Abstract
Background
The proportion of tumors of various histologies that may respond to drugs targeted to molecular alterations is unknown. NCI-MATCH, a collaboration between ECOG-ACRIN Cancer Research Group and the National Cancer Institute, was initiated to find efficacy signals by matching patients with refractory malignancies to treatment targeted to potential tumor molecular drivers regardless of cancer histology.
Methods
Trial development required assumptions about molecular target prevalence, accrual rates, treatment eligibility, and enrollment rates as well as consideration of logistical requirements. Central tumor profiling was performed with an investigational next-generation DNA–targeted sequencing assay of alterations in 143 genes, and protein expression of protein expression of phosphatase and tensin homolog, mutL homolog 1, mutS homolog 2, and RB transcriptional corepressor 1. Treatments were allocated with a validated computational platform (MATCHBOX). A preplanned interim analysis evaluated assumptions and feasibility in this novel trial.
Results
At interim analysis, accrual was robust, tumor biopsies were safe (<1% severe events), and profiling success was 87.3%. Actionable molecular alteration frequency met expectations, but assignment and enrollment lagged due to histology exclusions and mismatch of resources to demand. To address this lag, we revised estimates of mutation frequencies, increased screening sample size, added treatments, and improved assay throughput and efficiency (93.9% completion and 14-day turnaround).
Conclusions
The experiences in the design and implementation of the NCI-MATCH trial suggest that profiling from fresh tumor biopsies and assigning treatment can be performed efficiently in a large national network trial. The success of such trials necessitates a broad screening approach and many treatment options easily accessible to patients.
New anticancer agents are increasingly developed in biomarker-defined populations, requiring prospective molecular definition of the cancers hypothesized to be most responsive. Examples include receptor tyrosine-protein kinase erbB-2 (ERBB2) amplified breast cancer, where a 20–25% ERBB2 amplification rate (1,2) made prospective molecular screening essential for demonstration of trastuzumab efficacy (3). Likewise, for advanced lung adenocarcinoma containing activating epidermal growth factor receptor (EGFR) mutations, or anaplastic lymphoma kinase (ALK) and proto-oncogene tyrosine-protein kinase ROS (ROS) fusions, the benefit demonstrated with inhibitors of these pathways in most patients with the appropriate alterations was unequivocal (4–7).
Whether targeted therapies known to be effective in some tumor histologies might exhibit similar effectiveness in other tumor histologies in which they have not yet been evaluated is of great interest. Imatinib, for example, approved by the US Food and Drug Administration (FDA) for chronic myelogenous leukemia (defined by breakpoint cluster region protein (BCR)- abelson murine leukemia viral oncogene homolog 1 (ABL) translocation) and gastrointestinal stromal tumor (the majority have proto-oncogene c-KIT (KIT) mutations), has also been approved for systemic mastocytosis, chronic eosinophilic leukemia, and dermatofibroma protuberans (8–10) based on individual small trials in rare conditions defined by particular molecular alterations. More recently, the FDA approved drugs for certain rare molecular alterations based on trials that included several histologies. For example, the programmed cell death protein 1 (PD-1) inhibitor pembrolizumab was approved for any tumor characterized by mismatch repair deficiency (11), and larotrectinib was approved for any metastatic tumor with a tropomyosin receptor kinase (TRK) fusion (12). “Basket” trials, in which one drug or a combination is tested in several cohorts, each with a distinct histology in which the molecular alteration is reasonably frequent and sometimes including an “other” cohort for all cancers in which the molecular abnormality is less frequent, have been reported (13). However, for some tumors in which the targeted mutation is very rare, completing even a small study can be challenging (13).
In April 2013, the leaders of the ECOG-ACRIN Cancer Research Group (ECOG-ACRIN) and the National Cancer Institute (NCI) Division of Cancer Treatment and Diagnosis began development of a signal-finding clinical trial to be performed in the National Clinical Trials Network (NCTN) and the National Community Oncology Research Program involving a large screening effort to select patients for treatments targeted to somatic genomic alterations. The trial goal, involving over 1000 academic and community clinical sites in every state and US territory, was to investigate the activity of genomically targeted treatments across common and less common tumor types.
We incorporated as many targeted therapeutics directed to as many gene alterations as possible, though allowing only one treatment in the trial for each alteration. The initial 3000-patient screening accrual goal was calculated to be enough to complete accrual to subprotocols seeking patients with tumors harboring an alteration with prevalence of 1–2% given a projected match rate of 30%. Prevalence projections of somatic alterations were based on data from the Cancer Genome Atlas and the International Cancer Genome Consortium. However, because these sources focused on primary tumors before treatment and results might not be applicable to patients with refractory metastatic disease, we also reviewed mutation rates observed in various cancers from cBioPortal (http://www.cbioportal.org/) and the University of Texas MD Anderson Cancer Center precision medicine effort.
A fresh biopsy was considered essential, because genetic evolution and an altered genomic profile from the time of initial cancer diagnosis might be expected in a refractory population with advanced cancer. Central profiling assays were employed.
The feasibility of accrual, the mix of tumor histologies screened and/or assigned to a targeted treatment in a trial with broad eligibility criteria, the adequacy of biopsies obtained from diverse clinical sites, the rate of eligibility, and enrollment to subprotocols were all difficult to predict. Because of these uncertainties and the need to establish feasibility of the many steps required to enroll a patient on the NCI-MATCH trial in a national network, an interim analysis was planned following accrual of 500 patients to assess trial progress and to make any needed adjustments. Based on the insights from this analysis, study procedures were adjusted substantially so that upon activating additional subprotocols, we improved assignment efficiency and match rate.
