Abstract
In bacteria, transcription and translation are coupled processes, in which movement of RNA polymerase (RNAP) synthesizing mRNA is coordinated with movement of the first ribosome translating mRNA. Coupling is modulated by the transcription factors NusG--which is thought to bridge RNAP and ribosome--and NusA. Here, we report cryo-EM structures of Escherichia coli transcription-translation complexes (TTCs) containing different-length mRNA spacers between RNAP and the ribosome active-center P-site. Structures of TTCs containing short spacers show a state incompatible with NusG bridging and NusA binding (TTC-A; previously termed “expressome”). Structures of TTCs containing longer spacers reveal a new state compatible with NusG bridging and NusA binding (TTC-B) and reveal how NusG bridges and NusA binds. We propose that TTC-B mediates NusG- and NusA-dependent transcription-translation coupling.
One Sentence Summary:
Cryo-EM defines states that mediate NusG- and NusA-dependent transcription-translation coupling in bacteria
Bacterial transcription and bacterial translation occur in the same cellular compartment, occur at the same time, and are coordinated processes, in which the rate of transcription by the RNA polymerase (RNAP) molecule synthesizing an mRNA is coordinated with the rate of translation by the first ribosome (“lead ribosome”) translating the mRNA (1–9; see, however, 10). Data indicate that the coordination is mediated by transcription elongation factors of the NusG/RfaH family, which contain an N-terminal domain (N) that interacts with RNAP β’ and β subunits and a flexibly tethered C-terminal domain (C) that interacts with ribosomal protein S10, and which are thought to bridge, and thereby connect, the RNAP molecule and the lead ribosome (2, 5–9). Further data indicate that the coordination is modulated by the transcription elongation factor NusA (11).
Cramer and co-workers recently reported a 7.6 Å resolution cryo-EM structure of an Escherichia coli transcription-translation complex (TTC) termed the “expressome,” obtained by halting a transcription elongation complex (TEC) and allowing a translating ribosome to collide with the halted TEC (12). However, the mRNA molecule in the structure was not fully resolved, precluding determination of the number of mRNA nucleotides between the TEC and the ribosome active center in the structure (12), and the functional relevance of the structure has been challenged, due to its genesis as a collision complex, and due to its incompatibility with simultaneous interaction of NusG-N with RNAP and NusG-C with the ribosome (6–9). Demo et al. recently reported a ~7 Å resolution cryo-EM structure of a complex of E. coli RNAP and a ribosome 30S subunit (13). However, the structure did not contain mRNA, did not position RNAP close to the 30S mRNA-entrance portal, and was incompatible with simultaneous interaction of NusG-N with RNAP and NusG-C with the ribosome (13).
Here, we report cryo-EM structures of E. coli TTCs containing defined-length mRNA spacers between the TEC and the ribosome active-center product site (P site), both in the presence of NusG and in the absence of NusG (Figs. 1, S1–S5; Tables S1–S2). We prepared synthetic nucleic-acid scaffolds that contained (i) DNA and mRNA determinants that direct formation of a TEC upon interaction with RNAP, (ii ) an mRNA AUG codon that enables formation of a translation complex having the AUG codon positioned in the ribosome active-center P site upon interaction with a ribosome and tRNAfMet, and (iii) an mRNA spacer having a length, n, of 4, 5, 6, 7, 8, 9, or 10 codons (12, 15, 18, 21, 24, 27, or 30 nt) between (i) and (ii) (Fig. 1A). We then incubated the nucleic-acid scaffolds with RNAP, with ribosome and tRNAfMet, and optionally with NusG and/or NusA, and we determined structures by single-particle-reconstruction cryo-EM (see Supporting Information, Materials and Methods). With nucleic-acid scaffolds having short spacers (n = 4, 5, 6, 7, or 8), we obtained structures matching the “expressome” of 12 (TTC-A; Figs. 1B-left, 2, S1–S3; Table S1). However, with nucleic-acid scaffolds having longer mRNA spacers (n = 8, 9, or 10), we obtained structures of a new molecular assembly with features strongly suggesting it is the molecular assembly that functionally mediates NusG-dependent, NusA-dependent transcription-translation coupling in cells (TTC-B; Figs. 1B-center, 1B-right, 3–4, S3–S8; Table S1; Movies S1–S2).
Fig. 1. Structure determination: transcription-translation complexes (TTCs).
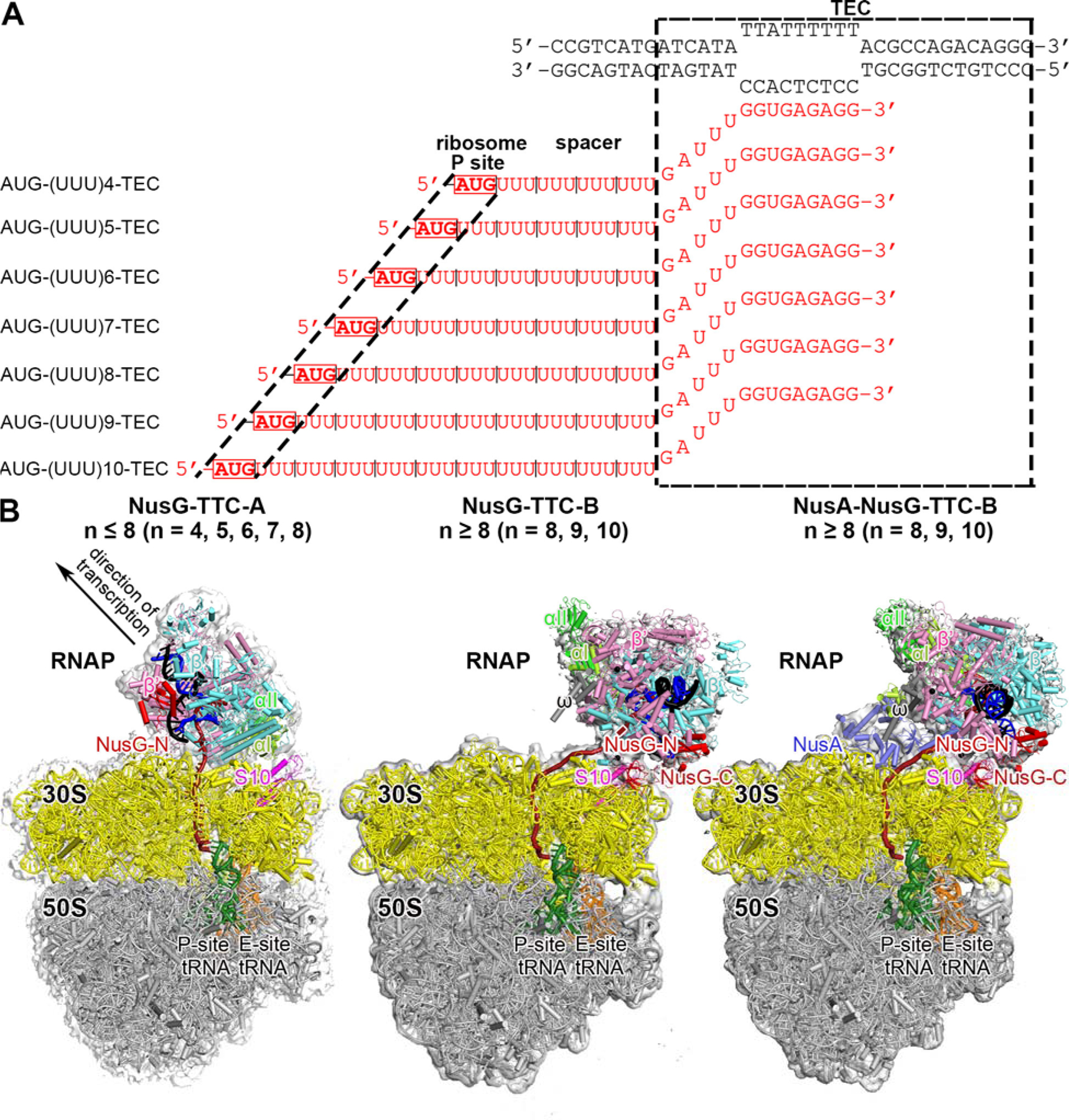
(A) Nucleic-acid scaffolds. Each scaffold comprises nontemplate- and template-strand oligodeoxyribonucleotides (black) and one of seven oligoribonucleotides having spacer length n of 4, 5, 6, 7, 8, 9, or 10 codons (red), corresponding to mRNA. Dashed black box labeled “TEC,” portion of nucleic-acid scaffold that forms TEC upon addition of RNAP (10 nt nontemplate- and template-strand ssDNA segments forming “transcription bubble,” 10 nt of mRNA engaged with template-strand DNA as RNA-DNA “hybrid,” and 5 nt of mRNA, on diagonal, in RNAP RNA-exit channel); dashed black lines labeled “ribosome P-site,” mRNA AUG codon intended to occupy ribosome active-center P site upon addition of ribosome and tRNAfMet; “spacer,” mRNA spacer between TEC and AUG codon in ribosome active-center P site.
