Abstract
Background
Antimicrobial resistance (AMR) is a rising health threat with 10 million annual casualties estimated by 2050. Appropriate treatment of infectious diseases with the right antibiotics reduces the spread of antibiotic resistance. Today, clinical practice relies on molecular and PCR techniques for pathogen identification and culture-based antibiotic susceptibility testing (AST). Recently, WGS has started to transform clinical microbiology, enabling prediction of resistance phenotypes from genotypes and allowing for more informed treatment decisions. WGS-based AST (WGS-AST) depends on the detection of AMR markers in sequenced isolates and therefore requires AMR reference databases. The completeness and quality of these databases are material to increase WGS-AST performance.
Methods
We present a systematic evaluation of the performance of publicly available AMR marker databases for resistance prediction on clinical isolates. We used the public databases CARD and ResFinder with a final dataset of 2587 isolates across five clinically relevant pathogens from PATRIC and NDARO, public repositories of antibiotic-resistant bacterial isolates.
Results
CARD and ResFinder WGS-AST performance had an overall balanced accuracy of 0.52 (±0.12) and 0.66 (±0.18), respectively. Major error rates were higher in CARD (42.68%) than ResFinder (25.06%). However, CARD showed almost no very major errors (1.17%) compared with ResFinder (4.42%).
Conclusions
We show that AMR databases need further expansion, improved marker annotations per antibiotic rather than per antibiotic class and validated multivariate marker panels to achieve clinical utility, e.g. in order to meet performance requirements such as provided by the FDA for clinical microbiology diagnostic testing.
Introduction
Antimicrobial resistance (AMR) is a rising health threat estimated to cause 700 000 annual deaths, with 10 million annual casualties expected by 2050.1 Appropriate antibiotic therapy improves patient outcomes and is a major factor in reducing the emergence of antibiotic resistance.2,3 Current clinical practice predominantly uses antibiotic susceptibility testing (AST) from bacterial culture, with long turnaround times ranging between 24 and 72 h, errors arising in inoculum preparation or culture conditions and limitations related to individual species–antibiotic combinations.3–5
With the reduction in WGS costs and runtimes, several researchers have assessed WGS-based AST (WGS-AST) for clinical practice, for example in Escherichia coli and Klebsiella pneumoniae, as well as using different computational approaches.3,6–9
The increasing availability of WGS data from clinical strains has helped to robustly identify antibiotic resistance determinants and to curate them in dedicated databases.10 Public databases like the Comprehensive Antibiotic Resistance Database (CARD),11 ResFinder12 and its companion database PointFinder,13 ARG-ANNOT,14 as well as others, have emerged as AMR repositories and software tools have been developed for WGS-AST based on these databases.
Stoesser et al.3 and Zankari et al.13 used ResFinder and its sister database PointFinder for WGS-AST on clinical isolates of E. coli, K. pneumoniae, Salmonella and Campylobacter jejuni. They showed that in silico AMR predictions may suffer from high false-susceptible (false-negative) and/or false-resistant (false-positive) rates. In resistance phenotyping, a false-negative result is considered a very major error (VME) and a false-positive result a major error (ME). A VME might result in use of an ineffective therapeutic agent for treatment, leading to treatment failure; an ME might limit therapeutic options and complicate treatment. The more serious effect of VMEs is reflected in FDA stipulations for diagnostic test approval, where the FDA requires VMEs <1.5% and MEs <3% for approval of a new AMR diagnostic test or device.15
As the coverage and quality of databases underlying WGS-AST are essential for clinically relevant AMR diagnostic tests,9,16 we evaluated the performance of public AMR marker reference databases for the prediction of resistance in bacterial isolates to spotlight current pitfalls and areas for improvement. Most studies that apply genotype to phenotype prediction focus on samples from a particular species or a single cohort. Here, we present a large-scale study encompassing bacterial isolates from multiple species and cohorts to investigate the utility of publicly available markers for AST prediction from WGS. To maximize the amount of data available for assessment, categorical phenotypes were used as provided by the public databases. A limitation may thus be that, with the increase of AMR, interpretive breakpoints have changed over time and isolates treated as susceptible may be considered resistant at present. As opposed to other studies that focus on smaller datasets or single pathogens together with expert manual curation of results,3,17 we evaluated the performance of AMR marker databases on a large and diverse dataset using default settings to assess the status quo and outline concrete steps to improve clinical utility of those databases without time-consuming result curation.
Materials and methods
Isolates and antibiotics collection
Assembled isolate genomes and categorical resistant/susceptible phenotypes as per the databases—filtered by antibiotics—were sourced from PATRIC (the Pathosystems Resource Integration Center) and NDARO (National Database of Antibiotic Resistant Organisms); accessed 31 January 2019. Details can be found in the Supplementary data (available as Supplementary data at JAC Online).
In silico phenotype prediction
CARD’s Resistance Gene Identifier (RGI) 4.2.2 with the CARD database 3.0.1 was run with default settings for all isolates to predict resistance phenotypes. ‘Perfect’ and ‘strict’ hits as per CARD publication were included according to per model curated similarity cut-offs where a perfect hit is an exact match to the curated reference sequences and a strict hit is a previously unknown variant of known AMR genes.18
ResFinder 4.0 (git commit 0007df1) was installed and run using the ResFinder database (git commit bd77b98) for resistance phenotype prediction for all isolates with default settings (minimum coverage 60%; minimum sequence identity 90%). PointFinder (git commit bd50f0b) was run for E. coli isolates using the PointFinder database (git commit d1413d2) with the scheme for E. coli.