Methods
Protocol Design
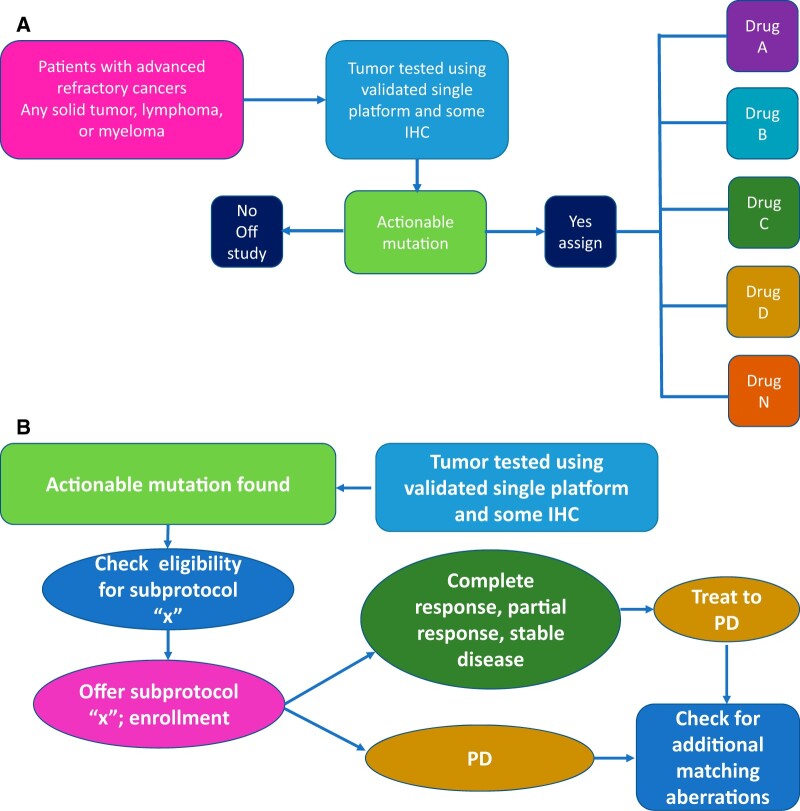
Several working committees (Supplementary Table 1, available online) developed an overall trial design and flow, chose appropriate eligibility molecular alterations, and evaluated targeted agents. These teams were overseen by a Steering Committee comprised of the chairpersons of each working committee. A parent master screening protocol, EAY131, housed an array of therapeutic Phase II subprotocols addressing gene alteration and drug pairs, with provision for amending all components as needed (Figure 1). The Agents/Genes Selection Working group recruited young investigators from across the NCTN to lead each subprotocol, each having its specific eligibility criteria and consent process. An interim analysis was planned, requiring screening suspension after 500 patients, to evaluate the trial structure, resources, and workflow, and to determine whether the initial projections for patient accrual, disease distribution, alteration detection, and treatment assignment were accurate. The protocol was reviewed and approved by NCI Central Institutional Review Board for Early Clinical Trials as the institutional review board of record (which has an assurance filed with and approved by the US Department of Health and Human Services) for all sites (NCT02465060).
Figure 1.
National Cancer Institute (NCI)-MATCH design and patient entry procedures. A) NCI -MATCH design (a type of platform trial with features of both umbrella and basket design). B) NCI -MATCH procedures for trial entry. IHC = immunohistochemistry; PD = progressive disease.
Selection of Drugs and Definition of Eligibility Variants
Single drugs or combinations were evaluated for inclusion if they targeted molecular alterations with an estimated prevalence of at least 1.5% of the population of refractory cancer patients, had a defined phase II dose and met criteria for clinical activity (ie, the drugs had either been approved or had shown clinically significant activity), or at least activity in a single patient with the relevant tumor molecular alteration. Actionable molecular alterations were those that, at minimum, were associated with preclinical evidence linking the alteration with drug activity.
Patient Selection
Patients eligible for screening were adults with any solid tumor, lymphoma, or myeloma who had progressed on prior treatment or for whom no curative treatment was available and who were willing to provide a tissue biopsy for profiling. Patients were required to have adequate hematopoietic, liver, and kidney function and an Eastern Cooperative Oncology Group (ECOG) performance status of grade 1 or better. Patients were excluded from eligibility to subprotocols if they had a tumor histology in which the drug was approved or in late clinical trial or was known not to be of benefit (4,14). All patients gave written informed consent for the trial.
Tumor Profiling
Patients underwent a biopsy for screening (with an estimated risk of an adverse event <2%), had a clinically indicated biopsy, or, added after the interim analysis, sent a tumor sample from a biopsy obtained within the prior 6 months and after which there had been no response to treatment. A preaddressed prepaid shipping kit with all required containers, fixatives, blood tubes, and instructions was provided for collection of specimens. Tumor profiling used a validated, targeted Next-generation sequencing (NGS) panel of 143 genes, which assayed single nucleotide variants, indels, amplifications, and selected fusions, and validated immunohistochemistry (IHC) assays for protein expression of phosphatase and tensin homolog (PTEN), mutL homolog 1, and mutS homolog 2 expression (15,16). All assays were performed as investigational assays under an abbreviated Investigational Device Exemption submitted to the NCI’s Investigational New Drug Application.
