Abstract
Changes in DNA methylation have been found to be highly correlated with aging in humans, but causes or consequences of these changes are not understood. We characterized the DNA methylomes of several hundred people in the Invecchiare in Chianti study to identify DNA sites in which percent methylation was systematically different with age. Then, we tested the hypothesis that changes of percent methylation in the same DNA sites occur longitudinally for the same DNA sites in the same subjects. We identified six differentially methylated regions in which percent methylation showed robust longitudinal changes in the same direction. We then describe functions of the genes near these differentially methylated regions and their potential relationship with aging, noting that the genes appear to regulate metabolism or cell type specificity. The nature of transcription factor binding sites in the vicinity of these differentially methylated regions suggest that these age-associated methylation changes reflect modulation of two biological mechanisms: the polycomb repressive complex 2, a protein complex that trimethylates histone H3 on lysine 27, and the transcriptional repressor CCCTC-binding factor or CTCF, both of which are regulators of chromatin architecture. These findings are consistent with the idea that changes in methylation with aging are of adaptive nature.
Keywords: Epigenetics, Epigenomics, DNA methylation, Epigenetics of aging, Longitudinal epigenomics
Recent research has identified a group of cytosines in the DNA of mammals that undergo foreseeable changes in methylation with aging (1). Weighted sums of percent methylation in these overlapping sets of DNA sites are considered “epigenetic clocks” that closely approximate chronological age and, independently of chronological age, predict adverse health outcomes, including mortality (2–4). While several methylation-based measures of biological aging have been developed and correlated with health dimensions and outcomes, the molecular mechanism behind these systematic changes in methylation are less understood, although hypotheses have been considered in the literature (5). For example, it has been proposed that errors during replication of the DNA methylation from the parent to the daughter strand, as well as during subsequent methylation maintenance, may lead to hypermethylation or hypomethylation of certain sites with aging (6). While this hypothesis is compatible with the observation of regression to the mean in the CG methylation status of the genome with aging (7), a phenomenon referred to as “epigenetic drift,” it does not explain why aging predictably affects methylation only in specific regions and not others (8,9). An alternative interpretation is that changes in methylation are induced by internal or environmental stressors and represent adaptive responses, that is, attempts to avoid the accumulation of damage and its consequences. According to this view, a faster epigenetic clock indicates that damage accumulation is accelerated and triggers an accelerated adaptive response. Of note, this hypothesis is consistent with evidence that deviations from the normal trajectory of DNA methylation with aging conveys information on health status and predicts adverse health outcomes (10).
Methods
Invecchiare in Chianti (InCHIANTI) is a prospective cohort study of aging based in the Chianti region of Italy. Study participants were enrolled in 1998 and 2000 and followed up for 15 years (Table S1). Sample collection procedures have been described elsewhere (11). The InCHIANTI protocols were approved by the Instituto Nazionale Riposo e Cura Anziani institutional review board in Italy and study participants provided informed consent.
DNA methylation was evaluated from 395 study participants in the InCHIANTI study, with ages ranging from 30 to 97 years, for the study timepoint at 2013. Briefly, buffy coats were collected from participant blood samples by standard protocols, and genomic DNA extracted and quantified in an automated manner. DNA was bisulfite converted and analyzed with an Illumina 450k methylation array according to standard protocols (Illumina, Inc., San Diego, CA). The resulting probe intensities were normalized according to the method of Triche and colleagues (12). The normalized intensities were then converted from methylated signal and unmethylated signal values to estimates of the proportion of methylated/unmethylated (beta values). Finally, probes were mapped to the genome. For discovery of regions of differential methylation, the assumption was made that changes in DNA methylation during the course of aging are small but highly reproducible, as described earlier. Therefore, aging was used as a continuous variable and the cutoff for differential methylation was set very low; precisely, the threshold for differential methylation was varied between .01% and .1% average differences over the differential region. The number of differentially methylated regions thus was found to increase exponentially with decreasing threshold. For significance testing, the methylation data were 5,000-fold bootstrapped to generate a null distribution of differentially methylated regions, and the p value calculated as the likelihood that the differentially methylated region would be discovered randomly (13). The differential methylation cutoff was therefore set at the highest level which produced a significant set of genes differentially methylated with age, to find the genes whose methylation state was most strongly, and most robustly, correlated with age. For functional analysis and annotation, the Reactome database and Lincipedia were used (14,15). For longitudinal validation, DNA methylation for 352 of the 395 was evaluated at 1999, 2007, and 2013 timepoints for the differentially methylated region loci found cross-sectionally, and the lmfit R package was used with a random intercept model.