Evaluation of prediction performance
CARD and ResFinder predictions were compared against in vitro phenotypic AST results. CARD and ResFinder reported predicted phenotypes by antibiotic class. Observed phenotypes against individual antibiotics, as reported by PATRIC and NDARO, were mapped to predicted phenotypes by antibiotic class affiliation to evaluate prediction performance. For example, observed phenotypes for amikacin, gentamicin and tobramycin were mapped to CARD’s predictions on aminoglycoside resistance.
Retrieved PATRIC and NDARO phenotypes were labelled as resistant/susceptible, in line with the resistance/susceptible prediction output from CARD and ResFinder. Evaluation of prediction performance was treated as a binary classification task; predicted phenotypes were compared with observed phenotypes. An ME was defined as a resistant prediction discrepant with an observed susceptible phenotype; a VME was defined as a susceptible prediction discrepant with an observed resistant phenotype. To evaluate prediction performance, balanced accuracy (bACC)—the average of sensitivity and specificity—was used, which can be understood as the average accuracy obtained for each class, thus avoiding inflated performance statistics in the case of dataset class imbalance.19–21
To evaluate PointFinder in combination with ResFinder for WGS-AST for the antibiotics ciprofloxacin, cefotaxime and ceftazidime in E. coli, an isolate was classified as resistant if either tool predicted resistance.
Evaluation of marker performance
We used the Antibiotic Resistance Ontology (ARO) provided by CARD to group detected CARD AMR markers, i.e. genes and variants, into AMR marker families. For example, all aminoglycoside nucleotide transferase (ANT) alleles were treated as a marker family, all TEM alleles as TEM β-lactamases and so on. The aim was to describe the effect of low-resolution annotations of marker-to-phenotype relationships on predictive performance. While considerable differences in activity exist for members of groups of, for example, β-lactamases, such as TEMs, SHVs and KPCs, those differences were not annotated conclusively and comprehensively for analysis on the allele level across evaluated databases.
For the evaluation, the predictive performance of every marker family for an associated antibiotic resistance listed by CARD ARO was independently evaluated on all isolates. Standard metrics such as sensitivity, specificity and positive predictive value (PPV) were calculated.
To assess the performance of entire AMR marker groups, marker families were further aggregated using resistance mechanism-related ontology terms that are close to the root term of CARD’s ARO. This aggregation resulted in four ‘marker groups’ that covered all markers detected: (i) efflux-related AMR genes and mutations; (ii) β-lactamases; (iii) aminoglycoside-modifying enzymes (AMEs); and (iv) fluoroquinolone resistance-associated genes and mutations. Predictive performance of each marker group was calculated as the average performance of the marker families represented by the marker group for all species–antibiotic combinations evaluated above.
Mapping of ResFinder compound class predictions to compounds
We downloaded marker–phenotype associations available from ResFinder at https://bitbucket.org/genomicepidemiology/resfinder_db. Associations were used to predict antibiotic resistance using detected markers as per ResFinder output in analysed isolates per antibiotic rather than antibiotic class. Markers without phenotype associations for individual antibiotics were excluded.
Data availability statement
Genotype and phenotype data were downloaded from the public repositories NDARO and PATRIC and were publicly available at the time of this study.
Results
Selection of species and antibiotics
A total of 4278 isolate genomes and their resistance phenotypes were collected from PATRIC22 and NDARO,23 two public repositories of microbial isolate data, for assessment of WGS-AST based on publicly available AMR markers from CARD11 and ResFinder12 (Table 1). Regarding antibiotic selection, 17 antibiotics were selected as representatives from penams, cephalosporins and other β-lactams, tetracyclines, quinolones and aminoglycosides. Details regarding selection criteria that resulted in a final dataset size of 2587 isolates are listed in the supplementary material.
Table 1.
