Abstract
Background
Chemokine signaling may contribute to progression of low-grade gliomas (LGGs) by altering tumor behavior or impacting the tumor microenvironment. In this study, we investigated the role of CX3C chemokine receptor 1 (CX3CR1) signaling in malignant transformation of LGGs.
Methods
Ninety patients with LGGs were genotyped for the presence of common CX3CR1 V249I polymorphism and examined for genotype-dependent alterations in survival, gene expression, and tumor microenvironment. A genetically engineered mouse model was leveraged to model endogenous intracranial gliomas with targeted expression of CX3C ligand 1 (CX3CL1) and CX3CR1, individually or in combination.
Results
LGG patients who were heterozygous (V/I; n = 43) or homozygous (I/I; n = 2) for the CX3CR1 V249I polymorphism had significantly improved median overall (14.8 vs 9.8 y, P < 0.05) and progression-free survival (8.6 vs 6.5 y, P < 0.05) compared with those with the wild type genotype (V/V; n = 45). Tumors from the V/I + I/I group exhibited significantly decreased levels of CCL2 and MMP9 transcripts, correlating with reduced intratumoral M2 macrophage infiltration and microvessel density. In an immunocompetent mouse model of LGGs, coexpression of CX3CL1 and CX3CR1 promoted a more malignant tumor phenotype characterized by increased microglia/macrophage infiltration and microvessel density, resulting in shorter survival.
Conclusions
CX3CR1 V249I polymorphism is associated with improved overall and progression-free survival in LGGs. CX3CR1 signaling enhances accumulation of tumor associated microglia/macrophages and angiogenesis during malignant transformation.
Keywords: CX3CR1, glioma, malignant transformation, polymorphism, tumor microenvironment
Key Points.
1. A common CX3CR1 polymorphism improves survival in low-grade glioma patients.
2. CX3CR1 signaling promotes a protumoral microenvironment.
Importance of the Study.
Recently, management of LGG has shifted away from watchful waiting to aggressive treatment based on molecular characterization, generating renewed interest for new therapeutic targets and prognostic markers. In this study we found that a common polymorphism in the chemokine receptor gene CX3CR1 was associated with altered tumor microenvironment and improved survival in LGG patients. On the other hand, induction of CX3CR1 signaling in a mouse glioma model promoted a more malignant tumor phenotype and shorter survival. These results suggest that CX3CR1 is both a potential prognostic marker and a therapeutic target in LGG.
High-grade gliomas (HGGs), including 5–10% of the most common primary brain tumor, glioblastoma (GBM), may arise from malignant transformation of low-grade gliomas (LGGs).1 While LGGs demonstrate a slow rate of growth, once they transform to HGGs, survival rates decline precipitously despite maximal therapy.2 The time frame of LGG progression—influenced by various factors including age, extent of resection, adjuvant therapy, isocitrate dehydrogenase 1 (IDH1) mutant status, and presence of 1p/19q codeletion—provides a potential window for therapeutic intervention.3 However, the mechanisms underlying malignant transformation of LGGs remain incompletely understood. Recent studies have demonstrated that immunosuppressive tumor microenvironment may play a critical role in glioma progression. Compared with LGGs, HGGs contain increased numbers of regulatory T cells, myeloid derived suppressor cells, and tumor-associated microglia/macrophages (TAMs).4–6 Reversing immunosuppression delays tumor progression and prolongs survival in mouse models.7–10
Accumulating data suggest that chemokines influence the glioma microenvironment.11 In particular, expression of CX3CL1 is increased in HGGs compared with LGGs, whereas its cognate receptor CX3CR1 is expressed by the tumor cells, suggesting a potential role for CX3CL1-CX3CR1 signaling in malignant transformation of LGGs.12 However, mechanistic studies have yielded conflicting results. Common CX3CR1 V249I polymorphism improved overall survival in GBM patients following surgical resection and decreased TAM infiltration.13 In contrast, genetic deletion of Cx3cr1 in a mouse model of GBM shortened survival and promoted accumulation of inflammatory monocytes.14 To complicate matters further, intratumoral CX3CL1-CX3CR1 signaling was shown to directly influence glioma behavior, including invasion and adhesion.15
We hypothesized that CX3CL1-CX3CR1 signaling influences malignant transformation of LGGs. Interestingly, previous studies have demonstrated that the common CX3CR1 V249I variant is associated with reduced number of CX3CL1 binding sites16 and impaired signaling,17 although exact mechanisms remain unclear. Therefore, we determined the effect of this polymorphism on survival, gene expression, and tumor microenvironment in a cohort of patients with malignantly transformed LGGs. Furthermore, we investigated the effect of CX3CL1 and CX3CR1 expression on survival, malignant transformation, and tumor microenvironment in an immunocompetent mouse model. We show that LGG patients homozygous or heterozygous for the common CX3CR1 V249I polymorphism have a significant survival advantage compared with those with the wild-type allele. In addition, CX3CR1 signaling promotes vascular proliferation and TAM infiltration during malignant transformation.
Materials and Methods
Patients
Ninety patients who met the following inclusion criteria were analyzed in the study: initial histologic diagnosis of LGG (World Health Organization [WHO] grade II) between 1994 and 2011, subsequent histology-confirmed malignant transformation to HGG (WHO grade III) or GBM (WHO grade IV), archived tumor tissue available, clinical data and follow-up available, age at diagnosis ≥18 years, and written informed consent from patients or their representatives. The MD Anderson institutional review board approved the study (protocol #PA14-0709).
Patient characteristics were derived from chart review. Presence of IDH1 mutation was determined by immunohistochemistry as described below, if not already determined at the time of diagnosis. Codeletion status of 1p/19q was available for only 42 patients. Overall survival (OS) was defined as the length of time from histologic diagnosis of LGG to death or last follow-up. Progression-free survival (PFS) was defined as the length of time from histologic diagnosis of LGG to pathologic confirmation of malignant transformation. Patients still alive at last follow-up were censored from the analysis.
