Abstract
Objective
The Unified Medical Language System (UMLS) integrates various source terminologies to support interoperability between biomedical information systems. In this article, we introduce a novel transformation-based auditing method that leverages the UMLS knowledge to systematically identify missing hierarchical IS-A relations in the source terminologies.
Materials and Methods
Given a concept name in the UMLS, we first identify its base and secondary noun chunks. For each identified noun chunk, we generate replacement candidates that are more general than the noun chunk. Then, we replace the noun chunks with their replacement candidates to generate new potential concept names that may serve as supertypes of the original concept. If a newly generated name is an existing concept name in the same source terminology with the original concept, then a potentially missing IS-A relation between the original and the new concept is identified.
Results
Applying our transformation-based method to English-language concept names in the UMLS (2019AB release), a total of 39 359 potentially missing IS-A relations were detected in 13 source terminologies. Domain experts evaluated a random sample of 200 potentially missing IS-A relations identified in the SNOMED CT (U.S. edition) and 100 in Gene Ontology. A total of 173 of 200 and 63 of 100 potentially missing IS-A relations were confirmed by domain experts, indicating that our method achieved a precision of 86.5% and 63% for the SNOMED CT and Gene Ontology, respectively.
Conclusions
Our results showed that our transformation-based method is effective in identifying missing IS-A relations in the UMLS source terminologies.
Keywords: Unified Medical Language System, biomedical terminologies, SNOMED CT, Gene Ontology, quality assurance
INTRODUCTION
Objectives
The Unified Medical Language System (UMLS) integrates over 15 million concept names from more than 200 source terminologies, including SNOMED CT and Gene Ontology, to enable interoperability between biomedical information systems.1–5 As the information about concepts and relations between concepts in source terminologies is preserved in the UMLS Metathesaurus, the quality issues existent in source terminologies (eg, missing relations between concepts) affect the qualities of the UMLS and UMLS-based information systems.
In this article, we introduce a novel transformation-based auditing method that leverages the knowledge in the UMLS to systematically identify missing hierarchical IS-A relations in the source terminologies. Quality improvement of the source terminologies will in turn enhance the qualities of the UMLS knowledge sources.
Background and Significance
Unified Medical Language System
The UMLS, developed by the U.S. National Library of Medicine, integrates various health and biomedical vocabularies and standards to enable interoperability between different applications and systems. It has been used in supporting a wide range of applications in biomedicine including information retrieval, natural language processing (NLP), deep learning, phenotyping, and clinical decision support.6–18
The UMLS consists of 3 knowledge sources: the Metathesaurus that contains concepts from many terminologies, the Semantic Network that contains semantic types and their relationships, and the SPECIALIST Lexicon and Lexical Tools to facilitate NLP.1–5
The UMLS Metathesaurus is organized by concept or meaning. Because a concept can have many different names, the UMLS Metathesaurus links all the names from different source terminologies that have the same meaning. Every occurrence of a concept name (or string) in each source terminology is the basic building block or “atom” of the UMLS Metathesaurus and is assigned a unique atom identifier (AUI). Atoms with the same meaning are mapped to a concept assigned a concept unique identifier (CUI). For example, consider a concept in the SNOMED CT with ID 282766005 and preferred name “Lower back injury.” It also has a synonym “Lumbar region injury.”19 In the UMLS Metathesaurus, the AUI for its preferred name is A3255024 and the AUI for its synonym is A3288211. These 2 atoms are both mapped to the same UMLS concept with CUI C0560632, which has a total of 14 atoms mapped from different source terminologies. The UMLS preserves the relations between concepts from its source terminologies. For instance, the IS-A relation between the atom “Superficial injury of lower back” with AUI A28900983 and the atom “Lower back injury” with AUI A3255024 comes from SNOMED CT.
In addition, each UMLS concept (CUI) is assigned at least 1 semantic type in order to provide a consistent categorization of all concepts. For example, the concept “Lower back injury” (CUI: C0560632) is assigned a semantic type “Injury or Poisoning.” There are 127 semantic types in the UMLS, such as “Disease or Syndrome” and “Therapeutic or Preventive Procedure.”
Related work on auditing UMLS
Given its wide use, quality defects of the UMLS will impact the qualities of all the downstream applications based on the UMLS. For instance, missing IS-A relations reduce the recall of UMLS-based information retrieval systems with valid results being missed from the query results.20 For example, suppose there is a need to identify a cohort of patients with “Arthritis of left subtalar joint” from a UMLS-based electronic health record system. However, “Osteoarthritis of left subtalar joint” is currently not listed as its subtype in any of the UMLS source terminologies (ie, a missing IS-A relation). Consequently, all the patients with “Osteoarthritis of left subtalar joint” would be missing from the cohort query result.