The distribution of analysed isolates across different species
| Pathogen/species | NDARO | PATRIC | Total | Selected |
|---|---|---|---|---|
| A. baumannii | 220 | 492 | 712 | 663 |
| Enterobacter cloacae complex | 83 | 8 | 91 | 39 |
| E. coli | 185 | 1517 | 1702 | 563 |
| K. pneumoniae | 295 | 820 | 1115 | 668 |
| P. aeruginosa | 144 | 514 | 658 | 654 |
| Total | 4278 | 2587 |
Resistance phenotype prediction
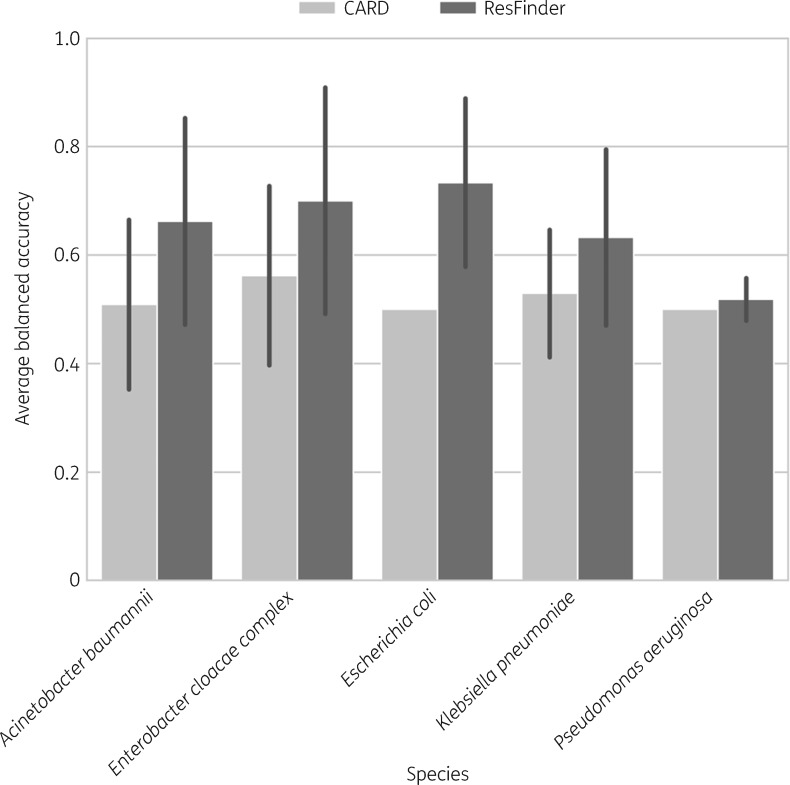
Despite the large number of isolates per species, the proportion of resistant to susceptible isolates, as deposited in PATRIC/NDARO, was often skewed to either extreme for a given antibiotic. This strong class imbalance (Figure S2) was reported before by Drouin et al.24 for PATRIC data. Therefore, averaged bACC was applied to assess resistance phenotype prediction for the five analysed species. Overall, bACC ranged from 0.50 to 0.73 (Figure 1).
Figure 1.
The bACC is shown for every species as the average of all bACCs for the species–antibiotic combinations mentioned in the main text. bACC was chosen as the evaluation criterion as it avoids performance inflation and provides a balanced representation of false-positive and false-negative rates even in the case of dataset class imbalance. Error bars indicate SD. CARD predicted all E. coli and P. aeruginosa isolates to be resistant to all tested antibiotics, resulting in a constant bACC of 0.5 and the absence of the error bars.
ResFinder showed slightly better performance than CARD. CARD predictions suffered from overclassification and failure to identify susceptible isolates, where bACC ranged from a minimum of 0.13 for aztreonam resistance in Acinetobacter baumannii to a maximum of 0.51 for tobramycin resistance in the same species. Notably, CARD predicted all E. coli and Pseudomonas aeruginosa isolates to be resistant to all tested antibiotics, resulting in a bACC measure of 0.50 across all tested antibiotics (n = 17 and n = 11, respectively). In contrast, ResFinder predictions had a bACC of 0.49 for piperacillin/tazobactam resistance in P. aeruginosa but in many instances achieved a bACC of 1.00 (Figure 2). Across all antibiotic–species combinations, CARD’s predictions showed an average bACC of 0.52 and ResFinder’s predictions an average bACC of 0.66. CARD’s predictions resulted in more MEs than ResFinder’s predictions, with CARD’s average ME rate at 0.42. ResFinder showed fewer MEs across all antibiotic–species combinations, where the average ME rate was 0.25.
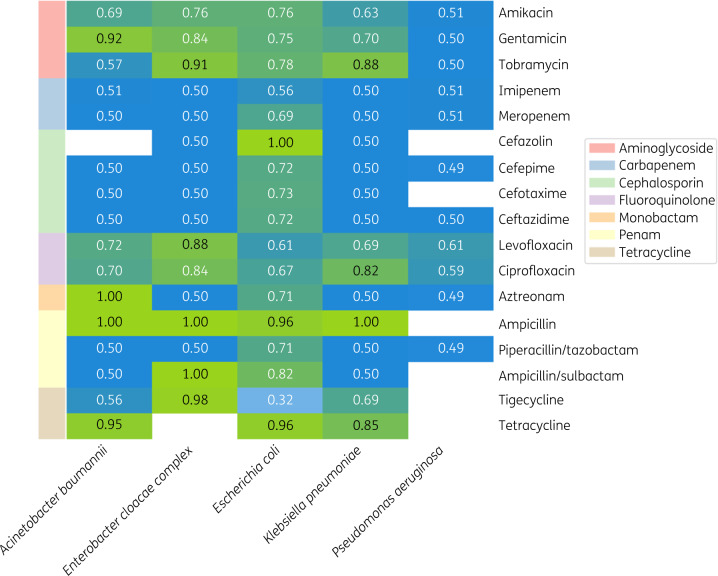
Figure 2.
bACC measures using ResFinder. The heatmap shows analysed antibiotics versus pathogens. White rectangles represent species–antibiotic pairs that were not analysed due to absent or insufficient AST data.