Assignment to Treatment
Patients whose tumors had an actionable alteration were assigned by a prospectively defined, NCI-designed computational platform (MATCHBOX) to one of the available treatment subprotocols (Figure 1). If multiple actionable variants were present, the patient was assigned by the variant with the highest level of evidence, followed by the variant with the highest allele frequency. If still equivalent, assignment was to the subprotocol with the fewest patients or random if accrual was equal. Patients who progressed after an initial response or had treatment for at least 6 months before progression could have a repeat biopsy for reassignment based on a different actionable alteration. Patients who progressed or stopped their assigned treatment for another reason within 6 months had their original sequencing results reviewed for additional molecular alterations for potential assignment to a second NCI-MATCH subprotocol.
Evaluation of Response
Response was evaluated every two cycles for drugs with a 28-day or 42-day cycle, or every three cycles for drugs with a 21-day cycle using appropriate response criteria (17–20).
Statistical Considerations
NCI-MATCH is a platform trial having features of both umbrella and basket trial designs, planned to screen 3000 patients to enroll 35 patients on subprotocols with variant prevalence of 1.2% or higher. The primary outcome for NCI-MATCH was the objective response rate for each subprotocol. An interim analysis to examine prevalence and enrollment rates and other assumptions was planned after 500 patients had been screened. In the interim analysis, actual rates computed from the patients already screened were used to project future subprotocol enrollment.
For each subprotocol, a response rate of 5 of 31 patients (16%) was considered promising. Although successful targeted agents often have a response rate of at least 50%, we expected some tumors may be refractory, and a heavily pretreated population may not be as responsive. Secondary objectives included progression-free survival at 6 months, progression-free survival, and toxicity assessment within each subprotocol. The number of subprotocols in the trial depended on the availability and scientific potential of therapies as well as feasibility of accrual, and funding. No single event would trigger the end of screening or accrual to the entire trial.
Results
Accrual
NCI-MATCH opened August 12, 2015, with 10 subprotocols (Figure 2). Patient demographics are shown in Table 1. Community oncology centers registered two-thirds of the patients. By October 2015, at least 500 patients had been accrued; new enrollment was suspended for interim analysis on November 11, 2015, resuming in May 2016. From August 12 to November 11, 2015, 739 tissue specimens were received from 795 registered patients. Screening was much more rapid than the predicted 50 patients per month, averaging 80 specimens per week after 8 weeks.
Figure 2.
Subprotocol activation timeline for National Cancer Institute (NCI)-MATCH. EGFR = epidermal growth factor receptor; HER2 = human epidermal growth factor receptor 2; ALK = anaplastic lymphoma kinase; ROS = proto-oncogene tyrosine-protein kinase ROS; BRAF = B-Raf proto-oncogene, serine/threonine kinase; NF2 = neurofibromatosis type 2; KIT = KIT proto-oncogene, receptor tyrosine kinase; PIK3CA = phosphatidylinositol 3-kinase catalytic subunit; PTEN = Phosphatase and tensin homolog; NF1 = neurofibromatosis type 1; GNAQ = G protein subunit alpha q; GNA11 = Guanine nucleotide-binding protein subunit alpha-11; PTCH1 = Patched1; SMO = Smoothened; DDR2 = DNA damage response 2; NRAS = Rat sarcoma virus GTPase, neuroblastoma; CCN = cyclin; MLH1 = mutL homolog 1; MSH2 = mutS homolog 2; MET = Mesenchymal Epithelial Transition, receptor tyrosine kinase; FGFR = Fibroblast Growth Factor Receptor; AKT = V-Akt Murine Thymoma Viral Oncogene Homolog.
Table 1.
Patient demographics at interim analysis of NCI-MATCH*
| Patient characteristic | Enrolled for screening | Assigned to treatment |
|---|---|---|
| No. (%) | No. (%) | |
| (n = 795) | (n = 33) | |
| Median age, y (Range) | 63 (24–93) | 68 (40–82) |
| Male | 305 (38.4) | 16 (48.5) |
| Female | 490 (61.6) | 17 (51.5) |
| White | 646 (81.27.9 | 29 (87.9) |
| Black | 88 (11.1) | 1 (3.0) |
| Asian | 27 (3.4) | 2 (6.1) |
| Native American | 4 (0.50) | 0 |
| Native Hawaiian | 1 (0.1) | 0 |
| Race not reported | 29 (3.6) | 1 (3.0) |
| Hispanic ethnicity | 36 (4.9) | 0 |
NCI = National Cancer Institute.
Although plans were made to restrict common tumors (non-small cell lung cancer, breast cancer, prostate cancer, and colorectal cancer) to 75% of the total, common cancers at interim analysis comprised only 35.5% of tumors, and 42.4% of those were assigned to subprotocol treatment. The most frequent tumors sequenced were colorectal cancer (13.0%), breast cancer (13.0%), ovarian cancer (11.2%), non-small cell lung cancer (7.4%), pancreatic cancer (5.3%), and head and neck cancer (5.3%) (Table 2).
Table 2.