Quality assurance or auditing of the UMLS and its sources has been an active research area. Cimino21 utilized the semantic information to detect ambiguous concepts, redundant concept pairs, inconsistent parent–child relationships, and missing relations between semantic types in the UMLS. Bodenreider22 investigated the problem of circular hierarchical relationships between concepts in the UMLS, identified potential causes and their corresponding treatments, and suggested prevention measures. Chen et al23 presented a structural method to group concepts with the same semantic type and partition the concepts into subgroups for the auditors to review and identify missing hierarchical relationships in the UMLS. He et al24,25 leveraged the mappings among different terminologies in the UMLS and developed a topological pattern–based method to enrich concepts in the SNOMED CT and National Cancer Institute (NCI) Thesaurus. Cui26 leveraged the UMLS mappings to identify inconsistent relationships between concepts across different terminologies. Abeysinghe et al27 leveraged the UMLS knowledge to identify supporting evidence for potential subtype inconsistencies detected in the Gene Ontology, NCI Thesaurus, and SNOMED CT. All these showed that the UMLS provides a promising environment for enhancing the qualities of its source terminologies.
Specific contribution
In this article, we leverage the UMLS knowledge to develop a novel, transformation-based method to automatically identify missing IS-A relations in the UMLS source terminologies. Our method takes full advantage of the rich knowledge provided by the UMLS for auditing and improving the qualities of its source terminologies, which in turn enhances the quality of the UMLS. Unlike the traditional terminology auditing methods that often rely on the knowledge within the terminology itself (ie, internal knowledge), our method leverages not only the terminology itself, but also the knowledge from other multiple terminologies in the UMLS (ie, both internal and external knowledge). This will result in newly identified missing IS-A relations that would not be uncovered by only looking into 1 or 2 individual terminologies.
In addition, unlike previous related work on auditing the UMLS that mainly focused on auditing high level views (eg, semantic types, concepts/CUIs, relations between concepts), this work intends to audit the UMLS source terminologies in the atom level.
MATERIALS AND METHODS
This work is based on the UMLS 2019AB release. A large proportion of concept names (or atoms) in the UMLS contain more than 1 noun chunk. The key idea of our transformation-based auditing method is to replace those noun chunks in a concept name with more general concept names. If a newly generated name after the replacement is an existing concept name in the same source terminology, then we consider that there is a potentially missing IS-A relation between the 2 concepts corresponding to the original and new concept names.
Our method consists of 4 main steps to identify potentially missing IS-A relations for each concept name in the UMLS: (1) parse the concept name and identify noun chunks, (2) generate replacement candidates for noun chunks, (3) perform concept name transformation and construct new potential concept names, and (4) map newly constructed concept names to atoms and identify potentially missing IS-A relations in the source terminologies.
Parsing concept names
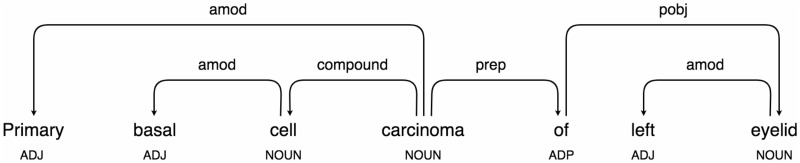
We first convert each concept name to lower case. We then use spaCy,28 an open-source library for advanced NLP, to perform dependency parsing and identify noun chunks within concept names. For example, Figure 1 shows the dependency graph of the concept name “Primary basal cell carcinoma of left eyelid,” in which 2 base noun chunks can be identified: “primary basal cell carcinoma” and “left eyelid.” Here, a base noun chunk consists of a head (eg, “carcinoma”) plus words describing the head (eg, “primary basal cell”).29 Note that “basal cell” is not a base noun chunk because it is used to modify or describe “carcinoma.” Instead, we consider such noun phrases describing the head as secondary noun chunks.
Figure 1.
Dependency graph of the concept name “Primary basal cell carcinoma of left eyelid.”
After the parsing, each concept name C can be represented as an ordered array of elements [c1, c2, …, cn], where ci can be a single word, a base noun chunk, or a secondary noun chunk. For instance, the concept name “Primary basal cell carcinoma of left eyelid” can be represented in 2 forms: (1) [primary basal cell carcinoma, of, left eyelid] and (2) [primary, basal cell, carcinoma, of, left eyelid].
Identifying replacement candidates
In this step, we identify replacement candidates that are more general than the noun chunks (base and secondary) in each concept name. If a noun chunk can be mapped to a UMLS atom (ie, the noun chunk is also a concept name in an existing source terminology), then we consider the concept names of this atom’s ancestors in its source terminology as replacement candidates for the noun chunk; otherwise, the noun chunk is considered as not having any replacement candidates. In other words, we leverage existing IS-A relations in the UMLS source terminologies to identify replacement candidates. To avoid replacement candidates being too general, we leveraged ancestors of the atom within a distance of 2 levels using depth-limited search.30
Take the concept name “Acute dacryoadenitis of left eye” in Table 1 as an example, it can be represented as an array [acute dacryoadenitis, of, left eye]. The noun chunk “acute dacryoadenitis” can be mapped to 9 atoms. For example, A2889158 is an atom sourced from the SNOMED CT (U.S. edition) with 7 level-2 ancestors. After going through all the 9 atoms, the following replacement candidates for “acute dacryoadenitis” can be obtained: “disorder of lacrimal gland,” “disorder of eyelid or lacrimal system,” “dacryoadenitis,” “inflammation of specific body systems,” “acute inflammatory disease,” “inflammatory disorder of head,” “acute disease,” and “inflammatory disorder.”