Results
A first attempt to examine the question of consistent methylation change with age is to look for areas of the genome that undergo predictable changes in methylation with aging, and identify genes and transcription factor binding sites in those areas that may be functionally affected by differential methylation. To this end, we analyzed methylation data from 395 human blood samples from participants of the InCHIANTI study, which enrolled representative samples of human subjects with a broad range of ages, and measured DNA methylation with the Illumina Infinium 450k chip at baseline and at the 9- and 13-year follow-up visits. In a cross-sectional discovery analysis of the third timepoint, we identified age-associated differentially methylated regions (aDMRs) with a pattern of methylation that changed linearly with aging. To validate these findings, we examined the methylation state of these same aDMRs in 352 of the original participants who had been followed longitudinally over a 14 years follow-up, with methylation assessed at baseline, Year 9, and Year 13. As these aDMRs were associated with age both cross-sectionally and longitudinally, we then annotated them as to their nearest gene. Finally, we discuss the involvement of these genes in processes involved in aging, suggesting that they may be involved in the maintenance of health with aging.
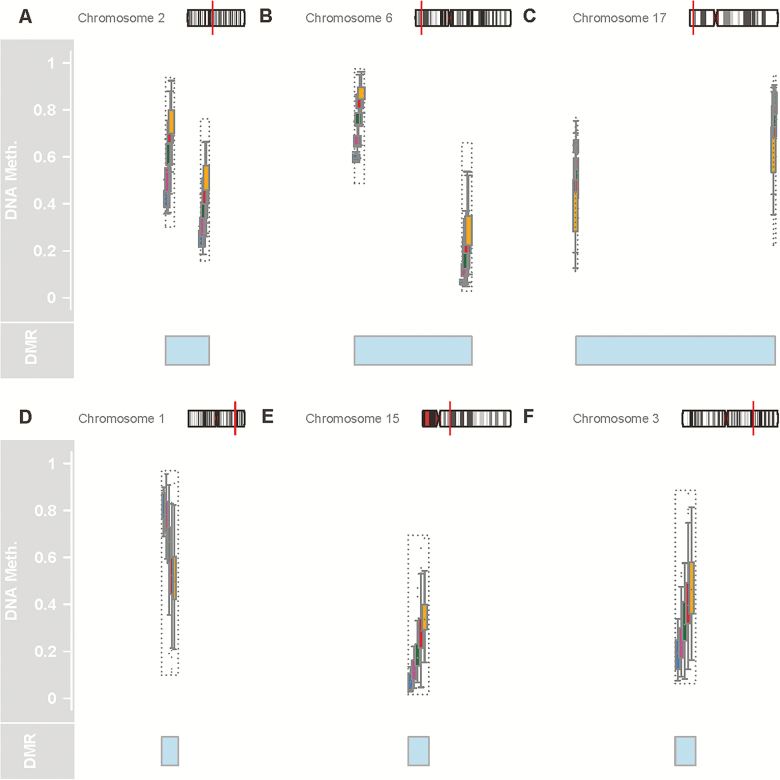
In the cross-sectional analysis, we searched for regions whose percent methylation was associated with age and are highly consistent between subjects. Analyses were adjusted for only technical variables (batch) and sex. To focus on overt systematic change in methylation well above random noise, a cutoff of .4% average differences per year was used to identify aDMRs with permutation-based p values (13). This resulted in six aDMRs associated with age with a p value of <.01 (Figure 1; Table 1). Figure 1 shows all six aDMRs, and their position in relation to start sites of neighboring genes. In Figure 1A, the aDMR maps to a CpG island just upstream from the transcription start site of several isoforms of the gene FHL2 (four and a half LIM domains 2), and differential methylation with age around this locus has been previously reported (16). The protein coded by FHL2 is a zinc-finger domain containing protein that inhibits the transcriptional activity of FOXO1 by facilitating its deacetylation through Sirtuin‐1 (SIRT1), two molecules that have been widely recognized as playing important roles in the biology of aging (17). In Figure 1B, the aDMR maps to a CpG island near the transcription start site of the protein ELOVL2 (elongase of very long chain fatty acids 2). Across multiple studies, the relation between percent methylation surrounding the ELOVL2 locus and aging is stronger than for any other loci, but in spite of significant research, the reason for this association is not understood. In Figure 1C, the aDMR maps to a CpG island near the TSS of one isoform of the gene aspartoacylase (ASPA). Previous