Sir,
The evolution of resistance that arises within individual patients remains an important clinical problem in certain clinical situations. In such cases, it is important to consider pharmacokinetic properties such as tissue penetration and half-life time of the drugs to avoid compartmentalization issues that can increase the likelihood of antibiotic resistance evolving.1 Here we describe a case of an acute Pseudomonas aeruginosa infection where compartmentalization within the CNS was associated with the in vivo evolution of resistance to multiple drugs, including piperacillin/tazobactam. We collected isolates from a single patient over the course of the infectious process, and sequenced and assembled their genome to identify the genomic changes leading to resistance.
At our institution, case reports are exempt from independent review. Individually identifying information, including patient age, sex, specific dates and comorbidities, was not disclosed to protect the patient’s identity.
We report a case of a patient in their early 20 s diagnosed with AML 3 months prior to admission. After a third cycle of high-dose Ara-C consolidation chemotherapy, the patient was brought to the emergency department with a 39.5°C fever and tachycardia but normal blood pressure. Admission blood cultures were positive for P. aeruginosa. Empirical treatment with piperacillin/tazobactam, vancomycin and micafungin was initiated (Figure 1a) which led to a rapid resolution of the bacteraemia. However, fevers persisted and the patient subsequently developed confusion and altered sensorium.
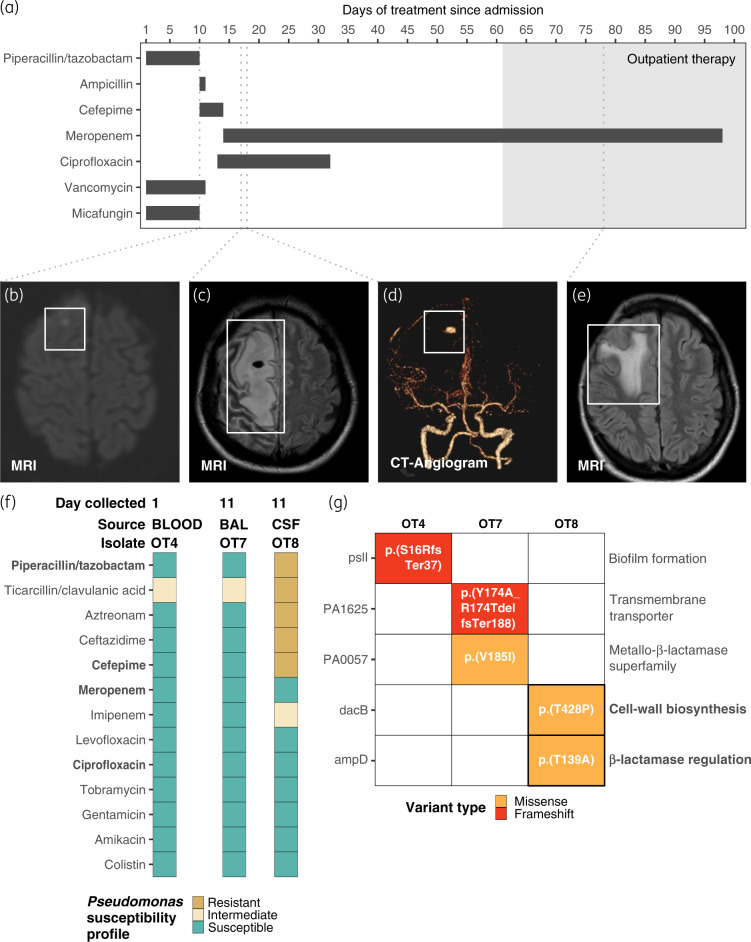
Figure 1.
