Supplemental Digital Content is available in the text.
Key Words: TIGIT, HCC, CD8+ T cell, immunosuppressive effect, immunotherapy
Abstract
The efficacy of adoptive cellular immunotherapy against cancer cells is limited due to the presence of immunosuppressive cells within the solid tumor microenvironment. The upregulation of certain coinhibitory receptors may lead to exhaustion of the immune effector cells. T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain (TIGIT) is an immune inhibitory receptor expressed by regulatory T cells and activated T cells and natural killer cells. The aim of this study was to determine the immunosuppressive effects of CD155/TIGIT signaling on CD8+ T cells of adoptive cellular immunotherapy in hepatocellular carcinoma (HCC). Our studies found that CD155 was overexpressed in HCC, and CD155hi HCC cells upregulated TIGIT on CD8+ T cells, which decreased the secretion of interferon-γ, tumor necrosis factor-α, and interleukin-17A and increased that of interleukin-10 from the effector cells. However, TIGIT blockade or CD155-knockdown reversed the inhibitory effect of HCC cells on CD8+ T-cell effector function. These results indicate that TIGIT can exert an immunosuppressive effect on CD8 T cells by modulating cytokine production through CD155, and is a promising target to optimize adoptive cellular immunotherapy against HCC.
Primary liver cancer is the sixth most commonly diagnosed malignancy, and the second most common cause of cancer-related deaths worldwide. In fact, the number of deaths recorded among liver cancer patients (745,500) every year is almost equal to that of the newly diagnosed cases (782,500).1 Hepatocellular carcinoma (HCC) is a prevalent primary liver cancer with poor long-term prognosis and high frequency of recurrence, despite therapeutic options such as surgery, transarterial chemoembolization, radiofrequency ablation, etc.2,3 Thus, it is imperative to develop novel strategies to improve survival of HCC patients, especially those in the advanced stages.
In recent years, plenty of studies have shown that the clinical benefits of immune-based therapies for HCC and other malignant diseases are emerging. Efforts to mobilize the body’s immune system to treat cancer, collectively known as cancer immunotherapy, have garnered the most attention so far, on the basis of new mechanisms and technology development in this area.4 Cancer immunotherapy is based on the activation and recruitment of antitumor immune cells.5–7 Adoptive cellular immunotherapy, a type of adjuvant therapy, induces a durable response against tumor cells8,9 through cytokine-induced killer (CIK) cells, tumor-infiltrating lymphocytes (TILs), expanding activated autologous lymphocytes, central memory T cells, and chimeric antigen receptor T cells, or natural killer (NK) cells. However, its therapeutic efficacy is limited within the solid tumor microenvironment10 due to the upregulation of immune coinhibitory receptors, such as cytotoxic T-lymphocyte–associated protein 4 (CTLA-4), lymphocyte activation gene-3, T-cell immunoglobulin-3, programmed cell death 1 (PD-1), and T-cell immunoglobulin (Ig) and immunoreceptor tyrosine-based inhibitory motif domain (TIGIT), which lead to exhaustion of the infused effector cells.11,12
TIGIT was first identified by Yu et al13 as a coinhibitory receptor consisting of an Ig variable domain, a transmembrane domain, and an immunoglobulin tail tyrosine (ITT)-like phosphorylation motif followed by an immunoreceptor tyrosine-based inhibitory motif domain. It is primarily expressed by activated and regulatory T cells (Tregs), as well as NK cells.13 The adhesion molecule CD155, also known as poliovirus receptor (PVR) or NECL5, is a high-affinity ligand for TIGIT binding. It is highly expressed on monocytes, dendritic cells (DCs), fibroblasts, endothelial cells, epithelial cells, platelets, and activated T and B cells.13–17 CD112 (nectin-2, also known as PRR2 or PVRL2), on the other hand, is a low-affinity ligand for TIGIT that is also expressed on monocytes, DCs, endothelial cells, hematopoietic cells, and activated T and B cells.18–20 Both CD155 and CD112 also bind to the costimulatory counterparts to TIGIT, such as CD226 (DNAM-1) or CD96, which act in concert with lymphocyte function-associated antigen 1 to activate T-cell responses.17,21 It is interesting to note that, CD155 and CD112 are also expressed by multiple solid tumors, including colorectal carcinoma,22 melanoma,23,24 ovarian cancer,25 etc.