(B) Cryo-EM structures of NusG-TTC-A (obtained with spacer lengths of 4–8 codons), NusG-TTC-B (obtained with spacer lengths of 8–10 codons), and NusA-NusG-TTC-B (obtained with spacer lengths of 8–10 codons). Structures shown are NusG-TTC-A (3.7 Å; n = 4; Table S1), NusG-TTC-B (4.7 Å; n = 9; Table S1), and NusA-NusG-TTC-B2 (3.5 Å; n = 8; Table S1). Images show EM density (gray surface) and fit (ribbons) for TEC, NusG and NusA (at top; direction of transcription, defined by downstream dsDNA, indicated by arrow in left panel and directly toward viewer in center and right panels) and for ribosome 30S and 50S subunits and P- and E-site tRNAs (at bottom). RNAP β’, β, αI, αII, and ω subunits are in pink, cyan, light green, and dark green, and gray; 30S subunit, 50S subunit, P-site tRNA, E-site tRNA are in yellow, gray, green, and orange; DNA nontemplate strand, DNA template strand, and mRNA are in black, blue, and brick-red (brick-red dashed line where modelled). NusG, NusA, and ribosomal protein S10 are in red, light blue, and magenta. Ribosome L7/L12 stalk omitted for clarity in this and all subsequent images.
Fig. 2. Cryo-EM structure of NusG-TTC-A.
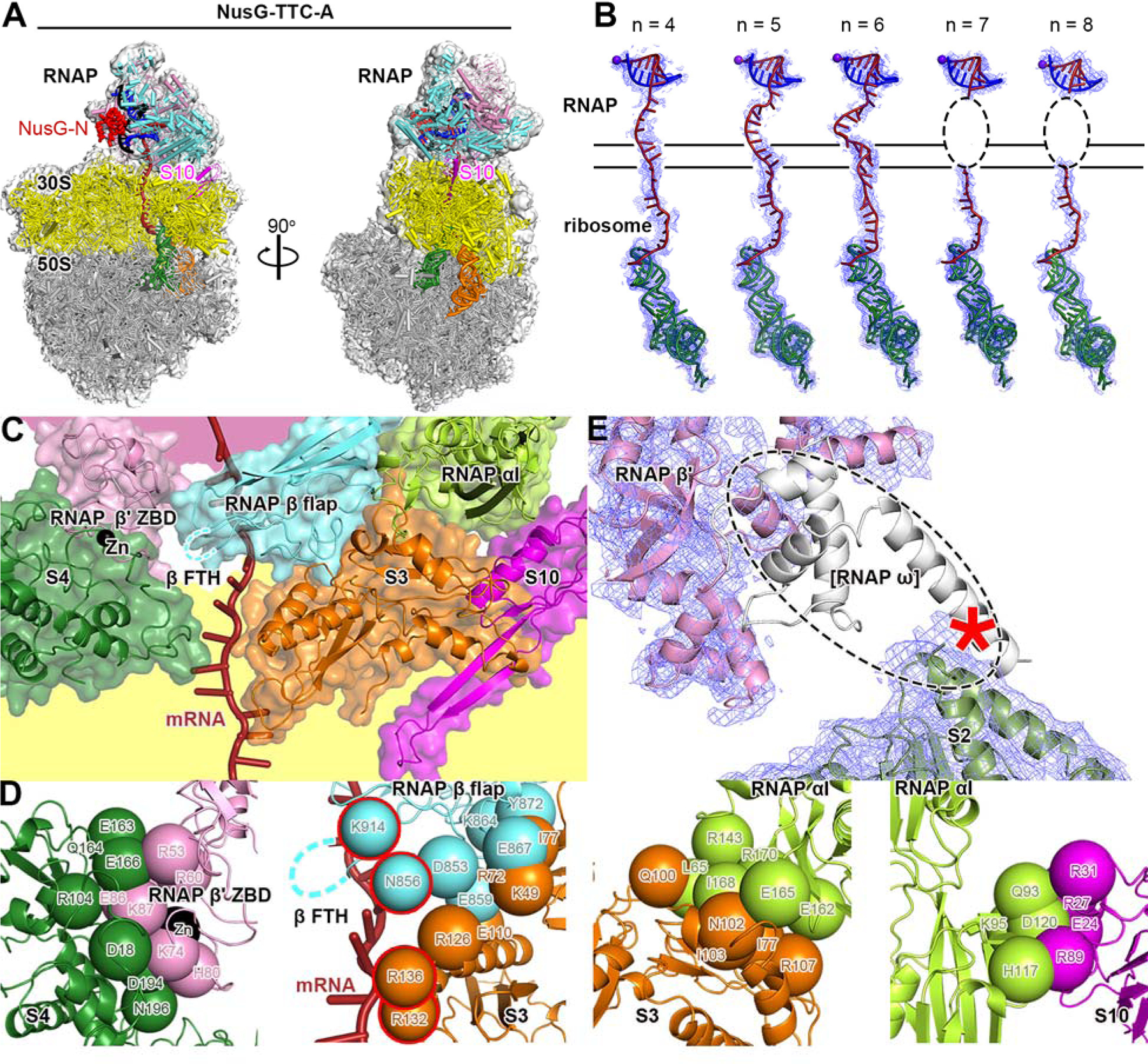
(A) Structure of NusG-TTC-A (3.7 Å; n = 4; Table S1). Two orthogonal views. Colors as in Fig. 1B.
(B) Accommodation of mRNA spacer lengths of 4, 5, 6, 7 and 8 codons in NusG-TTC-A. EM density, blue mesh; mRNA, brick-red (disordered mRNA nucleotides indicated by dashed oval); template-strand DNA in RNA-DNA hybrid, blue; RNAP active-center catalytic Mg2+, purple sphere; tRNA in ribosome P site, green. Upper and lower black horizontal lines indicate edges of RNAP and ribosome.
(C) RNAP-ribosome interface in NusG-TTC-A (n = 4; identical interface for n = 5, 6, 7, or 8), showing RNAP β’ zinc binding domain, (ZBD, pink; Zn2+ ion as black sphere), RNAP β flap, cyan, RNAP β flap tip helix (β FTH; disordered residues indicated by cyan dashed line), and RNAP αI (green) interacting with ribosomal proteins S4 (forest green), S3 (orange), and S10 (magenta) and with mRNA (brick red). Portions of RNAP β’ and ribosome 30S not involved in interactions are shaded pink and yellow, respectively.
(D) RNAP-ribosome interactions involving RNAP β’ ZBD and S4 (subpanel 1), RNAP β flap and S3 (subpanel 2; β FTH, dashed line; β and S3 residues that interact with mRNA, cyan and orange spheres with red outlines; mRNA, brick-red), RNAP αI and S3 (subpanel 3), and RNAP αI and S10 (subpanel 4). Other colors as in (C).
(E) Absence of EM density for RNAP ω subunit. EM density, blue mesh; atomic models for RNAP β’ and S2, pink ribbon and forest-green ribbon, respectively; location of missing EM density for ω, dashed oval; ω in TEC in absence of ribosome (PDB 6P19; 17), white ribbon.