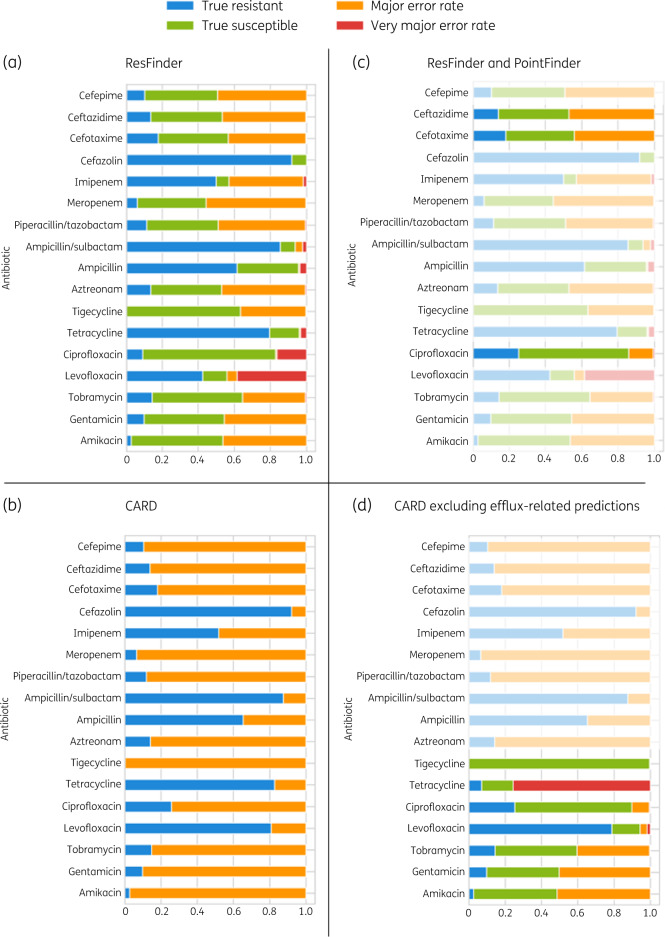
We chose E. coli (Figure 3) for a detailed comparison of ResFinder and CARD prediction performance.25 CARD and ResFinder results for the remaining species are in Tables S1 and S2. For E. coli, average bACC using ResFinder was 0.73 and average bACC using CARD was 0.50. Poor performance was mainly due to high ME rather than VME rates. ResFinder predictions had an average ME rate of 29.76% and an average VME rate of 3.99% whereas CARD predictions had an average ME rate of 65.4% and no VMEs. ResFinder’s predictions showed better overall performance than CARD except in the case of fluoroquinolones (Figure 3a and b).
Figure 3.
Evaluation of ResFinder and CARD (RGI) antibiotic resistance prediction performance on E. coli. (a) ResFinder prediction performance across 17 antibiotics. (b) CARD prediction performance across 17 antibiotics. (c) ResFinder and PointFinder prediction performance for ciprofloxacin, cefotaxime and ceftazidime. (d) CARD prediction performance excluding predictions based on markers related to efflux pump mechanism. ResFinder shows overall better prediction performance than CARD; PointFinder predictions improve ResFinder predictions and, excluding predictions based on efflux-related markers, improve CARD predictions except for tetracycline.
ResFinder’s bACC values of 0.67 for ciprofloxacin and 0.61 for levofloxacin were attributed to the fact that fluoroquinolone resistance is predominantly mediated by chromosomal mutations and ResFinder predictions are exclusively based on AMR genes.26,27 We combined results from ResFinder and PointFinder, the software developed by the ResFinder team to tackle chromosome mutation-mediated resistance. This approach could only be applied to E. coli as PointFinder’s database was implemented only for E. coli, Salmonella enterica and C. jejuni.13 Combining predictions from ResFinder and PointFinder for ciprofloxacin resulted in improved prediction performance and a reduced VME rate from 16.5% to 0.5%. Despite abundant literature on gyrA and parC mutations mediating resistance to fluoroquinolones in general,28 PointFinder exclusively associates gyrA and parC mutations with ciprofloxacin resistance; consequently levofloxacin resistance was not analysed (Figure 3c).
To further investigate CARD’s high ME rates, we repeated the calculation excluding all predictions based on markers with efflux-related resistance mechanisms. Efflux markers are highly affected by transcription regulation and detection by WGS is not always a sufficient indication of their contribution to AMR.29,30 This resulted in overall improvement in the performance of CARD’s RGI for aminoglycoside and fluoroquinolone compounds (Figure 3d). However, for tetracyclines, VME rates increased, where many resistant isolates were predicted to be susceptible, in agreement with efflux-mediated resistance being the main resistance mechanism against tetracyclines.31 For results of using CARD predictions without efflux markers for all species, see Table S3.
AMR marker performance
To provide an overview of contributions of entire marker groups to WGS-AST, four groups were built from CARD’s ARO: efflux-related markers, β-lactamases, AMEs and fluoroquinolone-resistance genes and mutations. For every group, we evaluated the contribution to the overall prediction of resistance phenotype using (i) PPV, i.e. the probability of a subject carrying a resistance marker being resistant, and (ii) specificity, i.e. the proportion of susceptible isolates that are correctly identified as such (Tables 2 and S4).
Table 2.