Primary disease sites of patients enrolled for screening at the interim analysis of NCI-MATCH
| Cancer type | Enrolled for screening (n = 795) No. (%) | Screened (n = 645) No. (%) | Assigned to treatment (n = 33) No. (%) |
|---|---|---|---|
| Common cancers | |||
| Colorectal | 104 (13.1) | 84 (13.0) | 6 (18.2) |
| Breast | 96 (12.1) | 84 (13.0) | 2 (6.1) |
| Non-small cell lung | 62 (7.8) | 48 (7.4) | 5 (15.2) |
| Prostate | 20 (2.5) | 17 (2.6) | 1 (3.0) |
| Common cancers subtotal | 282 (35.5) | 233 (36.1) | 14 (42.4) |
| Uncommon cancers | |||
| Ovarian | 89 (11.2) | 72 (11.2) | 6 (18.2) |
| Pancreas (adeno or NOS) | 43 (5.4) | 34 (5.3) | 0 |
| Head and neck* | 38 (4.8) | 34 (5.3) | 0 |
| Endometrial or uterine (nonsarcoma) | 34 (4.3) | 27 (4.2) | 0 |
| Esophageal, GE junction, or gastric | 31 (3.9) | 28 (4.3) | 4 (12.1) |
| Neuroendocrine† | 27 (3.4) | 20 (3.1) | 2 (6.1) |
| Cholangiocarcinoma | 24 (3.0) | 22 (3.4) | 1 (3.0) |
| Bladder or urinary tract | 21 (2.6) | 14 (2.2) | 1 (3.0) |
| Endometrial or uterine sarcoma‡ | 20 (2.5) | 16 (2.5) | 0 |
| Small cell lung | 16 (2.0) | 14 (2.2) | 0 |
| Other§ | 151 (19.0) | 116 (18.0) | 3 (9.1) |
| Primary site not specified | 19 (2.4) | 15 (2.3) | 2 (6.1) |
| Uncommon cancers subtotal | 513 (64.5) | 412 (63.9) | 19 (57.6) |
Salivary gland (three patients). NCI = National Cancer Institute; NOS = Not Otherwise Specified.
NOS (18 patients), pancreas (six patients), and carcinoid (three patients).
Uterine carcinosarcoma (seven patients).
Key other types: lymphoma (nine patients), brain tumor (nine patients), and melanoma (nine patients).
Molecular Profiling
Molecular profiling was evaluable in 87.3% of tumors (645 patients). Median time from registration to central biopsy receipt was 7 days. The robust accrual overwhelmed the initially planned resources. Median turnaround time from receipt of biopsy to results for assignment increased from 14 days in September 2015 to 36 days in October 2015.
We addressed these issues by three main revisions. First, we implemented a higher throughput NGS platform and increased sample processing personnel. Second, we improved tissue acquisition by allowing submission of clinical biopsies taken within 6 months of registration provided there was no clinical response to any treatment during those 6 months. We strongly recommended a fine needle aspirate simultaneous with the required biopsy—this rescued a proportion of those biopsy specimens that failed to yield usable material. These efforts increased the evaluable molecular profiling rate to 93.9%, decreased the median turnaround time to 14 days, and increased profiling capacity from an expected throughput of 50 per month to 100–150 per week. Finally, we implemented support desks at both the ECOG-ACRIN Operations office for clinical questions and at the central specimen receipt center (University of Texas MD Anderson Cancer Center) for laboratory issues. Educational efforts were increased to reinforce time and biopsy acquisition requirements, including reinforcing the use of provided detailed instructions for sample handling and fixation kits.
Assignment to Initial 10 Subprotocols
Based on initial assumptions, the anticipated match rate for the first 10 subprotocols was 9%. Of 645 patients’ tumors sequenced before interim analysis, 56 (8.7%) had an actionable molecular alteration (Table 3) for one of the open subprotocols. However, due to histology exclusions and other eligibility criteria, only 33 (5.1%) of all patients whose tumors were molecularly screened were eligible for assignment to a subprotocol (Supplementary Figure 1, available online), of which 16 (48.5%) enrolled. Of assigned patients who were not treated, five patients (15.2%) no longer met master protocol eligibility criteria; five patients (15.2%) did not meet subprotocol eligibility criteria; three patients (9.1%) progressed, deteriorated, or started other treatment; and four patients (12.1%) died.
Table 3.
Actionable alterations: 645 screened patients, initial 10 subprotocols*
| NCI-MATCH subprotocol | Assignment rate, % | Estimated prevalence of actionable mutation, % |
|---|---|---|
| Q: Ado-trastuzumab emtansine for HER2 amplification | 1.7 | 5 |
| U: Defactinib for NF2 mutations | 1.1 | 2 |
| B: Afatinib for HER2 mutations | 0.8 | 2-6 |
| H: Dabrefenib+trametinib for BRAF V600 mutations | 0.8 | 7 |
| R: Trametinib in BRAF non-V600 mutations | 0.3 | 2.8 |
| E: Osimertinib for EGFR T790M, rare EGFR mutations | 0.2 | 1–2 |
| F: Crizotinib for ALK translocations | 0.2 | <2 |
| V: Sunitinib for cKIT mutations (non-GIST) | 0.2 | 2 |
| A: Afatinib for EGFR mutations (nonlung) | 0 | 1–4 |
| G: Crizotinib for ROS1 translocations | 0 | <2 |
Table compares the actual assignment rate to the estimated prevalence during protocol design and illustrates that the actual assignment rate is much lower than the initial assumption. ALK = anaplastic lymphoma kinase; BRAF = B-Raf proto-oncogene, serine/threonine kinase; EGFR = epidermal growth factor receptor; HER2 = human epidermal growth factor receptor 2; GIST = gastrointestinal stromal tumor; KIT = KIT proto-oncogene, receptor tyrosine kinase; NCI = National Cancer Institute; NF2 = neurofibromatosis type 2; ROS = proto-oncogene tyrosine-protein kinase ROS.
