [5]. Moreover, Saccharomyces cerevisiae is one of the more promising organisms that reveal high tolerance to low pH-values. Interestingly, good LA productivities were achieved by genetically modified Candida spp [5].
Relatively to substrate sources, worldwide there is a lot of interesting agro-industrial waste or sub-products with a lower value, which can be fermented by several organisms. Molasses, juices waste, starchy biomass, agricultural residues, and forestry residues that is rich in mono and disaccharides, which in some cases need to be hydrolysed by pectinases to enhance the LA production. To use dairy wastes as a substrate, mainly whey, it is necessary to use an enriched mediums, due to insufficient proteolytic enzyme activity [5, 6, 7, 8]. In this paper, different bacterial groups that capable of producing lactic acid at different rates and under different conditions were discussed.
In this paper, different bacterial groups that capable of producing lactic acid at different rates and under different conditions were discussed. Moreover, chemical and physical pretreatment of substrates were explained.
2. LA producing microorganisms
2.1. Bacteria
2.1.1. Lactic acid bacteria
Lactic acid bacteria (LAB) are gram-positive microorganisms known as the main safe industrial-scale producers of lactic acid (LA). LA is produced by glycolysis pathway under anaerobic conditions, and this compound can be produced from hexoses and pentoses LAB metabolism pathways, as indicated in Figure 1. LA production yield and productivity depends on pH (3.5–9.6), temperature (5–45 °C), nutrients presence (such as amino acids, peptides, nucleotides and vitamins) and the LAB strain producers used (so far have been used strains belonging to the genus Leuconostoc, Lactococcus, Lactobacillus, Pediococcus, Enterococcus, Streptococcus, Vagococcus, Aerococcus, Carnobacterium, Tetragenococcus, Oenococcus and Weissella) [5, 6, 7, 8]. However, LAB species including Lactobacillus, Lactococcus, Leuconostoc, Streptococcus, and Pediococcus are also used as starter cultures in industrial food fermentations. Among LAB strains, Lactobacillus strains have great commercial importance due to high acid tolerance, high yield, and productivity, and can be engineered for the selective production of L/D-lactic acid [5]. However, there are some disadvantages when using the LAB for commercial LA production, such as the high requirement of complex nutrients (with increasing production costs) and the low fermentation temperature (that could result in contamination risks and prevention of simultaneous saccharification of starchy or cellulosic biomass and conversion to sugars by amylases enzymes and fermentation of sugars and lignocellulosic biomass) [9, 10]. However, the alkaliphilic LAB that includes Marinilactibacillus, Halolactibacillus, and Alkalibacterium spp. and other various strains from LAB genera, such as Microbacterium spp., Enterococcus spp., Alkalibacterium spp., Exiguobacterium spp., Oceanobacillus spp. and Bacillus spp., can produce LA at high pH-values (7.0–11.5), resulting in a contamination minimization during the fermentation process [9, 10, 11, 12]. For example, Exiguobacterium is a genus of bacilli, being the alkaliphile Exiguobacterium sp. strain 8-11-1 used to produce optically pure l-lactate, in nonsterile fed-batch fermentation with productivity of 8.15 g/L/h under glucose concentration of 80 g/L and using NaOH as a neutralizing agent [9].
Since the complex nutritional requirements of the LAB complicate industrial processes and enhance cost, genetic engineering methods by gene manipulation with plasmid transformation could improve the fermentation efficiency of LA production. Some microorganisms, such as Corynebacterium glutamicum (section 1-3), Escherichia coli (section 1-4) and yeasts lack activities for pyruvate-formate lyase and lactate dehydrogenase (LDH), and these genes can be inserted through gene sources of L-/D-LDH from LAB, bovine and fungi, to express the D(-)- LDH gene from LAB, producing D(-)-lactate in minimal medium with >99.9% optical purity.
Glucose fermentation by homofermentative LAB needs somewhat acidic to neutral pH. However, low pH, has an inhibitory impact on cellular metabolism, in turn lactic acid production. The large number of LAB cannot grow lower than pH 4. In order to maintain cell survival two solutions are used: i) lime is routinely introduced to the fermentors to keep a neutral pH, which cause to produce calcium lactate (>90% of the lactic acid). Subsequent fermentation, the broth containing calcium lactate would be acidified with sulfuric acid to generate lactic acid. High sulfuric acid consumption leads to form high content of insoluble calcium sulfate as gypsum compared to the amount of lactic acid produced, waste disposal concerns, further corrosion problems and a significant cost factor in the product recovery step of commercial operations. Ideally, microbial fermentation would take place in medium with a pH at or lower than the pKa of lactic acid (the pKa of lactic acid is 3.78), permitting direct purification of the acid form. ii) Metabolic engineering has been applied to modify for variants of Lactobacillus sp. with improved tolerance to the acidified medium generated during fermentation. Improved strains has been achieved after UV and nitrosoguanidine treatment, which they are capable to produce lactic acid at rates and yields like to those of the traditional, neutral-pH lactic acid processes. In order to maximize resistance to the acidic conditions inducing by lactic acid production, enzymes namely trehalose 6-phosphate phosphatase from Propionibacterium freudenreichii has been expressed in Lb. lactis, leading to 5- to 10-fold greater survivability at pH 3.0. Similarity, the enzymes in histidine decarboxylation pathway from Streptococcus thermophilus was expressed in Lb. lactis, making survival at pH levels as low as 3 in which the host cells were easily dying [1]. There are two fermentative LAB pathways:
-
A)
The homofermentative LAB
LAB possesses the aldolase enzyme and can convert glucose almost exclusively into LA. The homofermentative LAB usually uses hexose and pentose sugars via the Embden-Meyerhof (by using glycolysis pathway and pentose phosphate pathway). Homofermentative LAB produces two LA molecules as a major end-product per mole of consumed glucose, with a theoretical yield of 1 g.g−1 and experimental yields among being this related to the type of the carbon source used [11]. For LA commercial production (more than 100 g/L of lactic acid) only homofermentative LAB is available due to the high yield (near maximal theoretical value), productivity and a high optical purity of lactic acid (>99%). Homofermentative LAB includes Streptococcus, Lactococcus, Enterococcus, Pediococcus, and some Lactobacillus, as shown in Table 1. Homofermentative Lactobacillus spp. includes mainly Lb. delbruckii subsp. bulgaricus, Lb. acidophilus, Streptococcus salivarius subsp. thermophilus, and Lb. helveticus. Abdel-Rahman et al. [13, 14] reported that Enterococcus mundtii QU 25 and engineered Lactobacillus plantarum could also metabolize homofermentative pentoses to LA.
-
B)
The heterofermentative LAB
Table 1.
Compilation of organisms studied for lactic acid (LA) production, with respective LA concentration, yield, productivity, substrate source and reference.
| Organism | Lactic acid | Yield | Productivity | Source | Reference |
|---|---|---|---|---|---|
| g/L | g/g | g/(L/h) | |||
| Homo and Heterofermentative LAB | |||||
| Lb. delbruckii NCIMB 8130 | 90.0 | 0.97 | 3.8 | Molasses | [125] |
| Lb. delbrueckii sp. delbrueckii ATCC 9649 | 58 | 0.48 | Glucose | [13, 14] | |
| Lb. delbrueckii sp. lactis ATCC 8000 | 83 | 0.83 | Glucose | [13, 14] | |
| Lb. delbrueckii sp. lactis DSM 20073 | 82 | 0.82 | Glucose | [13, 14] | |
| Lb. delbrueckii mutant DP3 | 77 | 0.64 | Glucose | [13, 14] | |
| Lb. delbrueckii mutant DP3, 19 | 68 | 0.57 | Glucose | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus AU | 20 | 0.45 | Whey permeate | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus 5085 | 16 | 0.38 | Whey permeate | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus 5085 | 7.9 | 0.18 | Whey permeate | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus 5085 | 15 | 0.41 | 4 | Whey permeate | [13, 14] |
| Lb. delbrueckii sp. bulgaricus ATCC 11842 | - | - | - | Sorghum | [13, 14] |
| Lb. delbrueckii sp. lactis 447 | 55 | 0.85 | Lignocellulose hydrolysate | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus 5085 | 7.9 | 0.18 | Whey permeate | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus 5085 | 16 | 0.38 | Whey permeate | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus CRL 870 | 12 | - | - | Skim milk | [13, 14] |
| Lb. delbrueckii sp. delbrueckii ATCC 9649 | 106 | 0.82 | Hydrolysate wheat flour | [13, 14] | |
| Lb. delbrueckii IFO 3534 | 24 | 0.48 | Hydrolysate newspaper | [13, 14] | |
| 53 | 0.53 | Hydrolysate pure cellulose | |||
| Lb. delbrueckii sp. bulgaricus CBS 743.84 | 35 | 0.85 | Glucose | [13, 14] | |
| 37 | 0.82 | Lactose | |||
| Lb. delbrueckii sp. bulgaricus CNRZ 369 | 56 | 2.8 | Glucose | [13, 14] | |
| 32 | 1.6 | Cellobiose | |||
| 41 | 2.1 | Xylose | |||
| Lb. delbrueckii sp. delbrueckii | 87 | 0.87 | Glucose | [13, 14] | |
| 94 | 0.94 | Fructose + glucose | |||
| 85 | 0.85 | Sucrose | |||
| Lb. delbrueckii sp. delbrueckii ATCC 9649 | 58 | 0.85 | Glucose | [13, 14] | |
| 40 | 0.75 | Lactose | |||
| Lb. delbrueckii sp. bulgaricus ATCC 11842 | 18 | 0.11 | Hydrolysate of wheat flour | [13, 14] | |
| 26 | 0.18 | Hydrolysate wheat flour | |||
| Lb. delbrueckii sp. lactis ATCC 12315 | 100 | 1.0 | Hydrolysate potato | [13, 14] | |
| 93 | 0.78 | Hydrolysate potato waste | |||
| Lb. delbrueckii IFO 3534 | 83 | 0.83 | Glucose | [13, 14] | |
| 55 | 0.55 | Glucose | |||
| Lb. delbrueckii MIX several strains | 85 | 0.87 | Hydrolysate maize + barley | [13, 14] | |
| 71 | 0.73 | Hydrolysate maize + barley | |||
| Lb. delbrueckii NCIM-2365 | 90 | 0.9 | Glucose | [13, 14] | |
| 75 | 0.75 | Glucose | |||
| Lb. delbrueckii sp. bulgaricus | 44 | 0.95 | Whey | [13, 14] | |
| 13 | 0.28 | Whey | |||
| Lb. delbrueckii sp. bulgaricus ATCC 11842 | 50 | 1.0 | Whey | [13, 14] | |
| 9.5 | 0.19 | Whey | [13, 14] | ||
| Lb. delbrueckii sp. bulgaricus Ch H 2217 | 115 | 0.86 | Whey | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus NRRL B-548 | 45 | 0.90 | Lactose | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus ATCC 55163 | 50 | 0.64 | Whey | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus ATCC 11842 | - | - | Sorghum | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus CNRZ 369 | 25 | 0.48 | Whey | [13, 14] | |
| Lb. delbrueckii sp. bulgaricus NRRL B-548 | 52 | 0.58 | Cellulose | [13, 14] | |
| Lb. delbreuckii | 35.4 | 0.35 | 0.75 | Alfalfa fibers | [157] |
| Lb. delbrueckii NCIM 2025 | 81.9 | 0.94 | 1.36 | Cassava bagasse | [164] |
| Lb. delbrueckii subsp. delbrueckii IFO 3202 | 28.0 | 0.28 | 0.78 | Defatted rice bran | [13, 14] |
| Lb. delbrueckii mutant Uc-3 | 67.0 | 0.83 | 0.93 | Sugarcane bagasse waste | [174] |
| Lb.delbrueckii ssp. lactis DSM 20073 | 9.9 | Glucose | [24] | ||
|
Lb. delbrueckii sp. delbrueckii ATCC 9649 |
0.82 | 1.6 | Wheat | [13, 14] | |
|
Lb. delbrueckii sp. bulgaricus ATCC 11842 |
0.11 | 0.56 | Wheat | [13, 14] | |
| Lb. delbrueckii NCIM 2025 | 1.36 | Cassava bagasse | [164] | ||
| Homo and Heterofermentative LAB | |||||
| Lb. delbrueckii ZU-S2 | 0.92 | 0.93–5.75 | Corn cob residue | [206] | |
|
Lb. delbrueckii subsp.delbrueckii Mutant Uc-3 |
0.83 | 0.93 | Sugarcane bagasse | [174] | |
| Lb. delbrueckii UFV H2B20 | 0.99 | 0.82 | Brewer's spent grain | [207] | |
| Lb. delbrueckii NRRL B-445 | 108.0 | 0.9 | Wood | [155] | |
| Lb. delbrueckii | 79 | 0.81 | 3.58 | Broken rice | [208] |
| Lb. delbrueckii | Camel milk | [209] | |||
| Lb. delbrueckii | Cow milk | [209] | |||
| Lb. delbrueckii | Rice | [210] | |||
| Lb. delbrueckii | Grain cellulosic hydrolysate | [211] | |||
| Lb. delbrueckii | 88 | Molasses | [125] | ||
| Lb. delbrueckii | Yucca | [164] | |||
| Lb. delbrueckii sp. delbrueckii | 83.45–93.28 | 1.57–3.7 | Orange waste enzymatic hydrolysates | [216] | |
| Lb. delbrueckii subsp. delbrueckii Mutant Uc-3 | 166 | 4.15 | Molasses | [123] | |
| Lb. delbrueckii | 107 | 0.9 | 1.48 | Sugarcane molasses, sugarcane juice and sugar beet juice | [13, 14] |
| Lb. delbrueckii spp. delbrueckii | 4.2–6.72 | 0.94 | Orange peel wastes hydrolysates | [212, 213] | |
| Lb. delbrueckii and B. amyloliquefaciens | 40 | 0.96 | 0.42 | Cassava bagasse | [214] |
| Lb. delbrueckii | 16.15 | 0.5 | 0.9 | Cassava fibrous waste hydrolysis | [215] |
| Lb .delbrueckii subsp. delbrueckii NBRC3202 | 25.38 | 1.18 | 0.53 | Kodo millet bran residue | [216] |
| Lb. delbrueckii sp. bulgaricus CICC21101 | 18 | Corn stover | [217] | ||
| Lb. delbrüeckii spp. bulgaricus | 26.56 | 0.540 | 0.553 | Cheese whey | [177] |
| Lb. helveticus sp. milano | 18 | 0.36 | Glucose | [13, 14] | |
| 42 | 0.84 | Maltose | |||
| Lb. helveticus ATCC 15009 | 17 | 0.38 | Lactose | [13, 14] | |
| 8.9 | 0.20 | Whey | |||
| Lb. helveticus Milano | 40 | 0.83 | Whey permeate | [13, 14] | |
| Lb. helveticus sp. milano | 44 | - | - | Hydrolysate whey | [13, 14] |
| 41 | - | - | Hydrolysate clarified whey | ||
| 37 | - | - | Whey, Ultrafiltration (UF) | ||
| Lb. helveticus ATCC 15009 | 49 | 1.1 | Whey | [13, 14] | |
| Lb. helveticus L89 | Whey | [13, 14] | |||
| Lb. helveticus ATCC 15009 | 65.5 | 0.66 | 2.7 | Cheese whey | [218] |
| Lb. helveticus | 10.1 | 0.23 | 5.1 | Cheese whey | [219] |
| Lb. helveticus NCDO 1844 | 47 | 1.2 | Cheese Whey | [13, 14] | |
| Lb. helveticus R211 | 38.0 | - | 19–22 | Cheese whey | [218] |
| Lb. helveticus | 10.5 | Cheese whey | [218, 219, 220] | ||
| Lb. helveticus R211 | 66.0 | 1.4 | Cheese whey | [13, 14] | |
| Lb. helveticus&K. marxianus, Lb. helveticus (mixed culture) | 15.5 | 0.45 | 10.0 | Cheese whey | [219] |
| Lb. helveticus&Lb. bulgaricus (mixed culture) | 14.6 | 0.35 | 9.4 | Cheese whey | [219] |
| Lb. helveticus&Lb. bulgaricus& K. marxianus (mixed culture) | 19.8 | 0.47 | 12.8 | Cheese whey | [219] |
| Lb. rhamnosus ATCC 10863 | 68.0 | 0.76 | Glucose | [13, 14] | |
| Lb. rhamnosusATCC 7469 | 28 | 0.93 | Glucose | ||
| Lb. rhamnosusDSM 20024 | 22 | 0.74 | Glucose | ||
| Lb. rhamnosus ATCC 7469 | 24 | 0.80 | Glucose | ||
| Lb. rhamnosus CCM 1753 | 37 | 0.74 | Lignocellulose hydrolysate | ||
| Lb. rhamnosus ATCC 7469 | 18 | 0.40 | Molasses | ||
| Lb. rhamnosus ATCC 7469 | 30 | 0.71 | Whey permeate | ||
| Lb. rhamnosus ATCC 10863 | 30 | 0.71 | Whey permeate | ||
| Lb. rhamnosus ATCC 7469 | 21 | 0.38 | Lactose | ||
| Lb. rhamnosus ATCC 10863 | 17 | 0.86 | Glucose | ||
| 14 | 0.71 | Fructose | |||
| 16 | 0.81 | Glucose + fructose | |||
| 15 | 0.73 | Sucrose | |||
| Lb. rhamnosus ATCC 10863 | 45 | - | - | Alpha-cellulose | |
| Homo and Heterofermentative LAB | |||||
| 28 | Switch grass cellulose | ||||
| Lb. rhamnosus ATCC 10863 | 16 | 0.81 | Hydrolysate molasses | ||
| Lb. rhamnosus ATCC 10863 | 58 | 0.95 | Glucose | ||
| Lb. rhamnosus ATCC 10863 | 29 | 1.00 | Hydrolysate wood | ||
| Lb. rhamnosus ATCC 11443 | 53 | 0.66 | Glucose | ||
| Lb. rhamnosus ATCC 7469 | 34 | 1.1 | Glucose | ||
| Lb. rhamnosus ATCC 10863 | 80 | 0.74 | Sucrose | ||
| 80 | 0.89 | Glucose | |||
| 38 | 0.76 | Glucose | |||
| 32 | 0.80 | Glucose | |||
| 79 | 0.79 | Glucose | |||
| 25 | 0.91 | Glucose | |||
| 771 | - | Glucose | |||
| 45 | Cellulose | ||||
| Lb. rhamnosus ATCC 9595 (CECT288) | 32.5 | 0.88 | 5.41 | Apple pomace | [13, 14] |
| Lb. rhamnosus CECT-288 | 42.0 | 0.38 | 0.87 | Cellulosic biosludge | [170] |
| Lb. rhamnosus ATCC 7469 | 73.0 | 0.97 | 2.9 | Paper sludge | [175] |
| Lb. rhamnosus ATCC 10863 | 67 | 0.84 | 2.5 | Glucose | [13, 14] |
| Lb.rhamnosus IFO 3863 | 0.53–0.77 | 2.90–13.15 | Glucose | [221] | |
| Lb. rhamnosus ATCC 9595 (CECT288) | 0.36–0.88 | 0.82–5.41 | Apple pomace, cellulosic biosludge |
[13, 14] | |
| Lb. rhamnosus ATCC 7469 | 0.97 | 2.9 | Paper sludge | [175] | |
| Lb. rhamnosus and Lb. brevis (mixed culture) | 20.95 | 0.70 | 0.58 | Corn stover | [122] |
| Lb. rhamnosus ATCC 7469 | 18.58 | 0.73 | – | Liquid distillery stillage | [222] |
| Lb. rhamnosus LA-04-1 | 82 | 0.81 | 3.73 | White rice bran hydrolysate | [223] |
| Lb. rhamnosus ATCC 7469 | 34.7 | 0.81 | 0.66 | Liquid distillery stillage | [222] |
| 42.2 | 0.99 | 1.22 | Liquid distillery stillage | [222] | |
| Lb. rhamnosus | Date juice | [133] | |||
| Lb.rhamnosus | Glucose | [224] | |||
| Lb. rhamnosus ATCC 7469 | 73.2–179 | 0.81 | 0.76 | Recycled paper sludge | [225] |
| Lb. rhamnosus ATCC-10863 | 60 | Softwood pre-hydrolysate and paper mill sludge | [226] | ||
| Lb. rhamnosus | 41.65 | 0.83 | 0.87 | Cassava wastewater | [227] |
| L. rhamnosus ATCC 7469 | 97.1 | 1.80 | Bread stillage | [200] | |
| Lb.rhamnosus HG09F5-27 | 157.22 | 8.77 | Yam tuber starch | [228] | |
| Lb rhamnosus 6003 | 45.5 | Food waste | [229] | ||
| Lb. rhamnosus | 22–40 | 76.9 | 1.22 | Solid carob waste | [230] |
| Lb. rhamnosus PCM 489 | 27.5 | Cheese industry – whey | [231] | ||
| Lb. rhamnosus B103 | 143.7 | Dairy industry waste | [232] | ||
| L. rhamnosus ATCC 7469 | 58.01 | 1.19 | Brewer's spent grain | [233] | |
| Lb. bulgaricus NRRL B-548 | 38.7 | 0.90 | 3.5 | Lactose, glucose, and galactose | [234] |
| Lb. bulgaricus ATCC 8001, PTCC 1332 | 24.6 | 0.81 | - | Cheese whey | [235] |
| Lb. bulgaricus CGMCC 1.6970 | 70.70–113.18 | 1.47–2.36 | Cheese whey powder | [236] | |
| Lb. bulgaricus | 19.5 | 1.22 | Cheese whey | [182] | |
| Lb. bulgaricus & K. marxianus (mixed culture) | 16.2 | 0.41 | 10.5 | Cheese whey | [13, 14] |
| Lb. casei NRRL B-441 | 82.0 | 0.91 | 5.6 | Glucose | [13, 14] |
| 120 | 0.67 | - | Hydrolysate barley flour | [13, 14] | |
| Lb. casei SU No 22 | 16 | 0.32 | Whey | [13, 14] | |
| 20 | 0.39 | Deproteinised whey | [13, 14] | ||
| Lb. casei NRRL B-441 | 112 | 0.68 | Liquefied barley starch + glucoamylase | [13, 14] | |
| 162 | 0.87 | Liquefied barley starch + glucoamylase + alpha-amylase | |||
| 36 | 0.20 | Barley flour | |||
| Lb. casei L100 | 50 | 0.83 | Corn starch | [13, 14] | |
| Lb. casei Shirota | 94 82.6 |
0.92 2.5 |
2.61 2.50 |
Mixed food waste bakery waste | [237] |
| Lb.casei CICC 6056 | 55.1 | 0.835 | 0.574 | Sophora flavescens residues | [238] |
| Lb.casei | 21.3 | 0.63 | Sugarcane bagasse | [239] | |
| Lb. casei SU No 22 | 45 | 0.45 | 2.0 | Whey | [13, 14] |
| Homo and Heterofermentative LAB | |||||
| Lb. casei | 22 | 0.44 | Whey | [13, 14] | |
| Lb. casei NRRL B-441 | 80 | 0.89 | Glucose | [13, 14] | |
| Lb. casei | - | 0.10 | 0.13 | Banana wastes | [168] |
| Lb. casei | 39.1–63.3 | 0.51–0.91 | Food waste (mango, orange, green peas and) | [240] | |
| Lb. casei subsp. rhamnosus NRRL-B445 and Lc. lactis subsp. lactis ATCC19435 | 60.3 | - | 3.20 | Date juice | [133] |
| Lb. casei ATCC 10863 | 44 | 0.44 | 1.22 | Ram horn hydrolysate | [241] |
| Lb. casei NRRL B-441 | 96.0 | 0.93 | 2.2 | Cheese whey | [182] |
| Lb. casei SU No. 22 and Lb. lactis WS 1042 (mixed culture) | 22.5 | 0.48 | 0.93 | Cheese whey | [13, 14] |
| Lb.casei subsp. casei CRL 686 | 0.97 | Glucose | [13, 14] | ||
| Lb.casei NRRL B-441 | 0.74–1 | 3.5–5.6 | Glucose | [242] | |
| Lb.casei LA-04-1 | 0.90 | 2.14 | Glucose | [242] | |
| Lb. casei NRRL B-441 | 0.93 | 2.5–3.97 | Cheese whey | [13, 14] | |
| Lb. casei NCIMB 3254 | 1.40 | Cassava bagasse | [164] | ||
| Lb. casei NRRL B-441 | 162.0 | 3.4 | Barley | [13, 14] | |
| Lb. casei | 33.73 | Whey | [13, 14, 243] | ||
| Lb. casei | Molasses | [148] | |||
| Lb. casei A-8 | ~130 | Reuse of anaerobic digestion effluent | [244] | ||
| L. casei | Yucca | [164] | |||
| Lb. casei M-15 | Molasses | [129] | |||
| Lb. lactis ATCC 4797 | 12.5–24.3 | Casein whey permeate | [245] | ||
| L. lactis | Molasses | [246] | |||
| L. lactis | Pineapples syrup | [246] | |||
| L. lactis WS 1042 | 11 | 0.22 | Whey | [13, 14, 243] | |
| L. lactis sp. lactis 2432 | 8.3 | 0.21 | Whey permeate | ||
| L. lactis sp. cremoris 2487 | 37 | 0.88 | 4.6 | Whey permeate | |
| L. lactis sp. lactis 5085 | 37 | 0.88 | Whey permeate | ||
| L. lactis WS 1042 | 15 | 0.30 | Deproteinised whey | ||
| L. lactis sp. lactis 2432 | 9.0 | 0.20 | Whey permeate | ||
| L. lactis sp. cremoris SBT 1306 | 80 | 1.5 | Lactose | ||
| L. lactis sp. lactis ATCC 19435 | 96 | 0.76 | Hydrolysate wheat flour | ||
| L. lactis sp. lactis AS211 | 95 | 0.77 | Hydrolysate wheat flour | ||
| L. lactis sp. lactis NRRL B-4449 | 6.6 | 0.16 | Waste paper | ||
| L. lactis IO-l JCM 7638 | 23 | 0.45 | Xylose | ||
| 28 | 0.70 | Xylose + glucose | |||
| L. lactis sp. lactis ATCC 13673 | 36 | 1.0 | Glucose | ||
| 13 | 0.42 | Xylose | |||
| L. lactis sp. lactis ATCC 19435 | 4.9 | 0.86 | Glucose | ||
| 3.2 | 0.70 | Maltose | |||
| L. lactis sp. lactis NRRL B-4449 | 6.6 | 0.66 | Glucose | ||
| 2.8 | 0.28 | Galactose | |||
| 5.8 | 0.58 | Mannose | |||
| 1.8 | 0.18 | Xylose | |||
| 0.16 | Hydrolysate cellulose + glucose + mannose + xylose + galactose | ||||
| L. lactis IFO 12007 + Aspergillus awamori IFO 4033 | 25 | 0.50 | Potato starch | [13, 14] | |
| L. lactis IO-l JCM 7638 | 24 | 0.96 | Glucose | ||
| L. lactis sp. lactis AS211 | 107 | 0.91 | Hydrolysate wheat flour | ||
| L. lactis sp. lactis ATCC 19435 | 106 | 0.88 | Hydrolysate wheat flour | ||
| L. lactis sp. lactis ATCC 19435 | 90 | 0.98 | Hydrolysate wheat flour | ||
| 75 | 1.0 | Un hydrolysate wheat flour + glucose | |||
| 53 | Hydrolysate wheat flour | ||||
| L. lactis 65.1 | 39 | 0.75 | Glucose | ||
| L. lactis IFO 12007 | 25 | 0.50 | Potato starch | ||
| L. lactis sp. lactis ATCC 19435 | 65 | 1.5 | Glucose | ||
| L. lactis sp. lactis ATCC 19435 | 0.3 | 0.3 | Glucose | ||
| L. lactis IO-l JCM 7638 | 45 | 0.90 | Glucose | ||
| Homo and Heterofermentative LAB | |||||
| L. lactis 65.1 | 5.7 | 1.1 | Glucose | ||
| L. lactis IO-l JCM 7638 | 45 | 0.90 | Glucose | ||
| 66 | 0.88 | Glucose | |||
| L. lactis sp. lactis ATCC 19435 | 5.4 | 0.92 | Glucose | [149] | |
| 5.1 | 1.0 | Maltose | |||
| 96 | 0.76 | Hydrolysate wheat flour | [13, 14] | ||
| L. lactis sp. lactis biovar diacetylactis CNRZ 2125 | 38 | 0.73 | Lactose + citrate | ||
| L.lactis BME5-18 M | 0.97 | 2.2 | Glucose | [83] | |
| L. lactis IO-1 | 4.5 | Glucose | [247] | ||
| L. lactis sp. lactis | |||||
| ATCC 19435 | 0.76 | 3.0 | Wheat | [13, 14] | |
| L. lactis sp. lactis | |||||
| IFO 12007 | 0.76 | 0.6 | Cassava | [248] | |
| L. lactis sp. lactis | |||||
| AS211 | 0.77 | 1.7 | Wheat | [13, 14] | |
| L. lactis ATCC19435 | 92.5 | 0.68 | 0.5 | Artichoke hydrolysate | [249] |
| L. lactis IL 1403/pCUSαA | 15.6 | 0.89 | 1.57 | Soluble starch | [13, 14] |
| L. lactis IO-1 | 10.9 | 0.36 | 0.17 | Sugar cane baggage | [165] |
| Lb. lactis ssp. lactis IFO 12007 | 90.0 | 0.76 | 1.6 | [248] | |
| Lb. lactis NCIM 2368 | 17.01–72.24 | Glucose | [250] | ||
| Lb. plantarum NRRL B-787 | 17 | 0.42 | Solid waste | [13, 14] | |
| Lb. plantarum NRRL B-788 | 19 | 0.46 | Solid waste | ||
| Lb. plantarum NRRL B-813 | 18 | 0.43 | Solid waste | ||
| Lb. plantarum NRRL B-531 | 18 | 0.43 | Solid waste | ||
| Lb. plantarum | 17 | 0.70 | Corn syrup | [13, 14] | |
| Engineered Lb. plantarum NCIMB 8826 (GMO) | 73.2–141.9 | 0.9–0.93 | 2.95 | Glucose and xylose | [251] |
