Abstract
Background
Genetic analyses have identified many variants associated with the risk of inflammatory bowel disease (IBD) development. Among these variants, the ones located within the NOD2 gene have the highest odds ratio of all IBD genetic risk variants. Also, patients with Crohn’s disease (CD) have been shown to have an altered gut microbiome, which might be a reflection of inflammation itself or an effect of other parameters that contribute to the risk of the disease. Since NOD2 is an intracellular pattern recognition receptor that senses bacterial peptidoglycan in the cytosol and stimulates the host immune response (Al Nabhani et al., PLoS Pathog 13:e1006177, 2017), it is hypothesized that NOD2 variants represent perfect candidates for influencing host-microbiome interactions. We hypothesized that NOD2 risk variants affect the microbiome composition of healthy first degree relative (FDR) of CD patients and thus potentially contribute to an altered microbiome state before disease onset.
Methods
Based on this, we studied a large cohort of 1546 healthy FDR of CD patients and performed a focused analysis of the association of three major CD SNPs in the coding region of the NOD2 gene, which are known to confer a 15–40-fold increased risk of developing CD in homozygous or compound heterozygous individuals.
Results
Our results show that carriers of the C allele at rs2066845 was significantly associated with an increase in relative abundance in the fecal bacterial family Erysipelotrichaceae.
Conclusions
This result suggests that NOD2 polymorphisms contribute to fecal microbiome composition in asymptomatic individuals. Whether this modulation of the microbiome influences the future development of CD remains to be assessed.
Keywords: Fecal microbiota, Healthy human, Microbiome, rs2066844, rs2066845, rs2066847, NOD2, Inflammatory bowel disease
Background
Crohn’s disease (CD) is a complex disease with a significant public health impact [1]. Several factors are known to contribute to the risk of developing CD, but the exact cause(s) remains to be identified. It is generally thought that genetic and environmental factors play a crucial role in the development of CD. Also, alterations in the gut microbiome are described in patients with established CD [2]. NOD2 was the first susceptibility gene discovered for CD [3]. Since this discovery, genome-wide association studies (GWAs) have allowed the identification of many other genetic variants associated with CD [4], including several additional variants located in the NOD2 locus [4]. More precisely, three common variants located in the NOD2 leucine-rich repeat domain have been identified in the European population. These are R702W, G908R resulting from two cis-mutations in rs2066844, rs2066845 and L1007fs, a frameshift mutation identified by rs2066847 insC, respectively [5, 6]. Individuals who are homozygous or compound heterozygous for these mutations in NOD2 have a 15–40-fold increased risk of developing CD [5, 6].
Previous studies attempted to determine if NOD2 genotype is associated with any alteration in the composition of the intestinal microbiome in CD case-control studies. However, those studies fail to distinguish between the possibilities that microbial changes are the result of inflammation present in these patients with CD or drug therapies [7]. Several studies have performed such analysis using a GWAs approach in healthy individuals, but all failed to identify robust association looking at NOD2 genotype association with microbial composition [8–10]. The conservative threshold for association significance used in GWAs studies (p-value < 5 × 10− 8), may explain the absence of association. Alternatively, healthy individuals from the general population may have a relatively lower abundance of NOD2 variants associated with CD. For these two reasons, we addressed in a targeted fashion whether NOD2 CD risk alleles are associated with microbial composition in healthy first-degree relatives (FDR) of CD patients known to be genetically at higher risk to develop CD as compared to general population [11, 12]. We thus evaluated in a targeted fashion the association between three NOD2 CD risk variants known to have the largest effect size for CD (rs2066844, rs2066845 and rs2066847) with the fecal microbial composition in a large cohort of 1546 healthy FDRs of CD patients.
Methods
Patient recruitment
A total of 1546 subjects were recruited as part of the Crohn’s and Colitis Canada Genetic Environmental Microbial (CCC-GEM) project between the years 2008 and 2017. The study was approved by the Mount Sinai Hospital Research Ethics Board (Toronto - Managing center) and each participating recruitment center. All the participants or their guardians provided written consent before their enrollment. Participants provided blood and a stool sample for genetic and microbiome analysis, respectively.
Genotyping
DNA extraction from peripheral blood mononuclear cells (PBMC) was performed using the Gentra Puregene Blood Kit (Qiagen, CA, USA). Genomic DNA was quantified using Nanodrop®, and then dilutions were prepared at concentrations of 20 ng/μl and aliquoted into 96 well reaction plates. SNPs genotyping was performed using Illumina HumanCoreExome chip and ImmunoChip (Illumina, Inc. San Diego, CA) following the provider’s recommendations as previously described [8].
