Abstract
Nanomedicine involves the use of engineered nanoscale materials in an extensive range of diagnostic and therapeutic applications and can be applied to the treatment of many diseases. Despite the rapid progress and tremendous potential of nanomedicine in the past decades, the clinical translational process is still quite slow, owing to the difficulty in understanding, evaluating, and predicting nanomaterial behaviors within the complex environment of human beings. Microfluidics-based organ-on-a-chip (Organ Chip) techniques offer a promising way to resolve these challenges. Sophisticatedly designed Organ Chip enable in vitro simulation of the in vivo microenvironments, thus providing robust platforms for evaluating nanomedicine. Herein, we review recent developments and achievements in Organ Chip models for nanomedicine evaluations, categorized into seven broad sections based on the target organ systems: respiratory, digestive, lymphatic, excretory, nervous, and vascular, as well as coverage on applications relating to cancer. We conclude by providing our perspectives on the challenges and potential future directions for applications of Organ Chip in nanomedicine.
Keywords: Microfluidics, Nanoparticles, Nano-bio interaction, Body-on-a-chip, 3D cell culture model
Graphical abstract

Highlights
-
•
Microfluidics-based organ-on-a-chip (Organ Chip) techniques offer a promising way to understand, evaluate, and predict nanomedicine behaviors within the complex environment.
-
•
Organ Chip models for nanomedicine evaluations are categorized into seven broad sections based on the targeted body systems.
-
•
Limitations, challenges, and perspectives of Organ Chip for accelerating the assessment of nanomedicine are discussed, respectively.
1. Introduction
1.1. Conventional in vitro models for the evaluation of nanomedicine and their limitations
In the past two decades, there has been a surge in what is termed ‘nanomedicine’ research, which is now culminating in considerable commercialization efforts globally [1]. Nanomedicine involves the use of nanoscale or nanostructured materials in medical applications, such as biosensing/bioimaging, disease diagnosis, and therapeutic delivery [2]. Nanomaterials have unique medical effects according to their structures, and the continued development of nanomedicine has the potential to provide numerous benefits, including improved bioavailability, targeting ability, delivery efficacy, dose-response, and personalization compared to conventional medicines [3]. However, despite these potential benefits, essential data regarding the circulation, interaction, and toxicity of many nanomaterials are currently lacking, and there are evident challenges in terms of the safety assessment of nanomedicine in the human body [4,5]. To evaluate potential nanomedicine, animal models have been widely exploited [6]. Although animal models are useful for tracking the translocation of nanomedicine in vivo, the discrepancy in physiological responses between animals and humans may result in serious misunderstanding of their efficacy and toxicity [7]. In 2004, the US Food and Drug Administration (FDA) estimated that 92% of drugs that pass animal testing fail to proceed to the market due to efficacy and safety problems that were not predicted by animal tests [8]. More recent analyses suggest that, despite efforts to improve the predictability of animal testing, the failure rate has actually increased and is now closer to 96% [8]. Additionally, the increasing awareness of animal welfare has expedited efforts in generating in vitro human models that are more predictive of human responses, which may eventually replace animal models from both an accuracy as well as an ethical perspective [9].
In current in vitro studies, nanomedicine is most commonly tested in two-dimensional (2D) monolayer cell culture models [10]. Conventional 2D cell culture relies on adherence to a flat surface (e.g., glass or polystyrene) to provide mechanical support for single or multiple cell types that are either freshly isolated from tissues (primary cells) or are already immortalized (cell lines). Since the beginning of last century, 2D cell cultures have been well-established and became widespread and accepted by most researchers and scientists, due to their generally cost-effective nature and convenience for cell observation and measurements [11]. Although 2D cell cultures are still used for most research, the flat surface is not an accurate representation of how cells grow and keep their functions or how they are affected by diseases and injuries in nature, which in turn significantly limits their ability to recapitulate the appropriate levels of in vivo cellular responses [12]. For example, primary human hepatocytes are commonly used for in vitro studies of drug-induced liver injury [13]. However, when cultured as 2D monolayers, hepatocytes lose key phenotypic and hepatic characteristics within 7 days [14,15]. Even when various growth factors and other important constituents are provided in the medium in the 2D hepatocyte cultures, normal growth and differentiation do not occur due to the absence of a physiological matrix-like microenvironment [16,17]. Therefore, tests based on in vitro 2D cell culture models do not accurately predict in vivo toxicity and other biological effects of nanomedicine due to the absence of crucial physiological processes, such as the transport of nanomedicine through cell layers when they are brought in contact with the tissues [18].
Therefore, tremendous efforts in studying the efficiency and efficacy of nanomedicine have shifted from using static 2D models to three-dimensional (3D) cell culture models, which aim to mimic in vivo cell-cell or cell-ECM interactions, and thus provide better biomimetic platforms that are more physiologically relevant and predictive than 2D cultures [10,19]. Results from these 3D studies demonstrate that increasing the dimensionality of ECM around cells from 2D to 3D can significantly impact cell functions, including survival, proliferation, differentiation, and responses [20,21]. For example, Hsiao et al. compared the anti-cancer effect of different drugs on prostate cancer cells and showed that 3D-cultured cells were more resistant to 5-fluorouracil, which inhibits proliferation, whereas 2D cultures were more resistant to a different drug, tirapazamine, which is toxic in hypoxic conditions [22]. Various static 3D cell culture techniques have been introduced to establish in vivo-like conditions for the testing of nanomedicine, such as cell encapsulation in hydrogels [23], organoid/spheroid cultures [[24], [25], [26]], natural or customized scaffolds [[27], [28], [29]], and multilayered cell sheets [30]. However, despite providing valuable and important information, the static 3D method could result in rapid nanoparticle sedimentation due to the low rate of diffusion in static culture models, and then significantly alter transport kinetics and nanoparticle-cell interactions [31,32]. Although recent advances have been achieved in organoids and microtissues that are structurally sophisticated and functionally relevant compared with native tissues and organs, the dynamic transport of nanomedicine under these static conditions is still neglected [[33], [34], [35]]. Fortunately, the emerged microfluidics is an advanced technology that has been broadly applied to cell biology to develop devices and techniques for culturing, maintaining, analyzing, and experimenting with cells in dynamic microscale systems, which will be described in the following section [36].
1.2. Microfluidic organ-on-a-chip (Organ Chip) as a potential solution
Fluid flow, such as blood or interstitial flow, is crucially important for the functions of all tissues, with cells responding to flow through mechanisms such as differentiation and metabolic adaptation [37,38]. The microfluidic technology, developed in the 1990s, offers a unique opportunity for dynamic 3D cell culture, creating a platform for engineering highly complex microenvironments that are controllable, reproducible, and optimizable [[39], [172]]. In comparison to the static culture models, microfluidics permits continuous nutrient exchange, better oxygen perfusion, and physiological shear stress, which provide a better emulation of conditions in living organisms, including (1) readily mimicking the complex dynamic microscale 3D environments found in vivo; (2) ability to control features such as gas exchange, nutritional composition, and metabolite and waste removal; and (3) integration of multiple steps, such as cell culture, sampling, capture, lysis, imaging, and detection within a single device [40,41]. Hence, the combination of microfluidic technology with 3D cell culture, such as the emerging Organ Chip systems, offers great potential for in vivo-like tissue-based applications.
An Organ Chip is a microfluidic cell culture device created with microchip-manufacturing methods, which contains continuously perfused chamber(s) inhabited by living cells arranged to simulate tissue-level or organ-level physiology [42]. By recapitulating the multicellular architectures, tissue-tissue interfaces, physiological microenvironments, and vascular perfusion of the body, these devices produce levels of tissue and organ functionality not possible with conventional 2D or 3D culture systems [43]. Particularly, Organ Chip provides a novel platform for better predictive testing of nanotherapeutics by addressing issues that limit the pace of clinical translation of nanomedicine [[44], [45], [46]]. One of the most distinct advantages of using Organ Chip for nanomedicine evaluations is precise assessment of transport and translocation of nanomedicine across tissue-tissue interfaces under in vivo-relevant shear flow, which makes a considerable difference compared to the conventional methods. Other favorable features, such as the ability to explore the biological mechanisms of nanomedicine targeting effects in real time and revealing the adverse effects of nanomedicine via monitoring the slight changes in various parts of organs/tissues, provide Organ Chip with the potential to meet the demand of creating a robust preclinical screening in vitro model for the evaluations of nanomedicine. Due to these outstanding advantages, multiple companies have pursued the translation and commercialization of Organ Chip technology since the early 2010s, such as Emulate Inc. and Tissuse, as summarized in Table 1. Jodat et al. further cataloged the main research topics that have been reported by these companies, including Organ Chip research and development, molecular biology, disease modeling, and drug development [47]. This information suggests that the translation of Organ Chip for medicine evaluation is being investigated and further developed.
Table 1.
A partial list of companies involved in Organ Chip technologies and their selected publication from 2015 to 2020.
| Company | Product Image | Selected Products | References |
|---|---|---|---|
 |
 |
3D Cell Culture- on-a-Chip | [48] |
 |
 |
Standard/Triple Chamber Neuron Device | [49] |
 |
 |
MyrPlate MyrScreen |
[50] |
 |
 |
HUMIMIC Chip2 HUMIMIC Chip4 |
[51,52] |
 |
 |
Liver Bio-Kit Intestine Bio-Kit Kidney Bio-Kit |
[53,54] |
 |
 |
ParVivo™ Microfluidic-on-a-Chip | [55] |
 |
 |
OrganoPlate® 3-lane OrganoPlate® Caco-2 |
[56,57] |
 |
 |
3D InSight™ Liver 3D InSight™ Islet 3D InSight™ Tumor |
[[58], [59], [60]] |
 |
 |
SynTumor SynBBB SynTox |
[61] |
 |
 |
Multi-organ-on-a-Chip (2–4 organs) | [62,63] |
 |
 |
Nerve-on-a-Chip | [64] |
 |
 |
AXLung-on-a-Chip | [65,66] |
2. Modeling physiological functions of tissues/organs by Organ Chip platforms
Organ Chip models focus on the reconstitution of the 3D microstructures and tissue-tissue interfaces of human organs, by which they can also recapitulate primary physiological functions of the entire organs, including homeostasis and pathophysiological responses [[43], [67], [173]]. Thus, in recent years, Organ Chip has been expanded rapidly to encompass several organs as well as diseases, including the lung [68,69], liver [70,71], blood vessel [72,73], gut [74,75], heart [76,77], uterus [78,79], brain [[80], [81], [82]], bone marrow [83,84], and tumor/cancer [85,86], as illustrated in Scheme 1. The construction of these Organ Chip systems is guided by design principles based on a reductionist analysis of their target organs [47]. In general, the first step is to understand the anatomy of the target organ and reduce it to the basic elements essential for the physiological functions. These functional units are then examined and recapitulated to identify key features such as cell patterning, structural and architectural organizations, and organ-specific biophysical and biochemical microenvironments. To reduce or replace animal experiments and achieve more accurate and reliable preclinical data, the application of these Organ Chip models are numerous, such as developing personalized medicine, understanding disease etiology, and performing drug screening [87]. The final goal of the development of Organ Chip is personalized medicine, which helps researchers or doctors plan a more efficient treatment of the specimens and define appropriate medicines with the optimal doses for individual patients. Besides, Organ Chip can provide a more in-depth insight into disease etiology, which examines the reasons behind a disease by applying a situational microenvironment [47]. Moreover, the drugs or cosmetics being developed can be tested directly on Organ Chip models, which diminishes the development phase, reduces costs, and avoids adverse effects on animals and humans during the trials [88]. In particular, new microphysiological models of human organs have emulated physiological functions of the breathing motions in the lung, metabolism and excretion in the liver, blood cleansing functions in the spleen, reabsorption and transport in the kidney, blood-brain-barrier (BBB) in the brain, as well as microvascular networks and blood perfusion that can be used to potentially validate the efficacy, targeted delivery, pharmacokinetics and pharmacodynamics, functionality, and cytotoxicity of nanomedicine [89]. Hence, in the next section, we will discuss recent advancements in the development of the Organ Chip models, focusing on their applications for evaluating nanomedicine in a human-relevant manner.
Scheme 1.
Timeline of the development of microfluidics-based Organ Chip technology. Evolution of the field from the early concept of lung-on-a-chip pioneered by 2010 to the more complex model of multi-organ-on-a-chip reported by 2017.
3. Organ Chip platforms for the evaluation of nanomedicine
While the potential of converging the nanomedicine with Organ Chip microsystems is enormous, studying of nanoparticulates in the existing Organ Chip are nascent. Table 2 summarizes Organ Chip platforms that recapitulate different organs/tissues based on physiological systems, and each individual system and their applications in nanomedicine evaluation will be introduced in the following contents, respectively.
Table 2.