Studies indicate that TIGIT exerts its immunosuppressive effects by enhancing interleukin (IL)-10 production by DCs via CD155, which impedes CD4+ T-cell proliferation and function.13 In addition, TIGIT also inhibits CD4+ T cells by recruiting SHP phosphatases that suppress cytokine production and proliferation,26,27 and competes with CD226 for CD155.28 The TIGIT locus is demethylated in Tregs and may potentially bind to FOXP3.29 In fact, TIGIT+ Tregs are highly active and secrete fibrinogen-like protein 2, which selectively inhibits Th1 and Th17 responses.30 Recent gene expression analyses have detected TIGIT in the CD8+ TILs in several solid tumors, including lung, colon, breast, uterine, and renal cancers.31 Furthermore, elevated TIGIT expression in these TILs correlates with CD8 and PD-1 levels, and their immune function.31 However, the significance of TIGIT+ TILs in adoptive cellular immunotherapy is not completely clear. We found that TIGIT was upregulated in the HCC-targeting CD8+ T cells and exerted an immunosuppressive effect by binding to CD155.
MATERIALS AND METHODS
Cell Lines and Culture
The human HCC cell lines SNU423, Hep3B, and Bel-7402 were obtained from China Type Culture Collection (Shanghai, China). SNU423 and Bel-7402 cells were cultured in RPMI-1640 medium (Gibco) supplemented with 10% fetal bovine serum and 100 U/mL penicillin/streptomycin at 37°C under 5% CO2. The Hep3B cells were cultured in complete MEM (Gibco) under the same conditions.
Isolation of Peripheral Blood Mononuclear Cells (PBMCs)
Blood samples were collected from HCC patients and healthy volunteers. The PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation within 2 hours of sample collection. The immune cells were depleted by positive selection using EasySep Human CD45 Depletion Kit (STEMCELL Technologies Inc., Vancouver, BC, Canada).
Immunohistochemistry (IHC) and Immunofluorescence (IF)
Formalin-fixed, paraffin-embedded HCC and paratumor tissues were dehydrated, dewaxed, and rehydrated. For IHC, endogenous peroxidase activity was first blocked with 0.3% hydrogen peroxide in methanol, followed by antigen retrieval in boiling 10 mM citrate buffer (pH 6.0) for 10 minutes. After washing with phosphate buffer saline 3 times, the tissue sections were then incubated overnight with primary antibody, washed, and probed with a horseradish peroxidase–conjugated secondary antibody. The tissues were similarly processed for IF and probed with Alexa-Fluor 488-conjugated or Alexa-Fluor 555-conjugated secondary antibodies (1:1000; Thermo Scientific). An Ig control antibody conjugated to fluorophore: Alexa-Fluor 488 and Alexa-Fluor 555 were shown using adjacent tissue slides from the same fixed tissue block (Fig. S2, Supplemental Digital Content 1, http://links.lww.com/JIT/A560). The stained sections were visualized using a confocal laser scanning microscope.
CD8+ T-Cell Activation and Expansion
PBMCs were isolated from healthy volunteers as described, and T cells were isolated by immunomagnetic negative selection using EasySep Human T-Cell Isolation Kit (STEMCELL Technologies Inc.). The purified T cells were seeded into 25 cm2 flasks at a density of 1×106 cells/mL in 5 mL X-vivo 15 medium supplemented with 10% autologous serum, 100 IU/mL IL-2 and 100 U/mL penicillin/streptomycin, and cultured at 37°C under 5% CO2. The cells were activated by 25 µL/mL ImmunoCult Human CD3/CD28 T-Cell Activator (STEMCELL Technologies Inc.) for 3 days, and activation of variable CD3+ T cells was assessed in terms of surface expression of CD25 using flow cytometry. To expand the T cells, the cell density was adjusted to 1×106 cells/mL with the addition of fresh medium every 2–3 days. For long-term expansion, the cells were harvested and resuspended every 7–10 days in fresh medium and restimulated with ImmunoCult Human CD3/CD28 T-Cell Activator, in addition to adjusting the cell density every 2–3 days, as described. The CD8+ T cells were obtained by immunomagnetic positive selection using EasySep human CD8+ T-cell Isolation Kit (STEMCELL Technologies Inc.), and purity was verified to be >95%.
Flow Cytometry
Primary or cultured CD8+ T cells were stained with FITC-CD8, PE/cy7-TIGIT, and PE-CD25 antibodies, and the HCC cell lines with PE-CD155 antibody (Biolegend, San Diego, CA), as per standard protocols. The stained samples were analyzed by fluorescence-activated cell sorting and the data analyzed using FlowJo software. The gating strategies used in this study are shown in Figure S3 (Supplemental Digital Content 1, http://links.lww.com/JIT/A560). Isotype controls were used to set the gate.
Western Blotting
Total proteins were extracted from the cultured cells and analyzed by Western blotting with antibodies against TIGIT, β-actin, CD226, interferon (IFN)-γ, AKT, p-AKT, ERK, p-ERK, and p-IκBα (CST, Danvers, MA), as per standard protocols. When necessary, the gray value of the band is obtained by the Image J software; the gray value of the target protein is divided by the gray value of the internal reference protein, and then normalized for comparison.
Cytokine Measurement
The cytokines released by the CD8+ T cells were measured using the LEGENDplex Multianalyze Flow Assay Kit (Biolegend) according to the manufacturer’s instructions.