Abundance and WGS-AST performance metrics of the four main marker families from CARD
| Marker group | Average hits/sample | Average specificity | Average PPV |
|---|---|---|---|
| Efflux-related markers | 30.30 | 0.12 | 0.52 |
| β-Lactamases | 2.61 | 0.66 | 0.76 |
| AMEs | 2.51 | 0.65 | 0.55 |
| Fluoroquinolone resistance-associated genes and mutations | 0.72 | 0.94 | 0.82 |
Efflux-related AMR markers from CARD were the most abundant marker class in terms of hits per isolate (Table S5). On average, an isolate contained 30 hits related to ‘Antibiotic Efflux’ as per CARD ARO, significantly higher than the average hits of any other class. However, average specificity of an efflux-related AMR marker is lowest among marker families. P. aeruginosa isolates had the highest average number of efflux-related AMR determinants detected per isolate (mean = 44.66) compared with isolates from the remaining species (mean = 24.15), in agreement with studies demonstrating a major role of efflux pumps for P. aeruginosa resistance.32,33 In contrast, fluoroquinolone resistance genes and mutations show the highest specificity and PPV despite their low abundance (Figure S3). It should be noted that in addition to single-point mutations related to fluoroquinolone resistance, CARD also detects double mutations within the same gene, e.g. gyrA S83L/D87N, in line with scientific literature.34 Increased resistance due to the presence of more than one mechanism simultaneously was not accounted for.
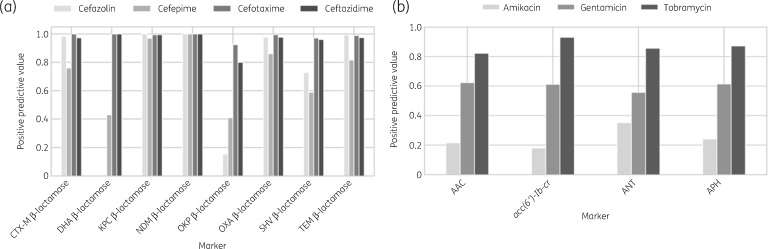
For the subsequent analysis, we focused on K. pneumoniae, another cause of highly threatening nosocomial infections.35 With the rise of β-lactam (mainly meropenems and cephalosporins) and aminoglycoside resistance in K. pneumoniae,36,37 accurate and precise detection of resistance is of high clinical relevance and value. Despite identical resistance profiles of some β-lactamases based on CARD ARO, we observed variability across some for resistance prediction against associated antibiotics (Figure 4).
Figure 4.
Differences in resistance profiles for β-lactamases and AMEs in K. pneumoniae. (a) KPC β-lactamases and NDM β-lactamases are consistently good predictors of resistance across all the analysed cephalosporins whereas OKP β-lactamases and CTX-M β-lactamases show variable resistance prediction performance. (b) AACs, aminoglycoside phosphotransferases (APHs) and ANTs consistently show lower PPVs for amikacin than tobramycin or gentamicin.
Annotation level and prediction performance
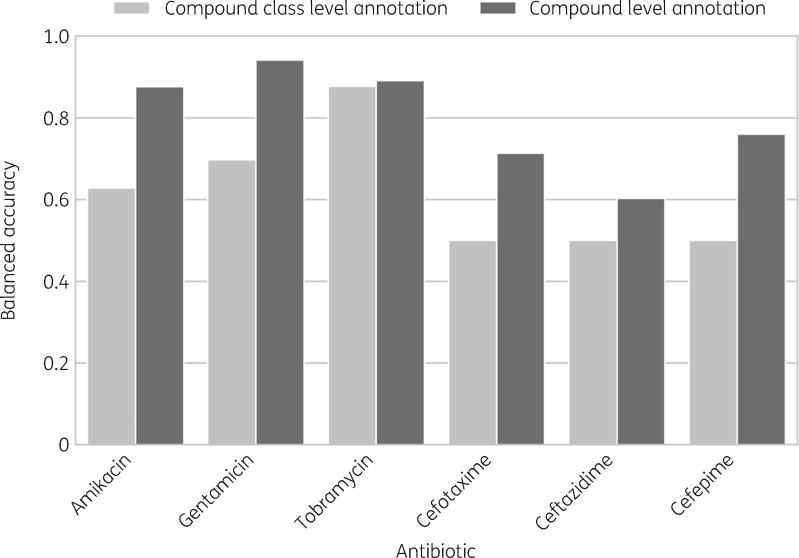
To investigate the effect of a higher level of annotation detail, i.e. marker–compound annotation, we compared the predictions of ResFinder with default settings against ResFinder predictions mapped to individual compounds (see the Materials and methods section). In Figure 5, we show that compound–level annotations improved prediction performance for cephalosporins as well as aminoglycosides in K. pneumoniae samples.
Figure 5.
bACC of ResFinder predictions based on ResFinder compound–class-level annotation versus compound-level annotation in K. pneumoniae samples. Compound-level prediction consistently performs better and the increase in bACC ranges between 0.01 for tobramycin resistance prediction and 0.26 for cefepime resistance prediction.