Genotyping of Polymorphism
Genomic DNA was purified from formalin-fixed paraffin-embedded (FFPE) tumor tissue or GBM stem cells (GSCs) using MasterPure kit (Epicentre). CX3CR1 V249I polymorphism (rs3732379) was genotyped using a commercially available TaqMan SNP Genotyping Assay (Applied Biosystems) on the 7500 Fast Real-Time PCR System (Applied Biosystems). Representative amplification plots for different genotypes are shown in Supplementary Figure 1.
RNA Sequencing and Heatmap Analysis
Total RNA was isolated from FFPE tumor tissue using MasterPure kit (Epicentre). Paired-end RNA sequencing was performed using HiSeq 2000 Sequencing System (Illumina) at the MD Anderson Sequencing and Microarray Facility. Raw reads were aligned to the human genome, counts quantified, and differential gene expression determined. Significantly differentially expressed genes were defined by a false discovery rate of 0.1 and log2 fold change threshold of 1 after filtering genes with low expression and illustrated by a heatmap. Samples with degraded RNA or low mapping rates were excluded. Additional details are provided in Supplementary Materials and Methods.
In Vitro Studies
GSCs were derived from acute cell dissociation of surgical GBM specimens from patients treated at MD Anderson, as described previously.18 Following overnight stimulation with 100 ng/mL recombinant human CX3CL1 (R&D Systems) or equivalent volume of phosphate buffered saline (PBS), RNA was extracted, reverse transcribed into cDNA, and amplified using TaqMan probes specific for chemokine C-C ligand 2 (CCL2) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Applied Biosystems). The difference in CCL2 transcript levels in response to CX3CL1 versus PBS stimulation was expressed as fold change using the delta delta cycle threshold (ddCt) method, with GAPDH as endogenous control.
For CX3CR1 immunofluorescence, GSCs were cultured overnight on chamber slides coated with Matrigel (Corning) and fixed in methanol prior to staining. Additional details are provided in Supplementary Materials and Methods.
Vector Construction
Replication-competent avian sarcoma-leukosis virus long terminal repeat with a splice (RCAS)—platelet-derived growth factor B (PDGFB) was generated as described previously.19 RCAS-CX3CL1 and RCAS-CX3CR1 were created by cloning human CX3CL1 and CX3CR1 (V249) cDNA into a Gateway-compatible RCAS vector using LR recombination (Invitrogen) and verified by sequencing.
Transfection of DF-1 Cells
Immortalized DF-1 chick fibroblast cells were grown in DMEM (Gibco) containing 10% fetal bovine serum at 37°C. Live virus was produced by transfecting RCAS-PDGFB, RCAS-CX3CL1, or RCAS-CX3CR1 into DF-1 cells using FuGENE-6 (Roche). Immunofluorescence with antibodies against human CX3CL1 and CX3CR1 were used to verify expression, with untransfected DF-1 cells as negative control.
Mouse Model
Generation of transgenic mice expressing the avian tumor virus receptor A under the Nestin promoter (Ntv-a) was described previously.20 DF-1 cells transfected with RCAS-PDGFB (1 × 104 cells in 1–2 μL of PBS), RCAS-CX3CL1 (1 × 105 cells), and/or RCAS-CX3CR1 (1 × 105 cells) were injected bilaterally into the frontal lobes of Ntv-a within 24–72 hours of birth when Nestin-positive cells are most proliferative. PDGFB (n = 31), PDGFB;CX3CL1 (n = 29), PDGFB;CX3CR1 (n = 23), and PDGFB;CX3CL1;CX3CR1 (n = 28) groups were generated. Mice were humanely euthanized by carbon dioxide asphyxiation between 90 and 105 days after injection, or sooner if symptoms related to tumor burden were present. The brains were removed, fixed in formalin, embedded in paraffin, and sectioned for immunohistochemical analysis. Animal experiments were approved by the MD Anderson Institutional Animal Care and Use Committee (protocol #00000900-RN01).
Tumor Grading
Histologic grades of mouse tumors were determined from hematoxylin and eosin–stained slides using WHO 2016 criteria by an investigator blinded to the group. LGGs were identified based on increased cellularity due to infiltrating tumor cells. HGGs were identified by the presence of microvascular proliferation, foci of necrosis, or brisk mitotic activity.
Immunohistochemistry and Immunofluorescence
Following heat-induced antigen retrieval and peroxide treatment, sections were stained with the following primary antibodies: mouse monoclonal anti-human IDH1 R132H (1:100, DIA-H09, Dianova), goat polyclonal anti-human tumor necrosis factor (TNF; 1:50, AF-410-NA, R&D Systems), rabbit polyclonal anti-human CD204 (1:100, orb100585, Biorbyt), rabbit polyclonal anti-human CD31 (1:50, ab28364, Abcam), or goat polyclonal anti-mouse CD31 (1:50, AF3628, R&D Systems).
For double immunofluorescence studies, sections were stained with the following combinations of primary antibodies: mouse monoclonal anti-human CX3CL1 (1:100, R&D Systems) and rabbit polyclonal anti-human CX3CR1 (1:100, ab8021, Abcam); goat polyclonal anti-human TNF (1:50) and rabbit polyclonal anti-human ionized calcium binding adaptor molecule 1 (Iba1: 1:500, Novus Biologicals); or mouse monoclonal anti-mouse Iba1 (1:500, MABN92, Millipore) and rabbit polyclonal anti-mouse oligodendrocyte transcription factor 2 (Olig2; 1:500, AB9610, Millipore).
Additional details are provided in Supplementary Material and Methods.
Analysis of Cell and Microvessel Density
The total number of cells and numbers of CD204 or Iba1-positive cells were counted in at least 5 non-overlapping high power fields from randomly selected, grade-matched human or mouse tumors. Density was calculated by dividing the number of positively stained cells by the total number of cells and expressed as a percentage.