Cell Coculture
The activated CD8+ T cells were sorted and cocultured with HCC cell lines either in regular culture plates or in transwells (Fig. S1, Supplemental Digital Content 1, http://links.lww.com/JIT/A560). In the transwell coculture format, the HCC cells were seeded in the bottom chambers and the effector CD8+ T cells in the lower chamber of the inserts. The functional anti-human TIGIT antibody (clone MBSA43, mouse IgG1; eBioscience, San Diego, CA) and recombinant human CD155/Fc protein (Sino Biological Inc., Beijing, China) were added to the culture medium, as appropriate. The cells were incubated for 2 days at 37°C under 5% CO2, harvested, and analyzed by Western blotting or flow cytometry. The culture supernatants were also collected and evaluated for cytokine levels, as described.
CD155 Knockout and Lentivirus Transduction
CRISPR/Cas9 was used for CD155 knockout. The sgRNA’s sequence (sgRNA-1 5′-AACTGGCATGGCCCGAGCCA-3′) was cloned into pLV vectors. HEK293T cells were cotransfected with pLV vector, pSPAX2, and pMD2.G plasmids, and the lentiviruses were precipitated using PEG8000-NaCl. The HCC cells were transduced with the lentivirus and selected 48 hours later using 3 μg/mL puromycin (MCE). Single-cell–derived clones were isolated through flow cytometric sorting, and CD155-knockdown (KD) was confirmed.
Statistical Analysis
A standard 2-tailed Student t test was used for comparing groups, and P-value<0.05 was considered statistically significant.
RESULTS
CD155 Was Overexpressed in HCC Tissues
The in situ expression of CD155 was significantly higher in the HCC tissues compared with the paired paratumor tissues by IHC (Fig. 1A) and in HCC cell lines by IF (Fig. 1B). Consistent with this, the CD155 positivity rates were 99.5%, 99.5%, and 99.1%, respectively, in the SNU423, Hep3B, and Bel-7402 cell lines (Fig. 1C).
FIGURE 1.
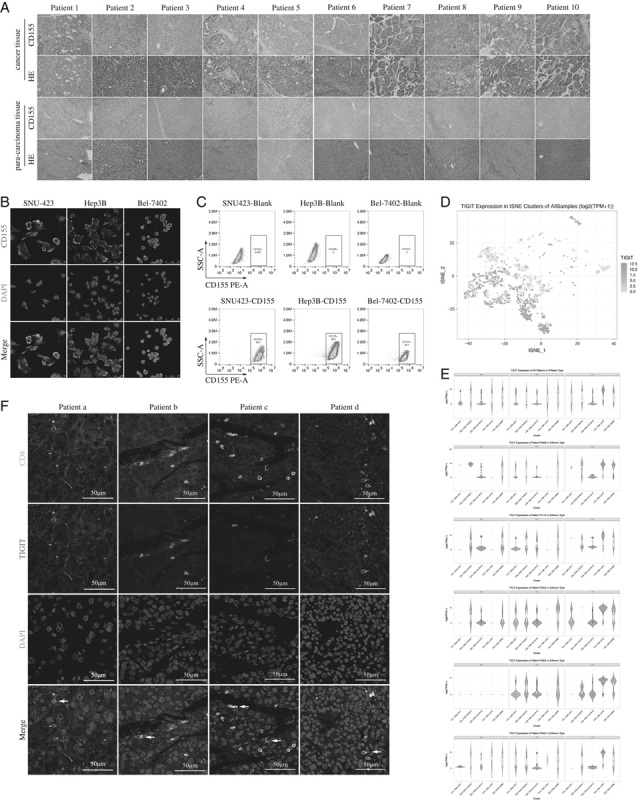
A, Representative immunohistochemical images showing in situ CD155 expression in hepatocellular carcinoma sections from 10 patients. B, Representative immunofluorescence images showing CD155 expression in SNU423, Hep3B, and Bel-7402 cells. C, Flow cytometry plots showing CD155 expression in SNU423, Hep3B, and Bel-7402 cells. D, The 2-dimensional visualization of CD8 T-cell clusters of 5 patients by t-SNE. Each dot corresponds to a single cell. The depth of each dot color indicates the level of T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain (TIGIT) expression. E, The violin plots showing the level of TIGIT expression in 5 CD8 T-cell clusters of all and individual patients. F, Representative immunofluorescence images showing the TIGIT+ CD8+ T cells (white arrows) in hepatocellular carcinoma sections.