Fig. 3. Cryo-EM structure of NusG-TTC-B.
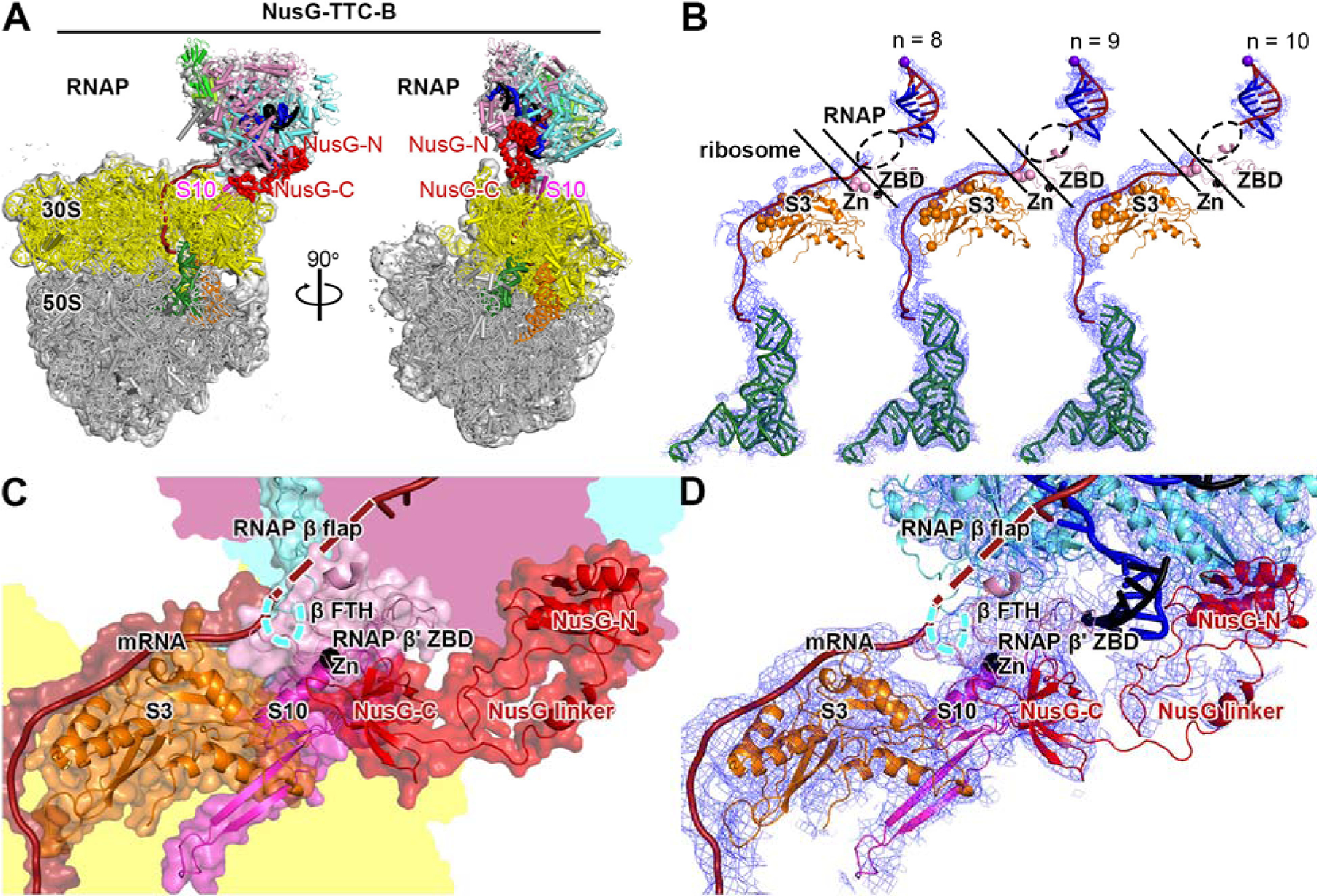
(A) Structure of NusG-TTC-B (4.7 Å; n = 9; Table S1). Views and colors as in Fig. 2A.
(B) Accommodation of mRNA spacer lengths of 8, 9, and 10 codons in NusG-TTC-B. EM density, blue mesh; mRNA, brick-red (disordered mRNA nucleotides indicated by dashed oval); template-strand DNA in RNA-DNA hybrid, blue; RNAP active-center catalytic Mg2+, purple sphere; tRNA in ribosome P site, green; ribosomal protein S3, orange (positively charged residues positioned to contact mRNA as orange spheres); RNAP β’ zinc binding domain (ZBD, pink; Zn2+ ion as black sphere; positively charged residues positioned to contact mRNA as pink spheres). Upper and lower black diagonal lines indicate edges of RNAP and ribosome.
(C) RNAP-ribosome interface and NusG bridging in NusG-TTC-B (n = 9; identical interface for n = 8, 9, or 10). RNAP β’ zinc binding domain, (ZBD, pink; Zn2+ ion as black sphere) interacts with ribosomal protein S3 (orange) and mRNA (brick red). NusG (red) bridges RNAP and ribosome, with NusG-N interacting with RNAP and NusG-C interacting with ribosomal protein S10 (magenta). Portions of RNAP β’, β, and ribosome 30S not involved in interactions are shaded pink, cyan, and yellow, respectively.
(D) As C, showing cryo-EM density as blue mesh.
Fig. 4. Cryo-EM structure of NusA-NusG-TTC-B.
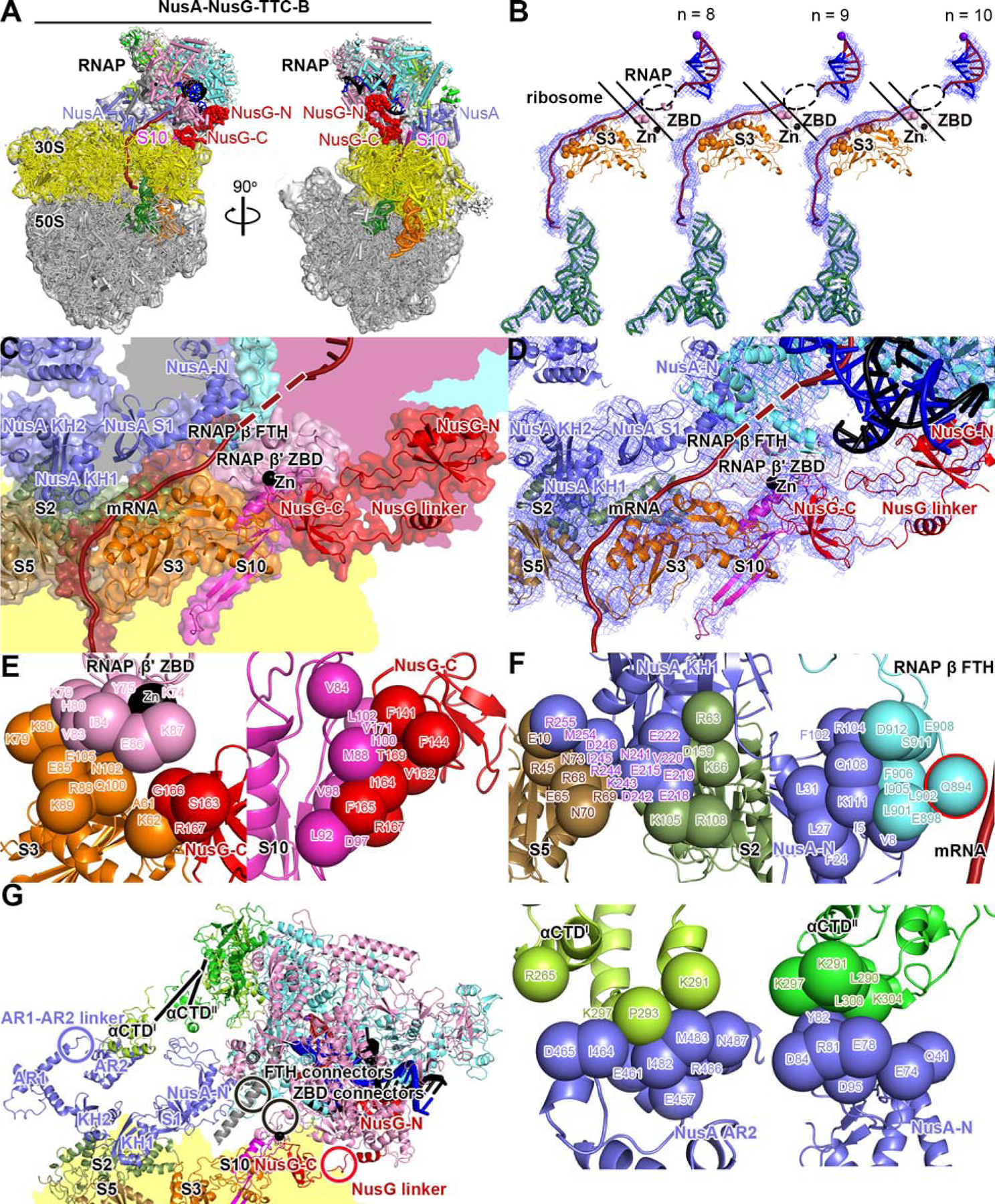
(A) Structure of NusA-NusG-TTC-B (NusA-NusG-TTC-B2; 3.5 Å; n = 9; Table S1). NusA, light blue. Views and other colors as in Figs. 2A and 3A.