In randomly selected, grade-matched mouse tumors, the number of distinct CD31-positive microvessels were counted from at least 5 non-overlapping high power fields in areas of highest microvessel density, and divided by the total quantified area in mm2. However, human tumors exhibited greater variability in overall cell density compared with mouse tumors, even with matching histologic grade. Therefore, CD31-positive cell density, rather than CD31-positive microvessel density, was compared in randomly selected human tumors using the same methods as above.
Statistical Analysis
Survival statistics were reported using Kaplan–Meier curves, which were compared using the log-rank test. Patient characteristics were compared using the chi-squared test, except for age at diagnosis, which was compared using the unpaired t-test. Unpaired t-test was used to compare GSC gene expression and CX3CR1 genotype or histology-dependent effects. Chi-squared test was used to compare mouse tumor grade and intratumoral hemorrhage. ANOVA with post hoc Tukey’s test was used to compare tumor size and microenvironment between mouse groups. Statistical analyses were performed using GraphPad Prism software.
Results
Effect of CX3CR1 V249I Polymorphism on Survival
Ninety LGG patients from an institutional repository with histological documentation of malignant transformation were included in the analysis. Genomic DNA was isolated from archived tumor samples and examined for the presence of the common germline CX3CR1 V249I polymorphism, associated with reduced number of CX3CL1 binding sites16 and decreased signaling,17 albeit with unclear mechanisms. Forty-five patients did not possess the polymorphism (V/V genotype), whereas 43 patients were heterozygous (V/I genotype) and 2 patients were homozygous for the I249 variant allele (I/I genotype). The calculated minor allele frequency of 0.27 in our cohort was comparable to what has been reported previously for patients with de novo GBMs13 and healthy controls.21
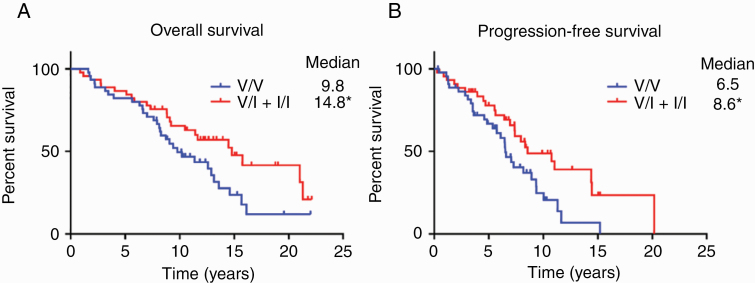
Patients with the germline CX3CR1 V249I polymorphism (V/I + I/I) had significantly longer OS than those without (V/V; 14.8 vs 9.8 y; P < 0.05, log-rank test; Fig. 1A). Similarly, patients in the V/I + I/I group had significantly longer PFS than those in the V/V group (8.6 vs 6.5 y; P < 0.05, log-rank test; Fig. 1B). Patient characteristics including sex, age at diagnosis, KPS, extent of resection, adjuvant therapy, and presence of IDH1 mutation and/or 1p/19q codeletion did not significantly differ between the genotypes (Table 1). Secondary analyses of OS in patients with IDH1 wild-type or 1p/19q codeleted tumors are shown in Supplementary Figure 2.
Fig. 1.
Overall (A) and progression-free survival (B) of patients with progressive LGGs, according to CX3CR1 genetic polymorphism. Kaplan–Meier survival curves were generated for patients heterozygous (V/I; n = 43) or homozygous (I/I; n = 2) for the V249I variant allele and compared against patients without the variant allele (V/V; n = 45). *P < 0.05, log-rank test.
Table 1.
Patient characteristics based on CX3CR1 genotype
| Characteristic | V/V, n (%) | V/I + I/I, n (%) |
|---|---|---|
| 45 | 45 | |
| Sex | ||
| Female | 19 (42) | 24 (53) |
| Male | 26 (58) | 21 (47) |
| Age at diagnosis, y, mean (SD) | 37.5 (12.2) | 38.1 (9.2) |
| KPS | ||
| 90 or 100 | 39 (87) | 42 (93) |
| <90 | 6 (13) | 3 (7) |
| Extent of resection | ||
| GTR | 11 (24) | 12 (27) |
| STR or biopsy | 34 (76) | 33 (73) |
| Adjuvant therapy | ||
| None | 12 (27) | 19 (42) |
| Radiation | 11 (24) | 12 (27) |
| Chemotherapy | 9 (20) | 9 (20) |
| Both | 13 (29) | 5 (11) |
| IDH1 mutation | ||
| No | 9 (20) | 10 (22) |
| Yes | 36 (80) | 35 (78) |
| 1p/19q codeletion* | ||
| No | 8 (30) | 10 (40) |
| Yes | 19 (70) | 15 (60) |
Abbreviations: GTR, gross total resection; IDH1, isocitrate dehydrogenase 1; SD, standard deviation; STR, subtotal resection; VV, homozygous for the common CX3CR1 V249 allele; V/I + I/I, heterozygous or homozygous for the variant CX3CR1 I249 allele.
*Only a subset of samples were analyzed for 1p/19q codeletion. None of the characteristics were significantly different between the 2 groups (unpaired t-test for age at diagnosis, chi-squared test for all others).