TIGIT Levels Were Higher in Activated CD8+ T Cells
Analysis of previously published single-cell sequencing GEO data of HCC immune cells32 revealed variations in TIGIT expression levels on CD8+ T cells from 5 HCC patients (Fig. 1D). Furthermore, the CD8+ TILs showed higher TIGIT expression compared with the CD8+ T cells in peripheral blood and adjacent tissues (Fig. 1E), and most TIGIT+ CD8+ TILs were distributed in the fourth C4_CD8-LAYN cluster that was predominantly composed of TILs expressing high levels of exhaustion markers such as CTLA-4, PDCD1, and HAVCR2. Consistent with these findings, the TILs in the HCC tissues of our cohort coexpressed CD8 and TIGIT (Fig. 1F). In addition, the percentage of TIGIT+ CD8+ T cells in the PBMCs was also significantly elevated following activation (Fig. 2A), as well as upon coculturing with the SNU423, Bel-7402, and Hep3B cells (Figs. 2B–D). Taken together, the CD155hi HCC cells upregulated TIGIT on CD8+ T cells.
FIGURE 2.
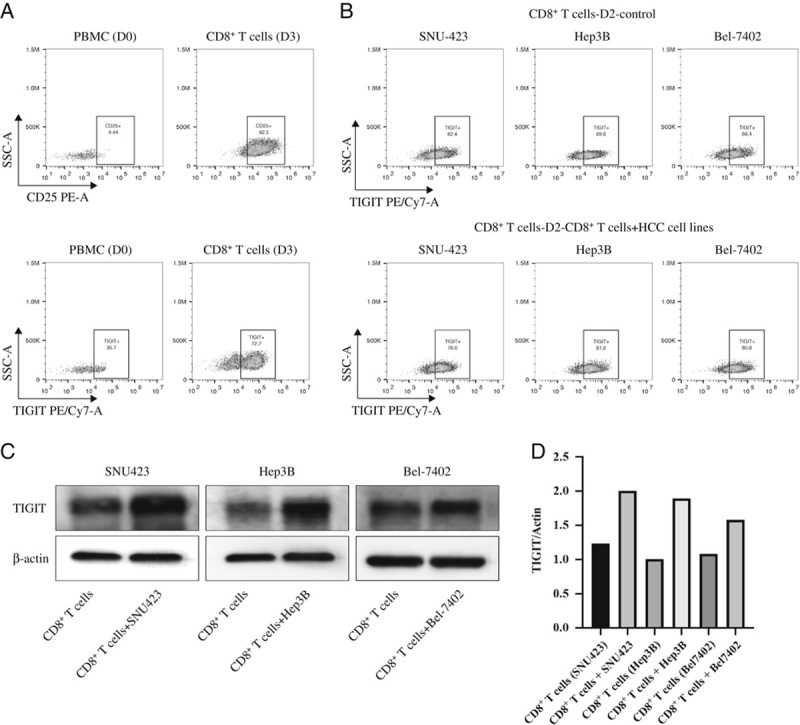
A, Flow cytometry plots showing peripheral blood TIGIT+ CD8 T cells stimulated with αCD3/CD28 for 3 days. B, Flow cytometry plots showing TIGIT+ CD8+ T cells following 3-day αCD3/CD28 stimulation and 2-day coculture with SNU423, Hep3B, and Bel-7402 cells. C, Representative immunoblots showing TIGIT expression levels in the CD8+ T cells treated as above. D, The gray value of the target protein is divided by the gray value of the internal reference protein, and then normalized for comparison. PBMC indicates peripheral blood mononuclear cell; TIGIT, T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain.
HCC Cells Inhibited CD8+ T-Cell Effector Function Via CD155/TIGIT Signaling
To determine whether the CD155/TIGIT signaling pathway inhibited CD8+ T-cell effector function, we treated them with recombinant human CD155. The cells cultured with CD155 secreted significantly less amount of IFN-γ, tumor necrosis factor (TNF)-α, and IL-17A, and higher levels of IL-10 compared with the unstimulated CD8+ T cells. Furthermore, blocking TIGIT reversed the secreted cytokine profile of T cells (Fig. 3A), indicating that the CD155/TIGIT axis relays inhibitory signals to these effector cells. Similarly, the CD8+ T cells cocultured with the CD155hi Bel-7402 cells (Figs. 1B, C) also released significantly lower amounts of IFN-γ, TNF-α, IL-17A, perforin, granzyme B and granulysin, and higher levels of IL-10 compared with the control cells (Fig. 3B). Antibody-mediated neutralization of TIGIT increased the production of effector cytokines and cytotoxic factors and decreased that of IL-10 in the cocultured CD8+ T cells (Fig. 3B). To further validate the immunosuppressive function of CD155, we generated a stable CD155-KD Bel-7402 cell line (Bel-7402-CD155 RNAi) using siRNA (Fig. 3C). CD8+ T cells cocultured with CD155hi Bel-7402 cells secreted lower levels of IFN-γ, TNF-α, IL-17A, perforin, and granulysin, and higher IL-10 compared with the control group with Bel-7402-CD155 RNAi (Fig. 3D), which was neutralized by blocking TIGIT (Fig. 3D). Taken together, CD155-expressing HCC cells inhibited CD8+ T-cell effector function through the CD155/TIGIT pathway.