Table 1.
An example of the transformation process for “Acute dacryoadenitis of left eye”
| Concept name | Acute dacryoadenitis of left eye |
|---|---|
| Representation ([c1, c2, c3]) | [acute dacryoadenitis, of, left eye] |
| Replacement candidates for “acute dacryoadenitis” (r1) | {dacryoadenitis, inflammation of specific body systems, acute disease, acute inflammatory disease, inflammatory disorder, inflammatory disorder of head, disorder of eyelid or lacrimal system, disorder of lacrimal gland} |
| Replacement candidates for “left eye” (r3) | {organ of special sense, eye, subdivision of face} |
| Combinatorial replacements | [{acute dacryoadenitis, dacryoadenitis, inflammation of specific body systems, acute disease, acute inflammatory disease, inflammatory disorder, inflammatory disorder of head, disorder of eyelid or lacrimal system, disorder of lacrimal gland}, of, {left eye, organ of special sense, eye, subdivision of face}] |
| Potentially missing IS-A relations detected in source terminologies | SNOMEDCT_US: “acute dacryoadenitis of left eye” IS-A “acute disease of eye” MEDCIN: “acute dacryoadenitis of left eye” IS-A “inflammatory disorder of eye” |
SNOMEDCT_US: SNOMED CT (U.S. version).
Concept name transformation
For each concept name with noun chunk(s) such that the replacement candidates have been identified already, we replace the original noun chunk(s) with their corresponding candidates to generate new potential concept names, which may serve as supertypes of the original concept name (since the replacement candidates are more general than the original noun chunk). Formally, given a concept name C represented by [c1, c2, …, cn], where there exists an i such that ci is a base or secondary noun chunk and ri is a set of replacement candidates for ci, then we replace ci with any candidate in ri and concatenate the array as a string to construct new concept names that may serve as C’s supertypes. If there are multiple such is, we will perform combinatorial replacements for multiple is.
Take the concept name “Acute dacryoadenitis of left eye” in Table 1 as an example. There are 3 elements in its array representation [c1, c2, c3], where c1 and c3 are base noun chunks. There are 8 replacement candidates for c1 and 3 for c3. A total of 35 new potential concept names can be obtained after the combinatorial replacements for c1 and c3, including “acute disease of eye” and “acute inflammatory disease of left eye.” Note that the total number 35 can be obtained by multiplying 9 (8 new noun chunks and 1 original noun chunk for c1) by 4 (3 new noun chunks and 1 original noun chunk for c3), and subtracting 1 (the original concept name) from it.
Identify missing IS-A relations in source terminologies
In this step, we check if the newly generated concept names exist in the UMLS (ie, exactly match the names of UMLS atoms) to identify potentially missing IS-A relations between atoms in source terminologies. Given a concept name C (mapped to an atom AUIC) and a potential concept name S serving as its supertype, if the following conditions hold: then we consider there is a potentially missing IS-A relation between AUIC and AUIS (ie, AUIC IS-A AUIS) in the terminology T. Note that missing IS-A relations between atoms from different source terminologies are beyond the scope of this work. The semantic type requirement of C and S is to avoid ambiguities caused by concept names that may have multiple meanings. For example, the concept name “cold” could refer to lower temperature (with a semantic type “Natural Phenomenon or Process”) or a kind of disease (with a semantic type “Disease or Syndrome”).
S can be mapped to a UMLS atom AUIS;
AUIS comes from the same source terminology T as AUIC;
Currently there is no IS-A relation (either direct or indirect) between AUIS and AUIC claimed in T; and
AUIC has the same semantic type as AUIS, or the set of semantic types of AUIC contains that of AUIS as a subset,
For “acute dacryoadenitis of left eye” in Table 1, after the transformation, “acute disease of eye” is one of its potential new concept names that can be mapped to atoms, while “acute inflammatory disease of left eye” cannot. By further mapping concept names to atoms, a potentially missing IS-A relation between “acute dacryoadenitis of left eye” with AUI A27761536 and “acute disease of eye” with AUI A3463187 can be identified in the SNOMED CT.