Genotype-Dependent Alterations in Gene Transcription and Tumor Microenvironment
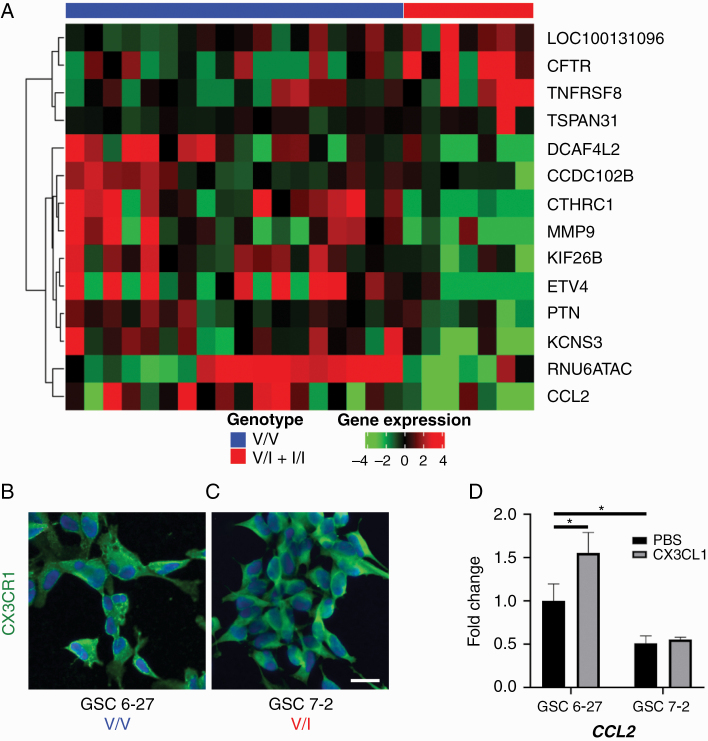
RNA isolated from HGG samples was sequenced and analyzed for differential gene expression based on genotype. HGGs from the V/I + I/I group demonstrated significantly decreased expression of CCL2, implicated in TAM recruitment,22 as well as significantly decreased expression of matrix metalloproteinase 9 (MMP9), implicated in tumor invasion and angiogenesis (Fig. 2A).23 Two separate GSC lines with proneural characteristics24 both exhibited membranous CX3CR1 expression despite different CX3CR1 genotypes (Fig. 2B, C). GSC6-27, derived from a V/V tumor, upregulated CCL2 in response to CX3CL1 stimulation; however, GSC7-2, derived from a V/I tumor, did not show this response (Fig. 2D). Interestingly, even in the absence of CX3CL1 stimulation, GSC7-2 exhibited reduced CCL2 transcript levels compared with GSC6-27. These results suggest that differential gene expression of CCL2 revealed by RNA sequencing was at least in part driven by altered genotype-dependent, intratumoral CX3CR1 signaling.
Fig. 2.
(A) Heatmap analysis of RNA sequencing data from HGGs, revealing differential expression of candidate genes CCL2 and MMP9 in tumors from V/I + I/I group compared with V/V group. Two separate GSC lines, 6-27 (B) and 7-2 (C), with similar proneural characteristics but different CX3CR1 genotypes were immunostained with antibody against CX3CR1 (green) and counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; blue), revealing robust membranous CX3CR1 expression. Stimulation of GSC6-27 (V/V) with CX3CL1 significantly increased CCL2 mRNA levels compared with treatment with PBS, whereas GSC7-2 (V/I) did not show this response (D; n = 3 for each condition). In addition, basal CCL2 mRNA levels were significantly lower in GSC7-2 compared with GSC6-27. *P < 0.05, unpaired t-test. Scale bar, 25 μm.
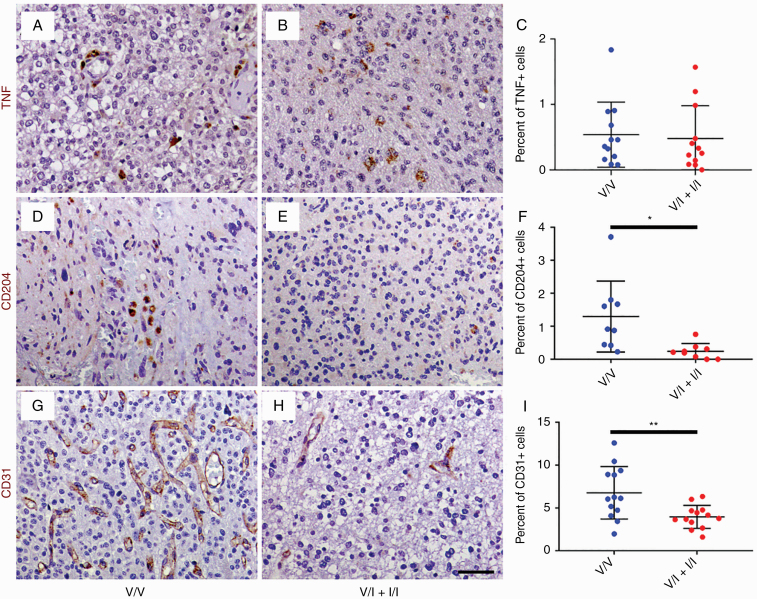
In order to determine the impact of altered CCL2 and MMP9 expression on the tumor microenvironment, histologic sections of randomly selected, grade-matched HGG samples from the V/V and V/I + I/I groups were immunostained with antibodies against the M1 macrophage marker TNF, M2 macrophage marker CD204, and endothelial marker CD31 (Fig. 3). HGGs from V/V and V/I + I/I genotypes exhibited sparse, punctate TNF immunoreactivity (Fig. 3A, B), predominantly within TAMs (Supplementary Figure 3). The mean density of TNF-positive cells was not significantly different between the groups (0.54% in V/V vs 0.48% in V/I + I/I; Fig. 3C). On the other hand, HGGs from the V/V genotypes demonstrated distinct areas of infiltration by CD204-positive TAMs with amoeboid morphology (Fig. 3D), which was largely absent in V/I + I/I HGGs (Fig. 3E). The mean density of CD204-positive cells in V/I + I/I tumors was 0.24%, which was significantly lower than 1.3% in V/V tumors (P < 0.05, unpaired t-test; Fig. 3F). In addition, V/I + I/I HGGs showed reduced numbers of CD31-positive microvessels compared with V/V HGGs (Fig. 3G, H). The mean density of CD31-positive cells in V/I + I/I tumors was 4.0%, which was significantly lower than 6.8% in V/V tumors (P < 0.01, unpaired t-test; Fig. 3I). Tumor histology did not significantly impact these results (Supplementary Figure 4).
Fig. 3.