Summary of clinical course, antibiotic treatment and genomic analysis of P. aeruginosa isolates. (a) Course of antibiotic treatment during hospitalization (white background) and as an outpatient (grey area). (b–e) MRI or CT angiogram images of the patient’s brain taken at different stages of disease progression. Grey dashed lines extending from panel (a) to (b–e) indicate the hospitalization day in which the images were taken. (f) Antibiotic susceptibility profiles of the P. aeruginosa isolates OT4, OT7 and OT8. BAL, bronchoalveolar lavage fluid. (g) Identified genomic variants in OT4, OT7 and OT8 over the entire closed genomes of the isolates. The functional annotation of each gene is given on the right side of the heat map. Annotation of each variant is indicated in Human Genome Variation Society (HGVS) format. Antibiotic doses in (a): piperacillin/tazobactam, 4.5 g IV q8h; ampicillin, 2 g IV q4h; cefepime, 2 g IV q8h; ciprofloxacin, 400 mg IV q8h; meropenem, 2 g IV q8h; vancomycin was not dosed consistently; and micafungin, 150 mg IV q24h.
On Day 10, the patient became more lethargic and developed acute left-sided weakness. On the same day, brain MRI revealed small foci of restricted diffusion in the right frontal lobe suggestive of an infiltrative inflammatory process (Figure 1b). The patient was transferred to the ICU and intubated for respiratory protection. The antibiotic treatment was then transitioned to cefepime, ampicillin and vancomycin (Figure 1a). On hospital Day 11 a bacterial culture from bronchoalveolar lavage fluid grew P. aeruginosa (representative isolate OT7) with similar antibiotic susceptibilities to the earlier blood culture collected on admission (representative isolate OT4; Figure 1f). A bacterial culture of the CSF taken on the same day grew P. aeruginosa (representative isolate OT8) resistant to multiple antibiotics, including piperacillin/tazobactam and cefepime (Figure 1f). Antibiotic therapy was changed to meropenem and ciprofloxacin (Figure 1a). A second brain MRI taken on hospital Day 17 demonstrated an increase of the inflammation in the right parasagittal frontal lobe with blood products around the area of restricted diffusion, consistent with the presence of an abscess (Figure 1c). A CT cerebral angiogram performed on hospital Day 18 demonstrated an 8 × 6 mm right middle cerebral artery aneurysm in the location of the brain abscess (Figure 1d). On the same day, the patient underwent frontal craniotomy for evacuation of subdural empyema and resection of the infected aneurysm.
Fevers resolved after surgery, and the patient improved clinically with resolution of left-sided weakness. The patient was treated with 2 more weeks of meropenem and ciprofloxacin, followed by an additional 6 weeks of meropenem monotherapy, partially as outpatient parenteral antimicrobial therapy. Brain MRI performed 8 weeks after surgery demonstrated sequelae of the infectious changes with minimal reactive changes (Figure 1e).
To identify the genetic changes between the three isolates taken from the patient (OT4, OT7 and OT8) we performed WGS using Illumina HiSeq 125 bp paired-end data, as well as long-read sequencing with the Oxford Nanopore MinION technology. Across the entire high-quality, closed genomes of all three isolates (assembled genomes are available at the NCBI database with the BioProject accession number PRJNA598709) we confidently identified just five variants in five different genes. To determine the ancestral and derived state of the five variants, we compared each with the reference strain P. aeruginosa PAO1 (Figure 1g). The OT4 isolate had a frameshift deletion in the gene pslI [p.(S16RfsTer37)], while the remaining isolates shared PAO1’s genotype. pslI is part of the polysaccharide psl locus, which encodes 15 co-transcribed genes predicted to synthesize Psl, a mannose- and galactose-rich exopolysaccharide required for the formation of structurally sound biofilms.2 Despite having a similar antimicrobial susceptibility profile to OT4, OT7 showed two additional variants in genes coding for an uncharacterized transmembrane protein [PA1625; p.(Y173A_R174delfsTer188)] and an uncharacterized protein belonging to the MBL superfamily, respectively [PA0057; p.(V185I)] (Figure 1g).
