Abstract
Background and Aim:
Salmonella spp. are one of the most important food-borne pathogens in the world, emerging as a major public health concern. Moreover, multidrug-resistant (MDR) strains have been isolated from salmonellosis outbreaks, which compromise its treatment success. This study was conducted to characterize the phenotypic and genotypic antibiotic resistance profile of Salmonella strains isolated from broilers and humans from the regions of Tolima and Santander (Colombia).
Materials and Methods:
Salmonella spp. strains (n=49) were confirmed through molecular detection by amplification of the invA gene. Phenotypic antibiotic resistance was determined by the automated method and the agar diffusion method, and the presence of resistance genes was evaluated by PCR. Genotypic characterization was conducted using the enterobacterial repetitive intergenic consensus (ERIC)-PCR method, from which a dendrogram was generated and the possible phylogenetic relationships were established.
Results:
Salmonella isolates were classified as MDR strains exhibiting resistance to four antibiotic classes, penicillins, aminoglycosides, sulfonamides, and cephalosporins, and the human strains were resistant to gentamicin. At the genotypic level, the isolates contained the genes blaCMY2, blaCTX-M, blaPSE-1, blaTEM, aadA1, srtB, dfrA1, sul2, and floR. The genotyping results obtained by ERIC-PCR allowed the grouping of strains according to the source of isolation.
Conclusion:
The Salmonella spp. strains exhibited resistance to multiple antibiotics, as well as multiple genes associated with them, and the ERIC-PCR method was a technique that was helpful in generating clusters with biological significance.
Keywords: broiler farm, genotyping, resistance genes
Introduction
Salmonella enterica is one of the major pathogenic bacteria that can be transmitted through food consumption [1]. Consumption of products such as milk, beef, pork, chicken meat, and eggs is considered as a transmission route, based on which salmonellosis can be classified as a disease of zoonotic origin [2]. In the United States, it has been estimated that this bacterium causes 1.2 million clinical cases per year, of which 1941 outbreaks have been documented [3]. In contrast, in Colombia, the clinical cases that were reported from during 2000-2013 were most frequently caused by the serotypes Typhimurium and Enteritidis [4].
Furthermore, several serotypes of Salmonella spp. have been reported to be antibiotic-resistant, which represents a public health problem due to the risk of transmission of resistance between bacterial populations. Due to the plasticity of these bacteria, they have adapted and developed mechanisms to resist the effects of antibiotics using genetic strategies such as gene mutations or acquisition of resistance genes by horizontal transfer [5]. One of the primary causes of this resistance is the use of antibiotics as growth promoters in animal diets or their direct use for prophylactic purposes [6].
In Salmonella and other bacteria, genotyping methods have been used for identifying the clonal and phylogenetic relationships between different isolates to generate control strategies and for surveillance of outbreaks caused due to multidrug-resistant (MDR) bacteria [7]. Repetitive element-based PCR (rep-PCR) is a simple and inexpensive method that can be used to discriminate between Salmonella strains through the analysis of band patterns. The enterobacterial repetitive intergenic consensus (ERIC) is a repetitive sequence that is highly conserved and located in the intergenic zones; it has a variable distribution along the bacterial chromosome, separated by different lengths of intragenic sequences, which allows these primers to offer different band profiles [8,9]. The REP-PCR technique has also been widely applied in Salmonella studies [8,10].
Therefore, the aim of this study was to characterize the phenotypic and genotypic antibiotic resistance profile of Salmonella strains isolated from broilers and human subjects from the regions of Tolima and Santander (Colombia).
Materials and Methods
Ethical approval
No ethical approval required for this study because samples were from Bacterial Strain Collection of the Laboratory of Immunology and Molecular Biology. All the procedures for the previous collection of the samples from animals and humans were approved by Bioethics Committee of the Central Office of Research from University of Tolima and complied with the guidelines for animal care and use in research and teaching.
Study period and location
The Salmonella strains from Tolima broiler farms were collected from March 2015 to March 2016. The strains from Santander broiler farms were collected from January 2015 to December 2015. In the case of the human strains, they were collected from August 2015 to December 2015 in local health care centers in Ibagué - Tolima. Finally, the study was conducted from October 2018 to June 2019 in the Laboratory of Immunology and Molecular Biology – LIBM of the University of Tolima.
Salmonella spp. strains
Salmonella spp. strains were obtained from the previous studies conducted by the Poultry Research Group of the University of Tolima [11,12]. A total of 39 strains of Salmonella spp. isolated from broiler farms were included, of which 15 strains were serotyped as Salmonella Heidelberg (Santander broiler farms), and 24 strains were serotyped as Salmonella Paratyphi B (Tolima broiler farms), according to the Kauffman–White–Le Minor scheme [13]. In addition, ten strains isolated from human subjects with gastroenteritis in Tolima region were included in the study, which belonged to the serotypes Newport (n=1), Enteritidis (n=4), Braenderup (n=1), Uganda (n=1), Typhimurium (n=2), and Grupensis (n=1).
Molecular confirmation
Genomic DNA (gDNA) was extracted from fresh colonies using the Invisorb® Spin Universal Kit (Stratec, Germany) and maintained at −20°C until use. All isolates were confirmed by PCR by the amplification of the invA gene (accession number NC003197.2) using the primers forward 5′-TGAAATTATCGCCACGTTCGGGCAA-3′ and reverse 5′-TCATCGCACCGTCAAAGGAACC-3′ with an amplicon size of 284 bp. S. enterica ATCC® 13076 strain (ATCC, USA) was used as a positive control. The PCR assay was conducted in a total volume of 25 μL consisting of 14.87 μL distilled deionized water, 5 μL of 5× colorless GoTaq® Flexi Buffer (Promega, USA), 1 μL dNTPs (1.5 mM) (Invitrogen, USA), 1 μL of each primer (forward and reverse) (10 pmol/μL), 1 μL MgCl2 (25 mM), 0.125 μL of 0.6 U GoTaq® Flexi DNA polymerase (Promega, USA), and 1 μL gDNA as the template. The amplification was performed in a T-100™ thermocycler (Bio-Rad, USA) with an initial denaturation step at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s, and a final step of extension at 72°C for 7 min. The amplicons were visualized on 2% agarose gel by electrophoresis (PowerPac™ HC, Bio-Rad, USA) using 100-bpDNA ladder Load Ready™ (Amplyus, USA). The gel was stained with HydraGreen™ (ACTGene, USA) and visualized under the UV light using the ENDURO™ GDS gel documentation system (Labnet International, Inc., USA).