Histological sections of WHO grade III gliomas from V/V (A, D, and G) and V/I + I/I patients (B, E, and H) were immunostained with antibodies against M1 macrophage marker TNF (brown, A and B), M2 macrophage marker CD204 (brown, D and E), and endothelial marker CD31 (brown, G and H), followed by counterstaining with hematoxylin (blue; A, B, D, E, G, and H). There were no significant differences in density of TNF-positive cells in HGGs from V/V (n = 12) and V/I patients (n = 12; C). On the other hand, HGGs from V/V patients (n = 9) demonstrated significantly increased density of CD204-positive macrophages compared with HGGs from V/I + I/I patients (n = 10; F). Similarly, V/V tumors (n = 13) showed significantly increased density of CD31-positive endothelial cells compared with V/I + I/I tumors (n = 13; I). *P < 0.05, **P < 0.01, unpaired t-test. Scale bar, 50 μm.
Effects of CX3CL1 and/or CX3CR1 Expression on Survival and Malignant Transformation in an RCAS/Ntv-a Mouse Model
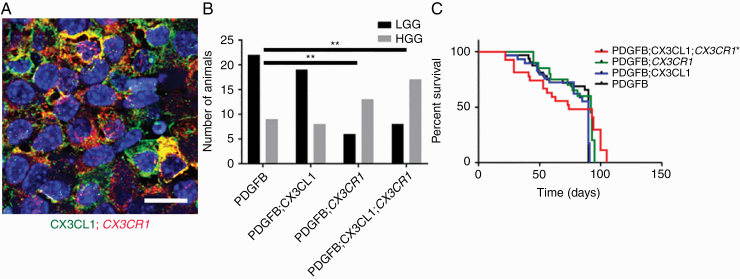
RCAS-driven expression of PDGFB ligand induces endogenous LGG formation from Nestin-positive glioneuronal precursors in immunocompetent Ntv-a mice.20 To further investigate the role of intratumoral CX3CL1-CX3CR1 signaling in vivo, CX3CL1 and CX3CR1 (V249) were expressed together with PDGFB in Ntv-a mice, separately or in combination. Tumor-specific, persistent expression of CX3CL1 and/or CX3CR1 was confirmed via immunofluorescence (Fig. 4A and Supplementary Figure 5). In the PDGFB group, all 31 mice formed tumors and 9 (29%) were HGGs. In the PDGFB;CX3CL1 group, 27 out of 29 mice formed tumors (93%) and 8 (30%) were HGGs. In the PDGFB;CX3CR1 group, 19 out of 23 mice formed tumors (83%) and 13 (68%) were HGGs. Finally, in the PDGFB;CX3CL1;CX3CR1 group, 25 out of 28 mice formed tumors (89%) and 17 (68%) were HGGs. The proportion of HGGs was significantly higher in the PDGFB;CX3CL1;CX3CR1 compared with the PDGFB group (P < 0.01, chi-squared test; Fig. 4B). Interestingly, the proportion of HGGs was also significantly higher in PDGFB;CX3CR1 compared with PDGFB group (P < 0.01, chi-squared test; Fig. 4B), suggesting cross-activation of the expressed human CX3CR1 by endogenous CX3CL1. In the PDGFB;CX3CL1;CX3CR1 group, the median survival was 74 days, which was significantly shorter than 90 days in the PDGFB group (P < 0.05, log-rank test; Fig. 4C). Median survival did not differ significantly in the PDGFB;CX3CL1 (90 days) or PDGFB;CX3CR1 groups (92 days), indicating that coexpression of ligand and receptor are required to fully promote the progression of PDGFB-induced gliomas in Ntv-a mice.
Fig. 4.
(A) Histological sections of murine gliomas induced by RCAS-PDGFB together with RCAS-CX3CL1 and RCAS-CX3CR1 were immunostained with antibodies against CX3CL1 (green) and CX3CR1 (red), followed by counterstaining with DAPI (blue), revealing membranous coexpression of CX3CL1 and CX3CR1. (B) Significantly higher proportion of HGGs were seen in mice injected with RCAS-PDGFB and RCAS-CX3CR1 (13/19) as well as in mice injected with RCAS-PDGFB, RCAS-CX3CL1, and RCAS-CX3CR1 (17/25) compared with mice injected with RCAS-PDGFB and RCAS-CX3CL1 (8/27) or with RCAS-PDGFB alone (9/31; **P < 0.01, chi-squared test). (C) Kaplan–Meier survival curves for PDGFB (n = 31), PDGFB;CX3CL1 (n = 29), PDGFB;CX3CR1 (n = 23), and PDGFB;CX3CL1;CX3CR1 (n = 28) groups show significantly shorter OS for PDGFB;CX3CL1;CX3CR1 group compared with PDGFB group (*P < 0.05, log-rank test). Scale bar, 25 μm.