It is worth noting that the potentially missing IS-A relations identified by our method may contain redundancy. Here, a missing IS-A relation (say “x IS-A y”) identified in a terminology T is considered redundant, if there exists another missing IS-A relation “x IS-A z” identified in T such that y is currently an ancestor of z in T. In this case, if “x IS-A z” is a valid missing IS-A relation, then “x IS-A y” can be implied as valid by “x IS-A z” and “z IS-A y.” Therefore, we further remove the potentially missing IS-A relations that are redundant from the result.
RESULTS
Identifying missing IS-A relations
We applied our method to the English-language concept names in the UMLS (2019AB release). In total, our method identified 42 362 potentially missing IS-A relations from 13 source terminologies in the UMLS. A total of 39 359 of 42 362 are nonredundant. Table 2 shows the number of potentially missing IS-A relations (nonredundant) detected in each source terminology. Table 2 also presents each terminology’s size including the number of concepts and the number of direct IS-A relations, as well as the number of existing IS-A relations that can be identified by our transformation-based method. In total 149 568 existing IS-A relations can be identified from 13 source terminologies, and 109 031 of them are direct IS-A relations.
Table 2.
The number of potentially missing IS-A relations detected in the UMLS source terminologies in English, as well as the terminology size and the number of existing IS-A relations that can be identified for each terminology
| Terminology size |
Existing IS-A relations identified |
Potentially missing IS-A relations identified | |||
|---|---|---|---|---|---|
| Source terminology | Concepts | Direct IS-A relations | Direct + indirect | Direct | |
| MEDCIN | 348 808 | 353 304 | 30 001 | 23 692 | 16 779 |
| UWDA | 61 127 | 62 285 | 34 564 | 24 594 | 10 865 |
| FMA | 102 595 | 104 341 | 54 644 | 39 274 | 7230 |
| SNOMEDCT_US | 401 832 | 994 499 | 19 859 | 14 529 | 3833 |
| NCI | 151 966 | 159 479 | 688 | 539 | 334 |
| GO | 49 907 | 77 067 | 9640 | 6246 | 250 |
| SNOMEDCT_VET | 36 527 | 40 689 | 82 | 81 | 23 |
| HPO | 16 222 | 18 313 | 37 | 30 | 11 |
| CPM | 3079 | 3853 | 7 | 7 | 10 |
| UMD | 27 309 | 12 889 | 0 | 0 | 8 |
| PDQ | 18 874 | 4298 | 43 | 36 | 8 |
| CPT | 40 892 | 14 072 | 1 | 1 | 7 |
| ATC | 5485 | 4969 | 2 | 2 | 1 |
GO: Gene Ontology; NCI: National Cancer Institute; SNOMEDCT_US: SNOMED CT (U.S. version).
Among 39 359 potentially missing IS-A relations identified, 36 997 were obtained from a single noun chunk replacement (1-replacement), 2338 from 2 noun chunk replacements (2-replacement), and 24 from 3 noun chunk replacements (3-replacement) (see Supplementary Appendix I for more details).
Evaluation
To assess the effectiveness of our method for identifying missing IS-A relations in the UMLS source terminologies, a sample of 200 IS-A relations from SNOMED CT (the “Clinical Finding” subhierarchy) and a sample of 100 from Gene Ontology were randomly selected. The samples were reviewed by domain experts (J.S. is a clinical expert familiar with SNOMED CT, and Y.Y. and W.J.Z. have expertise in systems biology and genomics). For each relation, we provided domain experts with the preferred names of the 2 concepts involved, as well as the links to the 2 concepts in their online browsers.
Domain experts verified that 173 of 200 potentially missing IS-A relations in SNOMED CT (a precision of 86.5%) and 63 of 100 in Gene Ontology (a precision of 63%) are valid (ie, true positives). Table 3 lists 15 valid examples (a complete list of evaluated samples can be found in Supplementary Appendix II for SNOMED CT and Supplementary Appendix III for Gene Ontology).
Table 3.
Examples of missing IS-A relations confirmed by domain experts
| Subtype concept | Supertype concept | Source terminology |
|---|---|---|
| Abrasion and/or friction burn of buttock with infection (disorder) | Superficial injury of buttock with infection (disorder) | SNOMEDCT_US |
| Camptodactyly of right hand (disorder) | Congenital deformity of right hand (disorder) | SNOMEDCT_US |
| Acute gastrojejunal ulcer with hemorrhage AND with perforation but without obstruction (disorder) | Peptic ulcer with hemorrhage AND with perforation but without obstruction (disorder) | SNOMEDCT_US |
| Malignant melanoma of skin of forearm (disorder) | Malignant neoplasm of skin of forearm (disorder) | SNOMEDCT_US |
| Deficiency of adenosylhomocysteinase (disorder) | Deficiency of hydrolase (disorder) | SNOMEDCT_US |
| Infestation caused by Boophilus (disorder) | Infestation caused by Ixodidae (disorder) | SNOMEDCT_US |
| Abscess of nasal septum (disorder) | Inflammatory disorder of cartilage (disorder) | SNOMEDCT_US |
| Obsessive compulsive disorder caused by cocaine (disorder) | Anxiety disorder caused by stimulant (disorder) | SNOMEDCT_US |
| Primary malignant neoplasm of frontal lobe (disorder) | Malignant neoplasm of cerebral cortex (disorder) | SNOMEDCT_US |
| Rupture of anterior cruciate ligament of left knee (disorder) | Injury of cruciate ligament of knee (disorder) | SNOMEDCT_US |
| negative regulation of testosterone biosynthetic process | negative regulation of steroid hormone biosynthetic process | GO |
| macrophage migration inhibitory factor binding | enzyme binding | GO |
| response to camptothecin | response to topoisomerase inhibitor | GO |
| formate dehydrogenase complex | oxidoreductase complex | GO |
| negative regulation of transmembrane | negative regulation of cellular process | GO |
GO: Gene Ontology; SNOMEDCT_US: SNOMED CT (U.S. version).