Organ Chip systems applied for the evaluations of nanomedicine.
| Physiological System | Recapitulated Organ/Tissue | Nanomedicine (Nanoparticulate) | Evaluation | References |
|---|---|---|---|---|
| Respiration System | Lung-on-a-chip | SiO2, ZnO, TiO2 | Studying of translocation and inflammatory effect of nanoparticles. | [68,90] |
| Digestive System | Liver-on-a-chip (integrated with intestine-on-a-chip) | Carboxylated polystyrene (PS) | Simulation of livery injury from ingested nanoparticles caused by gastrointestinal tract and liver tissue crosstalk. | [91] |
| Lymphatic System | Spleen- on-a-chip (a biospleen device) | Magnetic nanobeads | Cleaning pathogens from the blood of sepsis patients by using opsonin-coated nanobeads. | [92] |
| Excretory System | Kidney-on-a-chip | PS | Monitoring of in situ kidney injury via incorporated nanoparticles that introduced as an imaging adjuvant. | [93] |
| Nervous System | Blood–brain barrier (BBB)-on-a-chip | Angiopep-2 liposomes, gH625-functionalized PS | Evaluation of the efficiency of nanoparticles penetration by BBB. | [[94], [95], [96]] |
| Vascular System | Vascular-on-a-chip & Vascular-Tumor-on-a-chip | Poly (lactic-co-glycolic acid) (PLGA), tissue plasminogen activator (tPA)-coated PLGA, antibody-coated PS, functionalized liposomes. | Validation of extravasation, targeting specificity, and accumulating of nanoparticles. | [[97], [98], [99], [100], [101]] |
| Tumor/Cancer | Tumor/Cancer-on-a-chip | Gold nanoparticles (Au), PEGylated Au, cadmium telluride (CdTe)/Au, functionalized liposomes. | Evaluation of the efficiency of targeting, transport, and accumulation of nanoparticles. | [[102], [103], [104], [105]] |
3.1. Respiratory system
Limitation of conventional cell culture models for respiratory diseases. The respiratory system provides the main access point for nanoparticles to enter the human body [106]. In addition, the respiratory system is of great interest because various natural and engineered nanoparticles are known to be responsible for lung diseases, which represents three of the top five leading causes of death globally [107]. Currently, most approaches in studying the pulmonary system are highly dependent on animal models and in vitro 2D models [108]. However, the results from in vitro assays do not correspond well to in vivo results because these simplified 2D cell culture models lack the complex cellular architecture (e.g., alveolar-capillary barrier) and the dynamic microenvironment (e.g., gas exchange) that are found in the living lung, which are required for organ-level functionality and gene expressions [109]. Therefore, there is a necessity to develop in vitro models that facilitate the development of new nanomedicine for respiratory diseases [110].
The emergence of lung-on-a-chip (Lung Chip) and its application for nanoparticulates. The seminal paper that first used the term of Lung Chip was introduced by Huh et al. in which a biomimetic Lung Chip microsystem reconstituted the alveolar-capillary barrier, the smallest functional unit in the lung (Fig. 1A and B) [68]. To reproduce this air-liquid interface, a two-channel microfluidic device based on polydimethylsiloxane (PDMS) was used, with a collagen-coated porous membrane separating alveolar epithelial cells in contact with air in the top channel from microvascular endothelial cells in contact with perfused cell culture medium in the bottom channel. Another feature of this Lung Chip that is not available in traditional cell culture systems was the inclusion of a vacuum channel alongside the fluidic channels to generate cyclic strains of the alveolar-capillary interface to mimic the physiological breathing movements. This bioinspired microdevice recapitulated multiple complex and physiological organ-level responses, including pulmonary inflammation in response to bacteria and inflammatory cytokines introduced into the alveolar space. In addition, they also mimicked airborne exposure to toxic nanomaterials by exposing the epithelial cells of the Lung Chip to silica nanoparticles (diameter = 12 nm) for 5 h under the cyclic strains (Fig. 1C). The results showed that artificial respiration induced higher transport of nanoparticles from the epithelial to the endothelial channel and consequently higher uptake by the endothelial cells (Fig. 1D). This further led to significantly increased intercellular adhesion molecule-1 (ICAM-1) expression (Fig. 1E) and augmented production of reactive oxygen species (ROS) (Fig. 1F), suggesting that respiration enhances the pro-inflammatory behavior of nanoparticles and exacerbates the development of acute lung inflammation. The translocation behavior of the nanoparticles was then evaluated in a whole-mouse lung ventilation-perfusion model. The mouse studies confirmed that nanoparticles were transported across the alveolar-capillary barrier, with higher transport rates observed with cyclic breathing versus without, demonstrating the significance of using the in vitro Lung Chip model to predict physiological response in vivo. These results indicated that previous evaluations of nanoparticle toxicity using traditional cell culture systems likely underestimated the effect of the nanoparticles, especially in the context of lung damage after nanoparticle inhalation. This is not a general rule however, as the nanomaterial under consideration influences the differences in response observed.
Fig. 1.
An example of Lung Chip application in nanomedicine: the study of nanoparticle translocation and inflammatory effects. (A) Schematic of the Lung Chip integrated with mechanical stretching and an air-liquid interface. (B) Confocal image of the tissue-tissue interface consisting of a single layer of the alveolar epithelium (green) closely opposed to a monolayer of the microvascular endothelium (red), which express intercellular junctional structures that are visualized with antibodies to occludin or VE-cadherin. (C) Illustration of nanoparticle translocation across the alveolar-capillary interface of the lung. (D) Application of mechanical strain increased the rate of nanoparticle translocation across the alveolar-capillary interface compared with static controls or a transwell culture system. (E) Physiological mechanical strain and silica nanoparticles synergistically upregulate ICAM-1 expression. (F) Alveolar epithelial cells increased ROS production when exposed to silica nanoparticles in conjunction with cyclic strain, whereas nanoparticles or strain alone did not affect intracellular ROS levels. Reproduced from Ref. [68]. Copyright 2010 ©American Association for the Advancement of Science.
In a recent study, Zhang et al. compared the effects of 25-nm TiO2 and 40-nm ZnO nanoparticles on a Lung Chip containing co-cultured human lung primary epithelial cells with vascular endothelial cells [90]. While no significant cytotoxicity was observed with TiO2 nanoparticles up to 200 μg mL−1, at the same concentration, ZnO nanoparticles caused approximately 50% apoptosis in the epithelial cells compared to about 5% in endothelial cells. Since the nanoparticles were initially in contact with the epithelial cells, it is reasonable that the epithelial cells experienced more severe damage, including ROS generation and apoptotic cell death, than the endothelial cells. Furthermore, it was shown that the co-culture actually inhibited nanoparticle-induced apoptosis, as around 90% of monocultured epithelial cells underwent apoptosis when treated with the ZnO nanoparticles.
Outlook of using Lung Chip in the study of nanomedicine. In these pioneering Lung Chip models, nanoparticle-induced injuries on epithelial and endothelial cells were evaluated simultaneously under physiologically relevant microenvironments and stimuli within the context of the layered structure. It has been demonstrated that features such as mimicking cyclic breathing motions and cell-cell interactions are influential for determining nanoparticle-cell interactions. There is a great opportunity here beyond just toxicity testing to evaluate nanodiagnostics and nanotherapeutics within the Lung Chip. Future endeavors could also focus on integrating more cell types found in the alveoli, such as fibroblasts and alveolar macrophages, as well as including oxygen gradients to represent the microenvironment of the human lung to an even greater extent.
3.2. Digestive system
The digestive system, including the gastrointestinal (GI) tract, represents a likely route of entry for many nanomedicine, either directly through intentional ingestion or indirectly by secondary ingestion of inhaled particles [111]. The hollow organs that make up the GI tract are the mouth, esophagus, stomach, small intestine, large intestine, and anus, while the solid organs constituting the rest of the digestive system are the liver, pancreas, and gallbladder. Although several of these digestive organs have been recapitulated with Organ Chip, including stomach-on-a-chip (Stomach Chip) [112], small-intestine-on-a-chip (Intestine Chip) [113] or gut-on-a-chip (Gut Chip) [74,114], and pancreas-on-a-chip (Pancreas Chip) [115], only the liver-on-a-chip (Liver Chip) integrated with Intestine Chip has been specifically used for the study of nanomedicine so far [91].
Limitation of conventional cell culture models for liver diseases. The liver is the largest internal organ and gland in the human body and maintains a broad spectrum of vital functions, including drug metabolism and excretion [116]. Hepatocytes are the predominant cell type in the liver, which are epithelial cells that perform essential roles in metabolic, endocrine, and secretory functions of the body [117,118]. While the liver is important for performing these vital functions, it is also highly vulnerable to drug-induced damages [119]. Consequently, drug hepatotoxicity is one of the main concerns in identifying new medicines, making the ability to model hepatotoxicity highly crucial for the development of novel drug delivery systems [120]. Many studies have used hepatocytes as an in vitro model to investigate various liver functions [120,121]; however, isolated hepatocytes generally dedifferentiate when cultured in traditional 2D monolayers, resulting in the loss of phenotype and function [122]. This loss of morphology and function often causes failure to detect metabolism-mediated hepatotoxicity of medicine in vitro [123].
The emergence of Liver Chip and its application for nanoparticulates. The first Liver Chip model can be traced back to the last decade, with a microfluidic 3D Liver Chip designed primarily for drug toxicity screening [124,125]. For example, while not a pure “Organ Chip” model in the sense that the model only contained hepatocytes and no cell-cell interactions, Toh et al. demonstrated that a 3D microfluidic primary hepatocyte model was able to successfully maintain functional hepatocytes over a period of 72 h and also yield half-maximal inhibitory concentration (IC50) values that correlated well with in vivo median lethal concentration (LC50) toxicity data for different hepatotoxic drugs [126]. More recently, Du et al. developed a co-culture Liver Chip system integrating four major types of liver cells, including sinusoidal endothelial cells, Kupffer cells, hepatic stellate cells, and hepatocytes, into two adjacent channels separated by a permeable membrane, resulting in a microsystem recapitulating the key structures, configurations, and functions of the liver sinusoid [127]. As further consideration for nanoparticle toxicity evaluations in the digestive system, Esch et al. applied the Liver Chip in combination with an Intestine Chip (Fig. 2B) [91]. The liver compartment was represented using HepG2/C3A (hepatocellular carcinoma) cells cultured in a silicon chip between plexiglass layers (Fig. 2A), while the intestine was recapitulated using a co-culture of Caco-2 (colon carcinoma) and HT29-MTX (mucus-secreting colon epithelia) cells (Fig. 2C). The results showed that when only the liver compartment was exposed to 50 nm of carboxylated polystyrene nanoparticles, the HepG2/C3A cells released aspartate aminotransferase (AST), an indicator of sublethal cellular injury to the liver tissue (Fig. 2D). Interestingly, this phenomenon was exacerbated when the intestine compartment was linked upstream to the liver compartment via microfluidic channels to simulate first-pass metabolism, even though they also demonstrated that the Intestine Chip reduced nanoparticle exposure in the Liver Chip.
Fig. 2.
An example of Liver Chip and Intestine Chip application in nanomedicine: simulation of enhanced livery injury from ingested nanoparticles caused by GI tract and liver tissue cross-talk. Schematic of the (A) silicon chip with liver chamber and (C) GI tract module of the Body Chip system, and (B) the corresponding physiologically based pharmacokinetic modeling of the entire system. Cell culture medium was recirculated through both an apical circuit (green arrows) and a basolateral (systemic) circuit (black arrows). (D) Carboxylated polystyrene nanoparticles were added to the apical circuit at varying concentrations, and mean concentrations of AST released into systemic circulation were measured. Reproduced from Ref. [91]. Copyright 2014©Royal Society of Chemistry.
Outlook of using Liver Chip and liver-integrated multi-organ-on-a-chip (Multi-Organ Chip) in the study of nanomedicine. The above demonstrated that multi-organ in vitro devices are essential platforms for assessing nanomedicine toxicity, allowing the identification of feedback mechanisms where nanomedicine metabolism in the liver can affect the toxicity and/or efficacy of nanomedicine in other organ systems and vice versa. Therefore, Multi-Organ Chip and body-on-a-chip (Body Chip) platforms combining different organs are needed to investigate the interactions between multiple tissues following nanomedicine exposure. Although some investigations have shown proof-of-concept applications using Multi-Organ Chip systems consisting of immortalized cell lines [128,129], future endeavors must transition to primary cells, which can provide a more realistic physiological microenvironment for understanding nanomedicine delivery and effects.