Effect of CX3CL1 and/or CX3CR1 Expression on Tumor Microenvironment
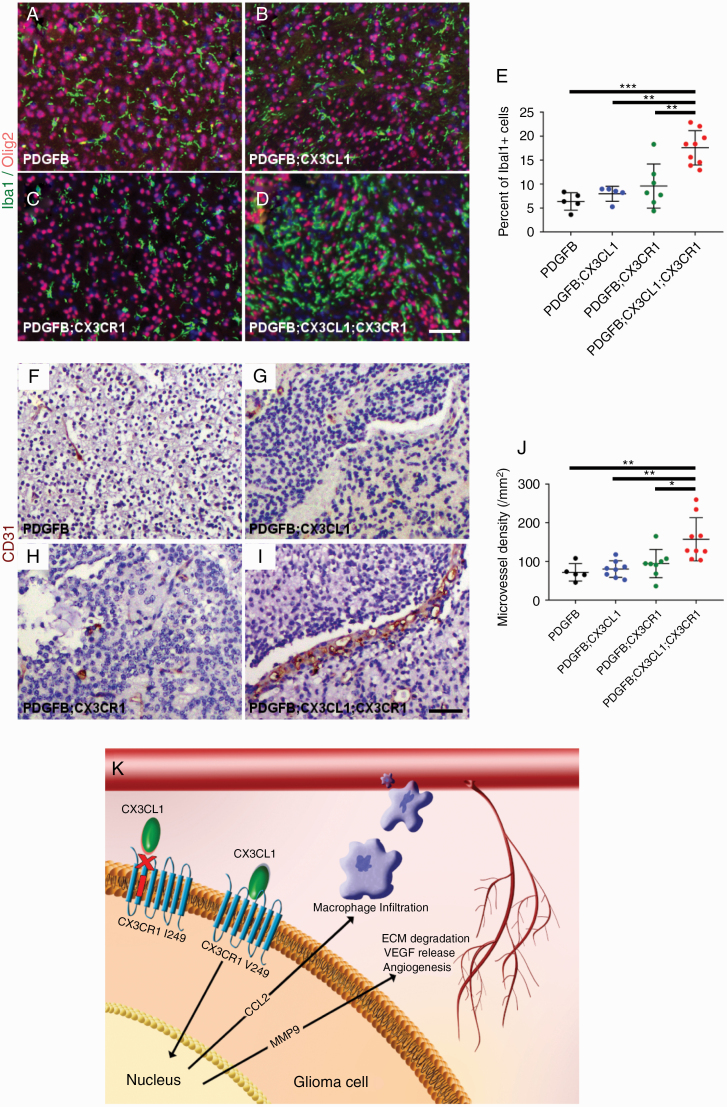
In order to characterize the tumor microenvironment, histologic sections of HGGs from PDGFB, PDGFB;CX3CL1, PDGFB;CX3CR1, and PDGFB;CX3CL1;CX3CR1 groups were immunostained with antibodies against the microglia/macrophage marker Iba1 and the oligodendrocyte lineage and tumor marker Olig2. All groups demonstrated more pronounced accumulation of Iba1-positive TAMs within Olig2-positive tumors compared with the surrounding normal brain (Fig. 5A–D). However, the mean density of Iba1-positive TAMs in HGGs from PDGFB;CX3CL1;CX3CR1 group (18%) was significantly higher compared with all other groups (6.4% in PDGFB, P < 0.001, ANOVA with post hoc Tukey’s test; 8.0% in PDGFB;CX3CL1, P < 0.01; and 9.6% in PDGFB;CX3CR1 groups, P < 0.01; Fig. 5E). TAM density did not correlate with tumor size, which was similar between groups (Supplementary Figure 6).
Fig. 5.
Histologic sections of murine HGGs from PDGFB (n = 5; A), PDGFB;CX3CL1 (n = 5; B), PDGFB;CX3CR1 (n = 7; C), and PDGFB;CX3CL1;CX3CR1 groups (n = 9; D) were immunostained with antibodies against microglia/macrophage marker Iba1 (green, A–D) and oligodendroglial lineage and tumor marker Olig2 (red, A–D), followed by counterstaining with DAPI (blue, A–D). Tumors from PDGFB;CX3CL1;CX3CR1 group exhibited significantly increased accumulation of microglia/macrophages, compared with tumors from all other groups (E). Histological sections of murine HGGs from PDGFB (n = 5; F), PDGFB;CX3CL1 (n = 8; G), PDGFB;CX3CR1 (n = 8; H), and PDGFB;CX3CL1;CX3CR1 groups (n = 9; I) were immunostained with antibody against endothelial marker CD31 (brown, F–I), followed by counterstaining with hematoxylin (blue, F–I). Tumors from PDGFB;CX3CL1;CX3CR1 group exhibited significantly increased density of CD31-positive microvessels, compared with tumors from all other groups (J). *P < 0.05, **P < 0.01, ***P < 0.001, ANOVA with post hoc Tukey’s test. Scale bar, 50 μm. (K) Working model. In gliomas, CX3CR1 signaling promotes CCL2-dependent TAM infiltration and MMP9-driven angiogenesis, which is abrogated by the I249 variant allele. ECM, extracellular matrix; VEGF, vascular endothelial growth factor.
In addition, tumor microvessel density was examined in HGGs from PDGFB, PDGFB;CX3CL1, PDGFB;CX3CR1, and PDGFB;CX3CL1;CX3CR1 groups via CD31 immunohistochemistry (Fig. 5F–I). Mean density of CD31-positive microvessels in HGGs from the PDGFB;CX3CL1;CX3CR1 group was 158/mm2, which was significantly higher compared with all other groups (72/mm2 in PDGFB, P < 0.01, ANOVA with post hoc Tukey’s test; 81/mm2 in PDGFB;CX3CL1, P < 0.01; and 95/mm2 in PDGFB;CX3CR1 groups, P < 0.05). Intratumoral hemorrhage occurred more frequently in HGGs from PDGFB;CX3CL1;CX3CR1 groups (16/17) compared with HGGs from all other groups (4/9 in PDGFB, P < 0.01, chi-squared test; 5/9 in PDGFB;CX3CL1, P < 0.05; and 5/13 in PDGFB CX3CR1 groups, P < 0.01; Supplementary Figure 7).
Discussion
In this study, we investigated the role of CX3CL1-CX3CR1 signaling in a retrospective cohort of patients with LGGs that ultimately progressed to HGGs. The germline CX3CR1 I249 variant allele had a profoundly beneficial impact, extending median OS by 5 years and PFS by nearly 2 years. Analysis of RNA sequencing data from tumor samples revealed reduced CCL2 and MMP9 transcript levels in association with the V249I polymorphism, consistent with reduced M2 TAM infiltration and microvessel density. In addition, GSC with the V/I genotype exhibited deficient response to CX3CL1 stimulation. Complementary to these findings, coexpression of CX3CL1 and CX3CR1 (V249) worsened survival and tumor grade in a PDGFB-driven mouse model of LGGs, by facilitating the accumulation of TAMs and increasing microvessel density. Taken together, our data suggest that CX3CL1-CX3CR1 signaling promotes malignant transformation of LGGs by upregulating CCL2-driven TAM infiltration and MMP9-driven angiogenesis, which is ameliorated by the I249 variant allele (Fig. 5K).