Genome imputation
Genotype imputation was performed on a merged HumanCoreEXOME chip and ImmunoChip hard genotype data. We performed two imputations, the first one based on the Haplotype Research Consortium (HRC) panel, and the second based on the 1000 Genomes imputation panel of October 2014. The former was performed by directly uploading the hard genotype to the HRC website (http://www.haplotype-reference-consortium.org/), while for the latter, IMPUTE2 (v. 2.3.0) [13, 14] was used. Markers were imputed in 1 M bp intervals with 750 K bp flanking regions with pre-phased haplotypes. Imputation results were quality controlled for minor allele frequency (MAF > 1%), missingness (< 5%), imputation certainty (> 0.4), and imputation information (> 0.9). The three NOD2 SNPs were identified based on their coordinates in the human genome build 37. The two SNPs rs2066844, rs2066845 were captured from the HRC imputation, whereas the rs2066847 was present as a hard-genotyped SNP in HumanCoreEXOME chip. PLINK v1.90b3.38 was used to extract the SNPs and to calculate their MAF [15].
Taxonomic profiling of the gut microbiota
Stool samples were collected in FB Commode Specimen Collector (Fisher Scientific, Waltham, MA) and put into polypropylene vials (Starplex Scientific Inc., Etobicoke, ON) and frozen prior their shipping to the local study sites where they were stored at − 80 °C. DNA extraction from fecal bacteria was performed using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). We added a step to the manufacturer’s recommendations, which consists of physical disruption of the bacterial cell wall using ceramic beads. The V4 hypervariable region of bacterial 16S ribosomal RNA (16S rRNA) was sequenced in paired-end mode (2 × 150 base pair) using MiSeq platform (Illumina Inc., San Diego, CA, USA). Paired reads were assembled using fastq-join software (ea-utils suite v1.1.2.537), resulting in 250 base pairs amplicons. The latter were then processed using the quantitative insights into microbial ecology (QIIME v1.9.0) pipeline using the default parameters. Operational taxonomic units (OTUs) were clustered at 97% sequence similarity against Greengenes database v.13_8 using a closed reference approach. Samples with less than 10,000 reads after quality filtering were removed from the analysis. Alpha diversity was expressed using the Shannon index after rarefaction at a depth of 10,000 reads per sample. 16S sequences are available by the study accession number PRJEB14839 [8].
Statistical considerations
To assess the association between the NOD2 polymorphisms and the fecal microbiome, we used two approaches. In the first approach, we evaluated the individual association of each of the three NOD2 risk SNPs with the microbiome. In the second approach, we stratified the cohort based on the presence of at least one NOD2 risk variant. Hence we had a new variable with two categories: subjects with at least one mutation (rs2066844, and/or rs2066845, and/or rs2066847) and subjects with no mutations.
In the two approaches, we tested first the association of the variables with the global composition of the microbiome at the genus level using permutational multivariate analysis of variance (PERMANOVA) on an OTU table rarefied to 10,000 reads and using a Bray-Curtis distance matrix. Then, the variables (NOD2 polymorphism and presence of at least one variant) were tested for their association with every member of the different taxonomy levels from phylum to genus using a two-part log-normal model as previously described [8, 16]. The model was applied to a non-rarefied OTU table and corrected for age, sex and the total number of reads per sample. All statistical analyses were performed using R (v3.3.3). PERMANOVA was performed using the Vegan package (V2.4–5), whereas the geeglm function of the geepack package (v1.2.0–2) was used to carry out the two-part log-normal model on each bacterial taxa.
The correction of multiple testing was performed by calculating the effective number of uncorrelated taxa using the approach described by Li and Ji [17]. Our taxonomy table contained 263 taxa from phylum to genus level. The application of the Li and Ji [17] method allowed us to identify 132 independent variables, bringing the raw p-value significance threshold to 3.8 × 10− 4.
Results
Frequency of NOD2 risk variants in healthy FDR of CD patients
A cohort of 1546 asymptomatic FDRs of CD patients recruited between 2008 and 2017 as part of the Crohn’s and Colitis Canada Genetic Environmental Microbial (CCC-GEM) project. All study subjects were of Caucasian descent with a mean age ± standard deviation of 19.4 ± 7.9 [6-35] were included in this study including 848 Female (See methods section). We calculated the MAF of the rs2066844 (R702W), rs2066845 (G908R) and rs2066847 (l1007fs) in our population of healthy FDRs of CD patients and compared this to those recorded in the 1000 genome project (self-reported healthy individuals) (see Table 1). The MAF of each of the three NOD2 risk SNPs were numerically higher in our cohort than reported in the 1000 genome project general healthy population. This increase in SNP frequency suggests that our cohort is enriched in NOD2 risk SNPs.
Table 1.
Minor allele frequency of NOD2 risk SNPs in healthy FDR of CD patients in comparison to the MAF in the 1000 genome project
| SNPs | MAF in healthy FDR of CD | MAF in the 1000 genomes project |
|---|---|---|
| rs2066844 | 8.9% | 1% |
| rs2066845 | 3.1% | < 1% |
| rs2066847 | 4.2% | < 1% |
MAF: Minor allele frequency, SNPs: single nucleotide polymorphisms. MAF from the 1000 genomes project was obtained using Ensembl database
Association of the individual NOD2 risk SNPs with the microbial composition and diversity of the microbiome in FDRs
To understand the associations between the three individual NOD2 variants and the significance in clustering patterns of the microbiome composition, we performed a PERMANOVA analysis; this showed no significant association (Table 2). Similarly, the group with at least one out of the three NOD2 variants compared with individuals without any NOD2 variant showed no significant difference in clustering patterns of stool microbiome composition. The alpha diversity, as represented by the Shannon index, also showed no significant association with each of the individual NOD2 variants, rs2066844, rs2066845 or rs2066847, respectively. The group with at least one mutation in the three NOD2 risk SNPs compared to the group with no variants also showed no significant difference in alpha diversity.