(B) Accommodation of mRNA spacer lengths of 8, 9, and 10 codons in NusA-NusG-TTC-B. Views and colors as in Fig 3B.
(C) RNAP-ribosome interface, NusG bridging, and NusA binding in NusA-NusG-TTC-B (n = 9; identical interface for n = 8, 9, or 10). RNAP β’ zinc binding domain, (ZBD, pink; Zn2+ ion as black sphere) interacts with ribosomal protein S3 (orange) and mRNA (brick red). NusG (red) bridges RNAP and ribosome, with NusG-N interacting with RNAP and NusG-C interacting with ribosomal protein S10 (magenta). NusA (light blue) KH1 domain interacts with ribosomal proteins S5 and S2 (brown and forest green). Portions of RNAP β’, β, ω, and ribosome 30S not involved in interactions are shaded pink, cyan, gray, and yellow, respectively.
(D) As C, showing cryo-EM density as blue mesh.
(E) RNAP-ribosome interactions involving RNAP β’ ZBD and S3 (subpanel 1) and NusG-ribosome interactions involving NusG-C and S10 (subpanel 2).
(F) NusA-ribosome interactions involving NusA KH1 and S5 and S2 (subpanel 1) and NusA-RNAP interactions involving NusA-N and RNAP β FTH (subpanel 2; β FTH residue that interacts with mRNA, cyan sphere with red outline; mRNA, brick-red), NusA AR2 and RNAP αCTDI (subpanel 3), and NusA-N and RNAP αCTDII (subpanel 4).
(G) Points of flexibility in NusA-NusG-TTC-B (NusA “coupling pantograph”): flexible linkage in NusA structure (AR1-AR2 linker; light blue circle), three flexible linkages between NusA and RNAP (αCTDI linker, αCTDII linker, and β FTH connectors; black lines and black circle), flexible linkage between RNAP and ribosome (β’ ZBD connectors; black circle), and flexible NusG bridging of RNAP and ribosome (NusG linker; red circle).
TTC-A was obtained with nucleic-acid scaffolds having mRNA spacers of 4, 5, 6, 7, or 8 codons--but not with longer mRNA spacers (Figs. 1B-left, 2, S1–S3; Table S1). TTC-A was obtained both in the absence of NusG and in the presence of NusG (Figs. S1–S3; Table S1). EM density maps of 3.7–6.3 Å resolution were obtained (~7 Å and ~3.5 Å local resolution for TEC and ribosome, respectively, in best maps), enabling unambiguous rigid-body docking of atomic structures of TEC, ribosome 30S subunit, ribosome 50S subunit with tRNA in active-center P site and exit site (E site), and, where present, NusG-N, followed by manual fitting of residues in the RNAP-ribosome interface and in DNA and mRNA (Figs. 1B-left, 2, S1–S3; Table S1).
Remarkably, in TTC-A, the spatial relationship of RNAP relative to the ribosome is identical in structures obtained with nucleic-acid scaffolds having mRNA spacer lengths of 4, 5, 6, 7, and 8 codons (Fig. S1F). High-resolution data for TTC-A reveal that differences in mRNA spacer length are accommodated through differences in extents of compaction of mRNA in the RNAP RNA-exit channel and RNAP-ribosome interface (Fig. 2B). As mRNA spacer length increases from 4 codons to 5 codons to 6 codons, the number of mRNA nucleotides in the RNAP RNA-exit channel and RNAP-ribosome interface increases from 7 nt (5 nt in exit channel; 2 nt in interface) to 10 nt (7 nt in exit channel; 3 nt in interface) to 13 nt (9 nt in exit channel; 4 nt in interface) (Fig. 2B, subpanels 1–3). When the mRNA spacer length increases to 7 or 8 codons, 16 or 19 nt of mRNA are accommodated in the RNAP RNA-exit channel and RNAP-ribosome interface, and the 16 or 19 nt of mRNA show disorder, indicating they adopt an ensemble of different conformations (Fig. 2B, subpanels 4–5). We point out that the volume of the RNAP RNA-exit channel and RNAP-ribosome interface cannot accommodate more than ~19 nt of mRNA without changing the conformation of the RNAP RNA-exit channel or disrupting the RNAP-ribosome interface, and we suggest that this accounts for our observations that TTC-A is obtained at relatively low particle populations with a nucleic-acid scaffold having an mRNA spacer length of 8 codons (18% vs. 91% for nucleic-acid scaffold having mRNA spacer length of 4 codons; Figs. S1, S3) and is not obtained with nucleic-acid scaffolds having mRNA spacer lengths >8 codons (Figs. S4–S5). The mRNA spacers analyzed in this work contained only U (Fig. 1A); because U is the RNA nucleotide having the smallest volume, the mRNA-spacer-length cut-off of 8 codons observed in this work is likely to represent an upper bound.
In TTC-A, the RNAP-ribosome interface is extensive (3,742 Å2 buried surface area) and involves contacts of RNAP β’ zinc-binding domain (ZBD), RNAP β flap, and RNAP αI with ribosomal proteins S4, S3, and S10, respectively (Figs. 2C–D).
In EM density maps of TTC-A, density is absent for RNAP ω subunit, indicating that RNAP ω subunit is either absent, or at a low occupancy level, or disordered (Fig. 2E). Molecular modelling suggests that, if RNAP ω were present and fully folded, the C-terminal α-helix of ω would clash with the ribosome (Fig. 2E).
In EM density maps of TTC-A obtained in the presence of NusG, EM density is present for NusG-N (residues 1–118) at its expected binding location on the RNAP β’ clamp helices and the RNAP β pincer tip (14; Figs. 1B-left, S1H), but is absent for the NusG linker and NusG-C, consistent with unrestricted motion of the linker and NusG-C relative to NusG-N (14; Fig. 1B-left). Density maps for TTC-A obtained in the absence of NusG are identical to those obtained in the presence of NusG, except that density for NusG-N is missing (Fig. S2). Model building indicates that the shortest sterically allowed distance between NusG-N bound to RNAP and NusG-C modelled as bound to its molecular target on the ribosome, ribosomal protein S10 (2, 5–9), is 160 Å in TTC-A--which is 1.9 times the maximum length of the NusG linker--indicating that TTC-A is incompatible with NusG bridging of RNAP and S10 (Fig. S9A).