Phenotypic resistance
The phenotypic resistance to ampicillin (AM) (4-16 μg/mL), piperacillin/tazobactam (4/4-64/4 μg/mL), gentamicin (GM) (2-8 μg/mL), trimethoprim/sulfamethoxazole (SXT) (1/19-4/76 μg/mL), ceftriaxone (CRO) (1-32 μg/mL), ceftazidime (CAZ) (1-16 μg/mL), cefepime (1-16 μg/mL), ertapenem (ETP) (0.25-4 μg/mL), imipenem (1-8 μg/mL), and meropenem (1-8 μg/mL) was evaluated using the automated BD Phoenix NMIC/ID-94 (Becton Dickinson, USA) through the minimum inhibitory concentration method following the recommendations of the CLSI [14]. The resistance to chloramphenicol (CHL, 30 μg), florfenicol (FFC, 30 μg), and streptomycin (STR, 10 μg) was determined using the Kirby–Bauer disk diffusion susceptibility test. A bacterial suspension was spread onto Mueller-Hinton agar (Oxoid, Germany), according to the McFarland turbidity scale of 0.5, and then, the bacterial growth inhibition was determined at 37°C for 18 h according to the CLSI [14] guidelines.
Genotypic resistance
The presence of antimicrobial resistance genes was determined by PCR using gene-specific primers described in Table-1. The gDNA extracted from the isolates was used as the template for the PCR assay that was conducted under the above-described conditions, except that the annealing temperature was adjusted according to the melting temperature of each primer set.
Table-1.
Primers used to evaluate the presence of resistance genes [42] in Salmonella spp. strains.
| Antibiotic | Target gene | Primer sequence | Amplicon size (bp) |
|---|---|---|---|
| Ampicillin | blaPSE-1 | F- GCAAGTAGGGCAGGCAATCA R- GAGCTAGATAGATGCTCACAA |
461 |
| blaTEM | F- ATCAGTTGGGTGCACGAGTG R- ACGCTCACCGGCTCCAGA |
608 | |
| Chloramphenicol | catA | F- CCAGACCGTTCAGCTGGATA R- CATCAGCACCTTGTCGCCT |
454 |
| cmlA | F- TGGACCGCTATCGGACCG R- CGCAAGACACTTGGGCTGC |
642 | |
| Florfenicol | floR | F- CACGTTGAGCCTCTATATGG R- ATGCAGAAGTAGAACGCGAC |
888 |
| Gentamicin | aadB | F-CTAGCTGCGGCAGATGAGC R-CTCAGCCGCCTCTGGGCA |
300 |
| Streptomycin | aadA1 | F- CTCCGCAGTGGATGGCGG R- GATCTGCGCGCGAGGCCA |
629 |
| aadA2 | F- CATTGAGCGCCATCTGGAAT R- ACATTTCGCTCATCGCCGGC |
501 | |
| strA | F- TGGCAGGAGGAACAGGAGG R- AGGTCGATCAGACCCGTGC |
404 | |
| strB | F- GCGGACACCTTTTCCAGCCT R- TCCGCCATCTGTGCAATGCG |
620 | |
| Trimethoprim | dfrA1 | F- CAATGGCTGTTGGTTGGAC R- CCGGCTCGATGTCTATTGT |
253 |
| dfrA10 | F- TCAAGGCAAATTACCTTGGC R- ATCTATTGGATCACCTACCC |
433 | |
| dfrA12 | F- TTCGCAGACTCACTGAGGG R- CGGTTGAGACAAGCTCGAAT |
330 | |
| Ceftriaxone | blaCMY2 | F- AAATCGTTATGCTGCGCTCT R- CCGATCCTAGCTCAAACAGC |
244 |
| blaCTX-M | F- TTCGCTAAATACCGCCATTC R- TATCGTTGGTTGTGCCGTAA |
236 | |
| Sulfamethoxazole | sul1 | F- CGGACGCGAGGCCTGTATC R- GGGTGCGGACGTAGTCAGC |
591 |
| sul2 | F- GCGCAGGCGCGTAAGCTGAT R- CGAAGCGCAGCCGCAATTC |
514 | |
| sul3 | F- GGGAGCCGCTTCCAGTAAT R- TCCGTGACACTGCAATCATTA |
500 |
ERIC-PCR
The 49 Salmonella spp. strains were fingerprinted using the primer set ERIC1 5′-ATGTAAGCTCCTGGGGATTCA-3′ and ERIC2 5′-AAGTAAGTGACTGGGGTGAGAGCG-3′ [9]. The PCR was performed in a total volume of 25 μL containing 11.85 μL distilled deionized water, 5 μL of 5× colorless GoTaq® Flexi Buffer (Promega, USA), 2 μL dNTPs (1.5 mM) (Invitrogen, USA), 1 μL of each primer (50 pmol/μL), 2 μL MgCl2 (25 mM), 0.15 μL of 0.7 U GoTaq® Flexi DNA polymerase (Promega, USA), and 2 μL gDNA as the template. The amplification was conducted in a T-100™ thermocycler (Bio-Rad, USA) with an initial denaturation step at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 1 min, extension at 72°C for 7 min, and a final step of extension at 72°C for 10 min.