studies have found that methylation around ASPA is affected by vitamin supplementation and especially vitamin B (18). In addition, ASPA is one of the genes whose methylation best discriminates monozygotic twins, suggesting that this gene is highly sensitive to environmental influences (19). In Figure 1D, the aDMR maps to a CpG island near the gene MIR29B2CHG (MicroRNA-29b2, c host gene). This gene is known to contain within its transcribed region two microRNAs, Mir-29b2 and Mir-29c, and so is possibly a so-called “host gene” for these microRNAs. Of note, hypomethylation in the Mir-29b2 neighbor with aging has been previously reported (20). Furthermore, corresponding to hypomethylation and increased gene expression, Mir-29b and Mir-29c gene expression was found to be significantly increased in a mouse model of Hutchinson–Gifford progeria syndrome (21). In Figure 1E, the aDMR maps to a CpG island near the TSS for ZIC4 (Zinc-finger protein family member 4). This protein is involved in organ specification and definition of cell identity during development. In Figure 1F, the aDMR maps to an intragenic CpG island in an exon of one isoform of the gene OTUD7A (OTU domain containing 7a), a deubiquitinase which may interact with the TRAF6 pathway, which may be involved in cell specification as well. Hypermethylation of OTUD7a with age was also reported previously (22). Overall, the genes identified function within the two broad categories of metabolism and cell and tissue type specification.
Figure 1.
Six age-associated differentially methylated regions (aDMRs) described in this study. Each panel shows, from top to bottom, the location of the aDMR within the chromosome; the normalized methylation data at each CpG along the aDMR; a highlight of the region considered the aDMR. The chromosomal location is represented in an ideogram showing the chromosome structure, in particular chromosome bands, with the aDMR location represented as a red line. Normalized methylation data at each CpG is shown with each box representing one age group as color-coded, with blue as ages 20–34, magenta as 35–49, green as 50–64, red as 65–79, and yellow for >80. The blue shaded region shows the region between the regulated CpGs, which we consider as a aDMR. In cases in which only one CpG is changing, the aDMR highlight marks only that CpG. In A, B, and C, two CpGs constitute the aDMR; in E, F, and G, one CpG represents the aDMR. For A, B, and C, the ends of the highlight denote the positions of the CpGs; for E, F, and G, the center of the highlight is position of the CpG.
Table 1.
All aDMRs Found Above the Threshold of .4% Average Methylation Change per Year, Including Those That Did Not Pass Significance Testing
| Chromosome | Start Index | End Index | Methylation Change | Length | p Value | FWER | Annotation | EZH2 Binding | SUZ12 Binding | CTCF Binding |
|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome 2 | 106015767 | 106015771 | 0.005309614 | 2 | 0 | 0 | FHL2 | Weak, displaced | None | Strong |