Molecular modelling indicates that TTC-A also is incompatible with other known functional properties of transcription elongation, pausing, and termination in E. coli. TTC-A is sterically incompatible with binding of NusA (15; Fig. S10A), formation of a 21 Q antitermination complex (16–17; Fig. S10B), and formation of pause and termination RNA hairpins (15,18–19; Fig. S10C–D). TTC-A also appears to be incompatible with ribosome 30S-head swivelling, the 21° rotation of the ribosome 30S head relative to the ribosome 30S body that occurs during ribosome translocation (20–22; Fig. S11A; Movie S3). The RNAP-ribosome interface in TTC-A spans the 30S head and 30S body in the unswivelled state (Figs. 2C, S11A-left) and is expected to be disrupted upon swivelling (loss of 1,972 A2 buried surface area; Fig. S11A-right). The finding that TTC-A--the “expressome” of 12-- lacks RNAP ω subunit, is incompatible with NusG bridging, and is incompatible with known functional properties of transcription and translation in E. coli indicates that TTC-A is unlikely to be functionally relevant to transcription-translation coupling under most conditions in E. coli. We propose that TTC-A is either: (i) a specialized complex that mediates transcription-translation coupling under specialized circumstances (e.g., transcription-translation coupling by RNAP deficient in ω or by ribosomes inactive in translocation), or (ii) an anomalous complex formed when the mRNA spacer between RNAP and ribosome is anomalously short (e.g. “collision-ome” or “crash-ome”).
TTC-B was obtained with nucleic-acid scaffolds having mRNA spacer lengths of 8, 9, or 10 codons--but not with shorter mRNA spacers (Figs. 1B, 3–4, S3–S7; Table S1). TTC-B was obtained only when NusG was present (Figs. S3–S8; Table S1) and was obtained both without bound NusA and with bound NusA (Figs. S3–S7; Table S1). TTC-B differs from TTC-A by translation of RNAP relative to the ribosome by ~70 Å and rotation of RNAP relative to the ribosome by ~180° (Fig. 1B; Movie S1). EM density maps at 3.1–12.6 Å resolution were obtained, (~7 Å and ~3 Å local resolution for TEC and ribosome, respectively, in best maps), enabling unambiguous rigid-body docking of atomic structures of components, followed by manual fitting (Figs. 3–4, S3–S7). TTC-B is identical to the NusG-bridged complex reported in a preprint by Weixlbaumer and co-workers (23) and is different from the NusA-containing complex reported in a preprint by Mahamid, Rappsilber, and co-workers (24).
In contrast to in TTC-A, where the RNAP RNA-exit channel is coupled directly to the ribosome mRNA entrance portal, in TTC-B, the RNAP RNA-exit channel is separated by ~60 Å from the ribosome mRNA entrance portal (Fig. 1B). In TTC-B, a ~60 Å, ~11 nt, mRNA segment connects the RNAP RNA-exit channel and the ribosome mRNA entry portal, running along the surface of the ribosome 30S head, making favorable electrostatic interactions with positively charged residues in ribosomal protein S3 and RNAP β’ ZBD (Figs. 3B, 4B, S3F, S4G). The requirement for this additional ~11 nt mRNA segment accounts for the fact that TTC-B is obtained only with nucleic-acid scaffolds having mRNA spacer lengths ≥8 codons.
In TTC-B, the spatial relationship of RNAP relative to the ribosome is identical in structures obtained with mRNA spacer lengths of 8, 9, and 10 codons (Figs. S4F, S5G). Analogously to in TTC-A, in TTC-B, differences in mRNA spacer length are accommodated through differences in extents of compaction of mRNA in the RNAP RNA-exit channel (Figs. 3B, 4B). As mRNA spacer length increases from 8 to 9 to 10 codons, the number of mRNA nucleotides in the RNAP RNA-exit channel increases from ~8 nt to ~11 nt to ~14 nt (disordered in each case; Figs. 3B, 4B). Assuming that the volume of the RNAP RNA-exit channel allows it to accommodate up to ~15 nt of mRNA (see above), it seems likely that mRNA spacer lengths up to ~10 codons could be accommodated in TTC-B. Noting that the mRNA segments in the RNAP-ribosome interface and near ribosomal protein S3 in TTC-B are solvent-accessible, it also seems possible that longer, possibly much longer, mRNA spacer lengths could be accommodated by looping of, or secondary-structure formation in, these mRNA segments.
In TTC-B, the interaction between RNAP and ribosome is small, involving only contact between the RNAP β’ ZBD sequence and ribosomal protein S3 (224 Å2 buried surface area; Figs. 3C–D, 4C–E).
In TTC-B, the RNAP-ribosome interaction is supplemented by bridging of RNAP and the ribosome by NusG, involving simultaneous binding of NusG-N to RNAP and binding of NusG-C to ribosomal protein S10 (1,409 Å2 buried surface area for NusG-C and S10; Figs. 1B, 3A,C–D, 4A,C–E, S3G, S4H, S9B–C). NusG-C interacts with S10 in the manner expected from published structures of a complex of NusG and S10 and of a complex of NusG and a ribosome (2,9; Figs. S3G, S4H). EM density maps show unambiguous density for NusG-N, NusG-C, and most residues of the NusG linker (Figs. 3D, 4D, S3G, S4H), and, at lower contour levels, show density for all residues of the NusG linker (Figs. 3C, 4C). Corresponding EM maps obtained in the absence of NusG do not show TTC-B (Fig. S8), indicating that NusG bridging is functionally important for the formation and/or the stability of TTC-B. The NusG bridging hypothesized in refs. 2 and 9 is unequivocally verified.
We first obtained structures of TTC-B in the presence of NusG and absence of NusA (NusG-TTC-B; Figs. 3, S3–S4; Table S1). Molecular modelling indicated that NusG-TTC-B potentially could accommodate binding of NusA (Fig. S10A). Therefore, we sought, and obtained, corresponding structures of TTC-B in the presence of both NusG and NusA (NusA-NusG-TTC-B; Figs. 4, S5; Table S1). As compared to structure determination of TTC-B in the absence of NusA, structure determination of TTC-B in the presence of NusA was associated with substantially higher particle populations (4% vs. 45% for n = 8, 18% vs. 28% for n = 9, and 17% vs. 40% for n = 10) and substantially higher resolutions (12.6 Å vs. 3.1 Å for n = 8, 4.7 Å vs. 4.2 Å for n = 9, and 5.0 Å vs. 3.7 Å for n = 10), indicating that NusA functionally stabilizes TTC-B. Three NusA-NusG-TTC-B subclasses were obtained: TTC-B1, TTC-B2, and TTC-B3, differing by up to 15° rotation of RNAP relative to NusA and ribosome (Figs. S5, S7A–B; Movie S2).
In all NusA-NusG-TTC-B subclasses, RNAP and NusG interact with the ribosome 30S head, with RNAP β’ ZBD contacting ribosomal protein S3 and NusG contacting ribosomal protein S10 (Fig. 4C–E, S6), essentially as in the absence of NusA (Fig. 3C–D).
In all NusA-NusG-TTC-B subclasses, NusA makes identical--and extensive--interactions with the surface of the ribosome S30 body, involving contacts between NusA KH1 domain and ribosomal proteins S2 and S5 (1,755 Å2 buried surface area; Figs. 4C–E, S6). The NusA-ribosome interactions observed here show no similarity to the putative NusA-ribosome interactions reported in 24; the orientation of NusA relative to the ribosome differs by ~180°, and the interactions involve a different module of the ribosome 30S subunit (body vs. head).