Table 2.
Association between the three NOD2 polymorphisms and the clustering patterns of the fecal microbiome composition
| Variants | PERMANOVA p value |
|---|---|
| rs2066844 | 0.44 |
| rs2066845 | 0.07 |
| rs2066847 | 0.10 |
| At least one NOD2 mutationa | 0.75 |
a in rs2066844 and/or rs2066845 and/or rs2066847
Association between NOD2 polymorphisms and specific fecal bacterial taxa and diversity
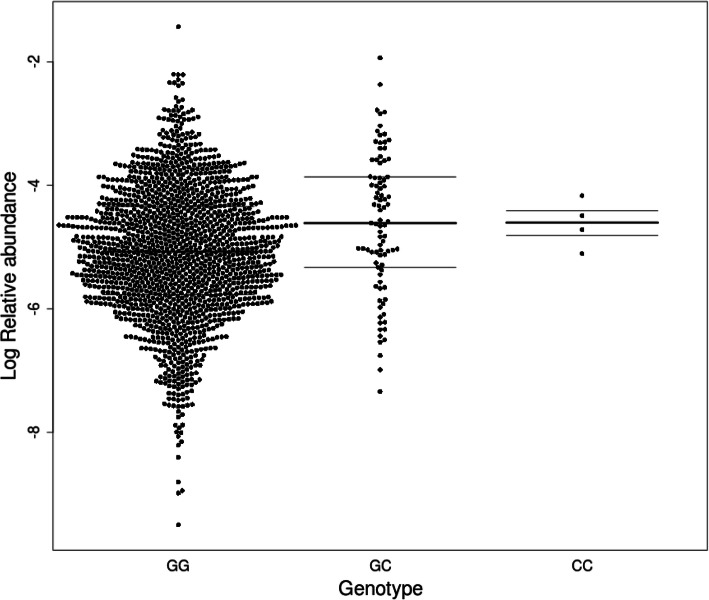
We assessed the association of each of the three NOD2 variants with each fecal bacterial taxon from phylum to genus level of the taxonomy. The rs2066845 was significantly associated with the family Erysipelotrichaceae (estimate ± standard deviation; 0.39 ± 0.10, p-value = 7.14 × 10− 5) (Fig. 1). The abundance of this taxon was significantly increased in the groups with at least one mutation in rs2066845 (the heterozygote and the minor homozygote) compared to groups with the null mutation. However, none of the genera of the Erysipelotrichaceae family appeared to significantly contribute to the effect observed at the family taxonomic level (Supplementary Table 1). No significant associations were observed for rs2066844 or rs2066847 with microbiome taxa (Supplementary Tables 2 and 3, respectively). When assessing the effect of binning the three NOD2 variants, no significant associations were observed between the presence of at least one mutation within the three NOD2 variants with any of the individual taxa (Supplementary Table 4).
Fig. 1.
Erysipelotrichaceae family is associated with rs2066845 genotype in 1546 healthy subjects. The x-axis corresponds to the rs2066845 genotype in the cohort. The y-axis corresponds to the log-transformed relative abundance of Erysipelotrichaceae. Each dot represents the value for a given individual. The lines represent the first, second, and third quartiles. Carrier of the C allele are enriched in the relative abundance of Erysipelotrichaceae (p-value = 7.14 × 10− 5)
Discussion
The NOD2 gene was the first found to be directly associated with CD risk through three main mutations occurring in the leucine-rich repeat region of the gene. Since this gene is a pattern recognition receptor and is involved in host-microbe interactions, a few studies have shown that NOD2 polymorphisms are involved in shaping the microbiome in IBD patients [18–21]. However, most of these studies were case-control studies, and as such, the inflammation process can affect microbiome composition regardless of the NOD2 genotype [19–21]. Our study cohort of healthy FDRs of CD patients permits to minimize some of these potential confounding factors as they are asymptomatic, do not take any anti-inflammatory or anti-bacterial medication, while having a high genetic risk of developing CD and carrying a higher frequency of CD associated mutations. As such, we assessed the association of three major NOD2 mutations with the composition of the gut microbiome in this cohort. We found that this cohort was enriched in NOD2 variants as reflected by higher MAFs of the CD risk SNPs, rs2066844 (R702W), rs2066845 (G908R), and rs2066847 (l1007fs) as compared to the general population (Table 2) confirming the findings of our previous study of healthy first-degree relatives [22]. Hence this cohort provides a higher prevalence of the NOD2 polymorphisms and thus is more powered than cohorts of healthy individuals from the general population in order to assess the potential association of NOD2 SNPs with gut microbiome composition in healthy individuals.