| Lb. plantarum | 15 | 0.30 | Hydrolysate soluble starch | [13, 14] | |
| Lb. plantarum | 15 | 0.30 | Hydrolysate tapioca starch | ||
| Lb. plantarum NRRL B-531 | 5.4 | 0.54 | Glucose | [13, 14] | |
| 3.7 | 0.37 | Galactose | |||
| 5.7 | 0.57 | Mannose | |||
| 0.43 | Hydrolysate cellulose: glucose, mannose, xylose, galactose | ||||
| Lb. plantarum NRRL B-787 | 6.2 | 0.62 | Glucose | ||
| 4.0 | 0.40 | Galactose | |||
| 6.6 | 0.66 | Mannose | |||
| 0.42 | Hydrolysate cellulose: glucose, mannose, xylose, galactose | ||||
| Lb. plantarum NRRL B-788 | 6.0 | 0.60 | Glucose | ||
| 4.9 | 0.49 | Galactose | |||
| 0.46 | Hydrolysate cellulose: glucose, mannose, xylose, galactose | ||||
| Lb. plantarum NRRL B-813 | 7.3 | 0.73 | Glucose | ||
| 4.7 | 0.47 | Galactose | |||
| 8.3 | 0.83 | Mannose | |||
| 0.43 | Hydrolysate cellulose: glucose, mannose, xylose, galactose | ||||
| Lb. plantarum USDA 422 | 5.2 | 0.52 | Glucose | ||
| 3.1 | 0.31 | Galactose | |||
| 6.2 | 0.62 | Mannose | |||
| 1.3 | 0.13 | Xylose | |||
| Lb. plantarum | 46.4 | 0.46 | 0.64 | Alfalfa fibers | [252] |
| Lb paracasei (NBRC 15889) | ~100 | Brown rice polish | [161] | ||
| Lb.uvarum | 139.71 | ||||
| Lb farraginis (NRIC 0676) | ~125 | ||||
| Lb brevis | 160.97 | ||||
| Lb plantarum (WCFS1) | 137.67 | ||||
| Homo and Heterofermentative LAB | |||||
| Lb plantarum (JCM 1149) | ~115 | ||||
| Lb. plantarum A6 | 8.41 | 0.98 | - | Mussel processing wastes | [13, 14] |
| Lb. plantarum ATCC 21028 | 41.0 | 0.97 | 1.0 | Synthetic lactose medium | [13, 14] |
| Lb. plantarum NCIMB 8826 | 73.2 | 0.85 | 3.86 | Corn starch | [253] |
| Lb. plantarum | Bamboo | [254] | |||
| Lb. plantarum A6 | 86.6 | 0.89 | 4.54 | Glucose | [255] |
| Lb. plantarum ΔldhL1 | 73.2 | 0.85 | 3.86 | Raw starch | [255] |
| Lb. plantarum ΔldhL1/pCU-CelA | 1.27 | - | - | Cellohexaose | [253] |
| Lb. plantarum ΔldhL1/pCU-CelA | 1.47 | - | - | β-glucan | |
| Lb. plantarum ΔldhL1-xpk1:tkt | 38.6 | 0.82 | 3.78 | Arabinose | |
| Lb. plantarumΔldhL1-xpk1: tkt-Δxpk2/pCU-PXylAB | 41.2 | 0.89 | 1.60 | Xylose | |
| Engineered Lb. Plantarum NCIMB 8826 (GMO) | 55.2–102.3 | 0.879 | 1.77–2.61 | Hardwood pulp, barley extract | [256] |
| Lb. plantarum | 28.45–34.19 39.72–42.34 |
0.87–0.94 0.93–0.99 |
4.57–14.22 7.56–9.93 |
Glucose Hydrolysate of microalga Chlorella vulgaris ESP-31 |
[257] |
| Lb plantarum BP04 | 57.5 | Dining-hall food waste | [201] | ||
| Lb. plantarum | 117.1 | 0.81 | Brown rice | [258] | |
| Lb. plantarum DldhL1: PxylABxpk1: tkt-Dxpk2: PxylAB | 39.7–74.2 | 0.78–0.79 | 1.53–2.85 | Glucose/xylose mixture | [259] |
| Lb. plantarum NCDC 414 | Vegetable juices | [260] | |||
| Lb. amylovorus ATCC 33620 | 4.2 | 0.1 | Potato | [140] | |
| Lb. amylophilus GV6 | 76.2 | 0.70 | 0.8 | [146] | |
| Lb. amylovorus ATCC 33622 | 93 | 0.52 | Hydrolysate barley flour | [13, 14] | |
| Lb. amylophilus ATCC 49845 | 21 | 0.95 | Glucose | ||
| 33 | 0.73 | Hydrolysate corn starch | |||
| Lb. amylovorus ATCC 33620 | 4.8 | 0.48 | Cassava starch | ||
| 10 | 1.0 | Corn starch | |||
| 4.2 | 0.42 | Potato starch | |||
| 7.9 | 0.79 | Rice starch | |||
| 7.8 | 0.78 | Wheat starch | |||
| Lb. amylovorus ATCC 33622 | 45 | 0.82 | Raw corn starch | ||
| Lb. amylovorus NRRL B-4542 | 114 | 0.63 | Barley flour + gluco amylase | ||
| Lb. amylophilus ATCC 49845 | - | - | Glucose | ||
| Lb. amylophilus ATCC 49845 | 30 | 0.60 | Starch | ||
| Lb. amylophilus GV6 | 27.3 | 0.3 | Barley | ||
| Lb. amylophilus BCRC 14055 | 21.62 | 0.98 | 0.31 | Starch | [261] |
| Lb. amylophilus | Corn | [146] | |||
| Lb. amylophilus | Potato | [146] | |||
| Lb. amylophilus | Wheat (bran or flour) | [143] | |||
| Lb. zeae ATCC 393 | 21 | 0.71 | Glucose | [13, 14] | |
| Lb. zeae ATCC 393 | 37 | 0.98 | 5.0 | Glucose | |
| Lb. salivarius sp. salivarius ATCC 11742 | 28 | 0.92 | Glucose | ||
| Str. thermophilus | 18 | 0.50 | Whey permeate | ||
| Str. thermophilus | 15 | 0.35 | Whey permeate | ||
| Str. thermophilus | 19 | 0.47 | Whey permeate | ||
| Str. thermophilus CRL 807 | 8.5 | Skim milk | |||
| Str. thermophilus | 40 | Lactose | |||
| Str. thermophilus | 24.18–39.71 | 0.55–0.80 | Magazine and office paper | [262] | |
| Lb. coryniformis ssp. torquens ATCC 25600 | 24.0 | 0.5 | Cellulose | [154] | |
| Lb. coryniformis ssp torquens ATCC | |||||
| 25600 | 23.1 | 0.51 | 0.48 | Cardboard waste | [154] |
| Lb. coryniformis ssp. torquens ATCC 25600 | 39 | 0.98 | 2.6 | Glucose | [13, 14] |
|
Lb. coryniformis Lb paracasei |
91.6–97.1 | 0.91–0.96 | 2.08–2.7 | Curcuma longa waste (food waste) | [263] |
| Lb. coryniformis subsp. torquens | 57.0 | 0.97 | 2.8 | Pulp mill residue | [264] |
| Lb. coryniformis sub. Torquens ATCC 25600 | 36.6 | 0.46 | 1.02 | Hydrodictyon reticulum | [199] |
| Lb. coryniformis sp. torquens ATCC 25600 | 23.4 | 0.51 | 0.49 | Waste cardboard | [154] |
| Lb. kefir | 9.8 | 0.20 | Paneer whey | [13, 14] | |
| Lb. acidophilus R | 8.6 | 0.17 | Paneer whey | ||
| Homo and Heterofermentative LAB | |||||
| Lb. acidophilus CRL 640 | 14 | Skim milk | [13, 14] | ||
| E. faecium | 11 | 0.45 | Hydrolysate cod + corn syrup | [13, 14] | |
| E. faecium | 27 | 0.91 | Alfalfa | [13, 14] | |
| E. faecalis RKY1 | 144.0 | 0.96 | 3.56–6.20 | Glucose | [136, 265] |
| E. faecium No. 78 | 3.04 | Sago | [266] | ||
| E. faecalis RKY1 | 0.93–1.04 | 0.5–4.8 | Corn, wheat, tapioca, potato | [136, 267] | |
| E. faecalis RKY1 | 1.7 | Wood | [268] | ||
| E. faecalis QU 11 | 55.3 | 0.991 | Glycerol | [269] | |
| E. faecalis RKY1 | 95.7 | 4.0 | Molasses | [140] | |
| E. faecalis RKY1 | 93.0 | 1.7 | Wood | [140] | |
| E. faecium No. 78 | 36.3 | 0.57 | 1.96 | Liquefied sago starch | [270] |
| E. faecalis RKY1 | 92–94 | – | 6.03–6.2 | Glucose | [136] |
| E. faecalis RKY1 | 48.0 | 0.92 | 4.0 | Wood hydrolyzate | [271] |
| E. durans BP130 | 28.8 | 0.85 | 0.24 | Food waste | [12] |
| E. mundtii QU 25 | 67.2–129 | 0.78–0.90 | 0.76–1.2 | Glucose/xylose mixture | [272] |
| E. faecium strain FW26 | 33.3 | 0.84 | Banana peels and food wastes mixture | [273] | |
| Ped. acidilacti | 13 | 0.51 | Hydrolysate cod + corn syrup | [13, 14] | |
| Engineered Pediococcus acidilactici | 87.8–104.5 | 1.22–1.45 | Corn stover feedstock | [236] | |
| Lb. plantarum NRRL | |||||
| B-4496, Lb. acidophilus NRRL B-4495, and L. reuteri B-14171 | Egg white hydrolysates | [274] | |||
| Lb. manihotivorans LMG18011 | 48.7 | 0.098 | 0.76 | Food wastes | [162] |
| Lb. pentosus NRRL B-227 | 21 | 0.51 | Solid waste | [13, 14] | |
| Lb. pentosus NRRL B-473 | 18 | 0.43 | Solid waste | ||
| Lb. pentosus | 46 | 0.92 | Glucose | ||
| 27 | 0.54 | Xylose | |||
| 90 | 1.8 | Glucose + xylose | |||
| 40 | 0.70 | Hydrolysate wood | |||
| Lb. pentosus NRRL B-473 | 6.9 | 0.69 | Glucose | ||
| 5.9 | 0.59 | Galactose | |||
| 7.4 | 0.74 | Mannose | |||
| 1.4 | 0.14 | Xylose | |||
| 0.43 | Hydrolysate cellulose: glucose + xylose + mannose + galactose | ||||
| Lb. pentosus ATCC 8041 | 21.8 | 0.77 | 0.84 | Vine-trimming wastes | [163] |
| Lb. sakei KTU05-06, Pediococcus acidilactici + KTU05-7 + P. pentosaceus KTU05-9 | 40.0–93.0 | 0.62–1.45 | 0.83–1.94 | Wheat bran | [275] |
| 28.4–54.6 | 0.50–0.97 | 0.59–1.14 | Spent distiller's grain with solids | ||
| 11.3–33.4 | 0.33–0.98 | 0.24–0.70 | Brewer's spent grain | ||
| Lb. pentosus ATCC-8041 | 23.0 | 0.93 | 0.45 | Nannochloropsis salina | [110] |
| Lb. pentosus CHCC 2355 | 0.88 | Wheat straw | [158] | ||
| Lb. pentosus ATCC 8041 | 0.65–0.77 | 0.1–0.9 | Vine-trimming wastes/Corn Stover |
[152, 158] | |
| Lb. pentosus | Grape marc | [276] | |||
| Lb. pentosus | Wheat straw | [158] | |||
| Lb. pentosus CECT4023T | 21 | 0.48–0.7 | Gardening lignocellulosic residues | [277] | |
| Lb. pentosus CECT-4023T (ATCC-8041) | 46 | 0.78 | 0.933 | Hemicellulosic hydrolyzates from trimming wastes of vine shoots | [278] |
| Lb. paracasei LA1 | 23.4 | 0.72 | 0.23 | Wastewater sludge | [176] |
| Lb. paracasei LA104 | 37.11 | 0.46 | 1.03 | Hydrodictyon reticulum | [199] |
| Lb. paracasei No. 8 | 81.5 | 2.7 | Sweet sorghum | [13, 14] | |
| Lb. paracasei No. 8 | 84.5 | 2.4 | Rye | [13, 14] | |
| Lb. paracasei No. 8 | 106.0 | 3.5 | Sweet sorghum | [13, 14] | |
| Lb. paracasei NCBIO01-M2 | 223.7 | 5.53 | Glucose | [279] | |
| Lb.paracasei | 169.9 | 1.42 | Molasses enriched potato stillage | [280] | |
| Lb. paracasei DSM 23505 | 123.7 | 0.91 | Chicory flour | [281] | |
| L. paracasei A-22 | 80.10 | 0.97 | 1.48 | Agro-industrial waste such as sunflower seed hull, brewers' spent grain, and sugar beet pulp | [282] |
| Lb. paracasei subsp. paracasei CHB2121 | 192 | 0.96 | 3.99 | Glucose | [283] |
| Lb. paracasei KCTC13169 | 92.5 | 0.9 | 8 1.2 | Artichoke tuber extract | [284] |
| Lb. sp. RKY2 | 129.0 | 2.9 | Rice | [140] | |
| Homo and Heterofermentative LAB | |||||
| Lb. sp. RKY2 | 3.1 | Rice and wheat bran | [140] | ||
| Lb.sp. strains A28a | ~52.4 | 0.07 | 0.27 | Mixed food waste | [285] |
| 0.22 | 0.27 | Starch | |||
| 0.14 | 0.27 | Sugar | |||
| Lb.sp. strains A59 | 0.14 | 0.53 | Mixed food waste | ||
| 0.43 | 0.53 | Starch | |||
| 0.29 | 0.53 | Sugar | |||
| Lb.sp. strains A211 | 0.14 | 0.37 | Mixed food waste | ||
| 0.41 | 0.37 | Starch | |||
| 0.24 | 0.37 | Sugar | |||
| Lb. brevis ATCC 14869 | 12.5 | 0.57 | 0.56 | Glucose, xylose or a glucose/xylose mixture | [286] |
| Lb. rhamnosus + L. brevis (mixed culture) | 14.8 | 0.73 | 0.4 | Glucose/xylose mixture | [287] |
| Lb. brevis | 15 | 0.22 | Cottonseed cake, wheat straw, sugarcane bagasse | [288] | |
| 10 | 0.49 | ||||
| 12.5 | 0.52 | ||||
| Lb. brevis and Lb. plantarum | ~15–35 | 0.52–0.8 | Lignocellulosic biomass | [289] | |
| Lb. brevis CHCC 2097 and Lb. pentosus CHCC 2355 | 7.1 | 0.95 | - | Wheat straw | [158] |
| Exiguobacterium sp. strain 8-11-1 | - | - | 8.15 | [290] | |
| Lb. bifermentans DSM 20003 | 0.83 | 1.17 | Wheat straw | [159] | |
| Halolactibacillus halophilus JCM 21694 | 65.8 | 0.83 | 1.1 | Sucrose | [291] |
| Lb. sp. G-02 and Aspergillus niger SL-09 (mixed culture) | 120.5 | 0.95 | 3.3 | Artichoke tubers | [91] |
| Sporolactobacillus sp. strain CASD | 207 | 0.93 | 3.8 | Peanut meal and glucose | [28] |
| Sporolactobacillus inulinus YBS1-5 | 107.2 | 0.85 | 1.19 | Corncob residues & cottonseed meal | [292] |
| Sporolactobacillus inulinus YBS1-5 | 87.3–99.5 | 0.65–0.89 | 0.81–1.94 | Wheat bran | [293] |
| Sporolactobacillus sp. strain CASD | 82.8 | 0.94 | 1.72 | Glucose | [40] |
| Sporolactobacillus inulinus | 93.4 | 1.37 | Glucose | [294] | |
| Sporolactobacillus inulinus YBS1-5 | 70.5 | 0.65 | Corn stover | [295] | |
| Sporolactobacillus laevolacticus DSM442 | 144.4 | 4.13 | Cotton seed | [296] | |
| Lb. sp. G-02 | 141.5 | 0.94 | 4.7 | Artichoke tubers | [297] |
| Lb. sp. RKY2 | 94.06 | 0.98 | 1.06 | Cheese whey | [184] |
| Lb. TY50 | 36.29 | ND | Kitchen waste | [298] | |
| Lb. sp. | 23.21 | Food waste + cu+2 | [201] | ||
| Lactobacillus sp. B2 | 19.5L | 0.81 | Crustacean waste | [299] | |
|
Lb. paracasei ATCC 334 |
1.2 | 1 | Chlorella | [300] | |
| Lb. lactis subsp. lactis NBRC 12007 | 0.8 | 1 | |||
| Lb. reuteri JCM 1112 | 1.02–4.29 | Glucose-sucrose | [301] | ||
| Lactococcus lactis JCM 7638 | Glucose-sucrose | ||||
| Lb. gasseri NCIMB 11718 | 8.42–18.7 | Glucose-sucrose | |||
| Lb. plantarum NCIMB 8826 | Glucose-sucrose | ||||
| Lb. paracasei ATCC 334 | 8.01–12.3 | Glucose-sucrose | |||
| 5.17–7.03 | |||||
| 7.77–9.60 | |||||
| Lb. paracasei 7B | 52.61 | 0.96 | 2.25–3.23 | Wood ligonocellulosic hydrolysate | [302] |
| Lb. paracasei h601 | 21.19 | ||||
| Lb. plantarum A1 | 41.91 | ||||
| Lb. plantarum K1 | 25.22 | ||||
| Lb. plantarum N14-2 | 36.95 | ||||
| Lb. fermentum h602 | 31.11 | ||||
| Lb. fermentum ATCC 14931 | 12.99 | ||||
| Lb. fermentum E1 | 5.91 | ||||
| Lb. brevis ATCC 8287 | 39.15 | ||||
| B. coagulans T10-2 | 13.44 | ||||
| B. coagulans T5-1 | 4.43 | ||||
| W. paramesenteroides H1-6 | 18.49 | ||||
| Homo and Heterofermentative LAB | |||||
| Lb. points (32%), Lb. frumenti (10%), Lb. acidophilus (8%), Lb. amylovorus and Bifidobacterium (mixed culture) | 10–20 | Acidogenic fermentation of fruit and vegetable wastes | [303] | ||
| Lb plantarum + Lb buchneri, + Lb rhamnosus; Lb. plantarum + Lb paracasei | 30.4–127.9 | Maize and amaranth | [304] | ||
| Lb. manihotivorans LMG18011 | 48.7 | 1.11 | Starch and food waste | [162] | |
| Lb. rhamnosus & B.coagulans | 112.5 | 0.88 | 2.74 | Cassava bagasse | [305] |
| Lb. delbrüeckii spp. bulgaricus | 31.70 | 0.645 | 0.660 | Hydrolysed cheese whey | [177, 275] |
| P. acidilactici KTU05-7 | 24.54 | 0.499 | 0.511 | ||
| P. pentosaceus KTU05-8 | 21.45 | 0.396 | 0.447 | ||
| P. pentosaceus KTU05-9 | 25.49 | 0.519 | 0.531 | ||
| P. pentosaceus KTU05-10 | 19.46 | 0.396 | 0.405 | ||
| P. acidilactici KTU05-7 | 27.86 | 0.567 | 0.580 | ||
| P. pentosaceus KTU05-8 | 25.21 | 0.513 | 0.525 | ||
| P. pentosaceus KTU05-9 | 28.06 | 0.571 | 0.584 | ||
| P. pentosaceus KTU05-10 | 22.82 | 0.464 | 0.475 | ||
| P. acidilactici | 97.3 | 0.95 | Corn stover | [306] | |
| P. acidilactici ZP26 | 77.66 | 1.06 | Corn stover | [307] | |
| Pediococcus acidilactici (DSM, 20284) | ~125 | Brown rice polish | [161] | ||
| Pediococcus pentosaceus (ATCC 25745) | ~65 | ||||
| Lb. buchneri NRRL B-30929 | 13.35 | Elephant grass | [308] | ||
| E. casseliflavus/Lb. casei (mixed culture) | 95 | 0.63 | 0.49 | Glucose/xylose mixture | [309] |
| Actinobacillus succinogenes | 183.4 | 0.97 | 1.53 | Glucose | [310] |
| Pediococcus acidilactici TM14 and Weissella paramesenteroides TA15 | Food waste composting | [311] | |||
| Weissella sp. S26/Bacillus sp.ADS3 | 13.2 | Xylose | [312] | ||
| Enterobacter aerogenes ATCC 29007 | 46.02 | 0.41 | Mannitol | [313] | |
| Thermoanaerobacterium aotearoense LA1002-G40 | 78.5 | 0.85 | 1.63 | Mixed bakery waste | [314] |
| Lb. sanfranciscensis MR29 | 2.85 | 0.057 | Wheat straw biomass | [315] | |
| Lb. rossiae GL14 | 0.96 | 0.0192 | |||
| Lb. frumenti H10 | 1.90 | 0.038 | |||
| Lb. rossiae M2 | 1.54 | 0.0308 | |||
| Lb. crustorum W19 | 2.94 | 0.058 | |||
| Lb. sanfranciscensis MW15 | 4.56 | 0.0988 | |||
| Lb. helveticus DSM 20075 | 2.03 | 0.0406 | |||
| Lb. delbrueckii subsp. bulgaricus MI | 4.74 | 0.0948 | |||
| Lb. delbrueckii subsp. bulgaricus DSM 20081 | 4.81 | 0.0962 | |||
| Leuconostoc mesenteroides NRRL B 512 | 60.2 | 1.25 | Sugarcane juice | [316] | |
| B. coagulans LA1507 and Lactobacillus rhamnosus LA-04-1 (Mixed culture) | 118 | 1.84 | Sweet sorghum juice | [317] | |
| Engineered Pediococcus acidilactici | 130.8 | 1.82 | Wheat straw | [318] | |
| Streptococcus sp.(indigenous consortium) | 50–69 | 1.27–2.93 | Highly viscous food waste | [319] | |
| Streptococcus sp. | 66.5 | 0.33 | 3.38 | Mixed food waste | [320] |
|
Bifidobacterium longum |
0.51 |
Cheese whey |
[321, 322] |
||
|
Bacillus strains | |||||
|
B. coagulans | |||||
| B. coagulans | 20.1 | 0.60 | 0.93 | Sucrose | [4] |
| B. coagulans 36D1 | 80 | 0 0.80 | 0.30 | Cellulose | [151] |
| B. coagulans strains 36D1 | 92.0 | 0.77 | 0.96 | Paper sludge | [20] |
| B. coagulans strains P4–102B | 91.7 | 0.78 | 0.82 | Paper sludge | [20] |
| B. coagulans SIM-7 DSM 14043 | 0.96 | 9.9 | Glucose | [24] | |
| B. coagulans DSM 2314 | 0.27 | Wheat straw | [323] | ||
| B. coagulans strain 36D1 | 103.6 | 0.93 | 0.71 | Glucose | [151] |
| B. coagulans strain 36D1 | 102.3 | 0.86 | 0.71 | Xylose | |
| B. coagulans NBRC 12583 | 2 | Sludge hydrolyzate | [324] | ||
| Alkaliphilic Bacillhilic | Sugars | [13, 14] | |||
| B. coagulans strain IPE22 | 46.12 | Wheat straw | [33] | ||
| B. coagulans C106 | 83.6–215.7 | 4–7.5 | Xylose | [325] | |
| B. coagulans NBRC12583 | Kitchen refuse | [27] | |||
| Bacillus strains | |||||
| B. coagulans | 60.7 | 0.71 | 2.68 | Municipal solid wastes |
[112] |
| B. coagulans DSM2314 | 58.7–70.4 | 0.83–0.73 | 1.14–1.81 | Sugarcane bagasse | [326] |
| B. coagulans | 79.4–93.7 | Glucose, xylose and cellobiose | [327] | ||
| B. coagulans BCS13002 | 11.75 | Gelatinized corn starch | [328] | ||
| 0.26 | Corn starch | ||||
| B. coagulans | 99.1 | 1.38 | Glucose | [329] | |
| B. coagulans | 145 | 1.5 | Glucose | [330] | |
| B. coagulans | 110 | 0.86 | 1.29 | Cassava bagasse | [304] |
| B. coagulans MA-13 | 29.7–33.7 | 0.92 | Lignocellulosic hydrolysate |
[331] | |
| B. coagulans JI12 | 0.97 | Oil palm empty fruit bunch hydrolysate | [332] | ||
| B. coagulans WCP 10-4 | 210 | 0.955 | 3.5 | Glucose or corn starch | [333] |
| B. coagulans C106 | 83.6 | 0.983 | 7.5 | Xylose | [334] |
| B. coagulans strainIPE22 | 38.73 | 0.813 | 0.39–0.65 | Pretreated wheat straw | [335] |
| B. coagulans | 0.94 | 0.33 | Corn stover hydrolysate | [336] | |
| B. coagulans | 165.7 168.3 |
0.92 0.88 |
1.6 2.1 |
Glucose Glucose/Cane molasses |
[337] |
| B. coagulans strain AD | 1.4 | 3.69 | Corn stover hydrolysate | [338] | |
| B. coagulans strain IPE 22 | 7.52–56.13 | 0.13–0.94 | 0.31–2.77 | Single sugar (glucose, xylose, arabinose) | [339] |
| 49.14–51.47 | 0.82–0.86 | 2.05–3.08 | Mixed sugar (glucose + xylose + arabinose) | ||
| 50.48–53.51 | 0.89–0.92 | 2.97–3.16 | Corn cob hydrolysate | ||
| B. coagulans L-LA 1507 | 78–97.5 | 0.325–0.406 | 1.25–3.25 | Corn stover | [340] |
| B. coagulans AT107 | 98.8 | 0.80–0.92 | 1.25–3.15 | Alfalfa green juices and clover green juice | [341] |
| B. coagulans | 79.1 | 0.76 | Lignocellulosic corncob residue | [342] | |
| B. coagulans | 92.5 | 0.578 | 2.01 | Dilute ethylediamine pre-treated rice straw | [343] |
| B. coagulans + B. thermoamylovorans. | 39.2 | 1.09 | Kitchen refuse medium | [118] | |
| B. coagulans IPE22 | 68.72 | 0.99 | 1.72 | Inedible starchy biomass | [344] |
| B. coagulans LA-15-2 | 117 | 2.79 | White rice bran | [345] | |
| B. coagulans A166 | 61.1 | 0.94 | Municipal solid waste | [346] | |
| B. subtilis ZM63, B. cereus, Paenibacillus polymyxa and B. cereus |
Glucose + Zn+2 |
[205] |
|||
|
B. licheniformis | |||||
| B. licheniformis TY7 | 40.0 | - | 2.50 | Kitchen refuse | [27] |
|
B. licheniformis TY7 |
24–40 |
1.29–1.35 |
Kitchen refuse |
[34, 347] |
|
|
B. subtilis | |||||
|
B. subtilis MUR1 (mutant) |
143.2 |
90.3 |
2.75 |
Glucose |
[36] |
|
B. sp. | |||||
| B. longum NCFB 2259 | 0.51–0.82 | 0.3–0.7 | Cheese whey | [181, 348] | |
| B. sp.36D1 | 0.60 | Sugar cane bagasse | [349] | ||
| B. sp. Na-2 | 106 | 0.94 | 3.53 | Glucose | [38] |
| B. sp. WL-S20 | 225 | 0.993 | 1.04 | Peanut meal and glucose | [16] |
| 180 | 0.98 | 1.61 | Peanut meal and glucose | [16] | |
| B. sp. 2-6 | 107 | 0.95 | 2.9 | Glucose | [40] |
| B. sp. Na-2 | 118 | 0.97 | 4.37 | Glucose | [39] |
|
B. sp. P38 |
180 |
0.96 |
2.4 |
Cellulosic hydrolysate |
[37] |
|
E. coli | |||||
| Engineered E. coli | 60–62.2 | 0.80–0.90 | Glucose | [348] | |
| Engineered E. coli | 45.5–51.8 | 0.91–0.99 | Glucose | [52] | |
| Engineered E. coli | 40 | 0.93 | Xylose | [57] | |
| Engineered E. coli | 56.8 | 0.88 | 0.94 | Glycerol | [350] |
| E. coli AC-521 | 85 | 0.85 | 1.0 | Sucrose | [54] |
| E. coli K12 strain | 32 | 0.85 | 0.44 | Glycerol | [59] |
| E. coli | 75 | 0.85 | 1.18 | Molasses | [351] |
| lactogenic Escherichia coli strain JU15 |
40 | 0.6 | Corn stover | [352] | |
| E. coli BW25113 (DpflA) (engineered) | 5.2 | 22.5 | 0.06 | cellobiose | [353] |
| 4.3–5 | 0.22–0.25 | ||||
| 5.3 | 29.6 | 0.11 | Glucose | ||
| E. coli | |||||
| E. coli MG1655-LA02Δdld (engineered) | 45 | 0.83 | 0.5 | Glycerol | [59] |
| E. coli strain CICIM B0013-070 (pUC-ldhA) (engineered) | 111.5 | 0.78 | 2.80 | Glycerol | [354] |
| Engineered E. coli | 50 | 0.90 | 0.60 | Glycerol | [53] |
| Engineered E. coli RR1 |
62.6 |
Glucose |
[13, 14] |
||
|
Corynebacteria glutamicum | |||||
| C. glutamicum | 120 | 0.865 | ~. 4.0 | Glucose | [48] |
| C. glutamicum | L-arabinose | [45] | |||
| C. glutamicum | Xylose | [46] | |||
| C. glutamicum | Glucose, fructose, sucrose, ribose | [355] | |||
| C. glutamicum | 60.27 | D-ribose | [51] | ||
|
Achromobacter denitrifleans NBRC 12669 |
3.9 |
0.41 |
– |
Glycerol |
[195] |
|
Fungi | |||||
|
Rhizopus sp. | |||||
|
R. oryzae | |||||
| R. oryzae ATCC 52311 | 83.0 | 0.88 | 2.6 | Glucose | [70] |
| R. oryzae | 62 | 72% | 2.5 | Glucose | [13, 14] |
| R. sp. MK-96-1196R. sp. MK-96-1196 | 33.3 | 0.93 | 1.80 | Cull potato glucose | [356] |
| R. oryzae | 83 | 65% | 1.6 | Glucose | [13, 14] |
| R. oryzae | 71.5 | 71% | - | Glucose | |
| R. oryzae | - | 70% | - | Glucose | |
| R. oryzae | 40 | 78% | 4.6 | Glucose | |
| R. oryzae | - | - | 6.2 | Glucose | |
| R. oryzae | - | 65% | - | Glucose | |
| R. oryzae | 112–173 | 78–94% | 2.8–5.6 | Glucose | [357] |
| R. oryzae | 104.6 | 87 | 1.8 | Glucose | [13, 14] |
| R. oryzae | 60 | 2.9–6.2 | Glucose | [13, 14] | |
| R. oryzae | - | - | 2.91 | Glucose | [72, 77] |
| R. oryzae NRRL 395 | 104.6 | 0.87 | 1.8 | Glucose | [153] |
| R. oryzae NRRL 395 | 0.87–0.90 | 1.8–2.5 | Glucose | [86] | |
| R. oryzae R1021 | 0.77 | Glucose | [83] | ||
| R. oryzae NRRL 395 | ≈1 | 1.65 | Corn | [86] | |
| R. oryzae RBU2-10 | 1.84 | Rice | [358] | ||
| R. arrhizus DAR 36017 | 1.3–1.6 | Potato | [172] | ||
| R. oryzae HZS6 | 0.80 | 0.99 | Corncob | [155] | |
| R. oryzae NRRL395 | 0.31 | Corncob | [65] | ||
| R. sp. MK-96-1196 | 24.0 | 0.3 | Corncob | [63] | |
| R. oryzae NRRL 395 | 49.1 | 0.7 | Waste paper | [153] | |
| R. oryzae GY18 | 115 | 0.81 | 1.6 | Glucose | [359] |
| R. oryzae GY18 | 80.1 | 0.89 | 1.67 | Sucrose | [359] |
| R. oryzae GY18 | 68.5 | 0.85 | 0.57 | Xylose | [359] |
| R. oryzae NBRC 5378 | 14.4 | – | 0.56 | Xylose | [69] |
| R. oryzae ATCC 9363 | 113 | 0.90 | 4.3 | Glucose | [360] |
| R. oryzae NRRL 395 | 91.0 | 0.76 | 2.02 | Corn starch | [13, 14] |
| R. oryzae | 103.7 | – | 2.16 | Glucose | [84] |
| 81–95 | – | 3.4–3.85 | Glucose | [84] | |
| R. oryzae NBRC 5384 | 145 | 0.95 | 1.42 | Glucose | [361] |
| 231 | 0.93 | 1.83 | Glucose | [361] | |
| R. oryzae | 51.7 | 0.68 | Oat | [362] | |
| R. oryzae | 173.5 | 0.86 | 1.45 | Tobacco waste water-extract and glucose | [363] |
| R. oryzae As3.819 | 80.2 | Glucose | [364] | ||
| R. oryzae | 463.18 | 0.83 | 2.76 | Cassava pulp | [365] |
| R. oryzae | 75.28 | 0.5 | 1.05 | Cassava pulp hydrolysates | [366] |
| R. arrhizus | 68.8 | 0.93 | 0.72 | Honeycomb matrix | [367] |
| R. arrhizus | 75.1 | 0.63 | 1.54 | Glucose | [368] |
| R. arrhizus | 1.2 | Pretreated dairy manure | [369] | ||
| R. arrhizus | 34–60.3 | 0.34–0.60 | Xylo-oligosaccharides manufacturing | [370] | |
| R. arrhizus UMIP 4.77 | 10 | 0.26 | 0.27 | Wheat straw | [371] |
|
Rhizopus sp. | |||||
| R. arrhizus | 46.78 | 0.97 | Animal feeds from Sophora flavescens residues | [372] | |
| R. microsporus | 84.3–119 | 0.84–0.93 | 1.25 | Liquefied cassava starch | [373] |
| R. arrhizus | 103.8 | Waste potato starch | [374] | ||
| Monascus ruber | 129–190 | 0.58–0.72 | 0.91–1.15 | Glucose | [375] |
| Engineered Aspergillus brasiliensis from Rhizopus oryzae | 13.1–32.2 | 0.26–0.47 | Glucose | [376] | |
|
Aspergillus niger |
7.7 |
0.13 |
Glucose |
[377] |
|
|
Yeast | |||||
| Engineered P. stipitis: LDH from L. helveticus (integrated, 1 copy) |
15–58 |
0.58 |
0.6 |
Glucose |
[100] |
|
Saccharomyces | |||||
| Engineered S. cerevisiae LDH from L. casei (multicopy vector) | 12 g/L | Glucose | [13, 14] | ||
| Engineered S. cerevisiae LDH from L. casei | 8.6 | 0.04 | Glucose | [13, 14] | |
| Engineered S. cerevisiae LDH from B. taurus (integrated, 1 copy) | 20 | Glucose | [13, 14] | ||
| Engineered S. cerevisiae LDH from B. taurus (multicopy plasmid) | 11.4 | Glucose | [13, 14] | ||