3.3. Lymphatic system
The lymphatic system consists of vessels, lymph nodes, and organs like the spleen, tonsils, and thymus gland. Delivery of medicine via the lymphatic system has several advantages, such as circumventing the first-pass metabolism in the liver and targeting drugs to diseases that spread through the lymphatic system [130]. Although lymph-node-on-a-chip is an established model and small molecule delivery to lymph nodes for improved immunotherapeutic treatments has attracted great attention [130,131], it has not yet been used as a platform for the study of nanomedicine. Alternatively, microfluidic devices recapitulating spleen functions, spleen-on-a-chip (Spleen Chip), have been applied in the field of nanomedicine.
The emergence of Spleen Chip and its application for nanoparticulates. Some initial work in designing a Spleen Chip was performed by Rigat-Brugarolas et al. [132]. In their design, they created a microengineered device to mimic the hydrodynamic forces and the physical properties of the splenon, the minimal functional unit of the red pulp, whose primary function is blood filtration. Although no spleen cells were cultured within the device, blood filtration was achieved purely based on fluid dynamics. They were able to distinguish human red blood cells (RBCs) and malaria-infected cells by demonstrating that infected RBCs were significantly more deformable than non-infected RBCs in their device, therefore providing the capacity for diagnostics directly through reproducing physiological conditions and observing the mechanical properties of cells within their chip.
The spleen is sometimes not enough to be able to completely remove pathogens on its own, especially when people are critically ill or have received traumatic injuries, which could result in potentially fatal sepsis [133]. To avert such an outcome in those situations, a microfluidic artificial spleen device has been invented to cleanse pathogens from the blood of sepsis patients, as shown in Fig. 3 [92]. In the device, to remove pathogens from blood, magnetic nanoparticles were employed and functionalized with mannose-binding lectin (MBL), a human blood opsonin that binds a wide variety of pathogens, such as bacteria, viruses, parasites, fungi, and toxins. Fig. 3A illustrates the designing of antibody fragments (FcMBL) through genetic engineering that retains the generic opsonin MBL's carbohydrate-recognition domain (CRD) and functionalizing magnetic nanobeads (diameter = 128 nm) with them through biotin-streptavidin interactions to produce magnetic opsonins. These opsonin-coated magnetic beads could capsulate various pathogens, such as bacteria Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) (Fig. 3B). On the other hand, the microfluidic device contains the architectural ingenuity of the spleen by incorporating a high-flow vascular arterial channel perfused with contaminated whole blood, a parallel low- or intermittent-flow venous sinusoid channel, and open slits interconnecting the two, which mimics the arterial red-pulp cord and venous systems that are separated by sinusoid slits (Fig. 3C). After mixing the magnetic nanobeads with contaminated blood and flowing the nanobead/blood mixture through the microchannels embedded in the chip, magnetic separation resulted in the removal of the pathogens through the venous sinusoid channel, leaving cleansed blood in the arterial channel. It was demonstrated that this artificial device could efficiently remove multiple Gram-negative and Gram-positive bacteria, fungi and endotoxins from whole human blood flowing through a single biospleen unit at up to 1.25 L per h in vitro.
Fig. 3.
An example of Spleen Chip application in nanomedicine: a biospleen device for blood cleansing. (A) Scheme for designing of magnetic nanobeads for pathogen capture. (B) Pseudocolored scanning electron micrographs showing multiple magnetic beads bound to the bacteria S. aureus and E. coli. (C) Schematic of a venous sinus in the red pulp of the spleen (left), longitudinal view of the biospleen (right), and a photograph of the engineered device (top right). Reproduced from Reference [92]. Copyright 2014©Nature Publishing Group.
Outlook of using Spleen Chip in the study of nanomedicine. In the future, it would be interesting to examine the role of the biological cell components of the spleen in homeostatic and pathological filtration processes. Additionally, since the inter-endothelial slits in the human spleen are approximately 200 nm, nanoparticles larger than 200 nm are expected to accumulate in the spleen [134,135]. There is, therefore, an opportunity to investigate the behavior of deformable nanoparticles and also to achieve a more comprehensive understanding of the interactions of accumulated nanoparticles with the spleen using dynamic chip models.
3.4. Excretory system
The human kidney is the main excretory organ that is exposed to drugs, and it is comprised of multiple cell types, including glomerular vascular endothelial cells and podocytes, performs many vital functions, including endocrine functions and cellular metabolism, and possesses a variety of critical structural components, including precisely arranged renal tubular segments and transcellular electrochemical and osmotic pressure gradients [136,137]. In particular, the epithelial cells of the proximal renal tubules in the kidney are the most susceptible targets for nanomedicine due to their capacity for drug metabolism [138].
The emergence of Kidney-on-a-chip (Kidney Chip). The Ingber Lab has created a Kidney Chip that mimics the in vivo renal tubular environment using human proximal tubular cells [139]. Under dynamic flow conditions mimicking living kidney tubules, superior primary cilia formation, epithelial cell polarization, albumin transport, glucose reabsorption, and brush border alkaline phosphatase activity were observed compared to cells cultured under static conditions. Additionally, the dynamic conditions revealed that exposure to apical fluid shear stress is important for facilitating cell recovery from cisplatin-induced damage and enhances P-glycoprotein (Pgp) efflux transporter activity, two features observed in vivo that are not reflected in conventional cell culture models. Hence, the human Kidney Chip is a platform that can meet the demand for mimicking human drug clearance and metabolism through precise control of drug concentrations and fluid flow rates [140].
Tracking nephrotoxicity using nanoparticulates upon Kidney Chip. To date, investigations of nanotherapeutic applications using the Kidney Chip have yet to be reported. Nevertheless, there has been one study in which nanoparticles were introduced as an imaging adjuvant for kidney injury [93]. γ-Glutamyl transpeptidase (GGT) is a protein presented on the apical membrane of proximal tubular cells (786-O) that is released upon cytotoxic insult. Therefore, introducing 500-nm fluorescent polystyrene nanoparticles conjugated with anti-GGT antibodies within the apical channel provided tracking capabilities of drug-induced nephrotoxicity due to agglutination of the nanoparticles upon immunocapture of released GGT, corresponding to increased fluorescence measured in the outflow (Fig. 4A). Interestingly, A smartphone-based fluorescence microscope was integrated as a handheld monitoring device attached to the chip (Fig. 4B), which provided a novel groundbreaking tool to enable the internal and external monitoring of the Kidney Chip. As such, this nanoparticle-based strategy overcomes the challenge of assessing cellular responses during chip experiments in a quick, real-time, non-disruptive, and in situ manner.
Fig. 4.
The incorporation of nanoparticles into Kidney Chip enables in situ monitoring of nephrotoxicity. (A) Schematic illustration of nephrotoxicity detection using nanoparticle-based strategy. (B) In situ monitoring Kidney Chip by a smartphone-based fluorescence microscope. Reproduced from Reference [93]. Copyright 2016©Elsevier B·V.
Outlook of using Kidney Chip in the study of nanomedicine. In recent years, highly sophisticated Kidney Chip platforms have been further developed using strategies such as 3D (bio)printing and induced pluripotent stem cells (iPSC)-derived podocytes [136,141]. As novel developments in Kidney Chip technology arise to integrate additional cell types and functions, there is an opportunity for more comprehensive research regarding the clearance of nanoparticles, particularly those smaller than 10 nm in diameter that will experience rapid clearance by the kidney [142], as well as the metabolism of drugs and nanomedicine, such that renal excretion and metabolism are not overlooked [143].
3.5. Nervous system
The development of new delivery strategies to penetrate the BBB has become a crucial goal of nanomedicine in treating the brain parenchyma [144,145]. The BBB is composed of tight junctions formed by brain endothelial cells lining the cerebral microvasculature, and it plays a vital role in maintaining homeostasis and protecting the brain from foreign substances; however, it also limits the accessibility of beneficial compounds [146]. A major issue that hinders the development of nanotherapeutics specifically targeted to the brain is the lack of reliable in vitro BBB models that mimic in vivo conditions and provide accurate predictions of nanomedicine transport and efficacy [147].
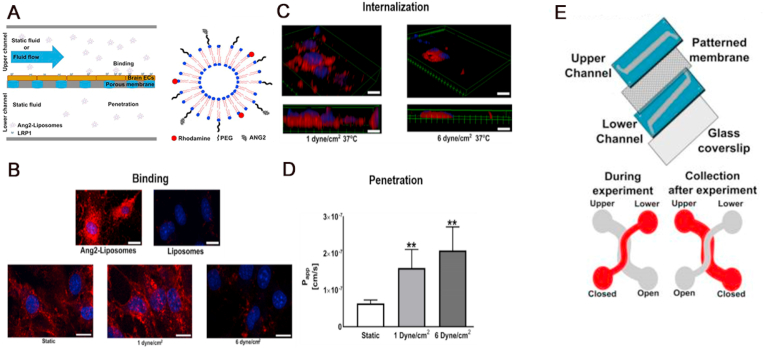
The emergence of BBB Chip and its application for nanoparticulates. The initial work in designing a BBB Chip was reported by Griep et al., using the human brain endothelial cell line hCMEC/D3 [81]. In their design, the BBB function was modulated straightforwardly by stimulation with shear stress, followed by exposure to the inflammation cytokine Tumor necrosis factor alpha (TNF-α). The results showed that the immortalized hCMEC/D3 cells could be cultured for up to 7 days in the BBB chip, and expressed BBB characteristics over time. Recently, Papademetriou et al. constructed a microfluidic BBB model (Fig. 5E) to evaluate binding and internalization by brain endothelial cells, as well as subsequent BBB penetration, of angiopep-2-coupled liposome nanocarriers (Ang2-Liposomes) [94]. Angiopep-2 is a peptide ligand of low-density lipoprotein receptor-related protein 1 (LRP1), and as such, confers the ability for the liposomes to shuttle across the BBB by utilizing LRP1 receptors on the luminal surface of the brain endothelial cells (Fig. 5A). Intriguingly, angiopep-2 conjugation not only significantly improved their binding compared to unconjugated liposomes, the efficiency of internalization and penetration was also regulated by the rate of flow. Ang2-Liposomes were internalized efficiently by brain endothelial cells under conditions of static flow or a lower fluid shear stress at 1 dyne cm−2 (Fig. 5B), while binding was reduced at a higher fluid shear stress, 6 dyne cm−2 (Fig. 5C). On the other hand, penetration of the BBB by Ang2-Liposomes was much more effective with applied shear stress, either at 1 dyne cm−2 or 6 dyne cm−2, rather than with static incubation (Fig. 5D). Their work highlights the relevance of dynamic flow for nanoparticle behavior within BBB in vitro models. Likewise, Falanga et al. developed a microfluidic BBB model to study the interaction of polystyrene nanoparticles coated with gH625, a virus-derived shuttling peptide, with brain endothelial cells in the dynamic flow conditions [95]. Their results demonstrated that these gH625-functionalized nanoparticles are an efficient platform for delivery to the brain, paving the way for future applications of peptide-mediated nanomedicine delivery to the BBB. However, it is crucial to consider that the two previous examples only utilized the mouse cerebral endothelial cell line bEnd.3, to model the BBB. Therefore, they are not human-specific models and likely do not reconstitute in vivo barrier strength.
Fig. 5.
An example of BBB Chip application in nanomedicine: evaluation of binding, internalization, and penetration of Ang2-Liposomes. (A) Left: schematic of the microfluidic BBB model and experimental design. Brain endothelial cells (bEnd.3) were grown in the upper channel of the device to enable barrier formation. Ang2-Liposomes were then added to the upper channel and incubated under static conditions or in the presence of flow, while the lower channel was kept static. Right: cartoon of the Ang2-Liposomes. Angiopep-2 was conjugated to the end of polyethylene glycol (PEG) chains. (B) Total binding of Ang2-Liposomes or non-functionalized liposomes in static fluid was visualized (upper row). Ang2-Liposomes binding to brain endothelial cells after incubation in static fluid or in the presence of flow (lower row). Cell nuclei were labeled in blue, while liposomes were labeled in red. (C) Internalization of Ang2-Liposomes in the presence of flow (D) Penetration of Ang2-Liposomes incubated in static fluid or the presence of flow. (E) Reproduced from Ref. [94]. Copyright 2018©Public Library of Science.
Recently, Park et al. published a method to use hypoxic conditions to enhance the barrier function of human iPSC-derived brain microvascular endothelial cells, a method in which trans-endothelial electrical resistance (TEER) values two orders of magnitude higher than previously reported could be achieved for up to a week [96]. Additionally, the endothelial cells were interfaced with primary human pericytes and astrocytes, which has been shown to be necessary for neurovascular function and inflammation, and the cells expressed high levels of functional efflux pumps, which allowed selective transcytosis of peptides and antibodies in a manner corresponding to previous in vivo observations [148]. The promise of this model for nanomedicine was demonstrated with a proof-of-concept experiment that 20-nm quantum dots conjugated with angiopep-2 were successfully shuttled across the BBB on the chip.