Our results showed that one of the NOD2 risk allele (rs2066845) is associated with the increased relative abundance of Erysipelotrichaceae family. Little is known regarding Erysipelotrichaceae family in terms of disease associations [23]. There is data to suggest that Erysipelotrichaceae is associated with metabolic disorders in humans [23]. Another study showed that species within Erysipelotrichaceae family are positively linked to inflammation in patients who had chronic HIV infection [19]. This taxon was decreased in early CD onset [20], and patients who experienced recurrence of CD had significantly lower levels of Erysipelotrichaceae [21]. Studies have shown inconsistent alterations in Erysipelotrichaceae family relative abundance in patients with established CD [7, 20, 21, 23]. In a recent report, Erysipelotrichaceae family was found to be increased in patients with CD patients with ileal disease compared to those with ileocolonic disease [24], which may be consistent with the fact that NOD2 is strongly associated with ileal CD [25]. Also, this taxon was found to be highly coated by IgA relative to other members of the gut microbiota in IBD patients [26]. It includes strictly anaerobic or aerotolerant organisms, and members of the Erysipelotrichaceae family can produce palmitic, stearic and oleic acid, fatty acids predominantly [27]. Over accumulation of tissue palmitic acid results in dyslipidemia, hyperglycemia, increased ectopic fat accumulation, increased inflammatory tone via toll-like receptor 4 [28] as well as pro-inflammatory adipokines mRNA expression in macrophages [29]. Also, stearic acid was shown to stimulate pro-inflammatory cytokine production [30]. It remained to be shown how the changes in this taxon contribute to the risk of CD in this population.
The other two NOD2 risk variants (rs2066844 and rs2066847) showed no significant associations with the fecal bacterial composition. A previous study detected an association of NOD2 risk allele dosages (comprised of combined rs104895431, rs104895467, rs2066844, rs2066845, rs5743277, rs5743293) with an increase of Enterobacteriaceae from biopsy sample in a cohort of 474 IBD affected patients [31]. However, the altered microbiome described in IBD patients is characterized by an increase in Proteobacteria species such as Escherichia coli (a species belonging to Enterobacteriaceae family) [2, 32]. Thus the increase of Enterobacteriaceae previously observed might be related to inflammation itself and not necessarily NOD2 genotype. Other studies of large cohorts of healthy individuals have failed to show any significant association of NOD2 genotype with microbiome composition [7–10, 33]. In these GWAS, only one study comprised of 1514 individuals showed that the NOD2 locus was associated with the enterobactin biosynthesis pathway, which is highly correlated with Escherichia coli abundance [10]. Our analysis showed that all the three tested NOD2 variants had a consistent direction of the association for which NOD2 variants had an increased Enterobacteriaceae despite not being significant. Thus the association of Enterobacteriaceae and NOD2 locus polymorphisms remains unclear.
The association between host genetics with microbial composition might be better detected in biopsy samples compared to fecal samples. In fact, in a study comparing the tissue associated bacteria in the ileum to the fecal bacteria in CD patients carrying the rs2066847 polymorphism or not showed that significant differences in microbial composition were only observed in the biopsy microbiome [34]. Recent GWAs studies of IBD subjects report that NOD2 variants are strongly associated in particular with ileal CD compared to colonic CD [25]. Thus, this may, in part, explain the weaker signal observed in fecal samples compared to biopsy samples in the ileum [35, 36].
Conclusion
In conclusion, this study is one of the largest studies of healthy subjects examining the specific association of the NOD2 genotype with the healthy human gut microbiota. We found that the NOD2 genotype has no significant influence on the overall composition of stool microbiota, suggesting that NOD2 polymorphism is not a major determinant of the overall composition of the microbiome in this healthy at-risk population. However, in our targeted analysis of the microbiota, we found that rs2066845 is significantly associated with an increased relative abundance of the Erysipelotrichaceae family. It remains possible that the effect of NOD2 polymorphisms on microbiome composition are limited under stable homeostatic conditions, and might be more enhanced after the disease onset, i.e. in the presence of active inflammation [31, 34, 37], or may be related to a specific location such as ileal CD. It remains possible that increased risk of developing CD contributed by NOD2 genotype is in part related to alteration of particular taxa such as Erysipelotrichaceae family in healthy individuals.
Supplementary information
Additional file 1 Supplementary Table 1. Association of rs2066845 with bacterial taxa in the GEM cohort. Supplementary Table 2. Association of rs2066844 with bacterial taxa in the GEM cohort. Supplementary Table 3. Association of rs2066847 with bacterial taxa in the GEM cohort. Supplementary Table 4. Cumulative NOD2 variants association with bacterial taxa in the GEM cohort.