New Estimate of Frequency of Actionable Alterations
Compared with initial assumptions, we observed a lower actionable alteration frequency in the initial 645 patients’ tumors (Table 3). The interim analysis allowed more accurate prediction of the potential assignment rate for subsequently added subprotocols (Table 4). After comparing the prevalence of actionable alterations in the 645 patients entered before the interim analysis to the eligibility requirements for the 24 future subprotocols, the expected assignment rate increased to 25.3%, allowing for ineligible histologies and other ineligibility criteria (Table 4). We projected that with 5000 screened patients, our upcoming subprotocols addressing alterations in phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA), cyclin D1 (CCND1), fibroblast growth factor receptor (FGFR), PTEN, ERBB2 (amplification), neurofibromatosis type 1 (NF1), and cyclin-dependent kinase 4 and 6 (CDK4/6)would each accrue at least 35 patients, and an additional four subprotocols would accrue nearly 30 patients. Eleven subprotocols addressing molecular alterations with a frequency of 1% or less would likely not reach the accrual goal. As a result, the screening accrual goal was increased from 3000 to 5000 and subsequently to approximately 6000 patients.
Table 4.
Anticipated assignment rate and expected subprotocol enrollment with screening accrual of 5000 patients based on mutation frequencies and tumor histology in NCI-MATCH interim analysis results in 645 screened patients*
| Target (subprotocol) | Expected assignment rate, % | Expected enrollment, No. |
|---|---|---|
| PIK3CA mutation (I) | 4.0 | 89 |
| CCND1 amplification (Z1B) | 3.6 | 79 |
| FGFR1/2/3 mutation or amplification or translocation (W) | 2.9 | 65 |
| PTEN expression loss (P) | 2.5 | 55 |
| ERBB2 amplification (Q) | 1.7 | 44 |
| NF1 (S1) | 1.9 | 41 |
| CDK4/6 amplification (Z1C) | 1.7 | 38 |
| TSC1/2 mutation (M) | 1.2 | 28 |
| AKT1 mutation (Y) | 1.2 | 28 |
| NRAS mutation (Z1A) | 1.2 | 28 |
| NF2 mutation (U) | 1.1 | 26 |
| PTEN mutation (N) | 1.1 | 24 |
| MET amplification (C1) | 0.9 | 21 |
| ERBB2 mutation (B) | 0.8 | 20 |
| BRAF V600 (H) | 0.8 | 19 |
| SMO/PTCH1 mutation (T) | 0.6 | 14 |
| MTOR mutation (L) | 0.3 | 7 |
| BRAF non V600 mutation (R) | 0.3 | 8 |
| EGFR T790M or other rare mutation (E) | 0.2 | 4 |
| ALK translocation (F) | 0.2 | 4 |
| cKIT mutation (V) | 0.2 | 3 |
| EGFR activating mutation (A) | — | — |
| ROS1 translocation (G) | — | — |
| GNAQ/GNA11 mutation (S2) | — | — |
— = the expected frequency is below 1% or 1 patient as indicated; ALK = anaplastic lymphoma kinase; AKT = V-Akt Murine Thymoma Viral Oncogene Homolog; BRAF = B-Raf proto-oncogene, serine/threonine kinase; CCN = cyclin; CDK4/6 = cyclin-dependent kinase 4 and 6; MLH1 = mutL homolog 1; EGFR = epidermal growth factor receptor; ERBB2 = receptor tyrosine-protein kinase erbB-2; FGFR = Fibroblast Growth Factor Receptor; GNAQ/GNA11 = G protein subunit alpha q/Guanine nucleotide-binding protein subunit alpha-11; KIT = KIT proto-oncogene, receptor tyrosine kinase; MET = Mesenchymal Epithelial Transition, receptor tyrosine kinase; NCI = National Cancer Institute; NF1 = neurofibromatosis type 1; NF2 = neurofibromatosis type 2; NRAS = Rat sarcoma virus GTPase, neuroblastoma; PIK3CA = phosphatidylinositol 3-kinase catalytic subunit; PTEN = Phosphatase and tensin homolog; SMO/PTCH1 = Smoothened/Patched1; ROS = proto-oncogene tyrosine-protein kinase ROS.
Changes made to the NCI-MATCH study after the interim analysis are detailed in Table 5.
Table 5.
Changes to NCI-MATCH after interim analysis and effect of changes, where known
| Design feature | Original protocol | Amended protocol |
|---|---|---|
| No. to screen | 3000 | 6000 |
| No. of subprotocols | 10 | 24 |
| Match rate, % | 8.7 | 25.3 |
| Treatment enrollment rate, % (of matched and eligible patients)* | 48.5 | To be determined |
| NGS platform throughput | 50/mo | 100–150/wk |
| Tumor tissue | Fresh biopsy | Fresh biopsy or archived tissue if obtained in previous 6 mo and patient had had no response to treatment during past 6 mo |
| NGS success rate, % | 87.3 | 93.9 |
| Turn-around time | Projected median: 14 d | Projected median: 14 d |
| Actual median: 36 d | Actual median: 14 d | |
| Education | — | Added support at ECOG-ACRIN and central laboratory to immediately field questions |
Matched and eligible patients possessed an actionable mutation and did not have any exclusionary characteristics (eg, tumor for which the drug was already approved, cooccurring mutations that would cause resistance to the treatment). ECOG-ACRIN = Eastern Cooperative Oncology Group-American College of Radiology Imaging Network; NGS = Next-generation sequencing; NCI = National Cancer Institute.