| Chromosome 6 | 11044877 | 11044888 | 0.004609445 | 2 | 0 | 0 | ELOVL2 | Strong | Weak | Strong |
| Chromosome 17 | 3379283 | 3379567 | −0.004233153 | 2 | 0 | 0 | ASPA | None | None | Weak |
| Chromosome 1 | 207997020 | 207997020 | −0.006858645 | 1 | 0 | 0 | MIR29B2CHG | None | None | Strong |
| Chromosome 15 | 31775895 | 31775895 | 0.005213145 | 1 | 0 | 0 | OTUD7a | Strong | Weak | Weak |
| Chromosome 3 | 147126753 | 147126753 | 0.005123596 | 1 | 0.007491 | 0.0004 | ZIC4 | Strong | Weak | Weak |
| Chromosome 2 | 145116633 | 145116633 | −0.004828198 | 1 | 0.037453 | 0.002 | ||||
| Chromosome 9 | 133908909 | 133908909 | −0.004801037 | 1 | 0.037453 | 0.002 | ||||
| Chromosome 5 | 16179633 | 16179633 | 0.004699174 | 1 | 0.048689 | 0.0026 | ||||
| Chromosome 2 | 164590272 | 164590272 | −0.004456642 | 1 | 0.146067 | 0.0078 | ||||
| Chromosome 14 | 94405681 | 94405681 | 0.00440554 | 1 | 0.17603 | 0.0092 | ||||
| Chromosome 5 | 95295740 | 95295740 | −0.004379721 | 1 | 0.198502 | 0.0104 | ||||
| Chromosome 2 | 66654644 | 66654644 | −0.004320193 | 1 | 0.277154 | 0.0142 | ||||
| Chromosome 19 | 16830749 | 16830749 | −0.004301742 | 1 | 0.303371 | 0.0156 | ||||
| Chromosome 20 | 2730488 | 2730488 | 0.004195198 | 1 | 0.430712 | 0.0218 | ||||
| Chromosome 6 | 110736772 | 110736772 | −0.004156555 | 1 | 0.509363 | 0.026 | ||||
| Chromosome 12 | 98151379 | 98151379 | −0.004086738 | 1 | 0.674157 | 0.0344 | ||||
| Chromosome 6 | 105388694 | 105388694 | 0.004028017 | 1 | 0.898876 | 0.0462 | ||||
| Chromosome 1 | 206685423 | 206685423 | −0.004024052 | 1 | 0.910112 | 0.0468 | ||||
| Chromosome 1 | 77907656 | 77907656 | 0.004003854 | 1 | 0.988764 | 0.0506 |
Note: Those that pass significance testing were annotated as to the most likely regulated gene and their transcription factor binding patterns.
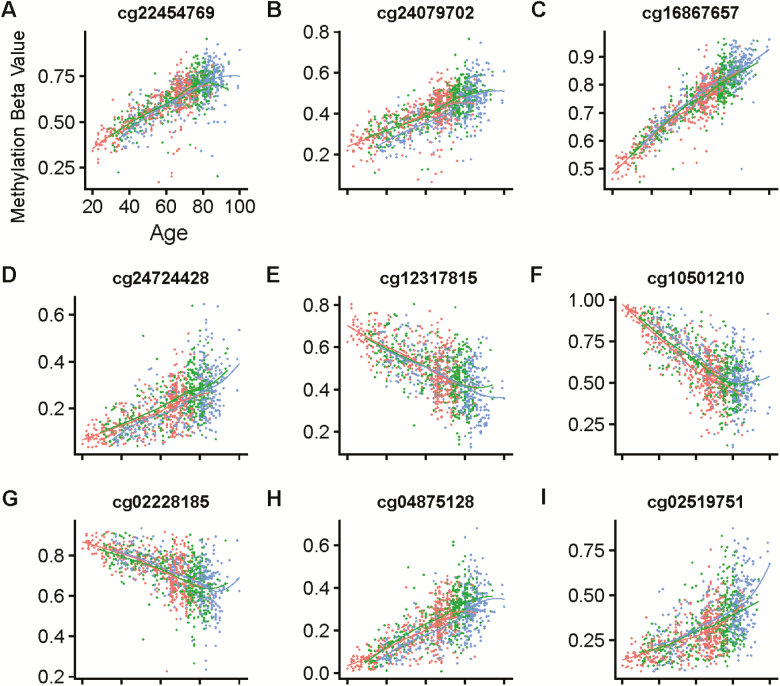
To validate our cross-sectional findings longitudinally, we extracted the methylation levels for all the CpG dinucleotides comprising our six aDMRs. As some of the aDMRs contain more than one CpG, this produces a list of nine CpGs. We then examined the longitudinal methylation levels of these CpG probes for 352 of the 395 subjects already described. Longitudinal trajectories of percent methylation over 3 timepoints that cover 13 years of follow-up are shown in Figure 2 for each of the nine CpGs considered. The FHL2 aDMR contains CpGs cg22454769 and cg24079702, which are shown in Figure 2A and B. The ELOVL2 aDMR contains CpGs cg16867657 and cg24724428, which are shown in Figure 2C and D. The ASPA aDMR contains CpGs cg12317815 and cg02228185, which are shown in Figure 2E and F. The MIR29B2CHG aDMR contains the CpG cg10501210, shown in Figure 2G. The ZIC4 aDMR contains the CpG cg02519751, shown in Figure 2H. Finally, the OTUD7a aDMR contains the CpG cg04875128, shown in Figure 2I. In a mixed-effects regression model adjusted for sex and methylation batch effects, changes in methylation in all sites considered were robustly associated with aging (p < .0001 for all models) Table S2.