NusA functions in this context as a large--70 Å x 50 Å--open rectangular frame that connects RNAP to the ribosome 30S body (Figs. 4G, S7C). One side of the NusA rectangular frame interacts with the ribosome 30S body, and three corners of the NusA rectangular frame interact with RNAP, contacting the RNA αI C-terminal domain (αCTDI), the RNA αII C-terminal domain (αCTDII), and the RNAP β flap-tip helix (FTH) (Figs 4F–G, S7C). The NusA rectangular frame contains an internal flexible linkage, the AR1-AR2 linker (light blue circle in Figs. 4G, S7C), and interacts with RNAP through three flexible linked modules: αCTDI and αCTDII, which are connected to the rest of RNAP through long, flexible linkers (25; lines in Figs. 4G, S7C), and β FTH, which is connected to the rest of RNAP through flexible connectors (15–18; black circle in Figs. 4G, S7C). The internal flexibility and flexible connections enable the NusA-RNAP subcomplex to maintain constant contact with the ribosome 30S body, despite differences in orientation of RNAP relative to the ribosome 30S body (Fig. S7C; Movie S2). We refer to the NusA rectangular frame as the “coupling pantograph,” analogizing it to an electric-railway coupling pantograph, the open rectangular frame, with internal flexibility and flexible connections, that enables a locomotive to maintain constant contact with a power cable, despite differences in orientation of the locomotive relative to the cable (26; Figs. S7C; Movie S2).
The separation between the RNAP RNA-exit channel and the ribosome mRNA entry portal in TTC-B, together with the open character of the NusA rectangular frame (“coupling pantograph”) in TTC-B, provides largely unrestricted access for transcriptional-regulatory factors to bind, and transcriptional-regulatory RNA secondary structures to form, at and adjacent to the mouth of the RNAP RNA-exit channel (Fig. S10). Molecular modelling indicates that TTC-B, unlike TTC-A, can accommodate formation of the 21 Q antitermination complex (16–17; Fig. S10B) and can accommodate formation of pause and termination RNA hairpins (15,18–19; Fig. S10C–D). In NusA-NusG-TTC-B, positively charged residues of NusA N and S1 domains are positioned to make favorable electrostatic interactions with the hairpin loop of a pause or termination RNA hairpin, and thereby potentially to nucleate formation of a pause or termination RNA hairpin (Fig. S7D; see 15). The different orientations of NusA N and S1 domains in NusA-NusG-TTC-B subclasses B1, B2, and B3 possibly enable interactions with different-length pause and termination RNA hairpins, with B1 accommodating shorter hairpins and B2 and B3 accommodating longer hairpins (Fig. S7D).
Molecular modelling also indicates that TTC-B, unlike TTC-A, is compatible with ribosome 30S-head swivelling, the rotation of the 30S head relative to the 30S body that occurs during ribosome translocation (20–22; Fig. S11B–C; Movies S4–S5). In NusG-TTC-B, all RNAP-ribosome and NusG-ribosome interactions involve the ribosome 30S head; accordingly, 30S-head swivelling can be accommodated by rotation of RNAP and NusG with the 30S head (Fig S11B, center) and/or by separate rotation of flexibly connected RNAP β’ ZBD and flexibly connected NusG-C with the 30S head (Fig S11B, right; Movie S4). In NusA-NusG-TTC-B, NusA-ribosome interactions involve the ribosome 30S body, and RNAP-ribosome and NusG-ribosome interactions involve the ribosome 30S head; nevertheless --exploiting the internal flexibility and flexible connections of the NusA-RNAP “coupling pantograph”-- 30S-head swivelling can be accommodated by rotation of RNAP and NusG with the 30S head (Fig S11B, center) and/or by separate rotation of flexibly connected RNAP β’ ZBD and flexibly connected NusG-C with the 30S head (Fig S11B, right; Movie S5).
Based on the observation that TTC-B is compatible with NusG bridging, NusA binding, known functional aspects of transcription, and known functional aspects of translation, we propose that TTC-B modulates NusG-dependent, NusA-dependent transcription-translation coupling in E. coli.
The structures presented were determined in the presence of CHAPSO, a non-ionic detergent that has been used extensively in cryo-EM structural analysis of RNAP and RNAP complexes to improve structural homogeneity by disrupting non-specific complexes and weak complexes, and by improving rotational-orientation distributions of particles by reducing interactions with the air-water interface (see 14–18). Analogous structure determination in the absence of CHAPSO yielded low-resolution maps of TTC-A for nucleic-acid scaffolds with mRNA spacer lengths of 4, 5, 6, and 7 codons (Figs. S12–S13; Tables S2; Movie S6) and yielded low-resolution maps of two additional complexes, TTC-C and TTC-D, for nucleic-acid scaffolds with mRNA spacer lengths of 7, 8, and 9 codons (Figs. S13–S16; Table S2; Movies S7–S11). The fact that TTC-C and TTC-D are observed only in the absence of CHAPSO suggests TTC-C and TTC-D may involve relatively weak interactions. In TTC-C and TTC-D, interactions between RNAP and ribosome are mediated by RNAP β sequence insert 2 (βSI2; also known as βi9; 26–27), a 60-Å long α-helical antiparallel coiled-coil flexibly tethered to the rest of RNAP, and the main interaction is an electrostatic interaction between the tip of βSI2 and the ribosome 30 S subunit (Figs. S15–S16). In TTC-C the orientation of RNAP relative to the ribosome is compatible with NusG bridging (Fig S15), and in TTC-D the orientation of RNAP relative to the ribosome is incompatible with NusG bridging (Fig S16). The structures suggest that TTC-C and TTC-D could play roles in NusG-dependent transcription-translation coupling and in NusG-independent transcription-translation coupling, respectively. The structural module that mediates RNAP-ribosome interaction in TTC-C and TTC-D--βSI2--is not essential for growth in rich media (26), but is essential for growth in minimal media (26), implying that TTC-C and TTC-D are unlikely to be important for transcription-translation coupling in general, but may be important for transcription-translation coupling in specific transcription units in specific regulatory contexts (see 6). Further analysis will be needed to determine whether, and, if so, in which contexts, TTC-C and TTC-D function in transcription-translation coupling in E. coli. The results presented define four structural classes of TTCs--TTC-A (the previously reported “expressome”; 12), TTC-B, TTC-C, and TTC-D--and show that TTC-B has structural properties indicating it mediates NusG-dependent, NusA-dependent transcription-translation coupling in E. coli.
The results presented reframe our understanding of the structural and mechanistic basis of transcription-translation coupling. The results provide high-resolution structures of the previously described “expressome” (12; TTC-A) that demonstrate the incompatibility of the previously described “expressome” with general transcription-translation coupling. In addition, the results provide high-resolution structures of a new structural state, TTC-B, with properties assignable to general, NusG-dependent, NusA-dependent transcription-translation coupling, show that NusG stabilizes TTC-B by bridging RNAP and the ribosome 30S head, show that NusA stabilizes TTC-B by bridging RNAP and the ribosome 30S body, and show that NusA serves as a “coupling pantograph” that bridges RNAP and the ribosome 30S body in a flexible manner that allows rotation of RNAP relative to the ribosome 30S body. Finally, the results provide testable new hypotheses regarding the identities of the RNAP and NusA structural modules crucial for transcription-translation coupling (RNAP β’ ZBD and NusA KH1) and regarding the interactions made by those structural modules (interactions with ribosomal protein S3 in the S30 head and interactions with ribosomal proteins S2 and S5 in the S30 body).
Supplementary Material
Movie shows the ~70 Å translation and ~180° rotation that relate NusG-TTC-A to NusG-TTC-B. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity).
Movie shows the up-to-15° rotation on one axis that relate NusG-TTC-B subclasses 1, 2, and 3. Flexible linkage between NusA AR1 and AR2 is shown as light blue circle; flexible linkers between RNAP αCTDI and αNTDI and between RNAP αCTDI and αNTDI are shown as white dashed lines; flexible connectors between RNAP β FTH and rest of RNAP and between RNAP β’ ZBD and rest of RNAP are shown as white circles; flexible linker between NusG-N and NusG-C is shown as red circle. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity).
Movie shows that NusG-TTC-A is incompatible with 30S-head swivelling, the ~21° rotation of the 30S head (brown) relative to the 30S body (yellow) that occurs during ribosome translocation (20–22). View and other colors as in Fig. S11 (ribosome 50S subunit and tRNAs omitted for clarity).