Outlook of using BBB Chip in the study of nanomedicine. As we achieve more advanced biomimetic models, future studies could inform the design of nanoparticle-based therapies with proper physicochemical properties that enable efficient binding and internalization by brain endothelial cells in the presence of dynamic flow, while maximizing BBB penetration [94]. Concurrently, we can investigate the behavior of the nanoparticles, how they interact with the constituents of the BBB, whether they have any inflammatory or toxic effects, and whether they influence neurovascular functions.
3.6. Vascular system
Intravenous injection is a common drug administration route because it is the most reliable method for systemic drug delivery, allows for 100% bioavailability, and bypasses efflux pumps and first-pass metabolism. Furthermore, no matter the route of administration, drugs and nanoparticles generally enter systemic circulation during distribution and clearance. It is, therefore, important to study the interactions of nanomedicine with the vasculature and consider vascular transport and toxicity [149].
The emergence of Vascular Chip and its application for nanoparticulates. One of the initial Vascular Chip was reported by Zheng et al., which was termed as a microfluidic flow stretch chip [150]. Their model chip could deliver fluid shear stress and cyclic stretch simultaneously or independently to vascular cells to mimic the hemodynamic microenvironment of the blood vessel in vivo. Hence, a key aspect of in vitro vascular models that use microfluidic systems is their ability to mimic the influence of shear stress, which is a primary factor in many aspects of vascular interactions and formation [151]. Shear stress is particularly crucial for nanoparticle delivery as it affects endocytic uptake of nanoparticles by endothelial cells [152]. Volkov et al. demonstrated that this shear stress effect was in fact even more critical than other factors that govern nanoparticle uptake, including treatment with mild detergents to increase permeability and treatment with TNF-α to increase endothelial cell activation, and the maximal uptake was registered at a relatively low shear stress of 0.05 Pa [153].
Aside from nanoparticle-endothelial cell interactions, another benefit of utilizing microfluidic models is their ability to validate the potential of shear-responsive nanoparticles for targeting and treating obstructed blood vessels (Fig. 6A) [97]. In this example, microscale aggregates of nanoparticles were created as thrombolytic delivery systems that were selectively delivered to regions of obstructed flow and then broken up into nanoscale components when exposed to abnormally high fluid shear stress, consequently concentrating and releasing drug specifically at the sites of thrombosis (Fig. 6B). These nanoparticles coated with tissue plasminogen activator (tPA) were able to rapidly dissolve preformed fibrin clots inside the microfluidic channel. This in vitro observation was then validated in an ex vivo mouse pulmonary embolism model, where a 100-fold lower dose of the shear-sensitive nanoparticle system was able to achieve the same lysing effect as free tPA. This microfluidic vascular stenosis model illustrates how nanoengineering approaches inspired by pathophysiological mechanisms can be used to develop safer and more effective therapeutic strategies.
Fig. 6.
An example of Vascular Chip application in nanomedicine: validation of the potential of shear-responsive nanoparticles for targeting and treating obstructed blood vessels. (A) Scanning electron micrographs of the microscale (~2–5 mm) shear-activated nanotherapeutics (SA-NTs) (left) and the poly (lactic-co-glycolic acid) (PLGA) nanoparticles (~180 nm) used to produce them (right). (B) Pathological shear-induced dissociation of SA-NTs and nanoparticle targeting under hemodynamic conditions in the microfluidic device. (1) A microfluidic vascular stenosis model showing how SA-NTs (large spheres) remain intact in the pre-stenotic region but then break up into individual nanoparticles (small spheres) when they flow through a constriction (90% lumen occlusion), which then accumulate in the endothelial cells lining the bottom of the channel. (2) A photograph of the PDMS-based microdevice that mimics vascular stenosis. (3) Computational fluid dynamics simulations of the microfluidic device shown in (2) demonstrating that a physiological level of inlet shear rate from the constriction increases to a pathological level in the post-stenotic region. Reproduced from Refs. [97]. Copyright 2012©American Association for the Advancement of Science.
As demonstrated above, on-chip vascular models are relevant when considering targeting vascular diseases using nanomedicine. They could, in turn, also contribute to the rational design of the ideal physicochemical properties of nanomedicine [154]. For example, Namdee et al. studied the differences in the accumulation of micro- and nano-sized spheres inside microchannels and observed that microspheres tended to localize more to the margins when compared to nanospheres [98]. This effect of size on accumulation, as well as hemodynamics and hemorheology, should be taken into account when designing nanomedicine targeted for vascular diseases and cancer. Moreover, some studies have highlighted the effect of particle shape on adhesion to microfluidic channel walls. In the case of an endothelialized channel, rod-shaped nanoparticles generally have higher targeting specificity and lower non-specific accumulation compared to sphere-shaped nanoparticles in these microfluidic models [99]. These observations were additionally corroborated by mouse experiments, which confirmed the shape-induced enhancement of vascular targeting in the lungs and brain under in vivo physiological conditions.
Outlook of using Vascular Chip in the study of nanomedicine. In summary, blood vessels are important to model due to their role in connecting the various other organs in the body and their contribution to the transport dynamics and kinetics of nutrients, oxygen, hormones, and nanomedicine. In this regard, future studies should incorporate the vasculature into other Organ Chip microfluidic models to mimic physiologically appropriate responses. Indeed, some studies have already pioneered the vascular-integrated Cancer Chip for nanomedicine evaluation [100,101]. For example, to mimic the tumor microenvironment and study nanoparticle extravasation through leaky tumor vasculature, Wang et al. linked a Vascular Chip in combination with a Cancer Chip [101]. The vascular structure with selective permeability was represented using human umbilical vein endothelial cells (HUVECs) cultured in the top channel of a PDMS chip, while the cancer was recapitulated using 3D tumor spheroids composed of SKOV3 (human ovarian cancer) cells in the bottom channel. The results showed that the vascular-tumor-on-a-chip model is capable of monitoring nanoparticle's extravasation and then tumor accumulation for hours up to days, suggesting that this combined model enable a more in-depth understanding about how nanoparticles take advantage of the enhanced permeability and retention effect and facilitate better prediction of the transport efficacy of nanoparticle formulations.
3.7. Tumor microenvironment
The tumor microenvironment (TME) is the environment around a tumor, and it is a hierarchical assembly of blood vessels, multiple cell types, signaling molecules, and ECM [155]. Such complexity can be partially recapitulated using the concept of Organ Chip platforms by replacing healthy cells and associated ECM with those of tumor or cancer origins; thus, tumor/cancer-on-a-chip (T/C Chip) models have been developed, including cancer cell intravasation/extravasation chip and orthotopic/metastatic cancer chip [68,[156], [157], [158]]. Besides, as a preclinical model, T/C Chips mimic the in vivo tumor architecture and dynamic flow conditions, which are essential factors for studying the interaction between nanomedicine and tumor cells [46,100,105].
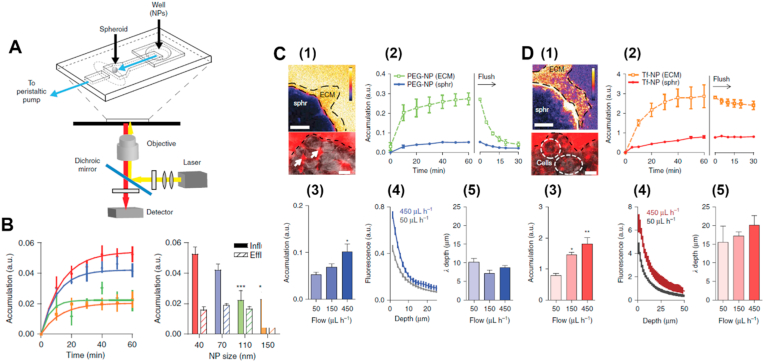
The T/C Chip and its application for nanoparticulates. Various prototype T/C Chip platforms have been developed for nanoparticle testing [159]. Yang et al. developed a breast T/C Chip using MCF-7 breast cancer cells and primary adipose-derived stromal cells for the evaluation of the efficiency of photodynamic therapy (PDT) with therapeutic agents, gold nanoparticles, under various irradiation conditions [102]. The results indicated that breast cancer cells in a 3D microfluidic culture environment exhibited more resistance to nanoparticle-based PDT than those in monolayer culture, demonstrating the suitability of the T/C Chip model for greater outcome reliability and physiological accuracy. In another study, Albanese et al. also customized a breast T/C Chip platform to investigate the transport behavior of PEGylated gold nanoparticles through tumor-like spheroids of the human melanoma cell line, MDA-MB-435, that have been immobilized in a PDMS-based chip (Fig. 7A) [103]. As have been observed in other studies [160,161], nanoparticle diameter (Fig. 7B) and surface functionalization (Fig. 7C and D) significantly affect their accumulation around the tumor site. In addition, the flow conditions in the TME also had a dramatic influence on nanoparticle accumulation. At a lower interstitial flow rate of 50 μL per h, the nanoparticles only accumulated in the periphery and failed to reach the tissue interface, while at a higher flow rate of 450 μL per h, the nanoparticles were able to accumulate to a greater extent both at the outer layer as well as within the tissue. However, Ran et al. showed an opposite trend for nanoparticle accumulation using a human ovarian T/C chip model, where lower flow rate of 0.25 μL per min resulted in improved anticancer efficacy of functioned liposomes [105]. Interestingly, they also demonstrated that the tumor spheroid size could influence the treatment outcome, in which smaller sized (200 μm) tumor spheroids showed a higher treatment efficacy compared to larger ones (250 and 300 μm).
Fig. 7.
An example of T/C Chip application in nanomedicine: investigation of transport behavior of gold nanoparticles with different sizes and functionalization through tumor spheroids under dynamic conditions. (A) Top: schematic of the microfluidic device on a microscope stage, with tumor spheroid immobilized at the end of the imaging chamber (B) The effect of nanoparticle size on accumulation in the spheroid measured. Left: tissue accumulation over time. Right: tissue accumulation after 1 h. (C) and (D) The effect of functionalization and flow rate on nanoparticle accumulation. Nanoparticles were functionalized with (C) PEG or (D) transferrin (Tf). (1) Intensity map (top) of fluorescence after 1 h. Image (bottom) of fluorescence in the interstitial spaces (arrows) and punctate fluorescence co-localizing with cell membranes (circles). (2) Mean fluorescence intensities of surrounding ECM and the tumor spheroids (sphr). (3) Mean spheroid fluorescence, (4) fluorescence distribution, and (5) penetration depth at various flow rates. Reproduced from Ref. [102]. Copyright 2013©Nature Publishing Group.
Outlook of using T/C Chip in the study of nanomedicine. Aside from testing the behavior and effect of nanoparticles on T/C models, it is important to consider toxicity effects on healthy tissues as well [162]. For example, Kotov et al. tested the effect of cadmium telluride/gold nanoparticles on a liver T/C model assembled with HepG2 hepatocellular carcinoma cells and demonstrated their toxicity to these tumor cells, but the toxicity to healthy liver tissue was not assessed [104]. Therefore, to unveil potential adverse effects of a nanomedicine while assessing its therapeutic efficacy, such as potential instigation of metastasis, future studies could connect T/C Chip containing cells from tumorous/cancerous tissues with healthy Organ Chip that are physiologically downstream [101]. Another consideration is the effect of other TME factors on the behavior of tumor cells. For example, it has been demonstrated that breathing motions suppress lung cancer growth and response to therapy in an orthotopic lung T/C Chip model in which H1975 human non-small-cell lung cancer cells were co-cultured with primary alveolar epithelial and endothelial cells [163]. These results matched the unique growth patterns and clinical responses to therapy observed in human patients. Cell-cell interactions were also shown to be critical determinants of tumor growth, as co-culture of H1975 with alveolar epithelial cells alone enhanced cancer cell growth, while co-culture with lung microvascular endothelial cells alone suppressed cell growth. Therefore, tumor activity and growth are highly dependent on both the mechanical and biological microenvironment, and evaluation of nanomedicine in T/C Chip models should take into account factors such as cyclic strain and cellular interactions, in both orthotopic as well as metastatic environments.
4. Challenges and conclusions of Organ Chip for the evaluation of nanomedicine
As an emerging candidate for in vitro screening of nanomedicine, Organ Chip is being developed with increasing complexity that allows direct control of biomechanical, biochemical, and biophysiological microenvironmental cues, which have already demonstrated their values and potentials for recapitulating key physiological conditions and characteristics that are not present in other in vitro systems. In the case of nanomedicine evaluations, we have seen several such examples. As introduced, in previous sections, the Lung Chip model demonstrated that damage from nanoparticles is strongly influenced by breathing motions and as a result, traditional cell culture systems likely have underestimated their effects. Similarly, the flow rate built in the BBB Chip model is found to be an essential regulator of the penetration of nanomedicine. Therefore, it is believed that there is a great opportunity to obtain more undiscovered information and immense value of nanomedicine by applying the Organ Chip technology.