FIGURE 3.
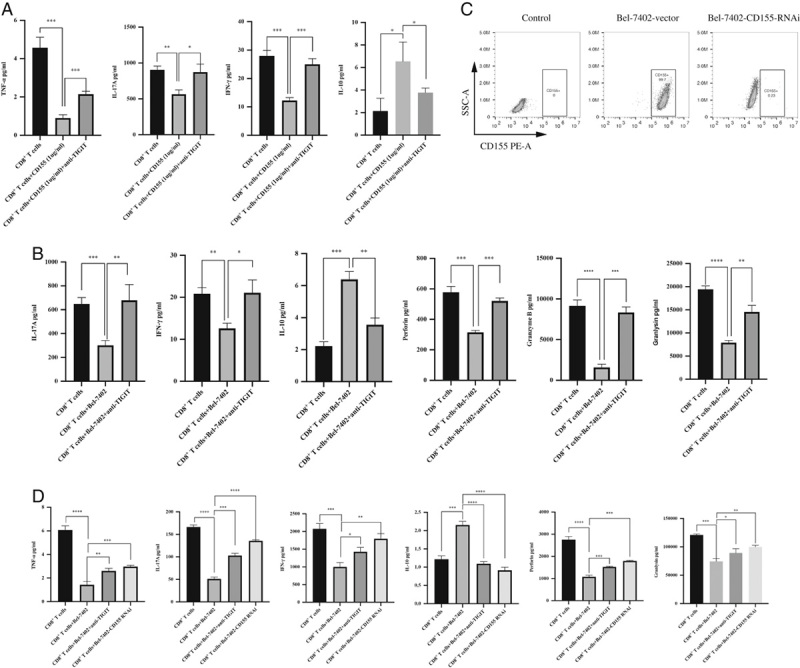
A, Amount of TNF-α, IL-17A, IFN-γ, and IL-10 secreted by CD8+ T cells cultured with 1 µg/mL CD155 with/out anti-TIGIT antibody. B, Amount of IL-17A, IFN-γ, IL-10, perforin, granzyme B, and granlysin produced by stimulated CD8+ T cells cocultured with Bel-7402 with/out anti-TIGIT antibody. C, Flow cytometry plots showing CD155 expression in Bel-7402-vector or Bel-7402-CD155 RNAi cells. D, Amount of TNF-α, IL-17A, IFN-γ, IL-10, perforin, and granlysin produced by stimulated CD8+ T cells cocultured with Bel-7402-vector or Bel-7402-CD155 RNAi cells with/out anti-TIGIT antibody. *P<0.05; **P<0.01; ***P<0.001, ****P<0.0001. IFN indicates interferon; IL, interleukin; TIGIT, T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain; TNF, tumor necrosis factor.
Mechanism of CD155/TIGIT Signaling Pathway
Studies have implicated phosphoinositide 3-kinase (PI3K), mitogen-activated protein kinase (MAPK) and nuclear factor-κB (NF-κB) signaling pathways in TIGIT/CD155-mediated immunosuppressive effects on NK cells33 (Fig. 4A). To this end, we analyzed the expression levels of related proteins in the CD8+ T cells cocultured with SNU423 and Bel-7402 cells. Blocking TIGIT during the coculture of T cells and wild-type HCC cells significantly increased the ratio of p-AKT/AKT and p-ERK/ERK and decreased p-IκBα levels (Figs. 4B, C). In addition, the p-AKT/AKT and p-ERK/ERK ratios were significantly lower and p-IκBα levels were higher in the CD8+ T cells cocultured with wild-type HCC cells compared with CD155 KD, which was reversed by blocking TIGIT (Fig. 4D). Taken together, the TIGIT/CD155 interaction suppresses effector functions of CD8+ T cells by inhibiting multiple signaling pathways.
FIGURE 4.