Biopsy Safety
Grade 3 biopsy-related toxicities were reported in three of 651 (0.5%) patients with data available at interim analysis (one each with abdominal pain and hypertension, and one patient with pneumothorax and cardiac dysrhythmia); no grade 4 or 5 toxicities were reported (Supplementary Table 2, available online).
Discussion
The implementation of NCI-MATCH provides important information to the field of precision oncology. Concerns regarding the ability to accrue for screening were allayed immediately: the rate of patient entry was almost 10-fold that anticipated, reflecting a large unmet patient need and interest in precision medicine in both academic and community centers.
Recently, several efforts to match patients with early-phase trials have reported that only a small minority of patients were able to enroll on relevant clinical studies due largely to lack of trial availability (21–24). NCI-MATCH partially remedied this limitation with the inclusion of subprotocols employing drugs addressing most of the genomic abnormalities that can presently be targeted in cancer and the ability to enroll patients at over 1000 clinical sites. The accrual rate for the initial 10 subprotocols was clearly too low to sustain a genomic trial with a clinically meaningful match rate. Many screened patients were ineligible for drugs in the initial 10 subprotocols because they had histologies for which the initial treatments were FDA approved. In addition, the time patients were required to be off treatment (4–6 weeks) for molecular profiling as well as processing delays due to inadequate laboratory resources likely allowed patient clinical deterioration and contributed to the low enrollment (48.5%).
Several lessons were learned in implementation of the NCI-MATCH study. First, the inclusion of an interim analysis was key to the success of the entire study. It allowed us to understand the demand for inclusion as well as the types of molecular alterations found in the population of patients to which NCI-MATCH was directed. This information assisted in forecasting which subprotocols would achieve the desired accrual of patients. Of note, when NCI-MATCH resumed accrual after the interim analysis, 24 subprotocols were available and the assignment rate increased from 5.1% to 25.3%, which was considered reasonable given that patients with actionable alterations for which drugs were approved or for which activity in that tumor was already known were excluded. Second, there is great demand for molecular profiling studies, and the volume of patients seeking these types of studies is larger than the number of patients seeking studies where molecular profiling for eligibility is not required. Adequate laboratory resources must be available and reliable. Third, education of clinical sites is critical. Medical personnel and patients need information to understand that profiling and matching takes some time and that patients must be able to be off therapy for at least 4 weeks. In addition, sites need information that generally more tissue is needed for profiling than the amount required for only diagnosis or confirmation of a malignant histology. Fourth, many actionable molecular alterations occur in 3% or less of patients with refractory malignancies. Trials addressing these alterations require access to a great number of clinical sites, and these sites must have as many treatments available as possible. Fifth, implementation in the NCTN and National Community Oncology Research Program, with investigators familiar with new drug administration and access to tens of thousands of patients with all types of malignancies, worked well. Sixth, the collaborative nature of the study, led by NCI and ECOG-ACRIN with participation of representatives from all the NCTN groups, was critical for success (Supplementary Table 1, available online). Furthermore, incorporation of expert input from across the several operational committees as well as from the principal investigators of the subprotocols and from drug developers in the pharmaceutical industry was essential. The trial necessitated a high level of ongoing involvement of all parties as well as project management support for trial development, protocol and amendment maintenance, and new subprotocol development. We implemented uniformity of approach for drug selection and incorporation and gained efficiencies by having all drugs and assays incorporated under an NCI-sponsored Investigational New Drug Application as well as by use of the NCI Central Institutional Review Board.
Success in precision medicine trials depends on the breadth and quality of the genomic assay(s) used for screening, the type of tumors and frequency of molecular alterations identified among registered patients, the number of treatment options available, and the drug target specificity, efficacy, and tolerability of the selected treatments. Addressing rarer molecular alterations remains a challenge. To accrue the Phase II cohorts of alterations in the prevalence range of 1–2%, NCI-MATCH increased the screening goal. Even then, it was clear that molecular subgroups with a prevalence of less than 1% require additional measures to achieve accrual goals. To complete enrollment on “rare variant” subprotocols, NCI-MATCH has now been modified to enroll patients who have an eligible alteration based on sequencing performed for clinical care at a CLIA-accredited laboratory external to the NCI-MATCH trial. This practicality must be considered in future trials addressing the “long tail” of less common, but still targetable, molecular alterations.
Funding
This study was conducted by the ECOG-ACRIN Cancer Research Group (Peter J. O'Dwyer, MD and Mitchell D. Schnall, MD, PhD, Group Co-Chairs) and the National Cancer Institute of the National Institutes of Health under the following award numbers: CA180820, CA180794, CA180858, CA180870, CA180867, CA180857, CA180853.