The PCR products were visualized by horizontal electrophoresis using 1% agarose gel (UltraPure™ Agarose, Invitrogen, USA) in 0.5× TBE, and the gel was stained with HydraGreen™ (ACTGene, Piscataway, USA). A 1-kb DNA ladder (Solis BioDyne, Estonia) was used in each gel as a molecular weight marker. The PCR products were run at 50 V for 3 h 30 min. For the cluster analysis, the banding patterns were analyzed using the BioNumerics version 7.5 software (Applied Maths, Sint-Martens-Latem, Belgium), and the genetic distances between strains were calculated according to the dice coefficient [15]. The dendrogram was constructed using the unweighted pair group method with arithmetic mean (UPGMA) in the BioNumerics version 7.5 software (Applied Maths, Sint-Martens-Latem, Belgium). In addition, the discriminatory index was calculated using the formula described by Hunter and Gaston [16] based on Simpson’s diversity index.
Results
Molecular confirmation
In all the 49 strains, the 284-bp fragment of the gene invA could be amplified, which indicated all the strains belonged to the genus Salmonella.
Phenotypic resistance
The 39 Salmonella spp. strains isolated from broiler farms were classified as MDR strains that were resistant to the four antibiotic classes of penicillins, aminoglycosides, sulfonamides, and cephalosporins (AM, GM, STR, SXT, CRO, and CAZ) (Table-2). Regarding the strains isolated from human subjects with gastroenteritis, the serotype Typhimurium (n=1) was classified as an MDR strain that exhibited resistance to GM, STR, chloramphenicol, and florfenicol. In total, 83.6% (41/49) of the strains were resistant to STR and 79.5% (39/49) were resistant to AM. In the case of cephalosporins, 75.5% (37/49) of the strains were resistant to CRO and CAZ. In addition, 71.4% (35/49) of the strains were resistant to SXT, and 65.3% (32/49) were resistant to GM. In contrast, all (100%; 49/49) the strains were susceptible to ETP, and 97.9% (48/49) of the strains were susceptible to amphenicols and carbapenems.
Table-2.
Phenotypic and genotypic profiles of resistance in Salmonella spp. strains.
| Strain code | Phenotypic antimicrobial resistance profile | Genotypic antimicrobial resistance profile |
|---|---|---|
| 1 | AM, STR, SXT, CRO, CAZ | aadA1, strA, strB, sul1, sul2, blaCMY2 |
| 2 | AM, STR, SXT, CRO, CAZ | strA, strB, sul1, sul2, blaCMY2 |
| 3 | AM, STR, SXT, CRO, CAZ | aadA1, strA, strB, sul1, sul2, blaCMY2 |
| 4 | AM, STR, SXT, CRO, CAZ | strA, strB, sul1, sul2, blaCMY2 |
| 5 | AM, STR, SXT, CRO, CAZ | aadA1, strA, strB, sul1, blaCMY2 |
| 6 | AM, STR, SXT, CRO, CAZ | aadA1, strA, strB, sul1, sul2, blaCMY2 |
| 7 | AM, STR, SXT, CRO, CAZ | strA, strB, sul1, blaCMY2 |
| 8 | AM, STR, SXT, CRO, CAZ | strA, strB, sul1, sul2, blaCMY2 |
| 9 | AM, STR, SXT, CRO, CAZ, FEP | aadA1, strA, strB, sul1, sul2, blaCMY2 |
| 10 | AM, STR, SXT, CRO, CAZ, FEP | aadA1, strA, strB, sul1, sul2, blaCMY2 |
| 11 | AM, STR, CRO, CAZ, FEP | aadA1, strA, strB, sul1, sul2, blaCMY2 |
| 12 | AM, TZP, STR, SXT, CRO, CAZ, FEP | aadA1, strA, strB, sul1, sul2, blaCMY2 |
| 13 | AM, STR, SXT, CRO, CAZ, FEP | strA, strB, sul1, sul2, blaCMY2 |
| 14 | AM, STR, SXT, CRO, CAZ, FEP | aadA1, strA, strB, sul1, sul2, blaCMY2 |
| 15 | AM, STR, SXT, CRO, CAZ, FEP | strA, strB, sul1, sul2, blaCMY2 |
| 16 | AM, GM, STR | blaPSE-1, blaTEM, aadA1, dfrA1, blaCMY2 |
| 17 | AM, GM, STR, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strB, dfrA1, sul2, blaCMY2 |
| 18 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strB, dfrA1, sul2, blaCMY2 |
| 19 | AM, STR, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strB, dfrA1, sul1, sul2, blaCMY2 |
| 20 | AM, GM, STR, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, dfrA1, sul2, blaCMY2 |
| 21 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, dfrA1, sul2, blaCMY2, blaCTX-M |
| 22 | AM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, dfrA1, sul2, blaCMY2, blaCTX-M |
| 23 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, dfrA1, sul1, sul2, blaCMY2 |
| 24 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, dfrA1, strA, strB, blaCMY2 |
| 25 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2, blaCTX-M |
| 26 | AM, GM, STR, SXT, CRO, CAZ, IPM | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2, blaCTX-M |
| 27 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2, blaCTX-M |
| 28 | GM, STR, SXT | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2 |
| 29 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strB, dfrA1, sul2, blaCMY2 |
| 30 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2 |
| 31 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2 |
| 32 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2, blaCTX-M |