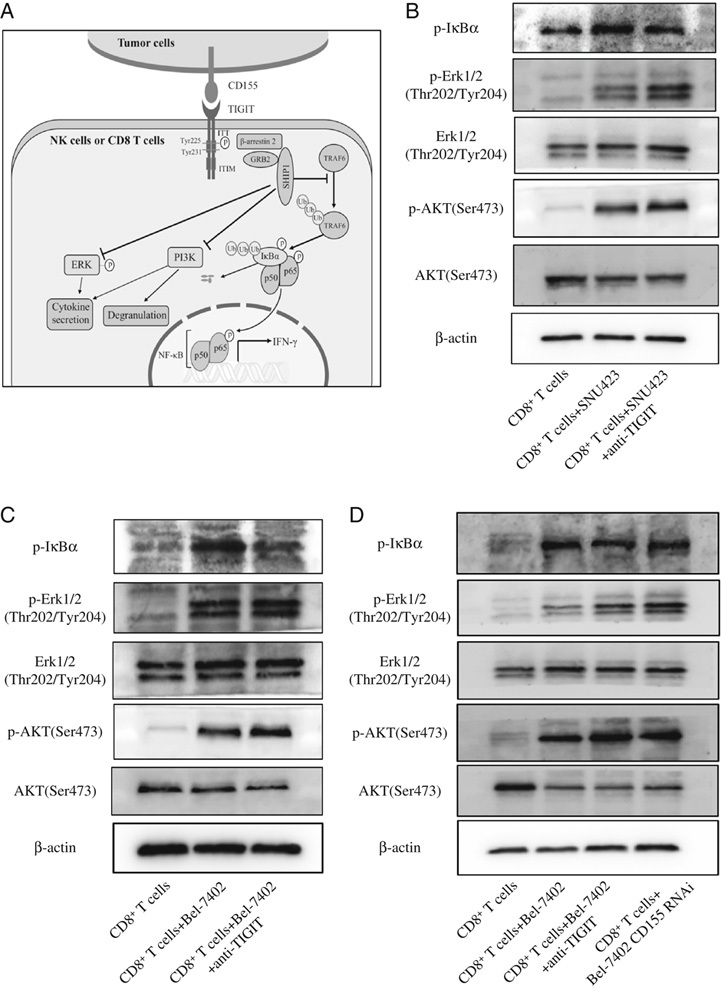
A, Mechanism of T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain (TIGIT)/CD155-mediated suppression effects in natural killer cells. Upon interaction with CD155, the immunoglobulin tail tyrosine (ITT)-like motif of TIGIT is phosphorylated on Tyr225 and binds the cytosolic adaptor growth factor receptor-bound protein 2 (GRB2), which can recruit SH2 domain–containing inositol-5-phosphatase 1 (SHIP1) to inhibit phosphoinositide 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) signaling. In addition, phosphorylated TIGIT recruits SHIP1 through β-arrestin 2 and impairs nuclear factor-κB (NF-κB) activation by blocking tumor necrosis factor receptor–associated factor 6 (TRAF6) autoubiquitylation. B–D, Representative immunoblot showing AKT, p-AKT, ERK, p-ERK, and p-IκBα expression in stimulated T cells cocultured with SNU423, Bel-7402, and Bel-7402-CD155 RNAi cells with/out anti-TIGIT antibody. ERK indicates extracellular-regulated kinase; IFN, interferon; NK, natural killer.
DISCUSSION
Adoptive cellular immunotherapy using NK cells, CIK cells or TILs, expanding activated autologous lymphocytes, central memory T cells, and chimeric antigen receptor T cells has gained considerable attention in recent years as a strategy against solid tumors. However, these cells function suboptimally within the tumor microenvironment10 on account of various immune inhibitory receptors such as PD-1, CTLA-1, T-cell immunoglobulin-3, and lymphocyte activation gene-3 that impair antitumor effects.10
TIGIT is a newly identified inhibitory receptor that is expressed on effector CD8+ T cells, the major effector cell type in antitumor immune responses and adoptive cellular immunotherapy. In addition, its ligand CD155 is overexpressed on human colon cancer,22 glioma,34 pancreatic cancer,35 and lung cancer36 cells. Not surprisingly, therefore, the TIGIT/CD155 pathway inhibits the tumor-infiltrating CIK, NK, and CD4+ T cells.37–39 However, CD155 expression in HCC cells has not been elucidated so far, and the role of CD155/TIGIT signaling on the CD8+ T cells infiltrating HCC tumors has also not been clearly defined. We found that CD155 was overexpressed in HCC compared with adjacent liver tissues, as well as in HCC cell lines such as SNU423, Hep3B, and Bel-7402. In addition, the HCC tissues had a high density of TIGIT and CD8 coexpressing T cells. This is consistent with the findings of Zhang et al32 who reported significantly higher proportion of TIGIT+ CD8+ T cells in the HCC tumors compared with that in paracarcinoma tissues and peripheral blood. In addition, most of these TIGIT+ CD8+ TILs were distributed in the C4_CD8-LAYN cluster, which predominantly consisted of cells expressing high levels of exhaustion markers such as CTLA-4, PDCD1, and HAVCR2. Therefore, the CD155/TIGIT signaling pathway likely plays an important role in immunosuppression in the HCC tumors.