However, despite the great promise, creating Organ Chip systems for nanomedicine evaluations is not a simple process, with a number of challenges to overcome. (1) While 3D Organ Chip systems represent higher levels of tissue organization, they still only embody some aspects of the real tissues, with some other tissue functions not being represented [164]. It is critical to consider what minimum functioning units are required to address specific applications for nanomedicine evaluations. (2) Using individual Organ Chip platforms limits the range of applications to a specific organ's functions. For systemic investigating of nanomedicine, reproducing the architectural complexity of human tissues and organs in a miniaturized fashion and interconnecting them in the right arrangement is a representative challenge. There have been recent advancements in constructing Body Chip or Multi-Organ Chip platforms, but more characterizations are still needed before they can be widely and accurately applied [91,128,129]. Similarly, building multiple experimental units arrayed on a single chip to achieve large(r)-scale, high (er)-throughput screening and analysis is also a challenge [165]. (3) While efforts are already underway to integrate biosensors within Organ Chip, this technique is facing many challenges [114,166]. One of the major challenges is to achieve real-time and continuous (e.g., more than a week) sensing and monitoring of the biological effects of nanomedicine. Although the current optical- and electrochemical-based methods can be used for real-time monitoring, sensor saturation and regeneration are still a remained problem [167]. Besides, it would be highly beneficial to be able to monitor multiple physicochemical parameters simultaneously in situ, such as oxygen level, pH, and even potential biomarkers, in order to track the effect of nanomedicine in a quantitative and real-time manner [168]. (4) Though PDMS and polystyrene are the most popular materials for chip fabrication due to several advantages, such as optical transparency and biocompatibility, other plastic materials have been used and a detailed comparison of their advantages and disadvantages in drug screening is missing [166,169]. Therefore, utilizing Organ Chip for nanomedicine studies should include consideration of any possible interactions between nanomedicine and the microfluidics systems. (5) Compared with animal experiments, Organ Chip constructed with primary human cells is closer to the real physiological and pathological conditions of the human body [170]. However, the challenge of cell sources and the difficulty of culturing primary cells in vitro are also current factors restricting the development of Organ Chip, which subsequently affects the reliability of testing nanomedicine on Organ Chip platforms. (6) Last but not least, Organ Chip platforms need to be standardized before their applications in the field of nanomedicine, with the hope that they could effectively address the core criteria of drug absorption, metabolism, and excretion [171]. Once standardized, we will certainly be able to eliminate numerous limitations and problems of nanomedicine evaluations; thus, more effective nanomedicine could be practiced using these advanced human-based in vitro models.
In conclusion, the process of nanomedicine evaluations is a long and costly procedure which is often practiced through animal models or other simplified in vitro models. However, these models always fail to accurately recapitulate the complexities of human physiology, resulting in probable failures of nanomedicine for human patients. There is realistic optimism that sophisticated in vitro human models will reduce or replace animal models in terms of accuracy and become more predictive of human responses. Therefore, as the potential candidate of in vitro model based on microfluidic technology, Organ Chip offers a novel platform to replace or complement the current nanomedicine study methods and expedite the nanomedicine assessment procedures by providing a more accurate replication of the microenvironments of native tissues and various tissue-tissue interactions. The present paper suggests that Organ Chip is improving at a rapid pace and becoming increasingly recognized among academic researchers, industry representatives, and regulatory agencies, which could be a powerful tool for elucidating the effect of nanomedicine on the human physiological systems and revealing additional insight for breakthroughs in the development of nanomedicine.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was primarily leveraged by National Natural Science Foundation of China for Innovative Research Groups (No. 51621002). Y.S.Z. was not supported by this fund; instead, support by the Brigham Research Institute is thanked. We acknowledge Dr. Amy Wen and Ms. Xuewei Zhang for helpful discussion. Any opinions, findings, conclusions, or recommendations expressed herein are those of the author(s).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Xi Chen, Email: chenxi@ecust.edu.cn.
Changsheng Liu, Email: liucs@ecust.edu.cn.
References
- 1.Duncan R. Nanomedicines in action. Pharm. J. 2004;273:485–488. [Google Scholar]
- 2.Wagner V., Dullaart A., Bock A.-K., Zweck A. The emerging nanomedicine landscape. Nat. Biotechnol. 2006;24:1211–1217. doi: 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]
- 3.Ventola C.L. The nanomedicine revolution: part 1: emerging concepts. Pharmacy and Therapeutics. 2012;37:512–525. [PMC free article] [PubMed] [Google Scholar]
- 4.Dickinson A.M., Godden J.M., Lanovyk K., Ahmed S.S. Assessing the safety of nanomedicines: a mini review. Applied in Vitro Toxicology. 2019;5:114–122. [Google Scholar]
- 5.Accomasso L., Cristallini C., Giachino C. Risk assessment and risk minimization in nanomedicine: a need for predictive, alternative, and 3Rs strategies. Front. Pharmacol. 2018;9:228. doi: 10.3389/fphar.2018.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shanks N., Greek R., Greek J. Are animal models predictive for humans? Philos. Ethics Humanit. Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moghimi S.M., Simberg D. Translational gaps in animal models of human infusion reactions to nanomedicines. Nanomedicine. 2018;13:973–975. doi: 10.2217/nnm-2018-0064. [DOI] [PubMed] [Google Scholar]
- 8.Akhtar A. The flaws and human harms of animal experimentation. Camb. Q. Healthc. Ethics. 2015;24:407–419. doi: 10.1017/S0963180115000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Resnik D.B., Tinkle S.S. Ethical issues in clinical trials involving nanomedicine. Contemp. Clin. Trials. 2007;28:433–441. doi: 10.1016/j.cct.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duval K., Grover H., Han L.H., Mou Y., Pegoraro A.F., Fredberg J., Chen Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology. 2017;32:266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 11.Imamura Y., Mukohara T., Shimono Y., Funakoshi Y., Chayahara N., Toyoda M., Kiyota N., Takao S., Kono S., Nakatsura T., Minami H. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015;33:1837–1843. doi: 10.3892/or.2015.3767. [DOI] [PubMed] [Google Scholar]
- 12.Fang Y., Eglen R.M. Three-dimensional cell cultures in drug discovery and development. SLAS DISCOVERY: Advancing the Science of Drug Discovery. 2017;22:456–472. doi: 10.1177/1087057117696795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmeichel K.L., Bissell M.J. Modeling tissue-specific signaling and organ function in three dimensions. J. Cell Sci. 2003;116:2377–2388. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., Chen B., Yan H., Zhao Y., Lou Z., Li J., Fu B., Zhu X., McManus D.P., Dai J., Jia W. Three-dimensional hepatocyte culture system for the study of Echinococcus multilocularis larval development. PLoS Neglected Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gómez-Lechón M.J., Tolosa L., Conde I., Donato M.T. Competency of different cell models to predict human hepatotoxic drugs. Expet Opin. Drug Metabol. Toxicol. 2014;10:1553–1568. doi: 10.1517/17425255.2014.967680. [DOI] [PubMed] [Google Scholar]
- 16.Ware B.R., Khetani S.R. Engineered liver platforms for different phases of drug development. Trends Biotechnol. 2017;35:172–183. doi: 10.1016/j.tibtech.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauschke V.M., Hendriks D.F., Bell C.C., Andersson T.B., Ingelman-Sundberg M. Novel 3D culture systems for studies of human liver function and assessments of the hepatotoxicity of drugs and drug candidates. Chem. Res. Toxicol. 2016;29:1936–1955. doi: 10.1021/acs.chemrestox.6b00150. [DOI] [PubMed] [Google Scholar]
- 18.Langhans S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018;9:6. doi: 10.3389/fphar.2018.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenda T., Kapałczyńska M., Przybyła W., Zajączkowska M., Teresiak A., Filas V., Ibbs M., Bliźniak R., Łuczewski Ł., Lamperska K. 2D and 3D cell cultures – a comparison of different types of cancer cell cultures. Arch. Med. Sci. 2016;14:910–919. doi: 10.5114/aoms.2016.63743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonnier F., Keating M.E., Wróbel T.P., Majzner K., Baranska M., Garcia-Munoz A., Blanco A., Byrne H.J. Cell viability assessment using the Alamar blue assay: a comparison of 2D and 3D cell culture models. Toxicol. Vitro. 2015;29:124–131. doi: 10.1016/j.tiv.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Baker B.M., Chen C.S. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tung Y.-C., Hsiao A.Y., Allen S.G., Torisawa Y.-s., Ho M., Takayama S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst. 2011;136:473–478. doi: 10.1039/c0an00609b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song H.-H.G., Park K.M., Gerecht S. Hydrogels to model 3D in vitro microenvironment of tumor vascularization. Adv. Drug Deliv. Rev. 2014;79–80:19–29. doi: 10.1016/j.addr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torisawa Y.S., Takagi A., Nashimoto Y., Yasukawa T., Shiku H., Matsue T. A multicellular spheroid array to realize spheroid formation, culture, and viability assay on a chip. Biomaterials. 2007;28:559–566. doi: 10.1016/j.biomaterials.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 25.Fennema E., Rivron N., Rouwkema J., van Blitterswijk C., de Boer J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013;31:108–115. doi: 10.1016/j.tibtech.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Pineda E.T., Nerem R.M., Ahsan T. Differentiation patterns of embryonic stem cells in two- versus three-dimensional culture. Cells Tissues Organs. 2013;197:399–410. doi: 10.1159/000346166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gauvin R., Chen Y.-C., Lee J.W., Soman P., Zorlutuna P., Nichol J.W., Bae H., Chen S., Khademhosseini A. Microfabrication of complex porous tissue engineering scaffolds using 3D projection stereolithography. Biomaterials. 2012;33:3824–3834. doi: 10.1016/j.biomaterials.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fierz F.C., Beckmann F., Huser M., Irsen S.H., Leukers B., Witte F., Degistirici Ö., Andronache A., Thie M., Müller B. The morphology of anisotropic 3D-printed hydroxyapatite scaffolds. Biomaterials. 2008;29:3799–3806. doi: 10.1016/j.biomaterials.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Suri S., Schmidt C.E. Cell-laden hydrogel constructs of hyaluronic acid, collagen, and laminin for neural tissue engineering. Tissue Eng. 2010;16:1703–1716. doi: 10.1089/ten.tea.2009.0381. [DOI] [PubMed] [Google Scholar]
- 30.Haraguchi Y., Shimizu T., Sasagawa T., Sekine H., Sakaguchi K., Kikuchi T., Sekine W., Sekiya S., Yamato M., Umezu M., Okano T. Fabrication of functional three-dimensional tissues by stacking cell sheets in vitro. Nat. Protoc. 2012;7:850–858. doi: 10.1038/nprot.2012.027. [DOI] [PubMed] [Google Scholar]