| 33 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2 |
| 34 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2, blaCTX-M |
| 35 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, sul1, sul2, blaCMY2, blaCTX-M |
| 36 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, aadA2, strA, strB, dfrA1, dfrA12, sul2, blaCMY2 |
| 37 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, aadA2, strA, strB, dfrA1, dfrA12, sul2, cmlA, blaCMY2 |
| 38 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, aadA2, strA, strB, dfrA1, sul2, blaCMY2 |
| 39 | AM, GM, STR, SXT, CRO, CAZ | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, dfrA12, sul2, blaCMY2 |
| 40 | GM | blaPSE-1, blaTEM, aadA1, strB, dfrA1, dfrA12, sul2, blaCMY2 |
| 41 | GM | blaPSE-1, blaTEM, aadA1, aadA2, strA, strB, dfrA1, dfrA12, sul2, blaCMY2 |
| 42 | GM | blaPSE-1, blaTEM, aadA1, aadA2, strB, dfrA1, dfrA12, sul2, blaCMY2 |
| 43 | GM | blaPSE-1, blaTEM, aadA1, strA, strB, dfrA1, dfrA12, sul2, blaCMY2 |
| 44 | GM | blaPSE-1, blaTEM, aadA1, aadA2, strB, dfrA1, dfrA12, sul2, blaCMY2 |
| 45 | GM | blaPSE-1, blaTEM, aadA1, aadA2, strA, strB, dfrA1, dfrA12, sul2, blaCMY2 |
| 46 | GM | blaPSE-1, blaTEM, aadA1, aadA2, strB, dfrA1, dfrA12, sul2, blaCMY2 |
| 47 | AM, GM, SXT | blaPSE-1, blaTEM, aadA1, aadA2, strB, dfrA1, dfrA12, sul2, sul3, cmlA, blaCMY2 |
| 48 | GM | blaPSE-1, blaTEM, aadA1, aadA2, strB, dfrA1, dfrA12, sul2, cmlA, blaCMY2 |
| 49 | GM, STR, CHL, FFC | blaPSE-1, blaTEM, aadA1, aadA2, strB, dfrA1, dfrA12, sul2, blaCMY2 |
AM=Ampicillin, TZP=Piperacillin/tazobactam, GM=Gentamicin, STR=Streptomycin, SXT=Trimethoprim/sulfamethoxazole, CHL=Chloramphenicol, FFC=Florfenicol, CRO=Ceftriaxone, CAZ=Ceftazidime, FEP=Cefepime, IPM=Imipenem. Poultry farms (Santander): Heidelberg (1-15); Poultry farms (Tolima): Paratyphi B (16-39); Human: Newport (40), Enteritidis (41-43), Braenderup (44), Uganda (45), Enteritidis (46), Typhimurium (47), Grupensis (48), Typhimurium (49)
Genotypic resistance
The gene blaCMY2 that confers resistance to CRO was detected in all the strains; however, some Salmonella spp. strains isolated from human subjects did not exhibit phenotypic resistance to this antibiotic (Table-2). Furthermore, in 93.9% of the strains that demonstrated the sul2 gene and in 69.4% of the strains showing the dfrA1 gene, these two genes conferred resistance to SXT. Moreover, other genes (sul1, sul3, dfrA10, and dfrA12) were evaluated, and it was observed that some strains contained one to three genes that can confer resistance to this antibiotic. However, some strains isolated from human subjects did not demonstrate phenotypic resistance. In addition, 69.4% of the strains demonstrated the presence of the genes blaPSE-1 and blaTEM that conferred resistance to AM, and 87.8% of the strains showed the presence of the genes aadA1 and strB that conferred resistance to STR. The gene aadB was not detected in the strains; however, the Salmonella spp. strains isolated from human subjects were phenotypically resistant to GM. Furthermore, the S. Typhimurium strain that was phenotypically resistant to amphenicols demonstrated the presence of the gene floR.
ERIC-PCR
Genotyping of Salmonella Heidelberg strains (n=15) isolated from the broiler farms in Santander region using the ERIC primers did not generate band patterns. However, in 34 of the 49 Salmonella spp. strains isolated from the broiler farms in Tolima and from human subjects with gastroenteritis, the molecular typing method ERIC generated 2-13 bands ranging in size from 200 to 4000 bp. The dendrogram revealed that the ERIC-PCR method could discriminate the strains according to their source of isolation from the broiler farms in Tolima and from human subjects with gastroenteritis. This typing method grouped the strains into six clusters at a Dice coefficient similarity of 90%, with a discriminatory index of 0.69 (Figure-1). The majority of isolates of the serotype S. Paratyphi B were grouped into cluster 6, and the four strains of the serotype S. Enteritidis were grouped into two clusters 1 and 2 (Figure-1). Furthermore, the strains grouped in each cluster had variable antibiotic resistance profiles.
Figure-1.
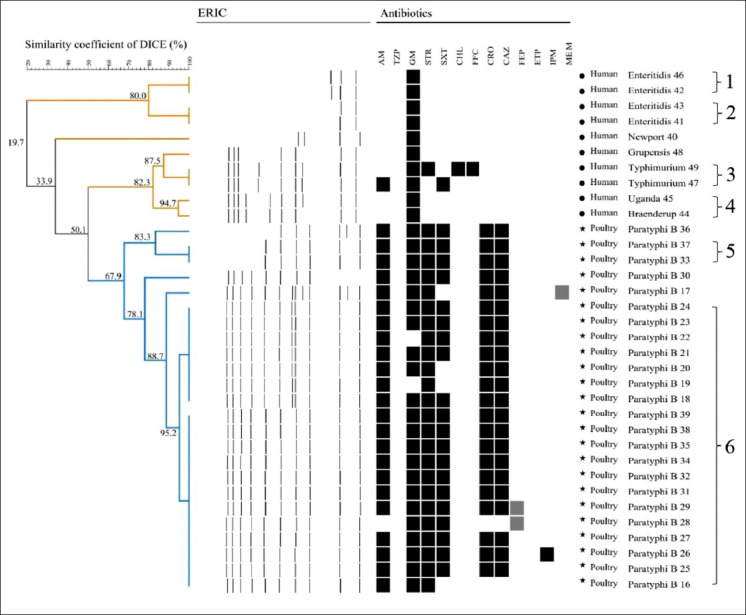
Dendrogram generated from enterobacterial repetitive intergenic consensus-PCR of 34 strains showing source, the serotype of Salmonella strains and phenotypic resistance of the strains. AM=Ampicillin, TZP=Piperacillin/tazobactam, GM=Gentamicin, STR=Streptomycin, SXT=Trimethoprim/sulfamethoxazole, CHL=Chloramphenicol, FFC=Florfenicol, CRO=Ceftriaxone, CAZ=Ceftazidime, FEP=Cefepime, ETP=Ertapenem, IPM=Imipenem, MEM=Meropenem, Black=Resistant, White=Susceptible, Gray=Intermediate.