It is interesting to note that, TIGIT levels were significantly upregulated on CD8+ T cells cocultured with HCC cells, indicating that the latter suppresses the tumor-infiltrating immune cells in an exocrine manner. This is consistent with previous studies showing that TIGIT can be rapidly induced by antigenic and other inflammatory stimuli.13,27,28,31 Furthermore, the immunosuppressive effects of HCC cells and recombinant CD155 on CD8+ T cells—measured in terms of the levels of effector cytokines released by the latter—was reversed after blocking TIGIT. Consistent with this, the CD155-KD HCC cells also failed to suppress the effector function of the cocultured CD8+ T cells. Studies indicate that TIGIT binding to CD155 induces phosphorylation at Tyr225 in the ITT-like motif and that it recruits SH2 domain–containing inositol-5-phosphatase 1 (SHIP1) through the cytosolic adaptor growth factor receptor-bound protein 2.40,41 TIGIT-mediated recruitment of SHIP1 inhibits NK cells by blocking the PI3K and MAPK signaling pathways. In addition, the ITT-like motif of phosphorylated TIGIT binds to β-arrestin 2 and recruits SHIP1 to limit NF-κB signaling.41 Consistent with this, we found that CD155/TIGIT blocked the PI3K, MAPK, and NF-κB signaling pathways in CD8+ T cells.
To summarize, we have shown, for the first time, that CD155 is overexpressed on HCC cells, and the latter upregulates TIGIT on the CD8+ TILs. TIGIT/CD155 binding then relays inhibitory signals into the effector cells by blocking multiple signaling pathways. Thus, tumors overexpressing CD155 likely escape the host immune response by upregulating TIGIT on the TILs, and inhibiting this blockade may improve the efficacy of adoptive cellular immunotherapy against HCC. In fact, the combination of targeted anticancer therapy and immune checkpoint inhibition has shown better clinical outcomes in HCC.9,42 Furthermore, a recent study reported coexpression of TIGIT and PD-1 in melanoma TILs.43 These data strongly support dual TIGIT and PD-1 blockade to optimize CD8+ T-cell-driven adoptive cellular immunotherapy in HCC, and will need further experimental and clinical validation.
Supplementary Material
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.immunotherapy-journal.com.
Acknowledgments
CONFLICTS OF INTEREST/FINANCIAL DISCLOSURES
Supported by the UMHS-PUHSC Joint Institute for Translational and Clinical Research (BMU2017JI006), National Natural Science Foundation of China (81570590, 81872508) and Project (RDY2018-03) supported by Peking University People’s Hospital Research and Development Funds. All authors have declared that there are no financial conflicts of interest with regard to this work.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.Poon D, Anderson BO, Chen LT, et al. Management of hepatocellular carcinoma in Asia: consensus statement from the Asian Oncology Summit 2009. Lancet Oncol. 2009;10:1111–1118. [DOI] [PubMed] [Google Scholar]
- 3.Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. [DOI] [PubMed] [Google Scholar]
- 4.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. [DOI] [PubMed] [Google Scholar]
- 5.Kean LS, Turka LA, Blazar BR. Advances in targeting co-inhibitory and co-stimulatory pathways in transplantation settings: the Yin to the Yang of cancer immunotherapy. Immunol Rev. 2017;276:192–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng X, Nakamura Y. Cancer precision medicine: from cancer screening to drug selection and personalized immunotherapy. Trends Pharmacol Sci. 2017;38:15–24. [DOI] [PubMed] [Google Scholar]
- 7.Barreira da Silva R, Laird ME, Yatim N, et al. Dipeptidylpeptidase 4 inhibition enhances lymphocyte trafficking, improving both naturally occurring tumor immunity and immunotherapy. Nat Immunol. 2015;16:850–858. [DOI] [PubMed] [Google Scholar]
- 8.Takayama T, Sekine T, Kondo Y, et al. Adjuvant adoptive immunotherapy against hepatocellular carcinoma. Hepatology. 1998;28:1436–1437. [DOI] [PubMed] [Google Scholar]
- 9.Fearon D. Combination immunotherapy for cancer. J Exp Med. 2016;213:1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon EK, Wang LC, Dolfi DV, et al. Multifactorial T-cell hypofunction that is reversible can limit the efficacy of chimeric antigen receptor-transduced human T cells in solid tumors. Clin Cancer Res. 2014;20:4262–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park HJ, Kusnadi A, Lee EJ, et al. Tumor-infiltrating regulatory T cells delineated by upregulation of PD-1 and inhibitory receptors. Cell Immunol. 2012;278:76–83. [DOI] [PubMed] [Google Scholar]
- 12.Abate-Daga D, Hanada K, Davis JL, et al. Expression profiling of TCR-engineered T cells demonstrates overexpression of multiple inhibitory receptors in persisting lymphocytes. Blood. 2013;122:1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Harden K, Gonzalez LC, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10:48–57. [DOI] [PubMed] [Google Scholar]