Notes
The study sponsors were the National Cancer Institute and ECOG-ACRIN Cancer Research Group, who were jointly responsible for the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication. In addition to the named authors above, the following contributed to the study either in support of its implementation, laboratory analysis, analysis and collection of data, or manuscript writing: NCI-MATCH Team: American College of Radiology Imaging Network (ACRIN), Philadelphia, PA: Mitch Schnall. Cancer Therapy Evaluation Program, Bethesda, MD: Tali Johnson. Dartmouth Laboratory, Bedford, NH: Francine B. de Abreu, Sophie J. Deharvengt, Torrey L. Gallagher, Kelley Godwin, Donald C. Green, Carolyn C. Hain, Brianna E. Houde, Jason D. Peterson, Heather B. Steinmetz, Gregory J. Tsongalis, Xiangjun Xiao. ECOG-ACRIN Cancer Research Group, Philadelphia, PA: Bruce Giantonio, Mary Lou Smith. Johns Hopkins University Sidney Kimmel Cancer Center, Baltimore, MD: Nilofer Azad. Massachusetts General Hospital Laboratory, Boston, MA: Michael Lambert, Long Phi Le, Amelia Raymond, Hayley Robinson, Kathy Vernovsky. Memorial Sloan Kettering Cancer Center, New York, NY: David Hyman. Moffitt Cancer Center, Tampa, FL: Christine Chung. NCI Center for Biomedical Informatics and Information Technology, Rockville, MD: Cathy Rowe, Frank Spina. NCI Division of Cancer Treatment and Diagnosis, Bethesda, MD: Sherry Ansher, Jason Cristofaro, Percy S. Ivy, Irina Lubensky, Hala Makhlouf, Lalitha Shankar, John Wright, Laura Yee, Liz Lison (contractor). The Frederick National Laboratory Biomedical Applications Development Center (BADC), Rockville, MD: Mary Anderson, Mark Benson, Nathan Birus, Lawrence Brem, Das Chebattina, Hector Donolo, Ericka Dumene, Mike Hammer, Matti Hendriksson, Sindu Kadiyala, Jennifer Lee, Anna Lu, Trevor O’Connor, Waleed Osman, Jeremy Pumphrey, Anjan Purkayastha, Vivek Ramani, Joshua Scibelli, Matt Smith, Anna Steinfeld, John Thompson, Subhash Vallala, Joseph Verbeck, Cindy Winter, Shan Yang, Qing Yu, Rick Zakharov. The Frederick National Laboratory Molecular Characterization Laboratory (MoCha), Frederick, MD: Courtney Bouk, Ting-Chia Chang, Ashley Fredlock, Kneshay Harper, Robin Harrington, Christopher Karlovich, Marvin Majano, Sean McDermott, Erin Mosher, Patricia Runge, Maria Saeed. University of Texas MD Anderson Cancer Center Laboratory, Houston, TX: Geeta S. Mantha, Divya Panditi, Keyur Pravinchandra Patel, Ryan Pepper, Luthra Rajyalakshmi, Neelima G. Reddy, Rajesh R. Singh, Debashish Tripathy, Johathan Yau. Yale Laboratory, New Haven, CT: Sandra Canosa, Karin Finberg, Jennifer Huang, Karyn Ronski, T. Zenta Walther.
Dr Flaherty reports personal fees and stock ownership from Loxo Oncology, Clovis Oncology, Strata Oncology, Vivid Biosciences, Checkmate Pharmaceuticals, X4 Pharmaceuticals, PIC Therapeutics, Fount Therapeutics, Shattuck Lab, Apricity, Oncoceutics, Fog Pharma, and Tvardi; personal fees from Sanofi, Amgen, Asana Biosciences, Aeglea, Array BioPharma, Tolero, Neon Therapeutics, Cell Signaling Technologies, Novartis, Genetech, Bristol Myers Squibb, Merck, Takeda, Verastem, Boston Biomedical, Pierre Fabre, Cell Medica, and Debiopharma, outside the submitted work. Dr Gray reports grants from National Cancer Institute, during the conduct of the study. Dr Chen reports employment from NCI, during the conduct of the study. Dr Hamilton reports grants from National Cancer Institute, during conduct of the study; grants from National Cancer Institute; consultant from Bristol-Myers Squibb Company, Guardant, and LOXO-Oncology; scientific or advisory committee member from Merck & Co, Inc. and Thermo-Fisher Scientific; advisory board member from Fred Hutchinson Cancer Research Center, Centers for Medicare and Medicaid Service, and Halio DX, outside the submitted work. Dr Mitchell reports grants and personal fees from Genentech; personal fees from Novartis, Janssen, and Bristol Myers Squibb, outside the submitted work. Dr Iafrate reports personal fees and equity holder from ArcherDx; grants from Sanofi, outside the submitted work. In addition, Dr. Iafrate has a patent USSN 13/793,564 issued to ArcherDx. Dr Catalano reports grants from the NIH, during the conduct of the study. Dr Alexander reports personal fees from Foundation Medicine, Inc., outside the submitted work; relationships with Global Coalition for Adaptive Research. Dr Kumar reports grants from Abbvie, Celgene, Janssen, Merck, Novartis, Roche, Sanofi, Takeda, KITE, and Medimmune/Astra Zeneca; personal fees from Oncopeptides and Adaptive, outside the submitted work. Dr Meric-Bernstam reports grants from Novartis/Aduro, Calithera, Bayer, Jounce, CytoMx, eFFECTOR, PUMA Biotechnology, Curis, Millennium, Glaxosmithkline, Daiichi Sankyo, Abbvie, Guardant Health, Takeda, and Aileron; personal fees from Inflection Biosciences, Pieris, Darwin Health, GRAIL, Clearlight Diagnostics, Dialectiva, Sumitomo Dainippon, Spectrum, Samsung Bioepis, Aduro, OrigiMed, Xencor, Jackson Laboratory, Mersana, Kolon Life Science, and Parexel International; grants and personal fees from Taiho, Genetech, Debio, Zymeworks, Pfizer and Seattle Genetics, outside the submitted work. Dr Caimi reports grants and personal fees from Genentech; grants from Millenium Pharmaceuticals, Rhizen Pharmaceuticals, TG Therapeutics, Roche, and ADC Therapeutics; personal fees from Fate Therapeutics, Kite Pharmaceuticals, and Celgene, outside the submitted work. Dr. Arteaga receives or has received research grants from Pfizer, Lilly and Takeda; holds minor stock options in Provista; serves or has served in an advisory role to Novartis, Merck, Lilly, Daiichi Sankyo, Taiho Oncology, OrigiMed, Puma Biotechnology, Immunomedics, AstraZeneca, Arvinas, and Sanofi; and reports scientific advisory board remuneration from the Susan G. Komen Foundation.