Discussion
In the present study, the strains corresponding to the isolates from broiler farms in Santander and Tolima regions were serotyped as Salmonella Heidelberg (n=15) and Salmonella Paratyphi B (n=24), respectively. S. Heidelberg represents a significant concern as it has been associated with food-borne infections in humans due to the consumption of poultry products [17]. Moreover, a high prevalence in chicken meat may indicate the risk associated with this product as a potential vehicle for the transmission of this bacterium [18]. The strains isolated from human subjects with gastroenteritis corresponded generally to Salmonella Enteritidis and Salmonella Typhimurium, which are the major serotypes that cause diseases in humans and have been reported previously from clinical cases in Colombia during the period 2005-2011 [19,20].
The results of the phenotypic resistance indicated that the Salmonella spp. strains isolated from broiler farms in Santander and Tolima could be categorized as resistant to MDR, that is, bacteria exhibiting resistance to one or more antibiotics from three or more classes of antibiotics [21]. These bacteria were resistant to β-lactams, aminoglycosides, and cephalosporins. The previous studies have reported that Salmonella spp. strains isolated from animal products were MDR strains in Colombia [22] and Brazil [23]. In the present study, we detected a high percentage of Salmonella strains that were phenotypically resistant to STR (79.5%); these findings are consistent with the previous studies that reported that strains isolated from broiler farms in Cundinamarca (Colombia) [22] and in the United States [24] exhibited a high percentage of resistance to STR. Regarding the resistance to AM (79.5%), previously studies from Santander [22] and Brazil [25] have reported high resistance rates. Moreover, 75.5% of the strains were found to be resistant to CRO and CAZ, which are higher than the results reported from Cuba (CRO, 10.7%; CAZ, 17.9%) [26], Cundinamarca (CRO, 0%; CAZ, 18.2%), and Santander (CRO, 4.5%; CAZ, 69.7%) in Colombia [22]. Resistance to third-generation cephalosporins exhibited by the strains isolated from broiler farms represents a concern because CRO and CAZ are the antibiotics used for salmonellosis treatment in adults and specifically in children, thus rendering the transmission of resistant bacteria a public health problem [26].
The strains isolated from human subjects with gastroenteritis were resistant to GM, which is one of the major antibiotics used in the treatment of urinary infections in humans [27]. On the other hand, all the Salmonella spp. strains were susceptible to ETP, which is similar to the result reported by Donado et al. [22]. Carbapenems are the final choice of antibiotics used in the treatment of salmonellosis when the bacteria exhibit resistance to antibiotics such as ciprofloxacin and CRO [28].
In the present study, the genotypic analyses revealed the presence of the gene blaCMY2 in all the strains. This gene encodes an extended-spectrum beta-lactamase that is responsible for hydrolyzing the β-lactam ring preventing it from combining with the penicillin-binding protein (PBP) [29]. This gene confers resistance to AM, ceftiofur, and cefoxitin and is associated with mobile elements, thereby increasing the probability of transmission between bacteria [30]. In our study, 69.4% of the strains demonstrated the presence of the genes blaPSE-1 and blaTEM that encode beta-lactamases that confer resistance to AM. In a study conducted in Ibagué (Colombia), 100% of the strains isolated from chicken carcasses had the gene blaTEM [31], a frequency that was higher than that found in the present study. Some strains that were phenotypically resistant to AM and CRO showed the presence of the genes blaPSE-1, blaTEM, blaCMY2, and blaCTX-M. However, in the case of Salmonella Heidelberg isolates, these strains were found to be phenotypically resistant to AM, but none of the genes evaluated in this study were found. It is possible that this resistance is mediated by other mechanisms, such as alterations in the target sites of beta-lactams (PBP) [32] or excessive expression of efflux pumps [33]. Regarding the strains isolated from human subjects and broiler farms in Tolima that had the gene but were phenotypically susceptible; Álvarez [34] mentioned that the hydrolysis generated by these ESBLs in the antibiotic differs between strains, although there is some hydrolysis performed by these enzymes, this is not sufficient to provide clinical resistance in the bacteria [35]. In addition, the gene sul2 was found in 93.9% of the strains; this gene encodes DHPS (dihydropteroate synthase) that has no affinity for the antibiotic [36]. Different results were found in strains isolated from chicken carcasses in Portugal (37%) [37]. In a previous study, the gene sul1 was reported to be the most prevalent [37], whereas in the present study, it was found in 57.1% of the strains. In contrast, trimethoprim resistance is mediated by the expression of the enzyme DHFR (dihydrofolate reductase) that has no affinity for the antibiotic and is encoded by the dfrA1 gene [38] that was detected in 69.4% of the strains. In general, the strains that were resistant to SXT showed the sul and/or dfrA resistance genes. However, in the case of strains isolated from human subjects, the sul2, dfrA1, and dfrA12 genes were present, but they were phenotypically susceptible, which may be due to the inactivation of these genes, as previously reported [39].
The STR resistance gene aadA1 was detected in 87.8% of the strains; this gene encodes a nucleotidyltransferase that catalyzes the transfer of a nucleotide monophosphate to a hydroxyl group in STR [40]. This result is higher than those reported in Salmonella strains isolated in Iran (45.6%) [41] and Thailand (17%) [42]. On the other hand, the gene strB also confers resistance to STR and encodes a phosphotransferase that catalyzes the ATP-dependent phosphorylation of hydroxyl groups in STR [43]. The strB gene was found in 87.8% of the strains, and similar results have been reported in Salmonella strains isolated from chicken carcasses in Ibagué [31]. Regarding the strains isolated from human subjects, it was observed that the strains contained three of four genes that conferred resistance to STR, whereas some strains were not resistant to this antibiotic. In a previous research, White et al. [44] reported Salmonella strains isolated from meat that had the aadA2 gene but were susceptible to STR, as the gene was silenced.