- 31.Cho E.C., Zhang Q., Xia Y. The effect of sedimentation and diffusion on cellular uptake of gold nanoparticles. Nat. Nanotechnol. 2011;6:385–391. doi: 10.1038/nnano.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leong D.T., Ng K.W. Probing the relevance of 3D cancer models in nanomedicine research. Adv. Drug Deliv. Rev. 2014;79–80:95–106. doi: 10.1016/j.addr.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Broutier L., Mastrogiovanni G., Verstegen M.M.A., Francies H.E., Gavarró L.M., Bradshaw C.R., Allen G.E., Arnes-Benito R., Sidorova O., Gaspersz M.P., Georgakopoulos N., Koo B.-K., Dietmann S., Davies S.E., Praseedom R.K., Lieshout R., Ijzermans J.N.M., Wigmore S.J., Saeb-Parsy K., Garnett M.J., van der Laan L.J.W., Huch M. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin N.Y.C., Homan K.A., Robinson S.S., Kolesky D.B., Duarte N., Moisan A., Lewis J.A. Renal reabsorption in 3D vascularized proximal tubule models. Proc. Natl. Acad. Sci. USA. 2019;116:5399–5404. doi: 10.1073/pnas.1815208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopper O., de Witte C.J., Lõhmussaar K., Valle-Inclan J.E., Hami N., Kester L., Balgobind A.V., Korving J., Proost N., Begthel H., van Wijk L.M., Revilla S.A., Theeuwsen R., van de Ven M., van Roosmalen M.J., Ponsioen B., Ho V.W.H., Neel B.G., Bosse T., Gaarenstroom K.N., Vrieling H., Vreeswijk M.P.G., van Diest P.J., Witteveen P.O., Jonges T., Bos J.L., van Oudenaarden A., Zweemer R.P., Snippert H.J.G., Kloosterman W.P., Clevers H. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 2019;25:838–849. doi: 10.1038/s41591-019-0422-6. [DOI] [PubMed] [Google Scholar]
- 36.Meyvantsson I., Beebe D.J. Cell culture models in microfluidic systems. Annu. Rev. Anal. Chem. 2008;1:423–449. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- 37.Kim K.M., Choi Y.J., Hwang J.-H., Kim A.R., Cho H.J., Hwang E.S., Park J.Y., Lee S.-H., Hong J.-H. Shear stress induced by an interstitial level of slow flow increases the osteogenic differentiation of mesenchymal stem cells through TAZ activation. PloS One. 2014;9 doi: 10.1371/journal.pone.0092427. e92427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonam S., Sathe S.R., Yim E.K., Sheetz M.P., Lim C.T. Cell contractility arising from topography and shear flow determines human mesenchymal stem cell fate. Sci. Rep. 2016;6:204–215. doi: 10.1038/srep20415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X.J., Valadez A.V., Zuo P., Nie Z. Microfluidic 3D cell culture: potential application for tissue-based bioassays. Bioanalysis. 2012;4:1509–1525. doi: 10.4155/bio.12.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehling M., Tay S. Microfluidic cell culture. Curr. Opin. Biotechnol. 2014;25:95–102. doi: 10.1016/j.copbio.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Halldorsson S., Lucumi E., Gómez-Sjöberg R., Fleming R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015;63:218–231. doi: 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 42.Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 43.Huh D., Hamilton G.A., Ingber D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi J.-H., Lee J., Shin W., Choi J.-W., Kim H.J. Priming nanoparticle-guided diagnostics and therapeutics towards human organs-on-chips microphysiological system. Nano Convergence. 2016;3 doi: 10.1186/s40580-016-0084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y.S., Khademhosseini A. Seeking the right context for evaluating nanomedicine: from tissue models in petri dishes to microfluidic organs-on-a-chip. Nanomedicine. 2015;10:685–688. doi: 10.2217/nnm.15.18. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y.S., Zhang Y.N., Zhang W. Cancer-on-a-chip systems at the frontier of nanomedicine. Drug Discov. Today. 2017;22:1392–1399. doi: 10.1016/j.drudis.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aliashrafi Jodat Y., Kang M., Kiaee K., Kim G., Flores Huidobro Martinez A., Rosenkranz A., Bae H., Shin S. Human-derived organ-on-a-chip for personalized drug development. Curr. Pharmaceut. Des. 2018;24:5471–5486. doi: 10.2174/1381612825666190308150055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uzel S.G.M., Amadi O.C., Pearl T.M., Lee R.T., So P.T.C., Kamm R.D. Simultaneous or sequential orthogonal gradient formation in a 3D cell culture microfluidic platform. Small. 2016;12:612–622. doi: 10.1002/smll.201501905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Au - Paranjape S.R., Au - Nagendran T., Au - Poole V., Au - Harris J., Au - Taylor A.M. JoVE; 2019. Compartmentalization of Human Stem Cell-Derived Neurons within Pre-assembled Plastic Microfluidic Chips. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cyganek L., Tiburcy M., Sekeres K., Gerstenberg K., Bohnenberger H., Lenz C., Henze S., Stauske M., Salinas G., Zimmermann W.H., Hasenfuss G., Guan K. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dehne E.-M., Hasenberg T., Horland R., Marx U. Multi-organ on a chip: human physiology-based assessment of liver toxicity. Toxicol. Lett. 2017;280:S75. [Google Scholar]
- 52.Spielmann H., Marx U. Human multi-organ-chips (MOCs) from vison to acceptance by industry and regulators. Toxicol. Lett. 2017;280:S30. [Google Scholar]
- 53.Jang K.J., Otieno M., Ronxhi J., Kodella K., Rubins J., Simic D., Singer M., Lim H.K., Lam W., Chen J., Petropolis D., Kulkarni G., Guzzie-Peck P., Karalis K., Hamilton G. Liver-chip identifies mitochondrial dysfunction, oxidative stress, and innate immune response as potential pathways of toxicity for the GPR40 agonist TAK-875. Toxicol. Lett. 2018;295:S129–S130. [Google Scholar]
- 54.Workman M.J., Gleeson J.P., Troisi E.J., Estrada H.Q., Kerns S.J., Hinojosa C.D., Hamilton G.A., Targan S.R., Svendsen C.N., Barrett R.J. Enhanced utilization of induced pluripotent stem cell–derived human intestinal organoids using microengineered chips. Cellular and Molecular Gastroenterology and Hepatology. 2018;5:669–677. doi: 10.1016/j.jcmgh.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber E.J., Chapron A., Chapron B.D., Voellinger J.L., Lidberg K.A., Yeung C.K., Wang Z., Yamaura Y., Hailey D.W., Neumann T., Shen D.D., Thummel K.E., Muczynski K.A., Himmelfarb J., Kelly E.J. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. 2016;90:627–637. doi: 10.1016/j.kint.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowman J., Wevers N., Spijkers X., Wilschut K., van Vught R., Trietsch S., Vulto P. BBB-on-a-chip: a 3D in vitro model of the human blood-brain barrier. Drug Metabol. Pharmacokinet. 2019;34:S54. [Google Scholar]
- 57.van Duinen V., Trietsch S.J., Joore J., Vulto P., Hankemeier T. Microfluidic 3D cell culture: from tools to tissue models. Curr. Opin. Biotechnol. 2015;35:118–126. doi: 10.1016/j.copbio.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Chiovaro F., Buschmann N., Strebel S., Guye P., Agarkova I. Development of an in vitro 3D model system for testing PD-1/PDL-1 immune checkpoint inhibitors. Eur. J. Canc. 2019;110:S18–S19. [Google Scholar]
- 59.Chiovaro F., Buschmann N., Strebel S., Wolf A., Agarkova I. 145P - immune-competent 3D InSightTM tumour models as novel platform to assess combinatorial biologics therapy. Ann. Oncol. 2019;30:xi52. [Google Scholar]
- 60.Rupp J., Strassfeld T., Steiert S., Guye P., Messner S., Kostadinova R. A novel microtissue-based 3D human liver NASH model for drug discovery. Toxicol. Lett. 2018;295:S79. [Google Scholar]
- 61.Deosarkar S.P., Prabhakarpandian B., Wang B., Sheffield J.B., Krynska B., Kiani M.F. A novel dynamic neonatal blood-brain barrier on a chip. PloS One. 2015;10 doi: 10.1371/journal.pone.0142725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Mello C.P.P., Rumsey J., Slaughter V., Hickman J.J. A human-on-a-chip approach to tackling rare diseases. Drug Discov. Today. 2019;24:2139–2151. doi: 10.1016/j.drudis.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oleaga C., Riu A., Rothemund S., Lavado A., McAleer C.W., Long C.J., Persaud K., Narasimhan N.S., Tran M., Roles J., Carmona-Moran C.A., Sasserath T., Elbrecht D.H., Kumanchik L., Bridges L.R., Martin C., Schnepper M.T., Ekman G., Jackson M., Wang Y.I., Note R., Langer J., Teissier S., Hickman J.J. Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system. Biomaterials. 2018;182:176–190. doi: 10.1016/j.biomaterials.2018.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen H., McCoy L., Willey H., Sharma A.D., Curley L., Moore M. Nerve-on-a-chip platform for assessing chemotherapy-induced peripheral neuropathy. J. Pharmacol. Toxicol. Methods. 2019;99:106595. [Google Scholar]
- 65.Roldan N., Rapet A., Stucki A.O., Raggi G., Fytianos K., Geiser T., Hobi N., Guenat O.T. Acute lung injury on-chip: building a disease predictive model that emulates alveolar biomechanics. Toxicol. Lett. 2018;295:S126–S127. [Google Scholar]
- 66.Stucki A.O., Raggi G., Sigrist S., Zamprogno P., Schneider-Daum N., Lehr C.M., Huwer H., Stucki J.D., Hobi N., Guenat O.T. Lung-on-a-Chip: the interplay of primary human epithelial and endothelial cells improves the alveolar barrier function. Toxicol. Lett. 2018;295:S67–S68. [Google Scholar]
- 67.Zheng F., Fu F., Cheng Y., Wang C., Zhao Y., Gu Z. Organ-on-a-chip systems: microengineering to biomimic living systems. Small. 2016;12:2253–2282. doi: 10.1002/smll.201503208. [DOI] [PubMed] [Google Scholar]
- 68.Huh D., Matthews B., Mammoto A., Montoya-Zavala M., Hsin H., Ingber D. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benam K.H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H.-H., Alves S.E., Salmon M., Ferrante T.C., Weaver J.C., Bahinski A., Hamilton G.A., Ingber D.E. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods. 2016;13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 70.Ho C.-T., Lin R.-Z., Chen R.-J., Chin C.-K., Gong S.-E., Chang H.-Y., Peng H.-L., Hsu L., Yew T.-R., Chang S.-F., Liu C.-H. Liver-cell patterning Lab Chip: mimicking the morphology of liver lobule tissue. Lab Chip. 2013;13:3578–3587. doi: 10.1039/c3lc50402f. [DOI] [PubMed] [Google Scholar]
- 71.Nakao Y., Kimura H., Sakai Y., Fujii T. Bile canaliculi formation by aligning rat primary hepatocytes in a microfluidic device. Biomicrofluidics. 2011;5:22212–22217. doi: 10.1063/1.3580753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franco C., Gerhardt H. Blood vessels on a chip. Nature. 2012;488:465–466. doi: 10.1038/488465a. [DOI] [PubMed] [Google Scholar]
- 73.Wang X., Phan D.T., Sobrino A., George S.C., Hughes C.C., Lee A.P. Engineering anastomosis between living capillary networks and endothelial cell-lined microfluidic channels. Lab Chip. 2016;16:282–290. doi: 10.1039/c5lc01050k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim H.J., Huh D., Hamilton G., Ingber D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 75.Kim H.J., Ingber D.E. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integrative Biology. 2013;5:1130–1140. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 76.Chen M.B., Srigunapalan S., Wheeler A.R., Simmons C.A. A 3D microfluidic platform incorporating methacrylated gelatin hydrogels to study physiological cardiovascular cell-cell interactions. Lab Chip. 2013;13:2591–2598. doi: 10.1039/c3lc00051f. [DOI] [PubMed] [Google Scholar]
- 77.Ma Z., Wang J., Loskill P., Huebsch N., Koo S., Svedlund F.L., Marks N.C., Hua E.W., Grigoropoulos C.P., Conklin B.R., Healy K.E. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat. Commun. 2015;6:7413. doi: 10.1038/ncomms8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang Y., Soroush F., Sheffield J.B., Wang B., Prabhakarpandian B., Kiani M.F. A biomimetic microfluidic tumor microenvironment platform mimicking the EPR effect for rapid screening of drug delivery systems. Sci. Rep. 2017;7:9359. doi: 10.1038/s41598-017-09815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li W., Liang G., Yan W., Zhang Q., Wang W., Zhou X., Liu D. Artificial uterus on a microfluidic chip. Chin. J. Anal. Chem. 2013;41:467–472. [Google Scholar]
- 80.Prabhakarpandian B., Shen M.C., Nichols J.B., Mills I.R., Sidoryk-Wegrzynowicz M., Aschner M., Pant K. SyM-BBB: a microfluidic blood brain barrier model. Lab Chip. 2013;13:1093–1101. doi: 10.1039/c2lc41208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Griep L.M., Wolbers F., de Wagenaar B., ter Braak P.M., Weksler B.B., Romero I.A., Couraud P.O., Vermes I., van der Meer A.D., van den Berg A. BBB on chip: microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed. Microdevices. 2013;15:145–150. doi: 10.1007/s10544-012-9699-7. [DOI] [PubMed] [Google Scholar]