Table 3 also contains 4 examples of missing IS-A relations in SNOMED CT that were obtained by multiple noun chunk replacements. For instance, the missing IS-A relation between “Obsessive compulsive disorder caused by cocaine (disorder)” and “Anxiety disorder caused by stimulant (disorder)” was obtained through the following 2 replacements: (1) “Obsessive compulsive disorder” IS-A “Anxiety disorder” in the NCI Thesaurus and (2) “Cocaine” IS-A “Psychostimulant” and “Psychostimulant” IS-A “Stimulant” in the SNOMED CT. The detailed replacements for the evaluated samples can be found in Supplementary Appendices II and III.
Analyses of false positive cases
Based on the evaluation results of the domain experts, we examined false positive cases (ie, invalid missing IS-A relations). More specifically, we looked into the noun chunks within the concept names and their replacement candidates (ie, existing IS-A relations) to find the potential causes.
Table 4 presents 7 invalid missing IS-A relations as well as the existing IS-A relations in the UMLS that were leveraged to obtain these invalid relations. We noted that the main cause of false positives is that the biomedical meanings of replacement candidates are not considered to be more general than their corresponding noun chunks. This could relate to either incorrect existing IS-A relations or different views of different terminologies. Take “cellular response to beta-carotene” IS-A “cellular response to vitamin A” detected in the Gene Ontology as an example. The domain experts believe that “beta-carotene” is an antioxidant that converts to vitamin A (which is not an IS-A relation), while SNOMED CT has an IS-A relation between “Beta-carotene (substance)” and “Retinol (substance)” (with a synonym “Vitamin A”), indicating that this is an incorrect IS-A relation in the SNOMED CT. Consider “Abscess of thumb of left hand (disorder)” IS-A “Abscess of finger of left hand (disorder)” detected in the SNOMED CT. It was obtained by leveraging an existing relation “Thumb” IS-A “Finger” in both UWDA and FMA. However, the detected missing IS-A relation is invalid, as in SNOMED CT “Finger” only includes the second to fifth digit of the hand (ie, “Thumb” is not a “Finger”).
Table 4.
Examples of false positives (or invalid missing IS-A relations) and the existing IS-A relations causing the false positives
| Subtype concept | Supertype concept | Source terminology | Existing IS-A relation(s) causing the false positive |
|---|---|---|---|
| Benign neoplasm of false vocal cord (disorder) | Benign neoplasm of vocal cord (disorder) | SNOMEDCT_US | “false vocal cord” IS-A “vocal cord” in the NCI Thesaurus |
| Deficiency of lysophospholipase (disorder) | Deficiency of triacylglycerol lipase (disorder) | SNOMEDCT_US | “lysophospholipase” IS-A “phospholipase” IS-A “triacylglycerol lipase” in the SNOMEDCT_US |
| Abscess of thumb of left hand (disorder) | Abscess of finger of left hand (disorder) | SNOMEDCT_US | “thumb” IS-A “finger” in the UWDA and FMA |
| Calculus of gallbladder with acute and chronic cholecystitis (disorder) | Calculus of gallbladder with acute cholecystitis (disorder) | SNOMEDCT_US | “acute and chronic cholecystitis” IS-A “acute cholecystitis” in the MEDCIN |
| cellular response to beta-carotene | cellular response to vitamin A | GO | “beta-carotene” IS-A “vitamin A” in the SNOMEDCT_US |
| caprolactam metabolic process | propylene metabolic process | GO | “caprolactam” IS-A “propylene” in the SNOMEDCT_US |
| cellular response to ammonium ion | cellular response to ammonia | GO | “ammonium ion” IS-A “ammonia” in the SNOMEDCT_US |
GO: Gene Ontology; SNOMEDCT_US: SNOMED CT (U.S. version).