Acknowledgments
We thank the members of the CCC GEM Global Project Office. The CCC IBD GEM Project research team is composed of: Maria Abreu, Paul Beck, Charles Bernstein, Kenneth Croitoru, Leo Dieleman, Brian Feagan, Anne Griffiths, David Guttman, Kevan Jacobson, Gilaad Kaplan, Denis O. Krause*(* deceased), Karen Madsen, John Marshall, Paul Moayyedi, Mark Ropeleski, Ernest Seidman*(* deceased), Mark Silverberg, Scott Snapper, Andy Stadnyk, Hillary Steinhart, Michael Surette, Dan Turner, Thomas Walters, Bruce Vallance, Guy Aumais, Alain Bitton, Maria Cino, Jeff Critch, Lee Denson, Colette Deslandres, Wael El-Matary, Hans Herfarth, Peter Higgins, Hien Huynh, Jeff Hyams, David Mack, Jerry McGrath, Anthony Otley, and Remo Panancionne. The CCC GEM Project recruitment site directors include Maria Abreu, Guy Aumais, Robert Baldassano, Charles Bernstein, Maria Cino, Lee Denson, Colette Deslandres, Wael El-Matary, Anne M. Griffiths, Charlotte Hedin, Hans Herfarth, Peter Higgins, Seamus Hussey, Hien Hyams, Kevan Jacobson, David Keljo, David Kevans, Charlie Lees, David Mack, John Marshall, Jerry McGrath, Sanjay Murthy, Anthony Otley, Remo Panaccione, Nimisha Parekh, Sophie Plamondon, Graham Radford-Smith, Mark Ropeleski, Joel Rosh, David Rubin, Michael Schultz, Ernest Seidman, Corey Siegel, Scott Snapper, Hillary Steinhart, and Dan Turner.
Abbreviations
- CD
Crohn’s disease
- IBD
Inflammatory bowel disease
- FDR
Healthy first degree relative
- GWAs
Genome-wide association studies
- CCC-GEM
Crohn’s and Colitis Canada Genetic Environmental Microbial
- PBMC
Peripheral blood mononuclear cells
- PERMANOVA
Permutational multivariate analysis of variance
- HRC
Haplotype Research Consortium
- OTUs
Operational taxonomic units
- MAF
Minor allele frequency
- SNP
Single nucleotide polymorphisms
- NOD2
Nucleotide-binding oligomerization domain-containing protein 2
- 16S rRNA
16S ribosomal RNA
- QIIME
Quantitative insights into microbial ecology
Authors’ contributions
Conceived and designed the analysis: WT, LB, WX, MIS, CCC IBD GEM Project research team, KC, MSS, DSG, AG, PM, RP, HH, HAS, GA, LAD, DT, ADP. Collected the data: MSS, DSG, AG, PM, RP, HH, HAS, GA, LAD, DT, CCC IBD GEM Project research team, KC. Contributed data or analysis tools: WT, LB, OEG, WX, MIS, JARG, SHL, PM, ADP, KC. Performed the analysis: WT, LB, OEG. Wrote the paper: WT, LB, OEG, JARG, SHL, KC. The authors read and approved the final manuscript
Funding
This study was supported by grants from Crohn’s and Colitis Canada Grant #CCC-GEMIII, Canadian Institutes of Health Research (CIHR) Grant #CMF108031 and the Helmsley Charitable Trust. Williams Turpin is a former recipient of a Postdoctoral Fellowship Research Award from the CIHR Fellowship/ Canadian Association of Gastroenterology (CAG)/ Ferring Pharmaceuticals Inc. and of a fellowship from the Department of Medicine, Mount Sinai Hospital, Toronto. Williams Turpin is partially supported by the Biocodex Microbiota Foundation. Sun-Ho Lee, Juan Antonio Raygoza Garay, are recipient of fellowship from the Department of Medicine, Mount Sinai Hospital, Toronto. Mark Silverberg is supported in part by the Gale and Graham Wright Chair in Digestive Diseases. Kenneth Croitoru is partially supported by a Canada Research Chair in Inflammatory Bowel Diseases. Osvaldo Espin-Garcia is a recipient of a CIHR STAGE fellowship.
Availability of data and materials
16S sequences are available by the study accession number PRJEB14839 (NCBI BioProject database); ENA-SUBMISSION: ERA675979 [9]. Requests for raw and analyzed data should follow the instruction given at http://www.gemproject.ca/data-access/. All submissions will be reviewed by the GEM Project Operating Committee to ensure that the requested samples/data will not interfere in any way with the intended GEM Project analysis of the nested cohort as per the original GEM Project Study Design and is not a duplication of analysis already ongoing. Those proposals meeting this evaluation will be distributed to all members of the GEM Project Steering Committee (GPSC) for review and open discussion. This review will focus on the global scientific merit of the proposal. This review will assess the basic scientific merit and the availability of requested samples and data, ensuring there is no compromise of the original intent of the GEM project. It would be of value to contact a member of the Steering Committee who could help sponsor your application. Those projects achieving majority vote of approval at the GPSC will be informed that the GEM Project will provide a letter of support stating that the requested samples or data will be made available to the applicants once the applicant receives funding from a granting agency that applies an independent peer review process to the proposal. The criteria to be used for review of all submissions will include the “scientific relevance” of the proposal and the judged availability of biological material requested. The budget to be requested from a funding agency, must allow for any expenses in processing samples or in setting up the appropriate queries of the database. The intent is to allow sufficient time for applicants to consider submission for funding opportunities.