Previous investigations of CX3CR1 signaling on glioma pathogenesis have yielded conflicting results. In contrast to our data, Cx3cr1 deletion in RCAS-based, PDGFB-driven, Ink4a-Arf and Pten deficient mouse GBM model worsened survival and increased tumor incidence, by indirectly promoting infiltration of interleukin (IL)-1β producing inflammatory monocytes into perivascular areas.14 On the other hand, Cx3cr1 deficiency had a negligible effect on survival as well as TAM or lymphocyte accumulation following intracranial implantation of murine GL261 glioma cells.25 However, both of these studies relied upon aggressive tumor models that do not allow sufficient time course for malignant transformation, which was the specific focus of our study. In particular, the Ink4a-Arf and Pten deficient background results in gliomas that are predominantly high grade, and GL261 is an already fully malignant tumor. Furthermore, PDGFB-driven murine GBMs, unlike their human counterparts, do not appear to express CX3CR1, given the absence of tumor-specific green fluorescent protein (GFP) labeling in Cx3cr1GFP/+ mice.14 Therefore, the intratumoral CX3CL1-CX3CR1 signaling could not be specifically assessed. Indeed, the presence of CX3CR1 V249I polymorphism, reported to reduce ligand binding and signaling,16,17 increased OS and diminished TAM accumulation in GBM patients,13 which is in agreement with our findings in patients with LGGs that progressed to HGGs.
By leveraging results from next-generation sequencing of tumor samples, we identified two key candidate genes, CCL2 and MMP9, in malignant transformation of LGGs, both with well-established tumor-promoting effects on the microenvironment. CCL2, produced by glioma cells, is a major chemoattractant for immunosuppressive myeloid cells and regulatory T cells. Intracranial implantation of CCL2-transfected rat CNS-1 glioma cells promoted TAM infiltration and angiogenesis, resulting in larger tumors and shorter survival.22Ccl2 deficiency decreased infiltration of regulatory T cells and myeloid derived suppressor cells in a GL261 mouse model.6 In human GBM patients, CCL2 expression was negatively correlated with survival and positively correlated with expression of myeloid derived suppressor cell markers. On the other hand, MMP9 plays important roles in tumor invasion and angiogenesis across a wide range of malignancies.23 In particular, several studies have highlighted the prognostic impact of MMP9 expression in GBM.26–28 Taken together, data from these previous studies suggest that increased TAM accumulation and angiogenesis observed in HGGs from V/V and PDGFB;CX3CL1;CX3CR1 groups are likely mediated by upregulation of CCL2 and MMP9.
In the normal CNS, CX3CR1 is expressed predominantly by microglia, whereas CX3CL1 is predominantly expressed by neurons.29,30 However, gliomas across the entire histologic spectrum and tumor-initiating GSCs demonstrate CX3CL1 and CX3CR1 expression.12 To complicate matters further, subsets of glioma-infiltrating peripheral macrophages and lymphocytes also express CX3CR1.31,32 In our study, we provide several lines of evidence that intratumoral rather than stromal CX3CL1-CX3CR1 signaling mediates the observed phenotypes. First, stimulation with CX3CL1 upregulated CCL2 transcript levels in V/V but not V/I GSCs. Second, tumor-specific, RCAS-driven coexpression of CX3CL1 and CX3CR1 (V249) in a mouse LGG model essentially phenocopied increased TAM accumulation and microvessel density seen in human tumors from V/V genotypes. Finally, intratumoral expression of CX3CR1, but not CX3CL1, promoted formation of more aggressive tumors in mice, suggesting that endogenous CX3CL1 cross-activates deleterious intratumoral CX3CR1 signaling.
Our study does have some limitations. While V/V and V/I + I/I groups do not show significant differences in demographic or major molecular characterizations, they were analyzed retrospectively, and our findings should be validated in a prospective cohort. In addition, our mouse model, while sufficient to examine the deleterious nature of wild-type CX3CR1 signaling, did not account for the beneficial effects of the polymorphism. Rescue experiments using RCAS-driven expression of variant CX3CR1 (I249) are currently ongoing. Furthermore, the contribution of CX3CR1 expressing stromal cells needs to be definitively ruled out in future investigations. Finally, despite similar incidence of HGGs between PDGFB;CX3CR1 and PDGFB;CX3CL1;CX3CR1 groups, survival was shortened only in the latter. This discrepancy is likely due to partial cross-activation of tumor expressed CX3CR1 by endogenous mouse CX3CL1. However, the effect on survival does not become manifest without coexpression of human CX3CL1, driving angiogenesis and intratumoral hemorrhage. Of note, PDGFB-driven tumors in mice do not express CX3CR114; therefore, ectopic expression of human CX3CL1 alone is insufficient to promote malignant transformation.
There has been a paradigm shift toward more aggressive treatment of LGGs, due to recent studies demonstrating improved survival with early surgery and concurrent adjuvant chemoradiation.33,34 In conjunction, classification of LGGs has transitioned toward molecular, rather than histologic, characterization, which is better at predicting tumor behavior.35 Within this context, the prognostic impact of CX3CR1 V249I polymorphism in LGGs may prove valuable in targeted therapy. Selective CX3CR1 inhibition also holds therapeutic promise to prevent malignant transformation of LGGs.
Funding
This work was supported by the National Institutes of Health (R01NS094615 to G.R. and R25NS070694 to S.L.); Tumor SPORE (P50CA127001 to G.R.); and Marnie Rose Foundation (G.R.). The Sequencing and Microarray Facility is funded by NCI Grant CA016672.
Conflict of interest statement. The authors report no conflict of interest.
Authorship statement. Conception and design: SL, KL, GR. Acquisition of data: SL, KL, YY, GR. Analysis and interpretation of data: SL, KL, GM, AR, GR. Writing, reviewing, editing: SL, KL, GM, YY, AR, GR.