Effect of restricting the IS-A source for noun chunk replacement
Relating to the subtle terminology difference, a natural question is whether restricting the IS-A relations leveraged for noun chunk replacement to be in the same terminology will have an effect on the performance of our method. To study this, we performed an experiment by restricting replacement candidates in the same terminology, which resulted in a total of 20 754 potentially missing IS-A relations, compared with 39 359 without applying the restriction (see Supplementary Appendix IV for more details).
We further looked into the evaluated samples regarding the performance comparison. For SNOMED CT, 173 of 200 evaluated relations (without applying the restriction) are valid, achieving a precision of 86.5%. Among 200 evaluated ones, 107 of them can be obtained by applying the restriction, and 103 of 107 are valid, achieving a precision of 96.26%. Therefore, the precision is increased by 9.76% with the restriction applied. However, the number of valid missing IS-A relations is decreased from 173 to 103, a 40.46% reduction. For Gene Ontology, 63 of 100 evaluated relations (without applying the restriction) are valid, achieving a precision of 63%. Among 100 evaluated ones, 21 of them can be obtained by applying the restriction, and 18 of 21 are valid, achieving a precision of 85.71%. Therefore, the precision is increased by 22.71%. However, the number of valid missing IS-A relations is decreased from 63 to 18, a 71.43% reduction. It can be seen that although restricting to the same source terminology for noun chunk replacement can improve the precision to some extent, leveraging multiple sources can identify more missing IS-A relations to a greater extent while still achieving acceptable precisions.
DISCUSSION
In this article, we introduced a transformation-based method to replace noun chunks in a concept name with more general concept names in order to detect potentially missing IS-A relations in the UMLS source terminologies. To find noun chunk replacement, we leverage abundant knowledge of IS-A relations between concept names provided by the UMLS.
Distinction with related work
Other auditing methods designed for a specific terminology including pattern-based, lexical-based and deep learning–based methods usually rely on the knowledge in the terminology itself and require transferring knowledge to features for representing concepts in order to identify missing IS-A relations between concepts.31–38 Therefore, the effectiveness of such methods to some extent relies on the terminology itself (ie, internal knowledge), while our method leverages both internal and external knowledge through the UMLS to perform the auditing. More importantly, our method enables the auditing of multiple source terminologies at the same time.
Exact vs normalized matching
For parsing and mapping concept names, we directly used the exact names without performing any normalization. We further tried normalizing concept names (after noun chunks were identified) using the UMLS lexical tool LuiNorm.39 We also utilized the normalized format for generating replacement candidates for noun chunks and mapping newly constructed concept names to atoms. As a result, the potentially missing IS-A relations identified using normalized matching contain all the 39 359 ones identified by exact matching as a subset. In addition, the normalized matching identified 10 627 extra potentially missing IS-A relations (see Supplementary Appendix V for more details).
Indeed, normalized matching helped identify extra valid missing IS-A relations. For example, a missing IS-A relation between “Malignant neoplasm of connective tissue” and “Neoplasm of connective tissues” in the SNOMED CT was identified by normalized matching, as “tissues” were normalized to “tissue.” However, there were also invalid cases identified. For instance, “Asymmetry” is a child of “Symmetries” in SNOMED CT. Performing normalization resulted in “Asymmetry” IS-A “Symmetry” and thus an invalid missing IS-A relation: “Asymmetry of mandible” IS-A “Symmetry of mandible.” Because the main focus of this article is the transformation-based method, it is beyond the scope of this work to thoroughly compare the actual performances of the exact matching and normalized matching, as it requires additional manual evaluation by domain experts.
Potential for concept enrichment
Because our focus in this work is to identify missing IS-A relations in the UMLS source terminologies, we require that the 2 atoms involved in a potentially missing IS-A relation be from the same terminology. For those ones with the 2 atoms coming from different source terminologies, missing concepts may be identified for concept enrichment in source terminologies. That is, if the supertype atom does not appear in the same source terminology with the subtype atom, then the supertype atom may be a potentially missing concept (ie, new concept) for the terminology or a missing synonym for an existing concept in the source terminology of the subtype atom.
Applicability to a specific terminology
Although our method was designed for auditing multiple source terminologies in the UMLS, it can be applied within a specific terminology such as SNOMED CT itself without using the UMLS. A question that may arise is: will this give the same results obtained for restricting the IS-A relations leveraged for noun chunk replacement to be in the UMLS SNOMED CT? The answer to this question depends on whether the IS-A relations in the UMLS SNOMED CT are identical to that in the original SNOMED CT. It is worth noting that relations in the UMLS are expressed in terms of CUIs (concepts) and AUIs (atoms or concept names). For SNOMED CT (U.S. 09/01/2019 edition) integrated in the UMLS (2019AB release), only IS-A relations between designated preferred names of SNOMED CT concepts are maintained. Therefore, if we only leverage such IS-A relations between preferred names of concepts when applying our method within SNOMED CT, then the same results will be obtained; however, if we leverage additional IS-A relations such as those between synonyms of concepts, then more results will be obtained and need further domain expert evaluation.