Discussion
Ruppé et al.16 reviewed studies performing genotype to phenotype comparisons from AMR determinants from public databases in an approach similar to the one presented here. These studies, however, focused on one or two species, with smaller datasets not exceeding 400 samples and were mainly derived from a single location.10,12,38–40 According to Su et al.,4 including large, diverse test sets for development and evaluation of WGS-AST tools is critical. However, most studies have used a sample with limited geographic and temporal variability. By using datasets from PATRIC41 and NDARO,23 comprised of genome assemblies and categorical resistant/susceptible phenotypes, we included a dataset from multiple clinically relevant species with diverse locations and collection times (Figure S1). A limitation may be that the study used categorical phenotypes as provided by the databases. With the increase of AMR, interpretive breakpoints have changed over time.42 For Enterobacteriaceae, for example, breakpoints (mg/L) for ciprofloxacin changed from susceptible (S) ≤1, intermediate (I) = 2, resistant (R) ≥4 when first published by CLSI to S ≤ 0.25, I = 0.5, R ≥ 1 in 2019. While S/I/R labels could have been recalculated, including only data with numerical MIC values, this would have drastically reduced the dataset. Moreover, breakpoint changes were expected not to affect the study’s main findings. This was validated exhaustively across all pathogen–compound pairs for which numeric MIC data could be retrieved. Database labels as per submission and labels recalculated with breakpoints from 2019 were 95.6% concordant, while bACC of WGS-AST only differed by an average of 0.7%.
In this study, as well as others,16 culture-based AST is treated as the gold standard, being the primary method employed by clinical laboratories.4 However, culture-based AST has its own limitations. For example, a lack of reproducibility and comparability across laboratories and countries due to biological variability, as well as differences in technical staff training and standards provided by EUCAST and CLSI.43,44 Adopting WGS-AST, backed by standardized and validated sequencing and bioinformatics pipelines, represents an opportunity to eliminate errors due to experimental procedure and differences in laboratory conditions in addition to achieving comprehensive and more accurate results.4,9
Our analysis highlighted factors that affect WGS-AST accuracy, starting at the databases, as shown by variable results from ResFinder and CARD. Poor performance reflected in our results was due to high rates of ME rather than VME. ResFinder prediction performance was improved by combination with PointFinder, in agreement with findings by Zankari et al.38 that high VME rates for fluoroquinolones, for example, were due to the non-consideration of mutational events. Compared with ResFinder, CARD showed worse overall performance with lower specificity and higher ME rates, a result in agreement with a comparison carried out on Enterobacteriaceae by Pesesky et al.8 However, CARD’s prediction performance for all antibiotic classes (except tetracyclines) improved upon removal of general AMR determinants such as efflux pumps and MEs were reduced significantly. These markers are likely to cause inaccurate predictions as they are affected by gene regulation where the presence of a gene detected by WGS is not indicative of its expression due to weak promoters or low copy numbers.29,40,45 The aforementioned findings highlight two of the main challenges facing WGS-AST, namely: (i) the impact of reference databases on downstream results and hence the need for an exhaustive and up-to-date knowledgebase of AMR determinants that encompasses AMR genes and variants (chromosomal and acquired), their prevalence in individual bacterial species and metadata on compound activity and reported intrinsic resistance;9,16 and (ii) the high false-positive rate due to an inadequate understanding of aspects such as gene regulation, mutations in intergenic regions and de novo rRNA mutations. In the latter case, false positives may arise because naive detection of an AMR-related gene without considering the aforementioned aspects may not be sufficient to causally assume resistance. For example, gene expression is not considered.43,45 It is clear that these challenges must be addressed to approach the FDA requirements for a diagnostic test of MEs <3% and VMEs <1.5%.
Our results also highlighted that P. aeruginosa presents a particularly challenging case for WGS-AST, with its unique and complex resistome being tightly regulated by gene expression.46 bACC ranged from 0.49 to 0.60, in agreement with Kos et al.47 who showed that using WGS-based detection of functional genetic targets alone failed to explain observed resistance phenotypes in that species.
Another important factor affecting WGS-AST accuracy was the similarity/identity cut-off used for resistance gene identification. As our analysis explored the applicability of ResFinder in a standard manner, we used its default settings. This resulted in much lower accuracies than other studies applying a more stringent cut-off. For instance, Zankari et al.38 used a more stringent cut-off in AMR gene identification (≥98%) compared with our ResFinder default settings (≥90%), which can explain the high rate of MEs (27.82%) in our results. In another study by Thomas et al.,40 applying a cut-off of ≥95% together with focusing on a chosen subset of ResFinder markers yielded specificity levels approaching 100% for all antibiotic classes except aminoglycosides. Clausen et al.39 claimed that the 98% sequence identity threshold, as recommended by Zankari et al.,38 was tested to be the ‘optimal threshold’. Our results here suggest that revisiting default parameters for ResFinder might be advisable.
A key factor determining WGS-AST accuracy was the annotation level of marker–phenotype relationships. Despite sharing an identical resistance profile on CARD, members of the same family of enzymes showed different performance in predicting resistance for different aminoglycosides in the case of AMEs or different cephalosporins in the case of β-lactamases. Individual members of these enzyme families interact differently with different members of the antibiotic class. For example, many members of the aminoglycoside acetyl transferase (AAC) class of AMEs confer resistance to gentamicin and tobramycin, but not amikacin.48 Similarly, some members of the TEM class of β-lactamases are ESBLs, affecting all cephalosporins, whereas others are broad-spectrum β-lactamases that only affect early cephalosporins.49,50 Allele-specific compound resistance annotations and statistics on marker prevalence and performance, such as sensitivity and specificity, comprehensively applied across all AMR markers, despite being a determining factor for accurate phenotype prediction, are not currently available from any public AMR database.51 In addition, resistance against antibiotic classes may be affected by the simultaneous presence of multiple resistance mechanisms. Accumulation of AMR point mutations or genes can cause incremental MIC shifts, e.g. if mutations in gyrA and parC are present in combination with qnr or aac(6′)-Ib-cr genes. Readily available data on those combinatorial marker–phenotype relationships would further improve WGS-AST accuracy.