The MDR OT8 isolate had missense variants in the genes dacB [p.(T428P)] and ampD [p.(T139A)] (Figure 1g). Complete knockouts of dacB and ampD have been associated with the deregulation of the β-lactamase AmpC, which led to high levels of resistance to almost all β-lactams, with the exception of the carbapenems.3 Several distinct variants in ampD have previously been reported to increase resistance to the β-lactams in clinical isolates of P. aeruginosa, highlighting the mutational diversity of this site.4–6 Indeed, 80 distinct non-synonymous SNPs in ampD were found in at least 1 of 99 clinical isolates of P. aeruginosa with varying levels of resistance to the β-lactams.7 Similarly, 36 non-synonymous SNPs in dacB were found in the same database.7
In humans, the effective concentration of piperacillin and tazobactam in the central nervous system is not commonly achieved as it can be highly variable among patients and usually lower than required for effective treatment.8 Previous studies have shown that piperacillin, alone and in combination with tazobactam, systematically leads to the derepression of AmpC. Thus, the variability in the concentration of both drugs within the CNS could have allowed selection for mutations in dacB and ampD that cause derepression of this β-lactamase. Additionally, the patient had a reduced capacity to fight the infection, which could have resulted in a larger population size, thereby increasing the probability of resistance emergence.9,10 Consistent with this interpretation, imaging showed P. aeruginosa established itself in high numbers within the CNS (visible in the MRI and CT scans in Figure 1b–e), subsequently acquiring two mutations leading to clinical levels of resistance to all β-lactams tested, except for meropenem (Figure 1g).
The link between theoretical principles and this patient’s clinical course was evident after the fact, but predicting evolution remains as challenging in patients as it is in all other aspects of biology.11 This case demonstrates the need to improve these efforts and suggests that integrating evolutionary principles into clinical risk prediction models may be fruitful.
Acknowledgements
We would like to thank Carol Young in the clinical microbiology laboratory of the University of Michigan Hospital for assistance acquiring bacterial samples.
Funding
This study was funded by the National Institutes of Health (NIH) (grant K08AI119182 from NIAID available to R.J.W.) and the German Research Foundation (DFG) (fellowship BA 6186/1-1 to C.B.).
Transparency declarations
None to declare.
References
- 1. Moreno-Gamez S, Hill AL, Rosenbloom DIS. et al. Imperfect drug penetration leads to spatial monotherapy and rapid evolution of multidrug resistance. Proc Natl Acad Sci USA 2015; 112: E2874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ma L, Lu H, Sprinkle A. et al. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J Bacteriol 2007; 189: 8353–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zamorano L, Moyá B, Juan C. et al. Differential β-lactam resistance response driven by ampD or dacB (PBP4) inactivation in genetically diverse Pseudomonas aeruginosa strains. J Antimicrob Chemother 2010; 65: 1540–2. [DOI] [PubMed] [Google Scholar]
- 4. Juan C, Maciá MD, Gutiérrez O. et al. Molecular mechanisms of β-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 2005; 49: 4733–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greipel L, Fischer S, Klockgether J. et al. Molecular epidemiology of mutations in antimicrobial resistance loci of Pseudomonas aeruginosa isolates from airways of cystic fibrosis patients. Antimicrob Agents Chemother 2016; 60: 6726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lister PD, Wolter DJ, Hanson ND.. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Antimicrob Agents Chemother 2009; 22: 582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hornischer K, Khaledi A, Pohl S. et al. BACTOME—a reference database to explore the sequence- and gene expression-variation landscape of Pseudomonas aeruginosa clinical isolates. Nucleic Acids Res 2019; 47: D716–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nau R, Kinzig-Schippers M, Sörgel F. et al. Kinetics of piperacillin and tazobactam in ventricular cerebrospinal fluid of hydrocephalic patients. Antimicrob Agents Chemother 1997; 41: 987–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Day T, Read AF.. Does high-dose antimicrobial chemotherapy prevent the evolution of resistance? PLoS Comput Biol 2016; 12: e1004689.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kouyos RD, Metcalf CJE, Birger R. et al. The path of least resistance: aggressive or moderate treatment? Proc R Soc B 2014; 281: 20140566.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lässig M, Mustonen V, Walczak AM.. Predicting evolution. Nat Ecol Evol 2017; 1: 77.. [DOI] [PubMed] [Google Scholar]