Movie shows that NusG-TTC-B is compatible with 30S-head swivelling, the ~21° rotation of the 30S head (brown) relative to the 30S body (yellow) that occurs during ribosome translocation (20–22). First clip models RNAP and NusG moving with 30S head; second clip models only RNAP β’ ZBD and NusG-C moving with 30S head. Flexible connectors between RNAP β’ ZBD and rest of RNAP are shown as white circle; f’lexible linker between NusG-N and NusG-C is shown as red circle. View and other colors as in Fig. S11 (ribosome 50S subunit, tRNAs, and mRNA omitted for clarity).
Movie shows that NusA-NusG-TTC-B is compatible with 30S-head swivelling, the ~21° rotation of the 30S head (brown) relative to the 30S body (yellow) that occurs during ribosome translocation (20–22). First clip for each subclass models RNAP and NusG moving with 30S head; second clip for each subclass models only RNAP β’ ZBD and NusG-C moving with 30S head. Flexible linkers between RNAP αCTDI and αNTDI and between RNAP αCTDI and αNTDI are shown as white dashed lines; flexible connectors between RNAP β FTH and rest of RNAP and between RNAP β’ ZBD and rest of RNAP are shown as white circles; flexible linker between NusG-N and NusG-C is shown as red circle. View and other colors as in Fig. S11 (ribosome 50S subunit, tRNAs, and mRMA omitted for clarity).
Movie shows the ~60° rotation that relates NusG-TTC-A subclasses A1 and A2. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the ~90° rotation on one axis and ~180° rotation on an orthogonal axis that relate NusG-TTC-A1 to NusG-TTC-C1. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the up-to-40° rotation that relates NusG-TTC-B subclasses C1, C2, C3, C4, C5, and CB6. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the ~45° rotation that relates NusG-TTC-A1 to NusG-TTC-D1. (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the ~180° rotation that relates NusG-TTC-C1 to NusG-TTC-D1. (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the up-to-20° rotation that relates NusG-TTC-D subclasses D1, D2, and D3. (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity)
Acknowledgements:
We thank the Rutgers University Cryo-EM Core facility, University of Michigan Life Sciences Institute Cryo-EM Facility, National Center for CryoEM Access and Training (supported by NIH grant GM129539, Simons Foundation grant SF349247, and New York state grants), and Pacific Northwest Center for Cryo-EM (supported by NIH grant GM129547 and Department of Energy Environmental Molecular Sciences Laboratory) for microscope access; K. Kuznedelov and K. Severinov for plasmids; and L. Minakhin, B. Nickels, and J. Winkelman for discussion; and E. Eng and H. Wei for assistance.
Funding: This work was supported by University of California discretionary funds to G.B., University of Michigan discretionary funds to M.S., and National Institutes of Health (NIH) grant GM041376 to R.H.E.
Footnotes
Data and Material Availability: Cryo-EM micrographs have been deposited in the Electron Microscopy Public Image Archive Resource (EMPIAR accession codes 10467 and 10468). Cryo-EM maps and atomic models have been deposited in the Electron Microscopy Database (EMDB accession codes 21386, 21468, 21469, 21470, 21471, 21472, 21474, 21475, 21476, 21477, 21482, 21483, 21485, 21486, 21494, 22082, 22084, 22087, 22107, 22141, 22142, 22181, 22192, and 22193) and the Protein Database (PDB accession codes 6VU3, 6VYQ, 6VYR, 6VYS, 6VYT, 6VYU, 6VYW, 6VYX, 6VYY, 6VYZ, 6VZJ, 6VZ2, 6VZ3, 6VZ5, 6VZ7, 6XDQ, 6XDR, 6XGF, 6XII, 6XIJ, 6X6T, 6X7F, 6X7K, and 6X9Q). Unique materials are available from the authors on request.
References:
- 1.Miller O, Hamkalo B, Thomas C, Visualization of bacterial genes in action, Science 169, 392–395 (1970). [DOI] [PubMed] [Google Scholar]
- 2.Burmann B, Schweimer K, Luo X, Wahl M, Stitt B, Gottesman M, Rösch P, A NusE-NusG complex links transcription and translation, Science 328, 501–504 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Proshkin S, Rahmouni A, Mironov A, Nudler E, Cooperation between translating ribosomes and RNA polymerase in transcription elongation., Science 328, 504–508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro-Roa D, Zenkin N, In vitro experimental system for analysis of transcription-translation coupling, Nucl. Acids Res 40, e45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGary K, Nudler E, RNA polymerase and the ribosome: the close relationship, Curr. Opin. Microbiol 16, 112–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Artsimovitch I, Rebuilding the bridge between transcription and translation, Mol. Microbiol 108, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena S, Myka K, Washburn R, Costantino N, Court L, Gottesman E, Escherichia coli transcription factor NusG binds to 70S ribosomes, Mol. Microbiol 108, 495–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevenson-Jones F, Woodgate J, Castro-Roa D, Zenkin N, Ribosome reactivates transcription by physically pushing RNA polymerase out of transcription arrest. Proc. Natl. Acad. Sci. USA 117, 8462–8467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Washburn R, Zuber P, Sun M, Hashem Y, Shen B, Li W, Harvey S, Knauer S, Frank J, Gottesman M, Escherichia coli NusG links the lead ribosome with the transcription elongation complex, www.biorxiv.org/content/10.1101/871962v1 (2019). [DOI] [PMC free article] [PubMed]
- 10.Chen M, Fredrick K, Measures of single- versus multiple-round translation argue against a mechanism to ensure coupling of transcription and translation, Proc. Natl. Acad. Sci. USA 115, 10774–10779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strauss M, Vitiello C, Schweimer K, Gottesman M, Roesch P, Knauer S, Transcription is regulated by NusA:NusG interaction, Nucl. Acids Res 44, 5971–5982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohler R, Mooney A, Mills J, Landick R, Cramer P, Architecture of a transcribing-translating expressome, Science 356, 194–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demo G, Rasouly A, Vasilyev N, Svetlov V, Loveland A, Diaz-Avalos R, Grigorieff N, Nudler E, Korostelev A, Structure of RNA polymerase bound to ribosomal 30S subunit, Elife 6, e28560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang J, Mooney R, Nedialkov Y, Saba J, Mishanina T, Artsimovitch I, Landick R, Darst S, Structural basis for transcript elongation control by NusG family universal regulators, Cell 173, 1650–1662 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo X, Myasnikov G, Chen J, Crucifix C, Papai G, Takacs M, Schultz P, Weixlbaumer A, Structural basis for NusA stabilized transcriptional pausing, Mol. Cell 69, 816–827 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, Gao X, Tian T, Yu Z, Gao B, Wen A, You L, Chang S, Zhang X, Zhang Y, Feng Y, Structural basis of Q-dependent transcription antitermination., Nature Commun 10, 2925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin Z, Kaelber J, Ebright R, Structural basis of Q-dependent antitermination, Proc. Natl. Acad. Sci. USA 116, 18384–18390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y, Mishanina V, Bellecourt J, Mooney A, Darst A, Landick R, RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing, Mol. Cell 69, 802–815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts J, Mechanisms of bacterial transcription termination, J. Mol. Biol 431, 4030–4039 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Schuwirth B. Borovinskaya M, Hau C, Zhang W, Vila-Sanjurjo A, Holton J, Doudna Cate J, Structures of the bacterial ribosome at 3.5Å resolution, Science 310, 827–834 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Ratje A, Loerke J, Mikolajka A, Brünner M, Hildebrand P, Starosta A, Dönhöfer A, Connell S, Fucini P, Mielke1 T, Whitford P, Onuchic J, Yu Y, Sanbonmatsu K, Hartmann R, Penczek P, Wilson D, Spahn C, Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites, Nature 486, 714–716 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Z, Noller H, Rotation of the head of the 30S ribosomal subunit during mRNA translocation, Proc. Natl. Acad. Sci. USA 109, 20391–20394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster M, Takacs M, Zhu C, Vidmar V, Eduljee A, Abdelkareem M, Weixlbaumer A, Structural basis of transcription-translation coupling and collision in bacteria, https://www.biorxiv.org/content/10.1101/2020.03.01.971028v1 (2020) [DOI] [PubMed]