- 14.Kamran N, Takai Y, Miyoshi J, et al. Toll-like receptor ligands induce expression of the costimulatory molecule CD155 on antigen-presenting cells. PLoS One. 2013;8:e54406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reymond N, Imbert AM, Devilard E, et al. DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J Exp Med. 2004;199:1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stengel KF, Harden-Bowles K, Yu X, et al. Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering. Proc Natl Acad Sci USA. 2012;109:5399–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibuya K, Shirakawa J, Kameyama T, et al. CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J Exp Med. 2003;198:1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ardolino M, Zingoni A, Cerboni C, et al. DNAM-1 ligand expression on Ag-stimulated T lymphocytes is mediated by ROS-dependent activation of DNA-damage response: relevance for NK-T cell interaction. Blood. 2011;117:4778–4786. [DOI] [PubMed] [Google Scholar]
- 19.Lopez M, Aoubala M, Jordier F, et al. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood. 1998;92:4602–4611. [PubMed] [Google Scholar]
- 20.Pende D, Castriconi R, Romagnani P, et al. Expression of the DNAM-1 ligands, Nectin-2 (CD112) and poliovirus receptor (CD155), on dendritic cells: relevance for natural killer-dendritic cell interaction. Blood. 2006;107:2030–2036. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs A, Cella M, Giurisato E, et al. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J Immunol. 2004;172:3994–3998. [DOI] [PubMed] [Google Scholar]
- 22.Masson D, Jarry A, Baury B, et al. Overexpression of the CD155 gene in human colorectal carcinoma. Gut. 2001;49:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bevelacqua V, Bevelacqua Y, Candido S, et al. Nectin like-5 overexpression correlates with the malignant phenotype in cutaneous melanoma. Oncotarget. 2012;3:882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casado JG, Pawelec G, Morgado S, et al. Expression of adhesion molecules and ligands for activating and costimulatory receptors involved in cell-mediated cytotoxicity in a large panel of human melanoma cell lines. Cancer Immunol Immunother. 2009;58:1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oshima T, Sato S, Kato J, et al. Nectin-2 is a potential target for antibody therapy of breast and ovarian cancers. Mol Cancer. 2013;12:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joller N, Hafler JP, Brynedal B, et al. Cutting Edge: TIGIT has T cell-intrinsic inhibitory functions. J Immunol. 2011;186:1338–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin SD, Taft DW, Brandt CS, et al. Vstm3 is a member of the CD28 family and an important modulator of T-cell function. Eur J Immunol. 2011;41:902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozano E, Dominguez-Villar M, Kuchroo V, et al. The TIGIT/CD226 axis regulates human T cell function. J Immunol. 2012;188:3869–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Maksimovic J, Naselli G, et al. Genome-wide DNA methylation analysis identifies hypomethylated genes regulated by FOXP3 in human regulatory T cells. Blood. 2013;122:2823–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joller N, Lozano E, Burkett PR, et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity. 2014;40:569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston RJ, Comps-Agrar L, Hackney J, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923–937. [DOI] [PubMed] [Google Scholar]
- 32.Zheng C, Zheng L, Yoo JK, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342.e16–1356.e16. [DOI] [PubMed] [Google Scholar]
- 33.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15:243–254. [DOI] [PubMed] [Google Scholar]
- 34.Gromeier M, Lachmann S, Rosenfeld MR, et al. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc Natl Acad Sci USA. 2000;97:6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishiwada S, Sho M, Yasuda S, et al. Clinical significance of CD155 expression in human pancreatic cancer. Anticancer Res. 2015;35:2287–2297. [PubMed] [Google Scholar]
- 36.Nakai R, Maniwa Y, Tanaka Y, et al. Overexpression of Necl-5 correlates with unfavorable prognosis in patients with lung adenocarcinoma. Cancer Sci. 2010;101:1326–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Zhao W, Li H, et al. Immunoreceptor TIGIT inhibits the cytotoxicity of human cytokine-induced killer cells by interacting with CD155. Cancer Immunol Immunother. 2016;65:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu F, Sunderland A, Zhou Y, et al. Blockade of CD112R and TIGIT signaling sensitizes human natural killer cell functions. Cancer Immunol Immunother. 2017;66:1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang FF, Wang Y, Wang L, et al. TIGIT expression levels on CD4+ T cells are correlated with disease severity in patients with psoriasis. Clin Exp Dermatol. 2018;43:675–682. [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Zhang H, Li M, et al. Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ. 2013;20:456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Xia P, Du Y, et al. T-cell immunoglobulin and ITIM domain (TIGIT) receptor/poliovirus receptor (PVR) ligand engagement suppresses interferon-gamma production of natural killer cells via beta-arrestin 2-mediated negative signaling. J Biol Chem. 2014;289:17647–17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes PE, Caenepeel S, Wu LC. Targeted therapy and checkpoint immunotherapy combinations for the treatment of cancer. Trends Immunol. 2016;37:462–476. [DOI] [PubMed] [Google Scholar]
- 43.Chauvin J-M, Pagliano O, Fourcade J, et al. TIGIT and PD-1 impair tumor antigen–specific CD8+ T cells in melanoma patients. J Clin Investig. 2015;125:2046–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.immunotherapy-journal.com.