A number of studies applied genotype to phenotype prediction while relying on a curated subset of markers from CARD, ResFinder and other resources and obtained better overall prediction performance. For instance, Stoesser et al.3 applied predictions relying on a curated in-house reference database that contained AMR determinants from ResFinder and other resources. Consequently, the sensitivity for predicting resistance for ∼140 E. coli and K. pneumoniae clinical isolates was 0.96 and specificity was 0.97.3 Another study by Gordon et al.17 on a dataset of ∼1000 Staphylococcus aureus clinical isolates predicted phenotype from genotype using a curated panel of AMR determinants based on a literature search that resulted in overall sensitivity and specificity of 0.97 and 0.99. A similar approach was applied in a recent study by Feldgarden et al.52 who used AMRFinder, a tool relying on a high-quality curated AMR gene reference database, combined with literature research on antibiotic resistance phenotypes. This resulted in predictions that are consistent with 98.4% of the actual phenotypes of 6242 isolates from four different species (mostly S. enterica).52 These studies highlight the effect of marker curation and high level of detail in phenotype–genotype annotation on WGS-based phenotype prediction.
Conclusions
To our knowledge, we have presented the first large-scale study to include a highly diverse dataset of thousands of bacterial isolates from multiple species, locations and times. Results illustrate that databases such as CARD and ResFinder serve as reference databases on resistance markers and offer functionality to predict resistance phenotypes from single markers, but may not yield optimal results using default settings. In this context, several challenges in genetic phenotype prediction still exist. To achieve FDA requirements for clinical microbiology diagnostic testing below 3% MEs and 1.5% VMEs, current AMR databases need to: (i) be further curated and expanded, (ii) provide AMR marker annotations per antibiotic rather than antibiotic class, and (iii) use combinations of individual AMR markers selected for optimal diagnostic performance in experimentally validated multivariate panels.
Funding
This work was supported by the Austrian Research Promotion Agency (FFG) (grants 863729, 866389, 874595, 879570) as well as the Vienna Business Agency (grant 2447823).
Transparency declarations
N.M., I.F. and S.B. are employees of Ares Genetics GmbH. A.E.P. is the Chief Executive Officer of Ares Genetics GmbH. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. All other authors: none to declare.
Author contributions
N.M., S.B., A.H. and A.E.P. designed the study. N.M. and I.F. analysed the data. N.M., S.B. and A.E.P. wrote the manuscript. All authors reviewed and approved the manuscript.
Supplementary Material
References
- 1. O’Neill J. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. The Review on Antimicrobial Resistance 2014. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf.
- 2. Lee C-R, Cho I, Jeong B. et al. Strategies to minimize antibiotic resistance. Int J Environ Res Public Health 2013; 10: 4274–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stoesser N, Batty EM, Eyre DW. et al. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 2013; 68: 2234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su M, Satola SW, Read TD.. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 2019; 57: e01405–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tamma PD, Fan Y, Bergman Y. et al. Applying rapid whole-genome sequencing to predict phenotypic antimicrobial susceptibility testing results among carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother 2018; 63: e01923–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hasman H, Saputra D, Sicheritz-Ponten T. et al. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J Clin Microbiol 2014; 52: 139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kleinheinz KA, Joensen KG, Larsen MV.. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 2014; 4: e27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pesesky MW, Hussain T, Wallace M. et al. Evaluation of machine learning and rules-based approaches for predicting antimicrobial resistance profiles in Gram-negative bacilli from whole genome sequence data. Front Microbiol 2016; 7: 1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Didelot X, Bowden R, Wilson DJ. et al. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 2012; 13: 601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moradigaravand D, Palm M, Farewell A. et al. Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLOS Comput Biol 2018; 14: e1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McArthur AG, Waglechner N, Nizam F. et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 2013; 57: 3348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zankari E, Hasman H, Cosentino S. et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zankari E, Allesøe R, Joensen KG. et al. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 2017; 72: 2764–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta SK, Padmanabhan BR, Diene SM. et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 2014; 58: 212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jorgensen JH, Ferraro MJ.. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 2009; 49: 1749–55. [DOI] [PubMed] [Google Scholar]
- 16. Ruppé E, Cherkaoui A, Lazarevic V. et al. Establishing genotype-to-phenotype relationships in bacteria causing hospital-acquired pneumonia: a prelude to the application of clinical metagenomics. Antibiotics 2017; 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gordon NC, Price JR, Cole K. et al. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 2014; 52: 1182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jia B, Raphenya AR, Alcock B. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017; 45: D566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bekkar M, Djemaa HK, Alitouche TA.. Evaluation measures for models assessment over imbalanced data sets. J Inf Eng Appl 2013; 3: 27–38. [Google Scholar]
- 20. Hicks AL, Wheeler N, Sánchez-Busó L. et al. Evaluation of parameters affecting performance and reliability of machine learning-based antibiotic susceptibility testing from whole genome sequencing data. PLOS Comput Biol 2019; 15: e1007349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brodersen KH, Ong CS, Stephan KE. et al. The balanced accuracy and its posterior distribution. Twentieth International Conference on Pattern Recognition, Istanbul, Turkey IEEE, 2010; 3121–4.