Supplementary Material
References
- 1. Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19(4):764–772. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5): 459–466. [DOI] [PubMed] [Google Scholar]
- 3. Tom MC, Park DYJ, Yang K, et al. Malignant transformation of molecularly classified adult low-grade glioma. Int J Radiat Oncol Biol Phys. 2019;105(5):1106–1112. [DOI] [PubMed] [Google Scholar]
- 4. Heimberger AB, Abou-Ghazal M, Reina-Ortiz C, et al. Incidence and prognostic impact of FoxP3+ regulatory T cells in human gliomas. Clin Cancer Res. 2008;14(16):5166–5172. [DOI] [PubMed] [Google Scholar]
- 5. Prosniak M, Harshyne LA, Andrews DW, et al. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin Cancer Res. 2013;19(14):3776–3786. [DOI] [PubMed] [Google Scholar]
- 6. Chang AL, Miska J, Wainwright DA, et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 2016;76(19):5671–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doucette TA, Kong LY, Yang Y, et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro Oncol. 2012;14(9):1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kong LY, Wu AS, Doucette T, et al. Intratumoral mediated immunosuppression is prognostic in genetically engineered murine models of glioma and correlates to immunotherapeutic responses. Clin Cancer Res. 2010;16(23):5722–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Latha K, Yan J, Yang Y, et al. The role of fibrinogen-like protein 2 on immunosuppression and malignant progression in glioma. J Natl Cancer Inst. 2019;111(3):292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu S, Wei J, Wang F, et al. Effect of miR-142-3p on the M2 macrophage and therapeutic efficacy against murine glioblastoma. J Natl Cancer Inst. 2014;106(8):dju162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sciumè G, Santoni A, Bernardini G. Chemokines and glioma: invasion and more. J Neuroimmunol. 2010;224(1-2):8–12. [DOI] [PubMed] [Google Scholar]
- 12. Erreni M, Solinas G, Brescia P, et al. Human glioblastoma tumours and neural cancer stem cells express the chemokine CX3CL1 and its receptor CX3CR1. Eur J Cancer. 2010;46(18):3383–3392. [DOI] [PubMed] [Google Scholar]
- 13. Rodero M, Marie Y, Coudert M, et al. Polymorphism in the microglial cell-mobilizing CX3CR1 gene is associated with survival in patients with glioblastoma. J Clin Oncol. 2008;26(36):5957–5964. [DOI] [PubMed] [Google Scholar]
- 14. Feng X, Szulzewsky F, Yerevanian A, et al. Loss of CX3CR1 increases accumulation of inflammatory monocytes and promotes gliomagenesis. Oncotarget. 2015;6(17):15077–15094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sciumè G, Soriani A, Piccoli M, Frati L, Santoni A, Bernardini G. CX3CR1/CX3CL1 axis negatively controls glioma cell invasion and is modulated by transforming growth factor-beta1. Neuro Oncol. 2010;12(7):701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moatti D, Faure S, Fumeron F, et al. Polymorphism in the fractalkine receptor CX3CR1 as a genetic risk factor for coronary artery disease. Blood. 2001;97(7):1925–1928. [DOI] [PubMed] [Google Scholar]
- 17. Faure S, Meyer L, Costagliola D, et al. Rapid progression to AIDS in HIV+ individuals with a structural variant of the chemokine receptor CX3CR1. Science. 2000;287(5461):2274–2277. [DOI] [PubMed] [Google Scholar]
- 18. Bhat KP, Salazar KL, Balasubramaniyan V, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25(24):2594–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15(15):1913–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25(1):55–57. [DOI] [PubMed] [Google Scholar]
- 21. Schaumberg DA, Rose L, DeAngelis MM, Semba RD, Hageman GS, Chasman DI. Prospective study of common variants in CX3CR1 and risk of macular degeneration: pooled analysis from 5 long-term studies. JAMA Ophthalmol. 2014;132(1):84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Platten M, Kretz A, Naumann U, et al. Monocyte chemoattractant protein-1 increases microglial infiltration and aggressiveness of gliomas. Ann Neurol. 2003;54(3):388–392. [DOI] [PubMed] [Google Scholar]
- 23. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161–174. [DOI] [PubMed] [Google Scholar]
- 24. Bhat KPL, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu C, Luo D, Streit WJ, Harrison JK. CX3CL1 and CX3CR1 in the GL261 murine model of glioma: CX3CR1 deficiency does not impact tumor growth or infiltration of microglia and lymphocytes. J Neuroimmunol. 2008;198(1–2):98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xue Q, Cao L, Chen XY, et al. High expression of MMP9 in glioma affects cell proliferation and is associated with patient survival rates. Oncol Lett. 2017;13(3):1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choe G, Park JK, Jouben-Steele L, et al. Active matrix metalloproteinase 9 expression is associated with primary glioblastoma subtype. Clin Cancer Res. 2002;8(9):2894–2901. [PubMed] [Google Scholar]
- 28. Wang M, Wang T, Liu S, Yoshida D, Teramoto A. The expression of matrix metalloproteinase-2 and -9 in human gliomas of different pathological grades. Brain Tumor Pathol. 2003;20(2):65–72. [DOI] [PubMed] [Google Scholar]
- 29. Jung S, Aliberti J, Graemmel P, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20(11):4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim KW, Vallon-Eberhard A, Zigmond E, et al. In vivo structure/function and expression analysis of the CX3C chemokine fractalkine. Blood. 2011;118(22):e156–e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Z, Feng X, Herting CJ, et al. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 2017;77(9):2266–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ren F, Zhao Q, Huang L, et al. The R132H mutation in IDH1 promotes the recruitment of NK cells through CX3CL1/CX3CR1 chemotaxis and is correlated with a better prognosis in gliomas. Immunol Cell Biol. 2019;97(5):457–469. [DOI] [PubMed] [Google Scholar]
- 33. Jakola AS, Myrmel KS, Kloster R, et al. Comparison of a strategy favoring early surgical resection vs a strategy favoring watchful waiting in low-grade gliomas. JAMA. 2012;308(18):1881–1888. [DOI] [PubMed] [Google Scholar]
- 34. Buckner JC, Shaw EG, Pugh SL, et al. Radiation plus procarbazine, CCNU, and vincristine in low-grade glioma. N Engl J Med. 2016;374(14):1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. The Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.