| Recombinant Saccharomyces cerevisiae CENPK2 | 2.22 | Food waste biomass | [378] | ||
| Engineered S. cerevisiae OC-2T T165R | ~45–50 | ~0.45–1.6 | Glucose | [379] | |
| Engineered S. cerevisiae LDH from B. taurus (multicopy plasmid) | 6.1 | Glucose | [13, 14] | ||
| Engineered S. cerevisiae LDH from L. plantarum (integrated, 1 copy) | 58 | Glucose | [380] | ||
| Engineered S. cerevisiae LDH from L. casei (integrated, 2 copy) | 1.6 mol/96h | Glucose | [92] | ||
| Engineered S. cerevisiae LDH from B. taurus (integrated, 2 copies) | 50.6 | Glucose | [381] | ||
| Engineered S. cerevisiae LDH from B. taurus (integrated, 6 copies) | 120 | Glucose | [381, 382] | ||
| Engineered S. cerevisiae LDH from L. mesenterioides (D-LDH, integrated, | |||||
| 2 copies) | 53.2 | Glucose | [383] | ||
| Engineered S. cerevisiae LDH from B. taurus (integrated, 2 copies) | 82.3 | Glucose | [95] | ||
| Engineered S. cerevisiae HDH from R. oryzae (multicopy plasmid) | 38 | Glucose | [96] | ||
| Engineered S. cerevisiae HDH from L. plantarum (multicopy plasmid) | 70 | 0.93 | Glucose | [98] | |
| Engineered S. cerevisiae LDH from B. taurus (integrated, 8 copies) | 80 | Glucose | [97] | ||
| Engineered S. cerevisiae LDH from B. taurus (integrated, 2 copies) | 74.1 | Glucose | [97] | ||
| Engineered S. cerevisiae LDH from B. taurus (integrated, 2 copies) | 71.8 | Glucose | [97] | ||
| Engineered S. cerevisiae | 122 | 0.61 | Cane juice | [67] | |
| S. cerevisiae | 117 | 0.58 | Glucose | [384] | |
| Recombinant Saccharomyces cerevisiae | 60.3 | 0.646 | 2.8 | [385] | |
| Engineered Issatchenkia orientalis: LDH from L. helveticus (integrated, 1 copy) | 66 | Glucose | [386] | ||
| Engineered Issatchenkia orientalis: LDH from L. helveticus (integrated, 1 copy) |
70 |
Glucose |
[387] |
||
|
Candida | |||||
|
Candida utilis | |||||
| Engineered Candida utilis: LDH from | 93.9 | 0.91 | 2.18 | Xylose | [388] |
| Engineered Candida utilis: LDH from B. taurus – optimised (integrated, 2 copies) |
103.3 |
[104] |
|||
|
Candida boidinii | |||||
| Engineered Candida boidinii: LDH from B. taurus – optimized (integrated, 1 copy) |
85.9 |
Glucose |
[99] |
||
|
Candida sonorensis | |||||
| Candida sonorensis | 92 | 0.94 | 4.9 | Glucose | [100] |
| Candida sonorensis | 40 | 0.60 | Glucose | [389] | |
| Engineered Candida glycerinogenes from Rhizopus oryzae |
Glucose |
[390] |
|||
|
Kluyveromyces | |||||
| K. marxianus | 8.8 | 0.24 | 4.3 | [219] | |
| Engineered K. marxianus from actobacillus plantarum | 122–130 | Jerusalem artichoke tuber powder | [391] | ||
| Engineered K. marxianus from Homo sapiens (HsLDH), Bacillus subtilis (BsLDH), Bacillus megaterium (BmLDH), Lactococcus lactis (LlLDH), Rhizopus oryzae (RoLDH), and Plasmodium falciparum (PfLDH) | 25–105 | Alkali-pretreated corncob | [392] | ||
| Engineered K. marxianus LDH from L. helveticus (integrated into PDC1 locus) | 99 | Glucose | [106] | ||
| Engineered K. marxianus LDH from L. helveticus (integrated into PDC1 locus) | 9.1 | Glucose | [106] | ||
| Engineered K. lactis LDH from B. taurus (low copy number plasmid, 5 copies) | 109 | 0.91 | Glucose | [13, 14] | |
| Engineered K. lactis LDH from B. taurus(multicopy plasmid) | 60 | 0.85 | Glucose | [93] | |
| Engineered K. lactis LDH from B. taurus |
0.58–1.00 |
Glucose |
[93] |
||
|
Schizosaccharomyces | |||||
| Engineered Schizosaccharomyces pombeLDH from R. oryzae | 80–100 | Glucose | [393] | ||
| Schizosaccharomyces pombe | 24.4 | 0.45 | Cellobiose | [394] | |
| Schizosaccharomyces pombe | 60.3 | 0.45 | Glucose | [395] | |
|
Schizosaccharomyces pombe |
112 |
2.2 |
Glucose |
[396] |
|
|
Microalgae and cyanobacteria | |||||
| Engineering Synechocystis sp. PCC 6803 | 3.31 | Glucose | [397] | ||
| Engineering of Schizosaccharomyces pombe |
24.4–25.2 |
0.68–0.81 |
Glucose and cellobiose |
[394] |
|
|
Consortia | |||||
| MAR compost | 34.2 | 0.54 | Kitchen refuse | [113] | |
| waste activated sludge (Bacillus, Clostridiaceae, Lactobacillus and Peptostreptococcaceae) | 26.63–29.77 | Food waste | [398] | ||
| Naturally inhabiting bacteria in garbage | 64 | 0.62 | Kitchen refuse | [114] | |
| Naturally inhabiting bacteria in garbage | 37.7 | 0.58 | Garbage | [399] | |
| Anaerobic digestion sludge | 4.17 | 0.429 | Glucose | [400] | |
| Anaerobic digestion sludge | 23 | 0.92 | Glucose | [401] | |
| Excess sludge | 8.5 | 1.06 | Sucrose | [402] | |
| Naturally inhabiting bacteria in garbage | ˂27.5 | Kitchen refuse | [298] | ||
| Microbial consortium CEE-DL15 Clostridium sensustricto (57.29%), Escherichia (34.22%), and Enterococcus (5.32%) | 112.3 18.5 |
0.81 | 4.49 | Sugarcane molasses | [403] |
| Anaerobic activated sludge as inocula | 28.4 | 0.46 | Methanogenic sludge and fresh food waste | [404] | |
Cases with no data indicate absence of results in the cited reference.
LAB can metabolize glucose into LA, acetic acid (AA), formate, ethanol, diacetyl, acetoin, and carbon dioxide (CO2 gas detection is a diagnostic test for heterofermentative from homofermentative fermentation) [14]. The heterofermentative LAB can use the phosphogluconate pathway (with a theoretical yield of 0.5 g/g) and phosphoketolase pathway (with a theoretical yield of 0.6 g/g), when metabolizing hexose and pentose sugars, respectively [13, 14].
The utilization of heterofermentative LAB as dairy starter cultures are not common due to CO2 release and simultaneous production of LA and other organic acids, considered as defects which induce several problems in the products, including bloated packaging and cracks in dairy products and hard cheeses, respectively. Heterofermentative LAB includes mainly Oenococcus, Leuconostoc, and some Lactobacillus spp., and the main heterofermentative Lactobacillus spp. are Lb. brevis, Lb. fermentum, and Lb. reuteri.
2.1.2. Bacillus strains
Bacillus also has metabolic capacity to produce LA. There are several advantages to the use of Bacillus spp. relatively to the LAB. The use of Bacillus spp., allows reducing the LA production cost, because: (1) they can grow and ferment in mineral salt media with inexpensive nitrogen sources such as steep corn liquor or (NH4)2SO4, temperature (50–55 °C) and pH (6–6.5); (2) media sterilization before the fermentation process can be avoided due to the high temperature of LA fermentation process (>50 °C) and so do not need also cooling after medium sterilization, with considerable costs reduction; (3) they can utilize all sugars from lignocellulose biomasses, due to the ability to metabolize pentose sugars via the pentose phosphate pathway and hexose sugars via the EMP pathway; (4) all strains of Bacillus produce only L-LA [15]; (5) they can convert substrates to LA with high yield or high productivity; (6) some strains namely B. coagulans JI12 was tolerant to both furfural (4 g/l) and acetate (20 g/l). Neither pre-detoxification nor separation of fermentable sugars from lignin was needed before the fermentation. Meng et al. [16] and Patel et al. [17] reported that the alkaliphilic Bacillus sp. WL-S20 and B. coagulans 36D1 produced L-LA at concentration and yield of (225 g/L and 0.993 g/g) and (92.0 and 0.96 g/g), respectively. Alkaliphilic Bacillus sp. WL-S20 generated L-lactic acid in fed-batch fermentation at pH 9.0, which would reduce a risk of the contamination during fermentation and also can produce lactic acid in thermal fermentation (≥50 °C) [16]. Bacillus spp. have been accredited by European Food Safety Authority (EFSA) and Food and Drug Administration (FDA) to the Qualified Presumption of Safety (QPS) list and Generally Recognized As Safe (GRAS) status for applications in livestock production [18]. Some Bacillus strains could produce LA, including B. coagulans [19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33], B. stearothermophilus [13, 14], B. licheniformis [34] thermophilic B. licheniformis [35], B. subtilis [36], Bacillus sp [37, 38, 39, 40]. and alkaliphilic bacilli such as B. circulans var. alkalophilus ATCC 21783, B. alcalophilus sp. halodurans ATCC 27557, B. alcalophilus ATCC 27647, alkaliphilic B. sp. WL-S20 and B. sp. 17-1 ATCC 31007 [16].
2.1.3. Corynebacterium glutamicum
Corynebacterium glutamicum is an aerobic Gram-positive bacterium that has been reported to be able to excrete amino acids (L-lysine and L-glutamate) and also small amounts of mix-organic acids (LA, succinic acid (SA), and AA) in industrial production. The organic acids production reported has occurred under oxygen deprivation conditions (anaerobic condition) due to cell growth inhibition and acceleration of mix-organic acids production from various sugars, including D-glucose [41, 42, 43]; L-arabinose [44]; D-glucose and L-arabinose [45] D-xylose and D-glucose [46] and D-xylose, D-cellobiose and D-glucose [44] in mineral salts medium [13]; C. glutamicum is engineered and has highly potential bacterium that can produce LA with high yield and productivity without requiring complex nutritional compounds. C. vitaeruminis MTCC 5488 produced 38.5 g/l LA in fed-batch fermentation [13]. Meanwhile, C. glutamicum, as well as E. coli (section 1-4), have extremely low tolerance to acidic condition; hence LA production needs to be performed at pH-values about 7.0.
However, the simultaneous production of LA and the formation of several organic acids such as SA and AA resulted in a low LA production yield which should be improved [47]. Several types of research strategies were attempted to increase the LA production by C. glutamicum fermentation, through the promotion of medium conditions changes or by using engineering methodologies, such as:
-
A)
Inui et al. [41] and Okino et al. [48] reported a novel system which consists in a reactor containing high-density cells (HDC) of C. glutamicum (the cell concentrations were almost 10-fold higher than those commonly used for batch fermentation) that could lead to the high volumetric productivity of LA. According to the results of Yukawa et al. [49], LA was produced by using the C. glutamicum R strain under an HDC condition.
-
B)
Manipulation of C. glutamicum could produce D-lactic acid at higher productivity and purity compared with the parental strain. Simultaneously knock out of the L-LDH gene, and over expression of the D-LDH encoding gene was performed by inserting this gene into C. glutamicum from Lb. delbrueckii [43] and Lb. Bulgaricus [42].
Song et al. [50] reported an engineered C. glutamicum strain that can produce D-lactyl-CoA (by D-LDH and propionyl-CoA transferase) and 3-hydroxybutyryl-CoA (by β-ketothiolase and NADPH-dependent acetoacetyl-CoA reductase) from glucose, under several enzymatic reactions. Copolymerization of 3-hydroxybutyryl-CoA and D-lactyly-CoA by using lactate polymerizing enzyme reaction resulted in the production of poly (LA-co-3HB) with high LA fractions (96.8 mol%) [50].
-
C)
On the other hand, some studies reported that an engineered C. glutamicum could utilize pentose sugars including xylose [46] and arabinose [45], as well as hexose sugars, such as galactose and glucose. Kawaguchi et al. [46] inserted the genes xylA and xylB from E. coli into the C. glutamicum R strain that encodes xylose isomerase and xylulokinase, respectively, using a multicopy plasmid under the controlled promoter condition. Both the expression of xylA and xylB genes with xylose utilization ability could enhance the growth rate and production pattern of organic acid including L-LA and SA with interesting productivities (29 and 17 mmol/l/h) and yields (0.53 and of 0.25 g/g), respectively [46]. Kawaguchi et al. [45] performed another study in order to gain arabinose utilization ability, throughout the expression of genes araA, araB and araD (encoding arabinose isomerase, ribulokinase, and ribulose-5-phosphate 4-epimerase, respectively) from E. coli into the C. glutamicum R strain. The results showed that the engineered C. glutamicum could consume arabinose, through successful arabinose genes expression, leading to the production of L-LA (3.4 mmol/h/g dry cell), SA and AA. This L-LA was produced using a mixture of sugars (arabinose and glucose), being the glucose consumption rate (0.76 g/h/g dry cell) significantly higher than the arabinose counterpart (0.06 g/h/g dry cell) [45].
-
D)
Pyruvate kinase (Pyk) plays a key role in the production of pyruvate and ATP in glycolysis pathway and, moreover, as an essential factor in controlling the carbon flux distribution. C. glutamicum only contains one Pyk (pyk1NCgl2008). Moreover, recently Chai et al. [51] found NCgl2809 as another novel pyruvate kinase (Pyk2) in C. glutamicum. These authors grew an engineered C. glutamicum containing Pyk1 or Pyk2 on D-ribose conditions, being the LA production enhanced by overexpression of either Pyk1 or Pyk2, due to the increase of the activity of the Pyk enzyme. They found that fermentation by the overexpression of Pyk2 in WTΔpyk1 C. glutamicum strain could increase LA production to 60.27 ± 1.40 g/L (about 47% higher than the parent strain) under oxygen deprivation condition.
2.1.4. Escherichia coli
Wild-type E. coli is capable of growing and producing LA using hexoses and pentoses sugars fermentation with production of a mixture of organic acids (AA, SA, and formic acid (FA)) and ethanol [47, 52]. Moreover, they can grow on broth with more straightforward nutrient requirements compared to the conventional LAB.
Engineered E. coli showed improved LA fermentation efficiency compared with wild E. coli [13, 14, 52]. These engineered strains were manipulated by (1) replacement of D-LDH with L-LDH from LAB, bovine and other sources [13, 14, 52].; (2) prevention synthesis of racemic mixtures of D- and L-lactates by omission of methylglyoxal bypass route and consequently its accumulation; (3) avoiding of the undesired utilization of L-lactate by blocking the aerobic L-LDH [53]. Engineered E. coli strains can grow and produce LA from several disaccharides including sucrose [54, 55] and monosaccharides (hexoses and pentoses) including glucose [13, 14, 52, 56, 57, 58], xylose [56], and also glycerol [13, 14, 59, 60]. Some researchers reported that engineered E. coli strains produce D-LA by the homofermentative substrate pathway that causes over-expressing of LA. However, engineered E. coli strains had shown several disadvantages, such as low productivity (≤1.04 g/L/h) and low tolerance to low pH conditions due to LA production, in comparison with LAB [13, 14, 57].
2.2. Filamentous fungi
Filamentous fungi are another microbial source that can produce LA. Numerous species of the genus Rhizopus such as R. oryzae and R. arrhizus can produce L-LA (as the main product) fumaric acid, and ethanol from different carbon sources [64]. Among carbon sources, they aerobically metabolize glucose to produce LA. However, there are several renewable carbon resources for LA production by Rhizopus strains, which include corncob hydrolysate [61, 62, 63, 64, 65]; xylose [66, 67], glucose [13, 14, 68], wheat straw [69], paper pulp sulfite liquor [70], chicken feather protein hydrolysate [71], molasses [71], cassava pulp hydrolysis [72], potato hydrolysate [73], and glycerol enriched with lucerne green juice and inorganic nutrients [74]. Media containing nitrogen sources lead to a fast growth that induces the production of chitin instead of LA [15]. On the other hand, lack of a nitrogen source leads to a decreased cell activity and product formation in long-term cultivation [15]. Two solutions to overcome this drawback was: 1) cells morphology affected LA productivity and yield (for example, fungal pellets instead of spores [73]; 2) medium composition manipulation by using low nitrogen sources and high content of carbon sources could enhance LA production [73]. Urea is one of the nitrogen sources used by genus Rhizopus that when added periodically within the production phase can avoid biofilm overgrowth, postpone sporulation, and retain high cell viability and LA productivity [72].
There are some advantages and disadvantages of using Rhizopus strains for LA production. Some benefits of Rhizopus strains in comparison to LAB include: 1) their amylolytic properties (containing amylolytic enzyme activity) that can convert various starchy biomasses directly to L-LA without prior saccharification process [75]; 2) simple medium requirements [76-78]; 3) their filamentous or pellet growth in fermentation medium facilitate their separation from fermentation broth, which can lead to lower-cost downstream process [79]; 4) fungal biomass is a worth fermentation by-product. On the other hand, R. oryzae is an obligate aerobe and requires vigorous aeration, usually above an oxygen transfer rate of 0.3 g O2/L/h [80, 81]. A disadvantage of using fungi is related with the different morphology of growth under fermentation, which includes extended filamentous appearances, pellets, mycelial mats, and clumps that significantly affect LA productivity and rheology of broth medium. Their morphology can affect the oxygen supply and mass transfer. In fungal fermentation, the low LA productivity (below 3 g/(L·h)) is a result of the low O2 mass transfer and synthetic route shift toward production of other by-products such as ethanol and fumaric acid. The preferable fungal morphology for industrial fermentations is small pellets by several reasons: 1) improved rheology of broth fermentation; 2) enhanced mass transfer in fermentation broth; 3) can be continuously utilized by using repeated batch fermentation for long operations [82].
Some researchers investigated fungi morphology that enhances the LA productivity. Abdel-Rahman et al. [13, 14] verified that high LA production was obtained by cotton-like mycelial flocs morphology, which was formed by the culture of R. oryzae in the air-lift bioreactor.
Several reports attempted to achieve high yield and productivity of pure L-LA with higher cell density by fungal fermentation [71, 83, 84], including the following:
-
1.
Immobilization techniques, being Rhizopus oryzae immobilized for L-LA production [13, 14, 85, 86], but entrapment of fungal cells on matrixes revealed to be time-consuming.
-
2.Controlling the production of undesirable by-products, mainly ethanol and fumaric acid leads to higher LA productivity [87, 88, 89].
-
2.1.Addition of alcohol dehydrogenase (ADH) inhibitor into the fermentation medium (i.e., 1,2-diazole and 2,2,2-trifluoroethanol) as an active inhibitor to decrease ethanol production and lactate dehydrogenase (LDH), as a useful promoter to increase LA and cell biomass production [90].
-
2.2.Metabolic engineering of the strain by deleting the alcohol dehydrogenase and malate dehydrogenase genes, thus shifting the metabolic flux, increasing LA production and yield [89].
-
2.1.
As far as we are aware, there are no reports that include other fungi to produce LA. The fungus, Aspergillus niger together with Lactobacillus sp. was used for LA production. The strategy, in this case, was that fungi enzymes would perform saccharification and de-polymerization of carbohydrate polymers to produce fermentable sugars to be used by the bacterium [10, 91].
2.3. Yeasts
Presently, LAB is the main microorganisms used to LA production. However, there is one problem associated to their use; their low pH sensitivity leads to the use of large amounts of neutralizing agents, including CaCO3 and results in the production of gypsum in fermentation medium [92]. Comparatively, yeasts versus bacteria, yeasts can tolerate low pH which leads to a reduction for the need of neutralizing agents and downstream processing cost. The worst important disadvantage of using wild-type yeasts is the reduced LA production as the main product. Nevertheless, engineered yeasts are the best solution to overcome this drawback.
Engineering yeast manipulation has been studied to obtain high LA productivity and yield, due to cancelation of pyruvate decarboxylase and/or pyruvate dehydrogenase activities, which results in the partial or full substitution of ethanol by LA production [93]. In order to improve the natural acid resistance of yeasts, lactic acid productivity has been enhanced by inserting the gene encoding L(+)-LDH from heterologous sources. The bovine gene encoding LDH has been successfully expressed in both Candida utilis and Saccharomyces cerevisiae, and the gene encoding LDH from Lb. helveticus has been expressed in Candida sonorensis [1]. Different research teams have been attempting to produce lactate from engineered yeasts genera including Saccharomyces cerevisiae [13, 14, 92, 94, 95, 96, 97, 98], Candida spp. [99], Kluyveromyces lactis [13, 14, 93], Torulaspora delbrueckii [13, 14], Pichia stipites [100] and Zygosaccharomyces bailii [101].
2.3.1. Saccharomyces cerevisiae
Saccharomyces cerevisiae is one of the more permissive organisms used for LA production due to a high intrinsic tolerance to low pH-values. This characteristic should give to S. cerevisiae several advantages over LAB and Bacillus spp. Firstly, it is a microorganism resistant to low pH and can grow aerobically on glucose sources with the basic anaerobic growth factors including oleic acid, nicotinic acid, and ergosterol.
Engineered S. cerevisiae can efficiently produce d-lactic acid due to its capability to grow fast under anaerobic and aerobic conditions. In transgenic strains, the coding section of pyruvate decarboxylase 1 (PDC1) was completely eliminated, and one or several copies of the d-lactate dehydrogenase (d-LDH) gene resources were inserted into the genome from mammalian LAB such as Leuconostoc mesenteroides subsp. mesenteroides strain NBRC3426. This study was for the first time performed by Porro et al., 1995, having achieved an LA production of 20 g/l and productivity up to 11 g/L/h using engineered S. Cerevisiae [13, 14].
2.3.2. Candida
2.3.2.1. Candida sonorensis
Candida sonorensis as a methylotrophic yeast that can ferment hexose (i.e., glucose) and pentose sugars (i.e., xylose and arabinose) to ethanol. They tolerate acid environments and require simple growth medium. C. sonorensis was manipulated by insertion of L-LDH genes from Lb. helveticus, B. megaterium, and R. oryzae. Multiple LDH gene copies were expressed to produce suitable mutants for LA production, which produced LA and ethanol. In order to increase the LA productivity, ethanol production was stopped by the elimination of two pyruvate decarboxylase genes (PDC) 1 and 2, being these the primary enzymes contributing to ethanol production. This modification (C. sonorensis expressing L. helveticus LDH) did not affect cell growth and resulted in the accumulation of lactate up to 92 g/l with a yield of 0.94 g/g glucose without ethanol production [102]. In another work, engineered C. sonorensis (L-lactic acid dehydrogenase (ldhL) from Lb. helveticus) was reported to produce 31 g/l LA from 50 g/l D-xylose free of ethanol [103].
2.3.2.2. Candida boidinii
Genetic engineering can be used to construct a crabtree-negative methylotrophic haploid of Candida boidinii that can efficiently produce high amounts of L-LA [99]. The ethanol production of C. boidinii was 17% reduced by knocking out of the PDC1 gene encoding pyruvate decarboxylase when compared with the wild-type strain and with simultaneous heterologous expression of the bovine L-LDH gene resulted in 85.9 g/l of LA with a productivity of 1.79 g/l/h [99].