Parts of the manuscript were presented in abstract form at the American Association for Cancer Research meeting in 2016 as well as at the 2017 EORTC-NCI-AACR Molecular Targets and Cancer Therapeutics meeting.
The authors thank the following, not named in committees (Supplementary Table 1, available online): Programs within the NCI Division of Cancer Treatment and Diagnosis: Margaret Mooney, Shannon Hatch, Richard Simon; University of Texas MD Anderson Cancer Center Clinical Trials Support Unit: Russell Broaddus, Andrew Say, Hyvan Dang, Donna Cole, Marilyn Dawlett; University of Texas MD Anderson Consulting Pathologists: Elizabeth Euscher, Greg Fuller, Michelle Williams, Victor Prieto, Annikka Weissferdt, Pheroze Tamboli, Asif Rashid; University of Texas MD Anderson Tissue Qualification Laboratory: Marlene Joya, Jasmine Saenz, Min Ye, Emily Hopper, Richard Rivas, Thomas Huynh, Aaron Belmares, Sameen Saleem, Alexis Villanueva, Anthony Herrera, Diem Tran, Sally Kahlil; University of Texas MD Anderson Image Analysis Laboratory: Mark Nwokocha; University of Texas MD Anderson Immunohistochemistry Laboratory: Suneetha Inampudi; Universtiy of Texas MD Anderson NCI MATCH Call Center: Paul Mills, Raquel Montelongo, Skariah Ancy; University of Texas MD Anderson Molecular Diagnostics Laboratory: Lynn Barron, Alexander Aheimer; Robert Brock, Paula Elizondo, Lisset Salazar, Hangnga Nguyen, Courtney Wunder, Roshen Jacob, Heather Vasquez, Somtochi Opara, Marcos Hernandez, Jennifer Newton Victor, Crystal Simien, Kristyn Jeter; University of Texas MD Anderson Cancer Center Cytology Laboratory: Surayya Siddiqui, Cicily Joseph; ECOG-ACRIN Central Biorepository and Pathology Facility at MD Anderson: Amanda Saladino, Julie Hodson, Jemimah Doma, Carlette Ledet.
Supplementary Material
References
- 1.Dawood S, Broglio K, Buzdar AU, Hortobagyi GN, Giordano SH.. Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment: an institutional-based review. J Clin Oncol. 2010;28(1):92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortesi L, De Matteis E, Cirilli C, Marcheselli L, Proietto M, Federico M.. Outcome evaluation in pre-trastuzumab era between different breast cancer phenotypes: a population-based study on Italian women. Tumori. 2012;98(6):743–750. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101(36):13306–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. [DOI] [PubMed] [Google Scholar]
- 6.Kwak EL, Bang YJ, Camidge R, et al. Anaplatic lymphoma kinase inhibition in non-small cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y-L, Yang JC-H, Kim D-W, et al. Phase II study of crizotinib in East Asian patients with ROS1 positive advanced non-small cell lung cancer. J Clin Oncol. 2018;36(14):1405–1411. [DOI] [PubMed] [Google Scholar]
- 8.Druker BJ.Imatinib as a paradigm of targeted therapies. Adv Cancer Res. 2004;91:1–30. [DOI] [PubMed] [Google Scholar]
- 9.Tefferi A, Pardanani A.. Imatinib therapy in clonal eosinophilic disorders, including systemic mastocytosis. Int J Hematol. 2004;79(5):441–447. [DOI] [PubMed] [Google Scholar]
- 10.Duffaud F, Le Cesne A.. Imatinib in the treatment of solid tumours. Targ Oncol. 2009;4(1):45–56. [DOI] [PubMed] [Google Scholar]
- 11.Le DT, Durham JN, Smith KN, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.US Food and Drug Administration. FDA approves larotrectinib for solid tumors with NTRK gene fusions. https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm626720.htm. Published December 14, 2018. Accessed January 14, 2019.
- 13.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373(8):726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. [DOI] [PubMed] [Google Scholar]
- 15.Lih CJ, Harrington RD, Sims DJ, et al. Analytical validation of the next-generation sequencing assay for a nationwide signal-finding clinical trial: molecular analysis for therapy choice clinical trial. J Mol Diagn. 2017;19(2):313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoury JD, Wang W-L, Prieto VG, et al. Validation of immunohistochemical assays for integral biomarkers in the NCI-MATCH EAY131 clinical trial. Clin Cancer Res. 2018;24(3):521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumors: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 18.Cheson BD, Fisher RI, Barrington SF, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol. 2014;32(27):3059–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Paiva B, Anderson KC, et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17(8):e328–e346. [DOI] [PubMed] [Google Scholar]
- 21.Le Tourneau C, Delord JP, Goncalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16(13):1324–1334. [DOI] [PubMed] [Google Scholar]
- 22.Stockley TL, Oza AM, Berman HK, et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016;8(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.TsimberIdou A-M, Hong DS, Ye Y, et al. Initiative for Molecular Profiling and Advanced Cancer Therapy (IMPACT): an MD Anderson Precision Medicine Study. J Clin Oncol Precis Oncol. 2017;(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zehir A, Benayed R, Shah RK, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.