- 82.Park J., Lee B., Jeong G.S., Hyun J.K., Lee C.J., Lee S.-H. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer's disease. Lab Chip. 2014;15:2524–2525. doi: 10.1039/c4lc00962b. [DOI] [PubMed] [Google Scholar]
- 83.Sun Q., Choudhary S., Mannion C., Kissin Y., Zilberberg J., Lee W.Y. Ex vivo construction of human primary 3D-networked osteocytes. Bone. 2017;105:245–252. doi: 10.1016/j.bone.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Torisawa Y.-s., Spina C.S., Mammoto T., Mammoto A., Weaver J.C., Tat T., Collins J.J., Ingber D.E. Bone marrow–on–a–chip replicates hematopoietic niche physiology in vitro. Nat. Methods. 2014;11:663–669. doi: 10.1038/nmeth.2938. [DOI] [PubMed] [Google Scholar]
- 85.Kong J., Luo Y., Jin D., An F., Zhang W., Liu L., Li J., Fang S., Li X., Yang X., Lin B., Liu T. A novel microfluidic model can mimic organ-specific metastasis of circulating tumor cells. Oncotarget. 2016;7:78421–78432. doi: 10.18632/oncotarget.9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Choi Y., Hyun E., Seo J., Blundell C., Kim H.C., Lee E., Lee S.H., Moon A., Moon W.K., Huh D. A microengineered pathophysiological model of early-stage breast cancer. Lab Chip. 2015;15:3350–3357. doi: 10.1039/c5lc00514k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reardon S. Organs-on-chips' go mainstream. Nature. 2015;523:266. doi: 10.1038/523266a. [DOI] [PubMed] [Google Scholar]
- 88.Kimura H., Sakai Y., Fujii T. Organ/body-on-a-chip based on microfluidic technology for drug discovery. Drug Metabol. Pharmacokinet. 2018;33:43–48. doi: 10.1016/j.dmpk.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 89.Prantil-Baun R., Novak R., Das D., Somayaji M.R., Przekwas A., Ingber D.E. Physiologically based pharmacokinetic and pharmacodynamic analysis enabled by microfluidically linked organs-on-chips. Annu. Rev. Pharmacol. Toxicol. 2018;58:37–64. doi: 10.1146/annurev-pharmtox-010716-104748. [DOI] [PubMed] [Google Scholar]
- 90.Zhang M., Xu C., Jiang L., Qin J. A 3D human lung-on-a-chip model for nanotoxicity testing. Toxicology Research. 2018;7:1048–1060. doi: 10.1039/c8tx00156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Esch M., Mahler G., Stokol T., Shuler M. Body-on-a-Chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip. 2014;14:3081–3092. doi: 10.1039/c4lc00371c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kang J., Super M., Yung C.W., Cooper R., Domansky K., Graveline A., Mammoto T., Berthet J., Tobin H., Cartwright M., Watters A., Rottman M., Waterhouse A., Mammoto A., Gamini N., Rodas M., Jiang A., Valentin T., Ingber D. An extracorporeal blood-cleansing device for sepsis therapy. Nat. Med. 2014;20:1211–1216. doi: 10.1038/nm.3640. [DOI] [PubMed] [Google Scholar]
- 93.Cho S., Islas Robles A., Nicolini A., Monks T., Yoon J.-Y. In situ, dual-mode monitoring of organ-on-a-chip with smartphone-based fluorescence microscope. Biosens. Bioelectron. 2016;86:697–705. doi: 10.1016/j.bios.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Papademetriou I., Vedula E., Charest J., Porter T. Effect of flow on targeting and penetration of angiopep-decorated nanoparticles in a microfluidic model blood-brain barrier. PloS One. 2018;13 doi: 10.1371/journal.pone.0205158. e0205158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Falanga A., Pitingolo G., Celentano M., Cosentino A., Melone P., Vecchione R., Guarnieri D., Netti P. Shuttle-mediated nanoparticle transport across an in vitro brain endothelium under flow conditions. Biotechnol. Bioeng. 2016;114:1087–1095. doi: 10.1002/bit.26221. [DOI] [PubMed] [Google Scholar]
- 96.Park T.-E., Mustafaoglu N., Herland A., Hasselkus R., Mannix R., Fitzgerald E., Prantil-Baun R., Watters A., Henry O., Benz M., Sanchez H., McCrea H., Goumnerova L., Song H., Palecek S., Shusta E., Ingber D. Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat. Commun. 2019;10:2621. doi: 10.1038/s41467-019-10588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Korin N., Kanapathipillai M., Matthews B.D., Crescente M., Brill A., Mammoto T., Ghosh K., Jurek S., Bencherif S.A., Bhatta D., Coskun A.U., Feldman C.L., Wagner D.D., Ingber D.E. Shear-activated nanotherapeutics for drug targeting to obstructed blood vessels. Science. 2012;337:738–742. doi: 10.1126/science.1217815. [DOI] [PubMed] [Google Scholar]
- 98.Namdee K., Thompson A.J., Charoenphol P., Eniola-Adefeso O. Margination propensity of vascular-targeted spheres from blood flow in a microfluidic model of human microvessels. Langmuir. 2013;29:2530–2535. doi: 10.1021/la304746p. [DOI] [PubMed] [Google Scholar]
- 99.Kolhar P., Anselmo A.C., Gupta V., Pant K., Prabhakarpandian B., Ruoslahti E., Mitragotri S. Using shape effects to target antibody-coated nanoparticles to lung and brain endothelium. Proc. Natl. Acad. Sci. Unit. States Am. 2013;110:10753–10758. doi: 10.1073/pnas.1308345110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vu M.N., Rajasekhar P., Poole D.P., Khor S.Y., Truong N.P., Nowell C.J., Quinn J.F., Whittaker M., Veldhuis N.A., Davis T.P. Rapid assessment of nanoparticle extravasation in a microfluidic tumor model. ACS Appl. Nano Mater. 2019;2:1844–1856. [Google Scholar]
- 101.Wang H.-F., Ran R., Liu Y., Hui Y., Zeng B., Chen D., Weitz D.A., Zhao C.-X. Tumor-vasculature-on-a-chip for investigating nanoparticle extravasation and tumor accumulation. ACS Nano. 2018;12:11600–11609. doi: 10.1021/acsnano.8b06846. [DOI] [PubMed] [Google Scholar]
- 102.Yang Y., Yang X., Zou J., Jia C., Hu Y., Du H., Wang H. Evaluation of photodynamic therapy efficiency using an in vitro three-dimensional microfluidic breast cancer tissue model. Lab Chip. 2015;15:735–744. doi: 10.1039/c4lc01065e. [DOI] [PubMed] [Google Scholar]
- 103.Albanese A., Lam A.K., Sykes E.A., Rocheleau J.V., Chan W.C.W. Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat. Commun. 2013;4:2718. doi: 10.1038/ncomms3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee J., Lilly G.D., Doty R.C., Podsiadlo P., Kotov N.A. In vitro toxicity testing of nanoparticles in 3D cell culture. Small. 2009;5:1213–1221. doi: 10.1002/smll.200801788. [DOI] [PubMed] [Google Scholar]
- 105.Ran R., Wang H.-F., Hou F., Liu Y., Hui Y., Petrovsky N., Zhang F., Zhao C.-X. A microfluidic tumor-on-a-chip for assessing multifunctional liposomes' tumor targeting and anticancer efficacy. Advanced Healthcare Materials. 2019;8 doi: 10.1002/adhm.201900015. [DOI] [PubMed] [Google Scholar]
- 106.Pacurari M., Lowe K., Tchounwou P., Kafoury R. A review on the respiratory system toxicity of carbon nanoparticles. Int. J. Environ. Res. Publ. Health. 2016;13:325. doi: 10.3390/ijerph13030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ferkol T., Schraufnagel D. The global burden of respiratory disease. Annals of the American Thoracic Society. 2014;11:404–406. doi: 10.1513/AnnalsATS.201311-405PS. [DOI] [PubMed] [Google Scholar]
- 108.Huh D. A human breathing lung-on-a-chip. Annals of the American Thoracic Society. 2015;12:S42–S44. doi: 10.1513/AnnalsATS.201410-442MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sayes C.M., Reed K.L., Warheit D.B. Assessing toxicity of fine and nanoparticles: comparing in vitro measurements to in vivo pulmonary toxicity profiles. Toxicol. Sci. 2007;97:163–180. doi: 10.1093/toxsci/kfm018. [DOI] [PubMed] [Google Scholar]
- 110.Gkatzis K., Taghizadeh S., Huh D., Stainier D., Bellusci S. Use of 3D organoids and lung-on-a-chip methods to study lung development, regeneration and disease. Eur. Respir. J. 2018;52:1800876. doi: 10.1183/13993003.00876-2018. [DOI] [PubMed] [Google Scholar]
- 111.Bergin I.L., Witzmann F.A. Nanoparticle toxicity by the gastrointestinal route: evidence and knowledge gaps. Int. J. Biomed. Nanosci. Nanotechnol. (IJBNN) 2013;3:163. doi: 10.1504/IJBNN.2013.054515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li L., Lieleg O., Jang S., Ribbeck K., Han J. A microfluidic in vitro system for the quantitative study of the stomach mucus barrier function. Lab Chip. 2012;12:4071–4079. doi: 10.1039/C2LC40161D. [DOI] [PubMed] [Google Scholar]
- 113.Kasendra M., Tovaglieri A., Sontheimer-Phelps A., Jalili-Firoozinezhad S., Bein A., Chalkiadaki A., Scholl W., Zhang C., Rickner H., Richmond C.A., Li H., Breault D.T., Ingber D.E. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018;8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jalili-Firoozinezhad S., Gazzaniga F.S., Calamari E.L., Camacho D.M., Fadel C.W., Bein A., Swenor B., Nestor B., Cronce M.J., Tovaglieri A., Levy O., Gregory K.E., Breault D.T., Cabral J.M.S., Kasper D.L., Novak R., Ingber D.E. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019;3:520–531. doi: 10.1038/s41551-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dishinger J.F., Reid K.R., Kennedy R.T. Quantitative monitoring of insulin secretion from single islets of Langerhans in parallel on a microfluidic chip. Anal. Chem. 2009;81:3119–3127. doi: 10.1021/ac900109t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Szabo G., Petrasek J. Inflammasome activation and function in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2015;12:387–400. doi: 10.1038/nrgastro.2015.94. [DOI] [PubMed] [Google Scholar]
- 117.Zhou Z., Xu M.-J., Gao B. Hepatocytes: a key cell type for innate immunity. Cell. Mol. Immunol. 2016;13:301–315. doi: 10.1038/cmi.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fausto N., Campbell J.S. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech. Dev. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 119.Björnsson E. Hepatotoxicity by drugs: the most common implicated agents. Int. J. Mol. Sci. 2016;17:224. doi: 10.3390/ijms17020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Beckwitt C.H., Clark A.M., Wheeler S., Taylor D.L., Stolz D.B., Griffith L., Wells A. Liver ‘organ on a chip’. Exp. Cell Res. 2018;363:15–25. doi: 10.1016/j.yexcr.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Godoy P., Hewitt N.J., Albrecht U., Andersen M.E., Ansari N., Bhattacharya S., Bode J.G., Bolleyn J., Borner C., Böttger J., Braeuning A., Budinsky R.A., Burkhardt B., Cameron N.R., Camussi G., Cho C.-S., Choi Y.-J., Craig Rowlands J., Dahmen U., Damm G., Dirsch O., Donato M.T., Dong J., Dooley S., Drasdo D., Eakins R., Ferreira K.S., Fonsato V., Fraczek J., Gebhardt R., Gibson A., Glanemann M., Goldring C.E.P., Gómez-Lechón M.J., Groothuis G.M.M., Gustavsson L., Guyot C., Hallifax D., Hammad S., Hayward A., Häussinger D., Hellerbrand C., Hewitt P., Hoehme S., Holzhütter H.-G., Houston J.B., Hrach J., Ito K., Jaeschke H., Keitel V., Kelm J.M., Kevin Park B., Kordes C., Kullak-Ublick G.A., LeCluyse E.L., Lu P., Luebke-Wheeler J., Lutz A., Maltman D.J., Matz-Soja M., McMullen P., Merfort I., Messner S., Meyer C., Mwinyi J., Naisbitt D.J., Nussler A.K., Olinga P., Pampaloni F., Pi J., Pluta L., Przyborski S.A., Ramachandran A., Rogiers V., Rowe C., Schelcher C., Schmich K., Schwarz M., Singh B., Stelzer E.H.K., Stieger B., Stöber R., Sugiyama Y., Tetta C., Thasler W.E., Vanhaecke T., Vinken M., Weiss T.S., Widera A., Woods C.G., Xu J.J., Yarborough K.M., Hengstler J.G. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen X., Jiang W., Ahmed A., Mahon C., Müllner M., Cao B., Xia T. Engineering protective polymer coatings for liver microtissues. Chem. Res. Toxicol. 2018;32:49–56. doi: 10.1021/acs.chemrestox.8b00120. [DOI] [PubMed] [Google Scholar]
- 123.Bachmann A., Moll M., Gottwald E., Nies C., Zantl R., Wagner H., Burkhardt B., Sánchez J.J.M., Ladurner R., Thasler W., Damm G., Nussler A.K. 3D cultivation techniques for primary human hepatocytes. Microarrays. 2015;4:64–83. doi: 10.3390/microarrays4010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim M.S., Lee W., Kim Y.C., Park J.-K. Microvalve-assisted patterning platform for measuring cellular dynamics based on 3D cell culture. Biotechnol. Bioeng. 2008;101:1005–1013. doi: 10.1002/bit.21962. [DOI] [PubMed] [Google Scholar]