2.3.2.3. Candida utilise
Candida utilis as crabtree-negative yeast is currently used for the production of several valuable chemicals, including glutathione, single cell protein, and RNA. The most pertinent advantage of C. utilise for LA production is the use of inexpensive substrates for growing, which includes pulping-waste liquors. In the study performed by Ikushima et al. [104], an engineered C. utilise strain produced L-LA with high efficiency. These authors reduce ethanol production (as a by-product of L-LA) by knocking out the gene encoding pyruvate decarboxylase (CuPDC1), and then two copies of the bovine L-lactate dehydrogenase (L-LDH) gene were inserted into the CuPdc1-null strain genome. The engineered C. utilise produced 103.3 g/l of L-LA with 95.1% conversion of basal medium and 99.9% purity.
2.3.3. Kluyveromyces
2.3.3.1. Kluyveromyces lactis
Kluyveromyces lactis is crabtree-negative yeast which was used for LA production after genetic modification. In comparison with some other yeasts strains, such as S. cerevisiae, which have a pyruvate decarboxylase (PDC) with two active structural genes (PDC1 and PDC5) [93], Kluyveromyces lactis has expressed PDC activity with a single gene, KlPDC1. The omission of KlPDC1 leads to production strains without PDC activity and increase LA production with free ethanol. The intense competition for pyruvate consumption by homologous PDC and heterologous LDH activities leads to a low LA yield, due to the simultaneous production of ethanol and LA. On the other hand, the elimination of pyruvate decarboxylase gene (KlPDC1), as a single gene with PDC activity in K. lactis, resulted in no ethanol production [93]. In this yeast, the bovine L-lactate dehydrogenase gene (LDH) insertion and decarboxylase gene deletion were sufficient to increase the LA production to 109 g.l−1, with a productivity of 0.91 g.l−1. h−1, and yield 1.19 mol.mol−1 of consumed glucose [13, 14]. In another study, the KlPDC1 and pyruvate dehydrogenase (PDH) genes were deleted, being the LDH gene inserted into a wild-type of K. lactis. The LA production improved by shifting of pyruvate flux toward homolactic fermentation with a yield level of 0.85 g g−1 (being the maximum theoretical yield 1 g.g−1) [93].
2.3.3.2. Kluyveromyces marxianus
Kluyveromyces marxianus has several advantages which make it economically attractive for commercial-industrial applications, including 1) proliferation occurs at high temperatures (up to 52 °C), reducing contamination control in commercial cultivation, whereas most organisms in an industrial environment cannot be cultivated well at this temperature [105]; 2) K. marxianus in enriched media conditions, can grow rapidly with doubling times of 0.75–1 h (37 °C) [105]; 3) Many K. marxianus strains can utilize various inexpensive carbon sources and require few additional nutrients [105]. In this yeast, the LA concentration was improved by the insertion of the LDH gene from B. megaterium [105]. Also, Hause et al. [106] transformed K. marxianus by insertion of the LDH gene (from Lb. helveticus and integrated into PDC1 locus) and verified an L-LA production at 9.1 g/L.
2.3.4. Zygosacchromyces
Zygosaccharomyces bailii has been suggested as another host for LA [107], due to its ability to tolerate environmental restrictions, including high sugar concentrations, acidic conditions, relatively high temperatures (higher than fermentation process) and LA production levels compared with S. cerevisiae. Z. bailii has a high growth rate and biomass yield which could improve the fermentation processes of LA production. An engineered Z. bailii was produced by heterologous LDH gene expression (from the bacterial L-LDH) to induce the shift of the glycolytic flux towards the lactate production [101, 107] to improve LA production efficiency.
2.3.5. Pichia stipitis
Pichia stipitis can utilize pentose and hexose sugars from lignocellulose hydrolysates as substrates to produce ethanol. The deletion of alcohol dehydrogenase 1 (ADH 1) and insertion of L-LDH (from heterologous Lb. helveticus) under the ADH1 promoter, led to an engineering P. stipitis producing 58 and 41 g/l of LA from 100 of xylose and 94 g/l glucose, respectively. Moreover, ethanol production was reduced by 15–30 % and 70–80 % compared with the wild-type strain, by xylose and glucose utilization, respectively [100].
2.4. Microalgae and cyanobacteria
Algae and cyanobacteria are included in the category of photosynthetic microorganisms, and they can grow almost anywhere, with a short harvesting cycle of about 1–10 days and produce various chemicals (including biofuels (H2), ethanol, lactic, AA and FA). Algal biomass can be proposed as an alternative candidate to LA production without carbohydrate feed medium costs, being induced in high content of carbohydrates and proteins and also lack lignin [15, 108].
A few reports have evaluated the content of LA production by microalgal species, such as:
-
1.
Scenedesmus obliquus strain D3 could produce d-LA as the main fermentation product [13, 14];
-
2.
Nannochlorum sp. 26A4 produced LA at 26 g/L with a yield of 70% and optical purity of 99.8% from starch (40% content per dry weight) under dark and anaerobic conditions [109];
-
3.
Biomass of Nannochloropsis salina contains 40% lipids, 20% carbohydrates, and 40% proteins. The neutralized and concentrated lipid-free residue has 64.3% of sugars (glucose and xylose). Co-fermentation of N. salina and L. pentosus under anaerobic fermentation could yield 10.1 g/l of LA with 92.8% of the conversion [110].
-
4.
Synechococcus elongates PCC7942 engineered with simultaneous genes expression encoding glucose; lactate and fructose-facilitated diffusion transporter; L-LDH (from E. coli) and invertase could produce 600 μM of LA. Similarly, engineered Synechocystis sp. PCC6803 by insertion L-LDH gene (derived from B. subtilis) could produce of 3.2 mM LA [111].
3. Substrates for lactic acid production
3.1. Food waste
Food waste can include any compounds from the food production process to the wastes formed by the final consumer. Food waste contain a high amount of carbohydrate which causing it suitable as a substrate for lactic acid fermentation. Regarding to Table 1, numerous studies stated food waste are suitable for lactic acid production such as kitchen residues/refuse and municipal solid wastes [112], model kitchen refuse medium contain water, vegetables, meat/fish and cereals [113], mixes of cooked rice, vegetables, meat, and bean curd [113, 114]; rice, noodles, meat, and vegetables [115, 116]; vegetables such as carrot peel, cabbage, and potato peel, fruit such as banana peel, apple peel, and orange peel, baked fish, rice, and used tea leaves [117, 118]; rice, noodles, meat and vegetables, and unsold bakery products including cakes, breads and pastries [119]; rice, vegetables, and meat [120]; coffee mucilage [119]; and coffee pulp [121].
3.2. Carbohydrates
3.2.1. Starchy biomass and sugar plant wastes (malt, molasses and sugar beet juice)
Lactic acid can be produced from sugar plant wastes (molasses and sugar beet juice), starchy, and lignocellulosic biomasses (Figure 2).
Figure 2.
Pathways of lactic acid production from pentose sugars obtained from lignocellulose hydrolysate. Genes AraA, AraB, and AraD encoding arabinose isomerase, ribulokinase, and ribulose-5-phosphate 4-epimerase, respectively. XylA, and xylB encodes xylose isomerase, and xylulokinase. (1) arabinose isomerase; (2) ribulokinase; (3) ribulose-5-phosphate 3-epimerase; (4) xylose isomerase; (5) xylulokinase; (6) phosphoketolase; (7) acetate kinase; (8) phosphotransacetylase; (9) aldehyde dehydrogenase; (10) alcohol dehydrogenase; (11) lactate dehydrogenase; (12) transketolase; (13) transaldolase; (14) 6-phosphofructokinase; (15) fructose-bisphosphate aldolase; and (16) triosephosphate isomerase. PK pathway and PP pathway are phosphoketolase and pentose phosphate pathway. GA3P: glyceraldehyde-3-P, DHAP: Dihydroxyacetone-P.
Disaccharides (lactose and sucrose) and monosaccharides hexoses (glucose, fructose, and galactose) and pentoses (xylose and arabinose) sugars can be fermented by LAB via EMP and/or the pentose PK pathway [122]. Molasses are waste products containing a large amount of sucrose and other essential nutrients, which can derive from sugar cane and sugar beet from sugar manufacturing plants. Several microorganisms can use molasses as a substrate including Lb. delbrueckii subsp. delbrueckii mutant Uc-3 [123], Lb. delbrueckii NCIM 2025 [124]; Lb. delbrueckii NCIMB 8130 [125]; Lb. delbrueckii C.E.C.T. 286 [13,14], Lb. delbrueckii IFO 3202 [13,14], Lb. delbrueckii [126], Lb. plantarum [127], Sporolactobacillus cellulosolvens [13, 14], Rhizopus arrhizus [128], Lb. casei M-15 [129], Bacillus sp. XZL9 [29] and E. faecalis [130]. Shukla et al. (2004) also reported that recombinant E. coli strain could produce D-lactic acid from molasses [131]. Raw sugar beet juice with a Brix of at least 60 was used for LA production by lactic acid-producing microorganisms including bacteria (lactobacilli and moderately thermophilic bacilli due to fermentation at relatively high temperature such as B. coagulans, B. thermoamylovorans, Geobacillus stearothermophylus and B. smithii, yeasts and fungi, such as, Rhizopus and Aspergillus [132]. Malt and date juice are another source for LA production by Lb. casei subsp. rhamnosus in batch and fed-batch cultures with a maximum LA production level of 89.2 g/L already achieved [133, 134].
There is a great interest to introduce cellulosic and starchy materials as substrates for LA production due to their abundance, low price and for being derived from renewable sources [135]. Amylolytic lactic acid bacteria (ALAB) such as Lb. plantarum, Lb. fermentum and Lb. manihotivorans, Lb. amylophilus and Lb. amylovorus can ferment starchy biomass into LA due to their α-amylases activity [13, 14, 136, 137]. Some ALAB were isolated from various amylaceous compounds, which include maize and maize-based fermented products [13, 14, 138], potato [13, 14, 73, 138, 139], cassava and cassava-based fermented products [13, 14], rice and rice-based fermented products [13, 14, 136, 140], sweet sorghum [13, 14], wheat [13, 14, 136, 141], rye [13, 14], oat [13, 14], barley [13, 14, 136] and other starchy substrates [134, 137, 142, 143, 144, 145, 146, 147].
3.2.2. Lignocellulosic biomass
Worldwide, there are abundant and cheap lignocellulosic materials, that include agricultural residues (corn stover, bagasse, and rice husk), forestry residues (sawdust), portions of municipal solid wastes (waste paper and brewer spent grains), herbs, switch-grass and shrubs (switchgrass and water hyacinth), woody plants (poplar trees), Stems, straws, leaves, stalks, shells, husks, and peels from cereals like wheat, rice, barley, corn, sorghum and various industrial wastes [Figures 3 and 4; [134,148]. Cellulosic materials are composed mainly by cellulose, xylan, arabinan, galactan, and lignin [13, 14, 149].
Figure 3.
Different modes of fermentative production of lactic acid.
Figure 4.
Lactic acid production from urban areas or the hospitality sector, and fruits and vegetables industry (Demichelis et al., 2017).
The addition of pectinases and cellulases in the fermentation medium can enhance LA production [150]. However, fermentation of lignocellulosic hydrolysates is prevented by the inhibitory effect of some compounds including acetic acid, furfural, and 5-hydroxymethylfurfural, which are formed during pre-treatment of lignocellulose [150]. To reduce this inhibition, studies were performed through physical and chemical detoxification of the hydrolysate, being this mentioned as the challenges that must be overcome for their efficient utilization [14]. For LA production, several cellulosic materials can be used as substrate, such as: pure cellulose [13, 14, 151], lignocellulosic pentoses including xylose and arabinose (Figure 2) [13, 14, 63, 65, 66, 152] corncob [63, 65] waste paper [13, 14, 153, 154], and wood [64, 130, 155].
Yadav et al. (2020) indicated that P. pentosaceus SKL-7, Lb. plantarum SKL-19, Lb. fallax SKL-15, Lb. plantarum SKL-22, and Lb. paracasei SKL-21grew well in presence of 1-Ethyl-3-methylimidazolium-acetate, 1-Butyl-3-methylimidazolium methane-sulfonate and 1-Butyl-3-methylimidazolium chloride. The L. plantarum SKL-22 demonstareted relatively high tolerance with greatest specific growth rates in presence of 0.5% and 1% 1-Butyl-3-methylimidazolium methane-sulfonate and 1-Butyl-3- methylimidazolium chloride. L. plantarum SKL-22 formed reasonable good content of lactic acid 34.26 g/l, so promising strain for production of lactic acid from lignocellulosic biomass [156].
Agricultural residues are another potential source of substrates for LA production. This category includes: alfalfa fiber [157], wheat bran and straw [158, 159], defatted rice bran [160, 161], food wastes [162], corn stover and cob [29, 65, 152, 157, 163], barley bran husks [163], sugarcane and cassava bagasse [164, 165, 166], trimming vine shoots [163], wine-trimming wastes [163], apple pomace [167], banana wastes [168], mango peel [169], mussel processing wastes [13, 14], cellulosic bio sludge [170], kitchen refuses and wastes [27, 171, 172], fish meal wastes [173], cardboard waste [154] and sugarcane bagasse waste [174]. Wastewater of paper sludges is another source that does not require pretreatment and have a high content of polysaccharide degradation products and short cellulose fibers [20, 68, 170, 175, 176].
3.3. Dairy wastes
3.3.1. Cheese whey
Whey is the primary by-product of the dairy industry, containing proteins, lactose, fats, water-soluble vitamins and minerals. Lactose can be hydrolyzed into glucose and galactose by entering the cell via a permease and β-galactosidase (Figure 1) and can produce four LA moles [122, 177]. LAB are fastidious microbes that require complex macro and micronutrients since they don't have enough proteolytic enzyme activity to utilize whey proteins [178]. For complete utilization of whey lactose and proteins, the addition of supplementary components with a nitrogen source such as yeast extract, peptone, and soy flour or steep corn liquor is necessary. Enriched whey showed a significant improvement in LA production [13, 108, 122, 177]. For instance, whey supplemented with whey protein hydrolysate or yeast extract enhanced the LA production and decreased the unused nutrients loss during bioprocessing [178, 179].
Several strains have been used for LA production from whey, including Lb. plantarum, Lb. helveticus, Lb. acidophilus, Lb. delbrueckii subsp. bulgaricus, Lb. casei, L. lactis, and K. marxianus. However, in conventional batch fermentation, there is a long lag phase in LA production from whey. To overcome this problem, a greater fermenter capacity is required, but this subsequently increases the operational costs [13, 14, 179]. On the other hand, continuous whey fermentation (without the requirement of high-volume) allowed obtaining a high LA productivity [13, 14, 180]. Semi-continuous fermentation conditions with nanofiltration membranes for recycling lactose and cells increased twice the LA production [181]. Lactobacillus and Lactococcus genus are the major LA producers who could efficiently utilize lactose and proteins, present in whey, with high conversion rates [13, 14, 179, 182, 183]. Lb. sp. RKY2, Lb. casei NRRL B-441 and L. lactis subsp. cremoris produced LA at 6.34, 3.97 and 4.6 g l−1 h−1; with a yield of 0.98, 0.93 and 0.88 g/g lactose, respectively [13, 14, 182, 184]. Also, B. longum NCFB 2259 could produce LA with a yield of 0.81 g/g whey lactose as a sole medium in a batch fermentation reactor [181]. On the other hand, LA initially present in whey could have an inhibitory effect in whey fermentation which can be reduced to a certain content by the application of mono or dipolar membranes in an electrodialysis system [185] or using a hollow fiber fermenter by a continuous dialysis process [13, 14].
3.3.2. Yogurt
There is a huge amount of damaged or expired yogurt as waste products, which could provide a good resource for LA production [186]. Sweetened yogurt contains additional sugars, including sucrose and glucose, which would lead to a higher LA production, in comparison to cheese whey, which has fewer sugars. From yogurt whey LA was obtained with a productivity of 0.76 g/L/h and a yield of 0.9 g/g by Lb. casei ATCC 393 with bioconversion of about 44% of total sugars, with increasing order of consumption glucose > sucrose > lactose [186].
3.4. Industrial waste
This category includes glycerol from biodiesel industry and petroleum-based polymers. Glycerol is a byproduct of biodiesel industry that can be produced at a weight ratio of 1:10 (glycerol:biodiesel) [187]. There is abundantly glycerol being a cheap raw material that could be utilized by several microorganisms, which can convert glycerol to LA, such as Klebsiella pneumonia [188], Clostridium pasteurianum [189], Lb. Reuteri [13, 14], Lb. Brevis [13, 14], Lb. Buchneri [13, 14], wild/engineered E. coli [53, 190, 191, 192, 193]. Engineered Enterococcus faecal [194], and Achromobacter denitrificans NBRC 12669 [195]. According to Mazumdar et al. (2010) [53] and Posada et al. (2012a, b) [59,187,196,197], the over expressing pathways in engineered E. coli strains via homofermentative route could convert glycerol to D-lactate [59, 187, 196].
3.5. Microalgae
Algal biomass is another source for LA production [15, 108, 134]. Some advantages of these substrates include: 1) the richness in carbohydrates, essential fatty acids, vitamins, and proteins; 2) the lignin absence in microalgae could simplify its conversion into fermentable sugars [198,199]; 3) the growth can be almost anywhere with extremely short harvest cycles of about 1–10 days [197]. 4) The use of microalgae and cyanobacteria is capable to decrease the feedstock cost, as a result of their ability to utilize light energy to fix CO2 [134]. The microalga Hydrodictyon reticulum has been utilized as a substrate for the production of L-LA by Lb. paracasei LA104 and Lb. coryniformis subsp. Torquens [198]. Lb. paracasei LA104 and L. coryniformis subsp. torquens, by simultaneous saccharification and co-fermentation, achieved values of 37.1 g/l and 36.6 g/l LA and D-LA, respectively, from 80 g Hydrodictyon reticulatum (47.5%) [198, 199]. Lipid-free microalgae are good sources for LA production, such as Nannochloropsis salina for Lb. Pentosus [199], Chlamydomonas reinhardtii, Chlorell pyrenoidosa, and Dunaliella tertiolecta for L. amylovorus [13, 14].
3.6. Feed stock pretreatment
Generally, three leading stages could be demonstrated for efficient fermentative LA production mainly (i) feedstock pretreatment, (ii) mixed and other substrates for LA production, (iii) ion requirement [10, 134, 147, 200].
The chemical composition of substrate mainly consist of carbon and nitrogen compounds. A lignocellulosic agricultural residue as worldwide resource is comprised of three main polymers: cellulose, hemicellulose and lignin, linked by covalent and non-covalent bonds. Not only, this organised structure cause to prevent cellulose and hemicelluloses hydrolysis into fermentable sugars, but also inhibit the valorisation of lignin into chemicals. The impacts of various pretreatment methods upon diverse lignocellulosic materials, e.g., wheat straw, corn stover, rice straw, switchgrass, and sugarcane bagasse have been demonstrated [10, 14, 134, 147, 200]. The pretreatment process is extremely crucial stage in lignocellulose bioconversion. If it is too intense, toxic compounds can be generated which prevent microbial metabolism and growth. In contrast, insufficient pretreatment will cause, the resultant residue is not easily saccharified through hydrolytic enzymes. Therefore, pretreatment has a great potential to affect the downstream costs due to enzymatic hydrolysis rates, enzyme loading, determining fermentation toxicity, mixing power, power generation, product purification, product concentrations, waste treatment demands, and other process variables. Numerous pretreatments for lignocellulosic materials are suggested as follow:
3.6.1. Physical pretreatment
-
1)
Milling is being conducted for approximately all solid feedstocks to decrease particle size and cause it more accessible to other treatments or hydrolysis.
In order to improve fermentation, hydrolysis of carbohydrates to fermentable sugars is performed to facilitate microorganisms growth and their accessibility for biochemical conversion to LA. The hydrolysis of starchy substrate is carried out by amylolytic enzymes upon gelatinization, liquefaction and saccharification. The optimization of hydrolysis could be conducted for numerous substrates, temperature, time and mixing conditions etc [10, 14, 134, 147, 200]. 2) Liquid hot water and emerging technologies including pulsed electric field, high hydrostatic pressure and high pressure homogenization, ionizing (X-ray, beam) and non-ionizing (microwaves) radiation and non-thermal plasma can be also suitable as pretreatments or co-treatments during hydrolysis in biorefinery processes, predominantly for lignocellulosic substrates and other substrates [10, 14, 134, 147, 200].
3.6.2. Chemical pretreatment
Combination of thermal pretreatments with alkaline, lime, organosolv, ammonia fiber explosion and ammonia recycle percolation, ionic liquid, natural deep eutectic solvents are “greener” method, and acids, making changes in all three portions of lignocellulose substrate [10]. Acid treatment was predominantly applied in the hydrolysis of lignocellulose. Dilute acid pretreatment reaction can cleave labile ester groups and catalyze the hydrolysis of the glycosidic bonds of hemicellulose and lignin. Hydrolysis of both hemicellulose and lignin, in turn, production of toxic by-products. Although, it minimizes the requirement for using hemicellulases, acid hydrolysis cannot be combined with further enzymatic steps. Moreover, thermo-chemical pretreatments are considered as energy demanding and not environment friendly. The major drawback in the production of LA on lignocelluloses is formation of numerous undesirable compounds including furfural, uronic acid, vanillic acid, 4-hydroxybenzoic acid lignin or salts can influence microbial growth during fermentation and slow-down the fermentation and increase purification costs [10, 14, 134, 147, 200].
3.6.3. Biological pretreatments
This category of pretreatment is greater eco-friendly method than others and consists of various methods including:
-
1)
Utilization of more productive species to decline time necessary for microbial growth and formation of enzymes and hence cause to increase efficiently and economically processes. For instance, basidiomycetes or their enzymes (lignin peroxidase, laccase and manganese peroxidase) to degrade lignocellulosic biomass [10, 14, 134, 147, 200].
-
2)
Enzymatic hydrolysis is the abundant method to produce fermentable sugars from pretreated lignocellulosic biomass via depolymerizes the polysaccharides in the water-insoluble solid fraction. Therefore, it is critical step to consume polysaccharides as a carbon source by LAB [14]. Cellulases and hemicellulases enzymes can convert cellulose and hemicellulose into soluble sugars, respectively. In order to enhance enzymatic hydrolysis efficiency, mixtures of these enzymes are required to improve hemicellulose hydrolysis and then rise the access of cellulase, inducing to a reduced hydrolysis time and process cost [14]. Effective degradation and saccharification of cellulose demand a synergistic reaction of the 3 categories of cellulolytic enzymes in order: (i) Endo-β-1,4-glucanases (EG; EC 3.2.1.3) can randomly dissociate accessible intramolecular β -1,4-glucosidic bonds of cellulose chains, generating a new reducing and non-reducing chain end pair. (ii) Exo- β -1,4-glucanases or cellobiohydrolases (CBH; EC 3.2.1.91) can hydrolyze cellulose chains at the ends of the polymer, forming soluble cellobiose or glucose. (iii) β -Glucosidases (β -G; EC 3.2.1.21) (cellobiases) are capable complete the hydrolysis by cleaving cellobiose into 2 glucose molecules. They are also active on cellooligosaccharides. Besides, there are accessory or “helper” enzymes that play a main role in hydrolysis by clearing the access of the leading enzymes to cellulose due to attack hemicellulose and lignin. Xylan does not generate tight crystalline structures, so the substrate is more easily accessible. However, in contrast to cellulose, xylans are chemically quite complex, and their hydrolysis needs multiple enzymes. Enzymatic hydrolysis of hemicellulose was performed by β -xylosidase, endo-1,4- β -xylanase, β - glucuronidase, α -l-arabinofuranosidase, galactomannanase, glucomannanase and acetylxylan esterase, which act on xylan cleavage and saccharification. β -mannanase and β -mannosidase, which cause to cleave the glucomannan polymer backbone [14]. The hydrolytic efficiency of lignocellulose substantially improved by utilizing combinations of the 3 enzymes, 2 cellulases, and 1 xylanase [10, 14, 134, 147, 200].
3.6.4. Mixed and other substrates for LA production
Wastes or by-products are main representatives of mixed substrates with different composition of carbohydrates and proteins. Meanwhile, they contain low nutritional values, so require additional fortification and often some treatment. Inhibitory or toxic components in these media have to be evaluated, also. Instead of consumption yeast extract or other Unconventional and expensive nitrogen sources, numerous agricultural residues or byproducts namely soya bean hydrolysate, corn steep liquor, corn meal and wheat bran hydrolysate, chicken feather hydrolysate, by-products from malting and brewing and oil production can be utilized as cheaper nitrogen sources [10, 14, 134, 147, 200] (Table 1). Substantial studies were demonstrated in the case of free amino nitrogen content such as amino acids, and phosphate to LA production. Complementary substrates in nitrogen and carbohydrate sources were combined for LA fermentation. For instance sugar beet molasses (rich in carbohydrates) and distillery stillage from bioethanol production from waste potato (rich in nitrogen source) were used for LA production by Lactobacillus paracasei. Many studies have shown that how to determine carbon to nitrogen ratio and correlate it with LA productivity. Carbon/nitrogen ratio significantly effects on LA yield and cell growth. When the carbon and nitrogen content are provided only from fermentable sugars and free amino nitrogen content, accurate optimization of media composition for LA production would be performed [10, 14, 134, 147, 200].
3.6.5. Ion requirement
It is obvious that metals play a key role in the biological processes, such as activating major enzymes in metabolisms as cofactor, improving the growth of microbial cells and activation of organic acid synthesis by fungal and bacterial species [201].
3.6.5.1. Copper
Copper (II) by far has acted as a cofactor within numerous copper-dependent enzymes [201]. Furthermore, the microbial populations including LAB are more affected in the presence of copper (II) [202, 203, 204]. There are several hypotheses to improve lactic acid production in the presence of copper: a) it was proved that copper (II) inhibited the conversion of D-lactic acid to pyruvate via preventing the activity of NAD independent D-lactate dehydrogenas (id-LDH) in the pure culture, b) carbohydrate hydrolysis and glycolysis pathway were both strengthened that resulted in the promoting of lactic acid production from organic waste. The amount of copper (Cu-15; 15 μM/g, Cu-30; 30 μM/g and Cu-70; 70 μM/g) influence on the production of lactic acid (23.21 g/L), (17.44 g/L) and (16.53 g/L), respectively. It is indicated that the maximum concentration of lactic acid increased in the presence of copper compared to that of Blank (13.11 g/L). Nevertheless, continuously raising the copper level gradually reduced the production of lactic acid imply that that 70 μM-Cu2+/g VSS might exceed the tolerance of Lactobacillus and variation of functional genes revealed that the suggested homeostatic system towards copper (II) was activated at pretty low content that cause to facilitate the membrane transport function as well as carbohydrate metabolism [201].
3.6.5.2. Zinc
Regarding to Mumtaz et al., 2019, ZnO solubilization was associated to the synthesis of specific organic acids like Lactic and acetic acids. The culture medium was acidified and then ZnO solubilized. Two Zn- and acid-tolerant strains. Rhizosphere isolate Bacillus sp. ZM20 and B. cereus culture-collection strain generated various organic acids at a remarkably greater content than less tolerant strains when cultured in the presence of inhibitory but non-lethal levels of ZnO. It is supposed that the enhanced synthesis of these acids is due to a generalized stress response [205].