- 22. Antonopoulos DA, Assaf R, Aziz RK. et al. PATRIC as a unique resource for studying antimicrobial resistance. Brief Bioinform 2019; 20: 1094–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NCBI. National Database of Antibiotic Resistant Organisms (NDARO)—Pathogen Detection—NCBI. https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/.
- 24. Drouin A, Letarte G, Raymond F. et al. Interpretable genotype-to-phenotype classifiers with performance guarantees. Sci Rep 2019; 9:4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blount ZD. The unexhausted potential of E. coli. Elife 2015; 4: e05826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wright GD. Molecular mechanisms of antibiotic resistance. Chem Commun 2011; 47: 4055. [DOI] [PubMed] [Google Scholar]
- 27. Munita JM, Arias CA.. Mechanisms of antibiotic resistance. Microbiol Spectr 2016; 4: 481–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morgan-Linnell SK, Becnel Boyd L, Steffen D. et al. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob Agents Chemother 2009; 53: 235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grkovic S, Brown MH, Skurray RA.. Transcriptional regulation of multidrug efflux pumps in bacteria. Semin Cell Dev Biol 2001; 12: 225–37. [DOI] [PubMed] [Google Scholar]
- 30. Yasufuku T, Shigemura K, Shirakawa T. et al. Correlation of overexpression of efflux pump genes with antibiotic resistance in Escherichia coli strains clinically isolated from urinary tract infection patients. J Clin Microbiol 2011; 49: 189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Munita JM, Arias CA.. Mechanisms of antibiotic resistance. Microbiol Spectr 2016; 4: VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rampioni G, Pillai CR, Longo F. et al. Effect of efflux pump inhibition on Pseudomonas aeruginosa transcriptome and virulence. Sci Rep 2017; 7: 11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aeschlimann JR. The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other Gram-negative bacteria. Pharmacotherapy 2003; 23: 916–24. [DOI] [PubMed] [Google Scholar]
- 34. Vila J, Ruiz J, Marco F. et al. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother 1994; 38: 2477–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bassetti M, Righi E, Carnelutti A. et al. Multidrug-resistant Klebsiella pneumoniae: challenges for treatment, prevention and infection control. Expert Rev Anti Infect Ther 2018; 16: 749–61. [DOI] [PubMed] [Google Scholar]
- 36. Xie Y, Tian L, Li G. et al. Emergence of the third-generation cephalosporin-resistant hypervirulent Klebsiella pneumoniae due to the acquisition of a self-transferable blaDHA-1-carrying plasmid by an ST23 strain. Virulence 2018; 9: 838–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liang C, Xing B, Yang X. et al. Molecular epidemiology of aminoglycosides resistance on Klebsiella pneumoniae in a hospital in China. Int J Clin Exp Med 2015; 8: 1381–5. [PMC free article] [PubMed] [Google Scholar]
- 38. Zankari E, Hasman H, Kaas RS. et al. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 2013; 68: 771–7. [DOI] [PubMed] [Google Scholar]
- 39. Clausen P, Zankari E, Aarestrup FM. et al. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J Antimicrob Chemother 2016; 71: 2484–8. [DOI] [PubMed] [Google Scholar]
- 40. Thomas M, Fenske GJ, Antony L. et al. Whole genome sequencing-based detection of antimicrobial resistance and virulence in non-typhoidal Salmonella enterica isolated from wildlife. Gut Pathog 2017; 9: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wattam AR, Davis JJ, Assaf R. et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 2017; 45: D535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Humphries RM, Abbott AN, Hindler JA.. Understanding and addressing CLSI breakpoint revisions—a primer for clinical laboratories. J Clin Microbiol 2019; 57: e00203–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boolchandani M, D’Souza AW, Dantas G.. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Genet 2019; 20: 356–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Khan ZA, Siddiqui MF, Park S.. Current and emerging methods of antibiotic susceptibility testing. Diagnostics 2019; 9: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moran RA, Anantham S, Holt KE. et al. Prediction of antibiotic resistance from antibiotic resistance genes detected in antibiotic-resistant commensal Escherichia coli using PCR or WGS. J Antimicrob Chemother 2017; 72: 700–4. [DOI] [PubMed] [Google Scholar]
- 46. Pang Z, Raudonis R, Glick BR. et al. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv 2019; 37: 177–92. [DOI] [PubMed] [Google Scholar]
- 47. Kos VN, Déraspe M, McLaughlin RE. et al. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 2015; 59: 427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramirez MS, Tolmasky ME.. Aminoglycoside modifying enzymes. Drug Resist Updat 2010; 13: 151–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 2001; 14: 933–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Palzkill T. Structural and mechanistic basis for extended-spectrum drug-resistance mutations in altering the specificity of TEM, CTX-M, and KPC β-lactamases. Front Mol Biosci 2018; 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McArthur AG, Wright GD.. Bioinformatics of antimicrobial resistance in the age of molecular epidemiology. Curr Opin Microbiol 2015; 27: 45–50. [DOI] [PubMed] [Google Scholar]
- 52. Feldgarden M, Brover V, Haft DH. et al. Validating the AMRFinder Tool and Resistance Gene Database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 2019; 63: e00483–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype and phenotype data were downloaded from the public repositories NDARO and PATRIC and were publicly available at the time of this study.