The aadB gene that confers resistance to GM was not found in any of the strains in the present study. However, the strains isolated from human subjects were resistant to this antibiotic. This resistance may be mediated by other resistance genes such as grm [45], which was not evaluated in this study. In contrast, the S. Typhimurium serotype isolated from human subjects that were phenotypically resistant to amphenicols did not present the genes catA and cmlA, which confer resistance to these antibiotics. However, this strain had the floR gene, which might explain the resistance of this strain. This gene encodes an efflux pump that confers resistance to amphenicols, which has been reported in the genomic island of Salmonella (SGI1) [46,47]. Earlier, this gene was reported in 90% of chloramphenicol-resistant strains isolated from clinical cases in Saudi Arabia [48].
The genotyping method ERIC-PCR is a technique that allows the typing of different Salmonella serotypes such as Typhimurium, Paratyphi B, Heidelberg, Pullorum, and Gallinarum [9,49-51]. However, in the present study, we could not type the Salmonella Heidelberg strains isolated from broiler farms in Santander using the ERIC-PCR method. Previously, Wilson and Sharp [52] described that mutations in the ERIC region as well as deletions of 60-75 bp can interfere with the site of primer annealing and hinder the generation of band patterns.
This molecular typing method generated 2-13 bands ranging in size from 200 to 4000 bp, which is different from the results reported by Fendri et al. [53] (150-1500 bp) and Oliveira et al. [7] (190-1430 bp). Moreover, the discriminatory index of this molecular typing method was 0.69, which is similar to that reported by Nath et al. [54] who found a discrimination index of 0.70 but lower than those reported by Fendri et al. [53] and Kim and Lee [55] who found discriminatory indexes of 0.85 and 0.97, respectively. However, in the present study, the ERIC-PCR method was able to discriminate the strains according to their source of isolation, and a greater discriminatory power does not always represent a grouping with biological significance [56].
The serotype Paratyphi B was grouped into cluster 6, but some strains were also not grouped in a cluster; these results are in agreement with those reported by Kim and Lee [55] who described that ERIC-PCR can be used to sub-typify within the serotype. In addition, S. Enteritidis strains were grouped into two clusters; this is possible due to the fact that S. Enteritidis strains could have divergent clonal lineages so that they can be grouped into a different cluster [49]. Finally, in the present study, no relationship was found between the clusters and the antibiotic resistance. Kim and Lee [55] and Oliveira et al. [7] also reported similar results in which the clusters did not exhibit the same pattern of antibiotic resistance.
Conclusion
Salmonella spp. isolated from broiler farms in Santander and Tolima were MDR strains and demonstrated the presence of resistance genes associated with such resistance. Furthermore, ERIC-PCR is a technique that allowed obtaining clusters with biological significance, although this genotyping method could not type the Heidelberg strains.
Authors’ Contributions
ISR and MPH were responsible for the design of the study. MPH performed the experiments and laboratory analyses. ISR administered the project. RR collected the strains. ISR and MPH wrote the manuscript. ISR, MPH and RR reviewed and edited the paper. ISRB revised the manuscript critically. All authors read and approved the final manuscript.
Acknowledgments
Financial support, facilities and reagents were provided by the Laboratory of Immunology and Molecular Biology – LIBM of the University of Tolima. The authors did not receive any funds for this study.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Gad A.H, Abo-Shama U.H, Harclerode K.K, Fakhr M.K. Prevalence, serotyping, molecular typing, and antimicrobial resistance of Salmonella isolated from conventional and organic retail ground poultry. Front. Microbiol. 2018;9:2653. doi: 10.3389/fmicb.2018.02653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu W, Zeng L. Current and emerging innovations for detection of food-borne Salmonella. In:Current Topics in Salmonella and Salmonellosis. Books on Demand, Rijeka, Croatia. 2017. Available from: https://www.intechopen.com/books/current-topics-in-salmonella-and-salmonellosis/current-and-emerging-innovations-for-detection-of-food-borne-salmonella? Retrieved on 20-01-2019.
- 3.Jackson B.R, Griffin P.M, Cole D, Walsh K.A, Chai S.J. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998-2008. Emerg. Infect. Dis. 2013;19(8):1239–1244. doi: 10.3201/eid1908.121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pérez C.C, Cardozo S.M. Reports of outbreaks and isolates of Salmonella sp. n Colombia. Cult. Cient. 2014;12:74–83. [Google Scholar]
- 5.Kapoor G.S, Saigal S, Elongavan A. Action and resistance mechanisms of antibiotics:A guide for clinicians. J Anaesthesiol. Clin. Pharmacol. 2017;33(3):300–305. doi: 10.4103/joacp.JOACP_349_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall B.M, Levy S.B. Food animals and antimicrobials:Impacts on human health. Clin. Microbiol. Rev. 2011;24(4):718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira S.D, Bessa M.C, Santos L.R, Cardoso M.R, Brandelli A, Canal C.W. Phenotypic and genotypic characterization of Salmonella Enteritidis isolates. Braz. J. Microbiol. 2007;38(4):720–728. [Google Scholar]
- 8.Bilung L.M, Pui C.F, Su'ut L, Apun K. Evaluation of BOX-PCR and ERIC-PCR as molecular typing tools for pathogenic. Leptospira. Dis. Markers. 2018;2018 doi: 10.1155/2018/1351634. Article ID 1351634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Versalovic J.T, Koeuth T, Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Souza A.I, Freitas-Neto O, Batista D, Estupinan A, Almeida A, Barrow P.A, Berchieri A. ERIC-PCR genotyping of field isolates of Salmonella enterica subspenterica (2015) serovar Gallinarum biovars Gallinarum and Pullorum. Avian Pathol. 44(6):475–479. doi: 10.1080/03079457.2015.1086975. [DOI] [PubMed] [Google Scholar]
- 11.Fandiño-de-Rubio L.C, Verjan N. A common sequence type of Salmonella Enteritidis of avian and human origin with gastroenteritis in Ibagué, Colombia. Biomédica. 2019;39(1):50–62. doi: 10.7705/biomedica.v39i1.4155. [DOI] [PubMed] [Google Scholar]
- 12.Castro R, Fandiño-de-Rubio L.C, Veja A, Rondon I.S. Phenotypic and genotypic resistance of Salmonella Heidelberg isolated from one of the largest poultry production region from Colombia. Int. J. Poult. Sci. 2019;18(12):610–617. [Google Scholar]
- 13.Grimont P.A, Weill F.X. White-Kauffmann-Le Minor scheme. In:Antigenic Formulae of the Salmonella Serovars WHO Collaborating Center for Reference and Research on. Salmonella Paris, France. 2007. Available from: https://www.pasteur.fr/sites/default/files/veng_0.pdf . Retrieved on 02?-12-2018.