Ethics approval and consent to participate
All subjects and/or their guardians gave written informed consent to participate in the study. The study was approved by the Mount Sinai Hospital Research Ethics Board (Toronto – Managing center) and local centers (07–0322-E). The local centers are comprised of: the Conjoint Health Research Ethics Board (Calgary), The Research Ethics Office (Edmonton), the Nova Scotia Health Authority (Halifax), the Hamilton Integrated Research Ethics Board (Hamilton), the Queen’s University Health Sciences and Affiliated Teaching Hospitals Research Ethics Board (Kingston), Western’s Research Ethics Boards (London, ON), the Research Centre of the Sainte-Justine University Hospital Research Ethics Board (Ste-Justine), McGill Research Ethics Boards (McGill), the Office of Research Ethics and Integrity (Ottawa), the Interdisciplinary Committee on Ethics in Human Research (St John’s), the Clinical Research Ethics and Regulatory Compliance Office (Sick Kids Hospital), the UBC Office of Research Ethics (Vancouver), the Biomedical Research Ethics Board (Winnipeg), the comité d’éthique de la recherche (Hôpital du St-Sacrement), the University of Saskatchewan Research Ethics Board (Saskatoon), the Sunnybrook’s research ethics board (Sunnybrook), the MUHC Centre for Applied Ethics (Hôpital Maisonneuve-Rosemont), the UBC Providence Health Care Research Ethics Board (Rhode Island), the University Health Network Research Ethics Board (Toronto General Hospital), Comité d’éthique de la recherche du CIUSSS de l’Estrie (Sherbrooke), the Institutional Review Board (Connecticut), The DHMC Clinical Ethics Committee (Dartmouth-Hitchcock Medical Center), the Helsinki Committee (Shaare Zedek), the Helsinki Committee of Rambam (Rambam), the Tel Aviv Sourasky Medical Center Institutional Review Board (Tel Aviv Sourasky Medical Center), the Helsinki Committee of Soroka (Soroka Medical Center), the Helsinki Committee of Baruch Padeh Medical Center, Poria (Poria Medical Center), Kaplan Medical Center Helsinki Committee, the Kaplan Medical Center, Rehovot (Kaplan), the Helsinki Committee of Tel Hashomer (Tel Hashomer Hospital), the Helsinki Committee of Emek (Emek), the Assaf Harofeh Medical Center Helsinki Committee, Assaf Harofeh Medical Center, Zerifin (Asaf Harofe), the Institutional Review Board (Boston), the Ethics Center at Cincinnati Children’s (Cincinnati), the UNC Office of Human Research Ethics (North Carolina), the UMOR Ethics & Compliance (Michigan), the Research Ethics Board (Winnipeg - John Buhler Research Centre), the Helsinki Committee of Barzilai (Barzilai Medical Center), the Helsinki Committee (Beilinson\Schneider), the Helsinki Committee of Maccabi (Maccabi Hashalom), the Helsinki Committee of Wolfson (Wolfson), the BSD/UCMC Institutional Review Boards at the University of Chicago (Chicago), the Miami Research Ethics and Integrity Program (Miami), the Lady’s Children’s Hospital Crumlin Research Ethics Committee (Ireland), the Hospital Research Ethics Committee (Ireland), the West of Scotland Research Ethics Committee 4 (Royal Hospital for Sick Children - Edinburgh), the Convener of the University Research Ethics Committee (Ninewells - Dundee), the West of Scotland Research Ethics Committee 4 (Western General Hospital - Edinburgh), the West of Scotland Research Ethics Committee 4 (Yorkhill - Glasgow), the West of Scotland Research Ethics Committee 4 (The Glasgow Royal Infirmary), the NRES Research Ethics Committee (Aberdeen Royal Infirmary the West of Scotland Research Ethics Committee 4 (Raigmore Hospital), the NHS Research Ethics Committee (The Hull University Teaching Hospitals), the Interior Health Research Ethics Board (The Royal Liverpool University Hospital), the South Birmingham Local Research Ethics Committee (Queen Elizabeth Hospital Birmingham), the HRA National Research Ethics Service (Birmingham Children’s Hospital), the University Hospitals of Coventry and Warwickshire Clinical Ethics Committee (University Hospital Coventry), the North West - Greater Manchester Central Research Ethics Committee (Queen’s Medical Centre), the Royal Devon and Exeter NHS Foundation Trust (The Royal Devon and Exeter Hospital), the Southampton & South West Hampshire local research ethics committee (Southampton General Hospital), the Central University Research Ethics Committee (The John Radcliffe Hospital), the National Research Ethics Service (Addenbrooke’s Hospital), the NHS Research Ethics Committee (Norfolk and Norwich University Hospitals), the Clinical Ethics Committee at Barts and the London NHS Trust (St Bartholomew’s Hospital), the West of Scotland Research Ethics Committee 4 (University Hospital Crosshouse), the Great Ormond Street Clinical Ethics Service (Great Ormond Street Hospital), t NHS Lothian (Alder Hey Children’s Hospital), the West of Scotland Research Ethics Committee 4 (Forth Valley Royal Hospital), the Human Research Protection Office (CHIP Pittsburgh), North West - Greater Manchester East Research Ethics Committee (Salford royal hospital), the Memorial Health Services Research Council (MemorialCare’s Long Beach Medical Center), the Bnai-Zion Medical Center Helsinki Committee (Bnai Zion Medecal Center), the Institutional Review Board (Children’s Hospital Los Angeles), the West of Scotland Research Ethics Committee 4 (Gartnavel General Hospital), the Southern District Health Board and the Dunedin School of Medicine (Dunedin Hospital), the Central University Research Ethics Committee (John Radcliffe Hospital), the Code of Ethics and