Limitations and future work
In this study, we only evaluated SNOMED CT and Gene Ontology because of the lack of expertise in other terminologies. Although the evaluation results showed that our transformation-based method is effective in identifying missing IS-A relations in SNOMED CT and Gene Ontology, additional evaluation by domain experts is still needed to assess the effectiveness of our method for auditing other source terminologies. Another limitation of this work is regarding the incorrect IS-A relations that can be further revealed through manual examination of the invalid missing IS-A relations identified by our method. It would be desirable to develop an automated approach to detect such incorrect IS-A relations in the source terminologies.
CONCLUSION
In this article, we introduced a transformation-based auditing method to detect potentially missing IS-A relations in the UMLS source terminologies. Leveraging rich knowledge in the UMLS (2019AB release), our method is able to audit multiple terminologies at the same time. Experts’ evaluation showed the effectiveness of our method (a precision of 86.5% for SNOMED CT and 63% for the Gene Ontology). Further analyses of invalid missing IS-A relations derived by our method revealed additional quality issues in the source terminologies. Because the source terminologies are regularly integrated into the UMLS, quality improvement of its source terminologies will directly enhance the quality of the UMLS itself.
FUNDING
This work was supported by National Science Foundation grants 1657306 and 1931134 received by LC; National Institutes of Health grants R21CA231904 and R01NS116287 received by LC, as well as 1UL1TR003167 and R01AG066749 received by WJZ; and Cancer Prevention and Research Institute of Texas grant RP170668 received by WJZ. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation, National Institutes of Health, or Cancer Prevention and Research Institute of Texas.
AUTHOR CONTRIBUTIONS
LC and FZ conceptualized and designed this study. FZ developed the auditing algorithms, generated the auditing results, and prepared the evaluation samples for SNOMED CT and Gene Ontology. JS reviewed and evaluated the samples for SNOMED CT. YY and WJZ reviewed and evaluated the samples for Gene Ontology. FZ and LC analyzed the evaluation results. FZ and LC wrote the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the anonymous reviewers for their valuable comments that help improve the quality of this article.
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare.
REFERENCES
- 1. Humphreys BL, Lindberg DAB. Building the Unified Medical Language System. Proc Annu Symp Comput Appl Med Care1989; 475-480; Washington, DC.
- 2. Lindberg DAB, Humphreys BL, McCray AT. The Unified Medical Language System. Yearb Med Inform 1993; 2 (1): 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Humphreys BL, Lindberg DAB, Schoolman HM, et al. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc 1998; 5 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32 (90001): 267D–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UMLS Reference Manual. Bethesda, MD: National Library of Medicine. https://www.ncbi.nlm.nih.gov/books/NBK9676/ Accessed January 20, 2020.
- 6. Chute CG, Yang Y, Evans DA. Latent Semantic Indexing of medical diagnoses using UMLS semantic structures. Proc Annu Symp Comput Appl Med Care1991; 185–9; Washington, DC. [PMC free article] [PubMed]
- 7. Nadkarni P, Chen R, Brandt C. UMLS concept indexing for production databases: a feasibility study. J Am Med Inform Assoc 2001; 8 (1): 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hersh W, Price S, Donohoe L. Assessing thesaurus-based query expansion using the UMLS Metathesaurus. Proc AMIA Symp2000; 34–8; Los Angeles, CA. [PMC free article] [PubMed]
- 9. Lu K, Mu X. Query expansion using UMLS tools for health information retrieval. Proc Am Soc Info Sci Technol 2009; 46 (1): 1–6; Vancouver, BC, Canada. [Google Scholar]
- 10. Martinez D, Otegi A, Soroa A, et al. Improving search over electronic health records using UMLS-based query expansion through random walks. J Biomed Inform 2014; 51: 100–6. [DOI] [PubMed] [Google Scholar]
- 11. McCray AT, Aronson AR, Browne AC, et al. UMLS knowledge for biomedical language processing. Bull Med Library Assoc 1993; 81 (2): 184–94. [PMC free article] [PubMed] [Google Scholar]
- 12. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp2001; 17–21. [PMC free article] [PubMed]
- 13. Chen L, Gu Y, Ji X, et al. Clinical trial cohort selection based on multi-level rule-based natural language processing system. J Am Med Inform Assoc 2019; 26 (11): 1218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yao L, Mao C, Luo Y. Clinical text classification with rule-based features and knowledge-guided convolutional neural networks. BMC Med Inform Decis Mak 2019; 19 (S3): 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maldonado R, Yetisgen M, Harabagiu SM. Adversarial learning of knowledge embeddings for the Unified Medical Language System. AMIA Jt Summits Transl Sci Proc 2019; 543–52. [PMC free article] [PubMed] [Google Scholar]
- 16. Adamusiak T, Shimoyama N, Shimoyama M. Next generation phenotyping using the Unified Medical Language System. JMIR Med Inform 2014; 2 (1): e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Achour SL, Dojat M, Rieux C, et al. A UMLS-based knowledge acquisition tool for rule-based clinical decision support system development. J Am Med Inform Assoc 2001; 8 (4): 351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee PJ, Lee YH, Kang Y, Chao CP. A medical decision support system using text mining to compare electronic medical records. In: proceedings of the International Conference on Human-Computer Interaction; 2019; 199–208; Orlando, FL.