- 14.Clinical and Laboratory Standards Institute. M100 Performance Standards for Antimicrobial Susceptibility. 27th ed. Pennsylvania (USA): CLSI; 2017. [Google Scholar]
- 15.Dice L.R. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 16.Hunter P.R, Gaston M.A. Numerical index of the discriminatory ability of typing systems:An application of Simpson's index of diversity. J. Clin. Microbiol. 1988;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.St-Amand J.A, Otto S.J, Cassis R, Annett-Christianson C.B. Antimicrobial resistance of Salmonella enterica serovar Heidelberg isolated from poultry in Alberta. Avian Pathol. 2013;42(4):379–386. doi: 10.1080/03079457.2013.811465. [DOI] [PubMed] [Google Scholar]
- 18.Donado P, Clavijo V, Leon M, Arevalo A, Castellanos R, Bernal J, Tafur M, Ovalle M, Alali Q, Hume M. Counts, serovars, and antimicrobial resistance phenotypes of Salmonella on raw chicken meat at retail in Colombia. J. Food Prot. 2014;77(2):227–235. doi: 10.4315/0362-028X.JFP-13-276. [DOI] [PubMed] [Google Scholar]
- 19.Rodríguez E.C, Díaz-Guevara P, Moreno J, Bautista A, Montaño L, Realpe M.E, della-Gaspera A, Wiesner M. Laboratory surveillance of Salmonella enterica from human clinical cases in Colombia 2005-2011. Enferm. Infecc. Microbiol. Clin. 2017;35(7):417–425. doi: 10.1016/j.eimc.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 20.Stevens M, Humphrey T.J, Maskell D.J. Molecular insights into farm animal and zoonotic Salmonella infections. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364(1530):2709–2723. doi: 10.1098/rstb.2009.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pintado V. Old and new drugs in the treatment of infection by multidrug-resistant bacteria. Rev. Esp. Quimioter. 2016;29(1):39–42. [PubMed] [Google Scholar]
- 22.Donado P, Gardner I, Byrne B.A, Leon M, Perez E, Ovalle M, Tafur M, Miller W. Prevalence, risk factors, and antimicrobial resistance profiles of Salmonella from commercial broiler farms in two important poultry-producing regions of Colombia. J. Food Prot. 2012;75(5):874–883. doi: 10.4315/0362-028X.JFP-11-458. [DOI] [PubMed] [Google Scholar]
- 23.Das-Neves G.B, Stefani L.M, Pick E, Araujo D, Giuriatti J, Percio C, Brisola M.C. Salmonella Heidelberg isolated from poultry shows a novel resistance profile. Acta Sci. Vet. 2016;44(1):1–6. [Google Scholar]
- 24.Liljebjelke K.A, Hofacre C.L, White D.G, Ayers S, Lee M.D, Maurer J.J. Diversity of antimicrobial resistance phenotypes in Salmonella isolated from commercial poultry farms. Front. Vet. Sci. 2017;4:96. doi: 10.3389/fvets.2017.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medeiros M.A, Oliveira D, Rodrigues D, Freitas D. Prevalence and antimicrobial resistance of Salmonella in chicken carcasses at retail in 15 Brazilian cities. Rev. Panam. Salud Publica. 2011;30(6):555–560. doi: 10.1590/s1020-49892011001200010. [DOI] [PubMed] [Google Scholar]
- 26.Rivera M, Granda A.E, Felipe L, Bonachea H. Antimicrobial resistance in Salmonella enterica subsp enterica isolated in imported poultry. Rev. Salud Anim. 2012;34(2):120–126. [Google Scholar]
- 27.Goodlet K.J, Benhalima F.Z, Nailor M.D. A systematic review of single-dose aminoglycoside therapy for urinary tract infection:Is it time to resurrect an old strategy? Antimicrob. Agents Chemother. 2019;63(1):e02165–e02218. doi: 10.1128/AAC.02165-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calayag A.M, Paclibare P, Santos P, Bautista C, Rivera W. Molecular characterization and antimicrobial resistance of Salmonella enterica from swine slaughtered in two different types of Philippine abattoir. Food Microbiol. 2017;65:51–56. doi: 10.1016/j.fm.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Morejón M. Extended spectrum betalactamases. Rev. Cubana Med. 2013;52(4):272–280. [Google Scholar]
- 30.Oladeinde A, Cook K, Lakin S.M, Woyda R, Abdo Z, Looft T, Herrington K, Zock G, Plumblee J, Thomas J, Beaudry M, Glenne T. Horizontal gene transfer and acquired antibiotic resistance in Salmonella enterica serovar Heidelberg following in vitro incubation in broiler ceca. Appl. Environ. Microbiol. 2019;85(22):e01903–e01919. doi: 10.1128/AEM.01903-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vélez D.C, Rodríguez V, Verjan N. Phenotypic and genotypic antibiotic resistance of Salmonella from chicken carcasses marketed at Ibague, Colombia. Braz. J. Poult. Sci. 2017;19(2):347–354. [Google Scholar]
- 32.Sun S, Selmer M, Andersson D. Resistance to ?-lactam antibiotics conferred by point mutations in penicillin-binding proteins PBP3, PBP4 and PBP6 in Salmonella enterica. PLoS One. 2014;9(5):e97202. doi: 10.1371/journal.pone.0097202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piddock L.J. Multidrug-resistance efflux pumps?Not just for resistance. Nat. Rev. Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 34.Álvarez D. Identification of extended spectrum beta-lactamases in Enterobacteriaceae. Rev. Haban. Cienc. Méd. 2010;9(4):516–524. [Google Scholar]