- 125.Sivaraman A., Leach J.K., Townsend S., Iida T., Hogan B.J., Stolz D.B., Fry R., Samson L.D., Tannenbaum S.R., Griffith L.G. A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr. Drug Metabol. 2005;6:569–591. doi: 10.2174/138920005774832632. [DOI] [PubMed] [Google Scholar]
- 126.Toh Y.C., Lim T.C., Tai D., Xiao G., van Noort D., Yu H. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip. 2009;9:2026–2035. doi: 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- 127.Du Y., Li N., Yang H., Luo C., Gong Y., Tong C., Gao Y., Lü S., Long M. Mimicking liver sinusoidal structures and functions using a 3D-configured microfluidic chip. Lab Chip. 2017;17:782–794. doi: 10.1039/c6lc01374k. [DOI] [PubMed] [Google Scholar]
- 128.McAleer C., Long C., Elbrecht D., Sasserath T., Bridges L., Rumsey J., Martin C., Schnepper M., Wang Y., Schuler F., Roth A., Funk C., Shuler M., Hickman J. Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aav1386. [DOI] [PubMed] [Google Scholar]
- 129.Edington C.D., Chen W.L.K., Geishecker E., Kassis T., Soenksen L.R., Bhushan B.M., Freake D., Kirschner J., Maass C., Tsamandouras N., Valdez J., Cook C.D., Parent T., Snyder S., Yu J., Suter E., Shockley M., Velazquez J., Velazquez J.J., Stockdale L., Papps J.P., Lee I., Vann N., Gamboa M., LaBarge M.E., Zhong Z., Wang X., Boyer L.A., Lauffenburger D.A., Carrier R.L., Communal C., Tannenbaum S.R., Stokes C.L., Hughes D.J., Rohatgi G., Trumper D.L., Cirit M., Griffith L.G. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci. Rep. 2018;8:4530. doi: 10.1038/s41598-018-22749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Darwis Y., Ali Khan A., Mudassir J., Mohtar N. Advanced drug delivery to the lymphatic system: lipid-based nanoformulations. Int. J. Nanomed. 2013;8:2733–2744. doi: 10.2147/IJN.S41521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moura Rosa P., Gopalakrishnan N., Ibrahim H., Haug M., Halaas Ø. The intercell dynamics of T cells and dendritic cells in a lymph node-on-a-chip flow device. Lab Chip. 2016;16:3728–3740. doi: 10.1039/c6lc00702c. [DOI] [PubMed] [Google Scholar]
- 132.Rigat-Brugarolas L.G., Elizalde-Torrent A., Bernabeu M., De Niz M., Martin-Jaular L., Fernandez-Becerra C., Homs-Corbera A., Samitier J., del Portillo H.A. A functional microengineered model of the human splenon-on-a-chip. Lab Chip. 2014;14:1715–1724. doi: 10.1039/c3lc51449h. [DOI] [PubMed] [Google Scholar]
- 133.Yung C.W., Fiering J., Mueller A.J., Ingber D.E. Micromagnetic-microfluidic blood cleansing device. Lab Chip. 2009;9:1171–1177. doi: 10.1039/b816986a. [DOI] [PubMed] [Google Scholar]
- 134.Moghimi S.M., Hunter A.C., Murray J.C. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol. Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 135.Almeida J.P., Chen A.L., Foster A., Drezek R. In vivo biodistribution of nanoparticles. Nanomedicine. 2011;6:815–835. doi: 10.2217/nnm.11.79. [DOI] [PubMed] [Google Scholar]
- 136.Musah S., Mammoto A., Ferrante T.C., Jeanty S.S.F., Hirano-Kobayashi M., Mammoto T., Roberts K., Chung S., Novak R., Ingram M., Fatanat-Didar T., Koshy S., Weaver J.C., Church G.M., Ingber D.E. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat. Biomed. Eng. 2017;1 doi: 10.1038/s41551-017-0069. 0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Allison S.J. Kidney glomerulus-on-a-chip. Nat. Rev. Nephrol. 2017;13:382. doi: 10.1038/nrneph.2017.79. [DOI] [PubMed] [Google Scholar]
- 138.Ashammakhi N., Wesseling-Perry K., Hasan A., Elkhammas E., Zhang Y.S. Kidney-on-a-chip: untapped opportunities. Kidney Int. 2018;94:1073–1086. doi: 10.1016/j.kint.2018.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jang K.-J., Mehr A.P., Hamilton G.A., McPartlin L.A., Chung S., Suh K.-Y., Ingber D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integrative Biology. 2013;5:1119–1129. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 140.Jeonghwan L., Sejoong K. Kidney-on-a-chip: a new technology for predicting drug efficacy, interactions, and drug-induced nephrotoxicity. Curr. Drug Metabol. 2018;19:577–583. doi: 10.2174/1389200219666180309101844. [DOI] [PubMed] [Google Scholar]
- 141.Homan K.A., Kolesky D.B., Skylar-Scott M.A., Herrmann J., Obuobi H., Moisan A., Lewis J.A. Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci. Rep. 2016;6:34845. doi: 10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Choi C.H.J., Zuckerman J.E., Webster P., Davis M.E. Targeting kidney mesangium by nanoparticles of defined size. Proc. Natl. Acad. Sci. USA. 2011;108:6656–6661. doi: 10.1073/pnas.1103573108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kim S., Takayama S. Organ-on-a-chip and the kidney. Kidney Res. Clinic. Pract. 2015;34:165–169. doi: 10.1016/j.krcp.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Saraiva C., Praça C., Ferreira R., Santos T., Ferreira L., Bernardino L. Nanoparticle-mediated brain drug delivery: overcoming blood–brain barrier to treat neurodegenerative diseases. J. Contr. Release. 2016;235:34–47. doi: 10.1016/j.jconrel.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 145.Shin Y., Choi S.H., Kim E., Bylykbashi E., Kim J.A., Chung S., Kim D.Y., Kamm R.D., Tanzi R.E. Blood–Brain Barrier dysfunction in a 3D in vitro model of Alzheimer's disease. Adv. Sci. 2019;6:1900962. doi: 10.1002/advs.201900962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stamatovic S.M., Keep R.F., Andjelkovic A.V. Brain endothelial cell-cell junctions: how to "open" the blood brain barrier. Curr. Neuropharmacol. 2008;6:179–192. doi: 10.2174/157015908785777210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Helms H.C., Abbott N.J., Burek M., Cecchelli R., Couraud P.-O., Deli M.A., Förster C., Galla H.J., Romero I.A., Shusta E.V., Stebbins M.J., Vandenhaute E., Weksler B., Brodin B. In vitro models of the blood–brain barrier: an overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cerebr. Blood Flow Metabol. 2016;36:862–890. doi: 10.1177/0271678X16630991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Herland A., van der Meer A.D., FitzGerald E.A., Park T.E., Sleeboom J.J., Ingber D.E. Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3D human Blood-Brain Barrier on a chip. PloS One. 2016;11 doi: 10.1371/journal.pone.0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bhise N.S., Ribas J., Manoharan V., Zhang Y.S., Polini A., Massa S., Dokmeci M.R., Khademhosseini A. Organ-on-a-chip platforms for studying drug delivery systems. J. Contr. Release. 2014;190:82–93. doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zheng W., Jiang B., Wang D., Zhang W., Wang Z., Jiang X. A microfluidic flow-stretch chip for investigating blood vessel biomechanics. Lab Chip. 2012;12:3441–3450. doi: 10.1039/c2lc40173h. [DOI] [PubMed] [Google Scholar]
- 151.Lamberti G., Tang Y., Prabhakarpandian B., Wang Y., Pant K., Kiani M.F., Wang B. Adhesive interaction of functionalized particles and endothelium in idealized microvascular networks. Microvasc. Res. 2013;89:107–114. doi: 10.1016/j.mvr.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 152.Castro-Núñez L.D.-V.I., Herczenik E., Mertens K., Meijer A.B. Shear stress is required for the endocytic uptake of the factor VIII-von Willebrand factor complex by macrophages. J. Thromb. Haemostasis. 2012;10:1929–1937. doi: 10.1111/j.1538-7836.2012.04860.x. [DOI] [PubMed] [Google Scholar]
- 153.Samuel S., Jain N., O'Dowd F., Paul T., Kashanin D., Gerard V., Gun'ko Y., Prina-Mello A., Volkov Y. Multifactorial determinants that govern nanoparticle uptake by human endothelial cells under flow. Int. J. Nanomed. 2012;7:2943–2956. doi: 10.2147/IJN.S30624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang J.-W., Shen Y.-C., Lin K.-C., Cheng S.-J., Chen S.-L., Chen C.-Y., Kumar P.V., Lin S.-F., Lu H.-E., Chen G.-Y. Organ-on-a-chip: opportunities for assessing the toxicity of particulate matter. Front. Bioengineer. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Joyce J.A., Pollard J.W. Microenvironmental regulation of metastasis. Nat. Rev. Canc. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zervantonakis I.K., Hughes-Alford S.K., Charest J.L., Condeelis J.S., Gertler F.B., Kamm R.D. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. Unit. States Am. 2012;109:13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jeon J.S., Zervantonakis I.K., Chung S., Kamm R.D., Charest J.L. In vitro model of tumor cell extravasation. PloS One. 2013;8 doi: 10.1371/journal.pone.0056910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bersini S., Jeon J.S., Dubini G., Arrigoni C., Chung S., Charest J.L., Moretti M., Kamm R.D. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials. 2014;35:2454–2461. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Farokhzad O., Khademhosseini A., Jon S., Hermmann A., Cheng J., Chin C., Kiselyuk A., Teply B., Eng G., Langer R. Microfluidic system for studying the interaction of nanoparticles and microparticles with cells. Anal. Chem. 2005;77:5453–5459. doi: 10.1021/ac050312q. [DOI] [PubMed] [Google Scholar]
- 160.Pirollo K.F., Chang E.H. Does a targeting ligand influence nanoparticle tumor localization or uptake? Trends Biotechnol. 2008;26:552–558. doi: 10.1016/j.tibtech.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 161.Sykes E.A., Chen J., Zheng G., Chan W.C. Investigating the impact of nanoparticle size on active and passive tumor targeting efficiency. ACS Nano. 2014;8:5696–5706. doi: 10.1021/nn500299p. [DOI] [PubMed] [Google Scholar]
- 162.Sung J.H., Wang Y.I., Narasimhan Sriram N., Jackson M., Long C., Hickman J.J., Shuler M.L. Recent advances in body-on-a-chip systems. Anal. Chem. 2019;91:330–351. doi: 10.1021/acs.analchem.8b05293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hassell B.A., Goyal G., Lee E., Sontheimer-Phelps A., Levy O., Chen C.S., Ingber D.E. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 2017;21:508–516. doi: 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 164.Reif R. The body-on-a-chip concept: possibilities and limitations. EXCLI J. 2014;13:1283–1285. [PMC free article] [PubMed] [Google Scholar]
- 165.Probst C., Schneider S., Loskill P. High-throughput organ-on-a-chip systems: current status and remaining challenges. Curr. Opin. Biomed. Engineer. 2018;6:33–41. [Google Scholar]
- 166.Shaegh S.A.M., Pourmand A., Nabavinia M., Avci H., Tamayol A., Mostafalu P., Ghavifekr H.B., Aghdam E.N., Dokmeci M.R., Khademhosseini A., Zhang Y.S. Rapid prototyping of whole-thermoplastic microfluidics with built-in microvalves using laser ablation and thermal fusion bonding. Sensor. Actuator. B Chem. 2018;255:100–109. [Google Scholar]
- 167.Kilic T., Navaee F., Stradolini F., Renaud P., Carrara S. Organs-on-chip monitoring: sensors and other strategies. Microphysiol. Syst. 2018;2:2. [Google Scholar]
- 168.Zhang Y.S., Aleman J., Shin S.R., Kilic T., Kim D., Mousavi Shaegh S.A., Massa S., Riahi R., Chae S., Hu N., Avci H., Zhang W., Silvestri A., Sanati Nezhad A., Manbohi A., De Ferrari F., Polini A., Calzone G., Shaikh N., Alerasool P., Budina E., Kang J., Bhise N., Ribas J., Pourmand A., Skardal A., Shupe T., Bishop C.E., Dokmeci M.R., Atala A., Khademhosseini A. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. USA. 2017;114:E2293–E2302. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pourmand A., Shaegh S.A.M., Ghavifekr H.B., Najafi Aghdam E., Dokmeci M.R., Khademhosseini A., Zhang Y.S. Fabrication of whole-thermoplastic normally closed microvalve, micro check valve, and micropump. Sensor. Actuator. B Chem. 2018;262:625–636. [Google Scholar]
- 170.Esch E.W., Bahinski A., Huh D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015;14:248–260. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nigam P. 2016. Lab-on-a-Chip Fabrication and Application || Cells and Organs on Chip—A Revolutionary Platform for Biomedicine. [DOI] [Google Scholar]
- 172.Xia Y., Whitesides G.M. Soft lithography. Annu. Rev. Mater. Sci. 1998;28:153–184. [Google Scholar]
- 173.Skardal A., Murphy S.V., Devarasetty M., Mead I., Kang H., Seol Y., Shrike Zhang Y. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017;7(1):8837. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]