- 19.SNOMED CT Browser. https://browser.ihtsdotools.org/ Accessed January 20, 2020.
- 20. Zhang GQ, Tao S, Zeng N, et al. Ontologies as nested facet systems for human–data interaction. Seman Web 2020; 11 (1): 79–86. [Google Scholar]
- 21. Cimino JJ. Auditing the Unified Medical Language system with semantic methods. J Am Med Inform Assoc 1998; 5 (1): 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bodenreider O. Circular hierarchical relationships in the UMLS: etiology, diagnosis, treatment, complications and prevention. Proc AMIA Symp2001; 57–61; Washington, DC. [PMC free article] [PubMed]
- 23. Chen Y, Gu HH, Perl Y, et al. Structural group-based auditing of missing hierarchical relationships in UMLS. J Biomed Inform 2009; 42 (3): 452–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He Z, Geller J, Chen Y. A comparative analysis of the density of the SNOMED CT conceptual content for semantic harmonization. Artif Intell Med 2015; 64 (1): 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He Z, Chen Y, de Coronado S, Piskorski K, Geller J. Topological-pattern-based recommendation of UMLS concepts for National Cancer Institute Thesaurus. AMIA Annu Symp Proc 2016; 2016: 618–27. [PMC free article] [PubMed] [Google Scholar]
- 26. Cui L. COHeRE: Cross-ontology hierarchical relation examination for ontology quality assurance. AMIA Annu Symp Proc 2015; 2015: 456–65. [PMC free article] [PubMed] [Google Scholar]
- 27. Abeysinghe R, Zheng F, Hinderer EW, et al. A lexical approach to identifying subtype inconsistencies in biomedical terminologies. In: proceedings of 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2018; 1982–9.
- 28. SpaCy: Industrial-Strength Natural Language Processing. https://spacy.io/ Accessed January 20, 2020.
- 29.SpaCy Linguistic Features. https://spacy.io/usage/linguistic-features Accessed January 20, 2020.
- 30. Hagberg A, Schult DA, Swart P. Exploring network structure, dynamics, and function using NetworkX. In: proceedings of the 7th Python in Science Conference (SciPy 2008); 2008: 11–6.
- 31. Liu H, Zheng L, Perl Y, Geller J, Elhanan G. Can a convolutional neural network support auditing of nci thesaurus neoplasm concepts? In proceedings of the International Conference on Biomedical Ontology; 2018.
- 32. Sun Q, Zhang GQ, Zhu W, et al. Validating auto-suggested changes for SNOMED CT in non-lattice subgraphs using relational machine learning. Stud Health Technol Inform 2019; 264: 378–82. [DOI] [PubMed] [Google Scholar]
- 33. Abeysinghe R, Brooks MA, Talbert J, et al. Quality assurance of NCI Thesaurus by mining structural-lexical patterns. In AMIA Annu Symp Proc 2018; 2017: 364–73; Washington, DC. [PMC free article] [PubMed]
- 34. Cui L, Zhu W, Tao S, et al. Mining non-lattice subgraphs for detecting missing hierarchical relations and concepts in SNOMED CT. J Am Med Inform Assoc 2017; 24 (4): 788–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bodenreider O. Identifying missing hierarchical relations in SNOMED CT from logical definitions based on the lexical features of concept names. https://mor.nlm.nih.gov/pubs/pdf/2017-snomedct_expo-ob-abstract.pdf Accessed June 30, 2020. [PMC free article] [PubMed]
- 36. Abeysinghe R, Hinderer EW, Moseley HN, Cui L. Auditing subtype inconsistencies among gene ontology concepts. In: proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2017; 1242–5; Kansas City, MO.
- 37. Cui L, Bodenreider O, Shi J, et al. Auditing SNOMED CT hierarchical relations based on lexical features of concepts in non-lattice subgraphs. J Biomed Inform 2018; 78: 177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abeysinghe R, Brooks MA, Cui L. Leveraging non-lattice subgraphs to audit hierarchical relations in NCI Thesaurus. AMIA Annu Symp Proc 2019; 2019: 982–91. [PMC free article] [PubMed] [Google Scholar]
- 39. LuiNorm. https://lexsrv3.nlm.nih.gov/LexSysGroup/Projects/lvg/2016/docs/userDoc/tools/luiNorm.html Accessed April 10, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.