- 24.O’Reilly F, Xue L, Graziadei A, Sinn L, Lenz S, Tegunov D, Blötz C, Hagen W, Cramer P, Stülke J, Mahamid J, Rappsilber J, In-cell architecture of an actively transcribing-translating expressome, https://www.biorxiv.org/content/10.1101/2020.02.28.970111v1 (2020). [DOI] [PMC free article] [PubMed]
- 25.Blatter E, Ross W, Tang H, Gourse R, Ebright R, (1994) Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell 78, 889–896. [DOI] [PubMed] [Google Scholar]
- 26. https://en.wikipedia.org/wiki/Pantograph_(transport)
- 27.Artsimovitch I, Svetlov V, Murakami K, Landick R, Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions, J. Biol. Chem 278, 12344–12355 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Lane W, Darst S, Molecular evolution of multisubunit RNA polymerases: sequence analysis, J. Mol. Biol 395, 671–685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belogurov G, Vassylyeva M, Svetlov V, Klyuyev S, Grishin N, Vassylyev D, Artsimovitch I, Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Biosyst 26, 117–129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molodtsov V, Sineva E, Zhang L, Huang X, Cashel M, Ades S, Murakami K, Allosteric effector ppGpp potentiates the inhibition of transcript initiation by DksA. Mol. Cell 69, 828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li K, Jiang T, Yu B, Wang L, Gao C, Ma C, Xu P, Ma Y, Escherichia coli transcription termination factor NusA: heat-induced oligomerization and chaperone activity. Sci. Rep 3, 2347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Artsimovitch I, Landick R, Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. USA 97, 7090–7095 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ederth J, Mandava C, Dasgupta S, Sanyal S, A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli. Nucl. Acids. Res 37, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan H, Conn A, Williams P, Diggs S, Hahm J, Gamper H, Hou Y, O’Leary S, Wang Y, Blaha G, Transcription-translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucl. Acids Res 45, 11043–11055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blaha G, Stelzl U, Spahn C, Agrawal R, Frank J, Nierhaus K, Preparation of functional ribosomal complexes and effect of buffer conditions on tRNA positions observed by cryoelectron microscopy. Meths. Enzymol 317, 292–309 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Robertson J, Paulsen H, Wintermeyer W, Pre-steady-state kinetic studies on ribosomal translocation. Meths. in Enzymol 164, 581–597 (1988). [DOI] [PubMed] [Google Scholar]
- 37.Zheng S, Palovcak E, Armache J, Verba K, Cheng Y, Agard D, MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Meths 14, 331–333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zivanov J, Nakane T, Forsberg B, Kimanius D, Hagen W, Lindahl E, Scheres S, New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang J, Mooney R, Nedialkov Y, Saba J, Mishanina T, Artsimovitch I, Landick R, Darst S, Structural basis for transcript elongation control by NusG family universal regulators. Cell 173, 1650–1662 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beckert B, Turk M, Czech A, Berninghausen O, Beckmann R, Ignatova Z, Plitzko JM, Wilson DN, Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat. Microbiol 3, 1115–1121 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Tian P, Steward A, Kudva R, Su T, Shilling PJ, Nickson AA, Hollins JJ, Beckmann R, von Heijne G, Clarke J, Best RB, Folding pathway of an Ig domain is conserved on and off the ribosome. Proc. Natl. Acad. Sci. USA 115, E11284–E11293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenner L, Demeshkina N, Yusupova G, Yusupov M, Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nature Structl. Mol. Biol 17, 555–U548 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Pettersen E, Goddard T, Huang C, Couch G, Greenblatt D, Meng E, Ferrin T, UCSF chimera - A visualization system for exploratory research and analysis. J. Comp. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Liebschner D, Afonine P, Baker M, Bunkoczi G, Chen V, Croll T, Hintze B, Hung L, Jain S, McCoy A, Moriarty N, Oeffner R, Poon B, Prisant M, Read RJ, Richardson J, Richardson D, Sammito M, Sobolev O, Stockwell D, Terwilliger T, Urzhumtsev A, Videau L, Williams C, Adams P, Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Cryst. D 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Cryst. D 66, 486 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter C, Carragher B, Automated molecular microscopy: the new Leginon system. J. Struct. Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Mastronarde D, Advanced data acquisition from electron microscopes with SerialEM. Microsc. Microanal 24, 864–865 (2018). [Google Scholar]
- 49.Cheng A, Eng E, Alink L, Rice W, Jordan K, Kim L, Potter C, Carragher B, High resolution single particle cryo-electron microscopy using beam-image shift. J. Structl. Biol 204, 270–275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su M, goCTF: Geometrically optimized CTF determination for single-particle cryo-EM. J. Struct. Biol 205, 22–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selmer M, Dunham C, Murphy F, Weixlbaumer A, Petry S, Kelley A, Weir J, Ramakrishnan V, Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Zhou J, Lancaster L, Donohue J, Noller H. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345, 1188–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie shows the ~70 Å translation and ~180° rotation that relate NusG-TTC-A to NusG-TTC-B. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity).
Movie shows the up-to-15° rotation on one axis that relate NusG-TTC-B subclasses 1, 2, and 3. Flexible linkage between NusA AR1 and AR2 is shown as light blue circle; flexible linkers between RNAP αCTDI and αNTDI and between RNAP αCTDI and αNTDI are shown as white dashed lines; flexible connectors between RNAP β FTH and rest of RNAP and between RNAP β’ ZBD and rest of RNAP are shown as white circles; flexible linker between NusG-N and NusG-C is shown as red circle. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity).
Movie shows that NusG-TTC-A is incompatible with 30S-head swivelling, the ~21° rotation of the 30S head (brown) relative to the 30S body (yellow) that occurs during ribosome translocation (20–22). View and other colors as in Fig. S11 (ribosome 50S subunit and tRNAs omitted for clarity).
Movie shows that NusG-TTC-B is compatible with 30S-head swivelling, the ~21° rotation of the 30S head (brown) relative to the 30S body (yellow) that occurs during ribosome translocation (20–22). First clip models RNAP and NusG moving with 30S head; second clip models only RNAP β’ ZBD and NusG-C moving with 30S head. Flexible connectors between RNAP β’ ZBD and rest of RNAP are shown as white circle; f’lexible linker between NusG-N and NusG-C is shown as red circle. View and other colors as in Fig. S11 (ribosome 50S subunit, tRNAs, and mRNA omitted for clarity).
Movie shows that NusA-NusG-TTC-B is compatible with 30S-head swivelling, the ~21° rotation of the 30S head (brown) relative to the 30S body (yellow) that occurs during ribosome translocation (20–22). First clip for each subclass models RNAP and NusG moving with 30S head; second clip for each subclass models only RNAP β’ ZBD and NusG-C moving with 30S head. Flexible linkers between RNAP αCTDI and αNTDI and between RNAP αCTDI and αNTDI are shown as white dashed lines; flexible connectors between RNAP β FTH and rest of RNAP and between RNAP β’ ZBD and rest of RNAP are shown as white circles; flexible linker between NusG-N and NusG-C is shown as red circle. View and other colors as in Fig. S11 (ribosome 50S subunit, tRNAs, and mRMA omitted for clarity).
Movie shows the ~60° rotation that relates NusG-TTC-A subclasses A1 and A2. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the ~90° rotation on one axis and ~180° rotation on an orthogonal axis that relate NusG-TTC-A1 to NusG-TTC-C1. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the up-to-40° rotation that relates NusG-TTC-B subclasses C1, C2, C3, C4, C5, and CB6. View and colors as in Fig. 1B (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the ~45° rotation that relates NusG-TTC-A1 to NusG-TTC-D1. (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the ~180° rotation that relates NusG-TTC-C1 to NusG-TTC-D1. (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity).
Movie shows the up-to-20° rotation that relates NusG-TTC-D subclasses D1, D2, and D3. (ribosome 50S subunit and tRNAs omitted for clarity; mRNA entering 30S subunit shown as in NusG-TTC-A1 for simplicity)