Figure 2.
The trajectories of the methylation levels (beta values) of all nine aDMR CpGs are plotted longitudinally, with each panel showing one CpG. Each dot represents one measure of CpG methylation, and dots are color-coded according to the timepoint, with red as timepoint 1 (time 0), green as timepoint 2 (time +9 years), and blue as timepoint 3 (time +15 years). Each line indicates a loess regression of each timepoint, to show batch effects. A–E: Each individual CpG considered.
Discussion
As these differentially methylated regions were found to be age-associated both cross-sectionally and longitudinally with very stringent criteria, we hypothesized that these methylation changes may reflect critical biological mechanism in aging, either indicating damage or compensatory/adaptive mechanisms. Hence, we sought to gain some understanding of the biological mechanism underneath these methylation changes by searching for transcription factor binding sites close by these aDMR loci using the catalog published by the ENCODE Factorbook project (23). Strikingly, all six of the aDMRs were found to have binding sites for proteins EZH2 and SUZ12, both of which are components of the polycomb repressive complex 2 (PRC2), a protein complex that trimethylates histone H3 on lysine 27, or bind the transcriptional repressor CTCF (Table 1). PRC2 is known to interact with all three DNA methyltransferase or DNMT enzymes and promote DNA methylation (24). Similarly, CTCF has recently been described to modulate so-called “hemimethylation” of genomic DNA, in which a CG dinucleotide on one strand of the genome is held unmethylated despite methylation of the opposite strand CG dinucleotide (25). Such hemimethylation could cause a progressive loss of methylation at these loci over the course of cell division. Consistent with this interpretation, the four aDMRs (FHL2, ELOVL2, OTUD7a, and ZIC4) that have binding sites for EZH2 or SUZ12 show increased methylation over aging. In contrast, the two aDMRs (ASPA, MIR29B2CHG) at which there is no EZH2 or SUZ12 binding, but where CTCF binding is prevalent, show decreased methylation during aging. It is possible that PRC2-signaled methylation is an active process, while loss of methylation by CTCF binding is stochastic, and occurs only when a PRC2 signal is absent. However, we admit that this mechanism is speculative and controlled experiments in a model system will be required to demonstrate it.
At least in part, these findings are consistent with the idea that epigenetic clock is related to cell division and proliferation during aging, as others have suggested (5). The input to trigger changes in methylation with aging are still undefined, but it is interesting to speculate that these changes are an adaptive mechanism aimed at maintaining normal cell function in spite of damage accumulation. Consistent with this hypothesis, the PRC2 pathway, has been implicated in hematopoietic stem cell aging (26) and in general in the maintenance of stem cell function. Indeed, our results are highly related those of Teschendorff and coworkers (27), who developed an epigenetic clock only from PRC2 targets which were hypermethylated with age. This clock was shown to correlate well with the rate of stem cell proliferation in a given tissue, especially in tissues with high stem cell proliferation. Thus, the relationship between PRC2 signaling, cellular proliferation, and the epigenetic clock deserves further study. The possibility that methylation changes with aging is signaling based, and compensatory in nature rather than reflecting random errors accumulation, drift or noise should be tested in future studies.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging. The InCHIANTI study baseline (1998–2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164; and 263 MD 821336); the InCHIANTI Follow-up 1 (2001–2003) was funded by the U.S. National Institute on Aging (Contracts: N.1-AG-1-1; and N.1-AG-1-2111); the InCHIANTI Follow-ups 2 and 3 studies (2004–2010) were financed by the U.S. National Institute on Aging (Contract: N01-AG-5-0002); supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Baltimore, MD.
Conflict of Interest
None reported.
Supplementary Material
Acknowledgments
L.F. designed the study and data collection, and R.T. and L.F. performed analyses. D.H. performed microarray experiments. C.U.-M., T.T., and A.Z.M. contributed to analysis and article.