- 35.Shaikh S, Fatima J, Shakil S, Rizvi S, Kamal M. Antibiotic resistance and extended-spectrum beta-lactamases:Types, epidemiology and treatment. Saudi J. Biol. Sci. 2015;22(1):90–101. doi: 10.1016/j.sjbs.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mąka Ł, Maćkiw E, Ścieżyńska H, Modzelewska M, Popowska M. Resistance to sulfonamides and dissemination of Sul genes among Salmonella spp. isolated from food in Poland. Foodborne Pathog. Dis. 2015;12(5):383–389. doi: 10.1089/fpd.2014.1825. [DOI] [PubMed] [Google Scholar]
- 37.Antunes P, Machado J, Sousa J, Peixe L. Dissemination of sulfonamide resistance genes (sul1, sul2 and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 2005;49(2):836–839. doi: 10.1128/AAC.49.2.836-839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alcaine S.D, Warnick L.D, Wiedmann M. Antimicrobial resistance in nontyphoidal Salmonellaa. J. Food Prot. 2007;70(3):780–790. doi: 10.4315/0362-028x-70.3.780. [DOI] [PubMed] [Google Scholar]
- 39.Olonitola O.S, Fahrenfeld N, Pruden A. Antibiotic resistance profiles among mesophilic aerobic bacteria in Nigerian chicken litter and associated antibiotic resistance genes. Poult. Sci. 2015;94(5):867–874. doi: 10.3382/ps/pev069. [DOI] [PubMed] [Google Scholar]
- 40.Vakulenko S.B, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 2003;16(3):430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doosti A, Mahmoudi E, Jami M.S, Mokhtari A. Prevalence of aadA1, aadA2, aadB, strA and strB genes and their associations with multidrug resistance phenotype in Salmonella Typhimurium isolated from poultry carcasses. Thai J. Vet. Med. 2016;46(4):691–697. [Google Scholar]
- 42.Chuanchuen R, Padungtod P. Antimicrobial resistance genes in Salmonella enterica isolates from poultry and swine in Thailand. J. Vet. Med. Sci. 2009;71(10):1349–1355. doi: 10.1292/jvms.001349. [DOI] [PubMed] [Google Scholar]
- 43.Pezzella C, Ricci A, DiGiannatale E, Luzzi I, Carattoli A. Tetracycline and streptomycin resistance genes, transposons, and plasmids in Salmonella enterica isolates from animals in Italy. Antimicrob. Agents Chemother. 2004;48(3):903–908. doi: 10.1128/AAC.48.3.903-908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White P.A, McIver C, Rawlinson W.D. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 2001;45(9):2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gebreyes W.A, Altier C. Molecular characterization of multidrug-resistant Salmonella enterica subsp enterica serovar Typhimurium isolates from swine. J. Clin. Microbiol. 2002;40(8):2813–2822. doi: 10.1128/JCM.40.8.2813-2822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004;28(5):519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Doublet B, Schwarz S, Kehrenberg C, Cloeckaert A. Florfenicol resistance gene floR is part of a novel transposon. Antimicrob. Agents Chemother. 2005;49(5):2106–2108. doi: 10.1128/AAC.49.5.2106-2108.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Tayeb M, Ibrahim A.S, Al-Salamah A.A, Almaary K.S, Elbadawi Y.B. Prevalence, serotyping and antimicrobials resistance mechanism of Salmonella enterica isolated from clinical and environmental samples in Saudi Arabia. Braz. J. Microbiol. 2017;48(3):499–508. doi: 10.1016/j.bjm.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson J.R, Clabots C, Azar M, Boxrud D.J, Besser J.M, Thurn J.R. Molecular analysis of a hospital cafeteria-associated salmonellosis outbreak using modified repetitive element PCR fingerprinting. J. Clin. Microbiol. 2001;39(10):3452–3460. doi: 10.1128/JCM.39.10.3452-3460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasschaert G, Houf K, Imberechts H, Grijspeerdt K, de Zutter L, Heyndrickx M. Comparison of five repetitive-sequence-based PCR typing methods for molecular discrimination of Salmonella enterica isolates. J. Clin. Microbiol. 2005;43(8):3615–3623. doi: 10.1128/JCM.43.8.3615-3623.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashemi A, Baghbani-Arani F. The effective differentiation of Salmonella isolates using four PCR-based typing methods. J. Appl. Microbiol. 2015;118(6):1530–1540. doi: 10.1111/jam.12805. [DOI] [PubMed] [Google Scholar]
- 52.Wilson L.A, Sharp P.M. Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli:Evolution and implications for ERIC-PCR. Mol. Biol. Evol. 2006;23(6):1156–1168. doi: 10.1093/molbev/msj125. [DOI] [PubMed] [Google Scholar]
- 53.Fendri I, Hassena A.B, Grosset N, Barkallah M, Khannous L, Chuat V, Gautier M, Gdoura R. Genetic diversity of food-isolated Salmonella strains through pulsed-field gel electrophoresis (PFGE) and enterobacterial repetitive intergenic consensus (ERIC-PCR) PLoS One. 2013;8(12):e81315. doi: 10.1371/journal.pone.0081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nath G.P, Maurya P, Gulati A.K. ERIC PCR and RAPD based fingerprinting of Salmonella Typhi strains isolated over a period of two decades. Infect. Genet. Evol. 2010;10(4):530–536. doi: 10.1016/j.meegid.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Kim J.E, Lee Y. Molecular characterization of antimicrobial-resistant non-typhoidal Salmonella from poultry industries in Korea. Ir. Vet. J. 2017;70:20. doi: 10.1186/s13620-017-0095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoke C.K, Teck-Ee K, Son R, Yoshitsugu N, Mitsuaki N. Molecular characterisation of Vibrio parahaemolyticus carrying tdh and trhg enes using ERIC-, RAPD-and BOX-PCR on local Malaysia bloody clam and lala. Int. Food Res. J. 2013;20(6):3299–3305. [Google Scholar]