Corporate Compliance Program of Atlantic Health System (New Jersey), the University Ethics of Research Committee (University Hospitals Bristol), the UC Irvine Interdisciplinary Center for the Scientific Study of Ethics and Morality (University of California Irvine), the NHS Research Ethics Committees (Royal Free Hospital), the Wirral University Teaching Hospital NHS (Arrowe Park Hospital), the Tallaght University Hospital / St. James’s Hospital Joint Research Ethics Committee (St James’s Hospital), the Auckland Health Research Ethics Committee (Auckland District Health Board), the Waikato DHB (Waikato District Health Board), the Helsinki Committee (Emek Medical Center), the Institutional Review Board (Children’s Hospital of Philadelphia), the Helsinki Committee (Karmel Medical Centre), the NHS Research Ethics Committees (Barnet and Chase Farm NHS Hospitals), the NHS Research Ethics Committees (New Victoria Hospital), the QIMR Berghofer Medical Research Institute-Human Research Ethics Committee (QIMR Berghofer Medical Research Institute), the Helsinki Committee (French Hospital Nazereth), the Regional Ethics Review Board in Stockholm (Örebro University Hospital), the Regional Ethics Review Board in Stockholm (Karolinska University Hospital), and Regional Ethics Review Board in Stockholm (Linköping University Hospital).
Consent for publication
Not applicable.
Competing interests
All authors disclose no potential conflicts (financial, professional, or personal) that are relevant to the manuscript.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kenneth Croitoru, Email: ken.croitoru@sinaihealth.ca.
CCC IBD GEM Project research team:
Maria Abreu, Paul Beck, Charles Bernstein, Kenneth Croitoru, Levinus Dieleman, Brian Feagan, Anne Griffiths, David Guttman, Kevan Jacobson, Gilaad Kaplan, Denis O. Krause, Karen Madsen, John Marshall, Paul Moayyedi, Ernest Seidman, Mark Silverberg, Andy Stadnyk, A. Hillary Steinhart, Michael Surette, Dan Turner, Thomas Walters, Bruce Vallance, Guy Aumais, Alain Bitton, Maria Cino, Jeff Critch, Lee Denson, Colette Deslandres, Wael El-Matary, Hans Herfarth, Peter Higgins, Hien Huynh, Jeff Hyams, David Mack, Jerry McGrath, Anthony Otley, Remo Panancionne, Robert Baldassano, Anne M. Griffiths, Charlotte Hedin, Seamus Hussey, Hien Hyams, David Keljo, David Kevans, Charlie Lees, Sanjay Murthy, Remo Panaccione, Nimisha Parekh, Sophie Plamondon, Graham Radford-Smith, Mark Ropeleski, Joel Rosh, David Rubin, Michael Schultz, Corey Siegel, and Scott Snapper
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-01115-w.
References
- 1.Rocchi A, Benchimol EI, Bernstein CN, Bitton A, Feagan B, Panaccione R, et al. Inflammatory bowel disease: a Canadian burden of illness review. Can J Gastroenterol = Journal canadien de gastroenterologie. 2012;26(11):811–817. doi: 10.1155/2012/984575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turpin W, Goethel A, Bedrani L, Croitoru Mdcm K. Determinants of IBD heritability: genes, bugs, and more. Inflamm Bowel Dis. 2018;24(6):1133–1148. doi: 10.1093/ibd/izy085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGovern DP, van Heel DA, Ahmad T, Jewell DP. NOD2 (CARD15), the first susceptibility gene for Crohn's disease. Gut. 2001;49(6):752–754. doi: 10.1136/gut.49.6.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411(6837):603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 6.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy NA, Lamb CA, Berry SH, Walker AW, Mansfield J, Parkes M, et al. The impact of NOD2 variants on fecal microbiota in Crohn's disease and controls without gastrointestinal disease. Inflamm Bowel Dis. 2018;24(3):583–592. doi: 10.1093/ibd/izx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. 2016;48(11):1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Thingholm LB, Skieceviciene J, Rausch P, Kummen M, Hov JR, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48(11):1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, et al. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48(11):1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 11.Joossens M, Van Steen K, Branche J, Sendid B, Rutgeerts P, Vasseur F, et al. Familial aggregation and antimicrobial response dose-dependently affect the risk for Crohn's disease. Inflamm Bowel Dis. 2010;16(1):58–67. doi: 10.1002/ibd.20985. [DOI] [PubMed] [Google Scholar]
- 12.Van Kruiningen HJ, Joossens M, Vermeire S, Joossens S, Debeugny S, Gower-Rousseau C, et al. Environmental factors in familial Crohn's disease in Belgium. Inflamm Bowel Dis. 2005;11(4):360–365. doi: 10.1097/01.MIB.0000158536.31557.90. [DOI] [PubMed] [Google Scholar]
- 13.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda, Md) 2011;1(6):457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L, Paterson AD, Turpin W, Xu W. Assessment and selection of competing models for zero-inflated microbiome data. PLoS One. 2015;10(7):e0129606. doi: 10.1371/journal.pone.0129606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 18.Al Nabhani Z, Dietrich G, Hugot JP, Barreau F. Nod2: the intestinal gate keeper. PLoS Pathog. 2017;13(3):e1006177. doi: 10.1371/journal.ppat.1006177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV. et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211(1):19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gevers D, Kugathasan S, Denson L, Vazquez-Baeza Y, Van Treuren W, Ren B. et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–92. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey N, Soergel DA, Repo S, Brenner SE. Association of gut microbiota with post-operative clinical course in Crohn's disease. BMC Gastroenterol. 2013;13:131. doi: 10.1186/1471-230X-13-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kevans D, Silverberg MS, Borowski K, Griffiths A, Xu W, Onay V, et al. IBD genetic risk profile in healthy first-degree relatives of Crohn's disease patients. J Crohns Colitis. 2016;10(2):209–215. doi: 10.1093/ecco-jcc/jjv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaakoush NO. Insights into the role of Erysipelotrichaceae in the human host. Front Cell Infect Microbiol. 2015;5:84. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, et al. A microbial signature for Crohn's disease. Gut. 2017;66(5):813–822. doi: 10.1136/gutjnl-2016-313235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleynen I, Boucher G, Jostins L, Schumm LP, Zeissig S, Ahmad T, et al. Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet (London, England) 2016;387(10014):156–167. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin a coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158(5):1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox LM, Sohn J, Tyrrell KL, Citron DM, Lawson PA, Patel NB, et al. Description of two novel members of the family Erysipelotrichaceae: Ileibacterium valens gen. Nov., sp. nov. and Dubosiella newyorkensis, gen. Nov., sp. nov., from the murine intestine, and emendation to the description of Faecalibaculum rodentium. Int J Syst Evol Microbiol. 2017;67(5):1247–1254. doi: 10.1099/ijsem.0.001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carta G, Murru E, Banni S, Manca C. Palmitic acid: physiological role, metabolism and nutritional implications. Front Physiol. 2017;8:902. doi: 10.3389/fphys.2017.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wisniewski PJ, Dowden RA, Campbell SC. Role of dietary lipids in modulating inflammation through the gut microbiota. Nutrients. 2019;11(1):117. doi: 10.3390/nu11010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao H, Chen L, Hao L, Zhang X, Chen Y, Ruan Z, et al. Stearic acid induces proinflammatory cytokine production partly through activation of lactate-HIF1alpha pathway in chondrocytes. Sci Rep. 2015;5:13092. doi: 10.1038/srep13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knights D, Silverberg M, Weersma R, Gevers D, Dijkstra G, Huang H, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6(12):107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Butcher J, Mack D, Stintzi A. Functional impacts of the intestinal microbiome in the pathogenesis of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21(1):139–153. doi: 10.1097/MIB.0000000000000215. [DOI] [PubMed] [Google Scholar]
- 33.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19(5):731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60(10):1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 35.Lesage S, Zouali H, Cezard JP, Colombel JF, Belaiche J, Almer S, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70(4):845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol. 2004;99(12):2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 37.Li E, Hamm CM, Gulati AS, Sartor RB, Chen H, Wu X, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One. 2012;7(6):e26284. doi: 10.1371/journal.pone.0026284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Supplementary Table 1. Association of rs2066845 with bacterial taxa in the GEM cohort. Supplementary Table 2. Association of rs2066844 with bacterial taxa in the GEM cohort. Supplementary Table 3. Association of rs2066847 with bacterial taxa in the GEM cohort. Supplementary Table 4. Cumulative NOD2 variants association with bacterial taxa in the GEM cohort.
Data Availability Statement
16S sequences are available by the study accession number PRJEB14839 (NCBI BioProject database); ENA-SUBMISSION: ERA675979 [9]. Requests for raw and analyzed data should follow the instruction given at http://www.gemproject.ca/data-access/. All submissions will be reviewed by the GEM Project Operating Committee to ensure that the requested samples/data will not interfere in any way with the intended GEM Project analysis of the nested cohort as per the original GEM Project Study Design and is not a duplication of analysis already ongoing. Those proposals meeting this evaluation will be distributed to all members of the GEM Project Steering Committee (GPSC) for review and open discussion. This review will focus on the global scientific merit of the proposal. This review will assess the basic scientific merit and the availability of requested samples and data, ensuring there is no compromise of the original intent of the GEM project. It would be of value to contact a member of the Steering Committee who could help sponsor your application. Those projects achieving majority vote of approval at the GPSC will be informed that the GEM Project will provide a letter of support stating that the requested samples or data will be made available to the applicants once the applicant receives funding from a granting agency that applies an independent peer review process to the proposal. The criteria to be used for review of all submissions will include the “scientific relevance” of the proposal and the judged availability of biological material requested. The budget to be requested from a funding agency, must allow for any expenses in processing samples or in setting up the appropriate queries of the database. The intent is to allow sufficient time for applicants to consider submission for funding opportunities.