4. Conclusions
The capacity of several microorganisms for production of LA was studied. Some of these microorganisms such as LAB require complex nutrients and low fermentation temperatures, which lead to increased costs and contamination risk. However, some of them like Bacillus spp., reduce the LA production cost due to fewer nutrition demands and a high temperature of fermentation. Agro-industrial waste or sub-products with a lower value such as molasses, juices waste, starchy biomass, agricultural residues, forestry residues that are rich in mono and disaccharides, which in some cases need to be hydrolysed by pectinases to enhanced the LA production. To use dairy wastes as a substrate, mainly whey, it is necessary to use an enriched mediums, due to insufficient proteolytic enzyme activity.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Upadhyaya B.P., DeVeaux L.C., Christopher L.P. Metabolic engineering as a tool for enhanced lactic acid production. 2014;32:637–644. doi: 10.1016/j.tibtech.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 2.de Oliveira R.A., Komesu A., Rossell C.E.V., Maciel Filho R. Challenges and opportunities in lactic acid bioprocess design – from economic to production aspects. Biochem. Eng. J. 2018;133:219–239. [Google Scholar]
- 3.Pleissner D., Demichelis F., Mariano S., Fiore S., Navarro Gutiérrez I.M., Schneider R., Venus J. Direct production of lactic acid based on simultaneous saccharification and fermentation of mixed restaurant food waste. J. Clean. Prod. 2017;143:615–623. [Google Scholar]
- 4.Payot T., Chemaly Z., Fick M. Lactic acid production by Bacillus coagulans—kinetic studies and optimization of culture medium for batch and continuous fermentations. Enzym. Microb. Technol. 1999;24:191–199. [Google Scholar]
- 5.Kylä-Nikkilä K., Hujanen M., Leisola M., Palva A. Metabolic engineering of lactobacillus helveticus CNRZ32 for production of purel-(+)-lactic acid. Appl. Environ. Microbiol. 2000;66:3835–3841. doi: 10.1128/aem.66.9.3835-3841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy G., Altaf M., Naveena B., Venkateshwar M., Kumar E.V. Amylolytic bacterial lactic acid fermentation—a review. Biotechnol. Adv. 2008;26:22–34. doi: 10.1016/j.biotechadv.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Singh S.K., Ahmed S.U., Pandey A. Metabolic engineering approaches for lactic acid production. Process Biochem. 2006;41:991–1000. [Google Scholar]
- 8.Zheng J., Wittouck S., Salvetti E., Franz C.M.A.P., Harris H.M.B., Mattarelli P., O’Toole P.W., Pot B., Vandamme P., Walter J., Watanabe K., Wuyts S., Felis G.E., Gänzle M.G., Lebeer S. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 9.Yokaryo H., Tokiwa Y. Isolation of alkaliphilic bacteria for production of high optically pure L-(+)-lactic acid. J. Gen. Appl. Microbiol. 2014;60:270–275. doi: 10.2323/jgam.60.270. [DOI] [PubMed] [Google Scholar]
- 10.John R.P., Anisha G.S., Nampoothiri K.M., Pandey A. Direct lactic acid fermentation: focus on simultaneous saccharification and lactic acid production. Biotechnol. Adv. 2009;27:145–152. doi: 10.1016/j.biotechadv.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Martinez F.A.C., Balciunas E.M., Salgado J.M., González J.M.D., Converti A., de Souza Oliveira R.P. Lactic acid properties, applications and production: a review. Trends Food Sci. Technol. 2013;30:70–83. [Google Scholar]
- 12.Hassan S.E.-D., Abdel-Rahman M.A., Roushdy M.M., Azab M.S., Gaber M.A. Effective biorefinery approach for lactic acid production based on cofermentation of mixed organic wastes by Enterococcus durans BP130. Biocataly. Agric. Biotechnol. 2019;20:101203. [Google Scholar]
- 13.Abdel-Rahman M.A., Tashiro Y., Sonomoto K. Recent advances in lactic acid production by microbial fermentation processes. Biotechnol. Adv. 2013;31:877–902. doi: 10.1016/j.biotechadv.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Abdel-Rahman M.A., Tashiro Y., Sonomoto K. Lactic acid production from lignocellulose-derived sugars using lactic acid bacteria: overview and limits. J. Biotechnol. 2011;156:286–301. doi: 10.1016/j.jbiotec.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Rahman M.A., Tashiro Y., Zendo T., Sonomoto K. Improved lactic acid productivity by an open repeated batch fermentation system using Enterococcus mundtii QU 25. RSC Adv. 2013;3:8437–8445. [Google Scholar]
- 16.Meng Y., Xue Y., Yu B., Gao C., Ma Y. Efficient production of L-lactic acid with high optical purity by alkaliphilic Bacillus sp. WL-S20. Bioresour. Technol. 2012;116:334–339. doi: 10.1016/j.biortech.2012.03.103. [DOI] [PubMed] [Google Scholar]
- 17.Patel M.A., Ou M.S., Harbrucker R., Aldrich H.C., Buszko M.L., Ingram L.O., Shanmugam K. Isolation and characterization of acid-tolerant, thermophilic bacteria for effective fermentation of biomass-derived sugars to lactic acid. Appl. Environ. Microbiol. 2006;72:3228–3235. doi: 10.1128/AEM.72.5.3228-3235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Authority E.F.S. The maintenance of the list of QPS microorganisms intentionally added to food or feed-Scientific Opinion of the Panel on Biological Hazards. EFSA J. 2008;6:923. [Google Scholar]
- 19.Bischoff K.M., Liu S., Hughes S.R., Rich J.O. Fermentation of corn fiber hydrolysate to lactic acid by the moderate thermophile Bacillus coagulans. Biotechnol. Lett. 2010;32:823–828. doi: 10.1007/s10529-010-0222-z. [DOI] [PubMed] [Google Scholar]
- 20.Budhavaram N.K., Fan Z. Production of lactic acid from paper sludge using acid-tolerant, thermophilic Bacillus coagulan strains. Bioresour. Technol. 2009;100:5966–5972. doi: 10.1016/j.biortech.2009.01.080. [DOI] [PubMed] [Google Scholar]
- 21.Hu J., Zhang Z., Lin Y., Zhao S., Mei Y., Liang Y., Peng N. High-titer lactic acid production from NaOH-pretreated corn stover by Bacillus coagulans LA204 using fed-batch simultaneous saccharification and fermentation under non-sterile condition. Bioresour. Technol. 2015;182:251–257. doi: 10.1016/j.biortech.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Ma K., Maeda T., You H., Shirai Y. Open fermentative production of l-lactic acid with high optical purity by thermophilic Bacillus coagulans using excess sludge as nutrient. Bioresour. Technol. 2014;151:28–35. doi: 10.1016/j.biortech.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Maas R.H., Springer J., Eggink G., Weusthuis R.A. Xylose metabolism in the fungus Rhizopus oryzae: effect of growth and respiration on L (+)-lactic acid production. J. Ind. Microbiol. Biotechnol. 2008;35:569–578. doi: 10.1007/s10295-008-0318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michelson T., Kask K., Jõgi E., Talpsep E., Suitso I., Nurk A. L (+)-Lactic acid producer Bacillus coagulans SIM-7 DSM 14043 and its comparison with Lactobacillus delbrueckii ssp. lactis DSM 20073. Enzym. Microb. Technol. 2006;39:861–867. [Google Scholar]
- 25.Ouyang J., Cai C., Chen H., Jiang T., Zheng Z. Efficient non-sterilized fermentation of biomass-derived xylose to lactic acid by a thermotolerant Bacillus coagulans NL01. Appl. Biochem. Biotechnol. 2012;168:2387–2397. doi: 10.1007/s12010-012-9944-9. [DOI] [PubMed] [Google Scholar]
- 26.Rosenberg M., Rebroš M., Krištofíková L., Malátová K. High temperature lactic acid production by Bacillus coagulans immobilized in LentiKats. Biotechnol. Lett. 2005;27:1943–1947. doi: 10.1007/s10529-005-3907-y. [DOI] [PubMed] [Google Scholar]
- 27.Sakai K., Ezaki Y. Open L-lactic acid fermentation of food refuse using thermophilic Bacillus coagulans and fluorescence in situ hybridization analysis of microflora. J. Biosci. Bioeng. 2006;101:457–463. doi: 10.1263/jbb.101.457. [DOI] [PubMed] [Google Scholar]
- 28.Walton S.L., Bischoff K.M., van Heiningen A.R., van Walsum G.P. Production of lactic acid from hemicellulose extracts by Bacillus coagulans MXL-9. J. Ind. Microbiol. Biotechnol. 2010;37:823–830. doi: 10.1007/s10295-010-0727-4. [DOI] [PubMed] [Google Scholar]
- 29.Wang L., Zhao B., Liu B., Yu B., Ma C., Su F., Hua D., Li Q., Ma Y., Xu P. Efficient production of L-lactic acid from corncob molasses, a waste by-product in xylitol production, by a newly isolated xylose utilizing Bacillus sp. strain. Bioresour. Technol. 2010;101:7908–7915. doi: 10.1016/j.biortech.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Deng W., Wang B., Zhang Q., Wan X., Tang Z., Wang Y., Zhu C., Cao Z., Wang G. Chemical synthesis of lactic acid from cellulose catalysed by lead (II) ions in water. Nat. Commun. 2013;4:2141. doi: 10.1038/ncomms3141. [DOI] [PubMed] [Google Scholar]
- 31.Ye L., Hudari M.S.B., Zhou X., Zhang D., Li Z., Wu J.C. Conversion of acid hydrolysate of oil palm empty fruit bunch to L-lactic acid by newly isolated Bacillus coagulans JI12. Appl. Microbiol. Biotechnol. 2013;97:4831–4838. doi: 10.1007/s00253-013-4788-y. [DOI] [PubMed] [Google Scholar]
- 32.Ye L., Zhao H., Li Z., Wu J.C. Improved acid tolerance of Lactobacillus pentosus by error-prone whole genome amplification. Bioresour. Technol. 2013;135:459–463. doi: 10.1016/j.biortech.2012.10.042. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Chen X., Luo J., Qi B., Wan Y. An efficient process for lactic acid production from wheat straw by a newly isolated Bacillus coagulans strain IPE22. Bioresour. Technol. 2014;158:396–399. doi: 10.1016/j.biortech.2014.02.128. [DOI] [PubMed] [Google Scholar]
- 34.Sakai K., Yamanami T. Thermotolerant Bacillus licheniformis TY7 produces optically active L-lactic acid from kitchen refuse under open condition. J. Biosci. Bioeng. 2006;102:132–134. doi: 10.1263/jbb.102.132. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q., Zhao X., Chamu J., Shanmugam K. Isolation, characterization and evolution of 2243 a new thermophilic Bacillus licheniformis for lactic acid production in mineral salts 2244 medium. Bioresour. Technol. 2011;102:8128–8152. doi: 10.1016/j.biortech.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Gao T., Wong Y., Ng C., Ho K. L-lactic acid production by Bacillus subtilis MUR1. Bioresour. Technol. 2012;121:105–110. doi: 10.1016/j.biortech.2012.06.108. [DOI] [PubMed] [Google Scholar]
- 37.Peng L., Wang L., Che C., Yang G., Yu B., Ma Y. Bacillus sp. strain P38: an efficient producer of L-lactate from cellulosic hydrolysate, with high tolerance for 2-furfural. Bioresour. Technol. 2013;149:169–176. doi: 10.1016/j.biortech.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 38.Qin J., Wang X., Zheng Z., Ma C., Tang H., Xu P. Production of L-lactic acid by a thermophilic Bacillus mutant using sodium hydroxide as neutralizing agent. Bioresour. Technol. 2010;101:7570–7576. doi: 10.1016/j.biortech.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 39.Qin J., Zhao B., Wang X., Wang L., Yu B., Ma Y., Ma C., Tang H., Sun J., Xu P. Non-sterilized fermentative production of polymer-grade L-lactic acid by a newly isolated thermophilic strain Bacillus sp. 2–6. PloS One. 2009;4:e4359. doi: 10.1371/journal.pone.0004359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao B., Wang L., Ma C., Yang C., Xu P., Ma Y. Repeated open fermentative production of optically pure L-lactic acid using a thermophilic Bacillus sp. strain. Bioresour. Technol. 2010;101:6494–6498. doi: 10.1016/j.biortech.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 41.Inui M., Murakami S., Okino S., Kawaguchi H., Vertès A.A., Yukawa H. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 2004;7:182–196. doi: 10.1159/000079827. [DOI] [PubMed] [Google Scholar]
- 42.Jia X., Liu P., Li S., Li S., Wen J. D-lactic acid production by a genetically engineered strain Corynebacterium glutamicum. World J. Microbiol. Biotechnol. 2011;27:2117–2124. [Google Scholar]
- 43.Okino S., Suda M., Fujikura K., Inui M., Yukawa H. Production of D-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl. Microbiol. Biotechnol. 2008;78:449–454. doi: 10.1007/s00253-007-1336-7. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki M., Jojima T., Inui M., Yukawa H. Simultaneous utilization of D-cellobiose, D-glucose, and D-xylose by recombinant Corynebacterium glutamicum under oxygen-deprived conditions. Appl. Microbiol. Biotechnol. 2008;81:691–699. doi: 10.1007/s00253-008-1703-z. [DOI] [PubMed] [Google Scholar]
- 45.Kawaguchi H., Sasaki M., Vertès A.A., Inui M., Yukawa H. Engineering of an L-arabinose metabolic pathway in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2008;77:1053–1062. doi: 10.1007/s00253-007-1244-x. [DOI] [PubMed] [Google Scholar]
- 46.Kawaguchi H., Vertes A.A., Okino S., Inui M., Yukawa H. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2006;72:3418–3428. doi: 10.1128/AEM.72.5.3418-3428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okano K., Tanaka T., Ogino C., Fukuda H., Kondo A. Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl. Microbiol. Biotechnol. 2010;85:413–423. doi: 10.1007/s00253-009-2280-5. [DOI] [PubMed] [Google Scholar]
- 48.Okino S., Inui M., Yukawa H. Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl. Microbiol. Biotechnol. 2005;68:475–480. doi: 10.1007/s00253-005-1900-y. [DOI] [PubMed] [Google Scholar]
- 49.Yukawa H., Omumasaba C.A., Nonaka H., Kos P., Okai N., Suzuki N., Suda M., Tsuge Y., Watanabe J., Ikeda Y. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology. 2007;153:1042–1058. doi: 10.1099/mic.0.2006/003657-0. [DOI] [PubMed] [Google Scholar]
- 50.Song Y., Matsumoto K.i., Yamada M., Gohda A., Brigham C.J., Sinskey A.J., Taguchi S. Engineered Corynebacterium glutamicum as an endotoxin-free platform strain for lactate-based polyester production. Appl. Microbiol. Biotechnol. 2012;93:1917–1925. doi: 10.1007/s00253-011-3718-0. [DOI] [PubMed] [Google Scholar]
- 51.Chai X., Shang X., Zhang Y., Liu S., Liang Y., Zhang Y., Wen T. A novel pyruvate kinase and its application in lactic acid production under oxygen deprivation in Corynebacterium glutamicum. BMC Biotechnol. 2016;16:79. doi: 10.1186/s12896-016-0313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou S., Causey T., Hasona A., Shanmugam K., Ingram L. Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl. Environ. Microbiol. 2003;69:399–407. doi: 10.1128/AEM.69.1.399-407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazumdar S., Blankschien M.D., Clomburg J.M., Gonzalez R. Efficient synthesis of L-lactic acid from glycerol by metabolically engineered Escherichia coli. Microb. Cell Factories. 2013;12:7. doi: 10.1186/1475-2859-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Tian T., Zhao J., Wang J., Yan T., Xu L., Liu Z., Garza E., Iverson A., Manow R. Homofermentative production of d-lactic acid from sucrose by a metabolically engineered Escherichia coli. Biotechnol. Lett. 2012;34:2069–2075. doi: 10.1007/s10529-012-1003-7. [DOI] [PubMed] [Google Scholar]
- 55.Zhou S., Yomano L., Shanmugam K., Ingram L. Fermentation of 10%(w/v) sugar to D (−)-lactate by engineered Escherichia coli B. Biotechnol. Lett. 2005;27:1891–1896. doi: 10.1007/s10529-005-3899-7. [DOI] [PubMed] [Google Scholar]
- 56.Dien B., Nichols N., Bothast R. Recombinant Escherichia coli engineered for production of L-lactic acid from hexose and pentose sugars. J. Ind. Microbiol. Biotechnol. 2001;27:259–264. doi: 10.1038/sj.jim.7000195. [DOI] [PubMed] [Google Scholar]
- 57.Zhou S., Shanmugam K., Ingram L. Functional replacement of the Escherichia coli D-(−)-lactate dehydrogenase gene (ldhA) with the L-(+)-lactate dehydrogenase gene (ldhL) from Pediococcus acidilactici. Appl. Environ. Microbiol. 2003;69:2237–2244. doi: 10.1128/AEM.69.4.2237-2244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Y., Eiteman M., DeWitt K., Altman E. Homolactate fermentation by metabolically engineered Escherichia coli strains. Appl. Environ. Microbiol. 2007;73:456–464. doi: 10.1128/AEM.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mazumdar S., Clomburg J.M., Gonzalez R. Escherichia coli strains engineered for homofermentative production of D-lactic acid from glycerol. Appl. Environ. Microbiol. 2010;76:4327–4336. doi: 10.1128/AEM.00664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q., Chamu Z.X.J., Shanmugam K. Isolation, characterization and evolution of 2243 a new thermophilic Bacillus licheniformis for lactic acid production in mineral salts 2244 medium. Bioresour. Technol. 2011;102:8128–8152. doi: 10.1016/j.biortech.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 61.Meussen B.J., de Graaff L.H., Sanders J.P., Weusthuis R.A. Metabolic engineering of Rhizopus oryzae for the production of platform chemicals. Appl. Microbiol. Biotechnol. 2012;94:875–886. doi: 10.1007/s00253-012-4033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bai D.-M., Li S.-Z., Liu Z.L., Cui Z.-F. Enhanced l-(+)-lactic acid production by an adapted strain of Rhizopus oryzae using corncob hydrolysate. Appl. Biochem. Biotechnol. 2008;144:79–85. doi: 10.1007/s12010-007-8078-y. [DOI] [PubMed] [Google Scholar]
- 63.Miura S., Arimura T., Itoda N., Dwiarti L., Feng J.B., Bin C.H., Okabe M. Production of L-lactic acid from corncob. J. Biosci. Bioeng. 2004;97:153–157. doi: 10.1016/S1389-1723(04)70184-X. [DOI] [PubMed] [Google Scholar]
- 64.Moldes A., Alonso J., Parajo J. Multi-step feeding systems for lactic acid production by simultaneous saccharification and fermentation of processed wood. Bioprocess Eng. 2000;22:175–180. [Google Scholar]
- 65.Ruengruglikit C., Hang Y. L (+)-lactic acid production from corncobs by Rhizopus oryzae NRRL-395. LWT-Food Sci. Technol. 2003;36:573–575. [Google Scholar]
- 66.Maas R.H., Bakker R.R., Eggink G., Weusthuis R.A. Lactic acid production from xylose by the fungus Rhizopus oryzae. Appl. Microbiol. Biotechnol. 2006;72:861–868. doi: 10.1007/s00253-006-0379-5. [DOI] [PubMed] [Google Scholar]
- 67.Saitoh S., Ishida N., Onishi T., Tokuhiro K., Nagamori E., Kitamoto K., Takahashi H. Genetically engineered wine yeast produces a high concentration of L-lactic acid of extremely high optical purity. Appl. Environ. Microbiol. 2005;71:2789–2792. doi: 10.1128/AEM.71.5.2789-2792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park E.Y., Anh P.N., Okuda N. Bioconversion of waste office paper to L (+)-lactic acid by the filamentous fungus Rhizopus oryzae. Bioresour. Technol. 2004;93:77–83. doi: 10.1016/j.biortech.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 69.Saito K., Hasa Y., Abe H. Production of lactic acid from xylose and wheat straw by Rhizopus oryzae. J. Biosci. Bioeng. 2012;114:166–169. doi: 10.1016/j.jbiosc.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 70.Taherzadeh M.J., Fox M., Hjorth H., Edebo L. Production of mycelium biomass and ethanol from paper pulp sulfite liquor by Rhizopus oryzae. Bioresour. Technol. 2003;88:167–177. doi: 10.1016/s0960-8524(03)00010-5. [DOI] [PubMed] [Google Scholar]
- 71.Taskin M., Esim N., Ortucu S. Efficient production of l-lactic acid from chicken feather protein hydrolysate and sugar beet molasses by the newly isolated Rhizopus oryzae TS-61. Food Bioprod. Process. 2012;90:773–779. [Google Scholar]
- 72.Thongchul N., Navankasattusas S., Yang S.-T. Production of lactic acid and ethanol by Rhizopus oryzae integrated with cassava pulp hydrolysis. Bioproc. Biosyst. Eng. 2010;33:407–416. doi: 10.1007/s00449-009-0341-x. [DOI] [PubMed] [Google Scholar]
- 73.Liu Y., Liao W. S.-l. Chen, Co-production of lactic acid and chitin using a pelletized filamentous fungus Rhizopus oryzae cultured on cull potatoes and glucose. J. Appl. Microbiol. 2008;105:1521–1528. doi: 10.1111/j.1365-2672.2008.03913.x. [DOI] [PubMed] [Google Scholar]
- 74.Vodnar D.C., Dulf F.V., Pop O.L., Socaciu C. L (+)-lactic acid production by pellet-form Rhizopus oryzae NRRL 395 on biodiesel crude glycerol. Microb. Cell Factories. 2013;12:92. doi: 10.1186/1475-2859-12-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin B., Huang L.P., Lant P. Rhizopus arrhizus–a producer for simultaneous saccharification and fermentation of starch waste materials to L (+)-lactic acid. Biotechnol. Lett. 2003;25:1983–1987. doi: 10.1023/b:bile.0000004389.53388.d0. [DOI] [PubMed] [Google Scholar]
- 76.Bulut S., Elibol M., Ozer D. Effect of different carbon sources on L (+)-lactic acid production by Rhizopus oryzae. Biochem. Eng. J. 2004;21:33–37. [Google Scholar]
- 77.Marták J., Schlosser Š., Sabolová E., Krištofíková L., Rosenberg M. Fermentation of lactic acid with Rhizopus arrhizus in a stirred tank reactor with a periodical bleed and feed operation. Process Biochem. 2003;38:1573–1583. [Google Scholar]
- 78.Oda Y., Yajima Y., Kinoshita M., Ohnishi M. Differences of Rhizopus oryzae strains in organic acid synthesis and fatty acid composition. Food Microbiol. 2003;20:371–375. [Google Scholar]
- 79.Zhang Z.Y., Jin B., Kelly J.M. Production of lactic acid from renewable materials by Rhizopus fungi. Biochem. Eng. J. 2007;35:251–263. [Google Scholar]
- 80.Liu T., Miura S., Yaguchi M., Arimura T., Park E.Y., Okabe M. Scale-up of L-Lactic acid production by mutant strain Rhizopus sp. Mk-96-1196 from 0.003 m3 to 5 m3 in airlift bioreactors. J. Biosci. Bioeng. 2006;101:9–12. doi: 10.1263/jbb.101.9. [DOI] [PubMed] [Google Scholar]
- 81.Liu Y., Liao W., Liu C., Chen S. Twenty-Seventh Symposium on Biotechnology for Fuels and Chemicals. Springer; 2006. Optimization of L-(+)-lactic acid production using pelletized filamentous Rhizopus oryzae NRRL 395; pp. 844–853. [PubMed] [Google Scholar]
- 82.Maneeboon T., Vanichsriratana W., Pomchaitaward C., Kitpreechavanich V. Optimization of lactic acid production by pellet-form Rhizopus oryzae in 3-L airlift bioreactor using response surface methodology. Appl. Biochem. Biotechnol. 2010;161:137–146. doi: 10.1007/s12010-009-8860-0. [DOI] [PubMed] [Google Scholar]
- 83.Bai D.-M., Wei Q., Yan Z.-H., Zhao X.-M., Li X.-G., Xu S.-M. Fed-batch fermentation of Lactobacillus lactis for hyper-production of L-lactic acid. Biotechnol. Lett. 2003;25:1833–1835. doi: 10.1023/a:1026276925649. [DOI] [PubMed] [Google Scholar]
- 84.Wu X., Jiang S., Liu M., Pan L., Zheng Z., Luo S. Production of l-lactic acid by Rhizopus oryzae using semicontinuous fermentation in bioreactor. J. Ind. Microbiol. Biotechnol. 2011;38:565–571. doi: 10.1007/s10295-010-0804-8. [DOI] [PubMed] [Google Scholar]
- 85.Efremenko E., Spiricheva O., Varfolomeyev S., Lozinsky V. Rhizopus oryzae fungus cells producing L (+)-lactic acid: kinetic and metabolic parameters of free and PVA-cryogel-entrapped mycelium. Appl. Microbiol. Biotechnol. 2006;72:480–485. doi: 10.1007/s00253-005-0297-y. [DOI] [PubMed] [Google Scholar]
- 86.Tay A., Yang S.T. Production of L (+)-lactic acid from glucose and starch by immobilized cells of Rhizopus oryzae in a rotating fibrous bed bioreactor. Biotechnol. Bioeng. 2002;80:1–12. doi: 10.1002/bit.10340. [DOI] [PubMed] [Google Scholar]
- 87.Litchfield J. Lactic acid, microbially produced. In: SchaechterMosel O., editor. Encyclopedia of Microbiology. Academic Press; Oxford: 2009. pp. 362–372. [Google Scholar]
- 88.Magnuson J.K., Lasure L.L. Springer; 2004. Organic Acid Production by Filamentous Fungi, Advances in Fungal Biotechnology for Industry, Agriculture, and Medicine; pp. 307–340. [Google Scholar]
- 89.Vink E.T., Davies S., Kolstad J.J. The eco-profile for current Ingeo® polylactide production. Ind. Biotechnol. 2010;6:212–224. [Google Scholar]
- 90.Thitiprasert S., Sooksai S., Thongchul N. In vivo regulation of alcohol dehydrogenase and lactate dehydrogenase in Rhizopus oryzae to improve L-lactic acid fermentation. Appl. Biochem. Biotechnol. 2011;164:1305–1322. doi: 10.1007/s12010-011-9214-2. [DOI] [PubMed] [Google Scholar]
- 91.Ge X.-Y., Qian H., Zhang W.-G. Improvement of l-lactic acid production from Jerusalem artichoke tubers by mixed culture of Aspergillus Niger and Lactobacillus sp. Bioresour. Technol. 2009;100:1872–1874. doi: 10.1016/j.biortech.2008.09.049. [DOI] [PubMed] [Google Scholar]
- 92.van Maris A.J., Winkler A.A., Porro D., van Dijken J.P., Pronk J.T. Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy-dependent lactate export. Appl. Environ. Microbiol. 2004;70:2898–2905. doi: 10.1128/AEM.70.5.2898-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bianchi M., Brambilla L., Protani F., Liu C.-L., Lievense J., Porro D. Efficient homolactic fermentation byKluyveromyces lactis strains defective in pyruvate utilization and transformed with the HeterologousLDH gene. Appl. Environ. Microbiol. 2001;67:5621–5625. doi: 10.1128/AEM.67.12.5621-5625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Colombié S., Sablayrolles J.-M. Nicotinic acid controls lactate production by K1-LDH: a Saccharomyces cerevisiae strain expressing a bacterial LDH gene. J. Ind. Microbiol. Biotechnol. 2004;31:209–215. doi: 10.1007/s10295-004-0138-5. [DOI] [PubMed] [Google Scholar]
- 95.Ishida N., Suzuki T., Tokuhiro K., Nagamori E., Onishi T., Saitoh S., Kitamoto K., Takahashi H. D-Lactic acid production by metabolically engineered Saccharomyces cerevisiae. J. Biosci. Bioeng. 2006;101:172–177. doi: 10.1263/jbb.101.172. [DOI] [PubMed] [Google Scholar]
- 96.Skory C.D. Lactic acid production by Saccharomyces cerevisiae expressing a Rhizopus oryzae lactate dehydrogenase gene. J. Ind. Microbiol. Biotechnol. 2003;30:22–27. doi: 10.1007/s10295-002-0004-2. [DOI] [PubMed] [Google Scholar]
- 97.Tokuhiro K., Ishida N., Nagamori E., Saitoh S., Onishi T., Kondo A., Takahashi H. Double mutation of the PDC1 and ADH1 genes improves lactate production in the yeast Saccharomyces cerevisiae expressing the bovine lactate dehydrogenase gene. Appl. Microbiol. Biotechnol. 2009;82:883–890. doi: 10.1007/s00253-008-1831-5. [DOI] [PubMed] [Google Scholar]
- 98.Valli M., Sauer M., Branduardi P., Borth N., Porro D., Mattanovich D. Improvement of lactic acid production in Saccharomyces cerevisiae by cell sorting for high intracellular pH. Appl. Environ. Microbiol. 2006;72:5492–5499. doi: 10.1128/AEM.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Osawa F., Fujii T., Nishida T., Tada N., Ohnishi T., Kobayashi O., Komeda T., Yoshida S. Efficient production of L-lactic acid by Crabtree-negative yeast Candida boidinii. Yeast. 2009;26:485–496. doi: 10.1002/yea.1702. [DOI] [PubMed] [Google Scholar]
- 100.Ilmén M., Koivuranta K., Ruohonen L., Suominen P., Penttilä M. Efficient production of L-lactic acid from xylose by Pichia stipitis. Appl. Environ. Microbiol. 2007;73:117–123. doi: 10.1128/AEM.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Branduardi P., Valli M., Brambilla L., Sauer M., Alberghina L., Porro D. The yeast Zygosaccharomyces bailii: a new host for heterologous protein production, secretion and for metabolic engineering applications. FEMS Yeast Res. 2004;4:493–504. doi: 10.1016/S1567-1356(03)00200-9. [DOI] [PubMed] [Google Scholar]