References
- 1. Koch CM, Wagner W. Epigenetic-aging-signature to determine age in different tissues. Aging (Albany NY). 2011;3:1018–1027. doi: 10.18632/aging.100395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weidner CI, Lin Q, Koch CM, et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15(2):R24. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bacalini MG, Deelen J, Pirazzini C, et al. Systemic age-associated DNA hypermethylation of ELOVL2 gene: in vivo and in vitro evidences of a cell replication process. J Gerontol A Biol Sci Med Sci. 2017;72:1015–1023. doi: 10.1093/gerona/glw185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim JY, Tavaré S, Shibata D. Counting human somatic cell replications: methylation mirrors endometrial stem cell divisions. Proc Natl Acad Sci U S A. 2005;102:17739–17744. doi: 10.1073/pnas.0503976102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maegawa S, Lu Y, Tahara T, et al. Caloric restriction delays age-related methylation drift. Nat Commun. 2017;8:539. doi: 10.1038/s41467-017-00607-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shipony Z, Mukamel Z, Cohen NM, et al. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature. 2014;513:115–119. doi: 10.1038/nature13458 [DOI] [PubMed] [Google Scholar]
- 9. Jenkinson G, Pujadas E, Goutsias J, Feinberg AP. Potential energy landscapes identify the information-theoretic nature of the epigenome. Nat Genet. 2017;49:719–729. doi: 10.1038/ng.3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY). 2016;8:1844–1865. doi: 10.18632/aging.101020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moore AZ, Hernandez DG, Tanaka T, et al. Change in epigenome-wide DNA methylation over 9 years and subsequent mortality: results from the inCHIANTI study. J Gerontol A Biol Sci Med Sci. 2016;71:1029–1035. doi: 10.1093/gerona/glv118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Triche TJ Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41(7):1090. doi: 10.1093/nar/gkt090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaffe AE, Murakami P, Lee H, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fabregat A, Jupe S, Matthews L, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–D655. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Volders PJ, Anckaert J, Verheggen K, et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019;47(D1):D135–D139. doi: 10.1093/nar/gky1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steegenga WT, Boekschoten MV, Lute C, et al. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age (Dordr). 2014;36:9648. doi: 10.1007/s11357-014-9648-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang Y, Hou H, Haller EM, Nicosia SV, Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005;24(5):1021–1032. doi: 10.1038/sj.emboj.7600570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Obeid R, Hübner U, Bodis M, Graeber S, Geisel J. Effect of adding B-vitamins to vitamin D and calcium supplementation on CpG methylation of epigenetic aging markers. Nutr Metab Cardiovasc Dis. 2018;28:411–417. doi: 10.1016/j.numecd.2017.12.006 [DOI] [PubMed] [Google Scholar]
- 19. Marqueta-Gracia JJ, Álvarez-Álvarez M, Baeta M, et al. Differentially methylated CpG regions analyzed by PCR-high resolution melting for monozygotic twin pair discrimination. Forensic Sci Int Genet. 2018;37:e1–e5. doi: 10.1016/j.fsigen.2018.08.013 [DOI] [PubMed] [Google Scholar]
- 20. Tserel L, Limbach M, Saare M, et al. CpG sites associated with NRP1, NRXN2 and miR-29b-2 are hypomethylated in monocytes during ageing. Immun Ageing. 2014;11:1. doi: 10.1186/1742-4933-11-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ugalde AP, Ramsay AJ, de la Rosa J, et al. Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. EMBO J. 2011;30:2219–2232. doi: 10.1038/emboj.2011.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Florath I, Butterbach K, Müller H, Bewerunge-Hudler M, Brenner H. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23:1186–1201. doi: 10.1093/hmg/ddt531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J, Zhuang J, Iyer S, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Viré E, Brenner C, Deplus R, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431 [DOI] [PubMed] [Google Scholar]
- 25. Xu C, Corces VG. Nascent DNA methylome mapping reveals inheritance of hemimethylation at CTCF/cohesin sites. Science. 2018;359:1166–1170. doi: 10.1126/science.aan5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beerman I, Bock C, Garrison BS, et al. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 2013;12:413–425. doi: 10.1016/j.stem.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 27. Yang Z, Wong A, Kuh D, et al. Correlation of an epigenetic mitotic clock with cancer risk. Genome Biol. 2016;17:205. doi: 10.1186/s13059-016-1064-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.