- 102.Ilmén M., Koivuranta K., Ruohonen L., Rajgarhia V., Suominen P., Penttilä M. Production of L-lactic acid by the yeast Candida sonorensis expressing heterologous bacterial and fungal lactate dehydrogenases. Microb. Cell Factories. 2013;12:53. doi: 10.1186/1475-2859-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koivuranta K.T., Ilmén M., Wiebe M.G., Ruohonen L., Suominen P., Penttilä M. L-lactic acid production from D-xylose with Candida sonorensis expressing a heterologous lactate dehydrogenase encoding gene. Microb. Cell Factories. 2014;13:107. doi: 10.1186/s12934-014-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ikushima S., Fujii T., Kobayashi O., Yoshida S., Yoshida A. Genetic engineering of Candida utilis yeast for efficient production of L-lactic acid. Biosci. Biotechnol. Biochem. 2009;73:1818–1824. doi: 10.1271/bbb.90186. [DOI] [PubMed] [Google Scholar]
- 105.Pecota D.C., Rajgarhia V., Da Silva N.A. Sequential gene integration for the engineering of Kluyveromyces marxianus. J. Biotechnol. 2007;127:408–416. doi: 10.1016/j.jbiotec.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 106.Hause B., Rajgarhia V., Suominen P. Methods and materials for the production of L-lactic acid in yeast. Google Patents. 2009 [Google Scholar]
- 107.Dato L., Branduardi P., Passolunghi S., Cattaneo D., Riboldi L., Frascotti G., Valli M., Porro D. Advances in molecular tools for the use of Zygosaccharomyces bailii as host for biotechnological productions and construction of the first auxotrophic mutant. FEMS Yeast Res. 2010;10:894–908. doi: 10.1111/j.1567-1364.2010.00668.x. [DOI] [PubMed] [Google Scholar]
- 108.Juturu V., Wu J.C. Microbial production of lactic acid: the latest development. Crit. Rev. Biotechnol. 2016;36:967–977. doi: 10.3109/07388551.2015.1066305. [DOI] [PubMed] [Google Scholar]
- 109.Hirayama S., Ueda R. Production of optically pure D-lactic acid by Nannochlorum sp. 26A4. Appl. Biochem. Biotechnol. 2004;119:71–77. doi: 10.1385/abab:119:1:71. [DOI] [PubMed] [Google Scholar]
- 110.Talukder M.M.R., Das P., Wu J.C. Microalgae (Nannochloropsis salina) biomass to lactic acid and lipid. Biochem. Eng. J. 2012;68:109–113. [Google Scholar]
- 111.Angermayr S.A., Paszota M., Hellingwerf K.J. Engineering a cyanobacterial cell factory for the production of lactic acid. Applied and Environmental Microbiology. AEM. 2012:1512–1587. doi: 10.1128/AEM.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lopez-Gomez J.P., Latorre-Sanchez M., Unger P., Schneider R., Coll Lozano C., Venus J. Assessing the organic fraction of municipal solid wastes for the production of lactic acid. Biochem. Eng. J. 2019;150:107251. [Google Scholar]
- 113.Tashiro Y., Matsumoto H., Miyamoto H., Okugawa Y., Pramod P., Miyamoto H., Sakai K. A novel production process for optically pure L-lactic acid from kitchen refuse using a bacterial consortium at high temperatures. Bioresour. Technol. 2013;146:672–681. doi: 10.1016/j.biortech.2013.07.102. [DOI] [PubMed] [Google Scholar]
- 114.Zhang B., He P., Ye N., Shao L. Enhanced isomer purity of lactic acid from the nonsterile fermentation of kitchen wastes. Bioresour. Technol. 2008;99:855–862. doi: 10.1016/j.biortech.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 115.Kwan T.H., Pleissner D., Lau K.Y., Venus J., Pommeret A., Lin C.S.K. Technoeconomic analysis of a food waste valorization process via microalgae cultivation and coproduction of plasticizer, lactic acid and animal feed from algal biomass and food waste. Bioresour. Technol. 2015;198:292–299. doi: 10.1016/j.biortech.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 116.Pleissner D., Lam W.C., Sun Z., Lin C.S.K. Food waste as nutrient source in heterotrophic microalgae cultivation. Bioresour. Technol. 2013;137:139–146. doi: 10.1016/j.biortech.2013.03.088. [DOI] [PubMed] [Google Scholar]
- 117.Kitpreechavanich V., Hayami A., Talek A., Chin C.F.S., Tashiro Y., Sakai K. Simultaneous production of L-lactic acid with high optical activity and a soil amendment with food waste that demonstrates plant growth promoting activity. J. Biosci. Bioeng. 2016;122:105–110. doi: 10.1016/j.jbiosc.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 118.Tashiro Y., Inokuchi S., Poudel P., Okugawa Y., Miyamoto H., Miayamoto H., Sakai K. Novel pH control strategy for efficient production of optically active L -lactic acid fromkitchen refuse using a mixed culture system. Bioresour. Technol. 2016;216:52–59. doi: 10.1016/j.biortech.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 119.Neu A., Pleissner D., Mehlmann K., Schneider R., Puerta-quintero G.I., Venus J. Fermentative utilization of coffee mucilage using Bacillus coagulans and investigation of down-stream processing of fermentation broth for optically pure L(+)-lactic acid production. Bioresour. Technol. 2016;211:398–405. doi: 10.1016/j.biortech.2016.03.122. [DOI] [PubMed] [Google Scholar]
- 120.Tang J., Wang X.C., Hu Y., Ngo H.H., Li Y. Dynamic membrane-assisted fermentation of food wastes for enhancing lactic acid production. Bioresour. Technol. 2017;234:40–47. doi: 10.1016/j.biortech.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 121.Pleissner D., Neu A.-K., Mehlmann K., Schneider R., Puerta-Quintero G.I., Venus J. Fermentative lactic acid production from coffee pulp hydrolysate using Bacillus coagulans at laboratory and pilot scales. Bioresour. Technol. 2016;218:167–173. doi: 10.1016/j.biortech.2016.06.078. [DOI] [PubMed] [Google Scholar]
- 122.Li Y., Cui F. Springer; Dordrecht: 2010. Microbial Lactic Acid Production from Renewable Resources, Sustainable Biotechnology; pp. 211–228. [Google Scholar]
- 123.Dumbrepatil A., Adsul M., Chaudhari S., Khire J., Gokhale D. Utilization of molasses sugar for lactic acid production by Lactobacillus delbrueckii subsp. delbrueckii mutant Uc-3 in batch fermentation. Appl. Environ. Microbiol. 2008;74:333–335. doi: 10.1128/AEM.01595-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bhatt S., Srivastava S. Lactic acid production from cane molasses by Lactobacillus delbrueckii NCIM 2025 in submerged condition: optimization of medium component by Taguchi DOE methodology. Food Biotechnol. 2008;22:115–139. [Google Scholar]
- 125.Kotzamanidis C., Roukas T., Skaracis G. Optimization of lactic acid production from beet molasses by Lactobacillus delbrueckii NCIMB 8130. World J. Microbiol. Biotechnol. 2002;18:441–448. [Google Scholar]
- 126.Rangaswamy V., Ramakrishna S. Lactic acid production by Lactobacillus delbrueckii in a dual reactor system using packed bed biofilm reactor. Lett. Appl. Microbiol. 2008;46:661–666. doi: 10.1111/j.1472-765X.2008.02362.x. [DOI] [PubMed] [Google Scholar]
- 127.Coelho L., De Lima C., Rodovalho C., Bernardo M., Contiero J. Lactic acid production by new Lactobacillus plantarum LMISM6 grown in molasses: optimization of medium composition. Braz. J. Chem. Eng. 2011;28:27–36. [Google Scholar]
- 128.Zhang Z.Y., Jin B., Kelly J.M. Production of L (+)-lactic acid using acid-adapted precultures of Rhizopus arrhizus in a stirred tank reactor. Appl. Biochem. Biotechnol. 2008;149:265–276. doi: 10.1007/s12010-007-8126-7. [DOI] [PubMed] [Google Scholar]
- 129.Chaisu K., Charles A.L., Guu Y.-K., Yen T.-B., Chiu C.-H. Optimization lactic acid production from molasses renewable raw material through response surface methodology with Lactobacillus casei M-15. APCBEE Proc. 2014;8:194–198. [Google Scholar]
- 130.Wee Y.-J., Yun J.-S., Park D.-H., Ryu H.-W. Biotechnological production of L (+)-lactic acid from wood hydrolyzate by batch fermentation of Enterococcus faecalis. Biotechnol. Lett. 2004;26:71–74. doi: 10.1023/b:bile.0000009464.23026.e0. [DOI] [PubMed] [Google Scholar]
- 131.Shukla V., Zhou S., Yomano L., Shanmugam K., Preston J., Ingram L. Production of d (−)-lactate from sucrose and molasses. Biotechnol. Lett. 2004;26:689–693. doi: 10.1023/b:bile.0000024088.36803.4e. [DOI] [PubMed] [Google Scholar]
- 132.Visser D., Van Breugel J., De Bruijn J.M., A'campo P. Lactic acid production from concentrated raw sugar beet juice. Google Patents. 2012 [Google Scholar]
- 133.Nancib A., Nancib N., Boudrant J. Production of lactic acid from date juice extract with free cells of single and mixed cultures of Lactobacillus casei and Lactococcus lactis. World J. Microbiol. Biotechnol. 2009;25:1423–1429. [Google Scholar]
- 134.de Oliveira R.A., Eduardo Vaz Rossell A.K.C., Filho R.M. Challenges and opportunities in lactic acid bioprocess design—from economic to production aspects. Biochem. Eng. J. 2018;15:219–239. [Google Scholar]
- 135.Vink E.T., Glassner D.A., Kolstad J.J., Wooley R.J., O’Connor R.P. The eco-profiles for current and near-future NatureWorks® polylactide (PLA) production. Ind. Biotechnol. 2007;3:58–81. [Google Scholar]
- 136.Oh H., Wee Y.-J., Yun J.-S., Han S.H., Jung S., Ryu H.-W. Lactic acid production from agricultural resources as cheap raw materials. Bioresour. Technol. 2005;96:1492–1498. doi: 10.1016/j.biortech.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 137.Vishnu C., Seenayya G., Reddy G. Direct fermentation of various pure and crude starchy substrates to L (+) lactic acid using Lactobacillus amylophilus GV6. World J. Microbiol. Biotechnol. 2002;18:429–433. [Google Scholar]
- 138.Oda Y., Saito K., Yamauchi H., Mori M. Lactic acid fermentation of potato pulp by the fungus Rhizopus oryzae. Curr. Microbiol. 2002;45:1–4. doi: 10.1007/s00284-001-0048-y. [DOI] [PubMed] [Google Scholar]
- 139.Huang L.P., Jin B., Lant P., Zhou J. Simultaneous saccharification and fermentation of potato starch wastewater to lactic acid by Rhizopus oryzae and Rhizopus arrhizus. Biochem. Eng. J. 2005;23:265–276. doi: 10.1007/s00449-005-0398-0. [DOI] [PubMed] [Google Scholar]
- 140.Yun J.-S., Wee Y.-J., Kim J.-N., Ryu H.-W. Fermentative production of dl-lactic acid from amylase-treated rice and wheat brans hydrolyzate by a novel lactic acid bacterium, Lactobacillus sp. Biotechnol. Lett. 2004;26:1613–1616. doi: 10.1023/B:BILE.0000045826.97010.82. [DOI] [PubMed] [Google Scholar]
- 141.Åkerberg C., Zacchi G. An economic evaluation of the fermentative production of lactic acid from wheat flour. Bioresour. Technol. 2000;75:119–126. [Google Scholar]
- 142.Naveena B., Altaf M., Bhadrayya K., Madhavendra S., Reddy G. Direct fermentation of starch to L (+) lactic acid in SSF by Lactobacillus amylophilus GV6 using wheat bran as support and substrate: medium optimization using RSM. Process Biochem. 2005;40:681–690. [Google Scholar]
- 143.Naveena B., Altaf M., Bhadrayya K., Reddy G. Screening and interaction effects of physical parameters, total N content and buffer on L (+) lactic acid production in SSF by Lactobacillus amylophilus GV6 using Taguchi designs. Indian J. Biotechnol. 2005;4:342–346. [Google Scholar]
- 144.Naveena B., Altaf M., Bhadriah K., Reddy G. Selection of medium components by Plackett–Burman design for production of L (+) lactic acid by Lactobacillus amylophilus GV6 in SSF using wheat bran. Bioresour. Technol. 2005;96:485–490. doi: 10.1016/j.biortech.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 145.Naveena B., Vishnu C., Altaf M., Reddy G. Wheat bran an inexpensive substrate for production of lactic acid in solid state fermentation by Lactobacillus amylophilus GV6—optimization of fermentation conditions. J. Sci. Ind. Res. (India) 2003;62:453–456. [Google Scholar]
- 146.Vishnu C., Seenayya G., Reddy G. Direct conversion of starch to L (+) lactic acid by amylase producing Lactobacillus amylophilus GV6. Bioprocess Eng. 2000;23:155–158. [Google Scholar]
- 147.Komesu A., de Oliveira J.A.R., da S Martins L.H., Wolf Maciel M.R., Maciel Filho R. Lactic acid production to purification: a review. BioResources. 2017;12:4364–4383. [Google Scholar]
- 148.Idrees M., Adnan A., Qureshi F.A. Optimization of sulfide/sulfite pretreatment of lignocellulosic biomass for lactic acid production. BioMed Res. Int. 2013;1:2013. doi: 10.1155/2013/934171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hofvendahl K., Hahn–Hägerdal B. Factors affecting the fermentative lactic acid production from renewable resources1. Enzym. Microb. Technol. 2000;26:87–107. doi: 10.1016/s0141-0229(99)00155-6. [DOI] [PubMed] [Google Scholar]
- 150.Wee Y., Yun J., Kim D., Ryu H. Batch and repeated batch production of L-(+)-lactic 2254 acid by RKY1 using wood hydrolyzate and corn steep liquor Enterococcus faecalis. J. Ind. Microbiol. Biotechnol. 2006;33:431–435. doi: 10.1007/s10295-006-0084-5. [DOI] [PubMed] [Google Scholar]
- 151.Ou M.S., Ingram L.O., Shanmugam K. L (+)-Lactic acid production from non-food carbohydrates by thermotolerant Bacillus coagulans. J. Ind. Microbiol. Biotechnol. 2011;38:599–605. doi: 10.1007/s10295-010-0796-4. [DOI] [PubMed] [Google Scholar]
- 152.Zhu Y., Lee Y., Elander R.T. Conversion of aqueous ammonia-treated corn stover to lactic acid by simultaneous saccharification and cofermentation. Appl. Biochem. Biotechnol. 2007;137:721–738. doi: 10.1007/s12010-007-9092-9. [DOI] [PubMed] [Google Scholar]
- 153.Park E.Y., Anh P.N., Okuda N. Bioconversion of waste office paper to L (+)-Lactic acid by filamentous fungus Rhizopus oryzae. Macro Rev. 2003;16:281–285. doi: 10.1016/j.biortech.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 154.Yáñez R., Alonso J.L., Parajó J.C. D-lactic acid production from waste cardboard. Journal of chemical technology & biotechnology: international research in process. Environ. Clean Technol. 2005;80:76–84. [Google Scholar]
- 155.Moldes A.B., Alonso J.K., Parajo J.C. Strategies to improve the bioconversion of processed wood into lactic acid by simultaneous saccharification and fermentation. J. Chem. Technol. Biotechnol. 2001;76:279–284. [Google Scholar]
- 156.Yadav N., Pranaw K., Khare S.K. Screening of lactic acid bacteria stable in ionic liquids and lignocellulosic byproducts for bio-based lactic acid production. Biores. Technol. Rep. 2020;11:100423. [Google Scholar]
- 157.Sreenath H.K., Moldes A.B., Koegel R.G., Straub R.J. Lactic acid production from agriculture residues. Biotechnol. Lett. 2001;23:179–184. [Google Scholar]
- 158.Garde A., Jonsson G., Schmidt A.S., Ahring B.K. Lactic acid production from wheat straw hemicellulose hydrolysate by Lactobacillus pentosus and Lactobacillus brevis. Bioresour. Technol. 2002;81:217–223. doi: 10.1016/s0960-8524(01)00135-3. [DOI] [PubMed] [Google Scholar]
- 159.Givry S., Prevot V., Duchiron F. Lactic acid production from hemicellulosic hydrolyzate by cells of Lactobacillus bifermentans immobilized in Ca-alginate using response surface methodology. World J. Microbiol. Biotechnol. 2008;24:745–752. [Google Scholar]
- 160.Tanaka T., Hoshina M., Tanabe S., Sakai K., Ohtsubo S., Taniguchi M. Production of D-lactic acid from defatted rice bran by simultaneous saccharification and fermentation. Bioresour. Technol. 2006;97:211–217. doi: 10.1016/j.biortech.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 161.Jukonyte R., Daiva Z., Bartkiene E., Lele V., Cernauskas D., Suproniene S., Juodeikiene G. A potential of brown rice polish as a substrate for the lactic acid and bioactive compounds production by the lactic acid bacteria newly isolated from cereal-based fermented products. LWT - Food Sci. Technol. 2018;97:323–331. [Google Scholar]
- 162.Ohkouchi Y., Inoue Y. Direct production of L (+)-lactic acid from starch and food wastes using Lactobacillus manihotivorans LMG18011. Bioresour. Technol. 2006;97:1554–1562. doi: 10.1016/j.biortech.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 163.Bustos G., Moldes A.B., Cruz J.M., Domínguez J.M. Production of fermentable media from vine-trimming wastes and bioconversion into lactic acid by Lactobacillus pentosus. J. Sci. Food Agric. 2004;84:2105–2112. [Google Scholar]
- 164.John R.P., Nampoothiri K.M., Pandey A. Simultaneous saccharification and fermentation of cassava bagasse for L-(+)-lactic acid production using Lactobacilli. Appl. Biochem. Biotechnol. 2006;134:263–272. doi: 10.1385/abab:134:3:263. [DOI] [PubMed] [Google Scholar]
- 165.Laopaiboon P., Thani A., Leelavatcharamas V., Laopaiboon L. Acid hydrolysis of sugarcane bagasse for lactic acid production. Bioresour. Technol. 2010;101:1036–1043. doi: 10.1016/j.biortech.2009.08.091. [DOI] [PubMed] [Google Scholar]
- 166.Oliveira R., Maciel Filho R., Rossel C.V. High lactic acid production from molasses and hydrolysed sugarcane bagasse. Chem. Eng. Trans. 2016;50:307–312. [Google Scholar]
- 167.Gullon B., Yanez R., Alonso J. L-Lactic acid production from apple pomace by sequential hydrolysis and fermentation 99. Bioresour. Technol. 2008;99:308–319. doi: 10.1016/j.biortech.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 168.Chan-Blanco Y., Bonilla-Leiva A., Velazquez A. Using banana to generate lactic acid through batch process fermentation. Appl. Microbiol. Biotechnol. 2003;63:147–152. doi: 10.1007/s00253-003-1374-8. [DOI] [PubMed] [Google Scholar]
- 169.Jawad A.H., Alkarkhi A.F., Jason O.C., Easa A.M., Norulaini N.N. Production of the lactic acid from mango peel waste–Factorial experiment. J. King Saud Univ. Sci. 2013;25:39–45. [Google Scholar]
- 170.Romaní A., Yáñez R., Garrote G., Alonso J.L. SSF production of lactic acid from cellulosic biosludges. Bioresour. Technol. 2008;99:4247–4254. doi: 10.1016/j.biortech.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 171.Kim K.I., Kim W.K., Seo D.K., Yoo I.S., Kim E.K., Yoon H.H. Biotechnology for Fuels and Chemicals. Humana Press; Totowa, NJ: 2003. Production of lactic acid from food wastes; pp. 637–647. [Google Scholar]
- 172.Zhang B. P.-j. He, N.-f. Ye, L.-m. Shao, Enhanced isomer purity of lactic acid from the non-sterile fermentation of kitchen wastes. Bioresour. Technol. 2008;99:855–862. doi: 10.1016/j.biortech.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 173.Huang L., Sheng J., Chen J., Li N. 2008 ICBBE 2008 the 2nd International Conference on. IEEE. 2008. Direct Fermentation of Fishmeal Wastewater and Starch Wastewater to Lactic Acid by Rhizopus Oryzae, Bioinformatics and Biomedical Engineering; pp. 3219–3222. [Google Scholar]
- 174.Adsul M.G., Varma A.J., Gokhale D.V. Lactic acid production from waste sugarcane bagasse derived cellulose. Green Chem. 2007;9:58–62. [Google Scholar]
- 175.Marques S., Santos J.A., Gírio F.M., Roseiro J.C. Lactic acid production from recycled paper sludge by simultaneous saccharification and fermentation. Biochem. Eng. J. 2008;41:210–216. [Google Scholar]
- 176.Nakasaki K., Adachi T. Effects of intermittent addition of cellulase for production of L-lactic acid from wastewater sludge by simultaneous saccharification and fermentation. Biotechnol. Bioeng. 2003;82:263–270. doi: 10.1002/bit.10573. [DOI] [PubMed] [Google Scholar]
- 177.Juodeikiene G., Zadeike D., Bartkiene E., Klupsaite D. Application of acid tolerant Pedioccocus strains for increasing the sustainability of lactic acid production from cheese whey. LWT - Food Sci. Technol. 2016;72:399–406. [Google Scholar]
- 178.Miller C., Fosmer A., Rush B., McMullin T., Beacom D., Suominen P. Industrial production of lactic acid. In: Moo-Young M., Butler M., Webb C., editors. second ed. Vol. 3. Elsevier; Burlington: 2011. pp. 179–188. (Comprehensive Biotechnology). [Google Scholar]
- 179.Fitzpatrick J., O'keeffe U. Influence of whey protein hydrolysate addition to whey permeate batch fermentations for producing lactic acid. Process Biochem. 2001;37:183–186. [Google Scholar]
- 180.Ghasemi M., Ahmad A., Jafary T., Azad A.K., Kakooei S., Wan Daud W.R., Sedighi M. Assessment of immobilized cell reactor and microbial fuel cell for simultaneous cheese whey treatment and lactic acid/electricity production. 2017;42:9107–9115. [Google Scholar]
- 181.Li Y., Shahbazi A., Coulibaly S., Mims M.M. Semicontinuous production of lactic acid from cheese whey using integrated membrane reactor. Appl. Biochem. Biotecnol. 2007:897–907. doi: 10.1007/s12010-007-9106-7. Springer. [DOI] [PubMed] [Google Scholar]
- 182.Büyükkileci A.O., Harsa S. Batch production of L (+) lactic acid from whey by Lactobacillus casei (NRRL B-441) J. Chem. Technol. Biotechnol. 2004;79:1036–1040. [Google Scholar]
- 183.Pauli T., Fitzpatrick J.J. Malt combing nuts as a nutrient supplement to whey permeate for producing lactic by fermentation with Lactobacillus casei. Process Biochem. 2002;38:1–6. [Google Scholar]
- 184.Kim H.-O., Wee Y.-J., Kim J.-N., Yun J.-S., Ryu H.-W. Twenty-Seventh Symposium on Biotechnology for Fuels and Chemicals. Springer; 2006. Production of lactic acid from cheese whey by batch and repeated batch cultures of Lactobacillus sp. RKY2; pp. 694–704. [PubMed] [Google Scholar]
- 185.Bazinet L. Electrodialytic phenomena and their applications in the dairy industry: a review. Crit. Rev. Food Sci. Nutr. 2005;45:307–326. doi: 10.1080/10408690490489279. [DOI] [PubMed] [Google Scholar]
- 186.Alonso S., Herrero M., Rendueles M., Díaz M. Residual yoghurt whey for lactic acid production. Biomass Bioenergy. 2010;34:931–938. [Google Scholar]
- 187.Posada J.A., Rincón L.E., Cardona C.A. Design and analysis of biorefineries based on raw glycerol: addressing the glycerol problem. Bioresour. Technol. 2012;111:282–293. doi: 10.1016/j.biortech.2012.01.151. [DOI] [PubMed] [Google Scholar]
- 188.Cheng K.-K., Zhang J.-A., Liu D.-H., Sun Y., Yang M.-D., Xu J.-M. Production of 1, 3-propanediol by Klebsiella pneumoniae from glycerol broth. Biotechnol. Lett. 2006;28:1817–1821. doi: 10.1007/s10529-006-9158-8. [DOI] [PubMed] [Google Scholar]
- 189.Biebl H. Fermentation of glycerol by Clostridium pasteurianum—batch and continuous culture studies. J. Ind. Microbiol. Biotechnol. 2001;27:18–26. doi: 10.1038/sj.jim.7000155. [DOI] [PubMed] [Google Scholar]
- 190.Dharmadi Y., Murarka A., Gonzalez R. Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnol. Bioeng. 2006;94:821–829. doi: 10.1002/bit.21025. [DOI] [PubMed] [Google Scholar]
- 191.Förster A.H., Gescher J. Metabolic engineering of Escherichia coli for production of mixed-acid fermentation end products. Front. Bioeng. Biotechnol. 2014;2:16. doi: 10.3389/fbioe.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gonzalez R., Murarka A., Dharmadi Y., Yazdani S.S. A new model for the anaerobic fermentation of glycerol in enteric bacteria: trunk and auxiliary pathways in Escherichia coli. Metab. Eng. 2008;10:234–245. doi: 10.1016/j.ymben.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 193.Murarka A., Dharmadi Y., Yazdani S., Gonzale R. Fermentative utilization of glycerol in 1882 Escherichia coli and its implications for the production of fuels and chemicals. Appl. Environ. Microbiol. 2008;74:1124–1135. doi: 10.1128/AEM.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Doi Y. L-Lactate production from biodiesel-derived crude glycerol by metabolically engineered Enterococcus faecalis: cytotoxic evaluation of biodiesel waste and development of glycerol-inducible gene expression system. Applied and Environmental Microbiology. AEM. 2015:3414–3418. doi: 10.1128/AEM.03418-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Norddahl B., Eriksen S., Pedersen F. 2004. Method for Producing Lactic Acid. Google Patents. No. 10/297,090. [Google Scholar]
- 196.Posada J.A., Cardona C.A., Gonzalez R. Analysis of the production process of optically pure D-lactic acid from raw glycerol using engineered Escherichia coli strains. Appl. Biochem. Biotechnol. 2012;166:680–699. doi: 10.1007/s12010-011-9458-x. [DOI] [PubMed] [Google Scholar]
- 197.Schenk P.M., Thomas-Hall S.R., Stephens E., Marx U.C., Mussgnug J.H., Posten C., Kruse O., Hankamer B. Second generation biofuels: high-efficiency microalgae for biodiesel production. Bioenergy Res. 2008;1:20–43. [Google Scholar]
- 198.Nguyen C.M., Kim J.-S., Hwang H.J., Park M.S., Choi G.J., Choi Y.H., Jang K.S., Kim J.-C. Production of L-lactic acid from a green microalga, Hydrodictyon reticulum, by Lactobacillus paracasei LA104 isolated from the traditional Korean food, makgeolli. Bioresour. Technol. 2012;110:552–559. doi: 10.1016/j.biortech.2012.01.079. [DOI] [PubMed] [Google Scholar]
- 199.Nguyen C.M., Kim J.-S., Song J.K., Choi G.J., Choi Y.H., Jang K.S., Kim J.-C. D-Lactic acid production from dry biomass of Hydrodictyon reticulatum by simultaneous saccharification and co-fermentation using Lactobacillus coryniformis subsp. torquens. Biotechnol. Lett. 2012;34:2235–2240. doi: 10.1007/s10529-012-1023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Djukić-Vuković A.P., Mojović L.V., Vukašinović-Sekulić M.S., Nikolić S.B., Pejin J.D. Integrated production of lactic acid and biomass on distillery stillage. Bioproc. Biosyst. Eng. 2013;36:1157–1164. doi: 10.1007/s00449-012-0842-x. [DOI] [PubMed] [Google Scholar]
- 201.Ye T., Li X., Zhang T., Su Y., Zhang W., Li J., Gan Y., Zhang A., Liu Y., Xue G. Copper (II) addition to accelerate lactic acid production from co-fermentation of food waste and waste activated sludge: understanding of the corresponding metabolisms, microbial community and predictive functional profiling. Waste Manag. 2018;76:414–422. doi: 10.1016/j.wasman.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 202.Ke X., Wang C., Li R., Zhang Y., Zhang H., Gui S. Biomethane production and dynamics of microflora in response to copper treatments during mesophilic anaerobic digestion. Waste Manag. Res. 2014;32:726–732. doi: 10.1177/0734242X14543816. [DOI] [PubMed] [Google Scholar]
- 203.Solioz M., Abicht H.K., Mermod M., Mancini S. Response of gram-positive bacteria to copper stress. J. Biol. Inorg. Chem. 2010;15:3–14. doi: 10.1007/s00775-009-0588-3. [DOI] [PubMed] [Google Scholar]
- 204.Solioz M., Mermod M., Abicht H.K., Mancini S. 2011. Responses of Lactic Acid Bacteria to Heavy Metal Stress. Springer, US; pp. 163–195. [Google Scholar]
- 205.Mumtaz M.Z., Barry K.M., Baker A.L., Nichols D.S., Ahmad M., Zahir Z.A., Britz M.L. Production of lactic and acetic acids by Bacillus sp. ZM20 and Bacillus cereus following exposure to zinc oxide: a possible mechanism for Zn solubilization. Rhizosphere. 2019;12:100170. [Google Scholar]
- 206.Shen X., Xia L. Lactic acid production from cellulosic material by synergetic hydrolysis and fermentation. Appl. Biochem. Biotechnol. 2006;133:251–262. doi: 10.1385/abab:133:3:251. [DOI] [PubMed] [Google Scholar]
- 207.Mussatto S.I., Fernandes M., Mancilha I.M., Roberto I.C. Effects of medium supplementation and pH control on lactic acid production from brewer's spent grain. Biochem. Eng. J. 2008;40:437–444. [Google Scholar]
- 208.Nakano S., Ugwu C.U., Tokiwa Y. Efficient production of d-(−)-lactic acid from broken rice by Lactobacillus delbrueckii using Ca(OH)2 as a neutralizing agent. Bioresour. Technol. 2012;104:791–794. doi: 10.1016/j.biortech.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 209.Gassem M., Abu-Tarboush H. Lactic acid production by Lactobacillus delbrueckii subsp. bulgaricus in camel's and cow's wheys. Milchwissenschaft. 2000;55:374–378. [Google Scholar]
- 210.Fukushima K., Sogo K., Miura S., Kimura Y. Production of D-lactic acid by bacterial fermentation of rice starch. Macromol. Biosci. 2004;4:1021–1027. doi: 10.1002/mabi.200400080. [DOI] [PubMed] [Google Scholar]
- 211.Roberto I., Mussatto S., Mancilha I., Fernandes M. The effects of pH and nutrient supplementation of brewer's spent grain cellulosic hydrolysate for lactic acid production by Lactobacillus delbrueckii. J. Biotechnol. 2007;2:S181–S182. doi: 10.1007/s10529-007-9494-3. [DOI] [PubMed] [Google Scholar]
- 212.de la Torre I., Ladero M., Santos V.E. D-lactic acid production from orange waste enzymatic hydrolysates with L. delbrueckii cells in growing and resting state. Ind. Crop. Prod. 2020;146:112176. [Google Scholar]
- 213.de la Torre I., Acedos M.G., Ladero M., Santos V.E. On the use of resting L. delbrueckii spp. delbrueckii cells for D-lactic acid production from orange peel wastes hydrolysates. Biochem. Eng. J. 2019;145:162–169. [Google Scholar]
- 214.John R.P., Sukumaran R.K., Nampoothiri K.M., Pandey A. Statistical optimization of simultaneous saccharification and L(+)-lactic acid fermentation from cassava bagasse using mixed culture of Lactobacilli by response surface methodology. Biochem. Eng. J. 2007;36:262–267. [Google Scholar]
- 215.Cingadi S., Srikanth K., Arun E.V.R., Sivaprakasam S. Statistical optimization of cassava fibrous waste hydrolysis by response surface methodology and use of dyrolysate based media for the production of optically pure D-lactic acid. Biochem. Eng. J. 2015;102:82–90. [Google Scholar]
- 216.Balakrishnana R., Tadib S.R.R., Sivaprakasamb S., Rajarama S. Optimization of acid and enzymatic hydrolysis of kodo millet (Paspalum scrobiculatum) bran residue to obtain fermentable sugars for the production of optically pure D (−) lactic acid. Ind. Crop. Prod. 2018;111:731–742. [Google Scholar]
- 217.Wang X., Wang G., Yu X., Chen H., Sun Y., Chen G. Pretreatment of corn stover by solid acid for D-lactic acid fermentation. Bioresour. Technol. 2017;239:490–495. doi: 10.1016/j.biortech.2017.04.089. [DOI] [PubMed] [Google Scholar]
- 218.Schepers A.W., Thibault J., Lacroix C. Continuous lactic acid production in whey permeate/yeast extract medium with immobilized Lactobacillus helveticus in a two-stage process: model and experiments. Enzym. Microb. Technol. 2006;38:324–337. [Google Scholar]
- 219.Plessas S., Bosnea L., Psarianos C., Koutinas A., Marchant R., Banat I. Lactic acid production by mixed cultures of Kluyveromyces marxianus, Lactobacillus delbrueckii ssp. bulgaricus and Lactobacillus helveticus. Bioresour. Technol. 2008;99:5951–5955. doi: 10.1016/j.biortech.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 220.Schepers A.W., Thibault J., Lacroix C. Lactobacillus helveticus growth and lactic acid production during pH-controlled batch cultures in whey permeate/yeast extract medium. Part I. Multiple factor kinetic analysis. Enzym. Microb. Technol. 2002;30:176–186. [Google Scholar]
- 221.Hirata M., Gao M.-t., Toorisaka E., Takanashi H., Hano T. Production of lactic acid by continuous electrodialysis fermentation with a glucose concentration controller. Biochem. Eng. J. 2005;25:159–163. [Google Scholar]
- 222.Djukić-Vuković A.P., Mojović L.V., Vukašinović-Sekulić M.S., Rakin M.B., Nikolić S.B., Pejin J.D., Bulatović M.L. Effect of different fermentation parameters on L-lactic acid production from liquid distillery stillage. Food Chem. 2012;134:1038–1043. doi: 10.1016/j.foodchem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 223.Li Z., Lu J., Yang Z., Han L., Tan T. Utilization of white rice bran for production of L-lactic acid. Biomass Bioenergy. 2012;39:53–58. [Google Scholar]
- 224.Zhao Z., Xie X., Wang Z., Tao Y., Niu X., Huang X., Liu L., Li Z. Immobilization of Lactobacillus rhamnosus in mesoporous silica-based material: an efficiency continuous cell-recycle fermentation system for lactic acid Production. J. Biosci. Bioeng. 2016;12:645–651. doi: 10.1016/j.jbiosc.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 225.Marques S., Gírio F., Santos J., Roseiro J. Pulsed fed-batch strategy towards intensified process for lactic acid production using recycled paper sludge. Biomass Conv. Bioref. 2017;7:127–137. [Google Scholar]
- 226.Lu Z., Wei M., Yu L. Enhancement of pilot scale production of l(+)-lactic acid by fermentation coupled with separation using membrane bioreactor. Process Biochem. 2012;47:410–415. [Google Scholar]
- 227.Coelho L.F., Bolner de Lima C.J., Bernardo M.P., Alvarez G.M., Contiero J. Improvement of L(+)-lactic acid production from cassava wastewater by Lactobacillus rhamnosus B 103. J. Environ. Sci. Health Part B. 2010;90:1944–1950. doi: 10.1002/jsfa.4039. [DOI] [PubMed] [Google Scholar]
- 228.Shi S., Kang L., Lee Y.Y. Production of lactic acid from the mixture of softwood pre-hydrolysate and paper mill sludge by simultaneous saccharification and fermentation. Appl. Biochem. Biotechnol. 2015;175:2741–2754. doi: 10.1007/s12010-014-1451-8. [DOI] [PubMed] [Google Scholar]
- 229.Wang X.Q., Wang Q.H., Ma H.Z., Yin W. Lactic acid fermentation of food waste using integrated glucoamylase production. J. Chem. Technol. Biotechnol. 2009;84:139–143. [Google Scholar]
- 230.Bahrya H., Abdalla R., Pons A., Taha S., Vial C. Optimization of lactic acid production using immobilized Lactobacillus Rhamnosus and carob pod waste from the Lebanese food industry. J. Biotechnol. 2019;306:81–88. doi: 10.1016/j.jbiotec.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 231.Lech M. Optimisation of protein-free waste whey supplementation used for the industrial microbiological production of lactic acid. Biochem. Eng. J. 2020;157:107531. [Google Scholar]
- 232.Bernardoa M.P., Coelhoa L.F., Sassa D.C., Contieroa J. L(+)-Lactic acid production by Lactobacillus rhamnosus B103 from dairy industry waste. Braz. J. Microbiol. 2016;47:640–646. doi: 10.1016/j.bjm.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Pejin J., Radosavljević M., Pribić M., Kocić-Tanackov S., Mladenović D., Djukić-Vuković A., Mojović L. Possibility of L-(+)-lactic acid fermentation using malting, brewing, and oil production by-products. Waste Manag. 2018;79:153–163. doi: 10.1016/j.wasman.2018.07.035. [DOI] [PubMed] [Google Scholar]
- 234.Burgos-Rubio C.N., Okos M.R., Wankat P.C. Kinetic study of the conversion of different substrates to lactic acid using Lactobacillus bulgaricus. Biotechnol. Prog. 2000;16:305–314. doi: 10.1021/bp000022p. [DOI] [PubMed] [Google Scholar]
- 235.Fakhravar S., Najafpour G., Heris S.Z., Izadi M., Fakhravar A. Fermentative lactic acid from deproteinized whey using Lactobacillus bulgaricus in batch culture. World Appl. Sci. J. 2012;17:1083–1086. [Google Scholar]
- 236.Liu P., Zheng Z., Xu Q., Qian Z., Liu J., Ouyang J. Valorization of dairy waste for enhanced D-lactic acid production at low cost. Process Biochem. 2018;71:18–22. [Google Scholar]
- 237.Kwan T.H., Hu Y., Lin C.S.K. Valorisation of food waste via fungal hydrolysis and lactic acid fermentation with Lactobacillus casei Shirota. Bioresour. Technol. 2016;217:129–136. doi: 10.1016/j.biortech.2016.01.134. [DOI] [PubMed] [Google Scholar]
- 238.Wang J., Gao M., Liu J., Wang Q., Wang C., Yin Z., Wu C. Lactic acid production from Sophora flavescens residues pretreated with sodium hydroxide: reutilization of the pretreated liquor during fermentation. Bioresour. Technol. 2017;241:915–921. doi: 10.1016/j.biortech.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 239.Oonkhanond B., Jonglertjunya W., Srimarut N., Bunpachart P., Tantinukul S., Nasongkl N., Sakdaronnarong C. Lactic acid production from sugarcane bagasse by an integrated system of lignocellulose fractionation, saccharification and fermentation, and ex-situ nanofiltration. J. Environ.Chem. Eng. 2017;5:2533–2541. [Google Scholar]
- 240.Mudaliyar P., Sharma L., Kulkarni C. Food waste management- lactic acid production by Lactobacillus species. Int. J. Adv. Biol. Biomed. Res. 2012;2:34–38. [Google Scholar]
- 241.Kurbanoglu E.B., Kurbanoglu N.I. Utilization for lactic acid production with a new acid hydrolysis of ram horn waste. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2003;225:29–34. doi: 10.1016/S0378-1097(03)00472-5. [DOI] [PubMed] [Google Scholar]
- 242.Hujanen M., Linko S., Linko Y.-Y., Leisola M. Optimisation of media and cultivation conditions for L (+)(S)-lactic acid production by Lactobacillus casei NRRL B-441. Appl. Microbiol. Biotechnol. 2001;56:126–130. doi: 10.1007/s002530000501. [DOI] [PubMed] [Google Scholar]
- 243.Panesar P.S., Kennedy J.F., Gandhi D.N., Bunko K. Bioutilisation of whey for lactic acid production. Food Chem. 2007;105:1–14. [Google Scholar]
- 244.Zhang C., Yang H.-Q., Wu D.-J. Study on the reuse of anaerobic digestion effluent in lactic acid Production. J. Clean. Prod. 2019;239:118028. [Google Scholar]
- 245.Prasad S., Srikanth K., Limaye A.M., Sivaprakasam S. Homo-fermentative production of D-lactic acid by Lactobacillus sp. employing casein whey permeate as a raw feed-stock. Biotechnol. Lett. 2014;36:1303–1307. doi: 10.1007/s10529-014-1482-9. [DOI] [PubMed] [Google Scholar]
- 246.Ueno T., Ozawa Y., Ishikawa M., Nakanishi K., Kimura T. Lactic acid production using two food processing wastes, canned pineapple syrup and grape invertase, as substrate and enzyme. Biotechnol. Lett. 2003;25:573–577. doi: 10.1023/a:1022888832278. [DOI] [PubMed] [Google Scholar]
- 247.Sirisansaneeyakul S., Luangpipat T., Vanichsriratana W., Srinophakun T., Chen H.H.-H., Chisti Y. Optimization of lactic acid production by immobilized Lactococcus lactis IO-1. J. Ind. Microbiol. Biotechnol. 2007;34:381. doi: 10.1007/s10295-007-0208-6. [DOI] [PubMed] [Google Scholar]
- 248.Roble N.D., Ogbonna J.C., Tanaka H. L-Lactic acid production from raw cassava starch in a circulating loop bioreactor with cells immobilized in loofa (Luffa cylindrica) Biotechnol. Lett. 2003;25:1093–1098. doi: 10.1023/a:1024192131343. [DOI] [PubMed] [Google Scholar]
- 249.Shi Z., Wei P., Zhu X., Cai J., Huang L., Xu Z. Efficient production of l-lactic acid from hydrolysate of Jerusalem artichoke with immobilized cells of Lactococcus lactis in fibrous bed bioreactors. Enzym. Microb. Technol. 2012;51:263–268. doi: 10.1016/j.enzmictec.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 250.Singhvi M., Zendo T., Iida H., Gokhale D., Sonomoto K. Stimulation of D- and L-lactate dehydrogenases transcriptional levels in presence of diammonium hydrogen phosphate resulting to enhanced lactic acid production by Lactobacillus strain. J. Biosci. Bioeng. 2017;124:674–679. doi: 10.1016/j.jbiosc.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 251.Tsuge Y., Kawaguchi H., Sasaki K., Tanaka T., Kondo A. Two-step production of D-lactate from mixed sugars by growing and resting cells of metabolically engineered Lactobacillus plantarum. Appl. Microbiol. Biotechnol. 2014;98:4911–4918. doi: 10.1007/s00253-014-5594-x. [DOI] [PubMed] [Google Scholar]
- 252.Sreenath H.K., Moldes A.B., Koegel R.G., Straub R.J. Lactic acid production by simultaneous saccharification and fermentation of alfalfa fiber. J. Biosci. Bioeng. 2001;92:518–523. doi: 10.1263/jbb.92.518. [DOI] [PubMed] [Google Scholar]
- 253.Okano K., Zhang Q., Shinkawa S., Yoshida S., Tanaka T., Fukuda H., Kondo A. Efficient production of optically pure D-lactic acid from raw corn starch by using a genetically modified L-lactate dehydrogenase gene-deficient and α-amylase-secreting Lactobacillus plantarum strain. Appl. Environ. Microbiol. 2009;75:462–467. doi: 10.1128/AEM.01514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Asada C., Nakamura Y., Kobayashi F. Waste reduction system for production of useful materials from un-utilized bamboo using steam explosion followed by various conversion methods. Biochem. Eng. J. 2005;23:131–137. [Google Scholar]
- 255.Okano K., Yoshida S., Yamada R., Tanaka T., Ogino C., Fukuda H., Kondo A. Improved production of homo-D-lactic acid via xylose fermentation by introduction of xylose assimilation genes and redirection of the phosphoketolase pathway to the pentose phosphate pathway in L-lactate dehydrogenase gene-deficient Lactobacillus plantarum. Appl. Environ. Microbiol. 2009;75:7858–7861. doi: 10.1128/AEM.01692-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hama S., Mizuno S., Kihara M., Tanaka T., Ogino C., Noda H., Kondo A. Production of D-lactic acid from hardwood pulp by mechanical milling followed by simultaneous saccharification and fermentation using metabolically engineered Lactobacillus plantarum. Bioresour. Technol. 2015;187:167–172. doi: 10.1016/j.biortech.2015.03.106. [DOI] [PubMed] [Google Scholar]
- 257.Chen P.-T., Hong Z.-S., Cheng C.-L., Ng I.-S., Lo Y.-C., Nagarajan D., Chang J.-S. Exploring fermentation strategies for enhanced lactic acid production with polyvinyl alcohol-immobilized Lactobacillus plantarum 23 using microalgae as feedstock. Bioresour. Technol. 2020;308:123266. doi: 10.1016/j.biortech.2020.123266. [DOI] [PubMed] [Google Scholar]
- 258.Okano K., Hama S., Kihara M., Noda H., Tanaka T., Kondo A. Production of optically pure D-lactic acid from brown rice using metabolically engineered Lactobacillus plantarum. Appl. Microbiol. Biotechnol. 2017;101:1869–1875. doi: 10.1007/s00253-016-7976-8. [DOI] [PubMed] [Google Scholar]
- 259.Yoshida S., Okano K., Tanaka T., Ogino C., Kondo A. Homo-D-lactic acid production from mixed sugars using xylose-assimilating operon-integrated Lactobacillus plantarum. Appl. Biochem. Biotechnol. 2011;92:67–76. doi: 10.1007/s00253-011-3356-6. [DOI] [PubMed] [Google Scholar]
- 260.Sharma V., Mishra H.N. Unstructured kinetic modeling of growth and lactic acid production by Lactobacillus plantarum NCDC 414 during fermentation of vegetable Juices. LWT - Food Sci. Technol. 2014;59:1123–1128. [Google Scholar]
- 261.Yen H.-W., Kang J.-L. Lactic acid production directly from starch in a starch-controlled fed-batch operation using Lactobacillus amylophilus. Bioproc. Biosyst. Eng. 2010;33:1017–1023. doi: 10.1007/s00449-010-0426-6. [DOI] [PubMed] [Google Scholar]
- 262.Yang S., Yu H., You Y., Li X., Jiang J. Effective lactic acid production from waste paper using Streptococcus thermophilus at low enzyme loading assisted by gleditsia saponin. Carbohydr. Polym. 2018;200:122–127. doi: 10.1016/j.carbpol.2018.07.063. [DOI] [PubMed] [Google Scholar]
- 263.Nguyen C.M., Kim J.-S., Nguyen T.N., Kim S.K., Choi G.J., Choi Y.H., Jang K.S., Kim J.-C. Production of L- and D-lactic acid from waste Curcuma longa biomass through simultaneous saccharification and cofermentation. Bioresour. Technol. 2013;146:35–43. doi: 10.1016/j.biortech.2013.07.035. [DOI] [PubMed] [Google Scholar]
- 264.de Oliveira Moraes A., Ramirez N.I.B., Pereira N. Evaluation of the fermentation potential of pulp mill residue to produce d(-)-lactic acid by separate hydrolysis and fermentation using Lactobacillus coryniformis subsp. torquens. Appl. Biochem. Biotechnol. 2016;180:1574–1585. doi: 10.1007/s12010-016-2188-3. [DOI] [PubMed] [Google Scholar]
- 265.Yun J.-S., Wee Y.-J., Ryu H.-W. Production of optically pure L (+)-lactic acid from various carbohydrates by batch fermentation of Enterococcus faecalis RKY1. Enzym. Microb. Technol. 2003;33:416–423. [Google Scholar]
- 266.Shibata K., Flores D.M., Kobayashi G., Sonomoto K. Direct L-lactic acid fermentation with sago starch by a novel amylolytic lactic acid bacterium, Enterococcus faecium. Enzym. Microb. Technol. 2007;41:149–155. [Google Scholar]
- 267.Wee Y.J., Reddy L., Ryu H.W. Fermentative production of L (+)-lactic acid from starch hydrolyzate and corn steep liquor as inexpensive nutrients by batch culture of Enterococcus faecalis RKY1. Journal of Chemical Technology & Biotechnology: international Research in Process. Environ. Clean Technol. 2008;83:1387–1393. [Google Scholar]
- 268.Ding S., Tan T. L-lactic acid production by Lactobacillus casei fermentation using different fed-batch feeding strategies. Process Biochem. 2006;41:1451–1454. [Google Scholar]
- 269.Murakami N., Oba M., Iwamoto M., Tashiro Y., Noguchi T., Bonkohara K., Abdel-Rahman M.A., Zendo T., Shimoda M., Sakai K., Sonomoto K. L-Lactic acid production from glycerol coupled with acetic acid metabolism by Enterococcus faecalis without carbon loss. J. Biosci. Bioeng. 2016;121:89–95. doi: 10.1016/j.jbiosc.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 270.Nolasco-Hipolito C., Zarrabal O.C., Kamaldin R.M., Teck-Yee L., Lihan S., Bujang K.B., Nitta Y. Lactic acid production by Enteroccocus faecium in liquefied sago starch. AMB Express. 2012;2:53. doi: 10.1186/2191-0855-2-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Wee Y.-J., Ryu H.-W. Lactic acid production by Lactobacillus sp. RKY2 in a cell-recycle continuous fermentation using lignocellulosic hydrolyzates as inexpensive raw materials. Bioresour. Technol. 2009;100:4262–4270. doi: 10.1016/j.biortech.2009.03.074. [DOI] [PubMed] [Google Scholar]
- 272.Abdel-Rahman M.A., Xiao Y., Tashiro Y., Wang Y., Zendo T., Sakai K., Sonomoto K. Fed-batch fermentation for enhanced lactic acid production from glucose/xylose mixture without carbon catabolite repression. J. Biosci. Bioeng. 2015;119:153–158. doi: 10.1016/j.jbiosc.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 273.Abdel-Rahman M.A., Hassan S.E.-D., Roushdy M.M., Aza M.S., Gaber M.A. Free-nutrient supply and thermo-alkaline conditions for direct lactic acid production from mixed lignocellulosic and food waste materials. Biores. Technol. Rep. 2019;7:100256. [Google Scholar]
- 274.Solvala K.M., Chouljenkob A., Chotikod A., Sathivel S. Growth kinetics and lactic acid production of Lactobacillus plantarum NRRL B-4496, L. acidophilus NRRL B-4495, and L. reuteri B-14171 in media containing egg white hydrolysates. LWT - Food Sci. Technol. 2019;105:393–399. [Google Scholar]
- 275.Juodeikiene G., Klupsaite D., Zadeike D., Cizeikiene D., Vidziunaite I., Bartkiene E., Cernauskas D. Bioconversion of agro-industrial by-products to lactic acid using Lactobacillus sakei and two Pediococcus spp. strains. Int. J. Food Sci. Technol. 2016;51:2682–2691. [Google Scholar]
- 276.Rivera O.M.P., Menduiña A.B.M., Agrasar A.M.T., González J.M.D. Biosurfactants from grape marc: stability study. J. Biotechnol. 2007;2:S136. [Google Scholar]
- 277.Cubas-Cano E., González-Fernández C., Ballesteros I., Tomás-Pejó E. Efficient utilization of hydrolysates from steam-exploded gardening residues for lactic acid production by optimization of enzyme addition and pH control. Waste Manag. 2020;107:235–243. doi: 10.1016/j.wasman.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 278.Bustos G., Moldes A.B., Manuel C.J., Domínguez J.M. Influence of the metabolism pathway on lactic acid production from hemicellulosic trimming vine shoots hydrolyzates using Lactobacillus pentosus. Biotechnol. Prog. 2005;21:793–798. doi: 10.1021/bp049603v. [DOI] [PubMed] [Google Scholar]
- 279.Tian X., Wang Y., Chu J., Mohsin A., Zhuang Y. Exploring cellular fatty acid composition and intracellular metabolites of osmotic-tolerant mutant Lactobacillus paracasei NCBIO-M2 for highly efficient lactic acid production with high initial glucose concentration. J. Biotechnol. 2018;286:27–35. doi: 10.1016/j.jbiotec.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 280.Mladenović D., Pejin J., Kocić-Tanackov S., Radovanović Ž., Djukić-Vuković A., Mojović L. Lactic acid production on molasses enriched potato stillage by Lactobacillus paracasei immobilized onto agro-industrial waste supports. Ind. Crop. Prod. 2018;124:142–148. [Google Scholar]
- 281.Petrova P., Velikova P., Popova L., Petrov K. Direct conversion of chicory flour into L(+)-lactic acid by the highly effective inulinase producer Lactobacillus paracasei DSM 23505. Bioresour. Technol. 2015;186:329–333. doi: 10.1016/j.biortech.2015.03.077. [DOI] [PubMed] [Google Scholar]
- 282.Mladenović D., Pejin J., Kocić-Tanackov S., Djukić-Vuković A., Mojović L. Enhanced lactic acid production by adaptive evolution of Lactobacillus paracasei on agroindustrial substrate. Appl. Biochem. Biotechnol. 2018;187:753–769. doi: 10.1007/s12010-018-2852-x. [DOI] [PubMed] [Google Scholar]
- 283.Moon S.-K., Wee Y.-J., Choi G.-W. A novel lactic acid bacterium for the production of high purity L-lactic acid, Lactobacillus paracasei subsp. paracasei CHB2121. J. Biosci. Bioeng. 2012;114:155–159. doi: 10.1016/j.jbiosc.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 284.Choi H., Ryu H., Park K., Lee E., H L., SW K., ES C. Direct lactic acid fermentation of Jerusalem artichoke tuber extract using Lactobacillus paracasei without acidic or enzymatic inulin hydrolysis. Bioresour. Technol. 2012;114:745–747. doi: 10.1016/j.biortech.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 285.Pleissner D., Demichelis F., Mariano S., Fiore S., Gutiérrez I.M., Schneider R., Venus J. Direct production of lactic acid based on simultaneous saccharification and fermentation of mixed restaurant food waste. J. Clean. Prod. 2017;143:615–623. [Google Scholar]
- 286.Kim J.H., Shoemaker S.P., Mills D.A. Relaxed control of sugar utilization in Lactobacillus brevis. Microbiology. 2009;155:1351–1359. doi: 10.1099/mic.0.024653-0. [DOI] [PubMed] [Google Scholar]
- 287.Cui F., Li Y., Wan C. Lactic acid production from corn stover using mixed cultures of Lactobacillus rhamnosus and Lactobacillus brevis. Bioresour. Technol. 2011;102:1831–1836. doi: 10.1016/j.biortech.2010.09.063. [DOI] [PubMed] [Google Scholar]
- 288.Grewal J., Khare S.K. One-pot bioprocess for lactic acid production from lignocellulosic agro-wastes by using ionic liquid stable Lactobacillus brevis. Bioresour. Technol. 2018;251:268–273. doi: 10.1016/j.biortech.2017.12.056. [DOI] [PubMed] [Google Scholar]
- 289.Zhang Y., Vadlani P.V. Lactic acid production from biomass-derived sugars via co-fermentation of Lactobacillus brevis and Lactobacillus plantarum. J. Biosci. Bioeng. 2015;119:694–699. doi: 10.1016/j.jbiosc.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 290.Jiang X., Xue Y., Wang A., Wang L., Zhang G., Zeng Q., Yu B., Ma Y. Efficient production of polymer-grade L-lactate by an alkaliphilic Exiguobacterium sp. strain under nonsterile open fermentation conditions. Bioresour. Technol. 2013;143:665–668. doi: 10.1016/j.biortech.2013.06.049. [DOI] [PubMed] [Google Scholar]
- 291.Calabia B.P., Tokiwa Y., Aiba S. Fermentative production of L-(+)-lactic acid by an alkaliphilic marine microorganism. Biotechnol. Lett. 2011;33:1429–1433. doi: 10.1007/s10529-011-0573-0. [DOI] [PubMed] [Google Scholar]
- 292.Bai Z., Gao Z., Sun J., Wu B., He B. D-Lactic acid production by Sporolactobacillus inulinus YBS1-5 with simultaneous utilization of cottonseed meal and corncob residue. Bioresour. Technol. 2016;207:346–352. doi: 10.1016/j.biortech.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 293.Li J., Sun J., Wu B., He B. Combined utilization of nutrients and sugar derived from wheat bran for d-Lactate fermentation by Sporolactobacillus inulinus YBS1-5. Bioresour. Technol. 2017;229:33–38. doi: 10.1016/j.biortech.2016.12.101. [DOI] [PubMed] [Google Scholar]
- 294.Zheng H., Gong J., Chen T., Chen X., Zhao X. Strain improvement of Sporolactobacillus inulinus ATCC 15538 for acid tolerance and production of Dlactic acid by genome shuffling. Appl. Microbiol. Biotechnol. 2010;85:1541–1549. doi: 10.1007/s00253-009-2243-x. [DOI] [PubMed] [Google Scholar]
- 295.Bai Z., Gao Z., He B., Wu B. Effect of lignocellulose-derived inhibitors on the growth and D-lactic acid production of Sporolactobacillus inulinus YBS1-5. Bioproc. Biosyst. Eng. 2015;38:1993–2001. doi: 10.1007/s00449-015-1440-5. [DOI] [PubMed] [Google Scholar]
- 296.Li Y., Wang L., Ju J., Yu B., Ma Y. Efficient production of polymer-grade d-lactate by Sporolactobacillus laevolacticus DSM442 with agricultural waste cottonseed as the sole nitrogen source. Bioresour. Technol. 2013;142:186–191. doi: 10.1016/j.biortech.2013.04.124. [DOI] [PubMed] [Google Scholar]
- 297.Ge X.-Y., Qian H., Zhang W.-G. Enhancement of L-lactic acid production in Lactobacillus casei from Jerusalem artichoke tubers by kinetic optimization and citrate metabolism. J. Microbiol. Biotechnol. 2010;20:101–109. [PubMed] [Google Scholar]
- 298.Wang W.-M., Wang W.-H., Wang X.C.-Q., Ma H.-Z. Effect of different fermentation parameters on lactic acid production from kitchen waste by Lactobacillus TY50. Chem. Biochem. Eng. Q. 2011;25:433–438. [Google Scholar]
- 299.Flores-Albino B., Arias L., Gomez J., Castillo A., Gimeno M., Shirai K. Chitin and L (+)-lactic acid production from crab (Callinectes bellicosus) wastes by fermentation of Lactobacillus sp. B2 using sugar cane molasses as carbon source. Bioproc. Biosyst. Eng. 2012;35:1193–1200. doi: 10.1007/s00449-012-0706-4. [DOI] [PubMed] [Google Scholar]
- 300.Kawai M., Harada R., Yoda N., Yamasaki-Yashiki S., Fukusaki E., Katakura Y. Suppression of lactate production by using sucrose as a carbon source in lactic acid bacteria. J. Biosci. Bioeng. 2020;129:47–51. doi: 10.1016/j.jbiosc.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 301.Kawai M., Tsuchiya A., Ishida J., Yoda N., Yashiki-Yamasaki S., Katakura Y. Suppression of lactate production in fed-batch culture of some lactic acid bacteria with sucrose as the carbon source. J. Biosci. Bioeng. 2020;129:535–540. doi: 10.1016/j.jbiosc.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 302.Kuo Y.-C., Yuan S.-F., Wang C.-A., Huang Y.-J., Guo G.-L., Hwang W.-S. Production of optically pure L-lactic acid from lignocellulosic hydrolysate by using a newly isolated and D-lactate dehydrogenase gene-deficient Lactobacillus paracasei strain. Bioresour. Technol. 2015;198:651–657. doi: 10.1016/j.biortech.2015.09.071. [DOI] [PubMed] [Google Scholar]
- 303.Wu Y., Ma H., Zheng M., Wang K. Lactic acid production from acidogenic fermentation of fruit and vegetable wastes. Bioresour. Technol. 2015;191:53–58. doi: 10.1016/j.biortech.2015.04.100. [DOI] [PubMed] [Google Scholar]
- 304.Haag N.L., Nägele H.-J., Fritz T., Oechsner H. Effects of ensiling treatments on lactic acid production and supplementary methane formation of maize and amaranth – an advanced green biorefining approach. Bioresour. Technol. 2015;178:217–225. doi: 10.1016/j.biortech.2014.08.048. [DOI] [PubMed] [Google Scholar]
- 305.Chen B., Chen Z., Su K., Wang B., Wang Y., Wang Z., Si Y., Wu D., Cai P Qin. Efficient lactic acid production from cassava bagasse by mixed culture of Bacillus coagulans and lactobacillus rhamnosus using stepwise pH controlled simultaneous saccharification and co-fermentation. Ind. Crop. Prod. 2020;146:112175. [Google Scholar]
- 306.Qiu Z., Gao Q., Bao J. Constructing xylose-assimilating pathways in Pediococcus acidilactici for high titer D-lactic acid fermentation from corn stover feedstock. Bioresour. Technol. 2017;245:1369–1376. doi: 10.1016/j.biortech.2017.05.128. [DOI] [PubMed] [Google Scholar]
- 307.Yi X., Zhang P., Sun J., Tu Y., Gao Q., Zhang J., Bao J. Engineering wild-type robust Pediococcus acidilactici strain for hightiter l- and d-lactic acid production from corn stover feedstock. J. Biotechnol. 2016;217:112–121. doi: 10.1016/j.jbiotec.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 308.Montipó S., Ballesteros I., Fontana R.C., Ayrton S., Martins F., Ballesteros M., Camassol M. Integrated production of second generation ethanol and lactic acid from steam-exploded elephant grass. Bioresour. Technol. 2018;249:1017–1024. doi: 10.1016/j.biortech.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 309.Taniguchi M., Tokunaga T., Horiuchi K., Hoshino K., Sakai K., Tanaka T. Production of L-lactic acid from a mixture of xylose and glucose by co-cultivation of lactic acid bacteria. Appl. Biochem. Biotechnol. 2004;66:160–165. doi: 10.1007/s00253-004-1671-x. [DOI] [PubMed] [Google Scholar]
- 310.Wang C., Lia Q., Wang D., Xing J. Improving the lactic acid production of Actinobacillus succinogenes byusing a novel fermentation and separation integration system. Process Biochem. 2014;44:23–33. [Google Scholar]
- 311.Tran Q.N.M., Mimoto H., Koyama M., Nakasaki K. Lactic acid bacteria modulate organic acid production during early stages of food waste composting. Sci. Total Environ. 2019;687:341–347. doi: 10.1016/j.scitotenv.2019.06.113. [DOI] [PubMed] [Google Scholar]
- 312.Li Q., Hudari M.S.B. J.C. Wu.. Production of optically pure D-lactic acid by the combined use of Weissella sp: S26 and Bacillus sp. ADS3. Appl. Biochem. Biotechnol. 2016;178:285–293. doi: 10.1007/s12010-015-1871-0. [DOI] [PubMed] [Google Scholar]
- 313.Thapa L.P., Lee S.J., Park C., Kim S.W. Production of L-lactic acid from metabolically engineered strain of Enterobacter aerogenes ATCC 29007. Enzym. Microb. Technol. 2017;107:1–8. doi: 10.1016/j.enzmictec.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 314.Yang X., Zhu M., Huang X., Lin C.S.K., Wang J., Li S. Valorisation of mixed bakery waste in non-sterilized fermentation for L-lactic acid production by an evolved Thermoanaerobacterium sp. Strain Biores. Technol. 2015;198:47–54. doi: 10.1016/j.biortech.2015.08.108. [DOI] [PubMed] [Google Scholar]
- 315.Cizeikiene D., Juodeikiene G., Damasius J. Use of wheat straw biomass in production of L-lactic acid applying biocatalysis and combined lactic acid bacteria strains belonging to the genus Lactobacillus. Biocataly. Agric. Biotechnol. 2018;15:185–191. [Google Scholar]
- 316.Coelho L.F., De Lima C.J.B., Bernardo M.P., Contiero J. D(-)-lactic acid production by Leuconostoc mesenteroides B512 using different carbon and nitrogen sources. Appl. Biochem. Biotechnol. 2011;164:1160–1171. doi: 10.1007/s12010-011-9202-6. [DOI] [PubMed] [Google Scholar]
- 317.Wang Y., Chen C., Cai D., Wang Z., Qin P., Tan T. The optimization of L-lactic acid production from sweet sorghum juice by mixed fermentation of Bacillus coagulans and Lactobacillus rhamnosus under unsterile conditions. Bioresour. Technol. 2016;218:1098–1105. doi: 10.1016/j.biortech.2016.07.069. [DOI] [PubMed] [Google Scholar]
- 318.Qiu Z., Gao Q., Bao J. Engineering Pediococcus acidilactici with xylose assimilation pathway for high titer cellulosic L-lactic acid fermentation. Bioresour. Technol. 2018;249:9–15. doi: 10.1016/j.biortech.2017.09.117. [DOI] [PubMed] [Google Scholar]
- 319.Peinemann J.C., Demichelis F., Fiore S., Pleissner D. Techno-economic assessment of non-sterile batch and continuous production of lactic acid from food waste. Bioresour. Technol. 2019;289:121631. doi: 10.1016/j.biortech.2019.121631. [DOI] [PubMed] [Google Scholar]
- 320.Demichelis F., Pleissner D., Fiore S., Mariano S., Navarro Gutiérrez I.M., Schneider R., Venus J. Investigation of food waste valorization through sequential lactic acid fermentative production and anaerobic digestion of fermentation residues. Bioresour. Technol. 2017;241:508–516. doi: 10.1016/j.biortech.2017.05.174. [DOI] [PubMed] [Google Scholar]
- 321.Shahbazi A., Mims M.R., Li Y., Shirley V., Ibrahim S.A., Morris A. Lactic acid production from cheese whey by immobilized bacteria. Appl. Biochem. Biotechnol. 2005;122(1-3):529–540. doi: 10.1385/abab:122:1-3:0529. [DOI] [PubMed] [Google Scholar]
- 322.Li Y., Shahbazi A., Coulibaly S. Lactic acid production from cheese whey by Bifidobacterium longum. Trans. ASABE. 2006;49(4):1263–1267. [Google Scholar]
- 323.Maas R.H., Bakker R.R., Jansen M.L., Visser D., De Jong E., Eggink G., Weusthuis R.A. Lactic acid production from lime-treated wheat straw by Bacillus coagulans: neutralization of acid by fed-batch addition of alkaline substrate. Appl. Microbiol. Biotechnol. 2008;78:751–758. doi: 10.1007/s00253-008-1361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Cipro C.V.Z., Cherel Y., Caurant F., Miramand P., Méndez-Fernandez P., Bustamante P. Trace elements in tissues of white-chinned petrels (Procellaria aequinoctialis) from Kerguelen waters, Southern Indian Ocean. Polar Biol. 2014;37:763–771. [Google Scholar]
- 325.Ye L., Zhou X., Hudari M.S.B., Li Z., Wu J.C. Highly efficient production of L-lactic acid from xylose by newly isolated Bacillus coagulans C106. Bioresour. Technol. 2013;132:38–44. doi: 10.1016/j.biortech.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 326.van der Pol E.C., Eggink G., Weusthuis R.A. Production of l(+)-lactic acid from acid pretreated sugarcane bagasse using Bacillus coagulans DSM2314 in a simultaneous saccharification and fermentation strategy. Biotechnol. Biofuels. 2016;9:248. doi: 10.1186/s13068-016-0646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Glaser R., Venus J. Co-fermentation of the main sugar types from a beechwood organosolv hydrolysate by several strains of Bacillus coagulans results in effective lactic acid production. Biotechnol. Rep. 2018;18 doi: 10.1016/j.btre.2018.e00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Yao K., Zhou Q.-X., Liu D.-M., Chen S.-M., Yuan K. Comparative proteomics of the metabolic pathways involved in L-lactic acid production in Bacillus coagulans BCS13002 using different carbon sources. LWT - Food Sci. Technol. 2019;116:108445. [Google Scholar]
- 329.Wang Q., Ingram L.O., Shanmugam K.T. Evolution of D-lactate dehydrogenase activity from glycerol dehydrogenase and its utility for D-lactate production from lignocellulose. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108:18920–18925. doi: 10.1073/pnas.1111085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Zhang C., Zhou C., Assavasirijinda N., Yu B., Wang L., Ma Y. Non sterilized fermentation of high optically pure D-lactic acid by a genetically modified thermophilic Bacillus coagulans strain. Microb. Cell Factories. 2017;16:213. doi: 10.1186/s12934-017-0827-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Aulitto M., Fusco S., Bartolucci S., Franzen C.J., Contursi P. Bacillus coagulans MA-13: a promising thermophilic and cellulolytic strain for the production of lactic acid from lignocellulosic hydrolysate. Biotechnol. Biofuels. 2017;10:210. doi: 10.1186/s13068-017-0896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Ye L.D., Bin Hudari M.S., Li Z., Wu J.C. Simultaneous detoxification, saccharification and co-fermentation of oil palm empty fruit bunch hydrolysate for L-lactic acid production by Bacillus coagulans JI12. Biochem. Eng. J. 2014;83:16–21. [Google Scholar]
- 333.Zhou X., Ye L., Wu J.C. Efficient production of l-lactic acid by newly isolatedthermophilic Bacillus coagulans WCP 10-4 with high glucose tolerance. Appl. Microbiol. Biotechnol. 2013;97:4309–4314. doi: 10.1007/s00253-013-4710-7. [DOI] [PubMed] [Google Scholar]
- 334.Ye L., Zhou X., Hudari M.B., Li Z., Wu J.C. Highly efficient production of l-lacticacid from xylose by newly isolated Bacillus coagulans C106. Bioresour. Technol. 2013;132:38–44. doi: 10.1016/j.biortech.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 335.Zhang Y., Chen X., Luo J., Qi B., Wan Y. An efficient process for lactic acidproduction from wheat straw by a newly isolated Bacillus coagulans strainIPE22. Bioresour. Technol. 2014;158:396–399. doi: 10.1016/j.biortech.2014.02.128. [DOI] [PubMed] [Google Scholar]
- 336.Garrett B.G., Srinivas K., Ahring B.K. Performance and stability of AmberliteTMIRA-67 ion exchange resin for product extraction and pH control during homolactic fermentation of corn stover sugars. Biochem. Eng. J. 2015;94:1–8. [Google Scholar]
- 337.Xu K., Xu P. Efficient production of L-lactic acid using co-feeding strategy based on cane molasses/glucose carbon sources. Bioresour. Technol. 2014;153:23–29. doi: 10.1016/j.biortech.2013.11.057. [DOI] [PubMed] [Google Scholar]
- 338.Ahring B.K., Traverso J.J., Murali N., Srinivas K. Continuous fermentation of clarified corn stover hydrolysate for the production of lactic acid at high yield and productivity. Biochem. Eng. J. 2016;109:162–169. [Google Scholar]
- 339.Wang Y., Cao W., Luo J., Wan Y. Exploring the potential of lactic acid production from lignocellulosic hydrolysates with various ratios of hexose versus pentose by Bacillus coagulans IPE 22. Bioresour. Technol. 2018;261:342–349. doi: 10.1016/j.biortech.2018.03.135. [DOI] [PubMed] [Google Scholar]
- 340.Chen H., Suc Z., Wang Y., Wang B., Si Z., Lu J., Su C., Rene W., Chen H., Cai D., Qin P. Lactic acid production from pretreated corn stover with recycled streams. Process Biochem. 2020;9:132–140. [Google Scholar]
- 341.Dietz D., Schneider R., Papendiek F., Venus J. Leguminose green juice as an efficient nutrient for L(+)-lactic acid production. J. Biotechnol. 2016;236:26–34. doi: 10.1016/j.jbiotec.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 342.Jiang S., Xu P., Tao F. L-Lactic acid production by Bacillus coagulans through simultaneous saccharification and fermentation of lignocellulosic corncob residue. Biores. Technol. Rep. 2019;6:131–137. [Google Scholar]
- 343.Chen H., Huo W., Wang B., Wang Y., Wen H., Cai D., Zhang C., Wu Y., Qin P. L-lactic acid production by simultaneous saccharification and fermentation of dilute ethylediamine pre-treated rice straw. Ind. Crop. Prod. 2019;141:111749. [Google Scholar]
- 344.Wang Y., Cao W., Luo J., Qi B., Wan Y. One step open fermentation for lactic acid production from inedible starchy biomass by thermophilic Bacillus coagulans IPE22. Bioresour. Technol. 2019;272:398–406. doi: 10.1016/j.biortech.2018.10.043. [DOI] [PubMed] [Google Scholar]
- 345.Wang Y., Cai D., He M., Wang Z., Qin P., Tan T. Open fermentative production of L-lactic acid using white rice bran by simultaneous saccharification and fermentation. 2015;198:664–672. doi: 10.1016/j.biortech.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 346.López-Gómez J.P., Alexandri M., Schneider R., Latorre-Sánchez M., Coll Lozano C., Venusa J. Organic fraction of municipal solid waste for the production of L-lactic acid with high optical purity. J. Clean. Prod. 2020;247:119165. [Google Scholar]
- 347.Sakai K., Fujii N., Chukeatirote E. Racemization of L-lactic acid in pH-swingopen fermentation of kitchen refuse by selective proliferation of Lactobacillus plantarum. Journal of Bioscience and. Bioengineering. 2006;102:227–232. doi: 10.1263/jbb.102.227. [DOI] [PubMed] [Google Scholar]
- 348.Li Y., Ruan R., Chen P.L., Liu Z., Pan X., Lin X., Liu Y., Mok C., Yang T. Enzymatic hydrolysis of corn stover pretreated by combined dilute alkaline treatment and homogenization. Trans. ASAE. 2004;47:821. [Google Scholar]
- 349.Patel M.A., Ou M.S., Ingram L.O., Shanmugam K. Simultaneous saccharification and co-fermentation of crystalline cellulose and sugar cane bagasse hemicellulose hydrolysate to lactate by a thermotolerant acidophilic Bacillus sp. Biotechnol. Prog. 2005;21:1453–1460. doi: 10.1021/bp0400339. [DOI] [PubMed] [Google Scholar]
- 350.Hong A.A., Cheng K.K., Peng F., Zhou S., Sun Y., Liu C.M., Liu D.H. Strain isolation and optimization of process parameters for bioconversion of glycerol to lactic acid. J. Chem. Technol. Biotechnol. 2009;84:1576–1581. [Google Scholar]
- 351.Wang Y.Z., Li K.P., Huang F., Wang J.H., Zhao J.F., Zhao X., Garza E., Manow R., Grayburn S., Zhou S.D. Engineering and adaptive evolution of Escherichia coli W for L-lactic acid fermentation from molasses and corn steep liquor without additional nutrients. Bioresour. Technol. 2013;148:394–400. doi: 10.1016/j.biortech.2013.08.114. [DOI] [PubMed] [Google Scholar]
- 352.Parra-Ramírez D., Martinez A., Cardona C.A. Lactic acid production from glucose and xylose using the lactogenic Escherichia coli strain JU15: experiments and techno-economic results. Bioresour. Technol. 2019;273:86–92. doi: 10.1016/j.biortech.2018.10.061. [DOI] [PubMed] [Google Scholar]
- 353.Aso Y., Tsubaki M., Long B.H.D., Murakami R., Nagata K., Okano H., Dung N.T.P., Ohara H. Continuous production of D-lactic acid from cellobiose in cell recycle fermentation using b-glucosidase-displaying Escherichia coli. J. Biosci. Bioeng. 2019;127:441–446. doi: 10.1016/j.jbiosc.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 354.Tian K., Chen X., Shen W., Prior B.A., Shi G., Singh S., Wang Z. High-efficiency conversion of glycerol to D-lactic acid with metabolically engineered Escherichia coli. Afr. J. Biotechnol. 2012;11:4860–4867. [Google Scholar]
- 355.Michel A., Koch-Koerfges A., Krumbach K., Brocker M., Bott M. Anaerobic growth of Corynebacterium glutamicum by mixed-acid fermentation. Applied and Environmental Microbiology. AEM. 2015:2413–2415. doi: 10.1128/AEM.02413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Liu Y., Wen Z., Liao W., Liu C., Chen S. Optimization of the process for the production of L (+)-Lactic acid from cull potato by Rhizopus oryzae. Eng. Life Sci. 2005;5:343–349. [Google Scholar]
- 357.Efremenko E.N., Spiricheva O.V., Veremeenko D.V., Baibak A.V., Lozinsky V.I. L (+)-Lactic acid production using poly (vinyl alcohol)-cryogel-entrapped Rhizopus oryzae fungal cells. Journal of Chemical Technology & Biotechnology: international Research in Process. Environ. Clean Technol. 2006;81:519–522. [Google Scholar]
- 358.Ganguly R., Dwivedi P., Singh R. Production of lactic acid with loofa sponge immobilized Rhizopus oryzae RBU2-10. Bioresour. Technol. 2007;98:1246–1251. doi: 10.1016/j.biortech.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 359.Guo Y., Yan Q., Jiang Z., Teng C., Wang X. Efficient production of lactic acid from sucrose and corncob hydrolysate by a newly isolated Rhizopus oryzae GY18. J. Ind. Microbiol. Biotechnol. 2010;37:1137–1143. doi: 10.1007/s10295-010-0761-2. [DOI] [PubMed] [Google Scholar]
- 360.Yu M.-C., Wang R.-C., Wang C.-Y., Duan K.-J., Sheu D.-C. Enhanced production of L (+)-lactic acid by floc-form culture of Rhizopus oryzae. J. Chin. Inst. Chem. Eng. 2007;38:223–228. [Google Scholar]
- 361.Yamane T., Tanaka R. Highly accumulative production of L (+)-lactate from glucose by crystallization fermentation with immobilized Rhizopus oryzae. J. Biosci. Bioeng. 2013;115:90–95. doi: 10.1016/j.jbiosc.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 362.Koutinas A.A., Malbranque F., Wang R., Campbell G.M., Webb C. Development of an oat-based biorefinery for the production of L (+)-lactic acid by Rhizopus oryzae and various value-added coproducts. J. Agric. Food Chem. 2007;55:1755–1761. doi: 10.1021/jf0627120. [DOI] [PubMed] [Google Scholar]
- 363.Zheng Y., Wang Y., Zhang J., Pan J. Using tobacco waste extract in pre-culture medium to improve xylose utilization for L-lactic acid production from cellulosic waste by Rhizopus oryzae. Bioresource Technology. 2016;218:344–350. doi: 10.1016/j.biortech.2016.06.071. [DOI] [PubMed] [Google Scholar]
- 364.Zhao W., Huang J., Lv C., Hu S., Yao S., Mei L., Lei Y. pH stabilization of lactic acid fermentation via the glutamate 1 decarboxylation reaction: simultaneous production of lactic acid and γ-aminobutyric acid. Process Biochem. 2015;50:1523–1527. [Google Scholar]
- 365.Phrueksawan P., Kulpreecha S., Sooksai S., Thongchul N. Direct fermentation of L (+)-lactic acid from cassava pulp by solid state culture of Rhizopus oryzae. Bioproc. Biosyst. Eng. 2012;35:1429–1436. doi: 10.1007/s00449-012-0731-3. [DOI] [PubMed] [Google Scholar]
- 366.Pimtonga V., Ounaeba S., Thitiprasertb S., Toliengb V., Sooksaib S., Boonsombatb R., Tanasupawatc S., Assabumrungratd S., Thongchul N. Enhanced effectiveness of Rhizopus oryzae by immobilization in astatic bed fermentor for l-lactic acid production. Process Biochem. 2017;52:44–52. [Google Scholar]
- 367.Wang Z., Wang Y., Yang S.T., Wang R., Ren H. A novel honeycomb matrix for cell immobilization to enhance lactic acid production by Rhizopus oryzae, Bioresour. Technol. 2010;101:5557–5564. doi: 10.1016/j.biortech.2010.02.064. [DOI] [PubMed] [Google Scholar]
- 368.Coban H.B., Demirci A. Enhancement and modeling of microparticle-added Rhizopus oryzae lactic acid production. Bioproc. Biosyst. Eng. 2016;39:323–340. doi: 10.1007/s00449-015-1518-0. [DOI] [PubMed] [Google Scholar]
- 369.Sun J., Zhu J., Li W. L-(D) lactic acid production by Rhizopus oryzae using pretreated dairy manure as carbon and nitrogen source. Biomass Bioenergy. 2012;4(7):442–450. [Google Scholar]
- 370.Zhang L., Li X., Yong Q., Yang S.-T., Ouyang J., Yu S. Simultaneous saccharification and fermentation ofxylo-oligosaccharides manufacturing waste residue for l-lactic acidproduction by Rhizopus oryzae. Biochem. Eng. J. 2015;94:92–99. [Google Scholar]
- 371.Vially G., Marchal R., Guilbert N. l(+) lactate production from carbohydratesand lignocellulosic materials by Rhizopus oryzae UMIP 4.77. World J. Microbiol. Biotechnol. 2010;26:607–614. [Google Scholar]
- 372.Ma X., Gao M., Yin Z., Zhu W., Liu S., Wang Q. Lactic acid and animal feeds production from Sophora flavescens residues by Rhizopus oryzae fermentation. Process Biochem. 2020;92:401–408. [Google Scholar]
- 373.Trakarnpaiboon S., Srisuk N., Piyachomkwan K., Yang S.-T., Kitpreechavanich V. L-Lactic acid production from liquefied cassava starch by thermotolerant Rhizopus microsporus: characterization and optimization. Process Biochem. 2017;63(2017):26–34. [Google Scholar]
- 374.Zhang Z., Jin B. L (+)-lactic acid production using sugarcane molasses and waste potato starch: an alternative approach. Int. Sugar J. 2010;112:17. [Google Scholar]
- 375.A Weusthuis R., Mars A.E., Springer J., H Wolbert E.J., van der Wal H., de Vrije T.G., Levisson M., Leprince A., Houweling-Tan G.B., Moers A.P.H.A., A Hendriks S.N., Mendes O., Griekspoor Y., Werten M.W.T., Schaap P.J., van der Oost J., Eggink G. Monascus ruber as cell factory for lactic acid production at low pH. Metab. Eng. 2017;42:66–73. doi: 10.1016/j.ymben.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 376.Laud N., Rosso M.N., Fabre N., Crapart S., Herpoël-Gimbert I., Sigoillot J.C., Raouche S., Levasseur A. L-lactic acid production by Aspergillus brasiliensis overexpressing the heterologous ldha gene from Rhizopus oryzae. Microb. Cell Factories. 2015;14:66. doi: 10.1186/s12934-015-0249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Dave K.K., Punekar N.S. Expression of lactate hehydrogenase in Aspergillus niger for L-lactic acid production. PloS One. 2015;10 doi: 10.1371/journal.pone.0145459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Kim Y.S., Jang J.Y., Park S.J., Um B.H. Dilute sulfuric acid fractionation of Korean food waste for ethanol and lactic acid production by yeast. Waste Manag. 2018;74:231–240. doi: 10.1016/j.wasman.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 379.Gao M.-T., Shimamura T., Ishida N., Takahashi H. Fermentative lactic acid production with a metabolically engineered yeast immobilized in photo-crosslinkable resins. Biochem. Eng. J. 2009;47:66–70. [Google Scholar]
- 380.Colombié S., Dequin S., Sablayrolles J. Control of lactate production by Saccharomyces cerevisiae expressing a bacterial LDH gene. Enzym. Microb. Technol. 2003;33:38–46. [Google Scholar]
- 381.Ishida N., Saitoh S., Tokuhiro K., Nagamori E., Matsuyama T., Kitamoto K., Takahashi H. Efficient production of L-lactic acid by metabolically engineered Saccharomyces cerevisiae with a genome-integrated L-lactate dehydrogenase gene. Appl. Environ. Microbiol. 2005;71:1964–1970. doi: 10.1128/AEM.71.4.1964-1970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Ishida N., Saitoh S., Onishi T., Tokuhiro K., Nagamori E., Kitamoto K., Takahashi H. The effect of pyruvate decarboxylase gene knockout in Saccharomyces cerevisiae on L-lactic acid production. Biosci. Biotechnol. Biochem. 2006;70:1148–1153. doi: 10.1271/bbb.70.1148. [DOI] [PubMed] [Google Scholar]
- 383.Ishida C., Aranda C., Valenzuela L., Riego L., DeLuna A., Recillas-Targa F., Filetici P., López-Revilla R., González A. The UGA3-GLT1 intergenic region constitutes a promoter whose bidirectional nature is determined by chromatin organization in Saccharomyces cerevisiae. Mol. Microbiol. 2006;59:1790–1806. doi: 10.1111/j.1365-2958.2006.05055.x. [DOI] [PubMed] [Google Scholar]
- 384.Lee J.Y., Kang C.D., Lee S.H., Park Y.K., Cho K.M. Engineering cellular redox balance in Saccharomyces cerevisiae for improved production of L-lactic acid. Biotechnol. Bioeng. 2015;112:751–758. doi: 10.1002/bit.25488. [DOI] [PubMed] [Google Scholar]
- 385.Yamada R., Wakita K., Mitsui R., Ogino H. Enhanced D-lactic acid production by recombinant Saccharomyces cerevisiae following optimization of the global metabolic pathway. Biotechnol. Bioeng. 2017;114:2075–2084. doi: 10.1002/bit.26330. [DOI] [PubMed] [Google Scholar]
- 386.Miller M., Suominen P., Aristidou A., Hause B.M., Van Hoek P., Dundon C.A. Lactic acid-producing yeast cells having nonfunctional L-or D-lactate: ferricytochrome C oxidoreductase cells. Google Patents. 2012 [Google Scholar]
- 387.Dundon C.A., Suominen P., Aristidou A., Rush B.J., Koivuranta K., Hause B.M., McMullin T.W., Roberg-Perez K. Yeast cells having disrupted pathway from dihydroxyacetone phosphate to glycerol. Google Patents. 2009 [Google Scholar]
- 388.Tamakawa H., Ikushima S., Yoshida S. Efficient production of L-lactic acid from xylose by a recombinant Candida utilis strain. J. Biosci. Bioeng. 2012;113:73–75. doi: 10.1016/j.jbiosc.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 389.Ilmén M., Koivuranta K., Ruohonen L., Rajgarhia V., Suominen P., Penttilä M. Production of L-lactic acid by the yeast Candida sonorensis expressing heterologous bacterial and fungal lactate dehydrogenases. Microb. Cell Factories. 2013;12:53. doi: 10.1186/1475-2859-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Hou Q., He Q., Liu G., Lu X., Zong H., Chen W., Zhuge B. Identification and application of novel low pH-inducible promoters for lactic acid production in the tolerant yeast Candida glycerinogenes. J. Biosci. Bioeng. 2019;128:8–12. doi: 10.1016/j.jbiosc.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 391.Bae J.-H., Kim H.-J., Kim M.-J., Sung B.H., Jeon J.-H., Kim H.-S., Jin Y.-S., Kweon D.-H., Sohn J.-H. Direct fermentation of Jerusalem artichoke tuber powder for production of L-lactic acid and D-lactic acid by metabolically engineered Kluyveromyces Marxianus. J. Biotechnol. 2018;266:27–33. doi: 10.1016/j.jbiotec.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 392.Kong X., Zhang B., Hua Y., Zhu Y., Li W., Wang D., Hong J. Efficient L-lactic acid production from corncob residue using metabolically engineered thermo-tolerant yeast. Bioresour. Technol. 2019;273:220–230. doi: 10.1016/j.biortech.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 393.Pavlovich S.S., Mikhajlovic V.M., Vladimirovich J.T., Aleksandrovich R.J., Isaakovna R.E., Georgievna T.N., Aleksandrovna V.M., Mikhajlovna A.A., Georgievich D.V. 2006. Method for Microbiological Synthesis of Lactic Acid and Recombinant Strain of Yeast Schizosaccharomyces pombe for its Realization. Russian Patent Application RU000002268304. [Google Scholar]
- 394.Ozaki A., Konishi R., Otomo C., Kishid M., Takayama S., Matsumoto T., Tanaka T., Kondo A. Metabolic engineering of Schizosaccharomyces pombe via CRISPR-Cas9 genome editing for lactic acid production from glucose and cellobiose. Metabol. Eng. Commun. 2017;5:60–67. doi: 10.1016/j.meteno.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Yamada R., Wakita K., Mitsui R., Ogino H. Enhanced D-lactic acid production by recombinant Saccharomyces cerevisiae following optimization of the global metabolic pathway. Biotechnol. Bioeng. 2017;114:2075–2084. doi: 10.1002/bit.26330. [DOI] [PubMed] [Google Scholar]
- 396.Baek S.H., Kwon E.Y., Kim Y.H., Hahn J.S. Metabolic engineering and adaptive evolution for efficient production of d-lactic acid in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2016;100:2737–2748. doi: 10.1007/s00253-015-7174-0. [DOI] [PubMed] [Google Scholar]
- 397.Varman A.M., Yu Y., You L., Tang Y.J. Photoautotrophic production of D-lactic acid in an engineered cyanobacterium. Microb. Cell Factories. 2013;12:117. doi: 10.1186/1475-2859-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Li J., Zhang W., Li X., Ye T., Gan Y., Zhang A., Chen H., Xue G., Liu Y. Production of lactic acid from thermal pretreated food waste through the fermentation of waste activated sludge: effects of substrate and thermal pretreatment temperature. Bioresour. Technol. 2018;247:890–896. doi: 10.1016/j.biortech.2017.09.186. [DOI] [PubMed] [Google Scholar]
- 399.Akao S., Tsuno H., Cheon J. Semi-continuous L-lactate fermentation of garbage without sterile condition and analysis of the microbial structure. Water Resour. 2007;41:1774–1780. doi: 10.1016/j.watres.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 400.Itoh Y., Tada K., Kanno T., Horiuchi J.I. Selective production of lactic acid in continuous anaerobic acidogenesis by extremely low pH operation. J. Biosci. Bioeng. 2012;114:537–539. doi: 10.1016/j.jbiosc.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 401.Kim D.H., Lim W.T., Lee M.K., Kim M.S. Effect of temperature on continuous fermentative lactic acid (LA) production and bacterial community, and development of LA-producing UASB reactor. Bioresour. Technol. 2012;119:355–361. doi: 10.1016/j.biortech.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 402.Maeda T., Yoshimura T., Shimazu T., Shirai Y., Ogawa H.I. Enhanced production of lactic acid with reducing excess sludge by lactate fermentation. J. Hazard Mater. 2009;168:656–663. doi: 10.1016/j.jhazmat.2009.02.067. [DOI] [PubMed] [Google Scholar]
- 403.Sun Y., Xu Z., Zheng Y., Zhou J., Xiu Z. Efficient production of lactic acid from sugarcane molasses by a newly microbial consortium CEE-DL15. Process Biochem. 2019;81:132–138. [Google Scholar]
- 404.Tang J., Wang X.C., Hu Y., Zhang Y., Li Y. Effect of pH on lactic acid production from acidogenic fermentation of food waste with different types of inocula. Bioresour. Technol. 2017;224:544–552. doi: 10.1016/j.biortech.2016.11.111. [DOI] [PubMed] [Google Scholar]